Abstract
Introduction
Circulating DNA can be pro-inflammatory when detected by leukocytes via toll-like receptor 9 (TLR9). Cell-free fetal DNA (cff-DNA) of placental origin, circulates in pregnancy, and increased concentrations are seen in conditions associated with placental and maternal inflammation such as pre-eclampsia. However, whether cff-DNA is directly pro-inflammatory in pregnant women and what regulates cff-DNA levels in pregnancy are unknown.
Methods
Using a human term placental explant model, we examined whether induction of placental inflammation can promote cff-DNA release, and the capacity of this cff-DNA to stimulate peripheral blood mononuclear cells (PBMCs) from pregnant women.
Results
We demonstrate lipopolysaccharide (LPS)-mediated inflammation in placental explants and induced apoptosis after 24 h. However, this did not increase levels of cff-DNA generation compared to controls. Furthermore, the methylation status of the cff-DNA, was not altered by LPS-induced inflammation. Cff-DNA did not elicit production of inflammatory cytokines from PBMCs, in contrast to exposure to LPS or the TLR9 agonist CpG-ODN. Finally, we demonstrate that cff-DNA acquired directly from pregnant women did not differ in methylation status from placental extracted DNA, or from placental explant generated cell-free DNA, and that, unlike Escherichia coli DNA, this cff-DNA has a low level of unmethylated CpG sequences.
Discussion
Our data suggest that placental inflammation does not increase release of cff-DNA and that placental cff-DNA is not pro-inflammatory to circulating PBMCs. It thus seems unlikely that high levels of cff-DNA are either a direct consequence or cause of inflammation observed in obstetric complications.
Keywords: Cell-free fetal DNA, Placental inflammation, Pre-eclampsia, Preterm birth, Pathogenesis, DNA methylation, Toll-like receptor 9
Highlights
-
•
Cell-free fetal DNA was generated using a human placental explant model.
-
•
Lipopolysaccharide causes inflammation and cell death in placental explants.
-
•
Inflammation does not increase cell-free fetal DNA release from placental explants.
-
•
Generated DNA does not elicit inflammation from blood cells from pregnant women.
1. Introduction
Cell-free fetal DNA (cff-DNA) is found in the maternal circulation as early as 5 weeks of gestation [1,2]. It is predominantly of placental origin and is generated during cell-death, likely by apoptosis of the trophoblasts in the syncytiotrophoblast layer [[2], [3], [4], [5]]. Although the method of release is not completely clear, there is a strong association between cff-DNA and placental microparticles including exosomes. Multiple studies have found that placental microparticles can release cff-DNA and have demonstrated the presence of fetal DNA in, or bound to, placental microparticles [6,7]. The levels of circulating cff-DNA increase throughout gestation [8,9]. Interestingly, women who develop obstetric pathologies that are related to placental inflammation, such as pre-eclampsia and preterm labour, have higher levels of cff-DNA compared to uncomplicated pregnancies [3,[10], [11], [12], [13], [14], [15]]. Despite extensive research, the pathophysiology of these obstetric complications are not completely understood, resulting in an unmet need for effective treatments [16,17]. However, it is well established that inflammation plays a key role in preterm labour [18,19] and pre-eclampsia [14,17] and indeed there is some evidence that they share pathophysiological mechanisms [20,21].
cff-DNA can be discriminated from maternal cell-free DNA by identifying fetal DNA specific characteristics such as sex (if a male fetus is present) or various genes that are differentially methylated in the placenta [22]. Studies have indicated that cff-DNA may be less methylated overall than adult DNA [21,23]. Because the innate pattern recognition receptor Toll Like Receptor 9 (TLR9) is activated by unmethylated CpG oligonucleotide (ODN) sequences [24], cff-DNA may have the potential be a TLR9 ligand and thereby potentially be pro-inflammatory. cff-DNA has therefore been hypothesized to play a role in the pathophysiology of preterm labour and pre-eclampsia [11,13,14,23,25].
In a model of miscarriage in preterm birth, human fetal DNA caused TLR9-dependent fetal resorption in mice [21]. In a model of pre-eclampsia, injection of CpG-DNA, caused vascular inflammation, resulting in maternal hypertension and vasoconstriction in rats [26]. Furthermore, cell-free DNA of tumor origin can elicit an inflammatory response in epithelial cells through TLR9 [27,28]. Importantly, maternal circulatory cytokines produced upon TLR9 activation, such as IL-6, and chemokines, such as CXCL10, have been associated with obstetric complications, including pre-eclampsia and preterm birth [[29], [30], [31]]. However, it is unknown whether cff-DNA contributes to the inflammation seen in these pathologies, and whether this can be a directly pro-inflammatory stimulus for circulating leukocytes from pregnant women.
The mechanisms by which cff-DNA is released and why circulating levels of cff-DNA are higher in women who develop certain pregnancy complications remains elusive. In vitro studies have demonstrated that inflammation, induced by lipopolysaccharide (LPS), can increase programmed cell-death (apoptosis) in placental explants [32], and can increase cff-DNA release from murine placental explants [33]. We hypothesized that in utero inflammation would promote apoptosis and increase cff-DNA release from the placenta into the circulation resulting in an increase of cff-DNA, inducing systemic leucocyte-mediated inflammation, contributing to disease the pathogenesis in women with pre-eclampsia and/or preterm birth.
In this study, we examine the effect of LPS-mediated inflammation on cff-DNA release in a primary human placental explant model, and investigate the pro-inflammatory effects of cff-DNA on maternal peripheral blood mononuclear cells.
2. Materials and methods
2.1. Human samples
Placental tissues and whole blood (up to 30 ml) from pregnant women (N = 31) were collected as part of Edinburgh Reproductive Tissue Bio Bank (NHS Research ethics committee approval reference 13/ES/0126). Samples were donated by women with term pregnancies (37–40 weeks gestation) who were not in labour, had a BMI < 30, with no pregnancy complications, no blood borne diseases, no immunosuppressive disease or medication, and no current signs of infection.
2.2. Placental explant culture and stimulation
Within 30 min of delivery, 10 × 10 cm of placental tissue with no visible areas of calcification or hemorrhage was dissected and immediately immersed cold Dulbecco's phosphate-buffered saline (DPBS; Gibco). 1 × 1 cm pieces were dissected and snap frozen for use as sample controls (see below). Two washes were performed with DPBS, followed by a wash in culture medium (RPMI (Gibco) containing 10% Fetal Bovine Serum, 1% penicillin/streptomycin). While immersed in culture medium, placental pieces of approximately 150 mg were separated, using forceps and knife, into separate 6-well plates (one villous explant of 150 mg per well) and then cultured at 37 °C, ~18% PO2, 5% CO2. These placental explants were then stimulated with LPS (derived from E. coli 0111:B4 Sigma) at 2 ng/ml, 20 ng/ml or 200 ng/ml for 1, 6 or 24 h. Placental samples were stimulated with LPS in triplicate. Supernatants were collected for cff-DNA extraction and quantification and explants were snap frozen for further analysis.
2.3. Quantification of Caspase 3 and 7 in placental explants
Placental explants and placental sample controls were lysed with 350 μl luciferase cell lysis buffer (Promega E1531) using the Qiagen tissuelyser for 2 × 2 min at 250 Hz. Protein concentration was used for normalization between samples and was quantified using the Bio-rad protein quantification assay. All samples were eluted with cell lysis buffer to 1 mg/ml of protein. 50 μl of Caspase 3/7 Glo buffer (Promega G8090) was added to 50 μl of sample and incubated for 90 min in a clear 96-well plate. Samples were analysed in duplicate. Results were read at 490 nm in luminescence plate reader and quantified relative to sample controls. Results under zero were regarded as non-detected.
2.4. Cell-free fetal DNA extraction and quantification
Supernatant was harvested from placental explant cultures and cff-DNA extracted using the QIAamp Circulating Nucleic Acid Kit (Qiagen). Clinical cff-DNA was harvested in the same manner from plasma recovered from freshly drawn blood (less than hour after venepuncture) from EDTA (Sarstedt) tubes. The manufacturer's protocol was followed, using 1 μg carrier RNA and eluting in buffer AE (10 mM Tris-Cl, 0.5 mM EDTA; pH 9.0). Quantification of the cff-DNA isolated was performed with an absolute quantitation Real-Time multiplex PCR machine (ACB/SRY), over seven serial dilutions, using a standard curve of placental DNA. cff-DNA was quantified in pg/μl per mg of villous tissue in each placental explant culture. Clinical cff-DNA for methylation quantification was quantified using Nanodrop spectrometry.
2.5. Placental and adult DNA extraction
Human placental DNA was extracted from placental tissue, which had been snap frozen at time of collection, using the Qiagen Blood & Tissue DNA extraction kit with prolonged lysing period for increased fragmentation (2 × 2 min at 25 Hz). This DNA was used as a control for cff-DNA validation and quantification, and as a control stimulus in parallel to cff-DNA in in vitro cell stimulation experiments. Adult DNA was extracted from whole blood from a healthy non-pregnant woman (ERTBB 13/ES/0126) in sodium citrate tubes. The kit described above was used following manufactures protocol for whole blood DNA extraction. Adult DNA and placental DNA were quantified using Nano-spectrometry.
2.6. Peripheral blood mononuclear cells (PBMC) isolation and stimulation
Whole blood was acquired as part of Edinburgh Reproductive Tissue Bio Bank (NHS Research ethics committee approval reference 13/ES/0126) in sodium citrate tubes. PBMCs were harvested using Histopaque with a density gradient of 1.07 g/ml, washed twice with PBS, and finally washed with the culture medium (RPMI with 10% FCS and 5% Penicillin/Streptomycin). Cell number and viability was assessed using a haemocytometer and Trypan blue staining. PBMCs were plated at 2.0 × 106/ml in 12-well plates and maintained at 37 °C, 5% CO2 for 3 h, before stimulation with 1 μg/ml CpG oligodeoxynucleotides 2395 (ODN; Invivogen) [34], 1 μg/ml E. coli DNA (dsDNA E. coli Invivogen), 0.5 μg/ml cff-DNA, or vehicle (endotoxin-free LAL-water; Invivogen). Supernatants were collected after 18 h culture at 37 °C, 5% CO2.
2.7. Enzyme-linked immunosorbent assay (ELISA)
R&D Duoset ELISAs for IL-6, IL-8, TNF-α, CXCL-10 were used for cytokine analysis, according to the manufacturer's protocols. Supernatants from PBMC stimulation studies were used after being centrifuged for 1 min at 9391×g to spin down any residual cell debris.
2.8. Methylation quantification
Whole DNA methylation quantification was performed using Epigentek MethylFlash Global DNA Methylation (5-mC) ELISA Easy Kit (Colorimetric). The manufacturer's protocol was followed. Whole DNA was methylated using M.SssI methyltransferase (New England BioLabs), using manufacturer's protocol, with methylation halted by incubation at 65 °C for 20 min. This yielded the maximum amount of methylated CpGs. The level of unmethylated CpGs in starting materials was quantified by subtracting the amount of methylation quantified after use of methylation enzyme (maximum methylation) to the amount of methylation quantified without use of the methylation enzyme.
2.9. Statistics
Data in graphs are show as mean ± standard deviation (SD). In vitro stimulations were analysed using one-way or two-way ANOVA with Dunnet's or Tukey's multiple analysis post-test. Statistical analyses were performed with GraphPad Prism version 7.0 (GraphPad, San Diego, CA). p < 0.05 was considered statistically significant.
3. Results
3.1. LPS induced inflammation and apoptosis in placental explants
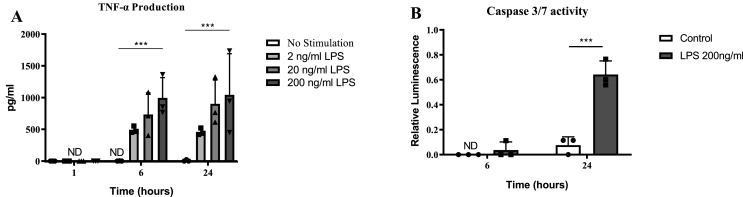
Placental explants, cultured with 2, 20 or 200 ng/ml of LPS or vehicle for 1, 6 or 24 h, showed a significant and concentration-dependent induction of TNF-α release, demonstrating LPS-induced inflammation (Fig. 1A).
Fig. 1.
LPS exposure induces inflammation and apoptosis in human placental explants. Placental explants were stimulated with different dosages of LPS. (A) TNF- α production increased in a concentration-dependent manner after 6 and 24 h (N = 3) exposure to LPS. ND = Not detected. Two-way ANOVA, Tukey's multiple comparisons test (B) Caspase 3/7 activity from placental explants (N = 3) is significantly increased after 24 h of 200 ng/ml LPS stimulation, two-way ANOVA, Tukey's multiple comparisons test. All bars represent mean with ±SD, * = p ≤ 0.05; ** = p ≤ 0.01, *** = p ≤ 0.001, **** = p ≤ 0.0001.
To determine the effect of LPS-induced inflammation on cell death, the induction of apoptosis was evaluated by measuring caspase 3/7 activation in the inflamed placental explants (treated with 200 ng/ml LPS for 6 or 24 h) relative to vehicle-treated control samples (N = 3, Fig. 1B). A significant increase in caspase 3/7 activation was seen in the LPS-treated placental explants at 24 h. These data demonstrate that LPS-mediated inflammation induced apoptosis by 24 h (Fig. 1B).
3.2. LPS-induced inflammation and cell death does not increase cff-DNA production by placental explants
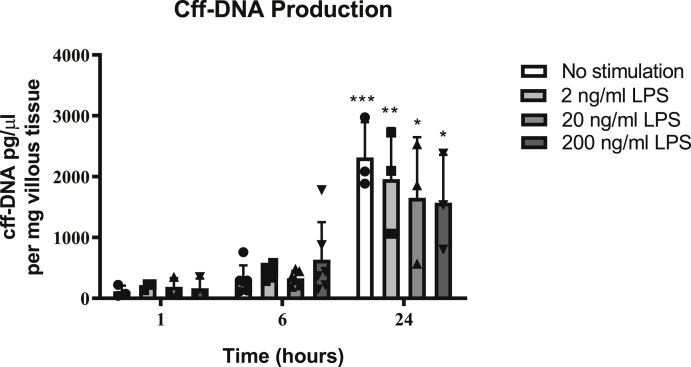
Unstimulated placental explants showed a significant increase in cff-DNA release at 24 h, compared to the 1-h samples (N = 3), but this was observed to be irrespective of LPS stimulation (Fig. 2). In order to more closely evaluate the effect of LPS at a time point prior to high spontaneous cff-DNA generation, concentration response experiments were repeated at 6-h (N = 6, Fig. 2). No significant cff-DNA release was seen at any of the concentrations of LPS used at 6 h, despite the pro-inflammatory cytokine response.
Fig. 2.
LPS-induced inflammation and cell death does not increase cff-DNA production in human placental explants. Placental explants were stimulated with various doses of LPS and supernatant was harvested for cff-DNA quantification. cff-DNA production (measured by quantifying the SRY gene) significantly increased at 24 h, irrespective of LPS stimulation, compared to 1 h non-stimulated samples, two-way ANOVA, Tukey's multiple comparisons test (N = 3 at 1/24 h and N = 6 at 6 h). NS = no stimulation. One-way ANOVA. Bars represent mean with ±SD, * = p ≤ 0.05; ** = p ≤ 0.01, *** = p ≤ 0.001, **** = p ≤ 0.0001.
Therefore, the cff-DNA generated in this model was produced independent of LPS-induced apoptosis.
3.3. Placental explant cff-DNA does not induce inflammation in human PBMCs from pregnant women
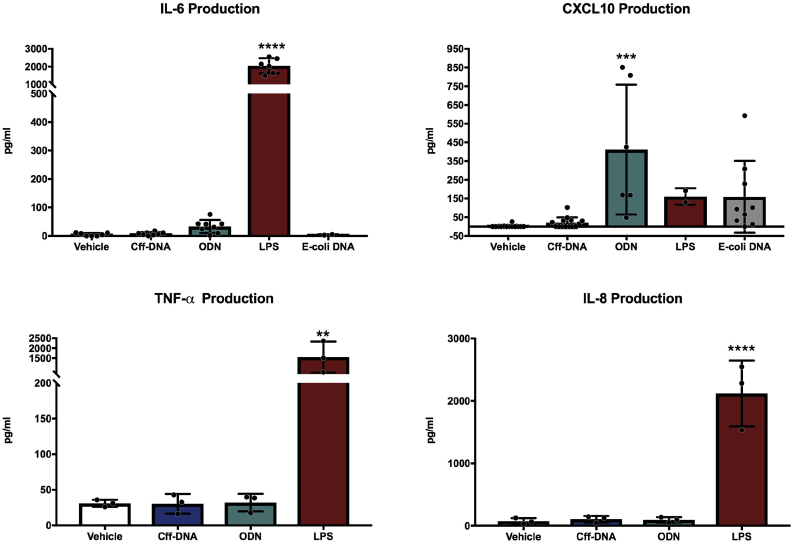
To assess the capacity of primary human placental explant cff-DNA to elicit an inflammatory response, PBMCs isolated from pregnant donors were cultured for 18 h in the presence of 500 ng/ml cff-DNA, generated ex-vivo from unstimulated cultured placental explants after 24 h of culture. Dosages above 500 ng/ml were not used because this would be not physiological in maternal blood [35], a dose response was performed for 5 ng/ml, 50 ng/ml and 500 ng/ml, where lower dosages did not produce an inflammatory response. TLR9-agonist CpG ODN (ODN; 1 μg/ml), E. coli DNA (1 μg/ml) or LPS (500 ng/ml) were used as positive controls. PBMCs stimulated with cff-DNA showed no difference in the levels of IL-6, CXCL10, TNF-α, or IL-8 produced, compared to vehicle-treated controls. In contrast, PBMCs stimulated with LPS showed a significant increase in IL-6, TNF-α and IL-8 production and ODN induced significant CXCL10 production (Fig. 3). This demonstrates that primary human placental explant cff-DNA does not induce an inflammatory response in PBMCs from pregnant women.
Fig. 3.
Human placental explant cff-DNA does not induce inflammatory responses in PBMCs from pregnant women. PBMCs from pregnant women were exposed to placental explant cff-DNA (500 ng/ml), CpG ODN (ODN; 1 μg/ml), E. coli DNA (1 μg/ml) or LPS (500 ng/ml) for 18 h before determination of cellular responses by ELISA assessment of supernatant. Sample size between 3 and 14, variable for different chemokines and stimulations. Data represent means ± SD. One-way ANOVA with Dunnett's multiple comparison test was performed for all cytokines and chemokines; **, p ≤ 0.01, ***, p ≤ 0.001, ****, p ≤ 0.0001 compared to vehicle.
3.4. Primary human placental explant cff-DNA is not hypomethylated
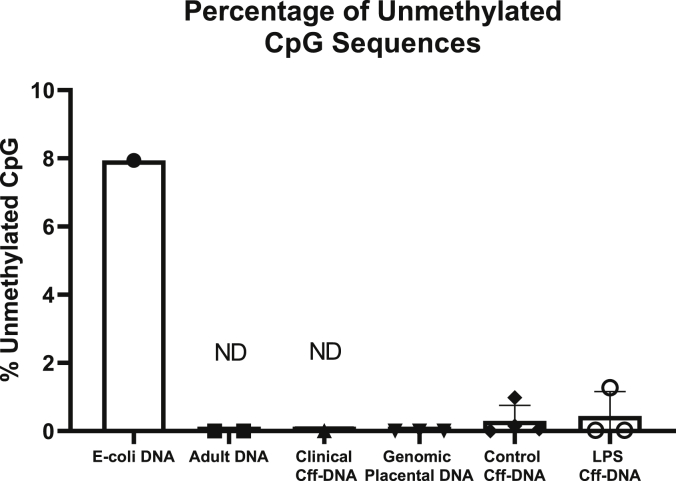
In order to determine the methylation status of the cff-DNA generated by our model, we quantified the proportion of CpG islands that were hypomethylated on the primary human placental explant cff-DNA, when compared to placental genomic DNA, adult human genomic DNA, clinically acquired cff-DNA (from three pregnant women between 39 and 40 weeks of gestation), and E. coli DNA (Fig. 4). This was calculated by quantifying total methylation of CpG sequences in the DNA samples after use of the M.SssI methylation enzyme, and subtracting total amount of pre-exisiting methylation quantified in the untreated sample. These analyses demonstrated that ~8% of the CpG motifs on E. coli DNA (a known TLR9 ligand) were unmethylated at baseline. In contrast, there was no evidence that cff-DNA (both from placental explant origin and pooled from the plasma of three pregnant women), or placental or adult genomic DNA, was significantly differentially methylated before or after treatment with M.SssI methylation enzyme. Thus, no evidence of hypomethylation was found on cff-DNA, making it unlikely to be an effective TLR9 agonst. These data are therefore compatible with our experiments demonstrating that cff-DNA is not proinflammatory. Furthermore, we demonstrate that cff-DNA acquired from LPS-exposed placental explants did not have ay significant alteration to the methylation composition. These data confirm that E. coli DNA is heavily unmethylated compared to human DNA, but failed to show cff-DNA being hypomethylated when compared to adult genomic DNA.
Fig. 4.
Primary human placental explant cff-DNA is not hypomethylatedDNA utilized was from E. Coli (E. coli DNA), genomic DNA from adult human blood leukocytes (Adult DNA), cff-DNA pooled from the blood of three pregnant women (clinical cff-DNA), genomic DNA extracted from primary placental explants (genomic placental DNA), or cff-DNA from placental explants, either untreated (control cff-DNA) or treated with 200 ng/ml LPS (LPS cff-DNA). Results are shown as percentage of total DNA CpG. Cff-DNA = cell-free Fetal DNA, ND = Not detected, values that were less than 0%.
4. Discussion
This study demonstrates that, in a primary human placental explant model; placental inflammation does not increase cff-DNA production, nor alter the methylation composition of cff-DNA, and that cff-DNA from human placental origin does not elicit a pro-inflammatory response from PBMCs from pregnant women. These findings suggest that placental-derived cff-DNA alone is not pro-inflammatory and that placental inflammation is unlikely to account for the increase in cff-DNA seen in obstetric complications associated with placental inflammation such as pre-eclampsia and preterm birth.
To our knowledge, this is the first study to use placental-derived cff-DNA generated from primary human placental explants as a stimulant on PBMCs from pregnant women. cff-DNA from term placentae were chosen, as mouse term placental DNA has been previously described to have the shortest telomere lengths compared to earlier in gestation [23] and telomere fragments have shown to affect the ability of DNA to elicit inflammation [23]. The cff-DNA used in our studies was released by placental explants and extracted using a specific cell-free DNA extraction kit. It was therefore proposed to be comparable to cff-DNA observed in systemic circulation during pregnancy [8,36]. The methylation profile of this placental explant cff-DNA was found to be comparable to cff-DNA collected directly from the blood of pregnant women. However samples had to be pooled from three different pregnant women to enable this analysis, and the yield was too low to enable testing of the inflammatory potential of cff-DNA from pregnant women in parallel with placental explant cffDNA; a limitation of our study. New techniques increasing the recovery of cff-DNA are being developed, and thus future studies may be able to use clinical cff-DNA in their experiments [37].
We examined the effects of LPS on inflammation, apoptosis and cff-DNA production using a placental explant culture. LPS was chosen as this is known to be a potent inducer of inflammation in placental explants by causing an increase in IL-6, IL-1β, IL-1α and TNF-α in term placental explants [38]. In placental explants, LPS has shown to cause a proliferation in placental resident macrophages, also known as Hofbauer cells [38], that can elicit a potent inflammatory response [39]. LPS has been used in various in vivo models for eliciting preterm birth [40,41] and recently, the experimental benefit of LPS in pre-eclampsia models has been emphasized, where LPS caused a pre-eclampsia-like phenotype in rats [42]. However, other clinically relevant inducers of inflammation and their effect on cff-DNA release, including bacteria or uric acid should be explored in future studies.
LPS-induced apoptosis was seen by an increase in caspase 3/7 activity. However, this method is limited due to the inability to demonstrate where apoptosis was taking place. cff-DNA is released primarily from the synctiotrophoblast layer of the placenta [2], and our methods cannot demonstrate the presence or absence of apoptosis in this site. This might explain partly, despite seeing general inflammation-induced apoptosis, we did not see an increase in cff-DNA release.
These experiments were limited to three individual placental samples as this sample size was sufficient to show a statistically significant increase in inflammation and apoptosis. However, no effect was seen on cff-DNA production. A subsequent power analysis demonstrated that 32 cases would be needed to demonstrate a difference between untreated placental explants and LPS treated explants, however, this number was not feasible to obtain in this study. Atmospheric oxygen was used for the placental explant culture, however, future studies should allow for adjusted oxygen levels as recent studies have shown that these levels of oxygen can favour pro-inflammatory and apoptotic transcriptomic responses [43]. In our study, all three samples were from women who gave birth to male babies, as this would ensure the ability to quantify cff-DNA by using the clinically established method of quantifying cff-DNA through qRT-PCR quantification of the SRY-gene. However, studies have demonstrated the LPS exposure to placental trophoblasts may have a sexually dismorphic effect [44], and this should be taken into account when interpreting these results. Finally, work using placental explant culture models has its limitations, as it is an ex vivo model, and future work should consider experiments using placental organoids, which is a promising model for future obstetric experimental work [45].
Our findings conflict with findings from Scharfe and colleagues who found that human genomic fetal DNA caused increased IL-6 production in PBMCs from pregnant women [21]. The different type of DNA used may explain this discrepancy. The choice for the DNA used in our experiments related to our hypothesis that this would be the most similar to the cff-DNA found in the maternal circulation. Whole genomic fetal DNA used by Scharfe and colleagues has less biological relevance, as whole genomic DNA is not readily found in the circulation in that composition [46] moreover, cff-DNA is predominantly from placental origin and not from the fetus itself [2]. In addition, the demographics (e.g. gestational age) of the pregnant women from which PBMC were obtained were not reported by Scharfe et al., and may be significant to outcomes. Gestational age and pregnancy complications can affect PBMC composition and the corresponding inflammatory response [47,48]. Furthermore, unlike the PBMCs from non-pregnant women, the effect of CpG on IL-6 production of the PBMCs from pregnant women was not reported [21]. To further analyse the response of DNA we also looked at the interferon-induced chemokine CXCL10, as 1 μg/ml of E. coli DNA can elicit a CXCL10 response from PBMCs [49]. Indeed, the CXCL10 response to cff-DNA is highly important in the context of placental inflammation, as CXCL10 is higher in the circulation of pregnant women with pre-eclampsia compared to uncomplicated pregnancies [29] and that CXC chemokines are involved in the pathogenesis of pre-eclampsia [30]. However, primary placental explant cff-DNA was found not to induce CXCL10.
The mechanisms that underpin the generation of cff-DNA in pregnancy remain to be determined, as does the contribution of placental inflammation, due to sterile inflammation or caused by infection, to this process [50,51]. LPS has been shown to increase apoptosis rates in placental trophoblasts [52] and mediate cff-DNA release from mouse placental explants [33]. In our placental explant model, using non-labouring term human placentas, we found that LPS-treated placental explants produced TNF-α in a concentration-dependent manner and significantly induced apoptosis by 24 h. However, we found no evidence that LPS-treated placental explants released higher amounts of cff-DNA compared to control samples. This suggests that LPS-induced inflammation and apoptosis alone may not be sufficient to cause a clinically relevant increases in cff-DNA. Indeed, pre-eclampsia is associated with significant amounts of placental necrosis, a cell death mechanism due to physical or chemical cell injury resulting in cell swelling and release of intracellular contents, compared to normal placentae [53]. Recently, pyroptosis, a form of cell death associated with inflammasome activation, has been found in decidual stromal cells from patients who developed preterm birth associated with inflammation and infection [54]. Future studies may therefore evaluate the role of necrosis and pyroptosis and their effect on cff-DNA release from placental explants.
Using methylation restriction enzymes, human fetal DNA [21] and mouse placental DNA [23], have been shown to have hypomethylated DNA segments present, unlike adult DNA. This difference has been proposed as a key factor in their ability to elicit inflammation [24,55]. It is notoriously difficult to quantify the percentage of hypomethylated DNA present in a sample and methylation restriction enzymes require high levels of DNA (often not achievable from clinical cff-DNA samples) and are unable to quantify the amount of methylation is present in a sample [56]. We sought to quantify the percentage of hypomethylated DNA present in various samples and compare this to a confirmed TLR9 ligand, E. coli DNA. We did this by using a method that can quantify the percentage of methylation present in a sample. However, the amount of methylation present in a sample does not represent the percentage of hypomethylation. Therefore, we quantified the amount of methylation present in our DNA after treatment with a methylation enzyme, used to methylate any hypomethylated sections of our DNA samples, and subtracted this with the untreated DNA samples. This yields an indication of the amount of hypomethylation present in a DNA sample, but has a limitation that it cannot detect very small amounts of hypomethylation. This may explain the fact that many samples in our study had no hypomethylated DNA detected (see Fig. 4). Neverthles, in keeping with data from whole genome bisulfite sequencing, we demonstrate that cff-DNA, generated from placental explants and from pregnant women, does not differ in methylation from adult genomic DNA [57]. Importantly, we demonstrate that, when compared to E. coli DNA, there are very few hypomethylated segments present, and that LPS-induced inflammation does not alter the methylation status of cff-DNA, thereby not promoting its pro-inflammatory properties in that manner. Together with the data from our PBMC stimulations, it is plausible that this cff-DNA is not sufficiently hypomethylated to be pro-inflammatory, and to stimulate TLR9.
In summary, cff-DNA is found in increasing amounts in the maternal circulation during pregnancy, but the regulation of this and the pro-inflammatory affects are not well understood. Based on our findings we propose that cff-DNA alone is not pro-inflammatory to PBMCs from pregnant women. This suggests that cff-DNA itself does not contribute to inflammation that can be seen in obstetric complications. Similarly, we show that the rise in cff-DNA measured in women with placental inflammation may not directly be caused by placental inflammation.
Contribution of authors
Sara R van Boeckel designed and conducted the experiments, interpreted the data and wrote the manuscript.
Heather MacPherson contributed to the experimental design, interpretation of data and writing of the manuscript.
Jane E Norman contributed to the writing of the manuscript.
Donald J Davidson contributed to experimental design, interpretation of data and writing of the manuscript.
Sarah J Stock designed the experiments, contributed to the interpretation of data and writing of the manuscript.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
This research was made possible by funding from Tommy's Baby Charity and the Medical Research Council Centre for Reproductive Health (MR/N022556/1).
DJD was funded by an MRC Senior non-clinical Fellowship (G1002046).
SJS is funded by Wellcome Trust Clinical Career Development Fellowships (209560/Z/17/Z).
References
- 1.Lo Y.M., Corbetta N., Chamberlain P.F., Rai V., Sargent I.L., Redman C.W. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 2.Bianchi D.W. Circulating fetal DNA: its origin and diagnostic potential-a review. Placenta. 2004;25(Suppl A):S93–S101. doi: 10.1016/j.placenta.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Hahn S., Huppertz B., Holzgreve W. Fetal cells and cell free fetal nucleic acids in maternal blood: new tools to study abnormal placentation? Placenta. 2005;26(7):515–526. doi: 10.1016/j.placenta.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Masuzaki H., Miura K., Yoshiura K.I., Yoshimura S., Niikawa N., Ishimaru T. Detection of cell free placental DNA in maternal plasma: direct evidence from three cases of confined placental mosaicism. J. Med. Genet. 2004;41(4):289–292. doi: 10.1136/jmg.2003.015784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy A., Zhong X.Y., Rusterholz C., Hahn S., Holzgreve W., Redman C.W. The effect of labour and placental separation on the shedding of syncytiotrophoblast microparticles, cell-free DNA and mRNA in normal pregnancy and pre-eclampsia. Placenta. 2008;29(11):942–949. doi: 10.1016/j.placenta.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 6.Orozco A.F., Jorgez C.J., Horne C., Marquez-Do D.A., Chapman M.R., Rodgers J.R. Membrane protected apoptotic trophoblast microparticles contain nucleic acids: relevance to preeclampsia. Am. J. Pathol. 2008;173(6):1595–1608. doi: 10.2353/ajpath.2008.080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong M., Chamley L.W. Placental extracellular vesicles and feto-maternal communication. Cold Spring Harb. Perspect. Med. 2015;5(3):a023028. doi: 10.1101/cshperspect.a023028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer M., Hutterer G., Eder M., Majer S., Leshane E., Johnson K.L. A prospective analysis of cell-free fetal DNA concentration in maternal plasma as an indicator for adverse pregnancy outcome. Prenat. Diagn. 2006;26(9):831–836. doi: 10.1002/pd.1513. [DOI] [PubMed] [Google Scholar]
- 9.Birch L., English C.A., O'Donoghue K., Barigye O., Fisk N.M., Keer J.T. Accurate and robust quantification of circulating fetal and total DNA in maternal plasma from 5 to 41 weeks of gestation. Clin. Chem. 2005;51(2):312–320. doi: 10.1373/clinchem.2004.042713. [DOI] [PubMed] [Google Scholar]
- 10.Farina A., LeShane E.S., Romero R., Gomez R., Chaiworapongsa T., Rizzo N. High levels of fetal cell-free DNA in maternal serum: a risk factor for spontaneous preterm delivery. Am. J. Obstet. Gynecol. 2005;193(2):421–425. doi: 10.1016/j.ajog.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 11.Dugoff L., Barberio A., Whittaker P.G., Schwartz N., Sehdev H., Bastek J.A. Cell-free DNA fetal fraction and preterm birth. Am. J. Obstet. Gynecol. 2016;215(2):e1–e7. doi: 10.1016/j.ajog.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Jakobsen T.R., Clausen F.B., Rode L., Dziegiel M.H., Tabor A. High levels of fetal DNA are associated with increased risk of spontaneous preterm delivery. Prenat. Diagn. 2012;32(9):840–845. doi: 10.1002/pd.3917. [DOI] [PubMed] [Google Scholar]
- 13.van Boeckel S.R., Davidson D.J., Norman J.E., Stock S.J. Cell-free fetal DNA and spontaneous preterm birth. Reproduction. 2018;155(3):R137–R145. doi: 10.1530/REP-17-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadeau-Vallée M., Obari D., Palacios J., Brien M., Duval C., Chemtob S. Sterile inflammation and pregnancy complications: a review. Reproduction. 2016;152(6):R277–R292. doi: 10.1530/REP-16-0453. [DOI] [PubMed] [Google Scholar]
- 15.Yu H., Shen Y., Ge Q., He Y., Qiao D., Ren M. Quantification of maternal serum cell-free fetal DNA in early-onset preeclampsia. Int. J. Mol. Sci. 2013;14(4):7571–7582. doi: 10.3390/ijms14047571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldenberg R.L., Culhane J.F., Iams J.D., Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371(9606):75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burton G.J., Redman C.W., Roberts J.M., Moffett A. Pre-eclampsia: pathophysiology and clinical implications. BMJ. 2019;366:l2381. doi: 10.1136/bmj.l2381. [DOI] [PubMed] [Google Scholar]
- 18.Lamont R.F. Infection in the prediction and antibiotics in the prevention of spontaneous preterm labour and preterm birth. BJOG. 2003;110(Suppl 20):71–75. doi: 10.1016/s1470-0328(03)00034-x. [DOI] [PubMed] [Google Scholar]
- 19.Romero R., Espinoza J., Gonçalves L.F., Kusanovic J.P., Friel L.A., Nien J.K. Inflammation in preterm and term labour and delivery. Semin. Fetal Neonatal Med. 2006;11(5):317–326. doi: 10.1016/j.siny.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasmussen S., Ebbing C., Irgens L.M. Predicting preeclampsia from a history of preterm birth. PloS One. 2017;12(7) doi: 10.1371/journal.pone.0181016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scharfe-Nugent A., Corr S.C., Carpenter S.B., Keogh L., Doyle B., Martin C. TLR9 provokes inflammation in response to fetal DNA: mechanism for fetal loss in preterm birth and preeclampsia. J. Immunol. 2012;188(11):5706–5712. doi: 10.4049/jimmunol.1103454. [DOI] [PubMed] [Google Scholar]
- 22.Peng X.L., Jiang P. Bioinformatics approaches for fetal DNA fraction estimation in noninvasive prenatal testing. Int. J. Mol. Sci. 2017;18(2) doi: 10.3390/ijms18020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillippe M. Cell-free fetal DNA, telomeres, and the spontaneous onset of parturition. Reprod. Sci. 2015;22(10):1186–1201. doi: 10.1177/1933719115592714. [DOI] [PubMed] [Google Scholar]
- 24.Vollmer J. TLR9 in health and disease. Int. Rev. Immunol. 2006;25(3–4):155–181. doi: 10.1080/08830180600743107. [DOI] [PubMed] [Google Scholar]
- 25.Phillippe M. Cell-free fetal DNA--a trigger for parturition. N. Engl. J. Med. 2014;370(26):2534–2536. doi: 10.1056/NEJMcibr1404324. [DOI] [PubMed] [Google Scholar]
- 26.Goulopoulou S., Wenceslau C.F., McCarthy C.G., Matsumoto T., Webb R.C. Exposure to stimulatory CpG oligonucleotides during gestation induces maternal hypertension and excess vasoconstriction in pregnant rats. Am. J. Physiol. Heart Circ. Physiol. 2016;310(8):H1015–H1025. doi: 10.1152/ajpheart.00834.2015. [DOI] [PubMed] [Google Scholar]
- 27.Fűri I., Kalmár A., Wichmann B., Spisák S., Schöller A., Barták B. Cell free DNA of tumor origin induces a 'Metastatic' expression profile in HT-29 cancer cell line. PloS One. 2015;10(7) doi: 10.1371/journal.pone.0131699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enninga E.A., Nevala W.K., Holtan S.G., Markovic S.N. Immune reactivation by cell-free fetal DNA in healthy pregnancies Re-purposed to target tumors: novel checkpoint inhibition in cancer therapeutics. Front. Immunol. 2015;6:424. doi: 10.3389/fimmu.2015.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gotsch F., Romero R., Friel L., Kusanovic J.P., Espinoza J., Erez O. CXCL10/IP-10: a missing link between inflammation and anti-angiogenesis in preeclampsia? J. Matern. Fetal Neonatal Med. 2007;20(11):777–792. doi: 10.1080/14767050701483298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darakhshan S., Hassanshahi G., Mofidifar Z., Soltani B., Karimabad M.N. CXCL9/CXCL10 angiostasis CXC-chemokines in parallel with the CXCL12 as an angiogenesis CXC-chemokine are variously expressed in pre-eclamptic-women and their neonates. Pregnancy Hypertens. 2019;17:36–42. doi: 10.1016/j.preghy.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Herrera-Muñoz A., Fernández-Alonso A.M., Fischer-Suárez N., Chedraui P., Pérez-López F.R. Maternal serum cytokine levels in pregnancies complicated with threatened preterm labour. Gynecol. Endocrinol. 2017;33(5):408–412. doi: 10.1080/09513590.2017.1284786. [DOI] [PubMed] [Google Scholar]
- 32.Ejima K., Koji T., Tsuruta D., Nanri H., Kashimura M., Ikeda M. Induction of apoptosis in placentas of pregnant mice exposed to lipopolysaccharides: possible involvement of Fas/Fas ligand system. Biol. Reprod. 2000;62(1):178–185. doi: 10.1095/biolreprod62.1.178. [DOI] [PubMed] [Google Scholar]
- 33.Phillippe M., Adeli S. Cell-free DNA release by mouse placental explants. PloS One. 2017;12(6) doi: 10.1371/journal.pone.0178845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Utaisincharoen P., Anuntagool N., Chaisuriya P., Pichyangkul S., Sirisinha S. CpG ODN activates NO and iNOS production in mouse macrophage cell line (RAW 264.7) Clin. Exp. Immunol. 2002;128(3):467–473. doi: 10.1046/j.1365-2249.2002.01866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taglauer E.S., Wilkins-Haug L., Bianchi D.W. Review: cell-free fetal DNA in the maternal circulation as an indication of placental health and disease. Placenta. 2014;35(Suppl):S64–S68. doi: 10.1016/j.placenta.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jain M., Balatsky A.V., Revina D.B., Samokhodskaya L.M. Direct comparison of QIAamp DSP Virus Kit and QIAamp Circulating Nucleic Acid Kit regarding cell-free fetal DNA isolation from maternal peripheral blood. Mol. Cell. Probes. 2019;43:13–19. doi: 10.1016/j.mcp.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 37.Stray J., Zimmermann B. Isolation of cell-free DNA from maternal plasma. Methods Mol. Biol. 2019;1885:309–323. doi: 10.1007/978-1-4939-8889-1_21. [DOI] [PubMed] [Google Scholar]
- 38.Duval C., Brien M.E., Gaudreault V., Boufaied I., Baker B., Jones R.L. Differential effect of LPS and IL-1β in term placental explants. Placenta. 2019;75:9–15. doi: 10.1016/j.placenta.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Reyes L., Golos T.G. Hofbauer cells: their role in healthy and complicated pregnancy. Front. Immunol. 2018;9:2628. doi: 10.3389/fimmu.2018.02628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rinaldi S.F., Makieva S., Frew L., Wade J., Thomson A.J., Moran C.M. Ultrasound-guided intrauterine injection of lipopolysaccharide as a novel model of preterm birth in the mouse. Am. J. Pathol. 2015;185(5):1201–1206. doi: 10.1016/j.ajpath.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyle A.K., Rinaldi S.F., Rossi A.G., Saunders P.T.K., Norman J.E. Repurposing simvastatin as a therapy for preterm labor: evidence from preclinical models. Faseb. J. 2018 doi: 10.1096/fj.201801104R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan M., Li X., Gao X., Dong L., Xin G., Chen L. LPS induces preeclampsia-like phenotype in rats and HTR8/SVneo cells dysfunction through TLR4/p38 MAPK pathway. Front. Physiol. 2019;10:1030. doi: 10.3389/fphys.2019.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brew O., Sullivan M.H.F. Oxygen and tissue culture affect placental gene expression. Placenta. 2017;55:13–20. doi: 10.1016/j.placenta.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 44.Yeganegi M., Watson C.S., Martins A., Kim S.O., Reid G., Challis J.R. Effect of Lactobacillus rhamnosus GR-1 supernatant and fetal sex on lipopolysaccharide-induced cytokine and prostaglandin-regulating enzymes in human placental trophoblast cells: implications for treatment of bacterial vaginosis and prevention of preterm labor. Am. J. Obstet. Gynecol. 2009;200(5) doi: 10.1016/j.ajog.2008.12.032. 532.e1-8. [DOI] [PubMed] [Google Scholar]
- 45.Turco M.Y., Gardner L., Kay R.G., Hamilton R.S., Prater M., Hollinshead M.S. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature. 2018;564(7735):263–267. doi: 10.1038/s41586-018-0753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hahn S., Giaglis S., Buser A., Hoesli I., Lapaire O., Hasler P. Cell-free nucleic acids in (maternal) blood: any relevance to (reproductive) immunologists? J. Reprod. Immunol. 2014;104–105:26–31. doi: 10.1016/j.jri.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 47.Mor G., Cardenas I. The immune system in pregnancy: a unique complexity. Am. J. Reprod. Immunol. 2010;63(6):425–433. doi: 10.1111/j.1600-0897.2010.00836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rajakumar A., Chu T., Handley D.E., Bunce K.D., Burke B., Hubel C.A. Maternal gene expression profiling during pregnancy and preeclampsia in human peripheral blood mononuclear cells. Placenta. 2011;32(1):70–78. doi: 10.1016/j.placenta.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGlasson S.L., Semple F., MacPherson H., Gray M., Davidson D.J., Dorin J.R. Human β-defensin 3 increases the TLR9-dependent response to bacterial DNA. Eur. J. Immunol. 2017;47(4):658–664. doi: 10.1002/eji.201646799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cappelletti M., Della Bella S., Ferrazzi E., Mavilio D., Divanovic S. Inflammation and preterm birth. J. Leukoc. Biol. 2016;99(1):67–78. doi: 10.1189/jlb.3MR0615-272RR. [DOI] [PubMed] [Google Scholar]
- 51.Gomez-Lopez N., StLouis D., Lehr M.A., Sanchez-Rodriguez E.N., Arenas-Hernandez M. Immune cells in term and preterm labor. Cell. Mol. Immunol. 2014;11(6):571–581. doi: 10.1038/cmi.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asagiri K., Nakatsuka M., Konishi H., Noguchi S., Takata M., Habara T. Involvement of peroxynitrite in LPS-induced apoptosis of trophoblasts. J. Obstet. Gynaecol. Res. 2003;29(1):49–55. doi: 10.1046/j.1341-8076.2003.00066.x. [DOI] [PubMed] [Google Scholar]
- 53.Roberts J.M., Escudero C. The placenta in preeclampsia. Pregnancy Hypertens. 2012;2(2):72–83. doi: 10.1016/j.preghy.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gomez-Lopez N., Romero R., Tarca A.L., Miller D., Panaitescu B., Schwenkel G. Gasdermin D: evidence of pyroptosis in spontaneous preterm labor with sterile intra-amniotic inflammation or intra-amniotic infection. Am. J. Reprod. Immunol. 2019 doi: 10.1111/aji.13184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bauer S. Toll-erating self DNA. Nat. Immunol. 2006;7(1):13–15. doi: 10.1038/ni0106-13. [DOI] [PubMed] [Google Scholar]
- 56.Kurdyukov S., Bullock M. DNA methylation analysis: choosing the right method. Biology. 2016;5(1) doi: 10.3390/biology5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jensen T.J., Kim S.K., Zhu Z., Chin C., Gebhard C., Lu T. Whole genome bisulfite sequencing of cell-free DNA and its cellular contributors uncovers placenta hypomethylated domains. Genome Biol. 2015;16:78. doi: 10.1186/s13059-015-0645-x. [DOI] [PMC free article] [PubMed] [Google Scholar]