Abstract
vanZ, a member of the VanA glycopeptide resistance gene cluster, confers resistance to lipoglycopeptide antibiotics independent of cell wall precursor modification by the vanHAX genes. Orthologs of vanZ are present in the genomes of many clinically relevant bacteria, including Enterococcus faecium and Streptococcus pneumoniae; however, vanZ genes are absent in Staphylococcus aureus. Here, we show that the expression of enterococcal vanZ paralogs in S. aureus increases the minimal inhibitory concentrations of lipoglycopeptide antibiotics teicoplanin, dalbavancin, oritavancin and new teicoplanin pseudoaglycone derivatives. The reduction in the binding of fluorescently labeled teicoplanin to the cells suggests the mechanism of VanZ-mediated resistance. In addition, using a genomic vanZ gene knockout mutant of S. pneumoniae, we have shown that the ability of VanZ proteins to compromise the activity of lipoglycopeptide antibiotics by reducing their binding is a more general feature of VanZ-superfamily proteins.
Keywords: Staphylococcus aureus, Streptococcus pneumoniae, antibiotic resistance, lipoglycopeptide antibiotics, VanZ
Introduction
Glycopeptide antibiotics are important for the treatment of multidrug-resistant infections caused by gram-positive bacteria. The emergence and spread of enterococcal strains resistant to vancomycin and teicoplanin (VRE) is a serious public health concern (Uttley et al., 1989). In VRE-resistant strains, cell wall biosynthesis is reprogramed to produce peptidoglycan precursors containing either D-alanine-D-lactate (D-Ala-D-Lac) or D-alanine-D-serine instead of the dipeptide D-alanine-D-alanine (D-Ala-D-Ala). As a result, the affinity of glycopeptide antibiotics to peptidoglycan dramatically decreases (Bugg et al., 1991; Lessard and Walsh, 1999). The three essential enzymes responsible for the precursor modification are encoded in vanHAX gene clusters. In the vanA-type gene cluster, two additional genes, vanY and vanZ, also contribute to glycopeptide resistance. VanY, a D, D-carboxypeptidase, eliminates D-Ala-D-Ala from peptidoglycan precursors, minimizing the number of primary binding sites for glycopeptide antibiotics (Arthur et al., 1994). VanZ decreases the sensitivity of Enterococcus faecalis to teicoplanin and oritavancin, but not vancomycin, independent of peptidoglycan modification (Arthur et al., 1995, 1999). The mechanism of VanZ-mediated resistance is not known. In addition to the vanA gene cluster, VanZ orthologs not associated with vancomycin resistance gene clusters are present in the genomes of clinically relevant bacteria, such as Bacillus, Streptococcus, Enterococcus, and Clostridium.
In this work, we compared the ability of vanZ from the vanA gene cluster encoded on Tn1546, which is present in various strains of Enterococcus faecium (vanZTei) (Arthur et al., 1995), and its paralog encoded in the chromosome of E. faecium Aus0004 (vanZg, locus tag: EFAU004_00030) to confer resistance to glycopeptide antibiotics in Staphylococcus aureus, which naturally does not encode vanZ. To further confirm the involvement of vanZ genes in lipoglycopeptide resistance, we studied the effect of the vanZ deletion in Streptococcus pneumoniae.
Materials and Methods
Strains
E. faecium Aus0004, S. aureus RN4220, S. aureus ATCC29213, S. pneumoniae R6, Escherichia coli XL1-Blue.
Antibiotics
Teicoplanin, vancomycin, oritavancin, and chloramphenicol (Sigma-Aldrich, Germany); dalbavancin (MedChemExpress, Sweden); MA79 (Csávás et al., 2015), ERJ390 (Pintér et al., 2009), and SZZS-12 (Szucs et al., 2017); carbenicillin, gentamicin and erythromycin (Duchefa Biochemie, Netherland); vancomycin BODIPY-FL conjugate (Thermo Fisher Scientific, Germany) and fluorescently labeled teicoplanin (Vimberg et al., 2019).
Preparation of Plasmids Expressing vanZTei and vanZg
The vanZTei and vanZg genes and their ribosome binding sites were amplified from the plasmid pAT398 (Arthur et al., 1995) and E. faecium Aus0004 chromosomal DNA, respectively, using the primers TecVanZ_SacI_F, TecVanZ_R, gVanZ_SacI_F, and gVanZ_R (Supplementary Table S1). The PCR products were cloned into the pRMC2 shuttle vector under the control of the anhydrotetracycline (AnhTet)-inducible promoter Pxyl/tetO using SacI and EcoRI restriction sites, resulting in the constructs pRMC2:vanZTei and pRMC2:vanZg. The constructs were confirmed by sequencing and then electroporated into S. aureus RN4220. Using the same procedure, we prepared the constructs pRMC2:vanZTei-His and pRMC2:vanZg-His encoding C-terminal His-tagged VanZ variants. However, primers TecVanZhis_R and gVanZhis_R replaced the reverse primers TecVanZ_R and gVanZ_R.
Construction of the S. pneumoniae R6 ΔvanZ and vanZ-Reverted Strains
Strain Sp539 (ΔvanZ) was constructed using a Sweet Janus cassette-based two-step negative selection strategy (Sung et al., 2001; Li et al., 2014). The Sweet Janus cassette contains the kanamycin resistance gene, the recessive rpsL gene and the sacB gene, which confers sucrose sensitivity (SuS), as counterselectable markers. In the first step, 1000 bp fragments corresponding to the upstream and downstream flanking regions of the vanZ gene (spr0050) were amplified from the wild-type chromosomal DNA with the primer pairs KB60/KB61 and KB62/KB63, respectively. The Sweet Janus cassette (2807 bp) amplified from the Sweet Janus cassette DNA fragment with the primers DP1/DP2 was attached to the regions flanking vanZ by fusion PCR using primers KB60 and KB63. The resulting PCR fragment was used for the transformation of the S. pneumoniae R6 strain, and KanR/SuS transformants (Sp537, vanZ:kan sacB) were selected. The PCR fragments consisting of the upstream and downstream flanking regions of the vanZ gene were amplified by the KB60/KB65 and KB64/KB63 primer pairs, respectively, and fused by overlap extension using primers KB60/KB63. The resulting fragment was transformed into the Sp537 strain to obtain Sp539 (SuR/KanS). To complement the vanZ deletion, we constructed strain Sp635 (SuR/KanS; ΔvanZ:vanZ) by transforming strain Sp537 (KanR/SuS) with the PCR fragment amplified with the primers KB60 and KB63 that contained wild-type loci using R6 chromosomal DNA as a template.
Minimal Inhibitory Concentration (MIC) Measurement
Minimal Inhibitory Concentrations were measured by the broth microdilution method according to ISO standard 20776-1 (EUCAST 2019). S. aureus strains with pRMC2, pRMC2:vanZTei and pRMC2:vanZg plasmids were cultured in the presence of 25 μg/ml chloramphenicol and 100 ng/ml AnhTet (Sigma-Aldrich, Germany) to induce vanZ gene expression. All measurements were performed twice in triplicate. S. aureus ATCC29213 was used as a control. MIC values of clinically accepted glycopeptide antibiotics were interpreted according to EUCAST clinical breakpoints (EUCAST, 2019).
Western Blot Analysis of VanZ Expression
Staphylococcus aureus RN4220 strains harboring plasmids pRMC2:vanZTei-His and pRMC2:vanZg-His were grown in 2 ml of brain heart infusion medium (Oxoid/Thermo Fisher Scientific, Germany) in the presence of chloramphenicol and AnhTet overnight at 37°C. Cells were harvested and lysed in 1 ml of 1× PBS (phosphate-buffered saline) buffer with 10 μg of lysostaphin (Sigma-Aldrich, Germany) for 15 min at 37°C. Cell debris was removed by centrifugation at 16,000 × g for 30 min. The supernatant was then transferred into fresh tubes and centrifuged at 30,000 × g for 30 min to separate the membrane and cytosolic fractions. Membrane sediment was resuspended in 50 μl of 1 M urea in 1× PBS. Supernatant proteins were precipitated with 10% TCA, washed twice with ice-cold acetone and resuspended in 50 μl of 1 M urea in 1× PBS buffer. Protein concentration was determined using a bicinchoninic acid (BCA)-based protein estimation kit (Thermo Fisher Scientific, Germany). Proteins were further denatured in SDS-loading buffer at 95°C for 10 min, and 20 μl aliquots were loaded on a 12% SDS-acrylamide gel. After separation by SDS-PAGE, proteins were transferred to a PVDF membrane (Immobilon-P, Merck Millipore, United States) at 15 V for 10 min with a BioRad SemiDry blotting system. His-tagged VanZ was detected with monoclonal anti-His antibody (Sigma-Aldrich, Germany) and subsequently with a secondary goat anti-mouse IgG antibody HRP conjugate (Sigma-Aldrich, Germany). Protein abundance was measured using Immobilon Western HRP Substrate (Merck Millipore, United States), and the signal was developed using the ChemiDoc MP Imaging System (Bio-Rad).
Binding of Fluorescent Vancomycin and Teicoplanin to S. aureus and S. pneumoniae Cells
Staphylococcus aureus pRMC2, pRMC2:vanZTei and pRMC2:vanZg, S. pneumoniae R6, S. pneumoniae R6, R6ΔvanZ, and R6ΔvanZ:vanZ cells were pregrown in Mueller Hinton medium (Oxoid/Thermo Fisher Scientific, Germany) to A600nm = 0.4. S. aureus was pregrown in the presence of chloramphenicol and AnhTet. Cells were harvested by centrifugation and resuspended to A600nm = 1 in 50 mM Tris–HCl buffer (pH = 7.4). Increasing amounts of Bodipy-Vancomycin (Thermo Fisher Scientific, Germany) or Fluorescent Teicoplanin (Vimberg et al., 2019) were added to 1 ml of resuspended cells. Cells were then incubated for 10 min at room temperature with the fluorescent antibiotics, harvested by centrifugation, washed two times with 50 mM Tris–HCl buffer (pH = 7.4), and finally resuspended in 100 μl of the same buffer. The fluorescence of 70 μl of resuspended cells was measured in automatic gain mode at Ex490nm/Em520nm in the case of fluorescent vancomycin or Ex530nm/Em580nm in the case of fluorescent teicoplanin in 96-well black plates (Thermo Fisher Scientific, Germany) by Tecan Infinite 200Pro. The experiment was repeated three times in duplicate.
Results
VanZ Reduces the Susceptibility of S. aureus and S. pneumoniae to Glycopeptide Antibiotics
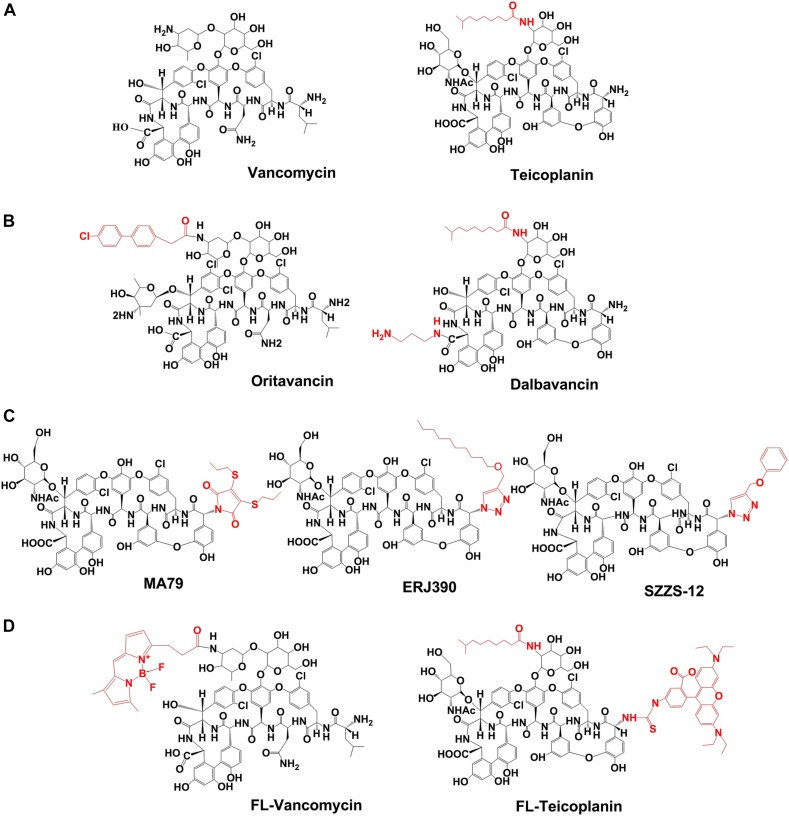
The ability of the E. faecium vanZTei and vanZg paralogs to confer resistance to glycopeptide antibiotics was tested in S. aureus, which naturally does not encode any proteins of the VanZ superfamily. In particular, we determined the susceptibility of S. aureus RN4220 expressing vanZTei and vanZg to the clinically used glycopeptide antibiotic vancomycin (VAN); the lipoglycopeptide antibiotics teicoplanin (TEI), oritavancin (ORI), and dalbavancin (DALB); and three experimental lipoglycopeptide antibiotics derived from teicoplanin pseudoaglycone: MA79 (Csávás et al., 2015), ERJ390 (Pintér et al., 2009) and SZZS-12 (Szucs et al., 2017; Figure 1). In addition, the non-glycopeptide antibiotics carbenicillin (CARB, cell wall-targeting) gentamicin (GEN, 30S ribosome-targeting) and erythromycin (ERY, 50S ribosome-targeting) were used as controls.
FIGURE 1.
Chemical structures of the glycopeptide antibiotics used in study. (A) Natural glycopeptide antibiotics, approved for the clinical use. (B) Semisynthetic lipoglycopeptide antibiotics, approved for the clinical use. (C) Semisynthetic derivatives of teicoplanin pseudoaglycone. (D) Fluorescently labeled vancomycin (FL-Vancomycin) and teicoplanin (FL-Teicoplanin). Lipophilic modifications of the glycopeptide antibiotics are shown in red.
As shown in Table 1, the expression of vanZg decreased the susceptibility of S. aureus to TEI and ERJ390 16-fold, to DALB four-fold, and to ORI and MA79 two times, and the expression of vanZg had no effect on the susceptibility of S. aureus to VAN, SZZS-12 or the control drugs. Similar to vanZg, the expression of vanZTei decreased the susceptibility of S. aureus to ERJ390 16-fold but had less or no activity against TEI, ORI and DALB. However, at the same time, cells expressing vanZTei were more active against MA79 and SZZS-12 (Table 1). To test whether different levels of protein expression cause different activities of VanZg and VanZTei, we performed Western blot analysis of the strains expressing C-terminal His-tagged versions of VanZ proteins. However, the analysis showed that both proteins were expressed at similar levels and that they were localized exclusively in the cell membrane (Supplementary Figure S1).
TABLE 1.
Summary of the MICs of glycopeptide and non-glycopeptide antibiotics against S. aureus RN4220 and S. pneumoniae R6, expressing or not expressing VanZ.
| MIC (μg/ml) | ATCC 29213 | RN4220 pRMC2 | RN4220 vanZTei | RN4220 vanZg | R6 | R6 ΔvanZ | R6 ΔvanZ:vanZ |
| VAN | 0.25 | 0.5 | 0.5 | 1 | 0.25 | 0.25 | 0.25 |
| TEI | 0.125 | 0.5 | 1 | 8 | 0.125 | 0.03125 | 0.125 |
| ORI | 0.125 | 0.125 | 0.125 | 0.25 | 0.0078 | 0.0039 | 0.0078 |
| DALB | 0.125 | 0.125 | 0.125 | 0.5 | 0.03125 | 0.0078 | 0.03125 |
| MA79 | 0.25 | 0.125 | 0.5 | 0.25 | 0.5 | 0.25 | 0.5 |
| ERJ390 | 0.125 | 0.0156 | 0.25 | 0.25 | 0.5 | 0.125 | 0.5 |
| SZZS-12 | 0.125 | 0.0156 | 0.0625 | 0.0156 | 0.5 | 0.125 | 0.5 |
| CARB | 0.25 | 0.25 | 0.25 | 0.25 | 0.0625 | 0.0625 | 0.0625 |
| GEN | 1 | 1 | 1 | 1 | 0.25 | 0.25 | 0.25 |
| ERY | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 |
VAN, vancomycin; TEI, teicoplanin; ORI, oritavancin; DALB, dalbavancin; CARB, carbenicillin; GEN, gentamicin; ERY, erythromycin. MIC values demonstrating resistance to clinically accepted glycopeptide antibiotics, according to EUCAST (2019) clinical breakpoints, are marked in bold. MIC values of the experimental glycopeptide antibiotics that increased due to vanZ expression are underlined. Decreased MIC values against S. pneumoniae R6ΔvanZ in comparison to wild-type S. pneumoniae R6 are in italics.
To further explore the effect of VanZ proteins on the resistance to glycopeptide antibiotics in the natural genetic background, we employed S. pneumoniae R6, which encodes the VanZ ortholog encoded by genome locus spr0050. We constructed a clean knockout of vanZ (ΔvanZ) and complemented vanZ in the S. pneumoniae R6 genome and tested the susceptibility of the strains to antibiotics. According to MIC measurements, the ΔvanZ mutant was up to four-fold more sensitive to lipoglycopeptide antibiotics, but not to VAN or non-glycopeptide antibiotics, than the wild-type strain (Table 1). Altogether, these data indicate that the transmembrane proteins VanZg and VanZTei, as well as VanZ from S. pneumoniae, decrease susceptibility to TEI and its derivatives, while they have no or a minor effect on VAN and its derivative ORI.
VanZ Decreases the Binding of FL-Teicoplanin to S. aureus and S. pneumoniae
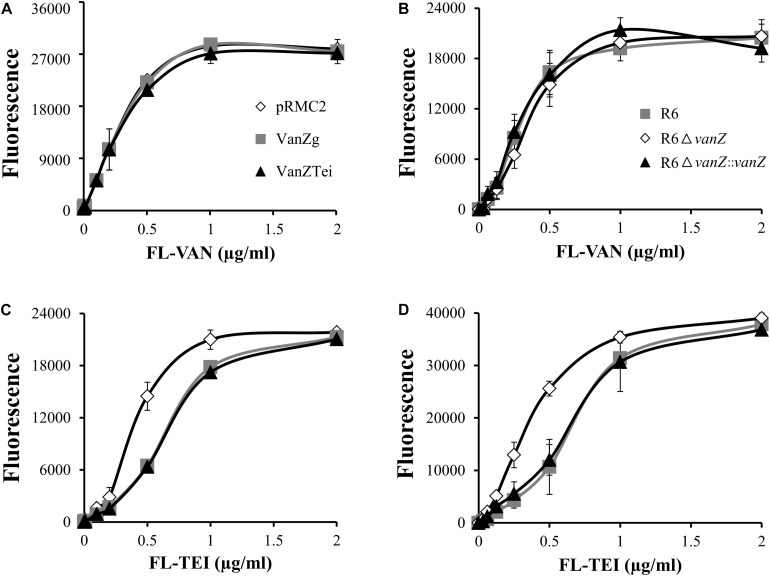
To determine whether the expression of VanZ might interfere with the binding of lipoglycopeptide antibiotics to the bacterial surface, we followed the binding of fluorescently labeled VAN and TEI (FL-VAN and FL-TEI) to S. aureus expressing vanZTei and vanZg, as well as to S. pneumoniae R6 wild-type, ΔvanZ and reverted strains. The titration curves of FL-VAN binding clearly showed that the presence of VanZ did not affected FL-VAN binding to S. aureus or S. pneumoniae cells (Figures 2A,B). On the other hand, FL-TEI bound less efficiently to S. aureus with VanZg or VanZTei than to cells without VanZ (Figure 2C). Similarly, S. pneumoniae R6 ΔvanZ was saturated with a lower amount of FL-TEI than the wild type (Figure 2D). Altogether, this experiment demonstrates that VanZ proteins might indeed affect the binding of lipoglycopeptide antibiotics to cells.
FIGURE 2.
Binding of fluorescent-vancomycin (FL-VAN) and fluorescent-teicoplanin (FL-TEI) to S. aureus RN4220, expressing vanZg and vanZTei (A,C) or to S. pneumoniae R6, R6ΔvanZ, and R6ΔvanZ:vanZ (B,D). FL-VAN and FL-TEI were titrated to the S. aureus and S. pneumoniae cells grown to exponential growth phase.
Discussion
Here, we showed that orthologous vanZ genes from E. faecium and S. pneumoniae decreased susceptibility to lipoglycopeptide antibiotics independent of their origins, genetic contexts, and host background. Surprisingly, when expressed in S. aureus, enterococcal genomic vanZg, which is not associated with any glycopeptide resistance gene cluster, was more efficient in conferring resistance to TEI than vanZTei (Table 1). In addition to E. faecium and S. pneumoniae, the involvement of genomic vanZ genes in the resistance to lipoglycopeptide antibiotics was reported for orthologs from Streptococcus suis and Clostridium difficile (Lai et al., 2017; Woods et al., 2018; Supplementary Figure S2). In addition to resistance, VanZ proteins might play a more general role in stress response and virulence, as it was observed that the expression of the streptococcal vanZ gene was induced by the epithelial antimicrobial peptide LL37 (Lai et al., 2017) and ribosome-targeting antibiotics (Ng et al., 2003) or was essential for lung infection (Hava and Camilli, 2002).
The VanZ-like family (PF04892) comprises a large number of transmembrane proteins of unknown function. Today, more than seven thousand VanZ family representatives can be found in the Pfam protein families database1. A phylogenetic tree constructed from 415 VanZ seed sequences, including five VanZ orthologs with activity against glycopeptides, showed that they belong to the same phylogenetic group (Supplementary Figure S3). We hypothesize that all these related VanZ proteins might mediate lipoglycopeptide antibiotic resistance.
Each of the vanZ genes conferred various levels of resistance to lipoglycopeptides but did not decrease susceptibility to VAN, and they had only a minor effect on the susceptibility to ORI (Table 1). Correspondingly, vanZ expression reduced the binding of FL-TEI but not FL-VAN to the cell surface (Figure 2). The hydrophobic moieties of TEI and DALB are thought to anchor the molecule to the bacterial membrane, thereby improving binding to the lipid II substrate (Beauregard et al., 1995; Kerns et al., 2000; Zeng et al., 2016). On the other hand, the hydrophobic substituent of ORI does not form a membrane anchor; instead, it is an essential part of the secondary binding to pentaglycyl bridge segments of the cell-wall peptidoglycan (Kim et al., 2013, 2017). Thus, VanZ proteins seem to affect the anchoring of lipoglycopeptides to a membrane rather than their binding to the peptidoglycan.
It is of great concern that VanZ orthologs were active against TEI pseudoaglycon derivatives, which represent the newest generation of lipoglycopeptides with promising in vitro activity against glycopeptide-resistant strains (Szucs et al., 2017). Similar to ORI, these derivatives show equal competition with FL-TEI and FL-VAN for binding to S. aureus cells, and this result correlates with their activity against vanHAX-mediated resistance (Vimberg et al., 2019). Nevertheless, whether the hydrophobic substituents interact with a membrane or with the peptidoglycan needs to be determined for these compounds.
The acquisition and spread of vanZ genes in S. aureus could become a critical problem. The vanZTei, gene, as a part of the vanA gene cluster, is occasionally transferred from enterococci into S. aureus, leading to highly vancomycin-resistant strains (VRSA) (Chang et al., 2003; Perichon and Courvalin, 2009; Saadat et al., 2014). Although the incidence of such an event remains low, apparently due to the high fitness cost of vanHAX-mediated resistance in S. aureus (Foucault et al., 2009), VRSA strains may represent progenitors for the generation of vanZTei-carrying mobile genetic elements.
Conclusion
In conclusion, our data indicate that VanZ family proteins protect bacteria from lipoglycopeptide antibiotics by affecting their binding to the cell surface. Considering that lipophilization of glycopeptides is an effective way to increase their activity, VanZ superfamily proteins commonly found in the genomes of relevant bacteria as well as the horizontal transfer of vanZ to vanZ-deficient strains represent a potential threat to the activity of the new generation of glycopeptide antibiotics.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
VV designed and performed the MIC measurements, fluorescence, and western-blot experiments. LZ constructed VanZ expressing plasmids and performed MIC measurements. KB constructed vanZ knockout in S. pneumoniae and its complementation and contributed to data interpretation and manuscript preparation. PB designed the S. pneumoniae experiments, interpreted the data, and contributed to the manuscript preparation. GB designed S. aureus experiments, interpreted data, and wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Pal Herczegh for the gift of the new semisynthetic antibiotics MA79, ERJ390, and SZZS-12. We also thank T. Foster for the gift of the pRMC2 plasmid.
Funding. This work was supported by the Czech Health Research Council (project no. NV15-28807A to GB), Czech Science Foundation (Grant 19-03269S to PB), the Institutional Research Concept (Grant RVO 61388971), and BIOCEV – Biotechnology and Biomedicine Center of the Academy of Sciences and Charles University (no. CZ.1.05/1.1.00/02.0109) through the European Regional Development Fund in the Czech Republic.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00566/full#supplementary-material
References
- Arthur M., Depardieu F., Molinas C., Reynolds P., Courvalin P. (1995). The vanZ gene of Tn1546 from Enterococcus faecium BM4147 confers resistance to teicoplanin. Gene 154 87–92. 10.1016/0378-1119(94)00851-i [DOI] [PubMed] [Google Scholar]
- Arthur M., Depardieu F., Reynolds P., Courvalin P. (1999). Moderate-level resistance to glycopeptide LY333328 mediated by genes of the vanA and vanB clusters in enterococci. Antimicrob. Agents Chemother. 43 1875–1880. 10.1128/aac.43.8.1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M., Depardieu F., Snaith H. A., Reynolds P. E., Courvalin P. (1994). Contribution of vanY D, D-carboxypeptidase to glycopeptide resistance in Enterococcus faecalis by hydrolysis of peptidoglycan precursors. Antimicrob. Agents Chemother. 38 1899–1903. 10.1128/aac.38.9.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauregard D., Williams D. H., Gwynn M. N., Knowles D. J. (1995). Dimerization and membrane anchors in extracellular targeting of vancomycin group antibiotics. Antimicrob. Agents Chemother. 39 781–785. 10.1128/aac.39.3.781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugg T. D. H., Wright G. D., Dutkamalen S., Arthur M., Courvalin P., Walsh C. T. (1991). Molecular-basis for vancomycin resistance in Enterococcus faecium BM4147 – biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30 10408–10415. 10.1021/bi00107a007 [DOI] [PubMed] [Google Scholar]
- Chang S., Sievert D. M., Hageman J. C., Boulton M. L., Tenover F. C., Downes F. P., et al. (2003). Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348 1342– 1347. [DOI] [PubMed] [Google Scholar]
- Csávás M., Miskovics A., Szcs Z., Röth E., Nagy Z. L., Bereczki I., et al. (2015). Synthesis and antibacterial evaluation of some teicoplanin pseudoaglycon derivatives containing alkyl-and arylthiosubstituted maleimides. J. Antibiot. (Tokyo) 68 579–585. 10.1038/ja.2015.33 [DOI] [PubMed] [Google Scholar]
- Foucault M. L., Courvalin P., Grillot-Courvalin C. (2009). Fitness cost of VanA-type vancomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53 2354–2359. 10.1128/AAC.01702-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hava D. L., Camilli A. (2002). Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45 1389–1406. 10.1046/j.1365-2958.2002.03106.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns R., Dong S. D., Fukuzawa S., Carbeck J., Kohler J., Silver L., et al. (2000). The role of hydrophobic substituents in the biological activity of glycopeptide antibiotics. J. Am. Chem. Soc. 122 12608–12609. 10.1021/ja0027665 [DOI] [Google Scholar]
- Kim S. J., Singh M., Sharif S., Schaefer J. (2017). Desleucyl-oritavancin with a damaged d -Ala- d -Ala binding site inhibits the transpeptidation step of cell-wall biosynthesis in whole cells of Staphylococcus aureus. Biochemistry 56 1529–1535. 10.1021/acs.biochem.6b01125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Tanaka K. S. E., Dietrich E., Rafai Far A., Schaefer J. (2013). Locations of the hydrophobic side chains of lipoglycopeptides bound to the peptidoglycan of Staphylococcus aureus. Biochemistry 52 3405–3414. 10.1021/bi400054p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L., Dai J., Tang H., Zhang S., Wu C., Qiu W., et al. (2017). Streptococcus suis serotype 9 strain GZ0565 contains a type VII secretion system putative substrate EsxA that contributes to bacterial virulence and a vanZ-like gene that confers resistance to teicoplanin and dalbavancin in Streptococcus agalactiae. Vet. Microbiol. 205 26–33. 10.1016/j.vetmic.2017.04.030 [DOI] [PubMed] [Google Scholar]
- Lessard I. A. D., Walsh C. T. (1999). VanX, a bacterial D-alanyl-D-alanine dipeptidase: resistance, immunity, or survival function? Proc. Natl. Acad. Sci. U.S.A. 96 11028–11032. 10.1073/pnas.96.20.11028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Thompson C. M., Lipsitch M. (2014). A modified Janus cassette (sweet Janus) to improve allelic replacement efficiency by high-stringency negative selection in Streptococcus pneumoniae. PLoS One 9:e100510. 10.1371/journal.pone.0100510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W. L., Kazmierczak K. M., Robertson G. T., Gilmour R., Winkler M. E. (2003). Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 185 359–370. 10.1128/jb.185.1.359-370.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perichon B., Courvalin P. (2009). VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53 4580–4587. 10.1128/AAC.00346-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintér G., Batta G., Kéki S., Mándi A., Komáromi I., Takács-Novák K., et al. (2009). Diazo transfer-click reaction route to new, lipophilic teicoplanin and ristocetin aglycon derivatives with high antibacterial and anti-influenza virus activity: an aggregation and receptor binding study. J. Med. Chem. 52 6053–6061. 10.1021/jm900950d [DOI] [PubMed] [Google Scholar]
- Saadat S., Solhjoo K., Norooz-Nejad M. J., Kazemi A. (2014). vanA and vanB positive vancomycin-resistant Staphylococcus aureus among clinical isolates in Shiraz, south of Iran. Oman Med. J. 29 335–339. 10.5001/omj.2014.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung C. K., Li H., Claverys J. P., Morrison D. A. (2001). An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67 5190–5196. 10.1128/aem.67.11.5190-5196.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szucs Z., Csávás M., Röth E., Borbás A., Batta G., Perret F., et al. (2017). Synthesis and biological evaluation of lipophilic teicoplanin pseudoaglycon derivatives containing a substituted triazole function. J. Antibiot. (Tokyo) 70 152–157. 10.1038/ja.2016.80 [DOI] [PubMed] [Google Scholar]
- Uttley A. H. C., George R. C., Naidoo J., Woodford N., Johnson A. P., Collins C. H., et al. (1989). High-level vancomycin-resistant enterococci causing hospital infections. Epidemiol. Infect. 103 173–181. 10.1017/s0950268800030478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimberg V., Gazak R., Szücs Z., Borbás A., Herczegh P., Cavanagh J. P., et al. (2019). Fluorescence assay to predict activity of the glycopeptide antibiotics. J. Antibiot. (Tokyo) 72 114–117. 10.1038/s41429-018-0120-5 [DOI] [PubMed] [Google Scholar]
- Woods E. C., Wetzel D., Mukerjee M., McBride S. M. (2018). Examination of the Clostridioides (Clostridium) difficile VanZ ortholog, CD1240. Anaerobe 53 108–115. 10.1016/j.anaerobe.2018.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D., Debabov D., Hartsell T. L., Cano R. J., Adams S., Schuyler J. A., et al. (2016). Approved glycopeptide antibacterial drugs: mechanism of action and resistance. Cold Spring Harb. Perspect. Med 6:a026989. 10.1101/cshperspect.a026989 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.