Abstract
N6-methyladenosine (m6A) is a well-known post-transcriptional modification that is the most common type of methylation in eukaryotic mRNAs. The regulation of m6A is dynamic and reversible, which is erected by m6A methyltransferases (“writers”) and removed by m6A demethylases (“erasers”). Notably, the effects on targeted mRNAs resulted by m6A predominantly depend on the functions of different m6A-binding proteins (“readers”) including YT521-B homology (YTH) domain family, heterogeneous nuclear ribonucleoproteins (HNRNPs), and insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs). Indeed, m6A readers not only participate in multiple procedures of RNA metabolism, but also are involved in a variety of biological processes. In this review, we summarized the specific functions and underlying mechanisms of m6A-binding proteins in tumorigenesis, hematopoiesis, virus replication, immune response, and adipogenesis.
Keywords: N6-methyladenosine, m6A-binding proteins, Cancer, Virus, Immunity, Adipogenesis
Introduction
Epigenetic abnormalities, such as DNA methylation, histone modification, genomic imprinting, and chromosome remodeling, mainly affect the characteristics and functions of genes through regulating the transcription or translation processes [1], without altering the DNA sequences. These changes in gene expression are stable during cell self-renewal, division, and differentiation. Over the past decades, more and more attention has been paid to RNA modification with the help of high-throughput sequencing. To date, more than 100 types of modifications have been confirmed in various RNAs, including messenger RNAs (mRNAs), transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), microRNAs (miRNAs), and long non-coding RNAs (lncRNAs). Notably, N6-methyladenosine (m6A), a well-known post-transcriptional modification first discovered and proposed in 1974, has been regarded as the most prevalent methylation in eukaryotic mRNAs [2, 3]. It is estimated that about 0.1–0.4% of all adenines are specially modified by m6A in mRNAs [4]. m6A usually appears at the RRACH sequences (R = A, G, or U; R = A or G; and H = A, C, or U) [5, 6], and mostly enriches in the 3′ untranslated regions (3’ UTRs) and near stop codons [7].
The regulation of m6A modification is dynamic and reversible (Table 1). It is established by m6A methyltransferases (also called “writers”), such as methyltransferase-like protein 3 (METTL3) [8, 9], METTL14 [8, 9] and Wilms tumor 1-associated protein (WTAP) [10]. And it is removed by m6A demethylases (also called “erasers”), containing fat-mass and obesity-associated protein (FTO) [16] and α-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5) [17]. More importantly, the effects of m6A modification on RNA metabolism predominantly depend on the recognition by different m6A-binding proteins (also called “readers”), including but not limited to YT521-B homology (YTH) domain family, heterogeneous nuclear ribonucleoproteins (HNRNPs), and insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs).
Table 1.
The specific function of each m6A-related enzyme
| Categories | m6A-related enzymes | Location | Mechanism | References |
| m6A writers | METTL3 | Nucleus | Catalyzing methyl-group transfer | [8, 9] |
| METTL14 | Nucleus | Forming a heterodimer with METTL3 and strengthening its catalytic activity | [8, 9] | |
| WTAP | Nucleus | Promoting METTL3-METTL14 complex localization to nuclear speckles and modulating their recruitment to RNA targets | [10] | |
| VIRMA | Nucleus | Preferentially mediating m6A modification in the 3’UTR and near stop codon, and affecting the selection of methylation sites | [11] | |
| RBM15/RBM15B | Nucleus | Mediating m6A methylation of lncRNA XIST | [12] | |
| ZC3H13 | Nucleus | Inducing the nuclear localization of Zc3h13-WTAP- Virilizer-Hakai complex | [13] | |
| METTL16 | Nucleus | Functioning as a conserved U6 snRNA methyltransferase and regulating the abundance of intracellular SAM | [14, 15] | |
| m6A erasers | FTO | Nucleus | Abrogating the m6A levels of targeted RNA via the oxidative demethylation activity | [16] |
| ALKBH5 | Nucleus | Removing the m6A modification of nuclear RNA | [17] | |
| m6A readers | YTHDF1 | Cytoplasm | Augmenting RNA translation through interacting with the translation initiation factor eIF3 | [18] |
| YTHDF2 | Cytoplasm | Promoting RNA degradation by recruiting the CCR4-NOT deadenylase complex | [19, 20] | |
| YTHDF3 | Cytoplasm | Not only promoting the translation of methylated RNA in cooperation with YTHDF1, but also strengthening RNA decay mediated by YTHDF2 | [21, 22] | |
| YTHDC1 | Nucleus | Mediating alternative splicing, facilitating m6A-methylated RNA nuclear export, and promoting X chromosome genes transcriptional silencing mediated by XIST | [12, 23, 24] | |
| YTHDC2 | Cytoplasm | Increasing the translation efficiency of RNA | [25, 26] | |
| HNRNPA2B1 | Nucleus | Accelerating the processing of primary miRNA, regulating alternative splicing, and acting as a “m6A-switch” | [27, 28] | |
| HNRNPC | Nucleus | Participating in the pre-mRNA processing and functioning as a “m6A-switch” | [29, 30] | |
| HNRNPG | Nucleus | Modulating pre-mRNA alternative splicing and acting as a “m6A-switch” | [31] | |
| IGF2BP1 | Cytoplasm | Fortifying RNA stability | [32] | |
| IGF2BP2 | Cytoplasm | Increasing the stability of RNA | [32] | |
| IGF2BP3 | Cytoplasm | Facilitating RNA stabilization | [32] |
The typical m6A methyltransferase complex mainly consists of METTL3, METTL14, and WTAP. METTL3, a core component with methyltransferase activity, can combine with S-adenosyl methionine (SAM) and then mediate RNA methylation in the nucleus [8, 9]. However, METTL14 is not a subunit to directly catalyze the methyl-group transfer. It functions as an RNA-binding platform to form a stable heterodimer with METTL3, eventually strengthening the catalytic effect of METTL3 [8, 9]. Interestingly, WTAP interacts with METTL3-METTL14 complex to not only promote their localization to nuclear speckles, but also modulate their recruitment to mRNA targets [10]. Moreover, there are a lot of regulatory factors binding to the catalytic complex, such as vir-like m6A methyltransferase associated (VIRMA, also termed as KIAA1429), RNA-binding motif protein 15/15B (RBM15/RBM15B), and zinc finger CCCH domain-containing protein 13 (ZC3H13). VIRMA preferentially mediates m6A modification in the 3′ UTR and near stop codon, affecting the selection of methylation sites [11]. RBM15/RBM15B play an important role in the X-inactivation and gene silencing through mediating the m6A methylation of lncRNA XIST [12]. ZC3H13 induces the nuclear localization of Zc3h13-WTAP-Virilizer- Hakai complex to regulate m6A methylation and trigger the self-renewal of embryonic stem cells [13]. It is noteworthy that novel methyltransferases are gradually discovered. For instance, METTL16, a conserved U6 snRNA methyltransferase, participates in the regulation of intracellular SAM abundance via methylating the SAM synthetase gene MAT2A and controlling its intron retention [14, 15].
Currently, only two demethylases have been identified, which both belong to AlkB family proteins. The first one is FTO that is located in the nucleus and abrogates the m6A levels via the oxidative demethylation activity [16]. Moreover, another demethylase ALKBH5 can also remove the m6A modification of nuclear RNA, and further modulate nuclear RNA export, RNA metabolism, and gene expression [17].
The YTH domain is found in 174 different proteins in eukaryotes [33], with a size from 100 to 150 amino acids. It is featured by 14 invariant and 19 highly conserved residues [34], and contains a structure of four α helices and six β strands [35]. Interestingly, the six β strands shape a β barrel and then stabilize the hydrophobic core through combining with α helices [35]. The recognized YTH domain family consists of YTH domain family protein 1-3 (YTHDF1-3, DF family) and YTH domain containing protein 1-2 (YTHDC1-2, DC family). YTHDF2, the first identified m6A reader, promotes mRNAs degradation and reduces the stability of targeted transcripts through recruiting the CCR4-NOT deadenylase complex [19, 20]. On the contrary, YTHDF1 augments mRNAs translation by interacting with the translation initiation factor eIF3 rather than by the m7G-cap-dependent manner [18]. Intriguingly, it has been reported that there are a great number of common targets that YTHDF3 shares with YTHDF1 and YTHDF2 [21]. Functionally, YTHDF3 not only cooperates with YTHDF1 to promote the translation of methylated mRNAs, but also strengthens mRNAs decay mediated by YTHDF2, showing a cooperative relationship among YTHDF proteins. YTHDF3 recognizes the m6A-containg mRNAs and increases their expression through combining with 40S and 60S ribosomal subunits [21, 22]. Moreover, YTHDF3 can remarkably facilitate protein synthesis of YTHDF1/3 common targets, but not YTHDF3 unique targets. Notably, YTHDF3 depletion decreases the binding of YTHDF1 and YTHDF2 to their target transcripts, while YTHDF1 or YTHDF2 loss reduces the amount of RNA that is bound by YTHDF3 [21]. It shows the important role of YTHDF3 in the RNA binding specificity and YTHDF1/2 in the RNA binding affinity. YTHDC1 is widely distributed in the nucleus and its YTH domain is in complex with the RNA 5-mer GG(m6A)CU to form a crystal structure [35]. The 3′ terminal nucleotides are stabilized via the interactions of cation-π-π; however, the 5′ terminal nucleotides remain flexible [36]. The function of YTHDC1 is to mediate alternative splicing by recruiting RNA splicing factor SRSF3 and blocking SRSF10 from binding to mRNAs [23]. YTHDC1 also interacts with SRSF3 and NXF1 to facilitate the m6A-methylated mRNAs nuclear export [24]. Furthermore, YTHDC1 plays a promoting role in the X chromosome genes transcriptional silencing mediated by XIST [12]. YTHDC2, a putative RNA helicase, contains YTH domain, helicase domain, R3H domain, and ankyrin repeats [37], which is of great significance to increase the translation efficiency of mRNAs [25, 26].
Besides, there are members of HNRNP family who have the potential to identify the m6A modification of mRNAs, including heterogeneous nuclear ribonucleoproteins A2/B1 (HNRNPA2B1), heterogeneous nuclear ribonucleoproteins C (HNRNPC) and heterogeneous nuclear ribonucleoproteins G (HNRNPG). The binding of HNRNP proteins to m6A sites can be enhanced via the mRNAs structural alteration induced by m6A methylation [31, 38, 39], which is called as “m6A-switch”. HNRNPA2B1 is a nuclear m6A-binding protein that accelerates the processing of primary miRNAs (pri-miRNAs) through interacting with DGCR8 protein in an m6A-dependent manner [27, 28]. Moreover, HNRNPA2B1 possesses a capability of modulating the alternative splicing of transcripts [27]. HNRNPC, a RNA-binding protein in nucleus, plays an important role in the pre-mRNAs processing [29, 30]. HNRNPG is an m6A reader that selectively binds RNAs via Arg-Gly-Gly (RGG) motifs in the low-complexity region [31]. HNRNPG modulates pre-mRNAs alternative splicing by interacting with both the phosphorylated carboxy-terminal domain of RNA polymerase II (RNAPII) and m6A-methylated pre-mRNAs in a RGG-region-dependent manner [31].
Additionally, IGF2BPs are the distinct and conserved m6A-binding proteins, whose RNA-binding domains consist of four K homology (KH) domains and two RNA recognition motif (RRM) domains [40]. However, only the third and fourth KH domains (KH3-4) are indispensable in recognizing the m6A sites of mRNAs. Functionally, IGF2BP1-3 fortify the stability and increase the translation efficiency of m6A-modified mRNAs [32]. And the mRNA stabilization mediated by IGF2BPs may be strengthened via recruiting the co-factors of IGF2BP1-3, including ELAV-like RNA binding protein 1 (ELAVL1, also called HuR) and matrin 3 (MATR3) [32].
In a word, m6A methylation participates in multiple procedures throughout the life cycle of mRNAs, such as alternative splicing, translation, translocation, and degradation. Recent years have witnessed a remarkable advance in understanding the roles of m6A modification in a variety of biological processes, particularly m6A readers. Herein, we provided an updated review to summarize the functions and mechanisms of m6A-binding proteins in tumorigenesis, hematopoiesis, viral replication, immunity, and adipogenesis.
The role of m6A-binding proteins in human solid cancers
Hepatocellular carcinoma (HCC)
Hypoxia is a common characteristic in various solid cancers, acting as a promoter in tumorigenesis via the regulation by hypoxia-inducible factor (HIF) [41]. A study reported by Zhong and colleagues showed that HIF1α-dependent hypoxia downregulated the expression of YTHDF2 in HCC cells, and that YTHDF2 depletion promoted tumor cells proliferation [42]. Mechanistically, EGFR, the upstream transcript of ERK/MAPK pathway, was modified by m6A and recognized by YTHDF2. Then YTHDF2 destabilized EGFR by facilitating mRNA decay, which in turn suppressed the ERK/MAPK signaling pathway. However, Hou et al. found that HIF2α-mediated hypoxia abrogated YTHDF2 expression in HCC cells, and that YTHDF2 deficiency facilitated inflammation, vasculature reconstruction, and metastasis through decreasing the degradation of m6A-marked interleukin-11 (IL-11) and serpin family E member 2 (SERPINE2) mRNAs which were two crucial factors in the processes of inflammation-induced malignancy and vascular remodeling [43]. SOCS2, a negative regulator of JAK/STAT pathway, was identified as a tumor suppressor of HCC. There was a direct binding between YTHDF2 and the m6A sites of SOCS2 mRNA in HCC [44]. Knockdown of YTHDF2 remarkably decreased the silencing of SOCS2 and increased its expression.
Notably, the function of YTHDF1 in the progression of HCC has also been reported. Epithelial-mesenchymal transition (EMT), a critical step of tumor cells metastasis, is a process in which epithelial tumor cells lose junction proteins and cell polarity [45, 46]. And Snail is a key transcription factor of EMT. In HCC cells, YTHDF1 could dramatically accelerate the translation of Snail mRNA via an m6A-dependent manner, thus contributing to tumor metastasis [47].
Additionally, Huang et al. demonstrated that IGF2BPs could preferentially recognize and bind to the m6A sites at the coding region instability determinant (CRD) of oncogene MYC in HCC [32]. Loss of IGF2BPs substantially reduced the expression of MYC mRNA and then inhibited HCC cells proliferation, migration, and colony formation ability. Furthermore, serum response factor (SRF) could promote cell proliferation, invasion, and metastasis of HCC. And IGF2BP1 increased SRF expression in a conservative manner, which was strictly dependent on the m6A modification at the 3′ UTR [48].
Colorectal cancer (CRC)
There was a study exploring the relationship between YTHDF1 and CRC. In CRC, YTHDF1 could promote cell proliferation and invasion [49]. Silencing of YTHDF1 significantly downregulated the expression of cancer stem cell markers but notably upregulated the enterocyte markers expression, suggesting that YTHDF1 was involved in modulating the stem cell-like activity in CRC. Mechanistically, YTHDF1 took advantage of the m6A-dependent mRNA translation to not only increase the β-catenin expression and its nuclear signaling activity, but also facilitate the expression of downstream targets of Wnt/β-catenin pathway, including WNT6 and FZD9 [49].
Interestingly, Ni et al. discovered a negative functional loop of lncRNA GAS5/YAP/YTHDF3 axis in CRC [50]. GAS5 could promote YAP to translocate from nucleus to cytoplasm, and facilitate YAP phosphorylation, ubiquitination and degradation, thus inhibiting CRC progression. And YAP targeted at the promoter of YTHDF3, leading to an increase of YTHDF3 expression. Then, YTHDF3 recognized m6A-containing GAS5 and accelerated its degradation, further affecting the activity of YAP signaling pathway in CRC [50].
Moreover, circNSUN2 is a circular RNA that facilitates CRC cells metastasis and aggressiveness, and contains m6A methylation at the exon 5-exon 4 junction site [51]. On the one hand, YTHDC1 promoted circNSUN2 export from nucleus to cytoplasm. On the other hand, the KH3-4 di-domain of IGF2BP2 interacted with circNSUN2 and then form a circNSUN2/IGF2BP2 complex to fortify the stability of HMGA2 that was a RNA inducing EMT and promoting CRC liver metastasis [51]. In addition, SOX2, an important gene to promote tumor initiation and metastasis, exhibited an elevated m6A level [52]. And IGF2BP2 could directly bind to the methylated SOX2 and dramatically enhance the stability of SOX2 mRNA by preventing it being degraded.
Gastric cancer (GC)
In GC, SEC62 overexpression promoted cell proliferation and inhibited cell apoptosis. And IGF2BP1 specially recognized the m6A-modified SEC62 mRNA and augmented its expression at both mRNA and protein levels [53]. Furthermore, another member of IGF2BP proteins IGF2BP3 could directly bind to the m6A sites of HDGF mRNA and then improved HDGF stability by enhancing the m6A-associated mRNA translation [54]. And HDGF has an ability of facilitating tumor angiogenesis. Knockdown of IGF2BP3 markedly reduced the expression of HDGF mRNA and suppressed tumor growth in GC.
Lung cancer
Pentose phosphate pathway (PPP) that is catalyzed by glucose-6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (6PGD) provides tumor cells the ribose-5-phosphate and NADPH. In lung cancer, YTHDF2 depletion could increase lactate production, and decrease oxidative PPP flux and NADPH/NADP+ ratio [55], indicating that YTHDF2 participated in lung cancer cells metabolism and affected tumor cells growth. Mechanistically, YTHDF2 significantly increased the translation of 6PGD, which was dependent on the m6A methylation sites of 6PGD mRNA.
Intriguingly, a recent research reported the different expression levels of YTHDF1 during disparate circumstances [56]. YTHDF1 was upregulated under normoxia but downregulated under hypoxia conditions. When non-small cell lung cancer (NSCLC) cells were in a normoxia state, overexpression of YTHDF1 promoted tumor cells proliferation and xenograft tumor formation by augmenting the translations of m6A-marked CDK2, CDK4, and cyclinD1, three key regulators in the G0/G1 cell cycle transition. However, when NSCLC cells were treated with cisplatin, a chemotherapy drug inducing the reactive oxygen species (ROS) accumulation, YTHDF1 was decreased and led to cisplatin resistance in tumor cells via regulating the Keap1-Nrf2-AKR1C1 axis [56].
Notably, m6A-binding proteins are observed to be associated with hippo signaling pathway effector YAP/TAZ. Jin et al. demonstrated that YTHDF1/3 complex, together with eIF3b, recognized m6A-containing YAP and upregulated its expression level through interacting with the translation initiation complex, thus boosting NSCLC cells growth, invasion, and migration [57]. Moreover, MALAT1 which was modified by m6A and then stabilized by the METTL3/YTHDF3 complex, sponged miR-1914-3p to further promote YAP expression [57]. Another important finding was that METTL3 facilitated lung cancer cells growth and invasion by functioning like an m6A reader [58]. METTL3 accelerated the translation of m6A-marked EGFR and TAZ through directly recruiting eIF3 to the translation initiation complex rather than relying on the methyltransferase activity or other m6A readers.
Bladder cancer (BC)
ITGA6 acted as a tumor promoter to facilitate BC cells growth and progression in vitro and in vivo. m6A enriched at ITGA6 mRNA, and was preferentially bound by YTHDF1 and YTHDF3 [59]. Knockdown of YTHDF1 and YTHDF3 abrogated the expression of ITGA6. Another study showed that BC-related oncogene CPCP1 could be marked by m6A and selectively recognized by YTHDF1 [60]. And YTHDF1 remarkably increased CDCP1 expression. Interestingly, in addition to catalyzing CDCP1 methylation, METTL3 also could bind to the m6A sites of CDCP1 and facilitate its translation, playing a role like an m6A-binding protein. Furthermore, METTL3 could accelerate the binding of YTHDF1 to CDCP1. Loss of METTL3 and YTHDF1 produced a cooperative effect on decreasing the expression of CDCP1 [60].
Endometrial cancer
In endometrial cancer, YTHDF1 and YTHDF2 regulated the expression of critical enzymes in the AKT signaling pathway via different ways [61]. PHLPP2 is a negative regulator of AKT pathway. YTHDF1 promoted the m6A-mediated translation of PHLPP2 through strengthening its binding to actively transcribing ribosomes. However, YTHDF2-mediated decay diminished the abundance of PRR5, PRR5L, and mTOR, which were three key components of mTORC2 complex, a positive regulator of AKT pathway.
Ovarian cancer
EIF3C is a subunit of EIF3 which is indispensable for protein synthesis. YTHDF1 was highly expressed in ovarian cancer and targeted at EIF3C to enhance its translation efficiency via an m6A-dependent manner; therefore, affecting the overall translational output and regulating tumor cells proliferation, migration, and invasion [62].
Cervical cancer (CC)
LncRNA GAS5-AS1 diminished the m6A level of GAS5 by interacting with m6A demethyltransferase in cervical cancer, and then decreased GAS5 degradation and increased GAS5 stability via reducing the YTHDF2-mediated decay, eventually resulting in the suppression of cancer tumorigenicity and metastasis [63].
Melanoma
In melanoma, depletion of YTHDF2 accelerated cell proliferation and migration by strikingly upregulating the mRNA levels of three key intrinsic pro-tumorigenic factors, including PD-1 (PDCD1), CXCR4, and SOX10, which was dependent on a reduced m6A-mediated mRNA decay [64]. Furthermore, YTHDF1 promoted the translation of tumor suppressor HINT2 mRNA which was methylated by m6A, thus playing a repressive role in ocular melanoma [65].
Breast cancer
BNIP3 is a pro-apoptosis gene. And YTHDF2 could recognize the m6A sites of BNIP3. As a result, the degradation of BNIP3 mRNA was increased and its expression was decreased, promoting the growth of tumor cells [66].
Pancreatic cancer
DANCR can facilitate cell proliferation and stemness-like properties. And IGF2BP2 targeted at m6A-containing DANCR to enhance its translation. Then, IGF2BP2 together with DANCR jointly boosted the tumorigenesis of pancreatic cancer [67].
The role of m6A-binding proteins in hematopoiesis
Hematopoiesis is an intricately detailed process in which immature hematopoietic stem cells (HSCs) differentiate into cellular components of peripheral blood, including leukocytes, erythrocytes, and platelets [68]. HSCs possess not only a capability of self-renewal but also a potential of multilineage differentiation, thus maintaining the homeostasis of hematological system. The sources of HSCs are commonly bone marrow (BM), umbilical cord blood (UCB), and mobilized peripheral blood (MPB) [69]. Moreover, HSCs are characterized by a variety of surface markers, such as CD34+, CD90+, Lin−, CD38−, and CD45RA− [70, 71].
Hematopoietic stem and progenitor cells (HSPCs) are initially derived from endothelial cells via a process called endothelial-to-hematopoietic transition (EHT) [72]. In EHT, Notch1 signaling pathway represses HSPCs programming and further influences the generation of HSPCs [73, 74]. A Chinese study first demonstrated that YTHDF2 regulated the production of earliest HSPCs during vertebrate embryogenesis [75]. YTHDF2 recognized the m6A peaks near the stop codons of arterial endothelial gene notch1a and promoted its mRNA decay, which in turn contributed to the inhibition of Notch1 signaling and modulated the development of HSPCs.
Besides, m6A-binding proteins are of great importance to HSCs expansion. Li et al. found that YTHDF2 knockout led to a dramatic increase in the numbers of both long-term HSCs and short-term HSCs, but not the progenitors and lineage cells [76]. Mechanistically, YTHDF2 facilitated the decay of m6A-modified mRNAs, such as TAL1, RUNX1, STAT5, and GATA2, which encoded transcription factors related with the self-renewal of stem cells. Furthermore, knockdown of YTHDF2 significantly promoted the stem cells maintenance and the human umbilical cord blood (hUCB) HSCs expansion in vitro [76]. Similarly, Wang et al. confirmed that the mRNAs clearance induced by YTHDF2 via an m6A-dependent manner was involved in the regeneration of HSCs [77]. Under transplantation and hematopoietic stress conditions, depletion of YTHDF2 increased the number of HSCs by reducing the degradation of Wnt signaling pathway downstream targets.
There is no doubt that the disorder of hematopoiesis is strongly associated with a lot of human diseases, including lymphoid and myeloid hematologic malignancies. Malignant hematopoiesis not only breaks the balance between self-renewal and differentiation of HSCs, but also puts the progenitor cells at a risk of developing leukemia [78]. So far, studies about the role of m6A readers in hematological neoplasms mainly focus on acute myeloid leukemia (AML). AML is regarded as a clonal hematopoietic dysregulation in which leukemic stem cells (LSCs) retain the self-renewal capacity, but lose the myeloid differentiation capacity [79, 80]. Paris and colleagues observed that YTHDF2 was overexpressed in AML and closely correlated with LSCs activity [81]. Functionally, YTHDF2 selectively compromised the initiation and propagation of AML, but does not impede normal hematopoiesis. Mechanistically, as a gene inhibiting the accumulation of leukemic cells, TNFR2 was marked by m6A. And its expression was regulated by the YTHDF2-mediated mRNA decay. In addition, silencing of YTHDF2 increased AML cells sensitivity to TNF-induced apoptosis. Su et al. revealed that YTHDF2 could recognize the m6A sites of MYC and CEBPA mRNAs in AML [82]. YTHDF2 loss facilitated the expression of MYC and CEBPA by increasing their stability, and also affected the sensitivity of leukemia cells to the tumor suppressor R-2-hydroxyglutarate (R-2HG).
The role of m6A-binding proteins in viruses
Kaposi’s sarcoma-associated herpesvirus (KSHV)
KSHV has two phases called latent phase and lytic replication phase. Under immunosuppressive conditions, KSHV in the latent phase is reactivated to undergo lytic replication, then producing new virus. Most transcripts encoded by KSHV were modified by m6A. Hesser et al. found that m6A modification regulated the lytic viral gene expression in a cell type-specific manner [83]. In KSHV-infected renal carcinoma cell lines, loss of YTHDF2 blocked virus lytic cycle progression and virion production through reducing the expression of m6A-modified ORF50. However, in the KSHV-positive B cell line, ORF50 expression was increased when YTHDF2 was depleted. Therefore, the same YTHDF2-mediated m6A machinery may lead to either pro- or anti-viral effects depending on the cell context, while the underlying mechanism remains unknown and demands further study.
In addition, the m6A peaks of replication transcription activator (RTA), a critical protein associating with KSHV lytic switch, could be recognized by YTHDC1 with its related splicing factors SRSF3 and SRSF10, therefore enhancing the pre-mRNA splicing of RTA [84]. Interestingly, RTA itself could conversely increase the m6A levels and promote its own pre-mRNA splicing in an m6A-dependent manner, which further facilitated lytic gene expression and replication. Tan et al. revealed that YTHDF2 could bind to RTA, ORF57, and ORF-K8 lytic transcripts in KSHV, and decrease their expression and half-life via the m6A-associated RNA decay [85].
Human immunodeficiency virus (HIV)
Tirumuru et al. have established a genome-wide mapping of m6A modification within HIV-1 RNA in several HIV-1-infected cells. The m6A peaks predominantly located at the 3′ UTR, 5′ UTR and some coding genes of the HIV-1 genome [86]. During the reverse transcription phase, YTHDF1-3 proteins could bind to the m6A-modified HIV-1 RNAs and facilitate their degradation. Therefore, YTHDFs suppressed HIV-1 infection through blocking the viral reverse transcription. Moreover, m6A methylation of HIV-1 RNA modulated by m6A writers or erasers was crucial for the effective HIV-1 protein synthesis [86]. Then, the underlying mechanisms of YTHDFs-induced HIV-1 infection inhibition were further explored [87]. In infected target cells, it was identified that YTHDFs preferentially interacted with m6A-marked viral genomic RNA (gRNA) at 5′ UTR and abolished the level of gRNA. As a consequence, both early and late reverse transcription products were decreased, which contributed to the impairment of HIV-1 replication and infectivity. Meanwhile, in virus-producing cells, endogenous YTHDFs and HIV-1 Gag protein constituted a complex with RNAs, and expression of Gag was strengthened by YTHDFs. It was noteworthy that the suppression of HIV-1 reverse transcription and promotion of Gag processing were both triggered by YTHDFs, which implied the complicated roles of m6A in viral infection [87].
Additionally, there was another study also revealing the m6A editing profiling of HIV-1 genome [88]. They found that m6A sites were mainly enriched in 3′ UTR and that m6A residues were sufficient to enhance mRNA expression via recruiting YTHDFs. Surprisingly, YTHDFs positively regulated the HIV-1 protein expression and virus replication, which was contradictory to previous conclusions. This may be caused by discrepant cells used or various time course of virus infection. Anyhow, the functional complexity of YTHDFs on HIV-1 production and infectivity requires further investigations.
Influenza A virus (IAV)
m6A modification enriched at the IAV mRNAs encoding major structural proteins, including HA, NA, M1/M2, and NP, but not at the virus mRNAs encoding viral polymerase proteins, such as PB2, PB1, and PA [89]. And the binding targets of YTHDF2 in IAV were mainly at the transcripts encoding viral structural proteins. Overexpression of YTHDF2 significantly increased the expression of IAV NS1, M2, and NP proteins, and facilitated IAV replication, infectious particle production and viral spread [89].
Hepatitis C virus (HCV)
In HCV infection, YTHDFs relocalized to lipid droplets where viruses assembled, and specially bound to m6A residues within the E1 region which were functionally relevant [90]. YTHDFs negatively modulated HCV particle production, showing a significant role of m6A in affecting the life cycle of HCV.
The role of m6A-binding proteins in immunity
Immune system has an ability to recognize and eliminate foreign antigenic substances via innate immune response and acquired immune response. Dendritic cells (DCs) are a class of professional antigen presenting cells (APCs) and play a key role in activating adaptive immune response and maintaining immune homeostasis. CCR7-induced DC migration was regulated by an lncRNA-associated feedback. To be specific, YTHDF2 could recognize the m6A-modified lnc-Dpf3 and promote its degradation. At the late stage after CCR7 stimulation, the m6A level of lnc-Dpf3 was diminished, which lead to a decreased YTHDF2-mediated mRNA decay of this lncRNA. Subsequently, enhanced lnc-Dpf3 in turn repressed DC migration by inhibiting HIF1α-dependent glycolysis, resulting in the suppression of immune response [91]. Moreover, it is well-characterized that CD40 and CD80 enhance DC antigen presentation and T cell activation, while Tirap induces TLR4/NF-κB signaling pathway. And they were all marked by METTL3-guided m6A modification. Then YTHDF1 interacted with CD40, CD80 and Tirap to facilitate their translation, thus contributing to the activation and maturation of DCs [92].
Recently, an emerging view of circRNAs-based innate immune system is proposed. Foreign circRNAs may function as powerful adjuvants to invoke antigen-specific T and B cell responses through activating RNA pattern recognition receptor RIG-I in the presence of K63-polyubiquitin (K63-Ubn). Nevertheless, endogenous human circRNAs would be modified by m6A methylation and bound by YTHDF2, which was capable of abrogating the activation of the RIG-I/K63-Ubn/circRNA complex. That is to say, YTHDF2 is crucial for the m6A-containing suppression of innate immunity [93].
Inflammatory response
Lipopolysaccharide (LPS) is the main component of the cell walls of gram-negative bacteria and can stimulate macrophages to secrete a number of inflammatory cytokines, such as IL-6, IL-1β, and TNF-α, through enhancing the activity of MAPK and NF-κB pathways [94]. YTHDF2 was upregulated in macrophages stimulated by LPS [95]. YTHDF2 silencing promoted the phosphorylation of p38 and ERK1/2 of MAPK signaling pathway and p65 of NF-κB signaling pathway by increasing MAP2K4 and MAP4K4 expression, which initiated the LPS-induced inflammatory response.
Antiviral immune response
Viruses have a feature of propagation, but it can be blocked by the type I interferon response and the expression of interferon-stimulated genes (ISGs) [96]. IFNB mRNA, which encoded interferon-β (IFN-β) to trigger type I interferon response, was methylated by m6A and bound by YTHDF2 [97]. YTHDF2 deficiency promoted the stability of IFNB and a constant IFN-β production, thereby leading to a strongly antiviral innate immune response and further decreasing the viral propagation. Similarly, a study conducted by Rubio et al. also showed that YTHDF1 and YTHDF2 play an antiviral role by modulating IFN in an m6A-recognition way [98]. During the DNA viruses infection, nucleus-localized HNRNPA2B1 recognized viral DNA and became dimerized. Then, it was demethylated at arginine-226 mediated by the arginine demethylase JMJD6 [99]. As a result, HNRNPA2B1 translocated from nucleus to cytoplasm and initiated the TBK1-IRF3 pathway, resulting in an increase of IFN-α/β production and the activation of type I interferon response. Interestingly, the constitutive association of hnRNPA2B1 and FTO was impaired after HSV-1 infection. Therefore, hnRNPA2B1 indirectly increased the m6A modification of bound transcripts including IFI16, cGAMP, cGAS, and STING. By regulating their nucleocytoplasmic trafficking and translation, hnRNPA2B1 further induced the activation of TBK1-IRF3 pathway.
Antitumor immune response
A few of antigens are considered to be expressed by tumor cells, mainly containing tumor-associated antigens (TAAs) and tumor-specific antigens (TSAs). Immune system can identify these foreign antigens to distinguish cancer cells from non-cancer cells and initiate antitumor immune responses. Recently, with the development of bioinformatic methods, the mysterious veil of tumor mutation-derived antigens (neoantigens) has been gradually uncovered. Neoantigens can induce antitumor immune responses and predict the responses to immunotherapy [100, 101]. Nevertheless, neoantigens stimulation does not produce a lasting T cell immune response to clear the tumor cells, thus resulting in the tumor immune escape.
m6A modification can modulate immune response induced by tumor neoantigens via YTHDF1. Functionally, absence of YTHDF1 specifically enhanced the cross-prime ability of DCs and further elicited CD8+ T cells responses [102]. Mechanistically, YTHDF1 recognized and bound to the m6A peaks of the transcripts encoding lysosomal proteases, markedly increasing the translation efficiency of lysosomal cathepsins in DCs but inhibiting the capability of cross-presentation of DCs. Notably, there were substantially increased neoantigen-specific CD8+ T cells when YTHDF1 was deficient. In addition, knockdown of YTHDF1 upregulated the expression of PD-L1 and improved the PD-L1 checkpoint blockade’s antitumor effect and the patients’ clinical outcomes [102].
The role of m6A-binding proteins in adipogenesis
Obesity is an important public health problem, which can increase the risk of cardiovascular diseases, diabetes mellitus, and cancers [103]. Adipogenesis is a sophisticated process of cell differentiation in which pre-adipocytes become adipocytes. It is closely associated with obesity and is regulated via transcriptional cascade and extracellular signals [104]. Notably, increasing evidence reveals that m6A-binding proteins play a critical role in adipogenesis.
Firstly, m6A reader YTHDF2 can influence the process of mitotic clonal expansion (MCE). CCNA2 and CDK2 are two pivot cell cycle regulators, promoting cells from S phase to G2 phase. During adipogenesis, the m6A levels of CCNA2 and CDK2 decreased, but their expression increased [105]. Mechanistically, YTHDF2 recognized the methylated sites of CCNA2 and CDK2 and subsequently decayed them. Interestingly, epigallocatechin gallate (EGCG), a catechin in green tea, plays an anti-adipogenesis role. EGCG could increase not only the m6A modification of CCNA2 and CDK2 but also the expression of YTHDF2 [106]. And YTHDF2-mediated decay reduced the expression of CCNA2 and CDK2 mRNAs, thereby impairing adipogenesis. Moreover, another key cell cycle regulator CCND1 is also involved in the adipocytes differentiation. YTHDF2 preferentially bound to m6A-methylated CCND1 mRNA and diminished its expression, partially arresting MCE and inhibiting adipogenesis [107].
Secondly, m6A reader YTHDF2 can modulate JAK/STAT signaling pathway. Wu et al. observed that there was a negative relationship between the expression of JAK2 mRNA and m6A level in preadipocytes [108]. Further study found that JAK2 was a direct target of YTHDF2, and that knockdown of YTHDF2 remarkably increased JAK2 mRNA and protein levels via a decreased m6A-based mRNA decay, which led to an inactivation of JAK2/STAT3/C/EBPβ pathway and thus hindered adipogenesis. Similarly, in the porcine bone marrow stem cells (pBMSCs), m6A methylation was enriched at JAK1 mRNA, and JAK1 knockdown could suppress adipocytes differentiation [109]. YTHDF2 recognized JAK1 and then diminish its expression through enhancing mRNA degradation, consequently inhibiting the phosphorylation of STAT5 and the initiation of JAK1/STAT5/C/EBPβ pathway, and preventing BMSCs from differentiating into adipocytes.
Thirdly, m6A reader YTHDF2 can regulate the expression of adipogenic-related genes. Bmal1 is an important part of mammalian circadian clock gene regulatory networks, mainly regulating metabolism. PPARα is a critical transcription factor that modulates the expression of genes involved in lipid metabolism. Loss of Bmal1 markedly increased the m6A levels of PPARα mRNA [110]. And YTHDF2 could mediate the mRNA decay of PPARα to decrease its stability and lead to a reduction of lipid accumulation, suggesting a close relationship between circadian clock regulation, m6A modification, and lipid metabolism. Furthermore, FAM134B is a positive regulator of lipid deposition in preadipocytes, whose overexpression causes the enhancement in the expression of adipogenic differentiating factors PPARγ, CEBPα, and FAS. And FAM134B was featured by m6A modification. Then, YTHDF2 interacted with FAM134B and drive its mRNA degradation via an m6A-based mechanism, leading to an inhibition of adipogenesis [111]. Song et al. found that Zfp217 could accelerate the expression of key adipogenic genes PPARγ, aP2, LPL, and Adiponectin [112]. Interestingly, Zfp217 could activate the transcription of FTO, while YTHDF2 was identified to inhibit the demethylase activity of FTO. And Zfp217 interacted with YTHDF2 to block its suppression of the m6A demethylase, finally induces the abolished m6A level of adipogenic genes. Thus, YTHDF2-mediated decay of these genes was decreased, which facilitated adipogenic differentiation [112].
Fourthly, m6A reader YTHDF2 can interact with autophagy-related genes. YTHDF2 targeted at m6A-modified Atg5 and Atg7, and modulated their expression via the mRNA decay [113]. Depletion of YTHDF2 accelerated autophagy and adipogenesis through enhancing the stability of Atg5 and Atg7, suggesting that m6A modification might regulate adipocytes differentiation by autophagy.
In addition to YTHDF2, m6A-binding protein YTHDF1 also participates in the process of adipogenesis. Jiang et al. determined the whole transcriptome-wide m6A profiles of the longissimus dorsi muscles (LDMs) in lean-type and obese-type pigs and found a unique m6A-methylated gene in the obese-type pigs called MTCH2 that was involved in lipid accumulation and oxidation [114]. And YTHDF1 targeted at the m6A motif of MTCH2 mRNA to improve its translation efficiency and further promote adipogenesis. Another study revealed that UCP2 and PNPLA2 were two distinguished m6A-containing genes in fat pig model [115]. After the depletion of m6A modification, mutated UCP2 suppressed adipogenesis while mutated PNPLA2 facilitated lipid accumulation. Mechanistically, m6A methylation promoted PNPLA2 expression by YTHDF1-mediated translation, whereas inhibited UCP2 expression seemingly dependent on neither YTHDF1 nor YTHDF2 [115].
Conclusions and perspective
With the advancements in the technique of high-throughput sequencing, m6A modification becomes an emerging focus of investigation. Increasingly, more researches have reported the roles of different m6A-binding proteins. YTHDF1, YTHDF3, and YTHDC2 promote mRNA translation, while YTHDF2 and YTHDF3 facilitate mRNA decay. YTHDC1 plays an important role in the alternative splicing, nuclear export and X chromosome silencing. IGF2BP1-3 have a function to increase the stability of targeted mRNAs. In addition, HNRNP family are involved in the alternative splicing, RNA processing and structure switching. The specific structures and functions of m6A-binding proteins are summed up in Fig. 1 and Fig. 2, respectively. More importantly, m6A readers act as a promoter or inhibitor in the multiple biological processes, such as tumorigenesis (Table 2), hematopoiesis, viral replication, immune response, and adipogenesis. However, until now, the understanding about m6A readers is just the tip of the iceberg. There remains a lot of space for exploring in the research on m6A-binding proteins.
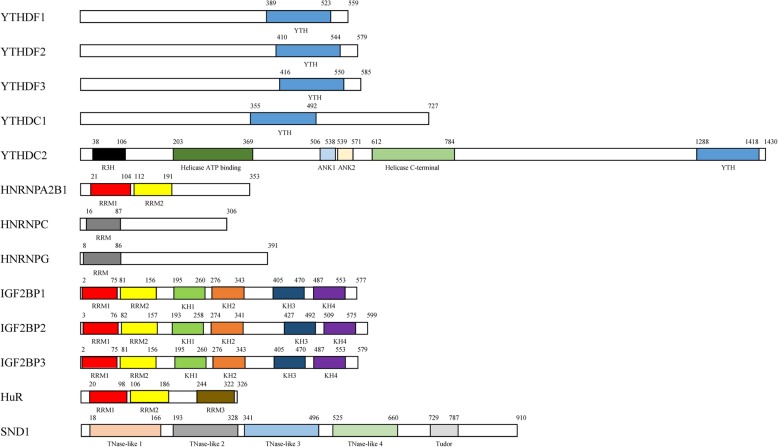
Fig. 1.
Domain architectures of m6A-binding proteins
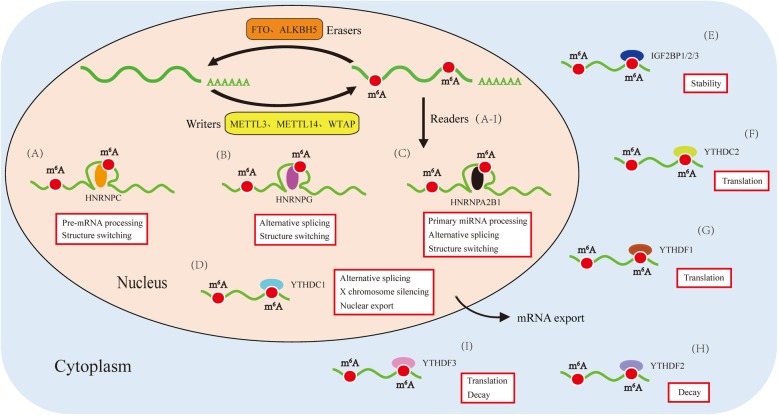
Fig. 2.
The regulation of m6A modification. m6A is established by m6A methyltransferases (“writers”) and removed by m6A demethylases (“erasers”). m6A readers are involved in multiple procedures of RNA metabolism through recognizing and binding to the m6A sites of RNAs. (a) HNRNPC plays an important role in the pre-mRNAs processing and structure switching. (b) HNRNPG modulates alternative splicing and structure switching. (c) HNRNPA2B1 accelerates primary miRNAs processing, alternative splicing, and structure switching. (d) YTHDC1 participates in the alternative splicing, nuclear export, and X chromosome silencing. (e) IGF2BP1/2/3 have a function to increase the stability of targeted mRNAs. (f) YTHDC2 promotes mRNAs translation. (g) YTHDF1 augments mRNAs translation. (h) YTHDF2 facilitates mRNAs decay. (i) YTHDF3 cooperates with YTHDF1 to increase mRNAs translation, and strengthens mRNAs decay mediated by YTHDF2
Table 2.
The role of m6A-binding proteins in human solid cancers and hematological malignancy
| Cancers | m6A readers | Target RNAs | Mechanism | Reference |
| Hepatocellular carcinoma | YTHDF1 | Snail | Accelerating the translation of Snail mRNA | [47] |
| YTHDF2 | EGFR | Destabilizing EGFR mRNA | [42] | |
| YTHDF2 | IL-11, SERPINE2 | Increasing the degradation of IL-11 and SERPINE2 mRNAs | [43] | |
| YTHDF2 | SOCS2 | Facilitating SOCS2 mRNA decay | [44] | |
| IGF2BP1/2/3 | MYC | Enhancing the expression of MYC mRNA | [32] | |
| IGF2BP1 | SRF | Promoting SRF mRNA translation | [48] | |
| Colorectal cancer | YTHDF1 | β-catenin, WNT6, FZD9 | Increasing the expression of β-catenin, WNT6 and FZD9A to activate Wnt/β-catenin pathway | [49] |
| YTHDF3 | GAS5 | Enhancing the degradation of lncRNA GAS5 | [50] | |
| YTHDC1 | circNSUN2 | Facilitating circNSUN2 export from nucleus to cytoplasm | [51] | |
| IGF2BP2 | HMGA2 | Forming a circNSUN2/IGF2BP2 complex to fortify the stability of HMGA2 | [51] | |
| IGF2BP2 | SOX2 | Stabilizing SOX2 mRNA | [52] | |
| Gastric cancer | IGF2BP1 | SEC62 | Augmenting SEC62 mRNA translation | [53] |
| IGF2BP3 | HDGF | Facilitating HDGF mRNA expression | [54] | |
| HuR | ZMYM1 | Fortifying the stability of ZMYM1 mRNA | [116] | |
| Lung cancer | YTHDF1 | CDK2, CDK4, cyclinD1 | Promoting the translations of CDK2, CDK4 and cyclinD1 | [56] |
| YTHDF1 | Keap1 | Leading to cisplatin resistance of tumor cells via modulating the Keap1-Nrf2-AKR1C1 axis | [56] | |
| YTHDF1/3 | YAP | Up-regulating YAP expression | [57] | |
| YTHDF2 | 6PGD | Facilitating 6PGD degradation | [55] | |
| YTHDF3 | MALAT1 | Increasing MALAT1 stability | [57] | |
| MELLT3 | EGFR, TAZ | Accelerating the translation of EGFR and TAZ | [58] | |
| Bladder cancer | YTHDF1/3 | ITGA6 | Promoting ITGA6 mRNA translation | [59] |
| YTHDF1 | CDCP1 | Enhancing the expression of CDCP1 mRNA | [60] | |
| MELLT3 | CDCP1 | Facilitating CDCP1 translation and strengthening the binding of YTHDF1 to CDCP1 | [60] | |
| Endometrial cancer | YTHDF1 | PHLPP2 | Increasing the expression of PHLPP2 | [61] |
| YTHDF2 | PRR5, PRR5L, mTOR | Diminishing the abundance of PRR5, PRR5L, and mTOR | [61] | |
| Ovarian cancer | YTHDF1 | EIF3C | Targeting at EIF3C to enhance its translation efficiency | [62] |
| Cervical cancer | YTHDF2 | GAS5 | Abrogating the GAS5 expression | [63] |
| Melanoma | YTHDF2 | PD-1 (PDCD1), CXCR4, SOX10 | Downregulating the mRNA and protein levels of three key intrinsic pro-tumorigenic factors, including PD-1 (PDCD1), CXCR4 and SOX10 | [64] |
| YTHDF1 | HINT2 | Promoting the translation of HINT2 mRNA | [65] | |
| Breast cancer | YTHDF2 | BNIP3 | Facilitating the degradation of BNIP3 mRNA | [66] |
| Pancreatic cancer | IGF2BP2 | DANCR | Enhancing the DANCR expression | [67] |
| Acute myeloid leukemia | YTHDF2 | TNFR2 | Reducing the TNFR2 expression | [81] |
| YTHDF2 | MYC, CEBPA | Accelerating the decay of MYC and CEBPA | [82] |
Firstly, a few of non-classic m6A readers are found through RNA sequencing, such as HuR and SND1. ZMYM1 mRNA, which could facilitate the process of EMT and the metastasis of GC, was modified by m6A and its stability was enhanced relying on the recognition by HuR [116]. Another novel m6A reader SND1 bound to the m6A sites of ORF50 transcript and promoted its stability. Depletion of SND1 inhibited the expression of KSHV early genes, affecting KSHV lytic replication [117]. Even so, the specific functions and mechanisms of new m6A-binding proteins are still not comprehensively elucidated.
Secondly, the inhibitors of m6A-related enzymes have been actively explored. For example, R-2HG [82] and meclofenamic acid (MA) [118] are two FTO inhibitors and have been confirmed to inhibit tumor cell growth and induce cell apoptosis. However, the inhibitors of m6A-binding proteins have not been identified. Actually, as the direct executers for m6A-dependent bioprocesses, m6A readers seem more indispensable for controlling multiple biological events including tumorigenesis. Therefore, further studies are needed to find the inhibitors of m6A readers in order to provide the novel and effective strategies for clinical therapy.
Thirdly, m6A exists in a broad range of viruses and is tightly associated with viral infectivity and replication. Surprisingly, the roles of m6A readers in the virus infections like KSHV, HIV, and IAV are sometimes contradicted with the well-recognized effects. A reasonable explanation may be that readers indirectly regulate the expression of viral genes via controlling the level of host antiviral transcripts. What is more, during the identical virus infection, even the same reader could function differentially relying on the cell context. It highlights the complexity of m6A-medaited impacts on virus processing which is host-dependent. Hence, it would be meaningful to systematically clarify the functions of m6A readers in the virus-host interaction.
Overall, m6A modification is emerging as a rising star in epigenetic research, and the evolving deciphering of m6A-binding proteins exposes a more comprehensive understanding of m6A methylation. The m6A readers are extensively responsible for multiple biological processes, and they extend the repertoire of m6A epitranscriptome, offering novel insights into its underlying molecular mechanisms. Apart from exploring more potential functional m6A-binding proteins, further investigations are required to not only exploit the complicated roles of m6A readers due to specific cell context, but also develop the efficient therapeutic interventions based on m6A readers.
Acknowledgements
Not applicable.
Abbreviations
- m6A
N6-methyladenosine
- YTHDF
YT521-B homology domain family
- YTHDC
YT521-B homology domain containing
- IGF2BP
Insulin-like growth factor 2 mRNA-binding protein
- HNRNPA2B1
Heterogeneous nuclear ribonucleoproteins A2/B1
- HNRNPC
Heterogeneous nuclear ribonucleoproteins C
- HNRNPG
Heterogeneous nuclear ribonucleoproteins G
- HCC
Hepatocellular carcinoma
- CRC
Colorectal cancer
- GC
Gastric cancer
- HSC
Hematopoietic stem cell
- AML
Acute myeloid leukemia
- KSHV
Kaposi’s sarcoma-associated herpes virus
- HIV
Human immunodeficiency virus
- IAV
Influenza A virus
- HCV
Hepatitis C virus
- DC
Dendritic cell
- IFN-β
Interferon-β
- MCE
Mitotic clonal expansion
Authors’ contributions
ZYC wrote and edited the manuscript; SYF collected the related literature; SHF finished the figures and tables; XWZ provided the feedback and guidance. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yanchun Zhao, Email: 21718093@zju.edu.cn.
Yuanfei Shi, Email: shiyuanfei0802@163.com.
Huafei Shen, Email: 464260761@qq.com.
Wanzhuo Xie, Email: xiewanzhuo@zju.edu.cn.
References
- 1.Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 2.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71(10):3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perry RP, Kelley DE. Existence of methylated messenger RNA in mouse L cells. Cell. 1974;1(1):37–42. doi: 10.1016/0092-8674(74)90153-6. [DOI] [Google Scholar]
- 4.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell. 1975;4(4):379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 5.Wei CM, Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977;16(8):1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- 6.Csepany T, Lin A, Baldick CJ, Jr, Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. J Biol Chem. 1990;265(33):20117–20122. [PubMed] [Google Scholar]
- 7.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534(7608):575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m6A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 2018;69(6):1028–38.e6. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169(5):824–35.e14. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, et al. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18(11):2004–2014. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27(3):315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27(3):444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61(4):507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 2017;6. 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed]
- 25.Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27(9):1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao Y, Dong L, Liu XM, Guo J, Ma H, Shen B, et al. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat Commun. 2019;10(1):5332. doi: 10.1038/s41467-019-13317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162(6):1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajagopalan LE, Westmark CJ, Jarzembowski JA, Malter JS. hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science. 2012;335(6076):1643–1646. doi: 10.1126/science.1218469. [DOI] [PubMed] [Google Scholar]
- 30.Zarnack K, König J, Tajnik M, Martincorena I, Eustermann S, Stévant I, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152(3):453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45(10):6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoilov P, Rafalska I, Stamm S. YTH: a new domain in nuclear proteins. Trends Biochem Sci. 2002;27(10):495–497. doi: 10.1016/S0968-0004(02)02189-8. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, et al. The YTH domain is a novel RNA binding domain. J Biol Chem. 2010;285(19):14701–14710. doi: 10.1074/jbc.M110.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10(11):927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Bedi RK, Wiedmer L, Huang D, Sledz P, Caflisch A. Flexible binding of m6A reader protein YTHDC1 to its preferred RNA motif. 2019;15(12):7004–14. [DOI] [PubMed]
- 37.Kretschmer J, Rao H, Hackert P, Sloan KE, Höbartner C, Bohnsack MT. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5’-3’ exoribonuclease XRN1. RNA. 2018;24(10):1339–1350. doi: 10.1261/rna.064238.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, et al. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun. 2018;9(1):420. doi: 10.1038/s41467-017-02770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70(15):2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schito L, Semenza GL. Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer. 2016;2(12):758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 42.Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, et al. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261. doi: 10.1016/j.canlet.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer. 2019;18(1):163. doi: 10.1186/s12943-019-1082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 45.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun. 2019;10(1):2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Müller S, Glaß M, Singh AK, Haase J, Bley N, Fuchs T, et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019;47(1):375–390. doi: 10.1093/nar/gky1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bai Y, Yang C, Wu R, Huang L, Song S, Li W, et al. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front Oncol. 2019;9:332. doi: 10.3389/fonc.2019.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer. 2019;18:143. doi: 10.1186/s12943-019-1079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10(1):4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, et al. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18(1):112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He H, Wu W, Sun Z, Chai L. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m6A-caused stabilization of SEC62. Biochem Biophys Res Commun. 2019;517(4):581–587. doi: 10.1016/j.bbrc.2019.07.058. [DOI] [PubMed] [Google Scholar]
- 54.Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. Gut. 2019. 10.1136/gutjnl-2019-319639 [Epub ahead of print].
- 55.Sheng H, Li Z, Su S, Sun W, Zhang X, Li L, et al. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinogenesis. 2019. 10.1093/carcin/bgz152 [Epub ahead of print]. [DOI] [PubMed]
- 56.Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10(1):4892. doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12(1):135. doi: 10.1186/s13045-019-0830-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin H, Ying X, Que B, Wang X, Chao Y, Zhang H, et al. N6-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019;47:195–207. doi: 10.1016/j.ebiom.2019.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang F, Jin H, Que B, Chao Y, Zhang H, Ying X, et al. Dynamic m6A mRNA methylation reveals the role of METTL3-m6A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene. 2019;38(24):4755–4772. doi: 10.1038/s41388-019-0755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20(9):1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020. 10.1093/nar/gkaa048 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 63.Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11(8):4909–4921. [PMC free article] [PubMed] [Google Scholar]
- 64.Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10(1):2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jia R, Chai P, Wang S, Sun B, Xu Y, Yang Y, et al. m6A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol Cancer. 2019;18(1):161. doi: 10.1186/s12943-019-1088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18(1):46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2019 Dec 5. 10.1038/s41418-019-0461-z [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 68.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2(7):502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 69.Sonoda Y. Immunophenotype and functional characteristics of human primitive CD34-negative hematopoietic stem cells: the significance of the intra-bone marrow injection. J Autoimmun. 2008;30(3):136–144. doi: 10.1016/j.jaut.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 70.Wognum AW, Eaves AC, Thomas TE. Identification and isolation of hematopoietic stem cells. Arch Med Res. 2003;34(6):461–475. doi: 10.1016/j.arcmed.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 71.Weissman IL, Shizuru JA. The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood. 2008;112(9):3543–3553. doi: 10.1182/blood-2008-08-078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ottersbach K. Endothelial-to-haematopoietic transition: an update on the process of making blood. Biochem Soc Trans. 2019;47(2):591–601. doi: 10.1042/BST20180320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang P, He Q, Chen D, Liu W, Wang L, Zhang C, et al. G protein-coupled receptor 183 facilitates endothelial-to-hematopoietic transition via Notch1 inhibition. Cell Res. 2015;25(10):1093–1107. doi: 10.1038/cr.2015.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gama-Norton L, Ferrando E, Ruiz-Herguido C, Liu Z, Guiu J, Islam AB, et al. Notch signal strength controls cell fate in the haemogenic endothelium. Nat Commun. 2015;6:8510. doi: 10.1038/ncomms9510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549(7671):273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 76.Li Z, Qian P, Shao W, Shi H, He XC, Gogol M, et al. Suppression of m6A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018;28(9):904–917. doi: 10.1038/s41422-018-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang H, Zuo H, Liu J, Wen F, Gao Y, Zhu X, et al. Loss of YTHDF2-mediated m6A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 2018;28(10):1035–1038. doi: 10.1038/s41422-018-0082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ye M, Zhang H, Yang H, Koche R, Staber PB, Cusan M, et al. Hematopoietic differentiation is required for initiation of acute myeloid leukemia. Cell Stem Cell. 2015;17(5):611–623. doi: 10.1016/j.stem.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Testa U. Leukemia stem cells. Ann Hematol. 2011;90(3):245–271. doi: 10.1007/s00277-010-1118-7. [DOI] [PubMed] [Google Scholar]
- 80.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 81.Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25(1):137–48.e6. doi: 10.1016/j.stem.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 2018;172(1-2):90–105.e23. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hesser CR, Karijolich J, Dominissini D, He C, Glaunsinger BA. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 2018;14(4):e1006995. doi: 10.1371/journal.ppat.1006995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ye F, Chen ER, Nilsen TW. Kaposi’s sarcoma-associated herpesvirus utilizes and manipulates RNA N6-adenosine methylation to promote lytic replication. J Virol. 2017;91(16). 10.1128/JVI.00466-17. [DOI] [PMC free article] [PubMed]
- 85.Tan B, Liu H, Zhang S, da Silva SR, Zhang L, Meng J, et al. Viral and cellular N6-methyladenosine and N6,2’-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat Microbiol. 2018;3(1):108–120. doi: 10.1038/s41564-017-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife. 2016;5. 10.7554/eLife.15528. [DOI] [PMC free article] [PubMed]
- 87.Lu W, Tirumuru N, St Gelais C, Koneru PC, Liu C, Kvaratskhelia M, et al. N6-methyladenosine-binding proteins suppress HIV-1 infectivity and viral production. J Biol Chem. 2018;293(34):12992–13005. doi: 10.1074/jbc.RA118.004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, et al. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. 2016;19(5):675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, et al. Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host Microbe. 2017;22(3):377–86.e5. doi: 10.1016/j.chom.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, et al. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe. 2016;20(5):654–665. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu J, Zhang X, Chen K, Cheng Y, Liu S, Xia M, et al. CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity. 2019;50(3):600–15.e15. doi: 10.1016/j.immuni.2019.01.021. [DOI] [PubMed] [Google Scholar]
- 92.Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, et al. Mettl3-mediated mRNA m6A methylation promotes dendritic cell activation. Nat Commun. 2019;10(1):1898. doi: 10.1038/s41467-019-09903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-methyladenosine modification controls circular RNA immunity. Mol Cell. 2019;76(1):96–109.e9. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao BB, Guo HJ, Liu Y, Luo XY, Yang SX, Wang YT, et al. K313, a novel benzoxazole derivative, exhibits anti-inflammatory properties via inhibiting GSK3β activity in LPS-induced RAW264.7 macrophages. J Cell Biochem. 2018;119(7):5382–5390. doi: 10.1002/jcb.26685. [DOI] [PubMed] [Google Scholar]
- 95.Yu R, Li Q, Feng Z, Cai L, Xu Q. m6A reader YTHDF2 regulates LPS-induced inflammatory response. Int J Mol Sci. 2019;20(6). 10.3390/ijms20061323. [DOI] [PMC free article] [PubMed]
- 96.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1(6):519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, et al. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol. 2019;20(2):173–182. doi: 10.1038/s41590-018-0275-z. [DOI] [PubMed] [Google Scholar]
- 98.Rubio RM, Depledge DP, Bianco C, Thompson L, Mohr I. RNA m6A modification enzymes shape innate responses to DNA by regulating interferon β. Genes Dev. 2018;32(23-24):1472–1484. doi: 10.1101/gad.319475.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang L, Wen M, Cao X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Sci. 2019;365(6454). 10.1126/science.aav0758. [DOI] [PubMed]
- 100.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 101.Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature. 2019;566(7743):270–4. [DOI] [PMC free article] [PubMed]
- 103.Haslam DW, James WP. Obesity. Lancet. 2005;366(9492):1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 104.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7(12):885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 105.Wu R, Liu Y, Yao Y, Zhao Y, Bi Z, Jiang Q, et al. FTO regulates adipogenesis by controlling cell cycle progression via m6A YTHDF2 dependent mechanism. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863(10):1323–1330. doi: 10.1016/j.bbalip.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 106.Wu R, Yao Y, Jiang Q, Cai M, Liu Q, Wang Y, et al. Epigallocatechin gallate targets FTO and inhibits adipogenesis in an mRNA m6A-YTHDF2-dependent manner. Int J Obes (Lond). 2018;42(7):1378–1388. doi: 10.1038/s41366-018-0082-5. [DOI] [PubMed] [Google Scholar]
- 107.Liu Q, Zhao Y, Wu R, Jiang Q, Cai M, Bi Z, et al. ZFP217 regulates adipogenesis by controlling mitotic clonal expansion in a METTL3-m6A dependent manner. RNA Biol. 2019;27:1–9. doi: 10.1080/15476286.2018.1557498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu R, Guo G, Bi Z, Liu Y, Zhao Y, Chen N, et al. m6A methylation modulates adipogenesis through JAK2-STAT3-C/EBPβ signaling. Biochim Biophys Acta Gene Regul Mech. 2019;1862(8):796–806. doi: 10.1016/j.bbagrm.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 109.Yao Y, Bi Z, Wu R, Zhao Y, Liu Y, Liu Q, et al. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway via an m6A-YTHDF2-dependent manner. FASEB J. 2019;33(6):7529–7544. doi: 10.1096/fj.201802644R. [DOI] [PubMed] [Google Scholar]
- 110.Zhong X, Yu J, Frazier K, Weng X, Li Y, Cham CM, et al. Circadian clock regulation of hepatic lipid metabolism by modulation of m6A mRNA methylation. Cell Rep. 2018;25(7):1816–28.e4. doi: 10.1016/j.celrep.2018.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cai M, Liu Q, Jiang Q, Wu R, Wang X, Wang Y. Loss of m6 A on FAM134B promotes adipogenesis in porcine adipocytes through m6 A-YTHDF2-dependent way. IUBMB Life. 2019;71(5):580–586. doi: 10.1002/iub.1974. [DOI] [PubMed] [Google Scholar]
- 112.Song T, Yang Y, Wei H, Xie X, Lu J, Zeng Q, et al. Zfp217 mediates m6A mRNA methylation to orchestrate transcriptional and post-transcriptional regulation to promote adipogenic differentiation. Nucleic Acids Res. 2019;47(12):6130–6144. doi: 10.1093/nar/gkz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang X, Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, et al. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2019;26:1–15. doi: 10.1080/15548627.2019.1659617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jiang Q, Sun B, Liu Q, Cai M, Wu R, Wang F, et al. MTCH2 promotes adipogenesis in intramuscular preadipocytes via an m6A-YTHDF1-dependent mechanism. FASEB J. 2019;33(2):2971–2981. doi: 10.1096/fj.201801393RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang X, Sun B, Jiang Q, Wu R, Cai M, Yao Y, et al. mRNA m6A plays opposite role in regulating UCP2 and PNPLA2 protein expression in adipocytes. Int J Obes (Lond). 2018;42(11):1912–1924. doi: 10.1038/s41366-018-0027-z. [DOI] [PubMed] [Google Scholar]
- 116.Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, et al. METTL3-mediated N6-methyladenosine modification is critical for epithelialmesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18(1):142. doi: 10.1186/s12943-019-1065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Baquero-Perez B, Antanaviciute A, Yonchev ID, Carr IM, Wilson SA, Whitehouse A. The Tudor SND1 protein is an m6A RNA reader essential for replication of Kaposi’s sarcoma-associated herpesvirus. Elife. 2019;8. 10.7554/eLife.47261. [DOI] [PMC free article] [PubMed]
- 118.Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43(1):373–384. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.