Centromeres ensure faithful chromosome partition upon cell division. This work reveals a novel role for centromeres in establishing/maintaining stem cell identity. Centromeric proteins are asymmetrically incorporated on sister chromatids before asymmetric division of Drosophila germline stem cells, which allows for selective chromosome segregation.
Abstract
Centromeres are epigenetically defined by CENP-A–containing chromatin and are essential for cell division. Previous studies suggest asymmetric inheritance of centromeric proteins upon stem cell division; however, the mechanism and implications of selective chromosome segregation remain unexplored. We show that Drosophila female germline stem cells (GSCs) and neuroblasts assemble centromeres after replication and before segregation. Specifically, CENP-A deposition is promoted by CYCLIN A, while excessive CENP-A deposition is prevented by CYCLIN B, through the HASPIN kinase. Furthermore, chromosomes inherited by GSCs incorporate more CENP-A, making stronger kinetochores that capture more spindle microtubules and bias segregation. Importantly, symmetric incorporation of CENP-A on sister chromatids via HASPIN knockdown or overexpression of CENP-A, either alone or together with its assembly factor CAL1, drives stem cell self-renewal. Finally, continued CENP-A assembly in differentiated cells is nonessential for egg development. Our work shows that centromere assembly epigenetically drives GSC maintenance and occurs before oocyte meiosis.
Introduction
Stem cells are fundamental for the generation of all tissues during embryogenesis and replace lost or damaged cells throughout the life of an organism. At division, stem cells generate two cells with distinct fates: (1) a cell that is an exact copy of its precursor, maintaining the “stemness,” and (2) a daughter cell that will subsequently differentiate (Betschinger and Knoblich, 2004; Inaba and Yamashita, 2012). Epigenetic mechanisms, heritable chemical modifications of the DNA/nucleosome that do not alter the primary genomic nucleotide sequence, regulate the process of self-renewal and differentiation of stem cells (Christophersen and Helin, 2010; Eun et al., 2010). In Drosophila male germline stem cells (GSCs), before division, phosphorylation at threonine 3 of histone H3 (H3T3P) preferentially associates with chromosomes that are inherited by the future stem cell (Xie et al., 2015). Furthermore, centromeric proteins seem to be asymmetrically distributed between stem and daughter cells in the Drosophila intestine and germline (García Del Arco et al., 2018; Ranjan et al., 2019). These findings support the “silent sister hypothesis” (Lansdorp, 2007), according to which epigenetic variations differentially mark sister chromatids driving selective chromosome segregation during stem cell mitosis (Dai et al., 2005; Lansdorp, 2007; Caperta et al., 2008; Tran et al., 2013; Xie et al., 2015). Centromeres, the primary constriction of chromosomes, are crucial for cell division, providing the chromatin surface where the kinetochore assembles (McKinley and Cheeseman, 2016). In turn, the kinetochore ensures the correct attachment of spindle microtubules and faithful chromosome partition into the two daughter cells upon division (Musacchio and Desai, 2017). Centromeric chromatin contains different kinds of DNA repeats (satellite and centromeric retrotransposons; Fukagawa and Earnshaw, 2014; Chang et al., 2019) wrapped around nucleosomes containing the histone H3 variant centromere protein A (CENP-A). Centromeres are not specified by a particular DNA sequence. Rather, they are specified epigenetically by CENP-A (Black and Cleveland, 2011; Allshire and Karpen, 2008; Fukagawa and Earnshaw, 2014; Karpen and Allshire, 1997). Centromere assembly, classically measured as CENP-A deposition to generate centromeric nucleosomes, occurs at the end of mitosis (between telophase and G1) in humans (Jansen et al., 2007; Hemmerich et al., 2008). Additional cell cycle timings for centromere assembly have been reported in flies (Mellone et al., 2011; Ahmad and Henikoff, 2001; Schuh et al., 2007). Interestingly, Drosophila spermatocytes and starfish oocytes are the only cells known to date to assemble centromeres before chromosome segregation, during prophase of meiosis I (Dunleavy et al., 2012; Swartz et al., 2019; Raychaudhuri et al., 2012). These examples show that centromere assembly dynamics can differ among metazoans and also among different cell types in the same organism.
A key player in centromere assembly in vertebrates is HJURP (holliday junction recognition protein), which localizes at centromeres during the cell cycle window of CENP-A deposition (Dunleavy et al., 2009; Foltz et al., 2009). Furthermore, centromere assembly is regulated by the cell cycle machinery. In flies, deposition of CID (the homologue of CENP-A) requires activation of the anaphase promoting complex/cyclosome (APC/C) and degradation of CYCLIN A (CYCA; Mellone et al., 2011; Erhardt et al., 2008). In humans, centromere assembly is antagonized by Cdk1 activity, while the kinase Plk1 promotes assembly (Silva et al., 2012; Stankovic et al., 2017; McKinley and Cheeseman, 2014). Additionally, the CYCLIN B (CYCB)/Cdk1 complex inhibits the binding of CENP-A to HJURP, preventing CENP-A loading at centromeres (Yu et al., 2015). To date, little is known about centromere assembly dynamics and functions in stem cell asymmetric divisions. Drosophila melanogaster ovaries provide an excellent model to study stem cells in their native niche (Yan et al., 2014). In this tissue, germline stem cells (GSCs) are easily accessible and can be manipulated genetically. Moreover, centromere assembly mechanisms in GSCs and their differentiated cells, cystoblasts (CBs), could be used to epigenetically discriminate between these two cell types. In Drosophila, CID binds to CAL1 (fly functional homologue of HJURP; Chen et al., 2014; Barnhart et al., 2011) in a prenucleosomal complex, and its localization to centromeres requires CAL1 and CENP-C (Erhardt et al., 2008; Mellone et al., 2011).
Here we investigated the dynamics of CENP-A deposition in Drosophila GSCs. We show that GSC centromeres are assembled after replication, but before chromosome segregation, with neural stem cells following the same trend. Centromere assembly in GSCs is tightly linked to the G2/M transition. Indeed, CYCA localizes at centromeres, and its knockdown is responsible for a marked reduction of centromeric CID and CENP-C, but not CAL1. Surprisingly, excessive CID deposition is prevented by CYCB, through the kinase HASPIN. Our superresolution microscopy analysis of GSCs at prometaphase and metaphase shows that CID incorporation on sister chromatids occurs asymmetrically, and chromosomes that will be inherited by the stem cell are loaded with more CID. Moreover, GSC chromosomes make stronger kinetochores, which anchor more spindle fibers. This asymmetric distribution of CID between GSC and CB is maintained also at later stages of the cell cycle, while it is not observed in differentiated cells outside of the niche. We also find that the depletion of CAL1 at centromeres blocks GSC proliferation and differentiation. Notably, overexpression of both CID and CAL1, as well as HASPIN knockdown, promotes stem cell self-renewal and disrupts the asymmetric inheritance of CID. Conversely, overexpression of CAL1 causes GSC-like tumors. Finally, CAL1 and CID knockdown at later stages of egg development have no obvious effect on cell division, suggesting that these cells inherit CID from GSCs. Taken together, our findings establish centromere assembly as a new epigenetic pathway that regulates stem cell fate.
Results
Nuclear distribution of centromeres in GSCs changes through the cell cycle
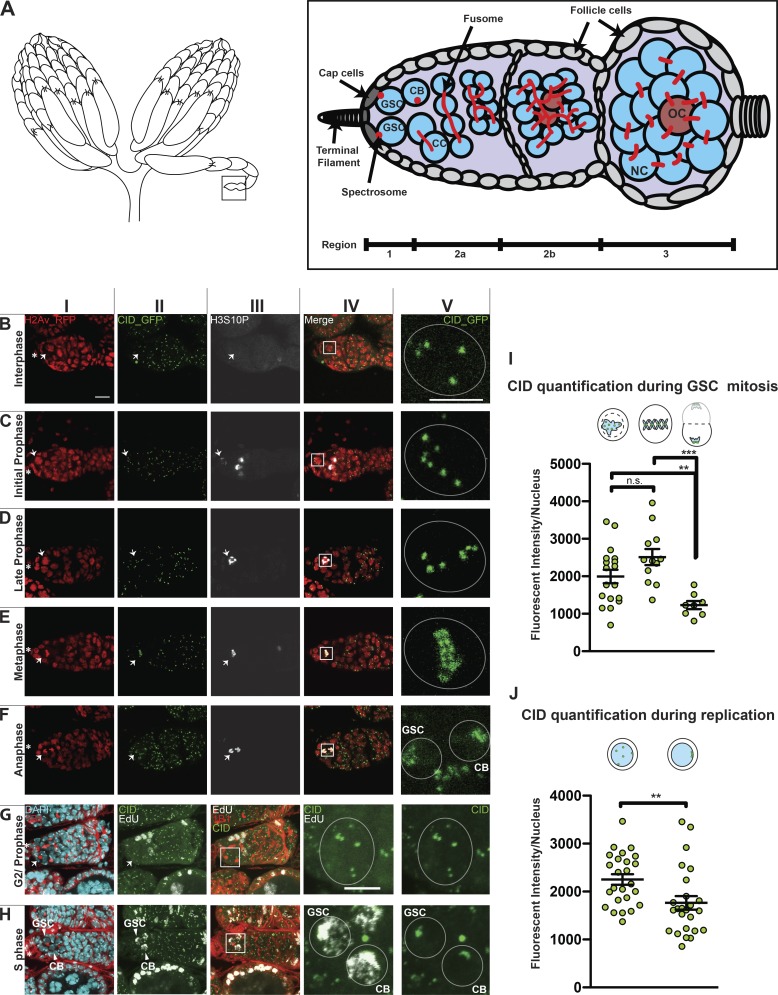
The cell cycle assembly time of centromeres in female GSCs is currently unknown. To elucidate this, we observed the distribution of centromeres throughout the cell cycle. The Drosophila female GSC niche is found at the apical end of the germarium, the anterior tip of the adult ovariole (Fig. 1 A, region 1). The niche comprises the terminal filament and the cap cells. A cytoplasmic roundish structure called the spectrosome connects two to three GSCs to the cap cells (Fig. 1 A). The spectrosome is present in both GSCs and CBs, and its shape can be used to define the cell cycle stage (Kao et al., 2015; Ables and Drummond-Barbosa, 2013). Upon asymmetric division, the daughter cell closer to the niche retains the stemness, while the other, the CB, differentiates and is detached from the niche together with its spectrosome. Each CB undergoes four rounds of mitosis with incomplete cytokinesis, giving rise to 16-cell cysts of cystocytes (CCs) interconnected to each other through the fusome, a branched spectrosome. After completion of S phase, 16-cell cysts start meiosis and form a synaptonemal complex. The oocyte originates from either of the two CCs with four fusome-bridges (Fig. 1 A, region 2a–b, brown cells; Rangan et al., 2011; Christophorou et al., 2013). In region 3, the 16-cell cysts mature to an egg chamber containing 15 nurse cells that provide for the oocyte (Fig. 1 A, region 3), which completes meiosis (McLaughlin and Bratu 2015; Hughes et al., 2018).
Figure 1.
Centromere assembly in GSCs occurs after replication but before chromosome segregation. (A) Diagram of Drosophila ovary (left) and germarium containing the germline stem cell (GSC) niche (right). NC, nurse cell; OC, oocyte; CB, cystoblast; CC, cystocyte. The spectrosome (red) connects GSCs to the cap cells (dark gray). (B–F) Confocal z-stack projection of a germarium expressing H2Av-RFP (red; I) and CID-GFP (green; II) and stained for H3S10P (white; III) showing centromere localization in GSC nuclei throughout the cell cycle; inset (V) marked by box in merged image (IV). Interphase (B), initial and late prophase (C and D), metaphase (E), anaphase (F). (G and H) Wild type germarium stained for DAPI (cyan), EdU (white), anti-CID (green), and anti-1B1 (red); G2/prophase GSCs (G) and EdU-positive (S phase) GSC and CB (H). (I and J) Quantification of CID-GFP fluorescence intensity observed at centromeres at prophase, metaphase, and anaphase (I) or antibody staining at replication and G2/prophase (J). Star indicates the terminal filament; arrows indicate GSCs; arrowheads indicate GSC and CB; <1-d-old heterozygous CID-GFP/H2Av-RFP and wild-type females; scale bar 10 µm (I–IV) or 5 µm (V). Cartoons indicate the cell cycle phase. Fluorescence Intensity is expressed as integrated density after background subtraction (see Materials and methods); data are represented as the mean ± SEM; **, P < 0.005; ***, P < 0.0005, n.s., not significant; calculated with unpaired t test with Welch’s correction.
To achieve our aim, we used transgenic flies expressing CID coupled to GFP to follow centromeres and H2Av coupled to RFP (Schuh et al., 2007) to follow chromatin condensation. To identify each phase of mitosis in GSCs, we used the phosphorylation at serine 10 of histone H3 (H3S10P; Matias et al., 2015; Hendzel et al., 1997). At interphase, chromatin is not condensed (Fig. 1 BI), centromeres are spread throughout the nucleus (Fig. 1, BII and BV), and H3S10P signal is absent (Fig. 1 BV and Fig. 3 BV). At prophase, H3S10P signal is present, chromosomes begin to condense, and centromeres start to align (Fig. 1, C and D). At this stage, we observed on average 5.7 centromere foci per cell. At metaphase, chromosomes and centromeres are completely aligned on the metaphase plate, and we observed an average of 6.9 centromere foci (Fig. 1, EI and EII). At this stage, it is possible to clearly distinguish the centromeres of each set of sister chromatids that will be inherited respectively by the new GSC and CB (Fig. 1, EII and EV). At anaphase, chromosomes and centromeres migrate to the opposite pole of each new daughter cell (Fig. 1, FI, FII, and FV), and an average of 3.1 centromere foci (despite the high level of clustering) are visible per cell. At telophase, the H3S10P signal is reduced, the chromatin starts to decondense, and centromeres remain located at the opposite side of each new nucleus (not depicted).
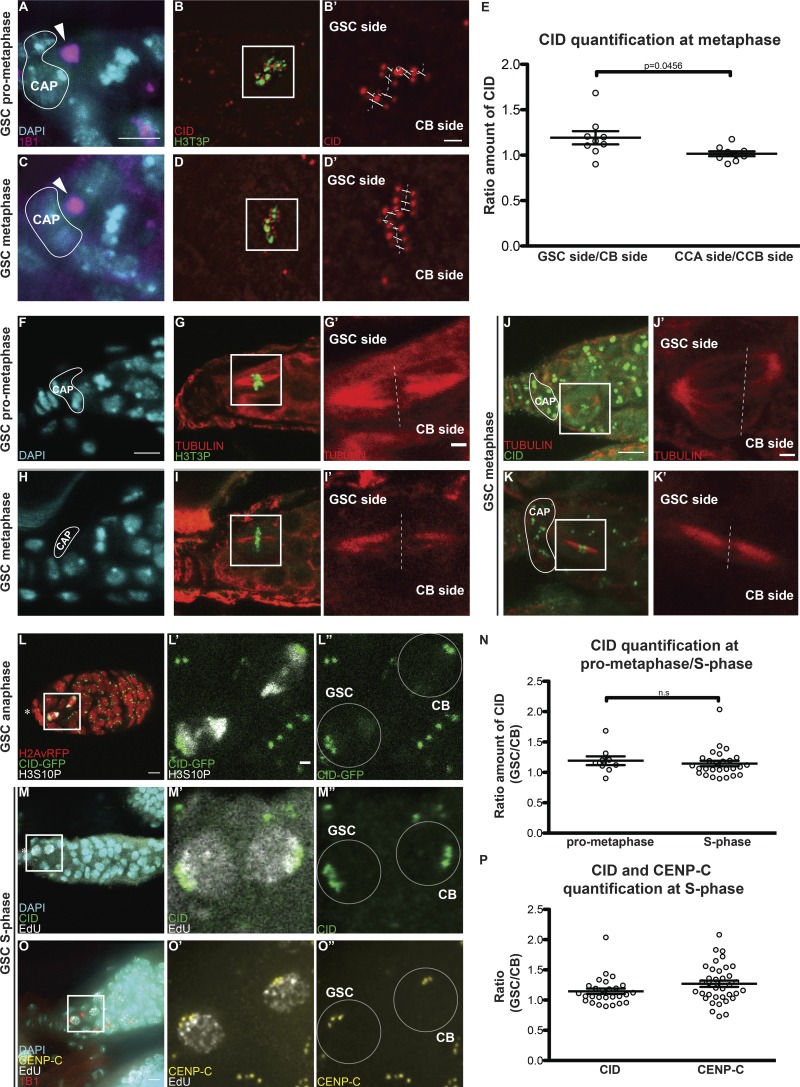
Figure 3.
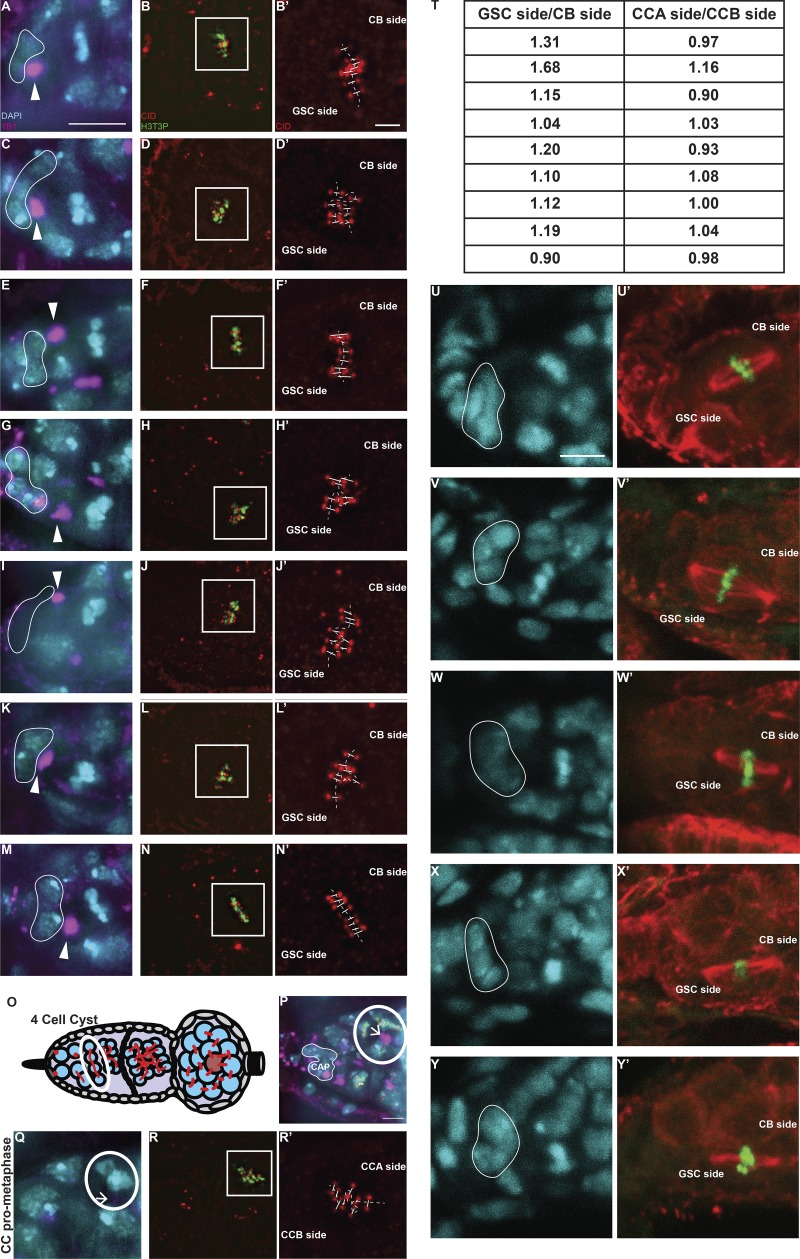
Sister chromatids of GSCs and CBs retain differential amounts of CID and CENP-C. (A–D′) Superresolution (N-SIM) z-stack projection of a GSC at prometaphase (A–B′) and metaphase (C–D′) stained for DAPI (cyan), anti-1B1 (spectrosome, magenta), anti-CID (red), and anti-H3T3P (green). (E) Ratio of the total amount of CID detected on the chromosomes of the GSC side and the total amount of CID detected on the chromosomes of the CB side, and similarly for the control CCA and CCB sides of cyst cells (CC). (F–I′) Confocal z-stack projection of a GSC at prometaphase (F–G′) and metaphase (H–I′) stained for DAPI (cyan), anti-TUBULIN (red), and anti-H3T3P (green). (J–K′) Confocal z-stack projection of a GSC at metaphase stained for anti-TUBULIN (red), anti-CID (green). (L–L″) Confocal z-stack projection of a H2Av-RFP/CID-GFP germarium, capturing a GSC and CB at anaphase. (M–M″) Confocal z-stack projection capturing a GSC and CB at S phase stained for DAPI (cyan), EdU (white), and anti-CID (green). (N) Comparison of the ratio of the total amount of CID detected on the chromosomes of the GSC side and the total amount of CID detected on the chromosomes of the CB side at prometaphase, and the amount of CID detected between GSCs and CBs at S phase. (O–O″) Confocal z-stack projection of a GSC and CB at S phase stained for DAPI (cyan), EdU (white), and anti-CENP-C (yellow). (P) Comparison of the ratio of CID and CENP-C between GSCs and CBs at S phase. White line, cap cells; arrowheads, spectrosome. Data are represented as the mean ± SEM; P value in E calculated through the use of different tests: unpaired t test with Welch’s correction (plotted); Mann–Whitney U test P value = 0.0244; Wilcoxon matched-pairs signed rank test P value = 0.0195; n.s., not significant. In A–L″, 30-min-old female flies; in N–O″, <1-d-old female flies; scale bar, 5 µm; inset, 1 µm.
To identify cells in S phase, we used 5-ethynyl-2′-deoxyuridine (EdU) to label nuclei with or without newly replicated DNA (Salic and Mitchison, 2008). After EdU incorporation, ovaries were antibody-stained to study centromere positioning in GSCs and CBs during replication (Fig. 1, G and H). In EdU-negative cells, the spectrosome is round, the DNA is not condensed, and on average 5.2 centromeres are scattered throughout the nucleus (Fig. 1, GI–GV), indicating that the cells are likely to be in G2 phase or early prophase. Interestingly, 100% of cells analyzed (50/50) show that GSCs and CBs were simultaneously positive for EdU staining (Fig. 1, HI–HV). In these cells, centromeres assumed a similar localization to that observed during anaphase and telophase, localizing to the opposite poles of the GSC and CB nuclei, with mostly four centromere foci (the exact number could not be detected due to clustering; Fig. 1, FV and HV). With the aid of several cell cycle markers (FUCCI, DACAPO) we did not succeed to isolate the G1 stage (not depicted), suggesting that it is very short in GSCs, as previously proposed (Ables and Drummond-Barbosa, 2013). In summary, our cell cycle analysis of centromere localization in M and interphase shows that centromeres are localized at the opposite poles of the new GSC and CB nuclei at anaphase, and that during DNA replication, centromeres retain this localization.
Centromeric recruitment of CID occurs after replication and before chromosome segregation in Drosophila GSCs and neuroblasts (NBs)
To assess the cell cycle timing of centromere assembly in GSCs, we quantified the CID fluorescent intensity (integrated density, Fig. S1, A–E; see Materials and methods) in mitosis and interphase. We first quantified the total amount of CID-GFP per nucleus at each phase of mitosis using the H3S10P marker (Fig. 1 I). No significant difference in CID level was detected between prophase (GSCp = 1,993 ± 180, n = 18 cells) and metaphase (GSCm = 2,512 ± 213, n = 12 cells). At anaphase, the CID level drops to about half the metaphase level (GSCa = 1,230 ± 109.4, n = 8 cells). Using antibody staining, we quantified CID in S phase and G2 phase/prophase cells (Fig. 1 J). The total amount of CID detected per nucleus in S phase cells was significantly lower than the value obtained for G2 phase/prophase: GSC+EdU = 1,764 ± 104.9 (n = 25 cells); GSCG2/prophase = 2,252 ± 108.6 (n = 25 cells). These results show that low levels of CID are observed at anaphase and replication, while considerably higher levels of CID are measured during G2 phase and prophase, suggesting that CID assembly in GSCs occurs after replication and before chromosome segregation. Furthermore, gradual deposition of CID might continue up to metaphase.
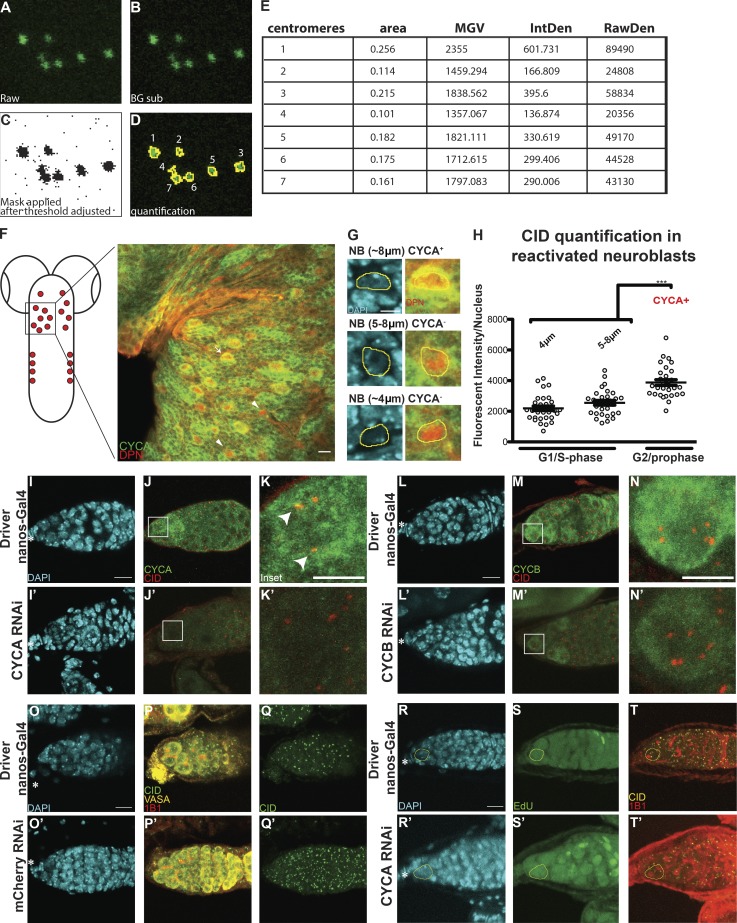
Figure S1.
Centromere assembly occurs after replication but before chromosome segregation. (A–E) Example image used for quantification (from Fig. 1 C, not oriented; see Materials and methods). (F) Diagram of Drosophila larval brain containing NBs (red) and confocal z-stack projection of a section of the tVNC stained with for DAPI (cyan), anti-CYCA (green), anti-DPN (red), and anti-CID (not depicted). (G) NBs in the tVNC are present in different sizes. (H) Quantification of fluorescence intensity of CID at centromeres in CYCA-negative and -positive NBs. Fluorescence intensity is expressed as integrated density after background subtraction (see Materials and methods); ***, P < 0.0005. (I–N′) Confocal z-stack projection of a nanos-Gal4 (I–N), CYCA RNAi (I′–K′), and CYCB RNAi (L–N′) germarium at 25°C, stained for DAPI (blue), anti-CID (red), and anti-CYCA and or CYC B (green). (O–Q′) Confocal z-stack projection of a nanos-Gal4 (O–Q), mCherry RNAi (O′–Q′) germarium. (R–T′) Confocal z-stack projection of a nanos-Gal4 (R–T), CYCA RNAi (R′–T′) germarium stained for DAPI (cyan), EdU (green), anti-CID (yellow), and anti-1B1 (spectrosome, red). Scale bar, 10 µm; inset, 5 µm.
To exclude the possibility that these dynamics were a specific feature of GSCs, we investigated CID deposition in neural stem cells of the thoracic ventral nerve chord (tVNC; Fig. S1 F) in larval brains. To isolate reactivated NBs in G2/prophase, we antibody stained with the NB marker Deadpan (Boone and Doe, 2008) and the G2 regulator CYCA that is degraded at metaphase (Lilly et al., 2000). Deadpan-positive NBs display different sizes, between 4 and 8 µm (Fig. 2 G; Chell and Brand, 2010). We quantified the total amount of CID per nucleus in these cells through antibody staining (Fig. 2 H). In the CYCA-negative NBs, the DNA is not condensed, indicating that they are neither in mitosis nor in G2/prophase. We therefore labeled them as G1/S phase NBs. Our quantification shows that CYCA-positive NBs have 65% and 90% more CID compared with the G1/S phase NBs (G2/prophase = 4,190 ± 364, n = 30 cells; G1/S phase (4 µm) = 2,191 ± 151, n = 31 cells; G1/S phase (5–8 µm) = 2,552 ± 155, n = 30 cells; 9 tVNC analyzed). Our results confirm that, similar to GSCs, neural stem cells likely also assemble centromeres during the G2/M transition.
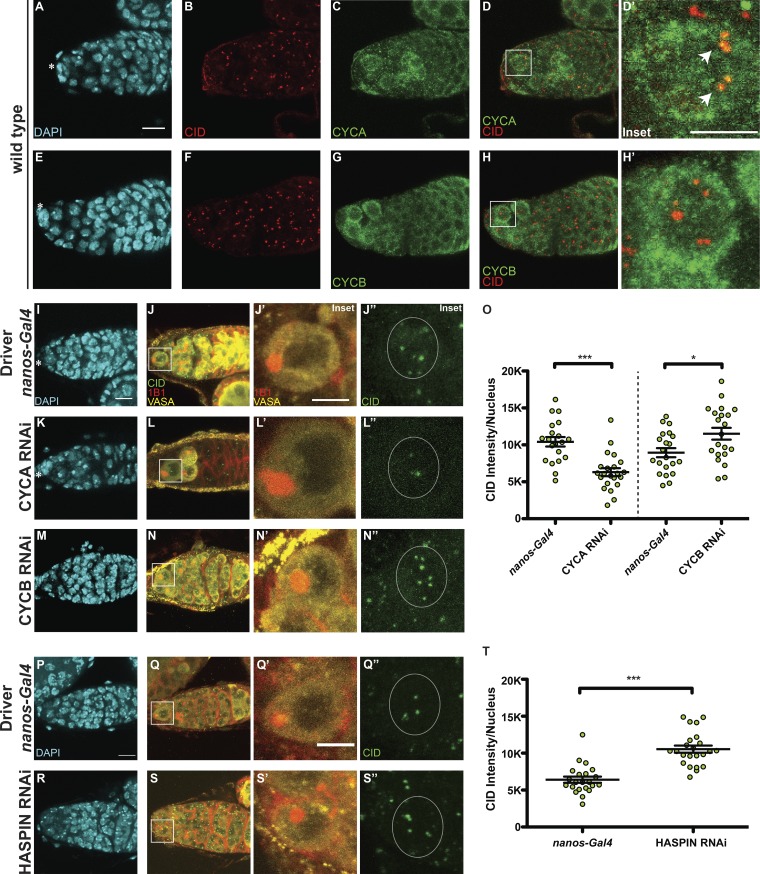
Figure 2.
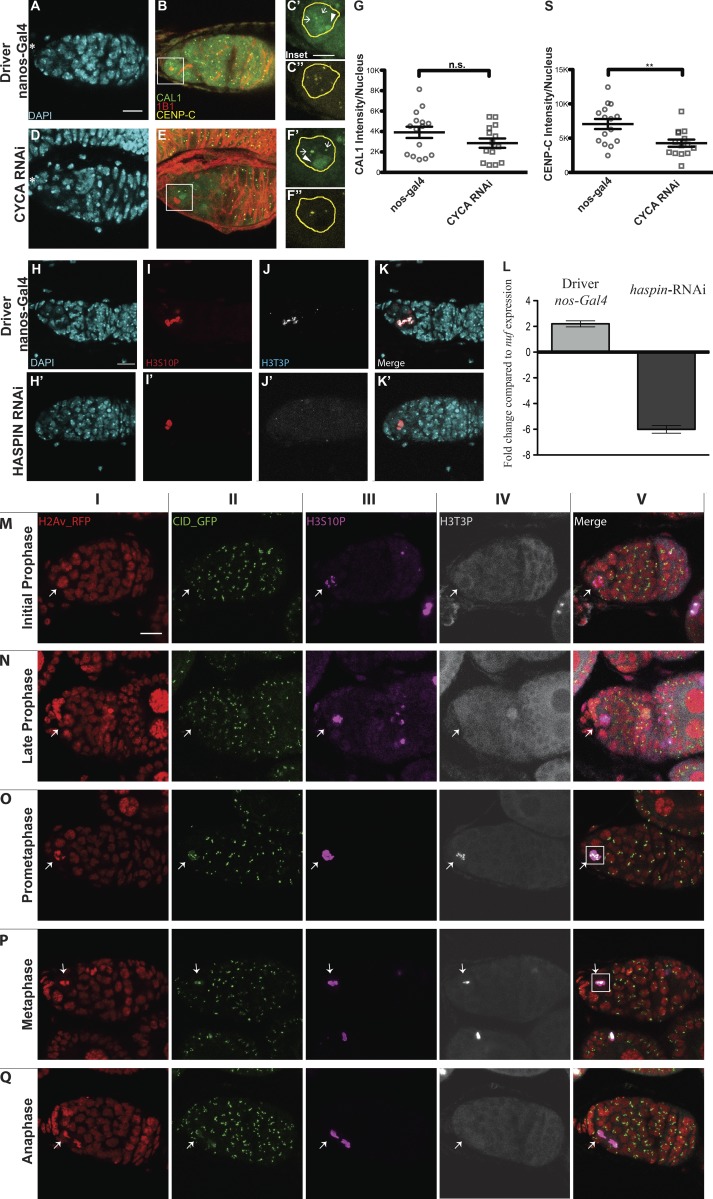
CID deposition in GSCs requires CYCA, CYCB, and HASPIN. (A–H) Wild-type germaria stained for DAPI (cyan), anti-CID (red), and anti-CYCA or anti-CYCB (green). (I–N″) Confocal z-stack projection of nanos-Gal4 (I–J″), CYCA RNAi (K–L″), CYCB RNAi (M–N″) germaria, stained for DAPI (cyan), anti-VASA (yellow), anti-CID (green), and anti-1B1 (spectrosome, red). (O) Quantification of CID fluorescence intensity at centromeres per nucleus (L). (P–S″) Confocal z-stack projection of nanos-Gal4 (P–Q″) and HASPIN RNAi germaria (R–S″), stained for DAPI (cyan), anti-VASA (yellow), anti-CID (green), and anti-1B1 (spectrosome, red). (T) Quantification of CID fluorescence intensity (MGVs) at centromeres per nucleus. Data are represented as the mean ± SEM; ***, P < 0.0005; *, P < 0.05, calculated with unpaired t test with Welch’s correction. Star indicates the terminal filament and arrows indicate centromeres; 3-d-old female flies; scale bar, 10 µm; inset, 5 µm.
Correct CID deposition at GSC centromeres requires both CYCA and CYCB
Previous work showed that CENP-A assembly into centromeric chromatin is tightly linked to key cell cycle regulators (Stankovic et al., 2017). For instance, in Drosophila, CYCA accumulation and degradation in G2 phase is crucial for CID assembly (Erhardt et al., 2008; Mellone et al., 2011). Our work shows that in GSCs, centromeric recruitment of CID initiates in early G2 phase and continues until at least prophase (Fig. 1), coinciding with CYCA and CYCB activities. Therefore, we characterized the localization pattern of CYCA and B in GSCs with respect to centromeres. CYCA was previously shown to have both cytoplasmic and nuclear localization, specifically colocalizing with CID at the centromeres in Kc167 cells (Erhardt et al., 2008). We confirm using antibody staining that this is the case also for GSCs (Fig. 2, A–D′). This is different from the CYCB localization pattern, as it shows both cytoplasmic and nuclear localization but fails to localize at centromeres (Fig. 2, E–H′). Next, we used the GAL4 upstream activating sequence (GAL4:UAS) system (Duffy, 2002) to induce the RNAi-mediated depletion of CYCA and B specifically in GSCs using the germline-specific driver nanos-Gal4 (Mathieu et al., 2013). To confirm both knockdowns, control nanos-Gal4 and CYCA/B RNAi ovaries were antibody stained against CYCA or CYCB (Fig. S1, I–N′). VASA staining (Yan et al., 2014) of ovaries showed that control germaria chambers are filled with germ cells (Fig. 2, I–J′), while CYCA depletion leads to a loss of germ cells (Fig. 2, K–L′). Furthermore, the few germ cells left appear to be as twice as big as the germ cells in the control (focus on inset in Fig. 2, J′ and L′). Similar to what has been previously described (Mathieu et al., 2013), we observed that CYCB-depleted germaria have more cells compared with the control, by counting the number of VASA-positive cells from a similar number of z-stack projections (nanos-Gal4 = 34.8 ± 2.3 cells, n = 21 germaria; CYCB RNAi = 50.6 ± 2.3, n = 23 germaria, not depicted; Fig. 2, M–N′). We did not observe these phenotypes in a nontarget mCherry RNAi control (Fig. S1, O–Q′). Given that CYCA knockdown can induce endo-reduplication (Rotelli et al., 2019), we performed EdU staining on control and CYCA RNAi germaria. GSCs with a round spectrosome and decondensed DNA are EdU negative and can therefore be considered in G2/prophase (Fig. S1, R–T′). We next quantified total centromeric CID in GSCs nuclei in G2/prophase (Fig. 2, J′, J″, L′, L″, N′, N″, and O). We first observed that nanos-Gal4 GSCs contain an average of 5.4 centromere foci detected with CID antibody, while GSCs depleted for CYCA show only 4. We found that in CYCA-knockdown GSCs, these levels are reduced by 40% compared with the control (Fig. 2 O, nanos-Gal4 = 10,411 ± 642, n = 20 cells; CYCA RNAi = 6,303 ± 538.5, n = 22 cells, reported as mean gray value [MGV] to not take into account differences measured in single centromere foci size [not depicted]). In the case of CYCB RNAi, quantitation of CID revealed that GSCs show a 28% increase in CID compared with the control (Fig. 2 O, nanos-Gal4 = 8,941 ± 610.8, 5.4 centromere foci, n = 21 cells; CYCB RNAi = 11,512 ± 801.8, 5.6 centromere foci, n = 21 cells, again reported as MGV). Taken together, our data show that CYCA and CYCB have opposite effects on CID intensity. Specifically, CYCA depletion is responsible for a 40% loss of centromeric CID, while CYCB RNAi causes a 28% increase in CID level.
Centromeric CAL1 level is not affected by CYCA deficiency, while CID incorporation is inhibited by the HASPIN kinase
To test whether the loss of CID observed in CYCA-deficient GSCs was due to a loss of CAL1 and/or CENP-C, we antibody stained control and knockdown germaria for both CAL1 and CENP-C (Fig. S2, A–F″). CAL1 is detectable not only at centromeres but also in nucleoli (Unhavaithaya and Orr-Weaver, 2013; Schittenhelm et al., 2010; Erhardt et al., 2008). In this case, we specifically quantified the centromeric CAL1 in GSCs at G2/prophase and found no significant difference between the nanos-Gal4 and CYCA RNAi samples (Fig. S2 R, nanos-Gal4 = 3,921 ± 546.4, n = 15 cells; CYCA RNAi = 2,865 ± 457.8, n = 14 cells). In contrast, CENP-C levels are reduced (Fig. S2 S, nanos-Gal4 = 7,060 ± 730.1, n = 15 cells; CYCA RNAi = 4,269 ± 525.6, n = 14 cells). These results suggest that the diminishment of CENP-C and CID observed in GSCs with reduced CYCA might be independent of CAL1. Because we found that CYCB has a role in CID deposition, we tested whether this occurs through its canonical pathway, which involves the activation of the kinase HASPIN that phosphorylates H3T3P (Moutinho-Santos and Maiato, 2014). Therefore, we performed HASPIN knockdown using the nanos-Gal4 driver (Fig. 2, O–R″). We first confirmed this knockdown by immunofluorescence (Fig. S2 H–K′) and real-time quantitative PCR (qPCR; Fig. S2 L). Next, we again measured the amount of CID in G2/prophase GSCs (Fig. 2 S). Interestingly, we found that GSCs in the HASPIN RNAi showed a 65% increase in CID level compared with the control (Fig. 2 S, nanos-Gal4, 6,408 ± 418, 4.1 centromere foci, n = 22 cells; HASPIN RNAi, 10,542 ± 479, 5.1 centromere foci, n = 22 cells, reported as MGV). This result indicates that regular CID deposition at centromeres involves HASPIN.
Figure S2.
CID deposition requires CYCA, CYCB, and HASPIN in Drosophila female GSCs. (A–F″) Confocal z-stack projection of nanos-Gal4 (A–C″), CYCA RNAi (D–F″) germaria stained for DAPI (blue), anti-CAL1 (green), anti-CENP-C (yellow), and anti-1B1 (spectrosome, red). (G) Quantification of fluorescence intensity of centromeric CAL1 per nucleus, using CENP-C as a centromeric marker based on two biological replicates. n.s., not significant. (S) Quantification of fluorescence intensity of CENP-C per nucleus, based on two biological replicates; **, P < 0.005. (H–K′) Confocal z-stack projection of nanos-Gal4 (H–K), HASPIN RNAi (H′–K′) germaria, stained for DAPI (blue), anti-H3S10P (red), and anti-H3T3P (gray). (L) HASPIN knockdown confirmation by real-time qPCR. (M–Q) Time course of the H3T3P (white) and H3S10P (magenta) signal appearance. Scale bar, 10 µm; inset, 5 µm.
Superresolution imaging reveals that GSC chromosomes are loaded with more CID, make stronger kinetochores, and capture more spindle fibers
To explore whether the timing of CID assembly might be linked to an asymmetric distribution of CID on chromosomes, we investigated CID distribution on sister chromatids in GSCs before division. Specifically, we used superresolution microscopy to examine CID intensity at sister centromeres at prometaphase and metaphase. To capture GSCs in this specific time window, we used the H3T3P marker (Xie et al., 2015). As expected, this marker first appears in GSCs at late prophase, while at anaphase the signal is lost (Fig. S2, M–Q). Importantly, superresolution microscopy allowed us to resolve eight individual sister chromatid pairs at these stages (16 centromere foci; Fig. 3). Using the position and orientation of the spectrosome, which has a round shape during mitosis (Ables and Drummond-Barbosa, 2013), we specifically identified centromeres that will be inherited by the GSCs (spectrosome proximal) and centromeres that will belong to the CBs (spectrosome distal, Fig. 3, A–D′; and Fig. S3, A–N′). Next, we measured the total amount of CID present on one set of chromosomes versus the other. For comparison, we conducted the same analysis on differentiated CCs of neighboring four-cell cysts that divide symmetrically (Figs. 3 E and S3, O–T). The ratio obtained shows that centromeres present on the GSC side incorporate ∼20% more CID, compared with centromeres of the CB side (ratio GSCside/CBside = 1.192 ± 0.072, n = 9 GSCs in prometaphase/metaphase; Fig. 3 E, values shown in Fig. S3 T). Importantly, this CID asymmetry is not observed in CCs at the same time window (ratio CCAside/CCBside = 1.016 ± 0.027, n = 9 CC in prometaphase/metaphase; Fig. 3 E, values shown in Fig. S3 T). To check whether bigger centromeres dock more spindle fibers, as already proposed (Drpic et al., 2018), we antibody stained GSCs in prometaphase and metaphase for tubulin (Fig. 3, F–I′; and Fig. S3 U–Y′). We observed more spindle fibers nucleated from the daughter centrosome, inherited by the GSC (Salzmann et al., 2014), compared with those nucleated from the mother centrosome on the CB side. This is detectable on bioriented spindles at both prometaphase and metaphase (Fig. 3, F–I′; and Fig. S3, U–Y′). To quantify this signal, we measured fluorescent intensity of the spindle on the GSC side versus the CB side (expressed as integrated density). The ratio of the two areas per cell analyzed show that GSC chromosomes display ∼48% more spindle microtubules compared with the CB chromosomes (ratio GSCside/CBside = 1.48 ± 0.2, n = 10 GSCs in prometaphase/metaphase). We also confirmed by performing costaining for both tubulin and CID that the microtubules nucleated from the centrosome were captured by centromeres (Fig. 3, J–K′). Next, we investigated whether the asymmetric distribution of CID is maintained later in the cell cycle. Specifically, we analyzed anaphase GSCs in transgenic flies expressing H2Av-RFP/CID-GFP and replicating GSC-CB couples (Fig. 3, L–O) in wild-type EdU-stained flies. As shown (Fig. 3, L–M″), both anaphase and S phase GSCs appear to retain the higher amount of CID. Quantitation revealed that at S phase, GSCs retain 14% more CID compared with CBs (Fig. 3 N; ratio GSC/CB = 1.14 ± 0.04, n = 27 couples GSC-CB analyzed; anaphase data not depicted because of the low number of cells analyzed). Furthermore, this value is not significantly different from the values found at prometaphase for GSC chromosomes (Fig. 3 N). Finally, we assessed whether centromeres that harbor more CID make bigger kinetochores, which could bias segregation (Drpic et al., 2018). For this, we quantified the amount of CENP-C in replicating GSC-CB couples (Fig. 3, O–O″), detecting a higher amount in GSCs (Fig. 3 P, ratio GSC/CB = 1.27 ± 0.05, n = 36 GSC-CB couples analyzed). These results suggest that chromosomes are labeled with a differential amount of CID upon centromere assembly. Moreover, chromosomes inherited by the GSCs harbor more CID and make bigger kinetochores that capture more spindle fibers compared with CB chromosomes.
Figure S3.
Chromosomes retain differential amounts of CID and CENP-C upon centromere assembly in Drosophila female GSCs. (A–N′) Superresolution SIM z-stack projection of a Drosophila GSC of a wild-type germarium in prometaphase and metaphase, stained for DAPI (blue), anti-CID (red), anti-H3T3P (green), and anti-SPECTROSOME (magenta). (O) Diagram of Drosophila germarium, highlighting the four-cell cyst stage containing four CCs. (P) Superresolution SIM z-stack projection of a germarium capturing four CCs in prometaphase/metaphase, which divide synchronously. (Q–R′) Superresolution SIM z-stack projection of a CC at prometaphase. (T) Table of the ratio values obtained for each cell analyzed. (U–Y′) Confocal z-stack projection of a GSC of a wild-type germarium in prometaphase and metaphase, stained for DAPI (cyan), anti-TUBULIN (red), and anti-H3T3P (green). White line highlights the cap cells; arrowheads, spectrosome; arrows, fusome (not yet visible in the z-stacks projected); scale bar, 5 µm; inset, 1 µm.
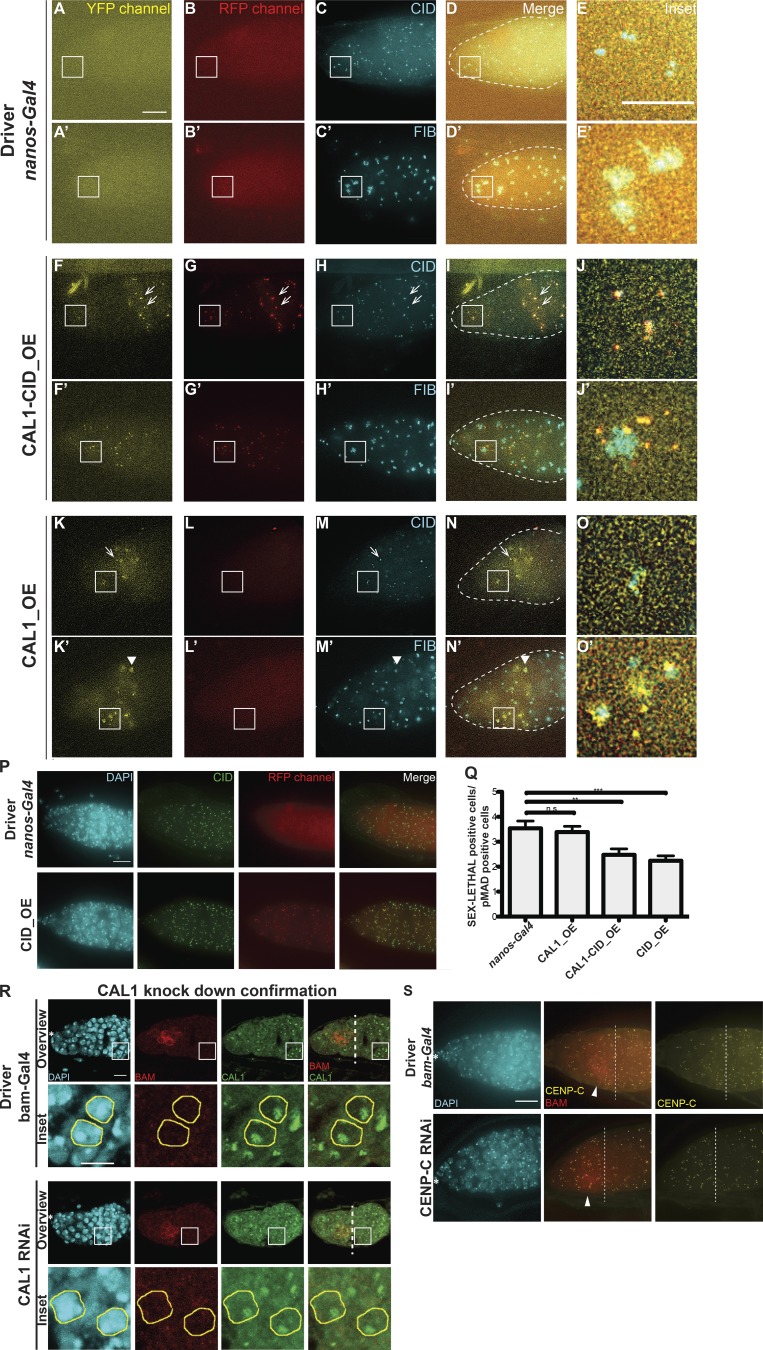
In GSCs, CAL1 is crucial for division and differentiation, as well as CID and CENP-C recruitment to centromeres
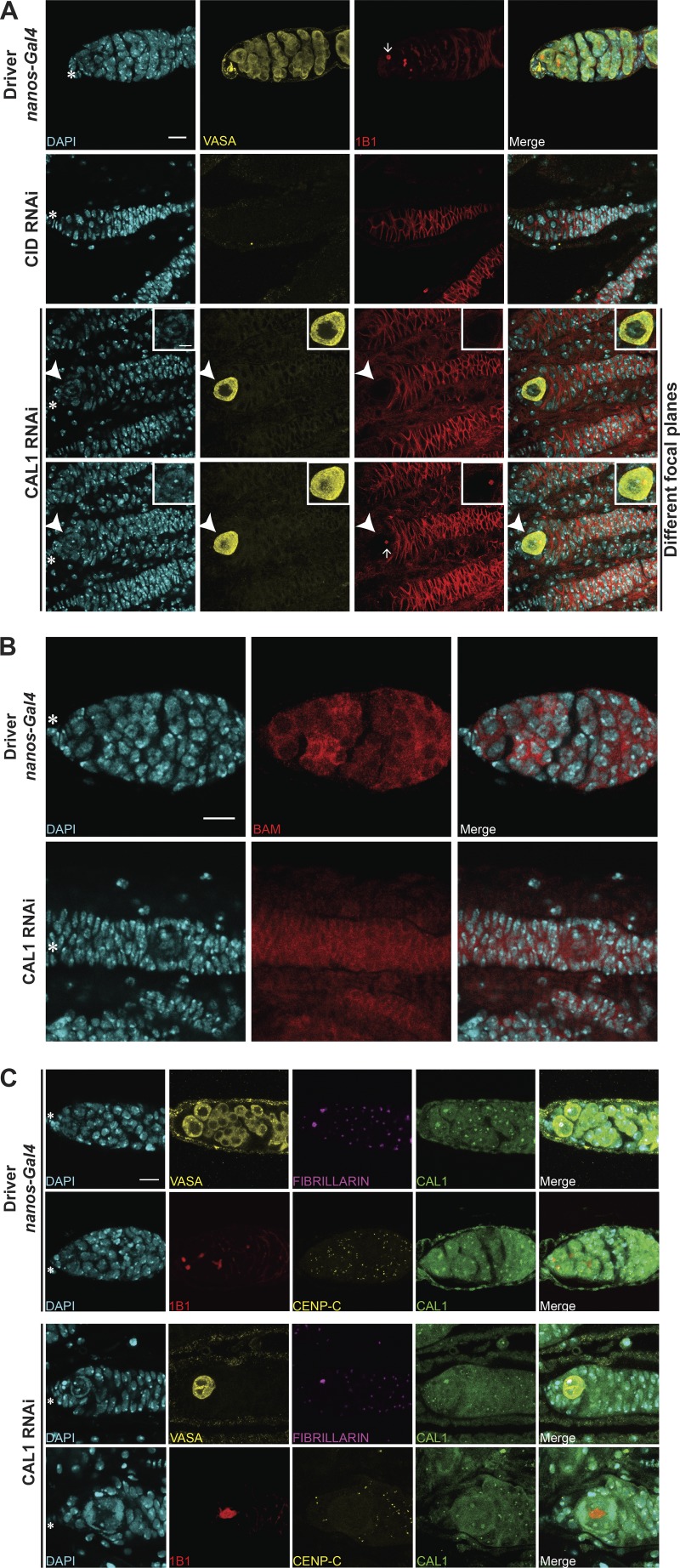
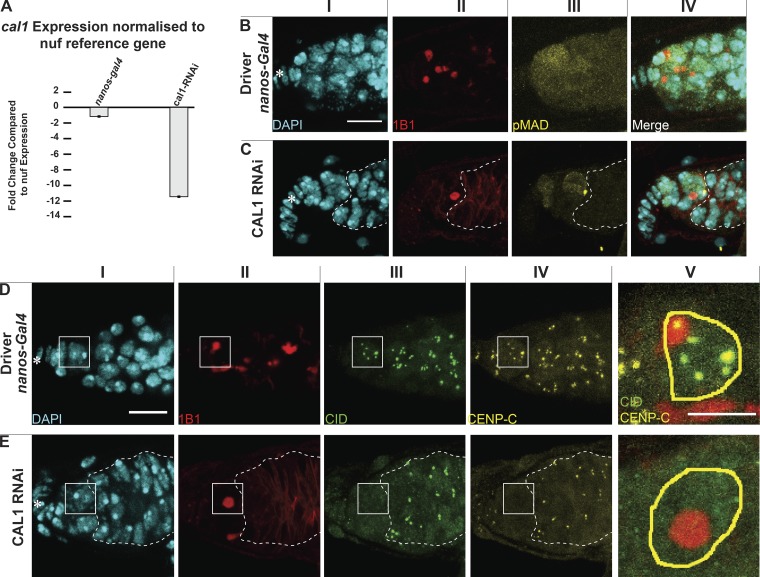
To test the role of centromeres in stem cell asymmetric division, we performed functional analyses knocking down CID and CAL1 in GSCs (Dietzl et al., 2007). Controls stained for VASA and the spectrosome (1B1) showed germaria filled with germ cells (Fig. S4 A) and GSCs having a round spectrosome attached to the cap cells (Fig. S4 A, arrows). As expected for an essential gene, CID knockdown resulted in empty ovaries with no VASA-positive cells, and therefore no germ cells (Fig. S4 A). For the CAL1 knockdown, we confirmed a >10-fold depletion of CAL1 expression through real-time qPCR (Fig. 4 A, see Materials and methods). Phenotypic analysis of CAL1-depleted ovaries showed they were largely empty (Fig. S4 A). However, 18% of germaria (3 of 16) showed the presence of a few cells (1–3) that were VASA positive with a round spectrosome and located 90% of the time at the apical end of germaria (Fig. S4 A). Older flies (7 d after eclosion) showed a higher frequency of this phenotype (∼30%, 5 of 16 germaria; not depicted). The 1–3 cells left show all the features of GSCs (Fig. 4, BI–BIV). They are VASA positive (Fig. S4 A) and located at the apical end of the germarium close to the terminal filament in the niche; have a round spectrosome (Fig. S4 A, compare Fig. 4, BII and CII); stain positive for phosphorylation of mothers against Dpp (pMAD), a BMP signaling indicator present in GSCs (Song et al., 2004; compare Fig. 4, BIII and CIII); and do not express the differentiation marker bag of marbles (BAM; Fig. S4 B). This analysis confirms that also in stem cells, CAL1 is crucial for cell division and therefore also for differentiation.
Figure S4.
CAL1 knockdown blocks cell proliferation. (A) Confocal z-stack projection of nanos-Gal4, CID RNAi, and CAL1 RNAi germaria, stained for DAPI (blue), anti-VASA (yellow), and anti-1B1 (spectrosome, red). (B) Confocal z-stack projection of bam-Gal4, CID RNAi, and CAL1 RNAi germaria, stained for DAPI (blue) and anti-BAM (red). (C) Confocal z-stack projection of a nanos-Gal4 (20°C) germarium stained for DAPI (blue) and anti-VASA (yellow), anti-FIBRILLARIN (magenta), and anti-CAL1 (green) and stained for DAPI (blue), anti-1B1 (red), anti-CENP-C (yellow), and anti-CAL1 (green). Confocal z-stack projection of a CAL1 RNAi (20°C) stained for DAPI (blue), anti-VASA (yellow), anti-FIBRILLARIN (magenta), and anti-CAL1 (green) and stained for DAPI (blue), anti-1B1 (red), anti-CENP-C (yellow), and anti-CAL1 (green). Star indicates the position of the terminal filament; 3-d-old female flies; scale bar, 10 µm.
Figure 4.
CAL1 knockdown blocks cell proliferation. (A) CAL1 knockdown confirmation by real time qPCR. (B and C) Confocal z-stack projection of nanos-Gal4 (B) and CAL1 RNAi (C) germaria, stained for DAPI (cyan), anti-1B1 (spectrosome, red), and anti-pMAD (labels GSCs, yellow). (D and E) Confocal z-stack projection of nanos-Gal4 (D) and CAL1 RNAi germaria (E), stained for DAPI (cyan), anti-1B1 (spectrosome, red), anti-CID (green), and anti-CENP-C (yellow). Star indicates the terminal filament; dotted lines represent follicle cells; 3-d-old female flies; scale bar, 10 µm; DV and EV, 5 µm.
Given that CAL1 is located at both centromeres and nucleoli, we investigated the depletion of both CAL1 pools in GSCs. In nanos-Gal4 flies (Fig. S4 C), CAL1 colocalizes with both CENP-C and the nucleolar marker FIBRILLARIN (Fig. S4 C). In the CAL1 knockdown samples, CAL1 is still present in the nucleolus of GSCs, colocalizing with FIBRILLARIN (Fig. S4 C); however, it is missing from the centromeres, as we could not distinguish any CAL1 signal outside of the nucleolus overlapping with CENP-C (Fig. S4 C). Indeed, neither CENP-C nor CID is detectable at centromeres in the knockdown GSCs (Fig. 4, D and E). From our observations, we conclude that (a) knockdown depletes the pool of CAL1 at centromeres, but not the one at the nucleolus; and (b) knockdown of centromeric CAL1 is responsible for the loss of functional centromeres in GSCs.
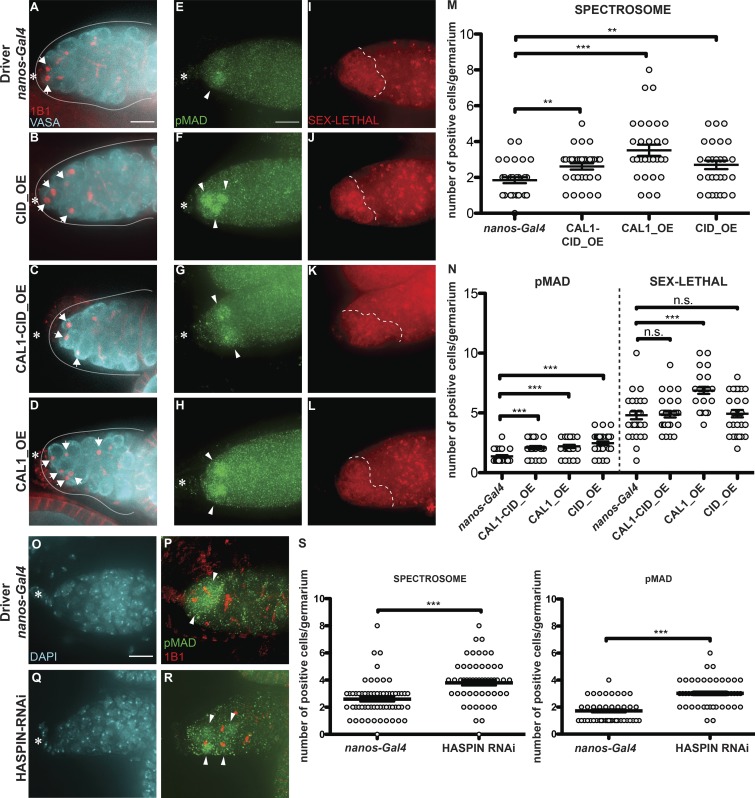
Overexpression of CAL1 and CID, as well as HASPIN knockdown, promotes stem cell self-renewal
To further explore centromere function in GSCs, we performed overexpression of CID, CAL1, or CAL1 together with CID. For this purpose, we crossed flies carrying CID-mCherry (CID_OE), CAL1-YFP (CAL1_OE), or both CAL1-YFP and CID-mCherry (CAL1-CID_OE) transgenes to a nanos-Gal4 driver line. Ovaries from F1 progeny were screened for correct localization of the fusion proteins, confirmed using antibody staining against CID and FIBRILLARIN (Fig. S5, A–O′). As expected, the nanos-Gal4 control does not show any YFP or RFP fluorescence (Fig. S5, A–E′). In the CAL1-CID_OE, CAL1-YFP colocalizes with CID-mCherry and with CID antibody (Fig. S5, F–I [arrow] and Fig. S5 J), but we could not detect any colocalization with FIBRILLARIN in nucleoli (Fig. S5, F′–I′ [arrow] and Fig. S5 J′). In the CAL1_OE, CAL1 localizes as expected at centromeres (Fig. S5, K–N, arrows, and Fig. S5 O) and at nucleoli (Fig. S5, K–N′ [arrowheads] and Fig. S5 O′). We also show that CID_OE colocalizes with CID antibody (Fig. S5 P). We next quantified the number of round spectrosomes, using antibody staining against 1B1, indicative of GSC and CB cells (Fig. 5, A–D and M). In nanos-Gal4, we found an average of two spectrosomes/germarium (Fig. 5 A [arrows] and Fig. 5 M; nanos-Gal41B1 = 1.84 ± 0.16, n = 30 germaria). In the CID_OE and CAL1-CID_OE, this number increases ∼1.4-fold (Fig. 5, B and C [arrows] and Fig. 5 M, CID_OE1B1 = 2.70 ± 0.24; CAL1-CID_OE1B1 = 2.61 ± 0.17, n = 30 germaria), while in the CAL1_OE, the number almost doubles (Fig. 5 D [arrows] and Fig. 5 M; CAL1_OE1B1 = 3.51 ± 0.31, n = 30 germaria). To measure the GSC/CB balance, we used the stem cell marker pMAD (Song et al., 2004) and SEX-LETHAL (SXL; Fig. 5, E–L and N), a marker that labels the GSC–CB transition. SXL is present from GSCs up to the two-cell cyst stage and can therefore be used to define the size of the germarium compartment containing GSCs and early differentiated cells (Chau et al., 2009). Our quantification shows that nanos-Gal4 germaria have approximately one pMAD-positive cell (nanos-Gal4pMAD = 1.36 ± 0.10, n = 30 germaria; Fig. 5, E and N), while this number doubles in all the overexpression lines (Fig. 5, F–H and N; CAL1-CID_OEpMAD = 2.10 ± 0.13; CAL1_OEpMAD = 2.20 ± 0.13 and CID_OE = 2.46 ± 0.15). SXL staining revealed that there is no difference in the number of SXL-positive cells between the nanos-Gal4 control, CID_OE, or CAL1-CID_OE (nanos-Gal4SXL = 4.80 ± 0.33, n = 30 germaria; CAL1-CID_OESXL = 4.86 ± 0.25; CID_OE = 4.93 ± 0.30; Fig. 5, I–K and N), while overexpression of CAL1 alone is responsible for an increase in the number of SXL-positive cells compared with the control (CAL1_OESXL = 6.86 ± 0.27, n = 30 germaria; Fig. 5, L and N). This analysis shows that CID, CAL1-CID, and CAL1 overexpression leads to an increase in the number of GSCs compared with the control. We next calculated the SXL/pMAD ratio as a measure for the number of GSCs, CBs, and two-cell cysts in the germarium. In control germaria, there are 3.5 SXL-positive cells for each pMAD-positive cell (nanos-Gal4SXL/pMAD = 3.53 ± 0.29). This ratio does not change in the CAL1_OE (CAL1_OESXL/pMAD = 3.38 ± 0.22), which indicates that differentiation occurs at an expected rate compared with the control. However, in the CAL1-CID_OE and CID_OE, ∼2 SXL-positive cells are present for each pMAD-positive cell (CAL1-CID_OESXL/pMAD = 2.47 ± 0.23; CID_OESXL/pMAD = 2.23 ± 0.19; Fig. S5 Q). This means that GSCs overexpressing CAL1-CID and/or CID self-renew rather than differentiate. To exclude the possibility that phenotypes were due to the genetic background of the responder flies, we conducted the same analysis on lines in which the overexpression is not induced and found similar values to the nanos-Gal4 control (not depicted). Altogether, our results suggest that the overexpression of CAL1 and CID together, as well as CID alone, promotes self-renewal, while CAL1 overexpression stimulates proliferation.
Figure S5.
CID and CAL1 overexpression and HASPIN knockdown promote stem cell self-renewal. (A–O′) Wide-field z-stack projection of nanos-Gal4, UAS_CAL1-YFP_UAS_CID-mCherry (CID-CAL1_OE), and UAS-CAL1-YFP (CAL1_OE) germaria, stained for anti-CID and or anti-FIB (cyan). Star, terminal filament; arrows, centromeres; arrowheads, nucleolus; 3-d-old female flies. (P) Wide-field z-stack projection of nanos-Gal4 and UAS_CID-mCherry germaria stained for DAPI (cyan) and anti-CID (green). (Q) Ratio of the number of SXL-positive cells to the number of pMAD-positive cells. n.s., not significant; **, P < 0.005; ***, P < 0.0005. (R) Confocal z-stack projection bam-Gal4 and CAL1 RNAi germaria stained for DAPI (blue), anti-BAM (red) and anti-CAL1 (green). Germ cells belonging to the 16-cell cyst chamber were selected based on the VASA marker (not depicted) and the lack of BAM signal in the control and in the CAL1 RNAi. (S) Confocal z-stack projection of bam-Gal4 and CENP-C RNAi germaria, stained for DAPI (blue), anti-BAM (red), and anti-CENP-C (yellow). Star, terminal filament; white dotted line in R and S, the end of the BAM-positive region; arrowheads, BAM-positive cells. Scale bar, 10 µm; inset, 5 µm.
Figure 5.
CID and CAL1 overexpression and HASPIN knockdown promote stem cell self-renewal. (A–D) Wide-field z-stack projection of nanos-Gal4 (A), UAS_ CID-mCherry (CID_OE; B) UAS_CAL1-YFP_UAS_CID-mCherry (CAL1-CID_OE; C), and UAS_CAL1-YFP (CAL1_OE; D) germaria, stained for VASA (cyan) and anti-1B1 (spectrosome, red). (E–L) Confocal z-stack projection of nanos-Gal4 (E and I), CID_OE (F and J), CAL1-CID_OE (G and K), and CAL1_OE (H and L) germaria, stained for anti-pMAD (green) and anti-SXL (red). (M) Spectrosome quantification. (N) pMAD and SXL quantification. (O–R) Wide-field z-stack projection of nanos-Gal4 (O and P) and HASPIN RNAi (Q and R) germaria stained for DAPI (cyan), anti-pMAD (green), and anti-1B1 (spectrosome red). (S) Spectrosome (left) and pMAD (right) quantitation in nanos-Gal4 and HASPIN RNAi. Data are represented as the mean ± SEM; ***, P < 0.0005; **, P < 0.005, n.s., not significant; calculated with unpaired t test with Welch’s correction. Star, terminal filament; arrows, spectrosome; arrowheads, pMAD-positive cells; dotted line, SXL regions; solid line, germarium; 3-d-old female flies; scale bar, 10 µm.
Because we found that HASPIN has a role in CID assembly, and given its proposed role in GSC asymmetric division in male flies (Xie et al., 2015), we tested whether HASPIN knockdown disrupts the GSC/CB balance in the germarium. We found that HASPIN knockdown germaria have a higher number of round spectrosomes, as well as a higher number of pMAD-positive cells, compared with the control (nanos-Gal41B1 = 2.6 ± 0.19, n = 60 germaria analyzed; nanos-Gal4pMAD = 1.72 ± 0.10, n = 54 germaria analyzed; HASPIN_RNAi1B1 = 3.80 ± 0.19, n = 60 germaria analyzed; HASPIN_RNAipMAD = 3.01 ± 0.13, n = 54 germaria analyzed; Fig. 5, O–S). These results confirm that also HASPIN regulates GSC/CB content in Drosophila germaria.
GSC self-renewal disrupts asymmetric inheritance of CID
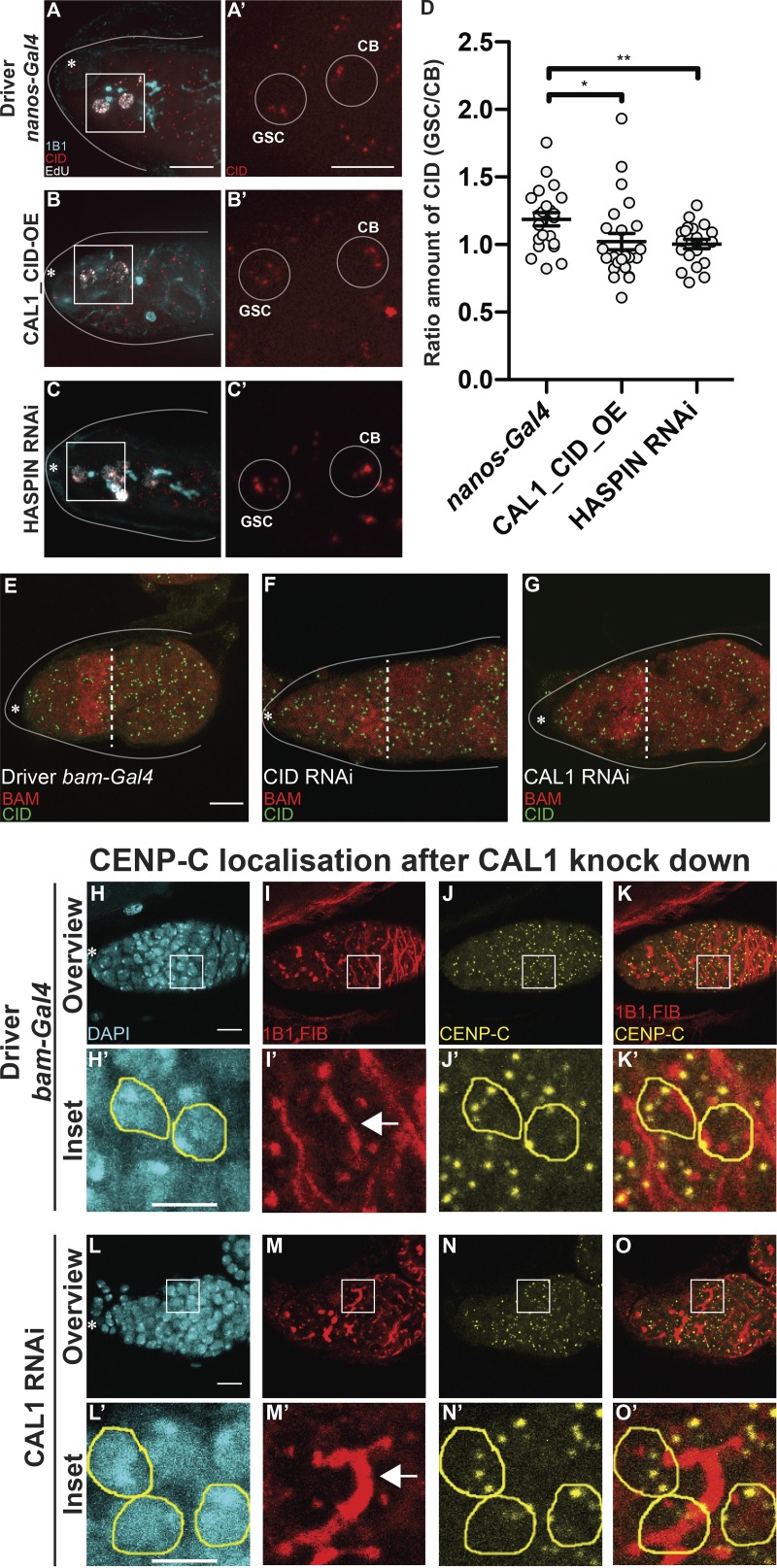
To investigate whether the asymmetric inheritance of CID between GSC and CB has a role in regulating the stem cell asymmetric division (Fig. 5), we quantified the amount of CID in replicating GSC-CB couples in controls, CAL1-CID_OE, and HASPIN knockdown germaria (Fig. 6, A–D). We again observed CID asymmetry in control couples (Fig. 6, A, A′, and D; ratio GSC/CB = 1.18 ± 0.04, n = 22 GSC-CB couples analyzed). Notably, CID asymmetry is lost in both CAL1-CID_OE or HASPIN knockdown couples (Fig. 6, B–D, ratio GSC/CB: CAL1-CID_OE = 1.02 ± 0.06, n = 24 GSC-CB couples analyzed; HASPINRNAi = 1.00 ± 0.03, n = 20 GSC-CB couples analyzed). This result indicates that in germaria enriched with GSCs at the expense of CBs, there is also a loss of asymmetric CID inheritance.
Figure 6.
GSC self-renewal disrupts CID asymmetric inheritance. (A–C′) Wide-field z-stack projection of nanos-Gal4 (A and A′), CAL1-CID_OE (B and B′), and HASPIN RNAi (C and C′) germaria stained for 1B1 (spectrosome, cyan), EdU (white), anti-CID (red). (D) Ratio of the total amount of CID detected in the GSC and the total amount of CID detected in the CB at S phase. (E–G) Confocal z-stack projection of bam-Gal4 (E), CID RNAi (F), and CAL1 RNAi (G) germaria, stained for anti-BAM (red) and anti-CID (green). (H–O′) Confocal z-stack projection of bam-Gal4 (H–K′) and CAL1 RNAi (L–O′) germaria, stained for DAPI (blue), anti-1B1 (red), anti-FIBRILLARIN (red), and anti-CENP-C (yellow). 16-cell cysts were selected based on the fusome morphology (arrow) in the control (H′–K′) and in the CAL1 RNAi (L′–O′). Data are represented as the mean ± SEM; *, P < 0.05; **, P < 0.005, calculated with unpaired t test with Welch’s correction. Star, terminal filament; dotted line, end of the BAM-positive region; arrow, fusome; solid line, germarium; 3-d-old female flies; scale bar, 10 µm; inset, 5 µm.
To examine CAL1 and CID requirements at later stages of egg development, we performed knockdown experiments for CENPs using the bam-Gal4 driver (active in 4–8-cell cysts). Ovaries were stained for VASA and BAM to mark 4–8-cell cysts. Surprisingly, we noticed that cell division past this stage is not impaired by depletion of either CID or CAL1 (Fig. 6, E–G). Remarkably, in the CID RNAi, we did not observe a significant diminishment of CID compared with the control (Fig. 6, E and F). In the CAL1 RNAi, CID levels appear to be reduced, but cell division proceeds normally (Fig. 6, E and G). In 16-cell cysts, after the BAM region, we confirmed a reduction of CAL1 in the CAL1 knockdown samples compared with the bam-Gal4 driver (Fig. S5 R). To check the impact of CAL1 reduction on centromere assembly, we antibody-stained samples against CENP-C. In the control cysts, identified through the fusome morphology (Fig. 6, H′–K′, arrows in Fig. 6 I′), two to four centromere foci closely opposed to the nucleolus are normally visible (Unhavaithaya and Orr-Weaver, 2013; Fig. 6, J′–K′). In the CAL1 knockdown sample (Fig. 6, L′–O′), we did not observe any change in the amount of CENP-C (Fig. 6, N′–O′). Since CID/CENP-C levels were not decreased after expression of CID/CAL1 RNAi using the bam-Gal4 driver, we sought to confirm this knockdown approach in germaria. Therefore, we tested the functionality of the driver on another centromere protein (CENP-C). Our results (Fig. S5 S) confirm effective CENP-C knockdown at this stage. In addition, since other drivers successfully knocked down CAL1 and CID, this observation supports the idea that at this stage CID and CENP-C are already assembled at centromeres and that CAL1 function is dispensable, at least for the cell division occurring after the eight-cell stage.
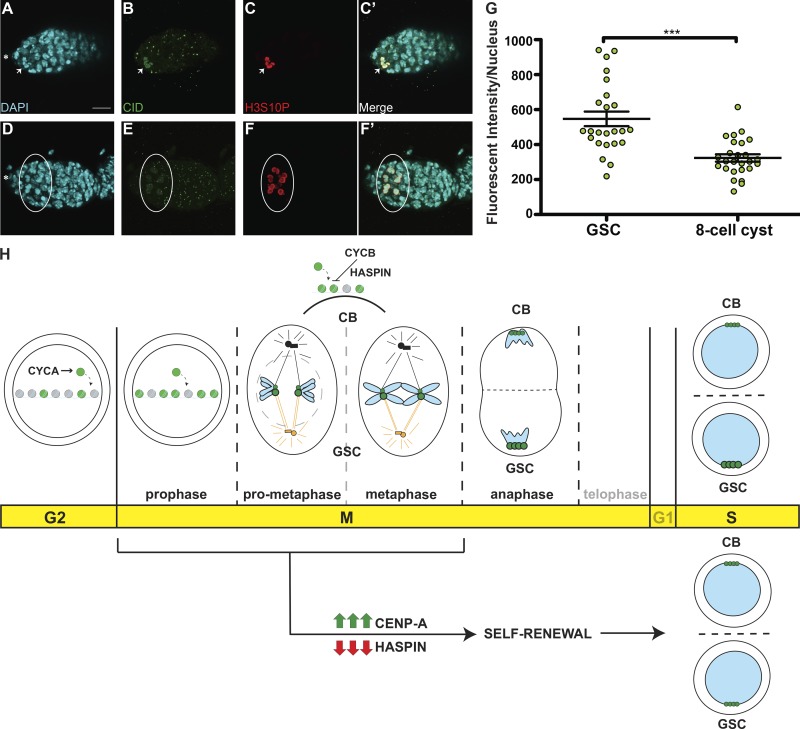
CID assembly dynamics differ between GSCs and cysts
To better understand the dynamics of CID assembly in GSCs and differentiated CCs, we measured the amount of CID per nucleus in both cell types to detect possible differences. We used H3S10P to mark GSCs and synchronously dividing eight-cell cysts at prophase (Fig. 7, A–F′). We noted that anti-CID staining at prophase labels centromeric CID but also shows a nuclear noncentromeric localization. As we did not observe this localization with CID-GFP, it is possible that it results from this specific antibody combination. Therefore, we focused our quantification on centromeric CID signals only. Compared with prophase GSCs, CC nuclei are smaller, yet centromeric foci are present in a similar number. From our quantification, we detected an ∼40% diminishment in the total amount of CID in CCs at the eight-cell stage (CC = 323.4 ± 20.94, n = 26 cell analyzed; Fig. 7 I), compared with GSCs (GSC = 547.2 ± 41.57, n = 24 cell analyzed; Fig. 7 I). This indicates a dramatic change in CID assembly dynamics, such that it is not replenished to 100% each cell division. Taken together with our observation of no significant reduction in CID after CID or CAL1 RNAi at this stage (Figs. 6 and S5), these data suggest that CID is inherited from the GSCs with little new CID loading occurring in cysts.
Figure 7.
Cyst cells incorporate less CID compared with GSCs. (A–F′) Confocal z-stack projection of a nanos-Gal4 germarium, stained for DAPI (cyan), anti-CID (green), and anti-H3S10P (red), to highlight a GSC (A–C′, arrow) and eight-cell cysts (D–F′, circle) in prophase. (G) Quantification of CID fluorescence intensity (integrated density) at centromeres in GSCs and eight-cell cysts at prophase obtained using wide-field microscopy. Data are represented as the mean ± SEM; ***, P < 0.0001. Star, terminal filament; 3-d-old female flies; scale bar, 10 µm. (H) Model for centromere assembly during the cell cycle. After replication, at early G2 phase, centromere assembly starts, promoted by CYCA, and centromeric nucleosomes (green) replace canonical nucleosomes (gray). This process continues until at least prophase. Excessive CID deposition is prevented by CYCB through HASPIN. At prometaphase, microtubules from centrosomes attach to centromeres through the kinetochore. At this point, sister chromatid pairs are loaded with differential amounts of CID (green) and CENP-C (not depicted) at centromeres. Chromosomes that retain more CID (bigger centromeres, figurative), make bigger kinetochores and attract more microtubules nucleating from the daughter centrosome (orange) and will be inherited by the GSC. At anaphase, and at replication, centromeres are clustered at the opposite sides of the two daughter nuclei. CID and CENP-C asymmetry is detected also at S phase. Telophase and G1 are shown as transparent because of the lack of data for these two phases. CID overexpression or HASPIN knockdown promotes GSC self-renewal and disrupts CID asymmetric inheritance.
Discussion
In this study, we performed a detailed characterization of centromere dynamics throughout the cell cycle in Drosophila female GSCs. Our analysis reveals that GSCs initiate CID incorporation after replication and that its deposition continues until at least prophase (Fig. 7 H). Drosophila neural stem cells follow the same trend. Notably, this timing is different from existing studies in other metazoans. We also found that CYCA, CYCB, and HASPIN are critically involved in CID (and CENP-C) loading at centromeres. According to our model (Fig. 7 H), CYCA promotes centromere assembly, while CYCB prevents excessive deposition of CID, through the HASPIN kinase. Moreover, chromosomes that will be inherited by GSCs are labeled with a higher amount of CID and capture more spindle microtubules (Fig. 7 J). Importantly, we show that overexpression of CAL1 and CID together, as well as HASPIN knockdown, promotes stem cell self-renewal, disrupting the asymmetric inheritance of CID. Depletion of CAL1 in stem cells blocks cell division, while CAL1 overexpression causes GSC-like tumors, highlighting its crucial role in cell proliferation. We raise three main points of discussion: (1) the biological significance of centromere assembly in G2-M phase; (2) CAL1 is a cell proliferation marker; and (3) CID incorporation into centromeric chromatin occurs before meiosis.
Biological significance of centromere assembly in G2-M phase
Cell cycle time
According to our data, CID deposition occupies a wide window of time from after replication and early G2 phase to prophase. The assembly of GSC centromeres during the G2/M transition could be due to the contraction of the G1 phase, a characteristic of stem cells (Pauklin and Vallier, 2013; White and Dalton, 2005; Becker et al., 2006). Yet, in fly embryonic divisions, G1 phase is missing, and instead CID loading occurs at anaphase (Schuh et al., 2007). Therefore, G2/M assembly might be a unique property of stem cells. This timing is also similar to the one found for Drosophila spermatocytes, which assemble centromeres in prophase of meiosis I (Dunleavy et al., 2012; Raychaudhuri et al., 2012). These spermatocytes undergo an arrest in prophase I for days, indicating a gradual loading of CID over a long period of time. Intriguingly, a similar phenomenon has been recently observed in G0-arrested human tissue culture cells and starfish oocytes (Swartz et al., 2019). Given that GSCs are mostly in G2 phase (Yamashita et al., 2003), Drosophila stem cells might show similar properties to quiescent cells. According to the most recent models, there is a dual mechanism for CENP-A deposition: (a) a rapid pulse during G1 in mitotically dividing cells; and (b) a slow but constant CENP-A deposition in nondividing cells to actively maintain centromeres (Swartz et al., 2019). Indeed, while previous studies in Drosophila NBs show a rapid pulse of CENP-A incorporation at telophase/G1 (Dunleavy et al., 2012), the majority of the loading could occur between G2 and prophase. Our new results also support this model.
Cell cycle regulation
Incorporation of CID before chromosome segregation might reflect a different CYCLIN-CDK activity in these cells. For instance, it has been already shown that in Drosophila GSCs CYCLIN E, a canonical G1/S cyclin, exists in its active form (in combination with Cdk2) throughout the cell cycle, indicating that some of the biological process commonly occurring in G1 phase might actually take place in G2 phase (Ables and Drummond-Barbosa, 2013). This is in line with our functional findings, where depletion of CYCA causes a decreased efficiency in CID and CENP-C assembly. We also found that this loss might be independent from CAL1. Surprisingly, correct CID deposition in GSCs also requires CYCB and HASPIN. Indeed, an inhibitory mechanism for CID deposition through CYCB has already been proposed in mammals (Stankovic et al., 2017). Interestingly, in Drosophila male GSCs, centromeric CAL1 is reduced between G2 and prometaphase (Ranjan et al., 2019), further suggesting a role for additional regulators of CID assembly, such as CYCA/B or HASPIN, at this time.
Epigenetic mechanism to drive cell fate during stem cell asymmetric division
According to our results, asymmetric cell division of GSCs is epigenetically regulated by differential amounts of centromeric proteins deposited at sister chromatids, which in turn can influence the attachment of spindle microtubules and can ultimately bias chromosome segregation. It is interesting to speculate on the temporal sequence of these events. Two scenarios can be proposed: (a) the nucleation of microtubules from the GSC centrosome requires bigger kinetochores; or (b) bigger kinetochores require a higher amount of spindle fibers to attach. Our results together with recent studies support the latter scenario. In fact, in Drosophila male GSCs, asymmetric distribution of centromeric proteins is established before microtubule attachment. Furthermore, microtubule disruption leaves asymmetric loading of CID intact, while it disrupts the asymmetric segregation of sister chromatids (Ranjan et al., 2019). Our data confirm this model, as we observe symmetric segregation of CID upon HASPIN knockdown. Indeed, in vertebrates HASPIN knockdown causes spindle defects (Wang et al., 2012; Yamagishi et al., 2010; Kelly et al., 2010). Specifically, we observed that a 1.2-fold difference in CID and CENP-C levels between GSC and CB chromosomes can bias segregation. While this difference is small, it fits with the observation that small changes in CENP-A level (on the order of 2–10% per day) impact on centromere functionality in the long run (Swartz et al., 2019). In Drosophila male GSCs, an asymmetric distribution of CID on sister chromatids >1.4-fold was reported (Ranjan et al., 2019). This higher value might reflect distinct systems in males and females or the quantitation methods used. Importantly, CID asymmetry in males is established in G2/prophase, in line with the time window we define for CID assembly. Further support for unexpected CID loading dynamics comes from our finding that GSCs in G2/prophase contain ∼30% more CID on average compared with S phase, indicating that CID is not replenished to 100% each cell cycle. Interestingly, the time course of H3T3P appearance during the GSC cell cycle closely follows the timing of CID incorporation, suggesting that the asymmetric deposition of CID might drive the differential phosphorylation of the histone H3 on sister chromatids. Finally, our results are in line with findings that the long-term retention of CENP-A in mouse oocytes has a role in establishing asymmetric centromere inheritance in meiosis (Smoak et al., 2016).
CAL1 is a cell proliferation marker
Our functional studies support a role for CAL1 in cell proliferation, with no apparent role in asymmetric cell division (Figs. 4 and 5). Indeed, centromeric proteins have been already proposed as biomarkers for cell proliferation (Swartz et al., 2019). Specifically, our functional analysis of centromeric proteins, as well as the HASPIN kinase, allowed us to discriminate between the classic role of centromeres in cell division and a role in asymmetric cell division. In our favorite scenario, CAL1 is needed to make functional centromeres crucial for cell division, while the asymmetric distribution of CID sister chromatids regulates asymmetric cell division and might depend on other factors, such as HASPIN. However, we cannot rule out that the effects on cell fate observed with our functional analysis might reflect alternative CAL1 functions outside of the centromere, for example due to changes in chromosome structure or gene expression.
CID incorporation into centromeric chromatin occurs before meiosis
Centromeres are crucially assembled in GSCs and therefore before meiosis of the oocyte takes place. Thus, it is possible that the 16-cell cysts inherit centromeric proteins synthesized and deposited in the GSCs, and the rate of new CID loading is reduced. This would explain why CAL1 function at centromeres is dispensable at this developmental stage.
Ultimately, our results provide the first functional evidence that centromeres have a role in the epigenetic pathway that specifies stem cell identity. Furthermore, our data support the silent sister hypothesis (Lansdorp, 2007), according to which centromeres can drive asymmetric division in stem cells.
Materials and methods
Generation of transgenic flies
Transgenic lines expressing either C-terminal tagged CAL1-YFP, or CID with the mCherry tag inserted between the N-terminus and the histone fold domain (CID-mCherry), or both (CAL1-YFP-CID-mCherry) under control of UASp sequences were generated by transposable (P) element transformation of pUASp vector (kind gift from X. Chen, Johns Hopkins University, Baltimore, MD) in w1118embryos (injection, selection, and balancing by BestGene). Specifically, CAL1-YFP and CID-mCherry constructs were placed in tandem following UASp sequences in the same plasmid. cid and cal1 cDNA were amplified from wild type (Oregon R). mCherry containing three codons for glycine residues at both sides was inserted in between cid N-terminal and cid C-terminal as described (Schuh et al., 2007). Cloning of pUASp_CID-mCherry and pUASp CAL1-YFP was performed through Gibson Assembly (NEB), combining multiple fragments including the gene of interest and the FP tag. Primer sequences used are as follows. (1) pUASp-CID-mCherry, pUASp-CID-Nt_Fw: 5′-AGGCCACTAGTGGATCTGGATCCTATGCCACGACACAGCAGAGCCAAGC-3′; CID-Nt_Rv-mCherry: 5′-ATCCTCCTCGCCCTTGCTCACCATACCACCACCGGTCTGGTTTTGCGCA-3′; mCherry_Fw-CID-Nt: 5′-TGCGCAAAACCAGACCGGTGGTGGTATGGTGAGCAAGGGCGAGGAGGAT-3′; CID-Ct_Fw: 5′-GCATGGACGAGCTGTACAAGGGTGGTGGTAGGCGGCGCAAAGCGGCCAA-3′; mCherry-Rv: 5′-TTGGCCGCTTTGCGCCGCCTACCACCACCCTTGTACAGCTCGTCCATGC-3′; CID-Ct-pUASp_Rv: 5′-TTAACGTTAACGTTCGAGGTCGACTCTAAAATTGCCGACCCCGGTCGCA-3′; (2) pUASp-CAL1-YFP, pUASp-CAL1_Fw: 5′-ATAGGCCACTAGTGGATCTGGATCCTATGGCGAATGCGGTGGTGGACGA-3′; CAL1-YFP_Rv: 5′-TCCTCGCCCTTGCTCACCATCTTGTCACCGGAATTATTCTCGAGTATGC-3′; CAL1-YFP_Fw: 5′-CAGCATACTCGAGAATAATTCCGGTGACAAGATGGTGAGCAAGGGCGAGGA-3′; pUASp-YFP_Rv: 5′-GTTAACGTTAACGTTCGAGGTCGACTTTACTTGTACAGCTCGTCCATGC-3′. pUASp-CAL1-YFP_UAS_CID-mCherry was performed through restriction cloning of the fragment CAL1 YFP and UASp sequence into the pUASp-CID-mCherry plasmid in front of CID-mCherry. Primers of the UASp sequence used are as follows. EcoRI-UASp_Fw: 5′-CCGAATTCTTACATACATACTAGAATTGGC-3′; UASp-NotI_Rv: 5′-CCGCGGCCGCTGCACTGAATTTAAGTGTATACTTC-3′.
Fly stocks and husbandry
Stocks were cultured on standard cornmeal medium (NUTRI-fly) preserved with 0.5% propionic acid and 0.1% Tegosept at 20°C under a 12-h light-dark cycle. UAS-RNAi lines were obtained from the Bloomington Stock Center (CYCA 35694; CYCB 38981; HASPIN 57787) and Vienna Drosophila RNAi Center (CAL1 45248; CENP-A/CID 102090). The germline-specific promoters nanos and bam were used to drive GAL4 expression (P{w[+mC] = UAS-Dcr-2.D}1, w[1118]; P{w[+mC] = GAL4-nos.NGT}40, provided by Bloomington Stock Center, #25751; bam-Gal4 was a kind gift from M.T. Fuller, Stanford University, Stanford, CA).
Crosses were performed at 20°C, 25°C, and 29°C, specifically: CAL1 and CID knockdown using the nanos-Gal4 driver were performed both at 25°C and 20°C, while CAL1 and CID knockdown using the bam-Gal4 driver were both conducted at 29°C. CYCA knockdown with nanos-Gal4 driver was performed at 25°C. CYCB knockdown using the nanos-Gal4 was performed at 25°C, HASPIN knockdown using the nanos-Gal4 driver was performed at 20°C, and then larvae were moved at 29°C for 8 d. Crosses for overexpression were performed using nanos-Gal4 driver at 25°C. Transgenic lines expressing GFP-tagged CENP-A/CID and RFP-tagged H2Av (heterozygotes; Schuh et al., 2007) under respective endogenous promoters were a kind gift from C. Lehner (University of Zurich, Zurich, Switzerland). Results obtained from each experiment rely on three biological replicates, unless otherwise specified.
Immunofluorescence
GSCs usually undergo mitotic division at very low frequency (Yamashita et al., 2003); therefore, to increase our chance to catch multiple cell cycle phases during cell division at once, we used young female flies (<1 d old) for centromere assembly quantifications and measurements of the asymmetry in the replication couples. To quantify metaphase GSCs in Fig. 3, we used young female flies 30 min old. For all the other experiments, we used 3-d-old female flies. Ovaries from young adult females were dissected in 1× PBS and fixed in 4% PFA. For quantification of CID in NBs, brains from third-instar larvae were dissected and fixed as described above. To carry out the tubulin staining, ovaries were fixed in ice-cold methanol for 20 min at 4°C, followed by acetone at −20°C for an additional 2 min. After fixation, samples were immediately washed in 1× PBS, 0.4% Triton X-100 (0.4PBT). Samples were then blocked in 0.4PBT with 1% BSA for 3–4 h at room temperature and incubated with primary antibodies (in blocking buffer) overnight at 4°C and with secondary antibodies (in blocking buffer) for 1 h at room temperature.
EdU assays
Ovaries from young female flies were dissected and incubated for 30 min with EdU (0.01 mM) in 1× PBS and then fixed as described. After washing in 0.4PBT, ovaries were incubated for 30 min in the dark with 2 mM CuSO4, 300 uM fluorescent azide and 10 mM ascorbic acid. Samples were then washed with 0.4PBT for 10 min and then blocked and stained as described above.
Antibodies
For immunostaining, the following antibodies were used: rabbit anti-CENP-A (CID) antibody (Active Motif 39719; 1:500), rat anti-CID (Active Motif 61735; 1:500), guinea pig anti-CENP-C (Erhardt et al., 2008; 1:1,000), sheep anti-CENP-C aa 1-732 (this study, 1:2,000), rabbit anti-H3S10P (Abcam ab5176, 1:1,000), mouse anti-H3S10P (Abcam ab14955; 1:1,000), rabbit anti-H3T3P (MERK 05-746R; 1:1,000), rabbit anti-VASA (Santa Cruz sc-30210; 1:250), goat anti-VASA (Santa Cruz sc-26877; 1:100), mouse anti-Fibrillarin (Abcam ab4566; 1:500), mouse anti-BAM (Developmental Studies Hybridoma Bank ab10570327; 1:10), mouse anti-CYCA (Developmental Studies Hybridoma Bank, A12 ab528188; 1:250 of the concentrated version), mouse anti-CYCB (Developmental Studies Hybridoma Bank, F2F4 ab2245815; 1:250 of the concentrated version), rabbit anti-pMAD (Abcam ab52903; 1:250), mouse anti-SPECTROSOME/1B1 (Developmental Studies Hybridoma Bank ab528070; 1:50), rat anti-DEADPAN (Abcam 195173; 1:100), mouse anti-tubulin (Abcam ab44928; 1:100), and rabbit anti-CAL1 (Bade et al., 2014; 1:1,000).
Confocal microscopy
Images of immunostained ovaries mounted in SlowFade Gold antifade reagent (Invitrogen S36936) were taken using an inverted Fluoview 1000 laser scanning microscope (Olympus) equipped with a 60× oil-immersion UPlanS-Apo objective (NA 1.2). The samples were excited at 404, 473, 559, and 635 nm, respectively, for DAPI and Alexa Fluor 488, 546, and 647. Light was guided to the sample via D405/473/559/635 dichroic mirror (Chroma). The emission light was guided via a size-adjustable pinhole, set at 115 µm. Fluorescence passed through a 430–455-, 490–540-, 575–620-, 655–755-nm bandpass filter for detection of DAPI and Alexa Fluor 488, 546, and 647, respectively, in sequential mode. Images were acquired as z-stacks with a step size of 0.5 µm.
Superresolution microscopy
Superresolution images of immunostained ovaries mounted in SlowFade Gold antifade reagent (Invitrogen S36936) were acquired using structured illumination microscopy (SIM). Samples were prepared on high precision cover glass (Zeiss). 3D SIM images were acquired on an N-SIM (Nikon Instruments) using a 100× 1.49-NA lens and refractive index–matched immersion oil (Nikon Instruments). Samples were imaged using a Nikon Plan Apo TIRF objective (NA 1.49, oil immersion) and an Andor DU-897X-5254 camera using 405-, 488-, 561-, and 640-nm laser lines. Z-step size for Z stacks was set to 0.120 µm as required by manufacturer software. For each focal plane, 15 images (five phases, three angles) were captured with the NIS-Elements software. SIM image processing, reconstruction, and analysis were performed using the N-SIM module of the NIS-Element Advanced Research software. Images were checked for artifacts using the SIM check software (http://www.micron.ox.ac.uk/software/SIMCheck.php). Images were reconstructed using NiS Elements software v4.6 (Nikon Instruments) from a z-stack comprising ≥1 µm of optical sections. In all SIM image reconstructions, the Wiener and Apodization filter parameters were kept constant.
Wide-field microscopy
Images of immunostained ovaries mounted in SlowFade Gold antifade reagent (Invitrogen S36936) were acquired using a DeltaVision Elite microscope system (Applied Precision) equipped with a 100× oil-immersion UPlanS-Apo objective (NA 1.4). Images were acquired as z-stacks with a step size of 0.2 µm. Fluorescence passed through a 435/48, 525/48, 597/45, 632/34 nm bandpass filter for detection of DAPI, Alexa Fluor 488, mCherry, and Alexa Fluor 647, respectively, in sequential mode.
Quantification
For each quantification, one cell/germarium was considered, unless specified otherwise. Images from a single cell (nucleus) were projected (maximum intensity) to capture all the centromeres present in the cell at a specific cell cycle phase. ImageJ software (National Institutes of Health; Schneider et al., 2012) was used to measure fluorescent intensity of CID in the following way (Fig. S1). The background was subtracted from the projected image. The threshold was adjusted, and the image was converted to binary. Overlapping centromeres were separated using the command “watershed.” Next, the command “analyze particles” was used to select centromeres. Size was adjusted to eliminate unwanted objects. Finally, integrated density (MGV*area) or MGVs from each centromere focus were extracted and used as fluorescent intensity to measure the total amount of fluorescence per nucleus. In Figs. 1, 2, 3, 6, and S1, we used the integrated density, because for stages such as replication, metaphase, and anaphase, centromeres are highly clustered and single centromere foci cannot be separated. In the quantification of CID in GSCs and cysts, we used the integrated density, because of the strong clustering observed after projection of the cysts. In the remaining quantifications of CID (Figs. 2 and S2), we used MGV. Quantification of pMAD- and SXL-positive cells was obtained by counting the positive cells for each signal through the z-stack of each image. Statistical analysis was performed using Prism software. Data distribution was assumed to be normal, but this was not formally tested. The P value in each graph shown was calculated with unpaired t test with Welch’s correction. In addition, for the graph shown in Fig. 3 E, we used Mann–Whitney U test and the Wilcoxon matched-pairs signed rank test.
RNA isolation and qPCR
Drivers (nanos-Gal4) and CAL1 knockdown flies were collected 7 d after hatching at 20°C and then dissected to extract ovaries. Drivers (nanos-Gal4) and HASPIN knockdown flies were collected after crossing was performed as described above. Total RNA from each sample was stored in TRIzol (Ambion, Life Technologies, 1559-6026) at −80°C until processing. RNA extraction and purification was performed with RNeasy MinElute clean up kit (Qiagen 74204). All the isolated RNAs were then standardized to the same concentration, and cDNA was synthesized (High Capacity RNA to cDNA, Thermo Fisher Biosciences, 4387406). qPCR was performed using the Applied Biosystem StepOnePlus Instrument and Power Up Sybr Green Master mix (Applied Biosystems A25780). The following genes, from Drosophila genome, were considered for this experiment: Glyceraldehyde 3-phosphate dehydrogenase (gapdh); Ribosomal Protein L32 (rpl32/rp49); nuclear-fall-out (nuf); and cal1. Primers for all the considered genes were designed using MacVector to amplify 75–150-bp fragments of the desired gene (Fig. S2 A). Before the qPCR experiment, these primers were tested with a mixture of cDNA from Drosophila ovaries to make sure they would amplify only a single region from the genome. Next, we checked primer efficiencies with a dilution curve (10−1 to 10−5) to make sure their range was within the negligible value of 1.9–2.0. Among the reference genes considered (gapdh; rpl32/rp49; nuf), nuf is highly stable at 20°C, between both control and the knockdown sample. Therefore, qPCR samples were standardized with nuf, and relative fold change values were calculated in Microsoft Excel and standardized against our reference gene based on published formulas (Livak and Schmittgen, 2001). Each qPCR experiment consisted of two biological replicates, and each sample was analyzed using three technical replicates per qPCR experiment. According to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Bustin et al., 2009), primer sequences used, relative efficiency, and amplification factor used in the calculation are as follows: gapdh, Fw 5′-GCTGGTGCCGAATACATCGTGG-3′; Rv 5′-CCAAGTTGACGCCGCAAACG-3′; efficiency, 90.7%, amplification factor, 1.91; rpl32/rpl49, Fw 5′-CCGCTTCAAGGGACAGTATCTGATGC-3′; Rv 5′-TTCTGCATGAGCAGGACCTCCAGC-3′; efficiency, 88.1%; amplification factor, 1.89; nuf, Fw 5′-TGGCGAAAATGAGTATCCCACCC-3′; Rv 5′-GGTTGTGTCCACTGTTGTTACCCACG-3′; efficiency, 105.8%; amplification factor, 2.06; cal1, Fw 5′-GTGAACGACAAGAGATTCCAGCGAC-3′; Rv 5′-AGTCCCTGCTCGGTCAGTGTGAAG-3′; efficiency, 102.9%; amplification factor: 2.03; haspin, Fw 5′-ACGTCGAAGCTCAATATGCCA-3′; Rv 5′-ACGGAAGTGGTGTACACTGATG-3′; efficiency, 109.2%; amplification factor, 2.09.
Online supplemental material
Fig. S1 illustrates the quantification strategy used in this study and shows the dynamics of CID assembly observed in Drosophila NBs as well as CYCA and CYCB knockdown confirmation. Fig. S2 shows the disrupted assembly dynamics of CENP-C and CAL1 on Drosophila GSC chromosomes upon CYCA knockdown. It also shows HASPIN knockdown confirmation and the temporal course of the H3T3P marker during Drosophila GSC mitosis. Fig. S3 shows the images used to determine asymmetric distribution of CID and spindle microtubules on GSC sister chromatids before chromosome segregation. Fig. S4 shows the consequences of CID and CAL1 knockdown using nanos-Gal4 driver on Drosophila germaria and centromere assembly. Fig. S5 shows the localization of overexpressed CAL1-YFP, CAL1-YFP_CID-mCherry, and CID-mCherry in the transgenic flies generated for this study. It also shows the ratio of SXL/pMAD-positive cells obtained for each transgenic line compared with the control and the CAL1 and CENP-C knockdown confirmation using bam-Gal4 driver.
Supplementary Material
Acknowledgments
We thank Gary Karpen and Sylvia Erhardt for antibodies. Antibodies obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the National Institutes of Health, are maintained at the University of Iowa, Department of Biology, Iowa City, IA.
E.M. Dunleavy, A.M. Kochendoerfer, C. Morgan, and A.E. Walsh are funded by Science Foundation Ireland PIYRA 13/YI/2187. A.A. Dattoli is funded by Government of Ireland Postdoctoral Fellowship 2017/1324 and Science Foundation Ireland PIYRA 13/YI/2187. B.L. Carty is funded by Government of Ireland Postgraduate Fellowship 2018/1208. The authors acknowledge the facilities and technical assistance of the National Centre for Bioengineering Science qPCR Facility, funded by National University of Ireland, Galway, and the Government of Ireland’s Program for Research in Third Level Institutions, Cycles 4 and 5, National Development Plan 2007-2013, the Centre for Microscopy & Imaging at the National University of Ireland, Galway (www.imaging.nuigalway.ie), and the Edinburgh Super-Resolution Imaging Consortium at the Institute of Genetics and Molecular Medicine at the University of Edinburgh (https://www.esric.org/). Stocks were obtained from the Bloomington Drosophila Stock Center (National Institutes of Health P40OD018537).
The authors declare no competing interests.
Author contributions: A.A. Dattoli and E.M. Dunleavy conceived and designed the study. A.A. Dattoli performed the molecular biological experiments, fluorescence microscopy imaging, and data analysis in the experiments presented in Figs. 1, 2 (A–O), 3, 4, and 6. B.L. Carty devised the GSC self-renewal assay (SXL/pMAD) in Fig. 5 and performed the molecular biological experiments, fluorescence microscopy imaging, and data analysis in the experiment presented in Figs. 3 (O and P) and 7 (A–I). A.A. Dattoli and A.M. Kochendoerfer performed the molecular biological experiments, fluorescence microscopy imaging, and data analysis in the experiments presented in Fig. 5 (A–N). A.A. Dattoli and C. Morgan performed the molecular biological experiments, fluorescence microscopy imaging, and data analysis in the experiments presented in Figs. 2 (P–T) and 5 (O–S). A.A. Dattoli and A.E. Walsh generated the transgenic fly lines. A.A. Dattoli and E.M. Dunleavy wrote the manuscript.
References
- Ables E.T., and Drummond-Barbosa D.. 2013. Cyclin E controls Drosophila female germline stem cell maintenance independently of its role in proliferation by modulating responsiveness to niche signals. Development. 140:530–540. 10.1242/dev.088583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K., and Henikoff S.. 2001. Centromeres are specialized replication domains in heterochromatin. J. Cell Biol. 153:101–110. 10.1083/jcb.153.1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire R.C., and Karpen G.H.. 2008. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9:923–937. 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bade D., Pauleau A.L., Wendler A., and Erhardt S.. 2014. The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev. Cell. 28:508–519. 10.1016/j.devcel.2014.01.031 [DOI] [PubMed] [Google Scholar]
- Barnhart M.C., Henning P., Kuich J.L., Stellfox M.E., Ward J.A., Bassett E.A., Black B.E., and Foltz D.R.. 2011. HJURP Is a CENP-A Chromatin Assembly Factor Sufficient to Form a Functional de Novo Kinetochore. J. Cell Biol. 194:229–243. 10.1083/jcb.201012017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K.A., Ghule P.N., Therrien J.A., Lian J.B., Stein J.L., van Wijnen A.J., and Stein G.S.. 2006. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell. Physiol. 209:883–893. 10.1002/jcp.20776 [DOI] [PubMed] [Google Scholar]
- Betschinger J., and Knoblich J.A.. 2004. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr. Biol. 14:R674–R685. 10.1016/j.cub.2004.08.017 [DOI] [PubMed] [Google Scholar]
- Black B.E., and Cleveland D.W.. 2011. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 144:471–479. 10.1016/j.cell.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone J.Q., and Doe C.Q.. 2008. Identification of Drosophila Type II Neuroblast Lineages Containing Transit Amplifying Ganglion Mother Cells. Dev. Neurobiol. 68:1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Caperta A.D., Rosa M., Delgado M., Karimi R., Demidov D., Viegas W., and Houben A.. 2008. Distribution patterns of phosphorylated Thr 3 and Thr 32 of histone H3 in plant mitosis and meiosis. Cytogenet. Genome Res. 122:73–79. 10.1159/000151319 [DOI] [PubMed] [Google Scholar]
- Chang C.H., Chavan A., Palladino J., Wei X., Martins N.M.C., Santinello B., Chen C.C., Erceg J., Belivea B.J., Wu C.T., et al. 2019. Islands of Retroelements Are Major Components of Drosophila Centromeres. PLoS. 17:e3000241 10.5061/dryad.rb1bt3j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau J., Kulnane L.S., and Salz H.K.. 2009. Sex-lethal facilitates the transition from germline stem cell to committed daughter cell in the Drosophila ovary. Genetics. 182:121–132. 10.1534/genetics.109.100693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chell J.M., and Brand A.H.. 2010. Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell. 143:1161–1173. 10.1016/j.cell.2010.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.C., Dechassa M.L., Bettini E., Ledoux M.B., Belisario C., Heun P., Luger K., and Mellone B.G.. 2014. CAL1 is the Drosophila CENP-A assembly factor. J. Cell Biol. 204:313–329. 10.1083/jcb.201305036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophersen N.S., and Helin K.. 2010. Epigenetic control of embryonic stem cell fate. J. Exp. Med. 207:2287–2295. 10.1084/jem.20101438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou N., Rubin T., and Huynh J.R.. 2013. Synaptonemal complex components promote centromere pairing in pre-meiotic germ cells. PLoS Genet. 9:e1004012 10.1371/journal.pgen.1004012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Sultan S., Taylor S.S., and Higgins J.M.G.. 2005. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19:472–488. 10.1101/gad.1267105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 448:151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Drpic D., Almeida A.C., Aguiar P., Renda F., Damas J., Lewin H.A., Larkin D.M., Khodjakov A., and Maiato H.. 2018. Chromosome Segregation Is Biased by Kinetochore Size. Curr. Biol. 28:1344–1356.e5. 10.1016/j.cub.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J.B. 2002. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis. 34:1–15. 10.1002/gene.10150 [DOI] [PubMed] [Google Scholar]
- Dunleavy E.M., Beier N.L., Gorgescu W., Tang J., Costes S.V., and Karpen G.H.. 2012. The cell cycle timing of centromeric chromatin assembly in Drosophila meiosis is distinct from mitosis yet requires CAL1 and CENP-C. PLoS Biol. 10:e1001460 10.1371/journal.pbio.1001460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., and Almouzni-Pettinotti G.. 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 137:485–497. 10.1016/j.cell.2009.02.040 [DOI] [PubMed] [Google Scholar]
- Erhardt S., Mellone B.G., Betts C.M., Zhang W., Karpen G.H., and Straight A.F.. 2008. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 183:805–818. 10.1083/jcb.200806038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun S.H., Gan Q., and Chen X.. 2010. Epigenetic regulation of germ cell differentiation. Curr. Opin. Cell Biol. 22:737–743. 10.1016/j.ceb.2010.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E.T., Bailey A.O., Yates J.R. III, Bassett E.A., Wood S., Black B.E., and Cleveland D.W.. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 137:472–484. 10.1016/j.cell.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., and Earnshaw W.C.. 2014. The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell. 30:496–508. 10.1016/j.devcel.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Del Arco A., Edgar B.A., and Erhardt S.. 2018. In Vivo Analysis of Centromeric Proteins Reveals a Stem Cell-Specific Asymmetry and an Essential Role in Differentiated, Non-proliferating Cells. Cell Reports. 22:1982–1993. 10.1016/j.celrep.2018.01.079 [DOI] [PubMed] [Google Scholar]
- Hemmerich P., Weidtkamp-Peters S., Hoischen C., Schmiedeberg L., Erliandri I., and Diekmann S.. 2008. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180:1101–1114. 10.1083/jcb.200710052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel M.J., Wei Y., Mancini M.A., Van Hooser A., Ranalli T., Brinkley B.R., Bazett-Jones D.P., and Allis C.D.. 1997. Mitosis-Specific Phosphorylation of Histone H3 Initiates Primarily within Pericentromeric Heterochromatin during G2 and Spreads in an Ordered Fashion Coincident with Mitotic Chromosome Condensation. Chromosoma. 106:348–360. [DOI] [PubMed] [Google Scholar]
- Hughes S.E., Miller D.E., Miller A.L., and Hawley R.S.. 2018. Female Meiosis: Synapsis, Recombination, and Segregation in Drosophila Melanogaster. Genetics. 208:875–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M., and Yamashita Y.M.. 2012. Asymmetric stem cell division: precision for robustness. Cell Stem Cell. 11:461–469. 10.1016/j.stem.2012.09.003 [DOI] [PubMed] [Google Scholar]
- Jansen L.E.T., Black B.E., Foltz D.R., and Cleveland D.W.. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176:795–805. 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S.H., Tseng C.Y., Wan C.L., Su Y.H., Hsieh C.C., Pi H., and Hsu H.J.. 2015. Aging and insulin signaling differentially control normal and tumorous germline stem cells. Aging Cell. 14:25–34. 10.1111/acel.12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G., and Allshire R.. 1997. The Case for Epigenetic Effects on Centromere Identity and Function. Trends Genet. 13:489–496. [DOI] [PubMed] [Google Scholar]
- Kelly A.E., Ghenoiu C., Xue J.Z., Zierhut C., Kimura H., and Funabiki H.. 2010. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 330:235–239. 10.1126/science.1189505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdorp P.M. 2007. Immortal strands? Give me a break. Cell. 129:1244–1247. 10.1016/j.cell.2007.06.017 [DOI] [PubMed] [Google Scholar]
- Lilly M.A., de Cuevas M., and Spradling A.C.. 2000. Cyclin A associates with the fusome during germline cyst formation in the Drosophila ovary. Dev. Biol. 218:53–63. 10.1006/dbio.1999.9570 [DOI] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Cauvin C., Moch C., Radford S.J., Sampaio P., Perdigoto C.N., Schweisguth F., Bardin A.J., Sunkel C.E., McKim K., et al. 2013. Aurora B and cyclin B have opposite effects on the timing of cytokinesis abscission in Drosophila germ cells and in vertebrate somatic cells. Dev. Cell. 26:250–265. 10.1016/j.devcel.2013.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias N.R., Mathieu J., and Huynh J.R.. 2015. Abscission is regulated by the ESCRT-III protein shrub in Drosophila germline stem cells. PLoS Genet. 11:e1004653 10.1371/journal.pgen.1004653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley K.L., and Cheeseman I.M.. 2014. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 158:397–411. 10.1016/j.cell.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley K.L., and Cheeseman I.M.. 2016. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17:16–29. 10.1038/nrm.2015.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J.M., and Bratu D.P.. 2015. Drosophila Melanogaster Oogenesis: An Overview In Drosophila Oogenesis: Methods in Molecular Biology. Vol. 1328 Bratu D and G McNeil, editors Humana Press, New York, NY: pp. 1–20. [DOI] [PubMed] [Google Scholar]
- Mellone B.G., Grive K.J., Shteyn V., Bowers S.R., Oderberg I., and Karpen G.H.. 2011. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 7:e1002068 10.1371/journal.pgen.1002068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho-Santos T., and Maiato H.. 2014. Plk1 puts a (Has)pin on the mitotic histone code. EMBO Rep. 15:203–204. 10.1002/embr.201438472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., and Desai A.. 2017. A Molecular View of Kinetochore Assembly and Function. Biology (Basel). 6:E5. https://doi.org/. 10.3390/biology6010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S., and Vallier L.. 2013. The cell-cycle state of stem cells determines cell fate propensity. Cell. 155:135–147. 10.1016/j.cell.2013.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P., Malone C.D., Navarro C., Newbold S.P., Hayes P.S., Sachidanandam R., Hannon G.J., and Lehmann R.. 2011. piRNA production requires heterochromatin formation in Drosophila. Curr. Biol. 21:1373–1379. 10.1016/j.cub.2011.06.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R., Snedeker J., and Chen X.. 2019. Asymmetric Centromeres Differentially Coordinate with Mitotic Machinery to Ensure Biased Sister Chromatid Segregation in Germline Stem Cells. Cell Stem Cell. 25:666–681.e5. 10.1016/j.stem.2019.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri N., Dubruille R., Orsi G.A., Bagheri H.C., Loppin B., and Lehner C.F.. 2012. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in Drosophila sperm. PLoS Biol. 10:e1001434 10.1371/journal.pbio.1001434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotelli M.D., Policastro R.A., Bolling A.M., Killion A.W., Weinberg A.J., Dixon M.J., Zentner G.E., Walczak E., Lilly M.A., and Calvi B.R.. 2019. A Cyclin A-Myb-MuvB-Aurora B Network Regulates the Choice between Mitotic Cycles and Polyploid Endoreplication Cycles. PLoS Genet. 15:e1008253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A., and Mitchison T.J.. 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA. 105:2415–2420. 10.1073/pnas.0712168105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann V., Chen C., Chiang C.-Y.A., Tiyaboonchai A., Mayer M., and Yamashita Y.M.. 2014. Centrosome-dependent asymmetric inheritance of the midbody ring in Drosophila germline stem cell division. Mol. Biol. Cell. 25:267–275. 10.1091/mbc.e13-09-0541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm R.B., Althoff F., Heidmann S., and Lehner C.F.. 2010. Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1. J. Cell Sci. 123:3768–3779. 10.1242/jcs.067934 [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M., Lehner C.F., and Heidmann S.. 2007. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 17:237–243. 10.1016/j.cub.2006.11.051 [DOI] [PubMed] [Google Scholar]
- Silva M.C.C., Bodor D.L., Stellfox M.E., Martins N.M.C., Hochegger H., Foltz D.R., and Jansen L.E.T.. 2012. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell. 22:52–63. 10.1016/j.devcel.2011.10.014 [DOI] [PubMed] [Google Scholar]
- Smoak E.M., Stein P., Schultz R.M., Lampson M.A., and Black B.E.. 2016. Long-Term Retention of CENP-A Nucleosomes in Mammalian Oocytes Underpins Transgenerational Inheritance of Centromere Identity. Curr. Biol. 26:1110–1116. 10.1016/j.cub.2016.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Wong M.D., Kawase E., Xi R., Ding B.C., McCarthy J.J., and Xie T.. 2004. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 131:1353–1364. 10.1242/dev.01026 [DOI] [PubMed] [Google Scholar]
- Stankovic A., Guo L.Y., Mata J.F., Bodor D.L., Cao X.J., Bailey A.O., Shabanowitz J., Hunt D.F., Garcia B.A., Black B.E., and Jansen L.E.T.. 2017. A Dual Inhibitory Mechanism Sufficient to Maintain Cell-Cycle-Restricted CENP-A Assembly. Mol. Cell. 65:231–246. 10.1016/j.molcel.2016.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz S.Z., McKay L.S., Su K.C., Bury L., Padeganeh A., Maddox P.S., Knouse K.A., and Cheeseman I.M.. 2019. Quiescent Cells Actively Replenish CENP-A Nucleosomes to Maintain Centromere Identity and Proliferative Potential. Dev. Cell. 51:35–48.e7. 10.1016/j.devcel.2019.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V., Feng L., and Chen X.. 2013. Asymmetric distribution of histones during Drosophila male germline stem cell asymmetric divisions. Chromosome Res. 21:255–269. 10.1007/s10577-013-9356-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y., and Orr-Weaver T.L.. 2013. Centromere proteins CENP-C and CAL1 functionally interact in meiosis for centromere clustering, pairing, and chromosome segregation. Proc. Natl. Acad. Sci. USA. 110:19878–19883. 10.1073/pnas.1320074110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Ulyanova N.P., Daum J.R., Patnaik D., Kateneva A.V., Gorbsky G.J., and Higgins J.M.. 2012. Haspin inhibitors reveal centromeric functions of Aurora B in chromosome segregation. J. Cell Biol. 199:251–268. 10.1083/jcb.201205106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J., and Dalton S.. 2005. Cell Cycle Control of Embryonic Stem Cells. Stem Cell Rev. 1:131–138. https://doi.org/SCR:1:2:131 [pii]\r. 10.1385/SCR:1:2:131 [DOI] [PubMed] [Google Scholar]
- Xie J., Wooten M., Tran V., Chen B.C., Pozmanter C., Simbolon C., Betzig E., and Chen X.. 2015. Histone H3 Threonine Phosphorylation Regulates Asymmetric Histone Inheritance in the Drosophila Male Germline. Cell. 163:920–933. 10.1016/j.cell.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., and Watanabe Y.. 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 330:239–243. 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- Yamashita Y.M., Jones D.L., and Fuller M.T.. 2003. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 301:1547–1550. 10.1126/science.1087795 [DOI] [PubMed] [Google Scholar]
- Yan D., Neumüller R.A., Buckner M., Ayers K., Li H., Hu Y., Yang-Zhou D., Pan L., Wang X., Kelley C., et al. 2014. A regulatory network of Drosophila germline stem cell self-renewal. Dev. Cell. 28:459–473. 10.1016/j.devcel.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Zhou X., Wang W., Deng W., Fang J., Hu H., Wang Z., Li S., Cui L., Shen J., et al. 2015. Dynamic phosphorylation of CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at centromeres. Dev. Cell. 32:68–81. 10.1016/j.devcel.2014.11.030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.