Mitochondria form a network that is constantly remodeled by fission and fusion. Abrisch et al. show that fission and fusion machineries converge at ER membrane contact sites (MCSs) to provide a unified platform where fission and fusion are coordinated to balance mitochondrial dynamics and morphology.
Abstract
The steady-state morphology of the mitochondrial network is maintained by a balance of constitutive fission and fusion reactions. Disruption of this steady-state morphology results in either a fragmented or elongated network, both of which are associated with altered metabolic states and disease. How the processes of fission and fusion are balanced by the cell is unclear. Here we show that mitochondrial fission and fusion are spatially coordinated at ER membrane contact sites (MCSs). Multiple measures indicate that the mitochondrial fusion machinery, Mitofusins, accumulate at ER MCSs where fusion occurs. Furthermore, fission and fusion machineries colocalize to form hotspots for membrane dynamics at ER MCSs that can persist through sequential events. Because these hotspots can undergo fission and fusion, they have the potential to quickly respond to metabolic cues. Indeed, we discover that ER MCSs define the interface between polarized and depolarized segments of mitochondria and can rescue the membrane potential of damaged mitochondria by ER-associated fusion.
Introduction
Mitochondrial morphology and dynamics are critical to normal cellular function. Mitochondrial morphology is maintained by a balance of constitutive fission and fusion reactions and by dynamic movements that occur along the cytoskeleton. Despite the fact that mitochondria rapidly fuse, divide, and move, they are somehow able to maintain a characteristic morphology (Twig et al., 2008; Youle and van der Bliek, 2012; Friedman et al., 2010). Disruption of this steady-state (dynamic equilibrium) morphology results in either a relatively fragmented or elongated network, both of which are associated with altered metabolic states and disease (Rambold et al., 2011; Wai and Langer, 2016). How fission and fusion machineries are coordinated to produce mitochondria of appropriate size is a fundamental question that is still unresolved.
Several aspects of mitochondrial dynamics and physiology are regulated by membrane contact sites (MCSs) with the ER. ER MCSs define the position of mitochondrial constriction and fission and provide a conduit for Ca2+ and lipid trafficking (Wu et al., 2018; Vance, 1990; Csordás et al., 1999; Rizzuto et al., 1998; Kornmann et al., 2009). Mitochondrial fission is regulated by members of the dynamin family of GTPases (including Drp1 in animal cells or its paralog Dnm1 in yeast; Smirnova et al., 2001; Lee et al., 2016; Bleazard et al., 1999). Fission dynamins are not membrane anchored, but rather are recruited from the cytoplasm to homo-oligomerize around the outer mitochondrial membrane (OMM) to constrict it in a process that is mechanically driven by GTP binding and hydrolysis (Ferguson and De Camilli, 2012). ER MCSs define the position where the constriction/division machineries assemble and where division occurs (Friedman et al., 2011; Lee et al., 2016; Cho et al., 2017). OMM and inner mitochondrial membrane (IMM) fusion is driven by integral membrane proteins that oligomerize in cis and trans to drive membrane fusion upon binding and hydrolysis of GTP (Santel and Fuller, 2001; Chen et al., 2003; Ferguson and De Camilli, 2012). OMM fusion is driven by the Mitofusin paralogs (Mfn1 and Mfn2), while IMM fusion is regulated by Opa1 (Ban et al., 2017; Meeusen et al., 2006; Legros et al., 2002; Misaka et al., 2002; Herlan et al., 2003; Lee et al., 2004). It is not known how the mitochondrial fusion machinery is positioned to determine the site of fusion. There have, however, been hints in the literature that fission and fusion can occur in rapid succession by an alternate mechanism termed “kiss-and-run” or “transient” fusion (Liu et al., 2009), where two mitochondria contact each other, mix contents, and subsequently divide while maintaining their original morphology. The existence of transient fusion events implies that fusion and fission could be spatially coordinated during transient fusion, but perhaps also more generally. Here we show that the fission and fusion machineries can assemble at the same ER MCS to modulate mitochondrial morphology in response to metabolic cues.
Results
Mitofusin is localized to ER MCSs
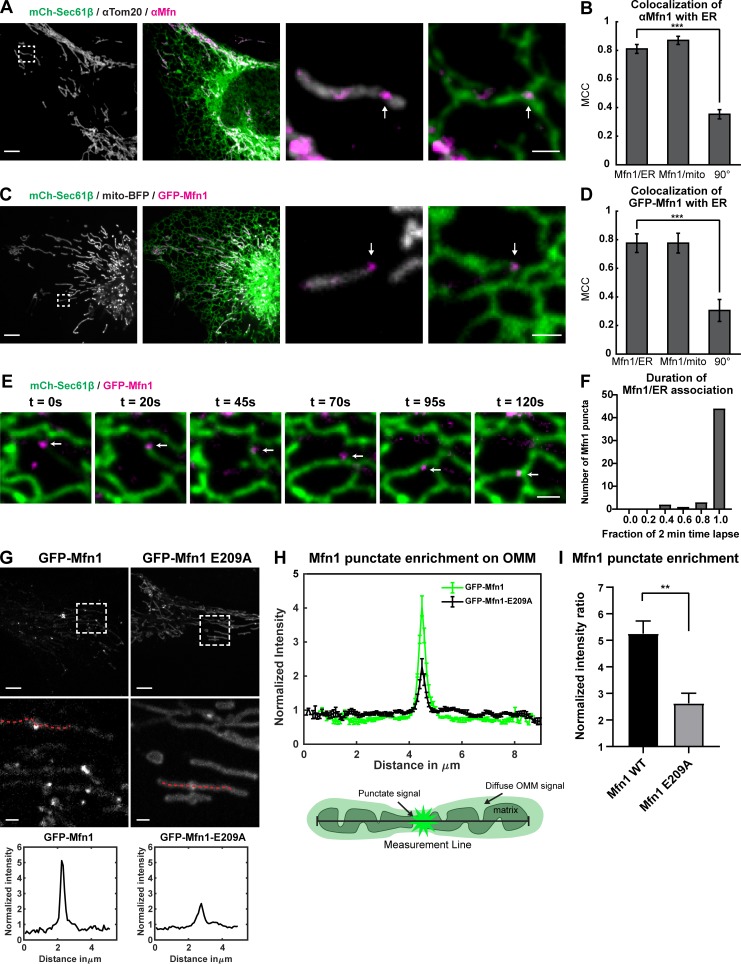
The position of mitochondrial fission is guided by ER MCSs that recruit the mitochondrial fission machinery (Friedman et al., 2011; Ji et al., 2017). We hypothesized that if fission and fusion are coordinated, then the mitochondrial fusion machinery might also assemble at ER MCSs. To test this, we transfected U-2 OS cells with a general ER marker (mCherry-Sec61β) and performed immunofluorescence with an antibody that is reported to recognize both Mfn1 and Mfn2, which will be referred to as Mfn (note that only Mfn1 was detected by quantitative RT-PCR [qPCR] in U-2 OS cells; Fig. S1 A), along with an antibody recognizing the OMM protein Tom20. Immunofluorescence staining revealed that, while Mfn weakly labeled the entire OMM like Tom20, Mfn also accumulated in punctate structures where ER tubules crossed the mitochondrial membrane (Fig. 1 A). We quantified the colocalization of endogenous Mfn puncta relative to the OMM marker Tom20 or the ER marker Sec61β by Manders colocalization coefficient (MCC; Manders et al., 1993; Dunn et al., 2011). Mfn puncta colocalized nearly as well with the ER as with the mitochondria (MCC = ∼0.8 vs. ∼0.9, respectively; Fig. 1 B). As a negative control, we rotated the Mfn image 90° with respect to the ER image and measured the random overlap, which was dramatically reduced to MCC = ∼0.35, indicating that the overlap of Mfn puncta with ER is significant and not due to chance.
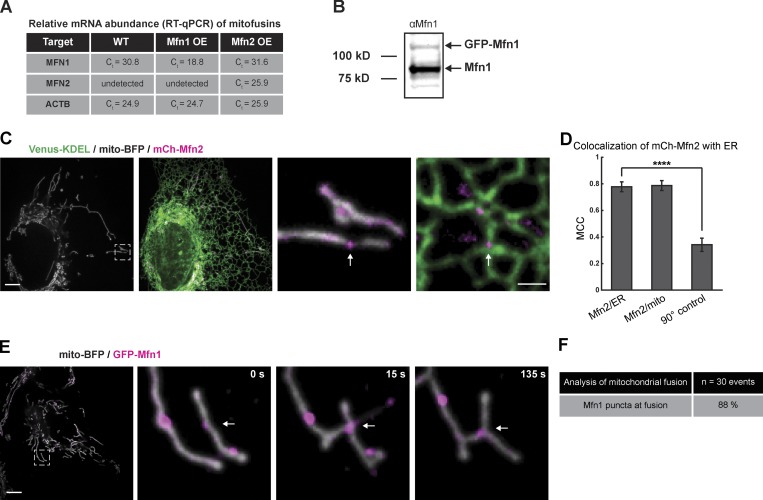
Figure S1.
Expression of endogenous and transfected Mitofusin. (A) Mean of triplicate threshold cycle (Ct) values for MFN1, MFN2, and ACTB mRNAs. U-2 OS cells were either not transfected (WT) or transfected with either GFP-Mfn1 or mCh-Mfn2 (OE). Target signifies the mRNA target of the primer set. (B) Western blot showing the signal from the anti-Mfn1 antibody in U-2 OS cells transfected with GFP-Mfn1 (molecular weight standards indicated on the left). (C) Representative image of a live cell expressing mCh-Mfn2 (magenta), Venus-KDEL (green), and mito-BFP (gray). Magnified merged images of inset show mCh-Mfn2 puncta relative to mitochondria and ER (right panels). (D) A graph of the MCC of GFP-Mfn2 relative to ER, mitochondria, or a 90° rotated Mfn2 relative to the original ER image. Significance determined as in Fig. 1 B. ****, P < 0.0001 by two-tailed Wilcoxon test; n = 30 regions. (E) Representative merged image of a cell expressing GFP-Mfn1 (magenta) and mito-BFP (gray) shows an Mfn1 punctum localized to the site of mitochondrial fusion event (at white arrow). (F) Table of fusion events scores 88% occur at a Mfn1 punctum (n = 42 events from 28 cells). Scale bars for whole cell, 5 µm; insets, 1 µm.
Figure 1.
Mitofusins form puncta that localize to ER tubules. (A) Representative image of a U-2 OS cell expressing mCh-Sec61β (ER, green) and immunostained with antibody against Mfn1/2 (magenta) and Tom20 (gray). Magnified merged image of insets show punctate distribution of Mfn1/2 (magenta) relative to mitochondria and ER (in right panels). (B) Graph of the MCC measured for Mfn1/2 relative to the ER, mitochondria, or a 90° rotated Mfn1/2 relative to the ER. Error bars represent SEM. ***, P < 0.0001 by two-tailed paired t test; normality was determined by Shapiro–Wilk test; n = 12 regions. (C) Representative live-cell image of a cell expressing GFP-Mfn1 (magenta), mCherry-Sec61β (green), and mito-BFP (gray). Magnified merged images of inset show GFP-Mfn1 puncta relative to mitochondria and ER (right panels). (D) Graph of the MCC of GFP-Mfn1 relative to ER, mitochondria, or a 90° rotated MFN1 relative to the ER. Significance determined as in B; ***, P = 0.0007, n = 10 regions. (E) Time-lapse images of live cells as in C reveals tracking of GFP-Mfn1 (magenta) puncta with ER tubules (green) over time. (F) Graph shows that the vast majority of Mfn1 puncta (84%) maintain their association with ER tubules during the entire 2-min video (n = 50 puncta from six cells). (G) Representative images of U-2 OS cells, GFP-Mfn1 or GFP-Mfn1-E209A. Insets and dashed lines correspond to the location of the linescans graphed below. (H) Linescan of Mfn1 signal on mitochondria showing the mean of five cells with seven linescans (as in Fig. 1 G) from each cell. Error bars represent SEM. Cartoon describes where measurements are taken and how signal is distributed on mitochondria. (I) Graph showing the difference in ratio of maximum punctate signal over the mean diffuse mitochondrial signal from GFP-Mfn1 and GFP-Mfn1-E209A. **, P = 0.0026 by two-tailed t test. Scale bars for whole cell image = 5 µm; insets = 1 µm. Error bars represent SEM.
To visualize Mfn1 localization relative to ER MCSs over time during live-cell imaging, we performed the same analyses as above on GFP-Mfn1. We cotransfected U-2 OS cells with markers for the ER (mCh-Sec61β) and mitochondria (mito-BFP) along with low levels of GFP-Mfn1 (immunoblot analysis confirms that exogenous GFP-Mfn1 is expressed at lower levels than endogenous Mfn1; Fig. S1 B). As expected, GFP-Mfn1 displayed the same pattern of localization as the endogenous protein (labeling of the OMM generally along with punctate enrichments at ER–mitochondria MCSs). The calculated MCC between GFP-Mfn1 puncta and the ER also recapitulated the behavior of the endogenous protein, being nearly identical to that with mitochondria (MCC = ∼0.77 and ∼0.78 respectively; Fig. 1, C and D). For comparison, we cotransfected cells with mCh-Mfn2, mito-BFP, and a general ER marker (Venus-KDEL) to measure whether Mfn2 puncta also localize to ER MCSs. Again, the calculated MCC revealed high overlap in signal between mCh-Mfn2 puncta and the ER, similar to mitochondria (MCC = ∼0.78 and ∼0.79, respectively; Fig. S1, C and D). To assess the dynamic localization of Mfn1 puncta relative to ER tubules, we tracked individual Mfn1 puncta and ER tubules over time by transfecting mito-BFP (not depicted), mCh-Sec61β, and GFP-Mfn1 and imaged live U-2 OS cells over 2-min time lapses (Fig. 1 E and Video 1). We found that of the 50 Mfn1 puncta analyzed, 84% remained ER associated for the entire 2-min time-lapse video (Fig. 1 F). Not only do Mfn1 puncta localize to ER MCSs, they also denote the location of mitochondrial fusion events (Fig. S1 E and Video 2); of 48 fusion events recorded, the vast majority (88%) are marked by an Mfn1 punctum (Fig. S1 F).
Video 1.
Mfn1 (magenta) tracking with an ER tubule (green) from Fig. 1 E. Two-minute time-lapse images of a live U-2 OS cell transfected with GFP-Mfn1 and mCh-Sec61β. Images were acquired every 5 s using a spinning-disk confocal microscope. Playback rate is 7.5 fps.
Video 2.
Mfn1 (magenta) at a mitochondrial fusion event (gray) from Fig. S1 E. Five-minute time-lapse image of a live U-2 OS cell transfected with mito-BFP and GFP-Mfn1. Images were acquired every 5 s using a spinning-disk confocal microscope. Playback rate is 7.5 fps.
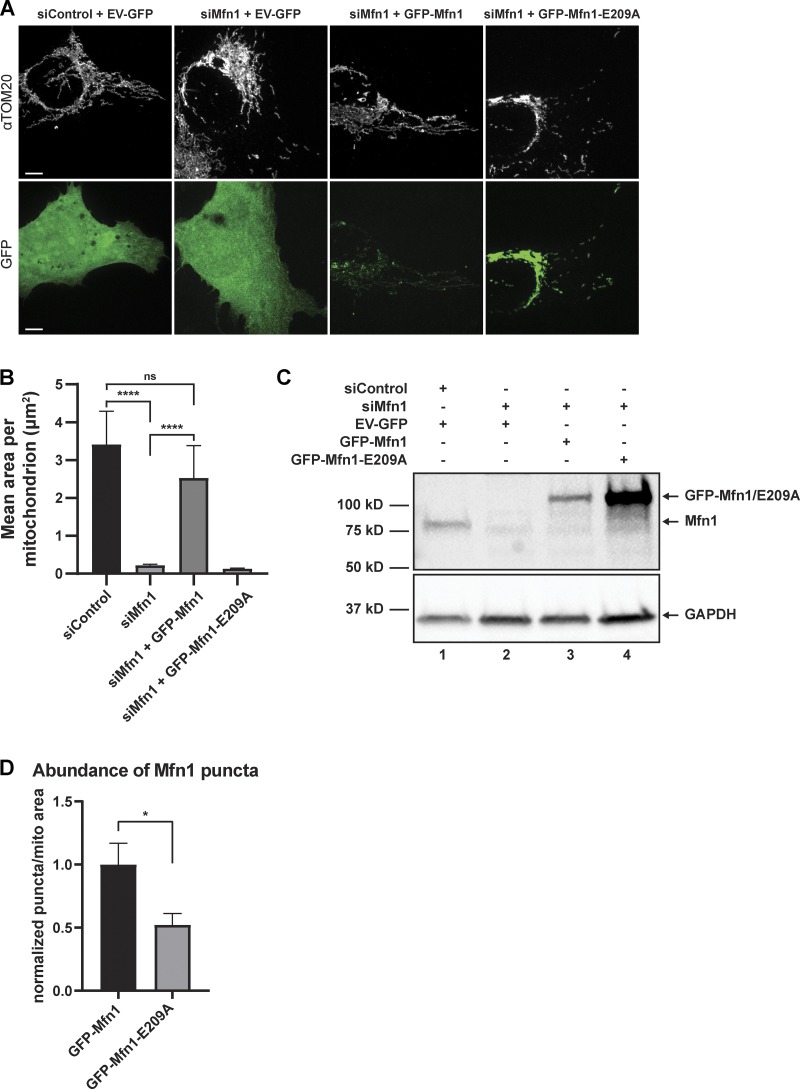
Mfn1 is a dynamin family member that forms oligomers in a manner that is dependent on nucleotide binding, which is critical for its fusogenic activity. We reasoned that the punctate accumulations of Mfn1 we observed may be dependent on its ability to dimerize. To test this, we generated the GTPase domain (G-domain) mutant Mfn1-E209A, which was reported to be deficient in three categories: (1) it is unable to hydrolyze GTP, (2) its G-domain is unable to homodimerize, and (3) it lacks fusogenic activity (Cao et al., 2017; Sloat et al., 2019). We confirmed that the fragmented mitochondrial morphology caused by depletion of Mfn1 by siRNA can be rescued by an siRNA-resistant GFP-tagged WT Mfn1 but not by the siRNA-resistant E209A GFP-Mfn1 mutant (Fig. S2, A–C; Cao et al., 2017). We then transfected either GFP-Mfn1 (WT) or GFP-Mfn1-E209A (E209A) along with mito-BFP in WT U-2 OS cells and assessed their ability to form punctate accumulations. We measured linescans over the length of mitochondria that appeared to have enrichments of signal in cells expressing either WT or E209A. Representative linescans revealed that E209A was more diffusely distributed throughout the OMM, while WT accumulated in puncta to a greater extent (Fig. 1 G). We then analyzed linescans of 35 mitochondria from five cells under each condition (WT versus E209A). Because punctate enrichments of GFP-Mfn1-E209A were much less obvious, we sought out the most prominent enrichments of signal in the images for analysis. Each linescan of Mfn1 signal was normalized to the same linescan of mito-BFP signal to account for changes in intensity due to differences in position relative to the focal plane. We computationally aligned the linescans from each cell by their maximum, and cell averages were computed to yield an overall average linescan (error bars represent SEM; Fig. 1 H). We then measured the maximum from both conditions and divided it by the corresponding mean intensity value of micrometers 2–4 on the x axis in Fig. 1 I to assess the fold increase in intensity between punctate enrichments and diffuse signal on the OMM. This analysis revealed that WT is approximately twofold more enriched than the most obvious enrichments of E209A in the cells we analyzed (Fig. 1 I). We also analyzed the number of Mfn1 puncta per mitochondrial area in WT and E209A-expressing cells and found that, in addition to being twofold more enriched over E209A, WT puncta were also twofold more abundant (Fig. S2 D). Taken together, these data support that Mfn1 puncta represent functional assemblies that localize to ER MCSs and mark the position of mitochondrial fusion.
Figure S2.
GFP-Mfn1, but not E209A, is capable of rescuing morphology. (A) Representative images of U-2 OS cells transfected with control siRNA and pAcGFP-C1 empty vector; anti-Mfn1 siRNA and pAcGFP-C1 empty vector; anti-Mfn1 siRNA and siRNA-resistant GFP-Mfn1; or anti-Mfn1 siRNA and siRNA-resistant GFP-Mfn1-E209A. Mitochondria were visualized by immunofluorescence using an anti-TOM20 antibody and native GFP fluorescence of transfected constructs. (B) Graph representing the mean mitochondrial area from cells treated and imaged as in A; ns and ****, P < 0.0001 as calculated by Kruskal–Wallis test. (C) Western blot showing relative protein levels of endogenous and exogenous Mfn1 after control or anti-Mfn1 siRNA treatment. (D) Graph showing the relative fold change of the number of puncta per mitochondrial area in cells transfected with either GFP-Mfn1 or GFP-Mfn1-E209A. *, P = 0.0377 by two-tailed t test; normality determined by Shapiro–Wilk test; n = 5 cells. Scale bar: 5 µm.
ER tubules mark sites of mitochondrial fusion
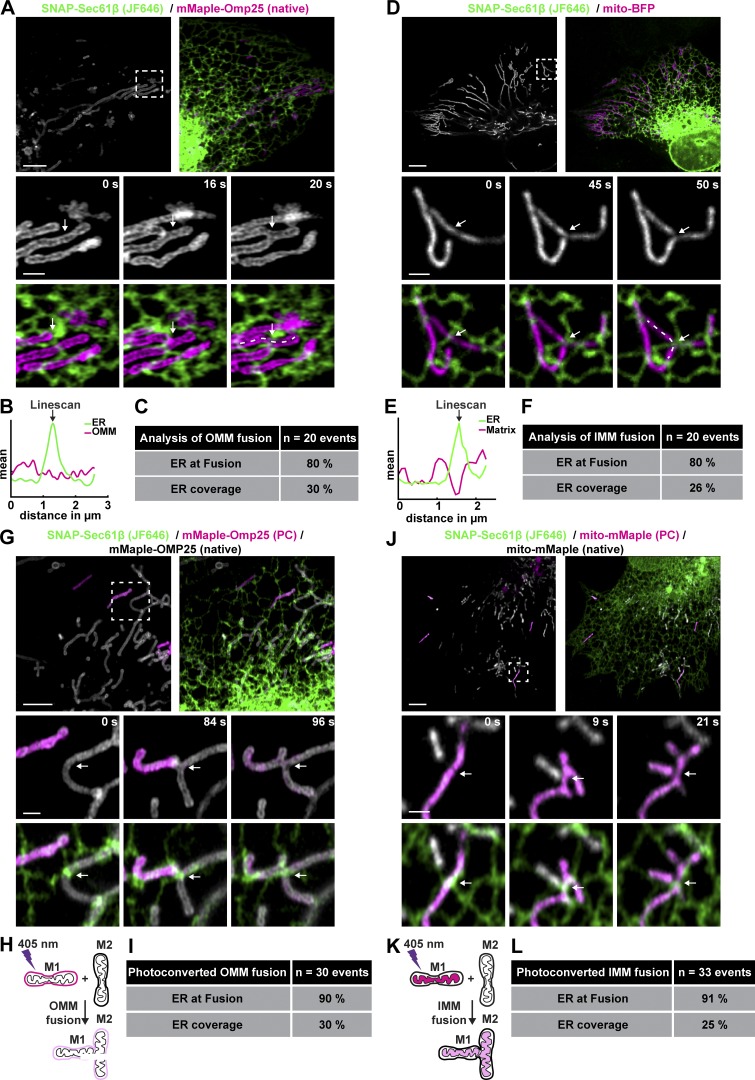
We next tested whether ER tubule crossings define the position of mitochondrial fusion as they do for mitochondrial fission. Cells were cotransfected with an OMM (mMaple-OMP25) and general ER marker (SNAP-Sec61β; JF646) and were imaged live to measure the percentage of apparent fusion events that occurred where ER tubules appear to contact mitochondria (Fig. 2, A–C; and Video 3). We performed a similar experiment with Snap-Sec61β and a soluble matrix marker (mito-BFP) to visualize apparent fusion events of the IMM (Fig. 2, D–F; and Video 4). We found that 80% of both OMM and matrix apparent fusion events are coincident with the location of a crossing ER tubule (Fig. 1, C and F). These numbers are much higher than would be expected based on the mitochondrial area that the ER overlaps (∼30%). A recent study using grazing-incidence structured illumination microscopy showed that ∼60% of apparent OMM mitochondrial fusion events in African green monkey kidney cells (COS7) occurred at ER tubules (the percentage of ER overlap with mitochondria was not reported for these experiments; Guo et al., 2018). Therefore, because scoring apparent fusion is subject to the limitations of even the best light microscopy, we sought a more accurate method that has been used by others to score the position and timing of fusion (Twig et al., 2006; Pham et al., 2012). We used a photoconvertible fluorescent protein, mMaple, to label the OMM or matrix, allowing us to photoconvert individual mitochondria and score fusion based on fluorescent protein transfer between photoconverted and native mitochondria upon a bona fide fusion event. The mMaple fusion protein has native excitation/emission maxima of ∼488/∼520 nm, respectively, and can be photoconverted to ∼550/590 nm upon stimulation with 405-nm light (McEvoy et al., 2012). We simultaneously converted approximately three mitochondria per cell from green to red by stimulating small regions encompassing each mitochondrion with low-power 405-nm laser and followed their fate for 8 min. First, we assessed fusion by fluorescent protein transfer between mitochondria upon fusion of the OMM and scored whether these events occurred at ER tubule crossings (Fig. 2, G and H; and Video 5). U-2 OS cells were cotransfected with an ER marker (SNAP-Sec61β, JF646) and a photo-convertible OMM marker (mMaple-OMP25). Using this more accurate method to score the time and position of OMM or IMM fusion events, we found that 90% of OMM fusion events occur at an ER tubule crossing (Fig. 2 I). Next, we assessed fusion by fluorescent protein transfer between mitochondria upon fusion of the matrix in cells cotransfected with an ER marker (SNAP-Sec61β, JF646) and a photo-convertible matrix marker (mito-mMaple; Fig. 2, J and K; and Video 6). Similarly, we found that 91% of matrix fusion events occur at an ER tubule crossing (Fig. 2 L).
Figure 2.
ER MCSs define the position of OMM and IMM fusion. (A) Representative image of a live U-2 OS cell expressing a marker of the ER (green) and OMM (gray or magenta). Magnified merged time-lapse images show an OMM fusion event (at arrow, middle and right panel) relative to a crossing ER tubule (at arrow, in bottom panels). (B) Linescan analysis (dashed line) at t = 20 s from A plots mean FI of an ER tubule that crosses over the location of fusion. (C) Percentage of OMM fusion events occurring at ER tubules relative to the mean coverage of ER along mitochondria (n = 20 events in seven cells). (D) As in A for cells expressing a marker of the ER (green) and mitochondrial matrix (magenta). Magnified merged time-lapse images show an IMM fusion event (at arrow, middle and bottom panel) relative to a crossing ER tubule (green, bottom panels). (E) As in B for 50-s panel in D. (F) As in C for D (n = 20 events in 17 cells). (G) Representative image of cells expressing an ER (green) and photoconvertible OMM marker (gray). Photoconversion of individual mitochondria converts gray mitochondria to magenta. Conversely, bona fide OMM fusion between magenta and gray mitochondria leads to content/color mixing (compare 84 s with 96 s). (H) Cartoon demonstrating content mixing of OMM upon fusion. (I) Percentage of OMM fusion events scored by content mixing that occur at a crossing ER tubule relative to the average percentage coverage of ER along mitochondria (n = 30 events in 14 cells). (J) As in G for a photoconvertible matrix marker. Time lapse images showing IMM fusion confirmed by content mixing (between 9 s and 21 s). (K) Cartoon demonstrating content mixing of matrix upon fusion. (L) As in I for matrix content mixing experiment in J (n = 33 events in 10 cells). Scale bars for whole cell = 5 µm; insets, 1 µm.
Video 3.
OMM (magenta) fusion at an ER tubule crossing (green) corresponding to Fig. 2 A. Time-lapse images over 2 min 16 s of a live U-2 OS cell transfected with SNAP-Sec61β and mMaple-OMP25. Frames were captured every 4 s using a laser-scanning confocal microscope. Playback rate is 7.5 fps.
Video 4.
Matrix (magenta) fusion at an ER tubule crossing (green) corresponding to Fig. 2 D. Time-lapse images over 8 min of a live U-2 OS cell transfected with SNAP-Sec61β and mito-BFP. Frames were captured every 5 s using a spinning-disk confocal microscope. Playback rate is 7.5 fps.
Video 5.
OMM fusion by content mixing (magenta and gray) at an ER tubule crossing (green) corresponding to Fig. 2 G. Time-lapse images showing 4 min 48 s of a live U-2 OS cell transfected with SNAP-Sec61β and mMaple-OMP25. Frames were captured every 3 s using a laser-scanning confocal microscope. Playback rate is 7.5 fps.
Video 6.
Matrix fusion by content mixing (magenta and gray) at an ER tubule crossing (green) corresponding to Fig. 2 J. Time-lapse images showing 1 min 30 s of a live U-2 OS cell transfected with SNAP-Sec61β and mito-mMaple. Frames were captured every 3 s using a laser-scanning confocal microscope. Playback rate is 7.5 fps.
Together, these data show that the vast majority (∼90%) of bona fide mitochondrial OMM and IMM fusion events occur at the intersection of ER tubules and mitochondria, statistics that are strikingly similar to the 88% of mitochondrial fission events scored to occur at ER MCSs (Friedman et al., 2011).
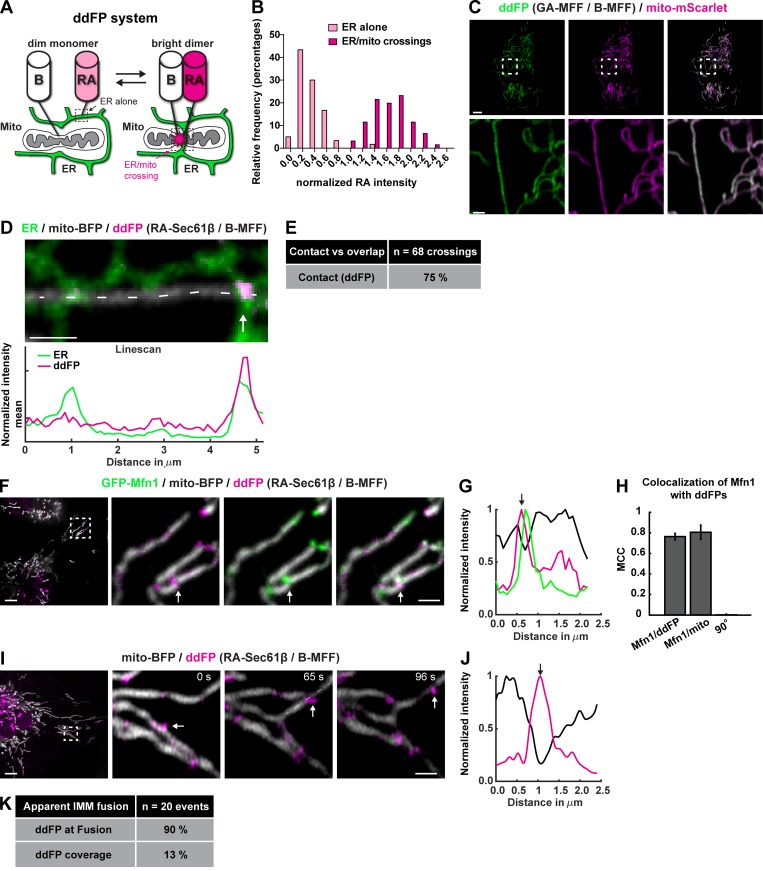
Dimerization-dependent fluorescent proteins (ddFPs) confirm ER contacts at Mfn1 puncta
We optimized a ddFP system to validate whether Mfn1 puncta and fusion events are localized to positions of close apposition between the ER and mitochondrial membrane (bona fide MCSs). The ddFP system consists of a heterodimeric fluorescent protein complex made up of a dim RFP or GFP (RA or GA, respectively) and protein binding partner (B) that lacks a chromophore. Upon dimerization of RA or GA with B, the brightness of RA or GA is increased (Ding et al., 2015). RA was fused to Sec61β to target it to the ER, and the B domain was fused to mitochondrial fission factor (MFF) to target it to the OMM. When both components are present in cells, the ER should display low-intensity RA signal in areas not contacting mitochondria. However, at ER MCSs with mitochondria, ER-localized RA should be close enough to dimerize with B on the mitochondrion, and RA should display a higher fluorescence intensity (FI) at that site (Fig. 3 A). This should manifest as a bimodal distribution of red signal in the image resulting from the contributions of monomeric RA (dim population) and dimeric B/RA (bright population). To validate the use of the ddFP system for measuring ER MCSs, we measured the distribution of RA signal intensity in cells cotransfected with RA-Sec61β, B-MFF, mito-BFP, and mNeonGreen-KDEL. RA FI was compared between 0.079-µm2 (8 × 8-pixel) regions along ER tubules where mitochondria were absent to 0.079-µm2 regions where ER tubules crossed over mitochondria. For each cell, we subtracted background and combined the values from all regions (ER with mitochondria and ER alone) and normalized to the mean of all RA intensity values in that cell, allowing us to compare relative intensities from different cells. This yielded two clear populations of RA signal intensity indicating the presence of a monomeric population and a heterodimeric population, which was approximately sevenfold brighter (Fig. 3 B). In other cell types, MFF has been reported to accumulate in puncta, and we wanted to rule out the possibility that MFF was forming puncta on its own and limiting the regions in which ER contact could be detected. This was addressed by testing whether B-MFF was accumulating in puncta on the OMM using transfected B-MFF and GA-MFF. Importantly, when we cotransfected B-MFF and GA-MFF in U-2 OS cells, the ddFP system diffusely labeled mitochondria and did not accumulate into puncta on its own (Fig. 3 C).
Figure 3.
Mfn1 puncta and fusion events localize to bona fide ER contact sites. (A) Cartoon demonstrating the ddFP system: the monomers are targeted to the mitochondria (B) and the ER (RA), and an increase in fluorescent signal is indicative of dimerization at MCSs. (B) Histogram of ddFP spot values taken from regions of ER alone (monomer) and regions where ER crossed mitochondria (dimer) validates signal increase upon dimerization (n = 120 spots). (C) Representative image of cells expressing GA-MFF (green dimerization partner), B-MFF, and mito-mScarlet (magenta) shows that B-MFF is not punctate without an ER partner. (D) Representative image of a cell expressing the ddFP system (RA-Sec61β + B-MFF) with a marker for ER (green) and mitochondria (mito-BFP; gray). Note that a bright ddFP signal (magenta) accumulates at a position where an ER tubule crosses the mitochondria. Linescan analysis of dashed line along the mitochondria shows relative FI of ddFP domains relative to ER tubule crossing. (E) Most but not all ER tubule crossings (75%) label positive for ddFP dimerization, indicating that ∼75% of ER tubule crossings are bona fide MCSs. (F) Representative image of cells expressing the ddFP system (magenta), GFP-Mfn1 (green), and mito-BFP (gray). (G) Magnified merged images and linescan analysis of F show the position of ddFP-positive domains relative to GFP-Mfn1 puncta along mitochondria. (H) Graph of MCC for GFP-Mfn1 puncta relative to ddFP-positive domains, mitochondria, or 90° rotated GFP-Mfn1 puncta shows that Mfn1 puncta and ddFP ER MCSs strongly overlap. (I) Representative merged time-lapse images of a ddFP-positive fusion event in a cell expressing the ddFP system (magenta) and mito-BFP (gray). (J) Linescan analysis of dashed line in I shows the relative FI of ddFP-positive domains along the length of a mitochondrion at the location of fusion (white arrow). (K) Percentage of fusion events that are ddFP positive for ER contact (90%) as in I relative to the mean coverage of ddFP (13%) on mitochondria. Scale bar: 5 µm in whole cell images; insets, 1 µm.
We then scored the percentage of apparent ER tubule–mitochondria crossings that were labeled with the ddFP system in cells cotransfected with an ER marker (mNeonGreen-KDEL), a mitochondrial matrix marker (mito-BFP), and the ddFP system (RA-Sec61β and B-MFF). We found that 75% of “apparent” ER tubule–mitochondria crossings (n = 68 crossings) are indeed bona fide ddFP ER MCSs that display a bright RA signal in foci where the ER tubule overlapped with the mitochondrion (Fig. 3, D and E). Next, we cotransfected cells with GFP-Mfn1, mito-BFP, and the ddFP system and asked whether Mfn1 puncta were colocalized to ER MCSs. Colocalization of Mfn1 puncta with ddFP ER MCS signal was high (MCC = ∼0.76) and similar to the mitochondria (MCC = ∼0.81; Fig. 3, F–H). If we rotate the Mfn1 image 90° to represent a scrambled position relative to the ddFP image, the overlap was lost (MCC = 0.0011; Fig. 3 H). Importantly, we scored that ∼90% of apparent IMM fusion events (n = 20 events in 11 cells) were spatially coincident with ddFP ER MCS signal, which strongly supports the notion that fusion happens at molecular distances between the ER and mitochondria (Fig. 3, I–K). Notably, the ddFP system is advantageous for the purpose of live-cell imaging of unaltered MCSs, because B has a relatively low binding affinity for RA (Kd = ∼7 µM; Ding et al., 2015).
Bidirectional membrane dynamics occur at ER–mitochondria MCSs
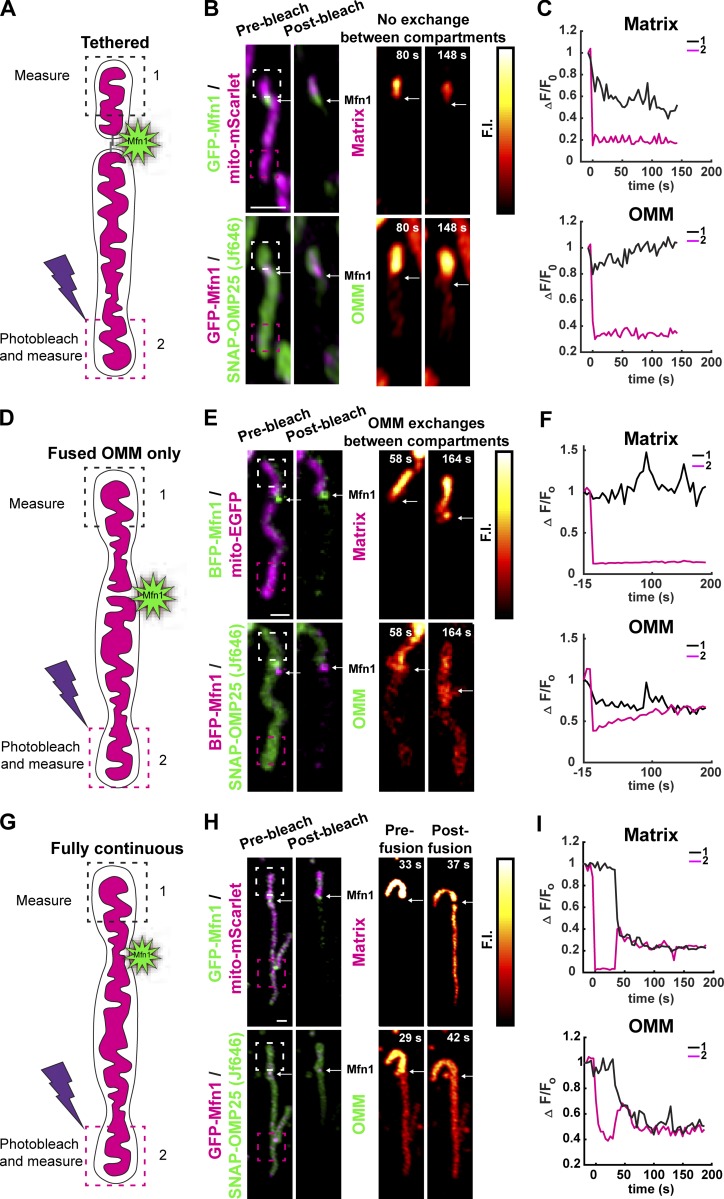
Mitochondrial membrane continuity cannot be conclusively determined by light microscopy because of its current resolution limitations. However, when light microscopy is coupled with techniques such as fluorescence loss in photobleaching (FLiP) and photoconversion, it is possible to probe the continuity of mitochondrial compartments using the diffusion of fluorescent proteins as a proxy. Mitochondria are known to exist in multiple membrane configurations: (1) tethered mitochondria where neither OMM nor IMM is continuous, (2) fused OMM and discontinuous IMM, and (3) continuous OMM and IMM. Visually continuous mitochondria can also modulate compartment continuity at points along their length in the absence of visually detectable morphological change (apparent fission or fusion events), as revealed by photoactivation and diffusion of photoactivatable GFP (Twig et al., 2008, 2006; Wong et al., 2019). Additionally, mitochondria undergo repeated constriction and relaxation of the IMM at locations defined by ER tubule crossings (Cho et al., 2017). Therefore, we hypothesized that ER MCSs at Mfn1 puncta may denote predefined branch points, or nodes, where IMM and OMM continuity can be reversibly modulated, and where we should be able to see all combinations of compartment continuity described above. To identify Mfn1-marked fusion intermediates, we performed FLiP experiments and measured the continuity of the OMM and IMM at Mfn1 puncta. Cells were cotransfected with a soluble matrix (mito-mScarlet) and OMM (SNAP-OMP25) marker, and with GFP-Mfn1 (Fig. 4 A, cartoon). We chose mitochondria to analyze that were solitary and contained an Mfn1 punctum that was located away from the tips (near the middle) to allow us to measure compartment continuity across a punctum from one side of the mitochondria to the other. A small region of the mitochondrion on one side of the Mfn1 punctum was irradiated long enough to bleach the entire continuous compartment, while simultaneously measuring FI on the bleached (box 2) and unbleached (box 1) side of the mitochondrion during recovery. This analysis revealed all three predicted membrane configurations at Mfn1 puncta: (1) mitochondria are tethered but IMM and OMM are not continuous (Fig. 4, A–C), (2) partially fused mitochondria where only OMM is continuous (Fig. 4, D–F), and (3) mitochondria that are initially tethered but later fuse resulting in OMM and matrix continuity (Fig. 4, G–I). Together, these data reveal that ER-associated Mfn1 puncta represent nodes where ER MCSs can regulate IMM and OMM continuity.
Figure 4.
Mfn1 puncta mark sites of reversible membrane continuity. (A) Cartoon description of a FLiP experiment designed to measure OMM and matrix continuity at Mfn1 puncta. (B) The matrix (mito-EGFP) and OMM (SNAP-OMP25) fluorescent markers were simultaneously photobleached (in box 2), which is located on one side of the Mfn1 punctum (Mfn1 in green or magenta, respectively, at white arrow). The FIs in box 1 and box 2 are imaged over time following photobleaching (PB). In snapshots shown, after t = 80 s and t =148 s after PB, there is still no diffusion of FI past the Mfn1 punctum (white arrow) for either the matrix (top images) or OMM signal (bottom images), which demonstrates that the matrix and OMM are tethered at the Mfn1 punctum but not continuous. (C) Graph of matrix and OMM FI measurements in box 1 and box 2 measured over time confirm that FI does not diffuse past the Mfn1 punctum for either marker. (D–F) Cartoon (D) and corresponding FliP experiment (E and F) where, after PB, the matrix FI does not diffuse past the Mfn1 punctum (Mfn1 in magenta with matrix, or green with OMM at white arrow) but the OMM fluorescent signal does. Thus, in this example, the OMM is continuous across the Mfn1 punctum, but the matrix is not. (G–I) Cartoon (G) and corresponding Flip experiment (H and I) for an example where for the first 37 s following PB the membranes are tethered but not continuous at the Mfn1 puncta (Mfn1 in green with matrix or magenta with OMM, white arrow). Then, at t = 37 s after PB, a fusion event restores OMM and matrix continuity, and fluorescent signal from box 1 diffuses past the Mfn1 punctum into box 2.
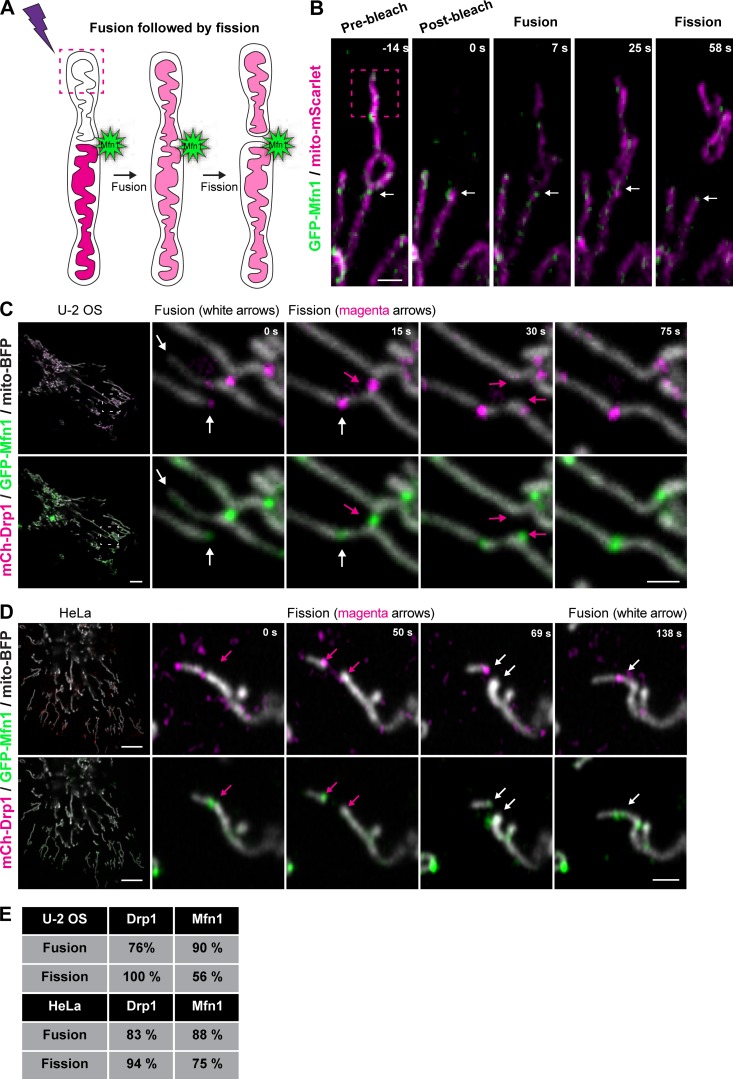
Because both mitochondrial fission (Friedman et al., 2011) and fusion (shown here) occur at ER MCSs, we asked whether fission and fusion were coordinated at the same ER MCS or whether they occur at separate locations. To test this, we first tracked single Mfn1 puncta over time to assess whether mitochondria can undergo both fusion and fission in successive events at these positions. Cells were cotransfected with GFP-Mfn1 and mito-mScarlet, and then we performed FLiP experiments (as in Fig. 4) to confirm fission and fusion events by content mixing at Mfn1 puncta. We show a representative example of an Mfn1-labeled fusion event followed by fission at the same Mfn1-labeled spot (Fig. 5, A and B). Based on this observation, we asked whether Drp1 and Mfn1 would localize to the same positions during mitochondrial fission and fusion events in two different human cell lines: U-2 OS and HeLa. Cells were cotransfected with mCherry-Drp1, GFP-Mfn1, and mito-BFP to visualize fission and fusion events relative to these opposing machineries. Mfn1 puncta colocalized with Drp1 at both fission and fusion events (Fig. 5, C and D, at arrows; and Video 7). We scored the percentage of apparent events marked by Drp1 and/or Mfn1 in both cell types cotransfected with either mCherry-Drp1 and mito-BFP or GFP-Mfn1 and mito-BFP (Fig. 5, C and D). Drp1 puncta were present at 76% and 83% of fusion events in U-2 OS and HeLa cells, respectively (and predictably at 100% and 94% of fission events; Fig. 5 E). Additionally, Mfn1 puncta were present at 56% and 75% of the fission events observed in U-2 OS and HeLa, respectively (and at 90% and 88% of fusion events; Fig. 5 E). These data demonstrate that ER MCSs, fission machinery, and fusion machinery converge at nodes that are hotspots for mitochondrial membrane dynamics.
Figure 5.
Fission, fusion, and respective machineries converge. (A and B) Cartoon description (A) and corresponding FLiP experiment (B) that reveals consecutive tethering, fusion, and then fission all occurring at the same Mfn1 punctum. A mitochondrion expressing GFP-Mfn1 and a matrix marker (mito-mScarlet, magenta) was photobleached (in box 2), which is located on one side of the Mfn1 punctum (Mfn1 in green, white arrow). Images shown are prebleach (t = −14 s), postbleach (t = 0 s), fusion at an Mfn1 punctum (at t = 7 s), prefission (25 s), and then postfission at an Mfn1 punctum (at t = 58 s). (C) A representative image of a U-2 OS cell expressing mCh-Drp1, GFP-Mfn1, and mito-BFP and magnified merged time-lapse images of the inset show the dynamics and colocalization of fission and fusion machineries over time. Note that both GFP-Mfn1 and mCh-Drp1 puncta colabel a fusion event (white arrow) and a neighboring fission event (magenta arrow). (D) Representative image of a HeLa cell expressing mCh-Drp1, GFP-Mfn1, and mito-BFP and magnified merged time-lapse images of the inset show the dynamics and colocalization of fission and fusion machineries over time. Note that both GFP-Mfn1 and mCh-Drp1 puncta co-label a fission event (magenta arrow) and remain labeled through a subsequent fusion event at the same spot (magenta arrow). (E) Table shows the percentage of fission or fusion events that are marked by GFP-Mfn1 and/or mCh-Drp1 puncta in U-2 OS and HeLa. Scale bars for whole cell = 5 µm; insets, 1 µm.
Video 7.
Mfn1 (green) and Drp1 (magenta) colocalize at fission and fusion events corresponding to Fig. 5 C. Two-minute time-lapse image of a live U-2 OS cell transfected with GFP-Mfn1, mCh-Drp1, and mito-BFP. Images were acquired every 5 s using a spinning-disk confocal microscope. Playback rate is 7.5 fps.
ER tubules and Dnm1 puncta are present at mitochondrial fusion and fission sites in yeast
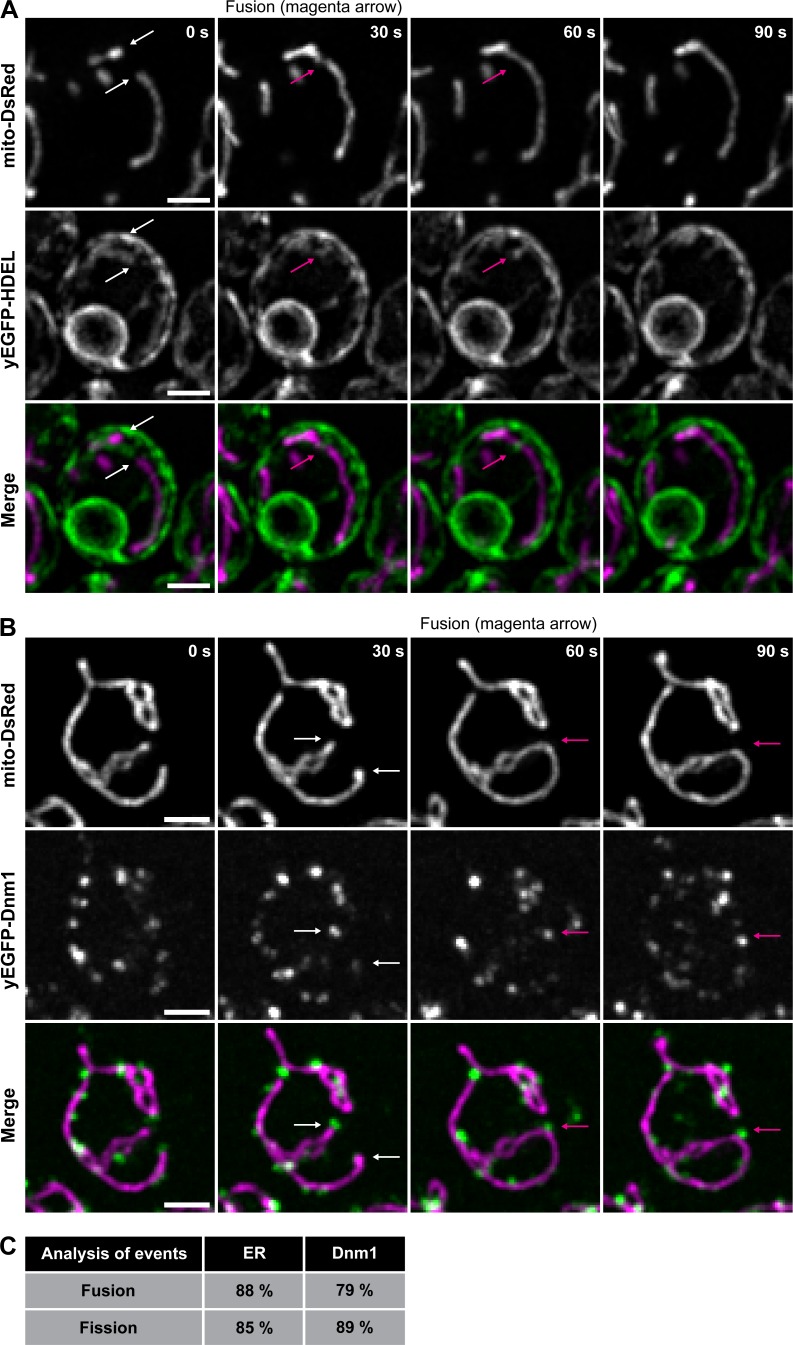
We performed similar experiments in Saccharomyces cerevisiae to determine if features of ER-associated fission and fusion are conserved in yeast. First, we monitored mitochondrial dynamics relative to ER tubules in live cells by confocal microscopy in a strain expressing fluorescent markers for ER (yEGFP-HDEL, green) and mitochondria (mito-DsRed, magenta). We assessed apparent fusion and fission events relative to ER tubules and found that the ER was present at 88% of fusion events and 85% of fission events (Fig. 6 A). This frequency is similar to our fusion statistics in human cells (Fig. 5) and is consistent with previously published data regarding ER-associated fission in yeast (Friedman et al., 2011). There is currently no functional fluorescent protein-tagged version of Fzo1 (the Mfn1 orthologue) for live yeast imaging. However, we can assess whether the yeast fission machinery Dnm1 marks ER MCS nodes where either fission or fusion can occur in yeast. We monitored mitochondrial dynamics relative to Dnm1 in cells expressing Dnm1-yEGFP and mito-DsRed. We scored that Dnm1 puncta localized to 79% of fusion and 89% of fission events in yeast (Fig. 6 B). These statistics are strikingly similar to those measured in animal cells (Fig. 5 E), supporting the notion that bidirectional ER MCS nodes are broadly conserved.
Figure 6.
ER tubules and Dnm1 puncta are present at mitochondrial fusion and fission sites in yeast. (A) Magnified time-lapse image set of a yeast cell expressing mito-DsRed to label mitochondria (top) and yEGFP-HDEL to label ER (middle). The merged images (bottom) show mitochondria in magenta and ER in green. White arrows indicate tips of mitochondria that fuse and corresponding location of ER. The magenta arrow indicates the position of fusion and corresponding location of the ER tubule, which marked the fusion event. Maximum-intensity projections of four focal planes are shown. (B) Magnified time-lapse image set of a yeast cell expressing mito-DsRed to label mitochondria (top) and Dnm1-yEGFP (middle). The merged images (bottom) show mitochondria in magenta and Dnm1 in green. White arrows indicate tips of mitochondria that fuse and the corresponding location of Dnm1. The magenta arrow indicates the position of fusion and corresponding position of Dnm1. Whole-cell, maximum-intensity projections are shown. (C) The percentage of mitochondrial fusion and fission events occurring at ER tubules (fusion n = 102 and fission n = 105 events), and at Dnm1 puncta (fusion n = 104 and fission n = 102 events). Scale bars = 2 µm.
Depolarized mitochondria are rescued (repolarized) at ER MCSs
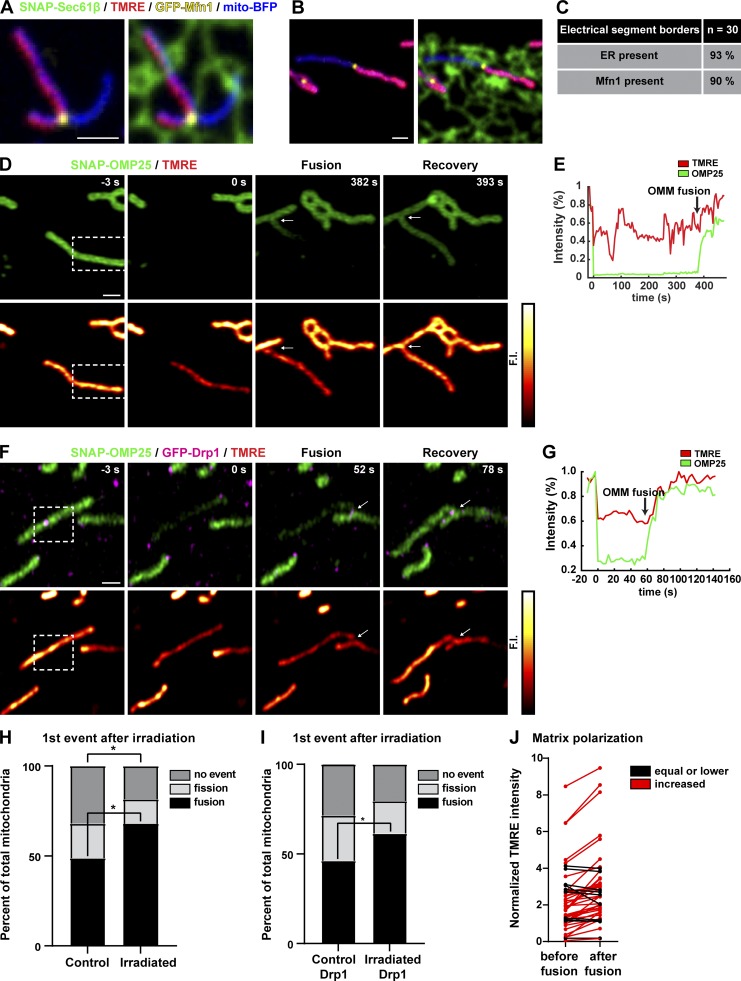
Because fission and fusion machinery accumulate together at the same ER-mitochondria MCS, we hypothesized that ER MCSs are poised to respond rapidly to stimuli and could be a site for rescuing damaged mitochondria. It has been proposed that transient or kiss-and-run type fusion (brief fusion events where the two fusing mitochondria subsequently divide and maintain their separate structures) could function to allow the exchange of proteins, metabolites, and ions for the purpose of rescuing “unhealthy” mitochondria (Liu et al., 2009; Rambold et al., 2011). One characteristic of “healthy” mitochondria is that they maintain a membrane potential across their IMM while mitochondria or mitochondrial segments that become depolarized are targeted for mitophagy (Twig et al., 2008). Therefore, we tested whether there is a relationship between ER MCSs and the spatial organization of differentially polarized mitochondrial segments. We cotransfected cells with a mitochondrial and ER marker (mito-BFP and SNAP-Sec61β, respectively), and GFP-Mfn1 and incubated them with [30 nM] tetramethylrhodamine ethyl ester (TMRE, a dye that accumulates in polarized mitochondria proportionally to the ΔΨm in a Nernstian fashion; O’Reilly et al., 2003). We found that the 405-nm laser exposure used to image the mito-BFP marker could trigger mitochondrial depolarization in segments along their length (see two examples in Fig. 7, A and B). Strikingly, the vast majority (90%) of boundaries between electrically uncoupled segments were marked by Mfn1 at ER MCSs (Fig. 7 C).
Figure 7.
Depolarized mitochondria are rescued by ER-associated fusion. (A) Representative image of a U-2 OS cell expressing GFP-Mfn1 (yellow), mito-BFP (blue), and SNAP-Sec61β (green) and loaded with a dye that accumulates within the matrix of mitochondria in proportion to membrane potential [30 nm] TMRE (red). Note the occurrence of an Mfn1 punctum at an ER tubule crossing at the border of electrically uncoupled mitochondrial segments. (B) Another example as in A. (C) Table of mitochondria imaged as in A and B shows that ER MCSs and Mfn1 puncta mark the boundaries between the vast majority of electrically uncoupled segments. (D) Cells expressing an OMM marker (SNAP-OMP25) were loaded with [30 nM] TMRE to indicate membrane polarization. Individual mitochondria were irradiated with 405 nm (in dashed boxes) to photobleach the OMM marker and depolarize the membrane. Depolarized mitochondria were imaged for 8 min following irradiation. In this example, a photobleached and depolarized mitochondrion fuses ∼382 s after irradiation, and this fusion leads to an increase in OMM FI and a subsequent increase in TMRE signal (indicating rescue of membrane potential). (E) Plot of relative FI of SNAP-OMP25 and TMRE signal for experiment in D. (F) Representative time-lapse image series of irradiated mitochondria in cells expressing GFP-Drp1 (to label ER MCSs), SNAP-OMP25, and loaded with [30 nM] TMRE. Dashed box indicates irradiated area. Arrows indicate location of fusion event. (G) Relative FI plots of SNAP-OMP25 and TMRE signal from the time-lapse in F. Note that the TMRE signal recovers shortly after the time of OMM FI increase by fusion (arrow). (H) Two groups of mitochondria in (D; control and irradiated) were imaged over time following irradiation and were assigned to three categories: fission first, fusion first, and no event. The graph shows the distribution of categories between control and irradiated. *, P < 0.05 by χ2 analysis (n = 78 mitochondria for control, n = 81 mitochondria for irradiated). (I) As in H except for the addition of transfected Drp1 (n = 91 mitochondria for control, n = 88 mitochondria for irradiated). *, P < 0.05 by χ2 analysis. (J) Fusion rescues mitochondrial membrane potential. The mean normalized TMRE FI was measured for the 10 frames directly preceding fusion and the 10 frames immediately after fusion for individual irradiated mitochondria. The two measurements were connected by lines. Red lines indicate an increase in TMRE intensity after fusion (mean FI increase = 65%), and black lines indicate no change or a decrease in TMRE intensity. Events are categorized from irradiation experiments in U-2 OS cells expressing SNAP-OMP25 alone and SNAP-OMP25 with GFP-Drp1. Scale bars, 1 µm.
Since ER MCSs define boundaries between electrically uncoupled mitochondrial segments, we tested whether individual depolarized mitochondria are more likely to undergo fission or fusion at ER MCSs. Cells were transiently transfected with the SNAP-OMP25 (JF646; to label the OMM) and loaded with 30 nM [TMRE] for 30 min, and then individual mitochondria were depolarized by stimulating them with 405-nm light. Mitochondria were selected for irradiation based on two criteria: (1) a positive TMRE signal and (2) an isolated position. Mitochondria that met these criteria were photobleached with 405-nm laser in a region roughly half to one third their size. In addition to markedly reducing TMRE fluorescence, 405-nm irradiation bleached the OMM marker, allowing the detection of fluorescent protein exchange upon a subsequent fusion event. We then tracked the individual depolarized mitochondria for 8 min after photobleaching and categorized changes in their morphology and ΔΨm by TMRE fluorescence. Most irradiated mitochondria were able to recover TMRE fluorescence upon fusion with an unirradiated neighboring mitochondrion (Fig. 7, D and E; and Video 8). We followed irradiated and control mitochondria and categorized their first event postirradiation as either fission, fusion, or no event. As we expected, the distribution of these categories was significantly changed in irradiated mitochondria compared with control. Irradiated mitochondria were 39% more likely to fuse directly after irradiation. Most of this increase seemed to be due to a decrease in the fraction of mitochondria that did nothing, while the likelihood of fission showed little change (Fig. 7 H).
Video 8.
Irradiation of a single mitochondrion followed by fusion and repolarization corresponding to Fig. 7 D. OMM is green (left) and TMRE intensity (right) is represented by red hot heatmap. Time-lapse images over 8 min of a live U-2 OS cell transfected with SNAP-OMP-25 and loaded with TMRE. Frames were captured every 3.5 s using a spinning-disk confocal microscope. Playback rate is 7.5 fps.
To assess whether fusion of depolarized mitochondria in response to irradiation occurs at ER MCSs, we asked whether fusion after depolarization occurred at Drp1 puncta (which are known to localize to ER MCSs; Friedman et al., 2011; Ji et al., 2017). Mfn1 was avoided as a proxy for ER MCSs in this analysis to prevent Mfn1 overexpression from artificially increasing the probability of fusion. Therefore, we performed the same experiment as above with cells transfected with GFP-Drp1 and SNAP-OMP25 and loaded with [30 nM] TMRE. Despite the transfection of fission machinery, this yielded nearly identical results as before where, following irradiation, mitochondria would fuse with a neighboring mitochondrion, which had the effect of rescuing membrane potential (Fig. 7, F and G; and Video 9). The probability that a depolarized mitochondrion will fuse following laser-induced depolarization was significantly increased (∼33%) compared with control mitochondria (Fig. 7 I). Additionally, the large majority of fusion events (88%) that occurred following irradiation and depolarization were labeled by GFP-Drp1. To quantify the extent of rescue after fusion of depolarized mitochondria, we took all the mitochondria from OMP25/Drp1-transfected cells and the OMP25-transfected cells that underwent fusion following depolarization and compared the mean normalized TMRE fluorescence during the 10 frames before fusion to the mean of the 10 frames after fusion. We found that 77% of these events resulted in an increase of TMRE fluorescence, indicating rescue of the membrane potential in the irradiated mitochondrion with a mean increase of 65% (Fig. 7 J). These data show that, on a short timescale, the initial response of individual mitochondria to depolarizing photodamage is to increase their likelihood of ER-associated fusion where Drp1 is accumulated. Further, this increase in fusion was able to restore the membrane potential of the majority of irradiated mitochondria. Together, these findings support the hypothesis that damaged mitochondria can be rescued by fusing and sharing contents with “healthy” mitochondria at ER MCS nodes.
Video 9.
Irradiation of a single mitochondrion (OMM labeled in green and Drp1 in magenta) followed by fusion and repolarization corresponding to Fig. 7 F. TMRE intensity is represented by red hot heatmap. Time-lapse images over 2 min 48 s of a live U-2 OS cell transfected with SNAP-OMP-25 and GFP-Drp1, and loaded with TMRE. Frames were captured every 3.5 s using a spinning-disk confocal microscope. Playback rate is 7.5 fps.
Discussion
Here, we discovered that the processes of fission and fusion are spatially coordinated and colocalized in a system of ER MCS nodes to regulate mitochondrial shape and health. We show that fusion (mitofusins) and fission (Drp1) machineries colocalize in puncta to regulate bidirectional membrane dynamics at bona fide ER–mitochondria MCSs. Thus, ER MCSs provide a series of nodes on mitochondria that are enzyme-like, whereby both the forward and reverse reactions (fusion vs. fission) can be catalyzed in response to metabolic cues. Indeed, we find that ER MCS nodes and their machineries will accumulate at the boundaries between mitochondrial segments with different membrane potentials. We show that depolarization of individual mitochondria pushes the reaction at ER MCS nodes toward fusion, resulting in the recovery of membrane potential. Clearly, other signals may push the reaction toward fission. These signals may guide a path to fission and fragmentation of mitochondria segments that are beyond repair to target these segments toward mitophagy.
The major question that remains is how do ER MCSs contribute to both the fission and fusion of mitochondria? There are several ways that we can imagine that ER could regulate these processes based on what activities are already known to occur at ER MCSs: for example, an ER MCS could have a specialized lipid composition that promotes high membrane curvature, which would be favorable for both membrane fission and fusion reactions. A specialized lipid environment could recruit fusion and fission machinery proteins to these locations or kinases/phosphatases that regulate fission and fusion machineries. It is also conceivable that release of high Ca2+ from the ER lumen onto mitochondria at MCSs could stimulate fission and fusion, although this mechanism would not be conserved in yeast, because yeast ER does not store high Ca2+. Future studies will investigate whether the forward and reverse rates could be determined at the ER MCS by the relative recruitment and/or posttranslational modifications of fission and fusion machineries, by small signaling molecules like Ca2+, or by the recruitment of activators or inhibitors of these machineries.
Materials and methods
No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
DNA plasmids
GFP–Sec61β, mCherry-Sec61β (Friedman et al., 2011), mito-BFP (Friedman et al., 2011), BFP-KDEL (Friedman et al., 2011), mCherry-KDEL (Zurek et al., 2011), mCherry–Drp1 isoform 3 (NM_005690; Friedman et al., 2011), GFP-MFF (Friedman et al., 2011), and mito-EGFP (Song et al., 2009) were previously described. KDEL-Venus was a gift from Eric Snapp (Janelia Research Campus, Ashburn, VA). mito-mMaple was a gift from Raquel Salvador-Gallego. pSNAP-C1 and pmMaple-C1 vectors were created by inserting SNAP or mMaple ORF into the NheI/BspEI site, to replace AcGFP, in the pAcGFP-C1 vector (Clontech). SNAP-OMP25, GFP-OMP25, and mMaple-OMP25 were created by amplifying OMP25 from photoactivatable GFP-OMP25 (Addgene 69598) and inserting it into the XhoI/BamHI sites of corresponding C1 vectors. SNAP-Sec61β was created by amplifying the Sec61β ORF from mCherry-Sec61β and inserting it into the XhoI/KpnI site of the pSNAP-C1. mito-mScarlet was created by amplifying the mScarlet from pmScarlet-i_C1 ORF (Addgene 85044), adding AgeI and a BsaI site with a NotI overhang, and inserting it into the AgeI/NotI site of mito-BFP, replacing BFP. Mitofusin-1 (NM_033540.3) was cloned from HeLa cDNA and inserted into the EcoR1/BamH1 site in pAcGFP-C1 (Clontech), mScarlet-C1, and SNAP-C1. GFP-Drp1 was created by subcloning Drp1 into pAcGFP-C1 using XhoI/BamHI sites. mCherry-Mfn2 was created by inserting mCherry into the NheI/XhoI site of pAcGFP-C1, replacing GFP, amplifying Mfn2 from HeLa cDNA, and inserting this into the XhoI/BamHI sites. RA was amplified from RA-nuclear export sequence (RA-NES) plasmid (Addgene 61019), and RA-C1 vector was derived from pAcGFP-C1 as above; Sec61β was inserted into the XhoI/KpnI sites. GA-MFF was created by amplifying the GA ORF from GA-NES (Ding et al., 2015; Addgene 61018) and replacing GFP from GFP-MFF using XhoI/BamHI sites. B-MFF was created by amplifying the B ORF from GB-NES (Ding et al., 2015) and replacing GFP from GFP-MFF as above. SNAP-Drp1 was made by inserting SNAP (gift from Luke Laevis, Janelia Research Facility, Ashburn, VA) into the AgeI/NotI site of pAcGFP-C1, replacing GFP, and then inserting Drp1 into the XhoI/BamHI site. GFP-Mfn1-E209A was created by site-directed mutagenesis of the GFP-Mfn1 construct described above.
Cell culture, transfection, and SNAP tag and TMRE labeling
U-2 OS cells (ATCC-HTB-96) were used for all experiments. Cell lines were tested for Mycoplasma contamination by ATCC at the time of purchase. Cells were grown in McCoy’s Medium 5A supplemented with 10% FBS and 1% penicillin/streptomycin. U-2 OS cells were plated directly onto 35-mm glass-bottomed microscope dishes (Cellvis) coated with fibronectin at ∼2.0 × 105 cells/35-mm dish ∼16 h before transfection. All imaging experiments were performed at 37°C in FluoroBrite DMEM (Invitrogen) supplemented with 10% FBS, 1× Glutamax (Gibco), 1× penicillin-streptomycin (Gibco), and 25 mM Hepes buffer (pH 7.4). Labeling SNAP constructs with [JF646 SNAP tag ligand] = 1.5 µM and incubation with [TMRE] = 30 nM were performed for 30 min in Fluorobrite medium supplemented as above but lacking serum.
Plasmid transfections were performed in OPTI-MEM (Invitrogen) for ∼5 h using 5 µl Lipofectamine 3000 (Invitrogen) per 35-mm dish with 2 µl/ug of P3000 reagent. Roughly 16–24 h after transfection, cells were transferred to imaging medium that was equilibrated in 5% CO2 at 37°C. Fluorobrite DMEM supplemented with 10% FBS, 1× Glutamax, 1% penicillin/streptomycin, and 25 mM Hepes buffer was used as imaging medium. For all experiments, the following amounts of DNA were transfected per 35-mm dish: 300–400 ng for all Sec61β constructs; 200–300 ng for all mito constructs; 100–200 ng for all Drp1 constructs; 200–300 ng for all KDEL plasmids; 250 ng for all Mfn1 and Mfn2 plasmids; 300 ng for all OMP25 plasmids; and 250–350 ng for RA-, B-, and GA-MFF.
Immunofluorescence
U-2 OS cells were seeded onto fibronectin-coated coverslips (neuVitro GG-18-1.5-Fibronectin) in 35-mm dishes and fixed 16–24 h later. Cells were washed three times quickly with 1× Dulbecco's Phosphate Buffered Saline (DBPS; Sigma-Aldrich D1408) and then incubated for 1 h at 4°C in 4% PFA (Polysciences, 00380-1), 1× DPBS that was prechilled to 4°C. Cells were then washed three times with 1× DPBS at 4°C before adding 1 ml of MeOH at −20°C for 1 min. Cells were immediately washed three times with 1× DPBS at 4°C and then allowed to equilibrate at RT. Coverslips were blocked for 1 h at RT in blocking solution (1% BSA/0.3 M [Glycine]/1% normal donkey serum). Blocking solution was replaced with blocking solution containing primary antibodies and incubated at RT for 1 h. Coverslips were then washed three times with 1× DPBS for 5 min each, and secondary antibodies were added in blocking solution as above for 1 h at RT. Coverslips were washed three times with 1× DPBS at RT for 5 min each, mounted on slides using ProLong Glass (Invitrogen, P36980) mounting medium, and imaged the next day. Antibodies used were mouse αMfn1/2 [3C9] (Abcam, ab57602) and pRb αTom20 (Santa Cruz, sc-11415). For mouse mAb αTom20 (Santa Cruz, sc-17764), staining in cells (Fig. S3) was processed as above except that MeOH was omitted, fixation for 15 min and washes were performed at RT, and cells were permeabilized with 0.1% Triton X-100.
Microscopy
Photoconversion and FLiP experiments were performed in part at the BioFrontiers Institute Advanced Light Microscopy Core using a Nikon A1R microscope with a 100× 1.45-NA Plan Apo I objective. Images were acquired using Nikon Elements software. Photoconversion and FLiP experiments were also performed in part on a Zeiss Axio Observer inverted fluorescence microscope equipped with a 63× 1.4-NA Plan Apo objective, LSM 880, and Airyscan detector. Images were acquired using Zeiss Zen software. All other live-cell imaging was performed on either the Zeiss Airyscan instrument as above or a Nikon eclipse Ti2 inverted microscope equipped with a 100× 1.45-NA Plan Apo objective, Yokagowa CSU-X1 spinning disk confocal scanner, an Andor iXon 897 electron-multiplying charge-coupled device camera, and OBIS LX/LS lasers (405/488/561/640 nm). Images were acquired using Micro-Manager software and ImageJ (National Institutes of Health). Immunofluorescence imaging was performed on the Nikon Ti2 described above.
Analyses of apparent mitochondrial dynamics relative to ER tubules, Mfn1, and Drp1
Apparent mitochondrial fusion was assessed by imaging U-2 OS cells expressing OMM or mitochondrial matrix markers (described above) every 5 s up to 8 min in a single focal plane. Apparent fusion was defined as two mitochondria coming into visual contact and then remaining and moving together. Fission was assessed using the same markers and defined as an apparently continuous mitochondrion that transitions to apparently discontinuous mitochondria that exhibit uncoupled movement. To identify fusion and fission events occurring at ER tubules, Mfn1, and/or Drp1, cells were imaged as above with the inclusion of the respective markers. “At” was defined in all experiments as the relevant marker in overlapping pixels or in pixels directly adjacent to the pixels where the mitochondrial marker merges or separates. ER coverage was measured in all images used for quantification and was determined by creating binary thresholded images of ER and mitochondria and calculating the percentage of pixels occupied by mitochondrial signal that were also occupied by ER signal.
Photoconversion and content mixing to confirm fusion events
Photoconversion and content mixing experiments were performed by imaging U-2 OS cells expressing mMaple-OMP25 to label the OMM or mito-mMaple to label the mitochondrial matrix along with an ER marker (described above). Fusion reported by content mixing was defined as a decrease in FI of photoconverted mMaple in one mitochondrion and a concomitant increase in a neighboring mitochondrion indicating free diffusion of mMaple and membrane continuity. Photoconversion was done on the Zeiss Airyscan microscope (described above). Parameters for photoconversion of mMaple in individual mitochondria were optimized slightly for each experiment and were always close to the following: Airyscan fast mode, four scans before bleaching, 15 iterations of 405-nm laser at 6% effective laser power (2-µs pixel dwell as in the imaging scans).
Analyzing colocalization of proteins
MCC between Mfn and ER or ddFP was calculated using Matlab according to the following equation:
where Mfni,colocal is the Mfn intensity in pixels that are above threshold in the ER image and Mfni is the sum of Mfn intensity values in all pixels above threshold in the Mfn image. All images were thresholded manually using the “imbinarize” function in Matlab. Mfn images were thresholded to exclude general mitochondrial shape and isolate puncta. MCC was calculated between Mfn and the general mitochondrial marker as described above. As a negative control, Mfn images were rotated 90° clockwise using the “imrotate” function in Matlab, and MCC with ER was calculated as above.
Tracking of Mfn1 puncta with ER signal over time
To track Mfn1 puncta relative to ER tubules, U-2 OS cells expressing GFP-Mfn1 and an ER marker (described above) were imaged over 2 min at 5-s intervals. Individual Mfn1 puncta were categorized as positive or negative for showing pixel overlap with an ER tubule during each frame of the time lapse.
ddFP analysis
As a positive control for ddFP system function, 3 × 3-pixel spots in the ddFP (red) channel were taken from U-2 OS cells expressing B-MFF, RA-Sec61β, a general ER marker, and a mitochondrial matrix marker for analysis. Two groups of spots were chosen: group 1 spots were positive for the general ER marker with no mitochondrial matrix marker present, and group 2 spots were positive for both the ER marker and the mitochondrial matrix marker. For background subtraction, a background spot was chosen for each experimental spot that was located as close as possible to its corresponding experimental spot and was negative for both ER marker and matrix marker. The mean pixel value of the background spot was subtracted from each corresponding experimental spot. For each cell, all spots from groups 1 and 2 were grouped into one set and were normalized by dividing by the mean spot value in the set. All normalized spot values from each cell were then combined and represented as a histogram.
To determine if ddFP signal was enriched at the site of mitochondrial fusion, we measured the maximum ddFP signal in three regions of interest (ROIs) for each fusion event: two 1-µm2 ROIs on an ER tubule that was not overlapping with a mitochondrion, and one 0.5-µm2 ROI at the site of fusion. We then took the ratio (maximum signal at the site of fusion)/(mean of the two maximum signals on an ER tubule without a mitochondrion). If the ratio was >1.5, meaning the signal at the site of fusion was ≥50% of the background signal on the ER with no mitochondrion, then the signal was counted as enriched at the site of fusion. We then compared ddFP signal at the site of fusion to ddFP coverage over mitochondria in the same image.
To determine the percentage of ER tubule crossings on mitochondria that are contact sites as reported by ddFP signal, we measured the ER and ddFP signal along a mitochondrion by linescan. To establish a threshold, we measured ddFP signal by linescan along a nearby ER tubule where there was no mitochondrion and took the mean of the values in the linescan. ER tubule crossings over mitochondria were categorized as contact sites if the ddFP signal at the ER crossing was greater than the threshold value. To determine colocalization of Mfn1 with ddFP signal, we manually thresholded images in Matlab and measured MCC as above.
Analysis of OMM and matrix continuity by FLiP
Individual mitochondria from U-2 OS cells expressing OMM, matrix, and Mfn1 markers were photobleached in a small region at the tip on one side of a Mfn1 punctum, and mean FI from regions on both sides of the Mfn1 punctum was measured throughout the time-lapse and plotted as ΔF/F0 versus time. Bleaching parameters were optimized for each experiment and were generally as follows: four scans before bleaching, 15 iterations of 405-nm laser at 6% effective laser power in Airyscan fast mode, with 2-µs pixel dwell as in the imaging scans.
Irradiation-induced depolarization of mitochondria
U-2 OS cells expressing an OMM marker were incubated with 30 nM [TMRE] for 30 min in serum-free FluoroBrite before imaging as described above. Irradiation parameters were optimized for each experiment and were generally as follows: four scans before bleaching, 15 iterations of 405-nm laser at 6% effective laser power in Airyscan fast mode, with 2-µs pixel dwell as in the imaging scans. Fusion was defined by content mixing as described above. TMRE and OMM FI of individual mitochondria was obtained by outlining the TMRE signal of the mitochondrion in each frame and taking the mean FI of both markers within the outline. To identify fusion events occurring at Drp1 puncta, cells were imaged, and individual mitochondria were irradiated as above except for the inclusion of a Drp1 marker. “At” was defined as Drp1 positive pixels overlapping or directly adjacent to pixels where the OMM marker of the fusing mitochondria meet.
Yeast strain construction and imaging
The yeast strain background used is W303 (ade2-1; leu2-3; his3-11, 15; trp1-1; ura3-1; can1-100). LLY522 (W303 ER-GFP/mito-DsRed::LEU/NAT) was created by transforming ClaI-digested pLL18 into W303. LLY3242 (W303 DNM1-yEGFP::HIS mito-DsRed::LEU/NAT) was created by transforming a PCR product amplifying the yEGFP::HIS cassette from pKT128 with homology to the DNM1 locus and transforming the resulting strain with EcoRI-digested pLL19 into W303.
The following plasmids were used in the study. pLL18 is pRS305 ER-GFP/mito-DsRed::LEU/NAT with a TPI promoter and CYC1 terminator driving HDEL-GFP expression, a TEF promoter and terminator driving NATNT2 expression, and a GPD promoter and ADH terminator driving expression of mito-DsRed (matrix-targeted DsRed amplified from pXY142-mito-DsRed; Friedman et al., 2011). pLL19 is pRS305 mito-DsRed::LEU/NAT with a TEF promoter and terminator driving NATNT2 expression and GPD promoter and ADH terminator driving expression of mito-DsRed. pKT128 is pFA6a–link–yEGFP–SpHIS5 (Sheff and Thorn, 2004).
For all S. cerevisiae strain imaging, cells were grown to midlog phase in synthetic complete medium + 2% (wt/vol) dextrose medium with 2× adenine, rotating at 24°C. Cells were concentrated by centrifugation and mounted on a 4% wt/vol agarose pad. Imaging was performed at 22°C. Z series of cells were imaged every 30 s over 20 min using a spinning disk confocal system (Leica) fitted with a spinning disk head (CSU-X1; Yokogawa), a PLAN APO 100× 1.44-NA objective (Leica), and an electron-multiplying charge-coupled device camera (Evolve 512 Delta; Photometrics). A step size of 0.4 µm was used over a range of 8 µm. Image capture was done using Metamorph (Molecular Devices). The images were deconvolved using AutoQuant X3’s (Media Cybernetics) iterative, constrained 3D deconvolution method. Fiji (National Institutes of Health) was used to make linear adjustments to brightness and contrast. Photoshop (Adobe) was used to crop images, separate channels, and add scale bars. Deconvolved images are shown.
Total mitochondrial fission and fusion events were quantified in S. cerevisiae from videos generated as described above using Fiji. Localization of Dnm1 and ER to mitochondrial fission and fusion sites was quantified in individual z-slices for each video.
Mitochondrial morphology analysis
U-2 OS cells were treated with either control siRNA (Silencer Negative Control siRNA, Invitrogen AM4635) or siRNA to deplete Mfn1 (target sequence 5′-GAAGAGCUCUGUUAUCAAU-3′, Dharmacon, siGenome D-010670-01-0005) and were transfected with empty vector (pAcGFP-C1, Clontech), siRNA-resistant GFP-Mfn1, or siRNA-resistant GFP-Mfn1-E209A. The cells were then prepared for immunofluorescence as described above and imaged on the Nikon Ti2 described above. Maximum-intensity projections were made from 2.2-µm Z-stacks sampled every 0.2 µm (11 z-slices) using Fiji. Peripheral mitochondria contained within a 230-µm2 ROI were analyzed using Matlab to report the number of mitochondria and their area.
Analysis of mitofusin puncta
U-2 OS cells were transfected with mito-BFP and either GFP-Mfn1 or GFP-Mfn1-E209A and imaged live as described above. For each condition, linescans were taken in Fiji of mitochondria that showed enrichments of mitofusin signal. Linescan analysis was performed using Matlab. Seven linescans were taken from each of five cells in both conditions, yielding 35 linescans from each condition. Linescans from individual cells were aligned to their maxima and averaged to yield a representative linescan for each cell. The average linescan for each cell was then normalized by its mean value, and all five normalized linescans were aligned to their maxima and averaged to yield the representative linescans shown in Fig. 1 H. Mfn1 punctate enrichment was calculated by measuring the mean value of Mfn1 maximum signal in puncta and dividing by the mean value of nonpunctate Mfn1 signal. Mitofusin puncta were counted in these images by manually thresholding the images to exclude the general mitochondrial signal and isolate puncta. Thresholded images were analyzed using the “Analyze particles” function in Fiji to return the number of puncta. Mitochondrial area was calculated by automatically thresholding the mito-BFP image using the Otsu method and summing the pixels that were positive for mito-BFP signal.
RT-qPCR
U2-OS cells were grown in six-well plates and were either transfected with GFP-Mfn1 or mCherry-Mfn2 or not transfected (WT). Three wells were grown for each condition above. Total RNA was isolated from each well using the RNeasy Mini Kit (Qiagen 74104) ∼17 h posttransfection. cDNA libraries (1 per 35-mm dish) were created using the SuperScript IV First-Strand Synthesis System with poly dT oligonucleotides to enrich for polyadenylated RNAs. qPCR was performed on an Applied Biosystems StepOnePlus thermal cycler in the standard mode using Power Sybr Green reagents (Applied Biosystems, 4367659). Threshold cycles were determined automatically. The sequences of the primers used for qPCR are as follows: MFN1_F (5′-GGAGCGAGCCTTTAAACAGCAG-3′), MFN1_R (5′-AGGCGAGCAAAAGTGGTAGC-3′), MFN2_F (5′-TGGCCAACTCAGAGTCCACC-3′), MFN2_R (5′-CCGCACCTCCTCCATGTACT-3′), ACTB_F (5′-TGGGCATGGGTCAGAAGGATTCC-3′), and ACTB_R (5′-GAAGGTGTGGTGCCAGATTTTCTCC-3′).
Western blot
Western blotting was performed with 4–12% Criterion TGX gels, 1-h wet transfer, blocking in milk for 30 min, primary antibody incubation for 1 h at RT, 3 × 5-min TBST washes, incubation with secondary antibody for 1 h at room temperature, and 3 × 5-min washes. Primary antibody against Mfn1 (rabbit polyclonal, Proteintech, 13798-1-AP) was used at 1:1,000 dilution. Goat anti-rabbit secondary antibody conjugated to HRP (Sigma-Aldrich, A6154) was used at 1:3,000, and signal was detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, 34577).
Data availability
The source data for all gels and blots are provided as a supplemental figure in the online version of the paper. All other data that support the findings of this study are available from the authors upon reasonable request.
Online supplemental material
Fig. S1 shows that Mfn2 enriches in puncta on mitochondria that colocalize with ER tubules and mark fusion events in the same fashion as Mfn1. Additionally, it shows that MFN1 mRNA is much more abundant that MFN2 mRNA in U-2 OS cells. Fig. S2 shows that GFP-Mfn1 can rescue mitochondrial fragmentation induced by siRNA silencing of MFN1 while GFP-Mfn1-E209A cannot. Video 1 shows an Mfn1 punctum tracking with an ER tubule over time. Video 2 shows two mitochondria fusing at an Mfn1 punctum. Video 3 shows an OMM fusion event at an ER tubule crossing. Video 4 shows a matrix fusion event at an ER tubule crossing. Video 5 shows a bona fide OMM fusion event at an ER tubule crossing that is confirmed by exchange of the photoconverted OMM marker (mMaple-OMP25) between the two mitochondria. Video 6 shows a bona fide matrix fusion event at an ER tubule crossing that is confirmed by exchange of the photoconverted OMM marker (mito-mMaple) between the two mitochondria. Video 7 shows a fusion event followed by a fission event, both labeled by Mfn1 and Drp1. Video 8 shows depolarization of an irradiated mitochondrion, fusion with another mitochondrion indicated by recovery of the OMM signal (SNAP-OMP25), and recovery of its membrane potential as indicated by TMRE fluorescence. Video 9 shows depolarization of an irradiated mitochondrion, fusion with another mitochondrion at a Drp1 punctum, and recovery of its membrane potential as indicated by TMRE fluorescence.
Acknowledgments
We thank E. Zamponi, J. Lee, R. Salvador-Gallego, and H. Wu in the Voeltz lab for helpful discussions and plasmids used here; Luke Lavis at the Janelia Research Facility for providing JF646 SNAP tag ligand; and J. Tay (University of Colorado Boulder, Boulder, CO) for Matlab assistance.
We thank the BioFrontiers Institute Advanced Light Microscopy Core for use of the Nikon A1R microscope, which was also supported by National Institute of Standards and Technology–University of Colorado Cooperative Agreement award number 70NANB15H226. L.L. Lackner is supported by a grant from the National Institutes of Health (GM120303). B.T. Wisniewski is supported by a National Institutes of Health Training Grant (T32GM008061). G.K. Voeltz is an investigator of the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
Author contributions: R.G. Abrisch and S.C. Gumbin designed, performed, did formal analysis and validated experiments in Figs. 1, 2, 3, 4, 5, 7, S1, and S2. B.T. Wisniewski and L.L. Lackner designed, performed, and did formal analysis on Fig 6. R.G. Abrisch and G.K. Voeltz wrote the original draft, and S.C. Gumbin, B.T. Wisniewski, and L.L. Lackner reviewed and edited it. G.K. Voeltz conceived the project, designed experiments, and provided funding, methodology, and supervision.
References
- Ban T., Ishihara T., Kohno H., Saita S., Ichimura A., Maenaka K., Oka T., Mihara K., and Ishihara N.. 2017. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat. Cell Biol. 19:856–863. 10.1038/ncb3560 [DOI] [PubMed] [Google Scholar]
- Bleazard W., McCaffery J.M., King E.J., Bale S., Mozdy A., Tieu Q., Nunnari J., and Shaw J.M.. 1999. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1:298–304. 10.1038/13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.-L., Meng S., Chen Y., Feng J.-X., Gu D.-D., Yu B., Li Y.-J., Yang J.-Y., Liao S., Chan D.C., and Gao S.. 2017. MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature. 542:372–376. 10.1038/nature21077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., and Chan D.C.. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160:189–200. 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B., Cho H.M., Jo Y., Kim H.D., Song M., Moon C., Kim H., Kim K., Sesaki H., Rhyu I.J., et al. 2017. Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat. Commun. 8:15754 10.1038/ncomms15754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordás G., Thomas A.P., and Hajnóczky G.. 1999. Quasi-synaptic calcium signal transmission between endoplasmic reticulum and mitochondria. EMBO J. 18:96–108. 10.1093/emboj/18.1.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Li J., Enterina J.R., Shen Y., Zhang I., Tewson P.H., Mo G.C.H., Zhang J., Quinn A.M., Hughes T.E., et al. 2015. Ratiometric biosensors based on dimerization-dependent fluorescent protein exchange. Nat. Methods. 12:195–198. 10.1038/nmeth.3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K.W., Kamocka M.M., and McDonald J.H.. 2011. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300:C723–C742. 10.1152/ajpcell.00462.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson S.M., and De Camilli P.. 2012. Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 13:75–88. 10.1038/nrm3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., and Voeltz G.K.. 2011. ER tubules mark sites of mitochondrial division. Science. 334:358–362. 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.R., Webster B.M., Mastronarde D.N., Verhey K.J., and Voeltz G.K.. 2010. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 190:363–375. 10.1083/jcb.200911024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Li D., Zhang S., Yang Y., Liu J.-J., Wang X., Liu C., Milkie D.E., Moore R.P., Tulu U.S., et al. 2018. Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell. 175:1430–1442.e17. 10.1016/j.cell.2018.09.057 [DOI] [PubMed] [Google Scholar]
- Herlan M., Vogel F., Bornhovd C., Neupert W., and Reichert A.S.. 2003. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 278:27781–27788. 10.1074/jbc.M211311200 [DOI] [PubMed] [Google Scholar]
- Ji W.-K., Chakrabarti R., Fan X., Schoenfeld L., Strack S., and Higgs H.N.. 2017. Receptor-mediated Drp1 oligomerization on endoplasmic reticulum. J. Cell Biol. 216:4123–4139. 10.1083/jcb.201610057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481. 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Jeong S.-Y., Karbowski M., Smith C.L., and Youle R.J.. 2004. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell. 15:5001–5011. 10.1091/mbc.e04-04-0294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Westrate L.M., Wu H., Page C., and Voeltz G.K.. 2016. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 540:139–143. 10.1038/nature20555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F., Lombès A., Frachon P., and Rojo M.. 2002. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol. Biol. Cell. 13:4343–4354. 10.1091/mbc.e02-06-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Weaver D., Shirihai O., and Hajnóczky G.. 2009. Mitochondrial ‘kiss-and-run’: interplay between mitochondrial motility and fusion-fission dynamics. EMBO J. 28:3074–3089. 10.1038/emboj.2009.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders E.M.M., Verbeek F.J., and Aten J.A.. 1993. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169:375–382. 10.1111/j.1365-2818.1993.tb03313.x [DOI] [PubMed] [Google Scholar]
- McEvoy A.L., Hoi H., Bates M., Platonova E., Cranfill P.J., Baird M.A., Davidson M.W., Ewers H., Liphardt J., and Campbell R.E.. 2012. mMaple: a photoconvertible fluorescent protein for use in multiple imaging modalities. PLoS One. 7:e51314 10.1371/journal.pone.0051314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., DeVay R., Block J., Cassidy-Stone A., Wayson S., McCaffery J.M., and Nunnari J.. 2006. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 127:383–395. 10.1016/j.cell.2006.09.021 [DOI] [PubMed] [Google Scholar]
- Misaka T., Miyashita T., and Kubo Y.. 2002. Primary structure of a dynamin-related mouse mitochondrial GTPase and its distribution in brain, subcellular localization, and effect on mitochondrial morphology. J. Biol. Chem. 277:15834–15842. 10.1074/jbc.M109260200 [DOI] [PubMed] [Google Scholar]
- O’Reilly C.M., Fogarty K.E., Drummond R.M., Tuft R.A., and Walsh J.V. Jr. 2003. Quantitative analysis of spontaneous mitochondrial depolarizations. Biophys. J. 85:3350–3357. 10.1016/S0006-3495(03)74754-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham A.H., McCaffery J.M., and Chan D.C.. 2012. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis. 50:833–843. 10.1002/dvg.22050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold A.S., Kostelecky B., Elia N., and Lippincott-Schwartz J.. 2011. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA. 108:10190–10195. 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R., Pinton P., Carrington W., Fay F.S., Fogarty K.E., Lifshitz L.M., Tuft R.A., and Pozzan T.. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 280:1763–1766. 10.1126/science.280.5370.1763 [DOI] [PubMed] [Google Scholar]
- Santel A., and Fuller M.T.. 2001. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114:867–874. [DOI] [PubMed] [Google Scholar]
- Sheff M.A., and Thorn K.S.. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670. 10.1002/yea.1130 [DOI] [PubMed] [Google Scholar]
- Sloat S.R., Whitley B.N., Engelhart E.A., and Hoppins S.. 2019. Identification of a mitofusin specificity region that confers unique activities to Mfn1 and Mfn2. Mol. Biol. Cell. 30:2309–2319. 10.1091/mbc.E19-05-0291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E., Griparic L., Shurland D.L., and van der Bliek A.M.. 2001. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 12:2245–2256. 10.1091/mbc.12.8.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Ghochani M., McCaffery J.M., Frey T.G., and Chan D.C.. 2009. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol. Biol. Cell. 20:3525–3532. 10.1091/mbc.e09-03-0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G., Elorza A., Molina A.J.A., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446. 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G., Graf S.A., Wikstrom J.D., Mohamed H., Haigh S.E., Elorza A., Deutsch M., Zurgil N., Reynolds N., and Shirihai O.S.. 2006. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. Am. J. Physiol. Cell Physiol. 291:C176–C184. 10.1152/ajpcell.00348.2005 [DOI] [PubMed] [Google Scholar]
- Vance J.E. 1990. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265:7248–7256. [PubMed] [Google Scholar]
- Wai T., and Langer T.. 2016. Mitochondrial Dynamics and Metabolic Regulation. Trends Endocrinol. Metab. 27:105–117. 10.1016/j.tem.2015.12.001 [DOI] [PubMed] [Google Scholar]
- Wong Y.C., Peng W., and Krainc D.. 2019. Lysosomal Regulation of Inter-mitochondrial Contact Fate and Motility in Charcot-Marie-Tooth Type 2. Dev. Cell. 50:339–354.e4. 10.1016/j.devcel.2019.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Carvalho P., and Voeltz G.K.. 2018. Here, there, and everywhere: The importance of ER membrane contact sites. Science. 361:eaan5835 10.1126/science.aan5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R.J., and van der Bliek A.M.. 2012. Mitochondrial fission, fusion, and stress. Science. 337:1062–1065. 10.1126/science.1219855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek N., Sparks L., and Voeltz G.. 2011. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 12:28–41. 10.1111/j.1600-0854.2010.01134.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The source data for all gels and blots are provided as a supplemental figure in the online version of the paper. All other data that support the findings of this study are available from the authors upon reasonable request.