Golenberg et al. report a citrullination-deficient zebrafish and demonstrate a role for Padi2 in fin wound responses and regeneration. This work identifies a distinct population of cells within the regenerative notochord bead that exhibits wound-induced histone citrullination.
Abstract
Calcium is an important early signal in wound healing, yet how these early signals promote regeneration remains unclear. Peptidylarginine deiminases (PADs), a family of calcium-dependent enzymes, catalyze citrullination, a post-translational modification that alters protein function and has been implicated in autoimmune diseases. We generated a mutation in the single zebrafish ancestral pad gene, padi2, that results in a loss of detectable calcium-dependent citrullination. The mutants exhibit impaired resolution of inflammation and regeneration after caudal fin transection. We identified a new subpopulation of cells displaying citrullinated histones within the notochord bead following tissue injury. Citrullination of histones in this region was absent, and wound-induced proliferation was perturbed in Padi2-deficient larvae. Taken together, our results show that Padi2 is required for the citrullination of histones within a group of cells in the notochord bead and for promoting wound-induced proliferation required for efficient regeneration. These findings identify Padi2 as a potential intermediary between early calcium signaling and subsequent tissue regeneration.
Introduction
Humans have limited regenerative capacity, which can present significant clinical challenges. Mammalian wound repair occurs through similar stages as regeneration in simple animal models (Yokoyama, 2008). In regenerative models, early wound signals activate a series of regenerative steps, including the recruitment of immune cells, generation of a wound epithelium (Roehl, 2018), and the subsequent formation of a blastema, a mass of stem cell–like cells, that mediates cell proliferation and repair (Whitehead et al., 2005). Although increased cytosolic calcium early in wound healing has been linked to later regenerative proliferation (Globus et al., 1987; Lagoudakis et al., 2010; Yoo et al., 2012a), there is limited understanding of how these early signals impact later regenerative events.
An attractive candidate to link calcium increase with subsequent regeneration is the family of calcium-dependent enzymes, peptidylarginine deiminases (PADs or PADIs), which catalyze the deimination of a peptidylarginine to the neutrally charged, noncoded amino acid, citrulline (Vossenaar et al., 2003). These enzymes were recently implicated in stem cell pluripotency and the regulation of gene expression (Christophorou et al., 2014; Wiese et al., 2019; Xiao et al., 2017). While PADs have been studied in mammalian models, the presence of multiple PAD isoforms and functional redundancy make it challenging to dissect the role of citrullination in normal development and wound healing.
Zebrafish, Danio rerio, have one highly conserved copy of a pad gene, padi2, that shares canonical mammalian PAD features, with conserved enzymatic activity and calcium dependence. We generated a padi2 mutant zebrafish line that lacks detectable calcium-dependent citrullination activity and displays normal developmental but impaired regenerative growth. This work provides insight into how calcium-dependent citrullination may integrate early signals induced by injury to mediate subsequent tissue repair.
Results and discussion
Characterization of zebrafish PAD
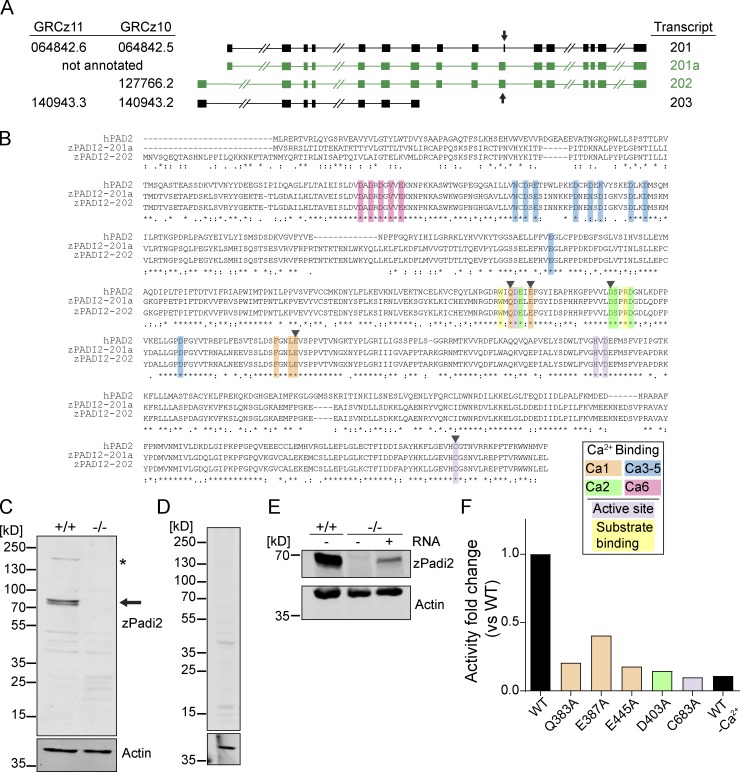
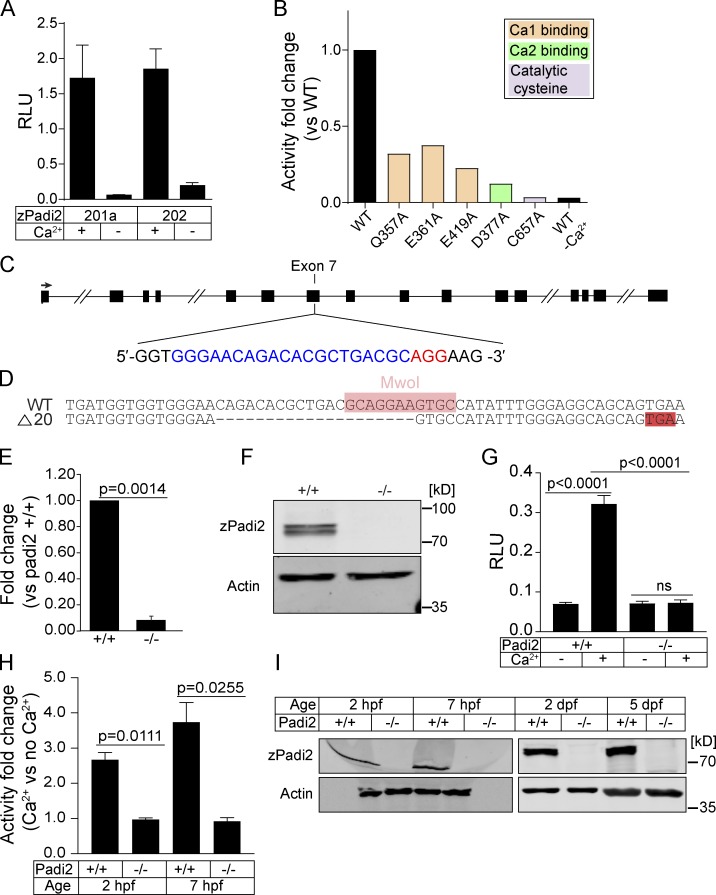
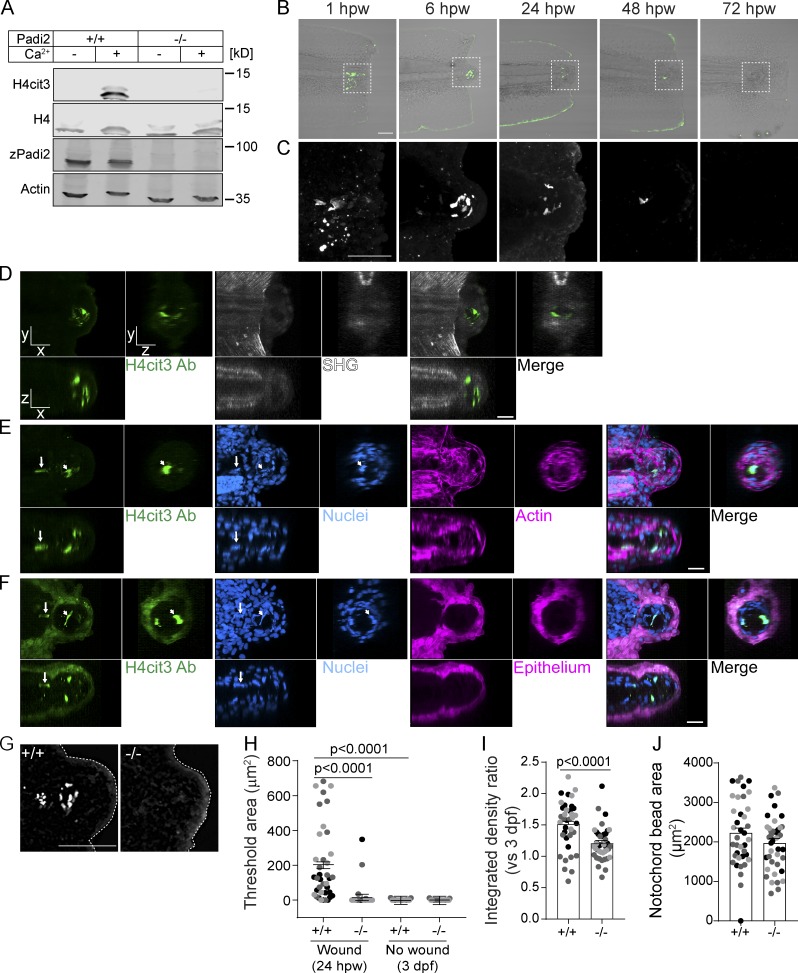
To examine the role of citrullination in zebrafish, we characterized the annotated zebrafish pad gene, padi2. A 7-exon transcript (203) and two 16-exon transcripts with alternative start sites (201 and 202) are annotated (Fig. S1 A). We cloned the transcripts that share exon 16 and identified two splice variants of padi2 (Fig. S1 A). These transcripts are highly conserved with human PADs and share 55% amino acid identity to human PAD2, with conserved catalytic and calcium-binding sites (Fig. S1 B; Smith and Waterman, 1981). Using a polyclonal antibody, immunoblotting showed a doublet at 75–80 kD, consistent with two full-length splice variants (Fig. S1 C). The absence of this doublet with preimmune serum and the detection of an appropriately sized protein in padi2-201a mRNA-injected larvae demonstrate the specificity of the antibody (Fig. S1, D and E). We used a colorimetric in vitro citrullination activity assay and detected citrullination activity with both Padi2 variants (Fig. 1 A; Nakayama-Hamada et al., 2005). Similar to mammalian PADs, zebrafish Padi2 activity was calcium dependent (Fig. 1 A), as alanine point mutations of predicted calcium binding amino acids (Arita et al., 2004) impaired in vitro citrullination activity (Fig. 1 B and Fig. S1 F). Mutation of the catalytic cysteine also abolished activity (Fig. 1 B and Fig. S1 F). These data indicate that zebrafish Padi2 is a canonical PAD with similar function to mammalian PADs.
Figure S1.
Characterization of zebrafish Padi2. (A) Schematics of padi2 transcripts, with exons represented by solid boxes and introns by connected lines (slashes indicate shortening of relative length for display purposes). Left: A list of the corresponding last seven digits of Ensembl ID from GRCz11 and GRCz10 genome assemblies (full Ensemble IDs listed in Materials and methods). Right: A list of the names based on GRCz10 used to reference the transcripts. Cloned transcripts discussed in this paper are in green, and arrows highlight exon 10. (B) Full amino acid sequences of human PAD2 and predicted zebrafish Padi2 splice variants (201a and 202). Amino acids are highlighted (as indicated in key) to demonstrate calcium binding, catalytic residues, and substrate-binding residues. Black arrowheads indicate amino acids referred to in F and Fig. 1 B. (C) Full Western blot (from Fig. 1 F) of pooled larvae probed with antibodies against zebrafish Padi2 and actin. WT and padi2−/− lysates were probed. Arrow demonstrates expected size of Padi2 transcripts at ∼75 and 80 kD, and asterisk marks ∼200 kD species. Notably, this antibody did not detect a protein of equivalent size to the predicted transcript 203 (∼35 kD). Representative blot from four replicates. (D) Western blot of pooled WT larvae probed with preimmune serum and actin antibody. (E) zPadi2 Western blot of pooled 2 dpf larvae. Lane 1, WT; lane 2, padi2−/−; lane 3, padi2 201a mRNA-injected padi2−/− larvae. (F) Citrullination activity of Padi2 202 and individual point mutations in select calcium-binding and catalytic amino acids (colors correspond to highlighted residues in B). Fold change of enzymatic activity normalized to WT Padi2 202. Data represent two independent experiments, and WT values are also represented in Fig 1 A.
Figure 1.
Characterization of zebrafish Padi2. (A) Citrullination activity of zebrafish Padi2 201a and 202 splice variants in total lysates with and without calcium. Absorbance of light was measured and expressed as mean (± SEM) relative light units (RLU), normalized for protein level. Data represent three independent replicates. (B) Citrullination activity of Padi2 201a and individual point mutations in calcium binding and catalytic amino acids. Fold change of enzymatic activity is shown relative to WT Padi2 201a. Data represent two independent replicates, and WT values are also represented in A. (C) Schematic of padi2 gene with exon 7 gRNA sequence highlighted for CRISPR/Cas9 mutagenesis. gRNA sequence is in blue and protospacer adjacent motif is in red. (D) Sequence alignment of WT and padi2−/− 20 bp mutation in exon 7. MwoI restriction site for genotyping highlighted in pink, early stop codon highlighted in red. (E) RT-qPCR of padi2 exon5/6 on individual larvae from a padi2+/− incross. Data from three pooled independent replicates with the means and SEM reported and a one-sample t test performed. (F) Representative Western blot for zebrafish Padi2 and Actin from pooled 2 dpf larvae (representative of four experiments). (G) Citrullination activity of pooled 2 dpf zebrafish lysates expressed as RLU. Data are from three independent replicates with the means and SEM reported and an ANOVA performed. (H) Citrullination activity of pooled embryo lysates during development. Fold change of enzymatic activity is shown as a ratio of calcium-treated to no calcium for each condition. Data are from three independent replicates. (I) Representative Western blot for zebrafish Padi2 and Actin from pooled zebrafish through stages of development (representative of three and two experiments).
Characterization of a padi2 zebrafish mutant
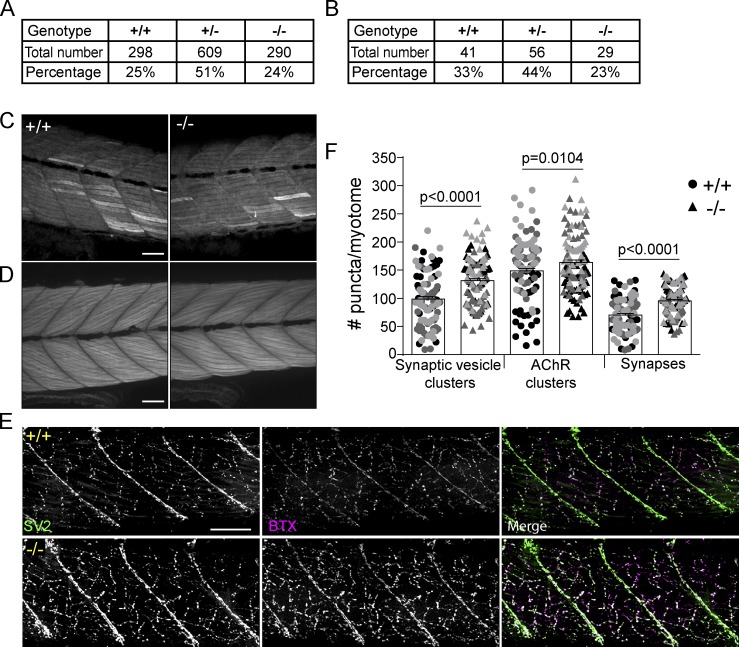
To characterize the role of citrullination in regeneration, we generated a zebrafish padi mutant using CRISPR/Cas9 gene editing, targeting exon 7 of padi2, as it preceded essential catalytic amino acids and resulted in a 20–base pair deletion and early stop codon (Fig. 1, C and D). Padi2 homozygous mutants (padi2−/−) had reduced levels of padi2 mRNA (Fig. 1 E) and loss of Padi2 protein (Fig. 1 F and Fig. S1 C) at 2 d post-fertilization (dpf). Lysates from mutant larvae lacked citrullination activity, even in the presence of excess calcium (Fig. 1 G), indicating that Padi2 is likely the PAD that mediates citrullination activity at 2 dpf. Although previous studies have indicated that mammalian PAD1 and PADI6 are necessary for normal development (Esposito et al., 2007; Kan et al., 2012; Zhang et al., 2016), we found that the Padi2-deficient zebrafish did not display any gross morphological defects and had normal viability (Fig. S2, A and B). A homozygous incross produces viable and developmentally normal maternal-zygotic embryos, indicating that maternal padi2 is not necessary for early embryonic development. To further assess citrullination in early development, we found that WT embryos showed citrullination activity and Padi2 protein expression during both pre- and post-maternal to zygotic transition that was absent in Padi2-deficient larvae (Fig. 1, H and I), providing further evidence that citrullination is not necessary for early zebrafish development. To characterize later phenotypes, we examined the muscles of Padi2-deficient larvae since mammalian PAD2 is the predominant isozyme in skeletal muscle and nervous system (Kubilus and Baden, 1983; Watanabe and Senshu, 1989). We found that both slow and fast-twitch skeletal muscles in the padi2−/− larvae appeared morphologically normal (Fig. S2, C and D). However, immunostaining of 5 dpf larvae for presynaptic vesicles (α-SV2) and acetylcholine receptors (AChR, α-BTX; Fig. S2, E and F) revealed that padi2−/− larvae formed more neuromuscular junctions than WT larvae (Fig. S2 F). This is interesting since Padi2 is expressed in central synapses (Bayés et al., 2017) and PAD2-deficient mice display behavioral defects (Falcão et al., 2019).
Figure S2.
Homozygous padi2 mutants are viable and have increased neuromuscular junctions. (A) Genotype frequency of 5 dpf larvae of incrossed padi2 heterozygotes. (B) Genotype frequency of adult offspring of incrossed padi2 heterozygotes. Data in A and B are from four and six clutches, respectively, and analyzed by χ2 tests. (C) Representative images of slow-muscle fibers immunostained with α-MyHC antibody in the trunk of 5 dpf larvae from three independent replicates. (D) Representative images of the trunk of phalloidin-stained 5 dpf larvae for visualization of F-actin in fast-muscle fibers. WT cousin (left) and padi2−/− (right) from three independent replicates. (E) Neuromuscular junctions are labeled with α-SV2 (green, presynaptic vesicles), α-BTX (red, postsynaptic AChRs), and merge (synapses) in WT cousins (top) and padi2−/− (bottom) larvae at 5 dpf. (F) Quantification of the number of SV2 puncta, AChR puncta, and synapses in a single myotome in the trunks of larvae. Data are from three pooled independent replicates with the lsmeans (±) SEM and P values calculated by ANOVA reported. Each symbol represents a single myotome, and measurements were taken from two myotomes per larva (n = 100 myotomes from 50 WT larvae, n = 114 myotomes from 57 padi2−/− larvae). Scale bars, 50 µm.
Padi2 is required for efficient epimorphic regeneration
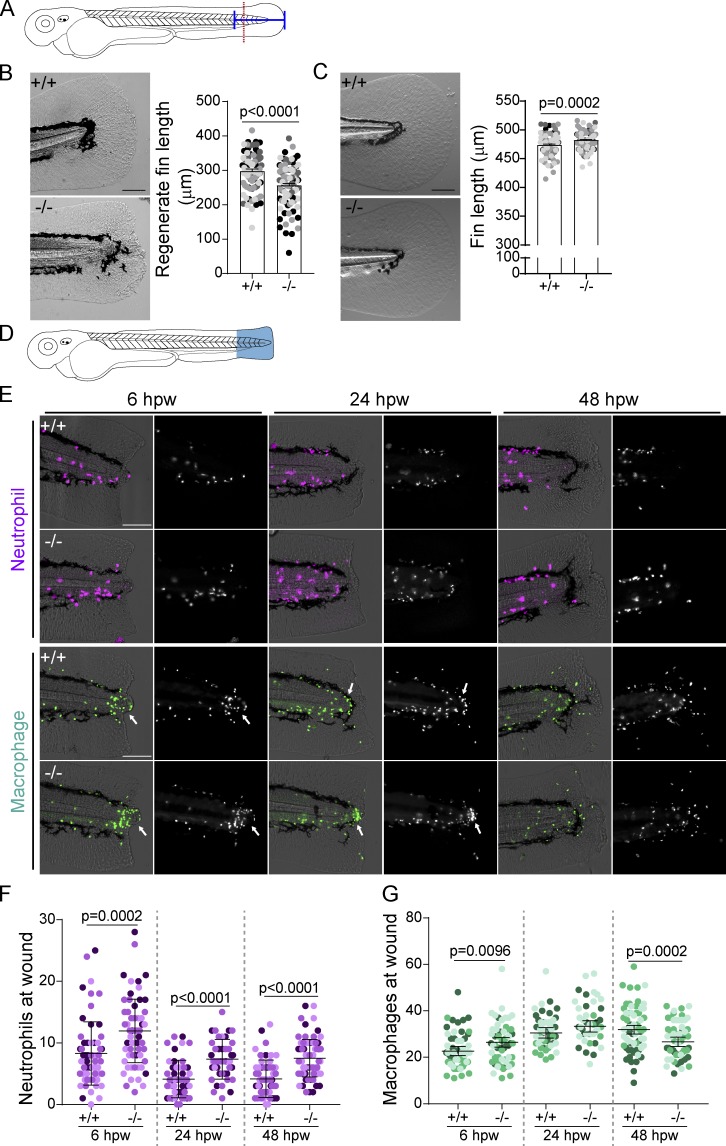
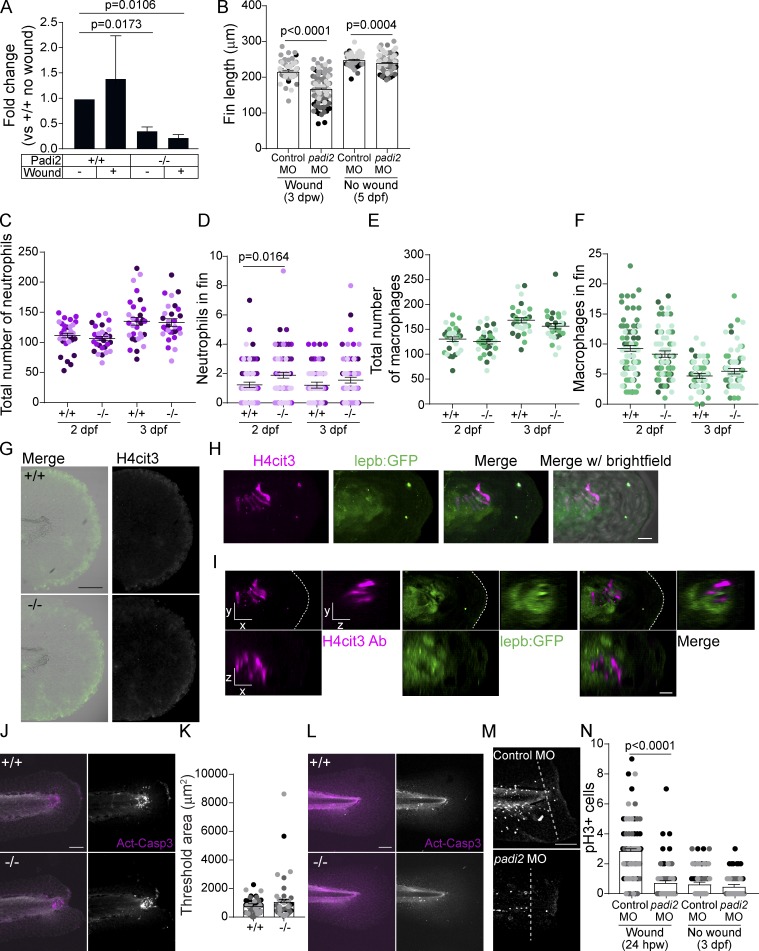
To determine the role of citrullination in regeneration, we performed a tail transection of 2.5 dpf larvae through the notochord as described by Rojas-Muñoz et al. (2009; Fig. 2 A). Analysis by quantitative RT-PCR (RT-qPCR) at 24 h post-wounding (hpw) showed that padi2 was expressed in the affected tissue during regeneration but was not detected in padi2−/− wounded fins (Fig. S3 A). Regeneration was impaired in the Padi2-decifient larvae compared with WT “cousins” (Fig. 2 B). Similar effects were observed with transient morpholino (MO) depletion of padi2 following a fin fold excision (Fig. S3 B). padi2−/− larvae had a slight, but statistically significant, increase in their developmental fin length at 5 dpf (Fig. 2 C). These findings suggest that Padi2 is necessary for efficient caudal fin regeneration and that different mechanisms mediate developmental and regenerative growth.
Figure 2.
Padi2 is required for proper regeneration and leukocyte recruitment. (A) Schematic of regeneration assay. Tail transections were performed through the notochord (red dotted line) at 2.5 dpf. Fin lengths were measured from the blood circulation to the end of the fin (blue solid line). (B) Representative bright field images of regeneration at 3 dpw and quantification of regenerate fin length from four independent replicates with n = 90 +/+, 95 −/−. (C) Representative images of 5 dpf developmental fins and quantification of developmental fin length from five independent replicates with n = 109 +/+, 108 −/−. (D) Schematic of leukocyte quantification region (in blue). (E) Representative images of leukocytes at a wound at 6, 24, and 48 hpw visualized with mCherry-labeled neutrophil nuclei (Tg(lyzC:H2B-mCherry)) and GFP-labeled macrophage nuclei (Tg(mpeg1:H2B-GFP)). Fluorescence image on the right, merge with bright-field on the left. Macrophage localization to the periphery of the notochord bead indicated with an arrow. (F) Quantification of neutrophil nuclei at a wound from three independent replicates (6 hpw, n = 62 +/+, 57 −/−; 24 hpw, n = 50 +/+, 47 −/−; 48 hpw, n = 63 +/+, n = 65 −/−). (G) Quantification of macrophage nuclei at a wound from three independent replicates (6 hpw, n = 61 +/+, 55 −/−; 24 hpw, n = 48 +/+, 44 −/−; and 48 hpw, n = 63 +/+, 57 −/−). All quantifications have lsmeans (± SEM) reported with P values calculated by ANOVA. Scale bars, 100 µm.
Figure S3.
Padi2-deficient larvae show regeneration defects. (A) RT-qPCR of padi2 exon5/6 on pooled fin extracts from 24 hpw and no wound controls (3 dpf) normalized to WT, no-wound fins. Data are from three pooled independent replicates with the means and SEM reported and a one-sample t test performed. (B) Quantification of regenerative and developmental fin length after MO knockdown of padi2. Data from five independent replicates with 3 dpw (n = 90 control MO, n = 113 padi2 MO) and 5 dpf (n = 104 control MO, n = 103 padi2 MO). (C) Quantification of neutrophils in whole larvae from three independent replicates, (n = 30 +/+, 29 −/− at 2 dpf; n = 30 +/+, 30 −/− at 3 dpf). (D) Quantification of neutrophil numbers in developmental, unwounded fins. Pooled from five independent replicates (n = 88 +/+, 78 −/− at 2 dpf and n = 79 +/+, 75 −/− at 3 dpf). (E) Quantification of macrophage numbers in whole larvae from three independent replicates (n = 30 +/+, 29 −/− at 2 dpf and n = 30 +/+, 30 −/− at 3 dpf). (F) Quantification of macrophage numbers in developmental, unwounded fins. Pooled from four independent replicates (n = 81 +/+, 74 −/− at 2 dpf and n = 70 +/+, 66 −/−). (G) Representative images of H4cit3 immunostaining in 3 dpf unwounded control larvae with H4cit3 antibody label on the right and merged with the bright-field on the left. (H) Representative multiphoton microscopy 3D reconstruction showing en face view of the notochord bead at 24 hpw in Tg(lepb:EGFP) expressing (green) larvae labeled with H4cit3 immunofluorescence (magenta). Last image in row includes bright-field overlay. (I) Section view of the notochord bead, showing en face (x, y view) and orthogonal (x, z view is below; y, z view is to the right) sections, with section thickness shown 2 µm for both x and y, 10 µm in z. Scale bars, 10 µm. (J and L) Representative images of active-Caspase3 labeled in 66 hpw fins (J) or developmental, unwounded fins (L). Merged images of active-Caspase3 (magenta) and DAPI (white) on the left, and single active-Caspase3 channel in white on the right. (K) Quantification of active-Caspase3 threshold area in padi2−/− and WT fins at 66 hpw from three independent replicates (n = 47 +/+, 47 −/−). (M and N) Representative images at 24 hpw (M) and quantification of mitotic cells (N) labeled with phosphorylated histone H3 in MO injected larvae past the notochord (white dotted line) from three independent replicates (24 hpw n = 68 control MO, 70 padi2 MO and 3 dpf n = 71 control MO, 64 padi2 MO). All quantifications have lsmeans and SEM reported, and P values were calculated by ANOVA. Scale bars, 100 µm.
Padi2 modulates leukocyte recruitment to a wound
Citrullination has been shown to affect the immune response in human disease (Li et al., 2010), with direct evidence for deimination of chemokines (Loos et al., 2009; Proost et al., 2008; Yoshida et al., 2014). To visualize leukocyte responses to a wound, we compared padi2−/− and WT cousin larvae with labeled neutrophils (Tg(lyzc:H2B-mCherry)) or macrophages (Tg(mpeg1:H2B-GFP); Fig. 2, D–G). padi2−/− larvae had a consistent increase in neutrophils at 6, 24, and 48 hpw (Fig. 2, E and F), with no change in total neutrophil numbers (Fig. S3 C), although there were slightly more neutrophils in unwounded fins (Fig. S3 D). We also found an early and small increase in macrophages at the wound in padi2−/− larvae, although this difference did not persist and there was no change in total macrophage numbers (Fig. 2, E and G; and Fig. S3, E and F). Interestingly, an aggregation of macrophages was observed around the notochord bead at 6 and 24 hpw (Fig. 2 E). Taken together, these findings suggest that resolution of neutrophil inflammation is impaired in Padi2-deficient larvae. It is unclear if this phenotype is due to a failure of wound resolution or potentially a direct effect of citrullination on leukocyte signaling pathways. Binding of the neutrophil chemokine, Cxcl8, to its receptors, Cxcr1 and Cxcr2, regulates neutrophil directional and reverse migration (Powell et al., 2017); interestingly, citrullination of Cxcl8 alters its binding to its receptors (Proost et al., 2008). Alternatively, citrullination of ECM components affects cell migration (Shelef et al., 2012; Sipilä et al., 2014; Yuzhalin et al., 2018), and could potentially regulate inflammation by altering the wound ECM.
Wounding induces localized histone citrullination in the notochord bead
We next considered whether wounding induces citrullination of histones in larval zebrafish due to the reported role of citrullinated histones in maintaining pluripotency (Christophorou et al., 2014; Wiese et al., 2019; Xiao et al., 2017). Whole WT larvae lysate showed calcium-dependent citrullination of histone H4 (H4cit3) that was not present in padi2−/− lysate (Fig. 3 A). Caudal fin transection results in increased calcium at a wound (Yoo et al., 2012a), which may promote citrullination. Visualization of H4cit3 upon caudal fin amputation in WT zebrafish revealed signal exclusively within a localized group of cells in the notochord bead (Fig. 3, B and C), a region previously described as the regeneration blastema (Rojas-Muñoz et al., 2009). Immunofluorescence microscopy revealed H4cit3 signal as early as 1 hpw, persisting to 24 hpw, and was absent by 72 hpw (Fig. 3, B and C). Histone H4 deimination is wound dependent, as no signal was observed in unwounded larvae (Fig. S3 G).
Figure 3.
Tail transection stimulates localized Padi2-dependent histone H4 citrullination. (A) Representative Western blot of padi2−/− and WT whole 2 dpf larvae lysates showing protein levels of citrullinated Histone4 (H4cit3), total Histone4 (H4), total Padi2 (zPadi2), and total actin (Actin; representative of two replicates). (B) Representative images of H4cit3 antibody staining in tail transected fins at 20× magnification. Merge images with bright-field are shown for orientation. Box denotes region imaged at 60× magnification in C showing H4cit3 immunolabel alone. (B and C) Representative images from three independent replicates. (D–F) Representative multiphoton microscopy enface (x, y view) and orthogonal (x, z view is below; y, z view is to the right) sections of 6 hpw WT caudal fins labeled with H4cit3 immunofluorescence (green) in conjunction with either SHG (D; white) or DAPI-labeled nuclei (E; blue) and rhodamine-phallodin–labeled actin (magenta) or DAPI-labeled nuclei (F; blue) and (Tg(krt-4:EGFP))-expressing epithelium (magenta). Note in F, fluorophore tag for epithelium crosses into H4cit3 antibody channel at the setting needed to detect the antibody signal. Arrow indicates nucleus with citrullinated histones in the notochord region, while arrowhead points to one example of a nucleus with citrullinated histones in the notochord bead. For image presentation, section thickness shown is 2 µm for both x and y, 10 µm in z. Representative images from two independent replicates. Videos 1, 2, 3, 4, 5, and 6 present z-stacks and 3D reconstructions of data in D-F. (G) Representative images of H4cit3 immunostaining in 24 hpw WT cousin (left) and padi2−/− (right). (H) Quantification of H4cit3 signal area at the notochord at 24 hpw and 3 dpf (no wound control). (I) Quantification of H4cit3 integrated density in 24 hpw larvae normalized to the average of 3 dpf for each genotype. (J) Quantification of the notochord bead area at 24 hpw. All quantifications are from three pooled independent replicates, have the lsmeans (±) SEM reported and P values calculated by ANOVA. (H–J) 24 hpw, n = 38 +/+, 41 −/−; 3 dpf n = 25 +/+, 25 −/−. Scale bars, 100 µm in B and G; 50 µm in C; 20 µm in D–F.
To further characterize this structure, we used multiphoton microscopy to understand the 3D localization of citrullinated histones within the context of the wounded fin. Using second harmonic generation (SHG) to visualize the collagen fiber network, in conjunction with H4cit3 immunostaining, we observed citrullinated histones in a region devoid of collagen fibers at 6 hpw (Fig. 3 D and Videos 1 and 2). The notochord bead containing this signal formed posterior to the original wound axis, demarcated by the end of the collagen network. Labeling F-actin to visualize cell borders demonstrated that the notochord bead is composed of multiple cells (Fig. 3 E and Videos 3 and 4). The H4cit3 signal colocalized with DAPI, but only a subset of nuclei in this region was positive for histone citrullination. Nuclei with histone citrullination can also be observed in cells outside the notochord bead; it is possible that these cells move into this region from the notochord (Fig. 3, E and F). Finally, visualization of the epithelium using Tg(krt4:EGFP) revealed that histone citrullination did not occur within these epithelial cells (Fig. 3 F and Videos 5 and 6). By 6 hpw, the wound epithelium has already formed and encompasses the cells with citrullinated histones, and the notochord bead (Fig. 3 F). Taken together, we have identified a new wound-induced structure within the notochord bead comprised of a subpopulation of cells with citrullinated histones. Future work will be needed to identify which cells or signals are necessary to promote citrullination only within this subpopulation of notochord bead cells.
Video 1.
Localization of nuclei exhibiting histone citrullination in the notochord bead, a region lacking collagen fibers. Scan through multiphoton z-stack of transected tail showing H4cit3 antibody labeled nuclei (green) in conjunction with SHG (white) imaging to show collagen fibers. Anterior is left. Sections are 2 microns apart. Frame rate is 5 fps. Corresponds to images in Fig 3 D.
Video 2.
3D localization of nuclei exhibiting histone citrullination, with SHG imaging. Rotations of 3D reconstruction (from z-stack shown in Video 1) of a transected tail at 6 hpw demonstrating the spatial relationship of nuclei with citrullinated histones (H4cit3 antibody, green) with respect to the collagen fiber structure (SHG, white). Anterior is left at start of video. Frame rate is 10 fps. Corresponds to images in Fig 3 D.
Video 3.
Cellular context of nuclei with citrullinated histones. Scan through multiphoton z-stack of transected tail illustrating colocalization of citrullinated histone label (H4cit3 antibody, green) with nuclear label (DAPI, blue) at 6 hpw. Cell outlines are identified by phalloidin-tagged actin (magenta). Anterior is left. Sections are 2 microns apart. Frame rate is 5 fps. Corresponds to images in Fig 3 E.
Video 4.
3D localization of nuclei with citrullinated histones in the cellular context of the wounded tail. Rotations of 3D reconstruction (from z-stack shown in Video 3) of a transected tail at 6 hpw showing the colocalization of H4cit3 antibody (green) with nuclear label (DAPI, blue) and demonstrating their spatial relationship to the actin outlines of cells (magenta) in the wounded tail, both in the notochord and in the bead. Anterior is left at start of video. Frame rate is 10 fps. Corresponds to images in Fig 3 E.
Video 5.
Nuclei with citrullinated histones are located within the bead, not in epithelium surrounding the bead. Scan through multiphoton z-stack of transected tail, further illustrating the colocalization of citrullinated histone label (H4cit3 antibody, green) with nuclear label (DAPI, blue) at 6 hpw and showing their relationship to the epithelial cells (krt4:GFP, magenta). Anterior is left. Sections are 2 microns apart. Frame rate is 5 fps. Corresponds to images in Fig 3 F.
Video 6.
3D localization of nuclei with citrullinated histones in the context of epithelium of the wounded tail. Rotations of 3D reconstruction (from z-stack shown in Video 5) of a transected tail at 6 hpw, again showing the colocalization of H4cit3 antibody (green) with nuclear label (DAPI, blue), both in the notochord and in the bead, as well as demonstrating their spatial relationship to the epithelial cells (magenta) surrounding the bead. Anterior is left at start of video. Frame rate is 10 fps. Corresponds to images in Fig 3 F.
Toward this goal, we found that wound-induced histone citrullination was absent in padi2−/− larvae at 24 hpw (Fig. 3 G). In the padi2−/− larvae, wounding did not induce histone H4 citrullination in the notochord bead above unwounded levels (Fig. 3, G–I). Morphologically, both WT and padi2−/− larvae formed similar-sized notochord beads early after wounding (Fig. 3 J). Previous reports suggest this region of cells act as a required wound-signaling center that orchestrates regeneration (Rojas-Muñoz et al., 2009; Romero et al., 2018). Importantly, this population of cells with citrullinated histones was associated with the blastema reporter, Tg(lepb:EGFP) (Fig. S3, H and I; Kang et al., 2016). Our data indicate that citrullination is necessary for efficient regeneration and that a localized population of cells within the blastema structure contains citrullinated histones. With a known role for histone citrullination in stem cell maintenance, we speculate that this population of cells has pluripotent features required for efficient tissue repair.
Padi2-deficient larvae have impaired wound-induced proliferation
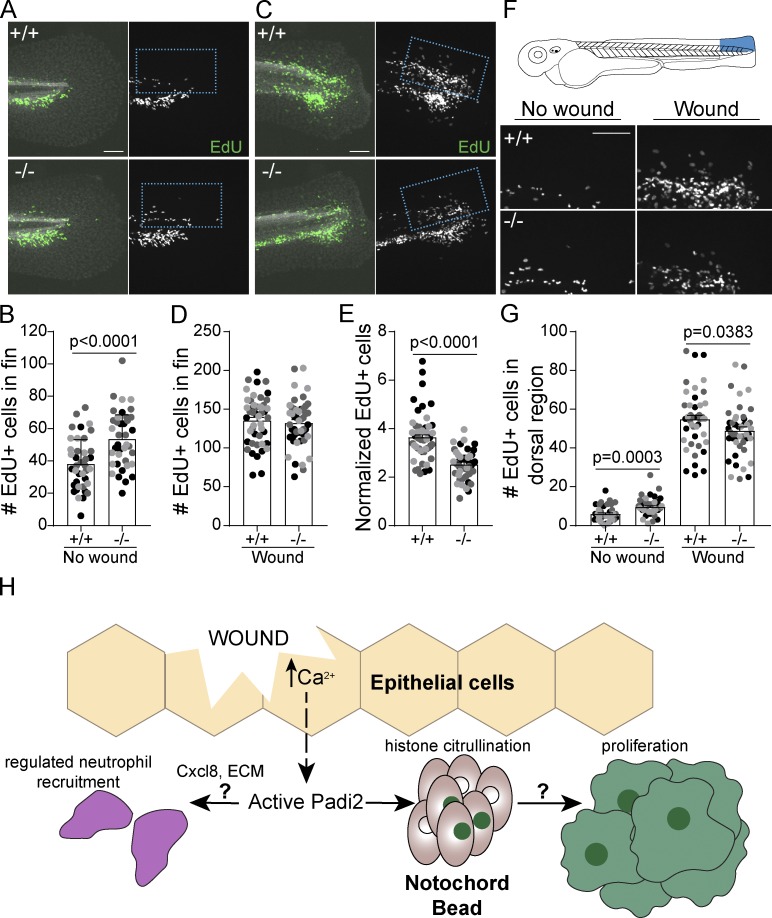
An essential aspect of epimorphic regeneration is remodeling by wound-induced apoptosis and proliferation (Gauron et al., 2013; Nechiporuk and Keating, 2002; Tseng et al., 2007). We did not observe a significant change in wound-stimulated apoptosis in mutant larvae (Fig. S3, J–L). Cell proliferation was assayed using 5-ethynyl-2’-deoxyurdine (EdU) incorporation, and while mutant larvae had a greater number of EdU-positive cells within the developing caudal fin (Fig. 4, A and B), induction of proliferation with wounding was impaired (Fig. 4, C–E). Similarly, padi2 MO knockdown resulted in decreased mitotic index at 24 hpw compared with control larvae (Fig. S3, M and N). To further quantify wound-induced proliferation, we focused on the dorsal region of the tail since much of the developmental proliferation is localized to the ventral fin. In this dorsal region we also observed impaired proliferation in the padi2−/− larvae compared with WT cousins (Fig. 4, F and G). These findings suggest opposing roles for Padi2 in developmental fins and wound-induced proliferation, supporting the idea that these two processes have distinct modes of regulation.
Figure 4.
Wound-induced proliferation is perturbed in Padi2-deficient larvae. (A and C) Representative images of 6 h EdU pulsed larvae in developmental, unwounded (A) or 66 hpw (C) fins. Merged images of EdU (green) and DAPI (white) on the left and single EdU (white) image on the right. (B and D) Quantification of EdU-positive cells in the fin. (E) Number of EdU-positive cells in the fin normalized to corresponding no wound conditions. (F) Representative images of the dorsal half of 6 h EdU-pulsed fins. (G) Quantification of EdU-positive cells within the dorsal region of the fin. All data are from three pooled independent replicates with the lsmeans and SEM reported and P values calculated by ANOVA (no wound, n = 39 +/+, 39 −/−; 66 hpw, n = 47 +/+, 47 −/−). Scale bars, 100 µm. (H) A proposed model depicting how the early wound epithelial calcium flux might activate (dashed arrow) Padi2 to catalyze citrullination events that, either directly or indirectly (left question mark), regulate neutrophil (purple) recruitment to the wound. Concomitantly, wound-dependent Padi2 citrullination of histones (green nuclei) within a subset of cells in the notochord bead (pink) potentially stimulates, through yet to be determined mechanisms (right question mark), regenerative proliferation.
In summary, we identified a new role for citrullination in wound healing and regeneration. Early calcium flux is a universal injury signal in organisms ranging in complexity, and while there are many citrullination-independent, calcium-induced wound pathways (Niethammer, 2016), this work identified one potential regenerative mechanism downstream of wound-induced calcium (Fig. 4 H). We showed that zebrafish Padi2 has conserved activity and calcium dependence and is necessary for calcium-mediated histone citrullination. The identification of a population of cells with wound-induced, Padi2-dependent histone citrullination in the notochord bead demarcates a novel signaling hub within a subset of blastemal cells that likely orchestrates efficient regenerative growth. Moreover, this citrullination-deficient vertebrate model provides a powerful tool for future studies to dissect the role of citrullination in development, disease, and wound healing, and will aid in the identification of in vivo Padi targets.
Materials and methods
Zebrafish maintenance and handling
All protocols in this study were approved by the University of Wisconsin-Madison Animal Care and Use Committee. Adult zebrafish were maintained on a 14 h/10 h light/dark schedule. Fertilized embryos were transferred and maintained in E3 buffer at 28.5°C. This study used adult AB and NHGRI-1 (LaFave et al., 2014) fish (obtained from the Zebrafish International Resource Center) as well as previously published transgenic lines Tg(mpeg1:H2B-GFP) (Miskolci et al., 2019), Tg(lyzc:H2B-mCherry) (Yoo et al., 2012b), Tg(krt4:EGFP) (Yoo et al., 2012a), and Tg(lepb:EGFP) (Kang et al., 2016).
Zebrafish and human PAD alignment
Sequence alignments were performed using the EMBOSS Water pairwise sequence alignment algorithm (Smith and Waterman, 1981). Predicted transcripts are listed in Table 1. Transcript annotations are from GRCz10 with transcript 201a indicating a sequence slightly divergent from GRCz10’s transcript 201. These transcripts have different first exons, but both splice variants share a complete exon 10, contrary to the genome assembly predictions. Due to these discrepancies, these transcripts are referred to here as 201a and 202 (Fig. S1 A).
Table 1. Annotated PAD transcripts used in this study.
| Name | Transcript ID | Genome assembly |
|---|---|---|
| zPadi2 202 | ENSDART00000140943.3 | GRCz11 |
| zPadi2 203 | ENSDART00000140943.2 | GRCZ10 |
| zPadi2 201 | ENSDART00000064842.6 | GRCz11 |
| zPadi2 201 | ENSDART00000064842.5 | GRCz10 |
| zPadi2 202 | ENSDART00000127766.2 | GRCz10 |
| hPADI2 202 | ENST00000375486.8 | GRCh38 |
Generation of a padi2 mutant line and genotyping
Zebrafish CRISPR/Cas9 injections were performed as previously described in our laboratory (LeBert et al., 2015, 2018). gRNA for zebrafish Padi2 (ENSDARG00000044167) was designed using CHOPCHOP (Montague et al., 2014). The exon 7 target sequence was 5′-GGGAACAGACACGCTGACGC-3′. The pT7 gRNA was prepared as previously described (LeBert et al., 2018). The gRNA and Cas9 protein (New England Biolabs) were injected into one-cell stage NHGRI-1 embryos in a 2 nl volume consisting of ∼50 ng/µl gRNA and ∼300 nM Cas9. To confirm genome editing by the gRNA, genomic DNA was extracted from 2 dpf embryos, amplified using the primers listed below, and separated on a 3% MetaPhor gel (Lonza): Padi2 F: 5′-CTGATACATGGCACAACCTACG-3′, and Padi2 R: 5′-GAAAACCAGCAAGCAGAGAAAGTT-3′.
Sequences of F0 mosaic cuts were confirmed by topoisomerase-based (TOPO) cloning (Invitrogen) and sequencing. Clutches of larvae with confirmed CRISPR cuts were grown to adulthood. Adult F0 CRISPR-injected fish were screened for germline mutations by testing their individual outcrossed offspring (2–5 dpf) using the primers listed above and Indel Detection and Amplicon Analysis (Yang et al., 2015). Sequences were analyzed using Peak Studio (McCafferty et al., 2012). Mutation sequence was confirmed by TOPO cloning and sequencing.
Heterozygous padi2 zebrafish were maintained by outcrossing the CRISPR mutants to AB WT background zebrafish and genotyped by genomic DNA isolated from fin clips and amplified using the primers listed above. PCR product was either separated on a 2% agarose gel for 3 h or digested overnight with MwoI (New England Biolabs) and separated on an agarose gel to determine individual fish genotypes. For experimental purposes, F2 or F3 heterozygotes were incrossed for the generation of adult homozygous mutant and WT siblings. These adults were then incrossed to produce padi2−/− and WT clutches, referred to as cousins, which were only used for experimentation and not for the maintenance of subsequent generations.
RT-qPCR
RNA and DNA were extracted from individual 2 dpf embryos from a padi2+/− incross using TRIZOL (Invitrogen) following the manufacturer’s protocol. Embryos were genotyped using GoTaq (Promega) as described above, and two or three embryos of each genotype were used for cDNA production using Superscript III First Strand Synthesis System with Oligo(dT) (Thermo Fisher Scientific). qPCR was performed using FastStart Essential Green Master (Roche) and a LightCycler96 (Roche). Primers for padi2 and ef1a are listed below. Data were normalized to ef1a using the ΔΔCt method (Livak and Schmittgen, 2001) and represented as fold change over WT embryos.
For evaluation of padi2 mRNA expression during wounding, incrosses of F3 or F4 adult WT and padi2−/− siblings were done to produce offspring cousins homozygous for the padi2 mutation or WT. Fin samples were amputated at the line of the blood circulatory loop, and 50–100 fins were pooled and flash frozen, with equivalent sample sizes used per replicate. RNA was extracted from fin tissue from 24 hpw and unwounded, 3 dpf, larvae, as described above. Primers for padi2 and rps11 (de Oliveira et al., 2013) are listed below. Data were normalized to rps11 and represented as fold change over WT, unwounded, age-matched control fins.
Primers were as follows: Padi2 exon5 RT-qPCR F: 5′-TAATGGCCATGGTGCAGTTC-3′, Padi2 exon6 RT-qPCR R: 5′-ATGGTCCATTAGTGCGCAAC-3′; Ef1a RT-qPCR F: 5′-TGCCTTCGTCCCAATTTCAG-3′, EF1a RT-qPCR R: 5′-TACCCTCCTTGCGCTCAATC-3′; and Rps11 RT-qPCR F: 5′-TAAGAAATGCCCCTTCACTG-3′, Rps11 RT-qPCR R: 5′-GTCTCTTCTCAAAACGGTTG-3′.
Generation of zebrafish padi2 clones and point mutations
Padi2 splice variants were amplified with Pfu Turbo DNA polymerase (Agilent) from cDNA using In-Fusion primers listed below. PCR products and a pCS2+8 vector (Gökirmak et al., 2012; Addgene) were digested with XbaI and BamHI (Promega) and ligated at RT using Takara ligation kit for long fragments. The 7-exon transcript (203) is predicted to lack the catalytic C terminus; therefore, we focused on cloning 16-exon transcripts, 201 and 202.
The primers were as follows: Padi2 cloning R, with XbaI: 5′-GGATCGTCTAGATTACAGCTCCAGGTTCCACC-3′, Padi2 cloning F transcript 201: 5′-CGATCCGGATCCATGGTGTCCCGTCGATCTCTTAC-3′, and Padi2 cloning F transcript 202: 5′-CGATCCGGATCCATGAATGTTTCGCAGGAGC-3′.
Both cDNA transcripts were cloned into pTRCHisA vector (Invitrogen) for N-terminal polyhistidine (his) tagging and expression in Escherichia coli (BL21(DE3)pLysS competent cells) using primers listed above. Constructs were inserted into the vector cut with BamHI and HindIII (Promega) using In-Fusion HD cloning kit (Clonetech). Point mutations were made with complementary primers (listed below) in pTRCHisA-padi2 vectors using QuikChange II Site-Directed Mutagenesis Kit (Agilent).
The primers were as follows: Catalytic C→A F: 5′-GTGAAGTTCACGCCGGGTCCAATGTTC-3′, Catalytic C→A R: 5′-GAACATTGGACCCGGCGTGAACTTCAC-3′; Ca1 binding Q→A F: 5′-ATCGCTGGATGGCGGATGAGCTTGAGTT-3′, Ca1 binding Q→A R: 5′-AACTCAAGCTCATCCGCCATCCAGCGAT-3′; Ca1 binding E→A F: 5′-GGATGAGCTTGCGTTTGGTTACATTG-3′, Ca1 binding E→A R: 5′-CAATGTAACCAAACGCAAGCTCATCC-3′; Ca1 binding E→A F: 5′-TTTCGGTAATCTGGCGGTCAGTCCACCA-3′, Ca1 binding E→A R: 5′-TGGTGGACTGACCGCCAGATTACCGAAA-3′; Ca2 binding D→A F: 5′-TGTTGTCCTGGCTTCTCCTCGTGAT-3′, and Ca2 binding D→A R: 5′-ATCACGAGGAGAAGCCAGGACAACA-3′.
Antibody production and Western blotting
The anti-zebrafish Padi2 antibody was generated in rabbits using combined full-length 201a and 202 variants fused to 6× poly-histidine in the pTRCHisA vector. Each immunogen was purified from BL21 E. coli lysates using a nickel-nitrilotriacetic acid superflow resin (Qiagen) and then combined and sent for anti-sera production (Covance). For Western blotting, 50–100 ∼2 dpf or 5 dpf larvae were pooled and deyolked in calcium-free Ringer’s solution via gentle disruption with a p200 pipette. Lysates from 2 hpf and 7 hpf larvae were not deyolked; samples were instead dechorionated on a Petri dish coated with 2% agarose and then rinsed with PBS. Larvae were washed twice with PBS and stored at −80°C until samples were lysed by sonication in 20 mM Tris, pH 7.6, 0.1% Triton X-100, 0.2 mM PMSF, 1 µg/ml Pepstatin, 2 µg/ml aprotinin, and 1 µg/ml Leupeptin at 3 µl per larva while on ice and clarified by centrifugation. Protein concentrations were determined using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Equal amounts of total protein were loaded on 6–20% gradient SDS-polyacrylamide gels and transferred to nitrocellulose. For citrullination analysis by Western blot of whole zebrafish lysates, methods for the citrullination colorimetric assay were followed, as described below, with the addition of dilution buffer in place of BAEE (Nα-benzoyl-L-arginine ethyl ester hydrochloride in 100 mM Tris, pH 7.4). The reaction was stopped after 90 min by boiling samples in SDS-PAGE sample buffer. zPadi2 rabbit anti-serum was used at 1:500 dilution, rabbit polyclonal anti-histone H4 (citrulline 3; 07–596, EMD-Millipore) at 1:50, mouse monoclonal anti-actin (ac15, A5441; Sigma-Aldrich) at 1:1,000, and rabbit polyclonal anti-histone H4 (07–108, EMD-Millipore) at 1:1,000. Western blots were imaged with an Odyssey Infrared Imaging System (LI-COR Biosciences).
padi2 mRNA reexpression
padi2-201a cloned into pCS2+8 (described above) was linearized using NotI restriction digest, and RNA was in vitro transcribed using the mMessage mMachine Sp6 kit (Ambion). RNA was cleaned up using an RNeasy Minikit column (Qiagen) and injected into single cell embryos (3 nl of 100 ng/μl). Embryo lysates were collected as described for Western blotting at 2 dpf and 5 dpf.
In vitro citrullination colorimetric assay
Zebrafish Padi2 constructs and point mutations were expressed in BL21 E. coli cells. Lysates were prepared on ice by sonication in 20 mM Tris, pH 7.6, 0.1% Triton X-100, 0.2 mM PMSF, 1 µg/ml Pepstatin, 2 µg/ml aprotinin, and 1 µg/ml Leupeptin and clarified by centrifugation. Bacterial lysates were aliquoted and frozen at −80°C. Lysates from zebrafish larvae were prepared as described above for Western blotting and used at equivalent amounts. The assay was performed as previously described (Nakayama-Hamada et al., 2005). In short, 12.5 µl lysate was incubated with 12.5 µl 4× reaction buffer (400 mM Tris, pH 7.4, ± 80 mM CaCl2, and 20 mM DTT), 12.5 µl 80 mM BAEE, and 12.5 µl dilution buffer (10 mM Tris, pH 7.6, 150 mM NaCl, and 2 mM DTT) for 1 h at 37°C. The reaction was stopped by the addition of 33 µM EDTA final concentration. Reactions were diluted 1:10 for an 8 mM BAEE final concentration, and 50 µl aliquots were done in triplicate in a 96-well plate. 150 µl colorimetric buffer, composed of 1 ml buffer A (80 mM diacetyl monoxime, 2 mM thiosemicarbazide) and 3 ml buffer B (3 M phosphoric acid, 6 M sulfuric acid, 2 mM ammonium iron [III] sulfate), were added to each well and incubated at 95°C for 15 min. Absorption was read at 540 nM. Relative light units were normalized to Western blot densitometry using the Odyssey Infrared Imaging System (LI-COR Biosciences).
MO injections
MO oligonucleotides (Genetools) were designed to the intron1/exon2 border of padi2. MOs were resuspended in water to a final concentration of 1 mM. MOs were diluted to a final concentration of 100 µM, and 3 nl injection mix was injected into one-cell stage embryos, which were subsequently maintained at 28.5°C. MO sequences used were as follows: padi2 MO: 5′-GAGCACATCTGGAATGGGAATATAT-3′; control MO: 5′-CCTCTTACCTCAGTTACAATTTATA-3′.
Regeneration assays
For larval regeneration assays, incrosses of F3 or F4 adult WT and padi2−/− siblings were done to produce offspring cousins homozygous for the padi2 mutation or WT. Dechorionated larvae were transferred to 35-mm milk-coated plates. Larvae were washed twice in E3 and wounded in a final 0.24 mg/ml tricaine (ethyl 3-aminobenzoate, Sigma-Aldrich)/E3 solution. Tail transections were performed on ∼2.5 dpf larvae with a surgical blade (feather no. 10) roughly four vacuolated cells from the posterior end of the notochord. Larvae were again washed three times with E3 and allowed to regenerate for 3 d post-wounding (dpw), at which point larvae were fixed with 4% PFA (Sigma-Aldrich) in PBS at 4°C overnight. Fins were imaged in PBS at RT on a Zeiss Zoomscope (EMS3/SyCoP3; Zeiss; 1× Plan-NeoFluor Z objective) with an Axiocam Mrm charge-coupled device camera using ZEN pro 2012 software (Zeiss). Regenerate length was measured from the edge of the blood vessel to the caudal edge of the tail fin using the FIJI image analysis software (Fig. 2 A; Schindelin et al., 2012). Unwounded, 5 dpf larvae fin lengths were measured as a developmental control. Fin transections were performed on MO-injected larvae similarly to as described above with amputation adjacent to the notochord, without causing damage to the notochord. Regenerated fins and developmental controls were measured from the caudal tip of the notochord to the caudal edge of the tail fin.
Immunofluorescence, microscopy, and analysis
Images are always shown with anterior to the left. All images were acquired at RT.
Neuromuscular labels
Immunostaining was performed on cousin offspring from incrossed adult F2 WT siblings and incrossed padi2−/− sibling zebrafish. 5 dpf larvae were fixed in 4% PFA, 0.125 M sucrose, and 1× PBS overnight at 4°C. For detection of slow muscles, larvae were washed three times with 0.1% PBS-Tween20 and incubated in 0.1% wt/vol collagenase type 1A (Sigma-Aldrich) in PBS at 37°C for 1.5 h, followed by three washes in PBSTD (0.3% Triton X-100 and 1% DMSO in PBS). Larvae were blocked for 2 h at RT in PBSTD with 2% BSA and 4% goat serum. Monoclonal mouse anti-myosin heavy chain antibody (F59; DSHB; Miller et al., 1985) was used at 1:20 in block buffer and incubated overnight in 4°C. Larvae were washed five times in PBSTD, and secondary DyLight 488 donkey anti-mouse IgG antibody (610–741-124, Rockland Immunochemicals) was used at 1:250 in block buffer overnight at 4°C. Five final washes were done in PBSTD. Zebrafish were stabilized in a zWEDGI (Huemer et al., 2017) in PBS during imaging acquisition. Images were acquired on a spinning disk confocal (CSU-X; Yokogawa) on a Zeiss Observer Z.1 inverted microscope and an electron multiplying charge-coupled device (EMCCD) evolve 512 camera (Photometrics) with a Plan-Apochromat NA 0.8/20× air objective and collected as z-stack of 1 µm optical sections at 512 × 512 resolution. Images were acquired using Zen 2 imagine software (Zeiss) and were z-projected using Zen 2.3 lite software (Zeiss).
For visualization of fast muscle, fixed fish were washed with PBS three times followed by three washes in PBS with 0.1% Tween20. Larvae were permeabilized with PBS 2% PBSTx (20% Triton X-100 in 1× PBS) for 1.5 h with gentle rocking. Fish were then incubated with rhodamin-phalloidin (R415, Invitrogen) diluted 1:100 in 2% PBSTx at 4°C overnight. Fish were rinsed in fresh 2% PBSTx followed by several washes in 0.2% PBSTx. Imaging was performed on the spinning disk microscope (described above) with a Plan-Apochromat NA 0.8/20× air objective (centered on cloaca) with 1 µm optical sections. Larvae in PBS were stabilized during imaging by use of a zWEDGI (Huemer et al., 2017).
For neuromuscular junction visualization, fix was washed off with three PBS washes. The skin was peeled with fine forceps (Dumont no. 55 dumostar, Fine Science Tools) starting above the swim bladder and removed down to the fin. Skinned larvae were incubated in 0.1% wt/vol collagenase type 1A at RT for 15 min with gentle rocking followed by three washes in PBS. For detection of AChRs, larvae were incubated for 30 min at RT 10 µg/ml Alexa Fluor 594–conjugated a-bungarotoxin (B13423, Thermo Fisher Scientific) diluted in incubation buffer (IB; 0.1% sodium azide, 2% BSA, and 0.5% Triton X-100 in PBS, pH 7.4). Embryos were rinsed three times in IB. Monoclonal mouse anti-synaptic vesicle glycoprotein 2A antibody (SV2, DSHB; Buckley and Kelly, 1985) was used at 1:50 in IB overnight at 4°C. Larvae were washed five times in IB and incubated with secondary DyLight 488 donkey anti-mouse IgG antibody (610–741-124, Rockland Immunochemicals) at 1:250 in IB for 4 h at RT or 4°C overnight. Final washes were done in IB before imaging on a spinning disk microscope (described above) with an EC Plan-NeoFluaR NA 0.75/40× air objective (Zeiss; centered around the cloaca with 2 × 1 tile images and 1 µm optical section z-stacks) using a zWEDGI (Huemer et al., 2017). To quantify colocalization of signal, maximum intensity projections were analyzed in FIJI using the plugin ComDet v3.7 for spot localization (https://github.com/ekatrukha/ComDet/wiki). Particles were threshold as approximate size being 5 pixels, intensity threshold for SV2 between 4 and 5, α-BTX between 2 and 3, and a 6-pixel maximum distance between particles.
Histone citrullination
Immunostaining was performed on offspring cousins from incrossed adult F3 WT siblings and incrossed padi2−/− siblings. To identify histone citrullination, larvae were fixed in a solution of 1% NP-40, 0.5% Triton-X, and 1.5% PFA in PBS at 4°C overnight. The following day, fix was replaced with a block solution of 2.5% BSA, 0.5% Tween-20, and 5% goat serum in PBS. Samples were blocked for at least 2.5 h at RT followed by the addition of polyclonal rabbit anti-histone H4 (citrulline 3) antibody (07–596, EMD Millipore) used at 1:100 and incubated overnight at 4°C. For time course experiments, samples were kept in block at 4°C until the final sample was prepared, at which time all samples were blocked at RT before the addition of the primary antibody. Samples were washed three times in PBS at RT for 5 min each, and secondary DyLight 488 donkey anti-rabbit (611–741-127, Rockland Immunochemicals) or Alexa Fluor 568 goat anti-rabbit IgG antibodies (A-11011, Invitrogen) were used at 1:250 in block buffer overnight at 4°C. When indicated, rhodamine-phalloidin (R415, Invitrogen) and 10 mg/ml DAPI (D9542, Sigma-Aldrich) were added with secondary antibodies at 1:100 and 1:10,000 dilutions, respectively. Finally, four washes were done in PBS. Caudal fins were isolated by removing larvae trunks with a scalpel blade and imaged in PBS in a glass-bottom dish. Images were acquired on a laser-scanning confocal microscope (FluoView FV1000; Olympus) with a NA 0.75/20× or PLANAPO NA 1.45/60× oil objective and FV10-ASW software (Olympus). 20× images used for quantification were acquired as z-stacks with 25, 1-µm optical slices at 640 × 640 resolution. Image analysis is discussed in a later section. Alternatively, images were acquired using multiphoton microscopy. For this, caudal fins of fixed, phenylthiourea (PTU)-treated, labeled larvae were removed from the trunk with a scalpel blade (Feather no. 15), then imaged in a 50-mm coverglass (no. 1.5) bottom dish (MatTek) in PBS, as previously described (LeBert et al., 2015, 2016). A second coverslip over the glass bottom depression minimized sample movement. The fins were imaged on a custom-built multiphoton microscope (Conklin et al., 2011; LeBert et al., 2016) at the Laboratory for Optical and Computational Instrumentation using a Nikon Eclipse TE2000U microscope with a Nikon 40× long working distance water immersion lens (1.2 NA, Nikon). All signals were detected sequentially using a H7422P-40 GaAsP Photomultiplier Tube (Hamamatsu). The backward SHG signal was collected with the multiphoton source laser (Chameleon UltraII, Coherent Inc.) tuned to 890 nm, with a 445/20-nm bandpass emission filter (Semrock). The fluorescent signal from H4cit3 antibody was collected using a either a 520/35-nm bandpass emission filter (Semrock) for the DyLight 488 donkey anti-rabbit secondary antibody (Rockland Immunochemicals) or a 615/20-nm bandpass emission filter (Semrock) for the Alexa Fluor 568 goat anti-rabbit secondary antibody (Invitrogen). The 615/20-nm emission filter was used to collect the fluorescent signal from the rhodamine-phalloidin, while the 520/35-nm emission filter was used to detect the krt4:EGFP and lepb:EGFP fluorescence. DAPI fluorescence was excited with the laser tuned to 740 nm and the emission collected using the 445/20-nm filter. Brightfield images were simultaneously collected using a separate photodiode-based transmission detector (Bio-Rad). Data were collected as z-stacks with optical sections 2 microns apart, at 512 × 512 resolution using WiscScan software (LOCI, University of Wisconsin-Madison).
Mitotic index
For evaluation of cells undergoing mitosis, 24 hpw and 3 dpf MO-injected larvae were fixed with 1.5% PFA in 0.1 M Pipes, 1.0 mM MgSO4, and 2 mM EGTA overnight at 4°C and immunolabeled with monoclonal mouse phosphorylated histone H3 (serine10) antibody (05–598, Millipore). To remove fixation solution, larvae were washed with PBS three times and placed in methanol at −20°C overnight. Samples were rehydrated in subsequent 5-min washes at ratios of 2:1, 1:1, and 1:2 methanol:PBSTx, and a final PBSTx wash. Larvae were incubated in 0.15 M glycine in PBS for 10 min at RT followed by three PBSTx washes. Fish were blocked in 1% BSA in PBSTx for 1 h at RT. Phosphorylated histone H3 (serine10) antibody diluted 1:300 in block was incubated overnight a 4°C. Samples were washed for 15–30 min in block, twice in PBSTx, and once more in block. Incubation with DyLight donkey anti-rabbit 488 secondary was used, followed by four washes in PBSTx. Samples in PBS on a glass-bottom dish were imaged and quantified on the laser-scanning confocal microscope with a NA 0.75/20×, as described above.
Leukocyte imaging
padi2+/− adults were crossed to AB WT zebrafish labeled with macrophage nuclei (Tg(mpeg1: H2B-GFP)) or neutrophil nuclei (Tg(lyzc:H2B-mCherry)) and subsequently incrossed to produce homozygous, fluorescently labeled adults. Experiments were performed on WT cousins and padi2−/− larvae resulting from incrossed adult transgenic siblings. Wounding was performed as described above, and larvae were fixed with 1.5% PFA in 0.1 M Pipes (Sigma-Aldrich), 1 mM MgSO4 (Sigma-Aldrich), and 2 mM EGTA (Sigma-Aldrich) overnight at 4°C. Caudal fins were imaged on a Zeiss Zoomscope, as described for regeneration assays. Leukocyte numbers were counted by hand in the region past the blood circulatory loop (Fig. 2 D) using Zen 2.3 lite software (Zeiss). Whole larvae were imaged in a zWEDGI (Huemer et al., 2017) and acquired on a spinning disk confocal (CSU-X; Yokogawa) on a Zeiss Observer Z.1 inverted microscope and an EMCCD evolve 512 camera (Photometrics) with a Plan-Apochromat NA 0.8/20× air objective (5 µm optical sections, 5 × 1 tiles, 2355 × 512 resolution).
EdU and apoptosis labeling
Immunostaining was performed on offspring cousins from incrossed adult F3 WT siblings and incrossed padi2−/− siblings. Proliferation in the fin was measured using Click-iT Plus EdU Imaging Kit (Life Technologies). Larvae were incubated in 10 µM EdU solution in E3 for 6 h with slight agitation. Wounded fish were incubated from 60 to 66 hpw along with age-matched unwounded controls. Larvae were fixed in 4% PFA in PBS overnight at 4°C and stored in methanol at −20°C until staining. Staining protocol was conducted according to the manufacturer’s instructions. EdU-stained larvae were also incubated with monoclonal rabbit anti-active Caspase3 antibody (559565, BD Biosciences) at 1:200 in block (PBS, 1% DMSO, 1% BSA, 0.05% Triton X-100, and 1.5% goat serum) followed by incubation with DyLight 550 donkey anti-rabbit secondary antibody (SA5-10039, Invitrogen) and 0.01 mg/ml DAPI (Sigma-Aldrich). Larvae were imaged in a zWEDGI bathed in PBS. Immunofluorescence images were acquired on a spinning disk confocal (CSU-X; Yokogawa) on a Zeiss Observer Z.1 inverted microscope with an EMCCD evolve 512 camera (Photometrics) and a Plan-Apochromat NA 0.8/20× air objective, as z-stacks, 3-µm optical sections, and with 512 × 512 resolution.
Image analysis/processing
Image analysis was performed on FIJI. For experiments where fluorescence intensity was quantified, no adjustments were made to the images before analysis. For histone H4cit3 analysis, a region of interest (ROI) 92 × 93 microns was centered around the notochord, as determined by the corresponding bright-field image. Immunostained images were z-projected as a maximum intensity projection, and the integrated density in the ROI was determined. Images were thresholded using the threshold plugin using auto-thresholding with the “Intermodes” method in FIJI (Prewitt and Mendelsohn, 1966), and the total area within the ROI was determined for particles larger than 8 pixels. For presentation purposes, images were processed to remove background using despeckling. Notochord bead area was determined in FIJI by outlining this structure as determined by examination of the optical bright-field slices.
Total neutrophil numbers were determined using Imaris (Bitplane) with the spots function as defined by a 10-µm diameter in the x, y plane and a z diameter of 20 µm. Total macrophage numbers were counted by hand using z-projected images in Zen 2.3 lite software. For total leukocyte quantifications, leukocytes within the yolk sac and heart were excluded.
For spatial assessment of nuclei with citrullinated histones, 3D reconstructions and slices were constructed using Imaris (Bitplane, Oxford Instruments). Videos of z-stack scans and 3D rotations were made in Imaris, annotated in FIJI using “Annotation_to_overlay1.3” plugin (Centre for Core Biotechnology Services, University of Leicester) and converted to MP4 using HandBrake (v1.2.2) software (The HandBrake Team).
For EdU analysis, images were 3D reconstructed using Imaris software (Bitplane). The number of EdU-positive cells were quantified in the fin region posterior of the blood circulatory loop with the spots function as defined by an x, y diameter of 7 µm and a z diameter of 14 µm. The level of apoptosis activation at the wound was determined by outlining the fin past the blood circulation using the corresponding bright-field image. In FIJI, total threshold area for active-Caspase3 signal in the wound was determined using the threshold plugin in FIJI by auto-thresholding with the “Yen Dark” method (Yen et al., 1995) for particles larger than 3 pixels.
Statistical analysis
For all statistical analyses, at least three independent replicates were conducted. For data in Fig. 1 G, analysis was done using one-way ordinary ANOVA with a Holm–Sidak multiple comparisons test. To examine mutant survival, Mendelian ratio was confirmed for both larvae and adult offspring from a heterozygous incross by χ2 tests. For all other quantitative experiments, data were pooled from the independent replicates, and results were summarized in terms of least-squared adjusted means (lsmeans) and standard errors (Vincent et al., 2016). Results were analyzed using ANOVA with a Tukey multiple comparisons test. Graphical representation shows individual data points color coded to reflect replicates. Statistical analysis and graphical representations were done using R version 3.4 and GraphPad Prism version 6.
Online supplemental material
Supplemental material includes additional data characterizing the zebrafish Padi2 transcripts and proteins (Fig. S1), additional characterization of the padi2 mutant (Fig. S2), and additional developmental phenotypes observed in the mutant and supporting MO data (Fig. S3). Videos 1, 2, 3, 4, 5, and 6 characterize the 3D context of the citrullinated histones after injury.
Supplementary Material
Acknowledgments
We thank members of the Huttenlocher laboratory for helpful discussions of the research, technical support, and zebrafish maintenance. We thank Emily Rosowski, Laurel Hind, and Francisco Barros-Becker for useful discussions, advice, and careful reading of the manuscript. We thank Jens Eickhoff for advice on statistical analysis.
This work was supported by National Institutes of Health grants R35 GM1 18027 (A. Huttenlocher), T32-GM07215 (N. Golenberg), and AHA16SDG30020001 (J. Kang), and National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases grant AR065500 (M.A. Shelef).
The authors declare no competing financial interests.
Author contributions: N. Golenberg, J.M. Squirrell, M.A. Shelef, K.W. Eliceiri, J. Kang, and A. Huttenlocher conceived and designed experiments. N. Golenberg, D.A. Bennin, J.M. Squirrell, J. Rindy, and P.E. Pistono conducted the experiments. N. Golenberg and D.A. Bennin performed the analysis. N. Golenberg, J.M. Squirrell, and A. Huttenlocher prepared the figures and wrote the manuscript.
References
- Arita K., Hashimoto H., Shimizu T., Nakashima K., Yamada M., and Sato M.. 2004. Structural basis for Ca(2+)-induced activation of human PAD4. Nat. Struct. Mol. Biol. 11:777–783. 10.1038/nsmb799 [DOI] [PubMed] [Google Scholar]
- Bayés À., Collins M.O., Reig-Viader R., Gou G., Goulding D., Izquierdo A., Choudhary J.S., Emes R.D., and Grant S.G.. 2017. Evolution of complexity in the zebrafish synapse proteome. Nat. Commun. 8:14613 10.1038/ncomms14613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley K., and Kelly R.B.. 1985. Identification of a transmembrane glycoprotein specific for secretory vesicles of neural and endocrine cells. J. Cell Biol. 100:1284–1294. 10.1083/jcb.100.4.1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou M.A., Castelo-Branco G., Halley-Stott R.P., Oliveira C.S., Loos R., Radzisheuskaya A., Mowen K.A., Bertone P., Silva J.C., Zernicka-Goetz M., et al. . 2014. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 507:104–108. 10.1038/nature12942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin M.W., Eickhoff J.C., Riching K.M., Pehlke C.A., Eliceiri K.W., Provenzano P.P., Friedl A., and Keely P.J.. 2011. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 178:1221–1232. 10.1016/j.ajpath.2010.11.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira S., Reyes-Aldasoro C.C., Candel S., Renshaw S.A., Mulero V., and Calado A.. 2013. Cxcl8 (IL-8) mediates neutrophil recruitment and behavior in the zebrafish inflammatory response. J. Immunol. 190:4349–4359. 10.4049/jimmunol.1203266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G., Vitale A.M., Leijten F.P., Strik A.M., Koonen-Reemst A.M., Yurttas P., Robben T.J., Coonrod S., and Gossen J.A.. 2007. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol. Cell. Endocrinol. 273:25–31. 10.1016/j.mce.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Falcão A.M., Meijer M., Scaglione A., Rinwa P., Agirre E., Liang J., Larsen S.C., Heskol A., Frawley R., Klingener M., et al. . 2019. PAD2-Mediated Citrullination Contributes to Efficient Oligodendrocyte Differentiation and Myelination. Cell Reports. 27:1090–1102.e10. 10.1016/j.celrep.2019.03.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauron C., Rampon C., Bouzaffour M., Ipendey E., Teillon J., Volovitch M., and Vriz S.. 2013. Sustained production of ROS triggers compensatory proliferation and is required for regeneration to proceed. Sci. Rep. 3:2084 10.1038/srep02084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globus M., Vethamany-Globus S., and Kesik A.. 1987. Control of blastema cell proliferation by possible interplay of calcium and cyclic nucleotides during newt limb regeneration. Differentiation. 35:94–99. 10.1111/j.1432-0436.1987.tb00155.x [DOI] [PubMed] [Google Scholar]
- Gökirmak T., Campanale J.P., Shipp L.E., Moy G.W., Tao H., and Hamdoun A.. 2012. Localization and substrate selectivity of sea urchin multidrug (MDR) efflux transporters. J. Biol. Chem. 287:43876–43883. 10.1074/jbc.M112.424879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huemer K., Squirrell J.M., Swader R., LeBert D.C., Huttenlocher A., and Eliceiri K.W.. 2017. zWEDGI: wounding and entrapment device for imaging live zebrafish larvae. Zebrafish. 14:42–50. 10.1089/zeb.2016.1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan R., Jin M., Subramanian V., Causey C.P., Thompson P.R., and Coonrod S.A.. 2012. Potential role for PADI-mediated histone citrullination in preimplantation development. BMC Dev. Biol. 12:19 10.1186/1471-213X-12-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Hu J., Karra R., Dickson A.L., Tornini V.A., Nachtrab G., Gemberling M., Goldman J.A., Black B.L., and Poss K.D.. 2016. Modulation of tissue repair by regeneration enhancer elements. Nature. 532:201–206. 10.1038/nature17644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubilus J., and Baden H.P.. 1983. Purification and properties of a brain enzyme which deiminates proteins. Biochim. Biophys. Acta. 745:285–291. 10.1016/0167-4838(83)90060-2 [DOI] [PubMed] [Google Scholar]
- LaFave M.C., Varshney G.K., Vemulapalli M., Mullikin J.C., and Burgess S.M.. 2014. A defined zebrafish line for high-throughput genetics and genomics: NHGRI-1. Genetics. 198:167–170. 10.1534/genetics.114.166769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagoudakis L., Garcin I., Julien B., Nahum K., Gomes D.A., Combettes L., Nathanson M.H., and Tordjmann T.. 2010. Cytosolic calcium regulates liver regeneration in the rat. Hepatology. 52:602–611. 10.1002/hep.23673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBert D.C., Squirrell J.M., Rindy J., Broadbridge E., Lui Y., Zakrzewska A., Eliceiri K.W., Meijer A.H., and Huttenlocher A.. 2015. Matrix metalloproteinase 9 modulates collagen matrices and wound repair. Development. 142:2136–2146. 10.1242/dev.121160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBert D.C., Squirrell J.M., Huttenlocher A., and Eliceiri K.W.. 2016. Second harmonic generation microscopy in zebrafish. Methods Cell Biol. 133:55–68. 10.1016/bs.mcb.2016.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBert D., Squirrell J.M., Freisinger C., Rindy J., Golenberg N., Frecentese G., Gibson A., Eliceiri K.W., and Huttenlocher A.. 2018. Damage-induced reactive oxygen species regulate vimentin and dynamic collagen-based projections to mediate wound repair. eLife. 7:e30703 10.7554/eLife.30703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Li M., Lindberg M.R., Kennett M.J., Xiong N., and Wang Y.. 2010. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 207:1853–1862. 10.1084/jem.20100239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Loos T., Opdenakker G., Van Damme J., and Proost P.. 2009. Citrullination of CXCL8 increases this chemokine’s ability to mobilize neutrophils into the blood circulation. Haematologica. 94:1346–1353. 10.3324/haematol.2009.006973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCafferty J., Reid R., Spencer M., Hamp T., and Fodor A.. 2012. Peak Studio: a tool for the visualization and analysis of fragment analysis files. Environ. Microbiol. Rep. 4:556–561. 10.1111/j.1758-2229.2012.00368.x [DOI] [PubMed] [Google Scholar]
- Miller J.B., Crow M.T., and Stockdale F.E.. 1985. Slow and fast myosin heavy chain content defines three types of myotubes in early muscle cell cultures. J. Cell Biol. 101:1643–1650. 10.1083/jcb.101.5.1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskolci V., Squirrell J., Rindy J., Vincent W., Sauer J.D., Gibson A., Eliceiri K.W., and Huttenlocher A.. 2019. Distinct inflammatory and wound healing responses to complex caudal fin injuries of larval zebrafish. eLife. 8:e45976 10.7554/eLife.45976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., and Valen E.. 2014. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 42(Web Server issue, W1):W401-7 10.1093/nar/gku410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama-Hamada M., Suzuki A., Kubota K., Takazawa T., Ohsaka M., Kawaida R., Ono M., Kasuya A., Furukawa H., Yamada R., and Yamamoto K.. 2005. Comparison of enzymatic properties between hPADI2 and hPADI4. Biochem. Biophys. Res. Commun. 327:192–200. 10.1016/j.bbrc.2004.11.152 [DOI] [PubMed] [Google Scholar]
- Nechiporuk A., and Keating M.T.. 2002. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 129:2607–2617. [DOI] [PubMed] [Google Scholar]
- Niethammer P. 2016. The early wound signals. Curr. Opin. Genet. Dev. 40:17–22. 10.1016/j.gde.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell D., Tauzin S., Hind L.E., Deng Q., Beebe D.J., and Huttenlocher A.. 2017. Chemokine Signaling and the Regulation of Bidirectional Leukocyte Migration in Interstitial Tissues. Cell Reports. 19:1572–1585. 10.1016/j.celrep.2017.04.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prewitt J.M., and Mendelsohn M.L.. 1966. The analysis of cell images. Ann. N. Y. Acad. Sci. 128:1035–1053. 10.1111/j.1749-6632.1965.tb11715.x [DOI] [PubMed] [Google Scholar]
- Proost P., Loos T., Mortier A., Schutyser E., Gouwy M., Noppen S., Dillen C., Ronsse I., Conings R., Struyf S., et al. . 2008. Citrullination of CXCL8 by peptidylarginine deiminase alters receptor usage, prevents proteolysis, and dampens tissue inflammation. J. Exp. Med. 205:2085–2097. 10.1084/jem.20080305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehl H.H. 2018. Linking wound response and inflammation to regeneration in the zebrafish larval fin. Int. J. Dev. Biol. 62:473–477. 10.1387/ijdb.170331hr [DOI] [PubMed] [Google Scholar]
- Rojas-Muñoz A., Rajadhyksha S., Gilmour D., van Bebber F., Antos C., Rodríguez Esteban C., Nüsslein-Volhard C., and Izpisúa Belmonte J.C.. 2009. ErbB2 and ErbB3 regulate amputation-induced proliferation and migration during vertebrate regeneration. Dev. Biol. 327:177–190. 10.1016/j.ydbio.2008.12.012 [DOI] [PubMed] [Google Scholar]
- Romero M.M.G., McCathie G., Jankun P., and Roehl H.H.. 2018. Damage-induced reactive oxygen species enable zebrafish tail regeneration by repositioning of Hedgehog expressing cells. Nat. Commun. 9:4010 10.1038/s41467-018-06460-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelef M.A., Bennin D.A., Mosher D.F., and Huttenlocher A.. 2012. Citrullination of fibronectin modulates synovial fibroblast behavior. Arthritis Res. Ther. 14:R240 10.1186/ar4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipilä K., Haag S., Denessiouk K., Käpylä J., Peters E.C., Denesyuk A., Hansen U., Konttinen Y., Johnson M.S., Holmdahl R., and Heino J.. 2014. Citrullination of collagen II affects integrin-mediated cell adhesion in a receptor-specific manner. FASEB J. 28:3758–3768. 10.1096/fj.13-247767 [DOI] [PubMed] [Google Scholar]
- Smith T.F., and Waterman M.S.. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195–197. 10.1016/0022-2836(81)90087-5 [DOI] [PubMed] [Google Scholar]
- Tseng A.S., Adams D.S., Qiu D., Koustubhan P., and Levin M.. 2007. Apoptosis is required during early stages of tail regeneration in Xenopus laevis. Dev. Biol. 301:62–69. 10.1016/j.ydbio.2006.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent W.J., Freisinger C.M., Lam P.Y., Huttenlocher A., and Sauer J.D.. 2016. Macrophages mediate flagellin induced inflammasome activation and host defense in zebrafish. Cell. Microbiol. 18:591–604. 10.1111/cmi.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossenaar E.R., Zendman A.J., van Venrooij W.J., and Pruijn G.J.. 2003. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. BioEssays. 25:1106–1118. 10.1002/bies.10357 [DOI] [PubMed] [Google Scholar]
- Watanabe K., and Senshu T.. 1989. Isolation and characterization of cDNA clones encoding rat skeletal muscle peptidylarginine deiminase. J. Biol. Chem. 264:15255–15260. [PubMed] [Google Scholar]
- Whitehead G.G., Makino S., Lien C.L., and Keating M.T.. 2005. fgf20 is essential for initiating zebrafish fin regeneration. Science. 310:1957–1960. 10.1126/science.1117637 [DOI] [PubMed] [Google Scholar]
- Wiese M., Bannister A.J., Basu S., Boucher W., Wohlfahrt K., Christophorou M.A., Nielsen M.L., Klenerman D., Laue E.D., and Kouzarides T.. 2019. Citrullination of HP1γ chromodomain affects association with chromatin. Epigenetics Chromatin. 12:21 10.1186/s13072-019-0265-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Lu J., Sridhar B., Cao X., Yu P., Zhao T., Chen C.C., McDee D., Sloofman L., Wang Y., et al. . 2017. SMARCAD1 Contributes to the Regulation of Naive Pluripotency by Interacting with Histone Citrullination. Cell Reports. 18:3117–3128. 10.1016/j.celrep.2017.02.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Steentoft C., Hauge C., Hansen L., Thomsen A.L., Niola F., Vester-Christensen M.B., Frödin M., Clausen H., Wandall H.H., and Bennett E.P.. 2015. Fast and sensitive detection of indels induced by precise gene targeting. Nucleic Acids Res. 43:e59 10.1093/nar/gkv126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen J.C., Chang F.J., and Chang S.. 1995. A new criterion for automatic multilevel thresholding. IEEE Trans. Image Process. 4:370–378. 10.1109/83.366472 [DOI] [PubMed] [Google Scholar]
- Yokoyama H. 2008. Initiation of limb regeneration: the critical steps for regenerative capacity. Dev. Growth Differ. 50:13–22. 10.1111/j.1440-169X.2007.00973.x [DOI] [PubMed] [Google Scholar]
- Yoo S.K., Freisinger C.M., LeBert D.C., and Huttenlocher A.. 2012a Early redox, Src family kinase, and calcium signaling integrate wound responses and tissue regeneration in zebrafish. J. Cell Biol. 199:225–234. 10.1083/jcb.201203154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.K., Lam P.Y., Eichelberg M.R., Zasadil L., Bement W.M., and Huttenlocher A.. 2012b The role of microtubules in neutrophil polarity and migration in live zebrafish. J. Cell Sci. 125:5702–5710. 10.1242/jcs.108324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Korchynskyi O., Tak P.P., Isozaki T., Ruth J.H., Campbell P.L., Baeten D.L., Gerlag D.M., Amin M.A., and Koch A.E.. 2014. Citrullination of epithelial neutrophil-activating peptide 78/CXCL5 results in conversion from a non-monocyte-recruiting chemokine to a monocyte-recruiting chemokine. Arthritis Rheumatol. 66:2716–2727. 10.1002/art.38750 [DOI] [PubMed] [Google Scholar]
- Yuzhalin A.E., Gordon-Weeks A.N., Tognoli M.L., Jones K., Markelc B., Konietzny R., Fischer R., Muth A., O’Neill E., Thompson P.R., et al. . 2018. Colorectal cancer liver metastatic growth depends on PAD4-driven citrullination of the extracellular matrix. Nat. Commun. 9:4783 10.1038/s41467-018-07306-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Liu X., Zhang M., Li T., Muth A., Thompson P.R., Coonrod S.A., and Zhang X.. 2016. Peptidylarginine deiminase 1-catalyzed histone citrullination is essential for early embryo development. Sci. Rep. 6:38727 10.1038/srep38727 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.