Abstract
Background:
A patient's infectivity is determined by the presence of the virus in different body fluids, secretions, and excreta. The persistence and clearance of viral RNA from different specimens of patients with 2019 novel coronavirus disease (COVID-19) remain unclear. This study analyzed the clearance time and factors influencing 2019 novel coronavirus (2019-nCoV) RNA in different samples from patients with COVID-19, providing further evidence to improve the management of patients during convalescence.
Methods:
The clinical data and laboratory test results of convalescent patients with COVID-19 who were admitted to from January 20, 2020 to February 10, 2020 were collected retrospectively. The reverse transcription polymerase chain reaction (RT-PCR) results for patients’ oropharyngeal swab, stool, urine, and serum samples were collected and analyzed. Convalescent patients refer to recovered non-febrile patients without respiratory symptoms who had two successive (minimum 24 h sampling interval) negative RT-PCR results for viral RNA from oropharyngeal swabs. The effects of cluster of differentiation 4 (CD4)+ T lymphocytes, inflammatory indicators, and glucocorticoid treatment on viral nucleic acid clearance were analyzed.
Results:
In the 292 confirmed cases, 66 patients recovered after treatment and were included in our study. In total, 28 (42.4%) women and 38 men (57.6%) with a median age of 44.0 (34.0–62.0) years were analyzed. After in-hospital treatment, patients’ inflammatory indicators decreased with improved clinical condition. The median time from the onset of symptoms to first negative RT-PCR results for oropharyngeal swabs in convalescent patients was 9.5 (6.0–11.0) days. By February 10, 2020, 11 convalescent patients (16.7%) still tested positive for viral RNA from stool specimens and the other 55 patients’ stool specimens were negative for 2019-nCoV following a median duration of 11.0 (9.0–16.0) days after symptom onset. Among these 55 patients, 43 had a longer duration until stool specimens were negative for viral RNA than for throat swabs, with a median delay of 2.0 (1.0–4.0) days. Results for only four (6.9%) urine samples were positive for viral nucleic acid out of 58 cases; viral RNA was still present in three patients’ urine specimens after throat swabs were negative. Using a multiple linear regression model (F = 2.669, P = 0.044, and adjusted R2 = 0.122), the analysis showed that the CD4+ T lymphocyte count may help predict the duration of viral RNA detection in patients’ stools (t = −2.699, P = 0.010). The duration of viral RNA detection from oropharyngeal swabs and fecal samples in the glucocorticoid treatment group was longer than that in the non-glucocorticoid treatment group (15 days vs. 8.0 days, respectively; t = 2.550, P = 0.013) and the duration of viral RNA detection in fecal samples in the glucocorticoid treatment group was longer than that in the non-glucocorticoid treatment group (20 days vs. 11 days, respectively; t = 4.631, P < 0.001). There was no statistically significant difference in inflammatory indicators between patients with positive fecal viral RNA test results and those with negative results (P > 0.05).
Conclusions:
In brief, as the clearance of viral RNA in patients’ stools was delayed compared to that in oropharyngeal swabs, it is important to identify viral RNA in feces during convalescence. Because of the delayed clearance of viral RNA in the glucocorticoid treatment group, glucocorticoids are not recommended in the treatment of COVID-19, especially for mild disease. The duration of RNA detection may relate to host cell immunity.
Keywords: COVID-19, 2019-nCoV, Nucleic acid detection, Glucocorticoid
Introduction
In January 2020, a new coronavirus was confirmed as the cause of unexplained pneumonia in a group of patients from Wuhan, Hubei, and was subsequently named the 2019 novel coronavirus (2019-nCoV).[1] Due to increasing numbers of cases reported out of China, the World Health Organization announced on January 30 that the emerging new coronavirus pneumonia epidemic constituted a public health emergency of international concern.[2] Up until February 11, 2020, there were 44,653 confirmed 2019 novel coronavirus disease (COVID-19) cases reported in China and 395 cases in 24 other countries.[3,4] The transmission capacity of 2019-nCoV was underestimated at first. Initial studies showed its regeneration number, R0, was 2.2 to 2.9,[5–7] meaning that each infector could transmit to another 2.2 to 2.9 people. Recently a novel study revealed that the R0 of 2019-nCoV is 3.77 based on clinical and epidemiological data from nearly 8866 patients in 30 provinces,[8] which is higher than the R0 of severe acute respiratory syndrome coronavirus (R0, 2–3).[9]
The patient's infectivity is determined by the presence of the virus in different body fluids, secretions, and excreta. All patients with positive viral RNA detection need to be isolated. As mentioned in the “Diagnosis and Treatment Scheme of New Coronavirus Infected Pneumonia” (trial version 5), only after the relief of symptoms and two successive (minimum 24 h sampling interval) negative viral nucleic acid results for respiratory specimens, the isolated cases can be disisolation. However, the persistence and clearance of viral RNA in different specimens of COVID-19 patients remains unclear. In this study, viral RNA detection was performed on throat swabs, and stool, urine, and serum specimens, which were analyzed based on different clinical conditions and lab results, to figure out the clearance time of the virus and factors which may influence this.
Methods
Ethical approval
This retrospective study was approved by the Shanghai Public Health Clinical Center Ethics Committee (No. YJ-2020-S015-01) and was exempted from the need for informed consent from patients.
Subjects
From January 20, 2020 to February 10, 2020, all confirmed patients with COVID-19 in the Shanghai region were admitted to the Shanghai Public Health Clinical Center. The convalescent patients refer to recovered non-febrile patients without respiratory symptoms who had two successive (minimum 24 h sampling interval) negative reverse transcription polymerase chain reaction (RT-PCR) results for viral RNA from oropharyngeal swabs.
Clinical measures
Clinical data and lab results were recorded at admission and last fecal viral RNA test, including sex, age, cluster of differentiation 4 (CD4)+ T lymphocyte counts, erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), and procalcitonin (PCT). The glucocorticoid treatment group included any patients who were ever treated with glucocorticoids, such as prednisolone or dexamethasone. The feces-positive and feces-negative groups were classified based on the detection of viral RNA in patients’ feces. The clinical conditions and lab results of study subjects, together with the viral RNA results from different specimens (oropharyngeal swab, stool, urine, and serum) from each day were collected retrospectively.
Detection of viral RNA in COVID-19
A magnetic bead-method nucleic acid extraction kit was applied in a fully automated nucleic acid extraction instrument (Master Biotechnology, China). The total RNA was extracted from a 200-μL sample and dual fluorescence PCR (Applied Biosystems 251658240 7500 Real-Time PCR Systems, Foster City, CA, USA) was performed according to the manufacturer's instructions.
Statistical analysis
Statistical analyses were performed using SPSS 20.0 (International Business Machines Corporation, IBM, Armonk, New York, USA). Normally distributed continuous variables are summarized as the mean and standard deviation, and non-normally distributed data are recorded as median and interquartile range (IQR) as appropriate. Categorical variables are expressed as counts and percentages for each category. The t-tests or Wilcoxon rank-sum tests were applied to test differences between two groups; Fisher exact tests or Chi-square tests were used for categorical variables. Multiple linear regression was applied to determine the relationship between outcomes and the exploratory factor. P < 0.05 was considered significant.
Results
Demographics and laboratory examination results
From January 20, 2020 to February 10, 2020, 292 patients with COVID-19 were admitted to the Shanghai Public Health Clinical Center. Sixty-six convalescent patients were included in our study. In total, 28 (42.4%) women and 38 men (57.6%) with a median age of 44.0 (34.0–62.0) years were analyzed. The oldest patient was 78 years old and the youngest was 16 years old. There was no difference in sex or age between those with or without glucocorticoid treatment (χ2 = 0.342, P = 0.599; t = 1.059, P = 0.294). On admission, the average high level of ESR was 70.0 (25.5–90.0) mm/h, high-sensitivity CRP was 8.4 (1.6–20.3) mg/L, and PCT was 0.03 (0.02–0.05) ng/mL, which decreased to 44.0 (29.5–81.3) mm/h, 0.5 (0.5–2.1) mg/L, and 0.02 (0.02–0.02) ng/mL, respectively, upon treatment [Table 1].
Table 1.
Clearance time of viral RNA with or without glucocorticoid treatment.
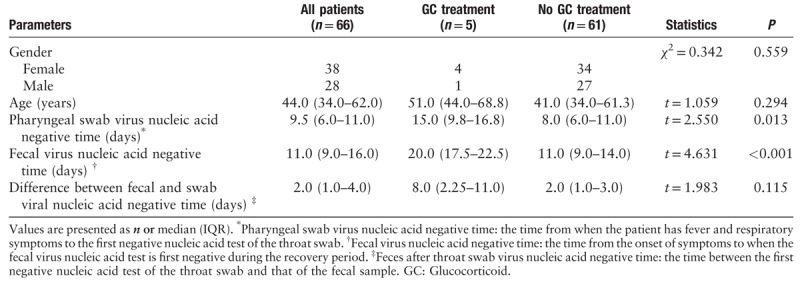
Virus RNA detection in different samples from patients with COVID-19
The median time from the onset of symptoms to first negative RT-PCR results for oropharyngeal swabs of convalescent patients was 9.5 (6.0–11.0) days with improvement in symptoms such as fever, cough, and dyspnea. This time varied greatly between patients, ranging from 2 to 22 days. The RT-PCR for viral RNA was performed using stool, urine, and blood specimens during convalescence. Until the end of the observation period (February 10, 2020), 11 convalescent patients (16.7%) still tested positive for viral RNA in stool specimens. The other 55 patients’ stool specimens were negative for 2019-nCoV following a median duration of 11.0 (9.0–16.0) days. Twelve patient (21.8% 12/55) Viral RNA in oropharyngeal swabs or fecal samples from was negative at the same time; 78.2% (43/55) of cases had longer duration until stool specimens were negative for viral RNA than throat swabs, with a median delay of 2.0 (1.0–4.0) days. Viral nucleic acid was found in urine in only four (6.9%) patients out of 58 cases [Table 1]; viral RNA was present in urine specimens after throat swabs were negative. Fourteen serum specimens were tested for 2019-nCoV and none of them showed positive results (Data not shown).
Factors related to virus clearance
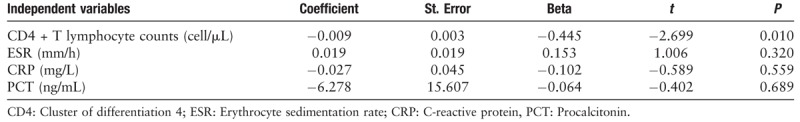
An analysis of the correlation between the absolute values of CD4+ T lymphocytes, CRP, red blood cell sedimentation rate, PCT, and the time of detoxification for feces during convalescence was performed. Using a multiple linear regression model (F = 2.699, P = 0.044, and adjusted R2 = 0.122), the analysis showed that the CD4+ T lymphocyte count may help predict the duration of viral RNA detection in patient stool samples (t = −2.699, P = 0.010) [Table 2].
Table 2.
Multiple linear regression analysis of immune and inflammatory parameters with respect to virus clearance.
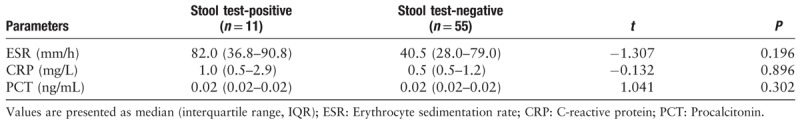
During hospitalization, five patients received glucocorticoid treatment. The duration of viral RNA detection in throat swabs and fecal samples in the glucocorticoid treatment group was longer than that in the non-glucocorticoid treatment group (15 days vs. 8.0 days, t = 2.550, P = 0.013; 20 days vs. 11 days t = 4.631, P < 0.001, respectively). We further analyzed the differences in the results of the last tests for inflammatory indicators upon positive results for viral RNA in fecal samples. There was no statistically significant difference in inflammatory indicators between patients with positive and negative fecal viral RNA test results (P > 0.05) [Table 3].
Table 3.
Viral RNA analysis of stool samples with the inflammatory indicators of patients.
Discussion
The novel coronavirus was firstly identified in respiratory specimens from patients with COVID-19 and viral nucleic acids were subsequently detected in patients’ stool, urine, and gastrointestinal mucosa.[10–12] Recently a neonatal infection was reported, indicating the possibility of fecal-oral and vertical transmission from mother to child, in addition to the currently confirmed droplet transmission and direct contact transmission. In this study, we found that the viral RNA can be detected in the stool of 81.8% (54/66) patients, even in those with negative results from throat swabs. The continuous detection of viral nucleic acids in feces suggests that the virus may be transmitted through the digestive tract or re-transmitted through aerosols containing viruses. Therefore, it is necessary urgently to standardize the stool transport process of COVID-19 patients to reduce risk of further transmission. Moreover, viral RNA detection in fecal samples should be applied regularly in patients with COVID-19, even during the recovery period. Transmission by urine or blood may occur less frequently because of the low rate of positive findings in patients.
Glucocorticoids have been widely used in the treatment of severe acute respiratory syndrome and Middle East respiratory syndrome, and are now also used in conjunction with other drugs to treat patients infected with 2019-nCoV. However, in the published clinical management opinions for the COVID-19, the application of glucocorticoids is not recommended unless there are other indications.[13,14] The use of glucocorticoids may delay the clearance of viral nucleic acids in patients and should be avoided during viral replication. Some bias exists in our study because the patients in the glucocorticoid treatment group had more severe disease and lower CD4+ T lymphocytes counts. Our point is that mild patients are not recommended glucocorticoid treatment, which may delay virus clearance. The randomized controlled double-blind experiments with expanded sample sizes will help clarify this issue. T cell immunity may play an important role in 2019-nCoV infection. The absolute values of CD4+ T lymphocytes, CRP, ESR, and PCT measured upon admission were analyzed with respect to virus clearance. The lower the absolute value of CD4+ T lymphocytes before treatment, the longer duration of virus clearance. The relationship between the fecal viral RNA results and inflammatory indicators of patients were analyzed and no statistical difference in ESR, CRP, or PCT during rehabilitation was found.
In brief, as the clearance of viral RNA from patients’ stools was delayed compared to that from oropharyngeal swabs, it is important to detect the viral RNA in feces during convalescence. Because of the delayed clearance of viral RNA in the glucocorticoid treatment group, glucocorticoids are not recommended in the treatment of COVID-19, especially for mild disease. The duration of RNA detection may be related to host cell immunity.
Funding
The work was supported by grants from the First-class university and first-class discipline building project of the Fudan University (No. IDF162005) and the Scientific research for special subjects on 2019 novel coronavirus (No. 2019-nCoV) of the Shanghai Public Health Clinical Center (No. 2020YJKY01).
Conflicts of interest
None.
Footnotes
How to cite this article: Ling Y, Xu SB, Lin YX, Tian D, Zhu ZQ, Dai FH, Wu F, Song ZG, Huang W, Chen J, Hu BJ, Wang S, Mao EQ, Zhu L, Zhang WH, Lu HZ. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J 2020;133:1039–1043. doi: 10.1097/CM9.0000000000000774
Yun Ling, Shui-Bao Xu, and Yi-Xiao Lin contributed equally to this work.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV). Geneva, World Health Organization; 2020. Available from: https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov). [Accessed February 20, 2020] [Google Scholar]
- 3.Text Transcription of Press Conference on February 11, 2020 (in Chinese). www.gov.cn, 2020. Available from: http://www.nhc.gov.cn/xcs/s3574/202002/53900c60791041e09070bca8f40f93ac.shtml. [Accessed February 20, 2020]. [Google Scholar]
- 4.Novel Coronavirus (2019-nCoV), Situation Report-22. Geneva: World Health Organization, 2 020. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200211-sitrep-22-ncov.pdf?sfvrsn=fb6d49b1_2. [Accessed February 20, 2020]. [Google Scholar]
- 5.Imai N, Cori A, Dorigatti I, Baguelin M, Donnelly CA, Riley S, et al. Transmissibility of 2019-nCoV. 2020; Geneva: World Health Organization, Available from: https://fpmag.net/wp-content/uploads/2020/01/Imperial-2019-nCoV-transmissibility.pdf. [Accessed February 20, 2020]. [Google Scholar]
- 6.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med 2020; 382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu T, Hu J, Kang M, Lin L, Zhong H, Xiao J, et al. Transmission dynamics of 2019 novel coronavirus (2019-nCoV). BioRixv 2020; doi: 10.1101/2020.01.25.919787. [Google Scholar]
- 8.Yang Y, Lu Q, Liu M, Wang Y, Zhang A, Jalali N, et al. Epidemiological and clinical features of the 2019 novel coronavirus outbreak in China. medRxiv 2020; doi: 10.1101/2020.02.10.20021675. [Google Scholar]
- 9.Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS). Geneva: World Health Organization, 2003. Available from: https://www.who.int/csr/sars/en/WHOconsensus.pdf. [Accessed February 20, 2020]. [Google Scholar]
- 10.Ren LL, Wang YM, Wu ZQ, Xiang ZC, Guo L, Xu T, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J 2020; 133:1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med 2020; 382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of 2019 novel coronavirus infection in China. N Engl J Med 2020; Feb 28. doi: 10.1056/NEJMoa2002032. [Google Scholar]
- 13.Clinical Management of Severe Acute Respiratory Infection When Novel Coronavirus (nCoV) Infection is Suspected. Geneva: World Health Organization, 2020. Available from: https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected. [Accessed January 28, 2020]. [Google Scholar]
- 14.Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 2020; 395:473–475. doi: 10.1016/s0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


