Abstract
Many receptors can be activated by bile acids (BAs) and their derivatives. These include nuclear receptors farnesoid X receptor (FXR), pregnane X receptor (PXR), and vitamin D receptor (VDR), as well as membrane receptors Takeda G protein receptor 5 (TGR5), sphingosine-1-phosphate receptor 2 (S1PR2), and cholinergic receptor muscarinic 2 (CHRM2). All of them are implicated in the development of metabolic and immunological diseases in response to endobiotic and xenobiotic exposure. Because epigenetic regulation is critical for organisms to adapt to constant environmental changes, this review article summarizes epigenetic regulation as well as post-transcriptional modification of bile acid receptors. In addition, the focus of this review is on the liver and digestive tract although these receptors may have effects on other organs. Those regulatory mechanisms are implicated in the disease process and critically important in uncovering innovative strategy for prevention and treatment of metabolic and immunological diseases.
Keywords: Bile acid receptor, Farnesoid X receptor (FXR), G protein-coupled bile acid receptor, Takeda G protein receptor 5 (TGR5), Sphingosine-1-phosphate receptor 2 (S1PR2), Acetylation, Methylation, Glycosylation
1. Introduction
Upon catalysis by hepatic and bacterial enzymes, cholesterol converts into bile acids (BAs).1,2 In addition, BAs have bacteriostatic effects. Thus, BAs are the intrinsic links that explain how foods, through gut microbiota, affect host metabolism and immunity. Hepatic enzymes generate free primary BAs such as chenodeoxycholic acid (CDCA) and cholic acid (CA). Hepatic conjugation of BAs increases the hydrophilicity of BAs and changes their binding affinity to their receptors. In the gut, bacterial enzyme, i.e., bile salt hydrolase deconjugates BAs. Moreover, bacterial enzyme 7α- dehydroxylase that can be found in Firmicutes converts primary BAs into secondary BAs such as deoxycholic acid (DCA) and lithocholic acid (LCA).3,4 Therefore, host and bacteria jointly produce various BAs, and eubiosis is essential for maintaining BA homeostasis. In contrast, dysregulated BA synthesis accompanied by dysbiosis is implicated in the development of metabolic diseases including obesity, steatosis, steatohepatitis, as well as liver and colon cancer.3,5–10
Free and conjugated primary as well as secondary BAs have differential binding affinities to various receptors including nuclear farnesoid X receptor (FXR) as well as membrane Takeda G protein receptor 5 (TGR5), and sphingosine-1-phosphate receptor 2 (S1PR2). Additionally, pregnane X receptor (PXR), vitamin D receptor (VDR), constitutive androstane receptor (CAR), and cholin-ergic receptor muscarinic 2 (CHRM2) can be activated by BAs or their precursors and metabolites (Table 1). Thus, BA receptors are essentially endobiotic and xenobiotic sensors. For an organism to adapt to constant environmental change, epigenetic mechanism is used to regulate host response. Epigenetic effects such as acetylation and methylation are ways to switch genes on and off without changing deoxyribonucleic acid (DNA) sequence. Thus, as nutrient and chemical sensors, epigenetic mechanisms should be important for regulating the expression and activity of BA receptors. This review article summarizes epigenetic regulation and post-transcriptional modification of BA receptors. The information is critically important to understand how these receptors are activated or silenced, thereby leading to metabolic or detoxification function or dysfunction. We focus on FXR, TGR5, and S1PR2 since the information available for other receptors in this area is limited. The search was done using combinations of following keywords: FXR, G protein-coupled bile acid receptor, TGR5, S1PR2, acetylation, methylation, glycosylation, epigenetics, and bile acid in the PubMed.
Table 1.
Bile acids and their derivatives as agonists for the listed receptors.
| Receptors | Ligands | References |
|---|---|---|
| FXR | CDCA > DCA > LCA > CA > UDCA | 11,12 |
| Bile alcohols, 6α-ethyl-CDCA | 13,14 | |
| 5β-cholanoic acid, 5β-norcholanoic acid, 5α-cholanoic acid | 15 | |
| TGR5 | LCA > DCA > CDCA > CA > UDCA | 16 |
| TLCA | 17 | |
| S1PR2 | Conjugated BAs (GCA, TCA, GDCA, TDCA, TUDCA) | 18 |
| PXR | 3-keto-LCA, LCA, CDCA, DCA, CA | 19,20 |
| 7α-hydroxy-4-cholesten-3-one | 21 | |
| VDR | LCA, 3-keto-LCA | 22 |
| CAR | CA, 6-keto-LCA, 12-keto-LCA | 23,24 |
| CHRM2 | TCA | 25 |
Note: Humans mainly make glycine conjugates of bile acids while mice make taurine conjugates.
Abbreviations: FXR, farnesoid X receptor; TGR5, Takeda G protein receptor 5; S1PR2, sphingosine-1-phosphate receptor 2; PXR, pregnane X receptor; VDR, vitamin D receptor; CAR, constitutive androstane receptor; CHRM2, cholinergic receptor muscarinic 2; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; LCA, lithocholic acid; CA, cholic acid; TLCA, taurolithocholic acid; UDCA, ursodeoxycholic acid; GCA, glycocholic acid; TCA, taurocholic acid; GDCA, glycodeoxycholic acid; TDCA, taurodeoxycholic acid; TUDCA, tauroursodeoxycholic acid.
2. FXR
2.1. FXR introduction
BAs regulate glucose and lipid metabolism as well as the inflammatory process. This paradigm shift was spurred by identification of the BA receptor FXR. The function of FXR has been extensively reviewed by recent articles.1,26–34 We only provide a general introduction here. FXR activation plays a key role in regulating BA homeostasis in the liver and intestine.3,5,35–39 The activation of FXR leads to the regulation of genes whose function is to decrease the concentrations of BAs. FXR increases the expression of hepatic small heterodimer partner (SHP) and intestinal fibroblast growth factor 15 (FGF15), which in turn inhibits hepatic cholesterol 7α-hydroxylase (CYP7A1) and sterol 12α-hydroxylase (CYP8B1), reducing BA synthesis. In addition, FXR activation increases the expression of canalicular transporters, such as the bile-salt export pump (BSEP), providing a pathway for excreting cholesterol and BAs. These regulatory pathways are important in part because accumulation of hydrophobic BAs leads to inflammation, injury, cirrhosis, and carcinogenesis.40,41 In contrast to the dysregulated BA synthesis found in metabolic disease patients, activation of FXR increases metabolism and insulin sensitivity, and FXR agonists are used to treat non-alcoholic steatohepatitis.42 In consistency, whole-body FXR knockout mice, which have dysregulated BA synthesis and dysbiosis, spontaneously develop non-alcoholic steatohepatitis and liver cancer.43–47 Moreover, reduced FXR is found in patients who have cirrhosis and colon or liver cancer as well as ulcerative colitis.48–51 Thus, it is important to understand the mechanism by which FXR is regulated. Taken together, FXR, which is mainly expressed in the liver and intestine, has a pivotal role in regulating BAs homeostasis leading to metabolic and anti-inflammatory beneficial outcomes (Fig. 1).
Fig. 1. Schematic overview of the functions of bile acid receptors.
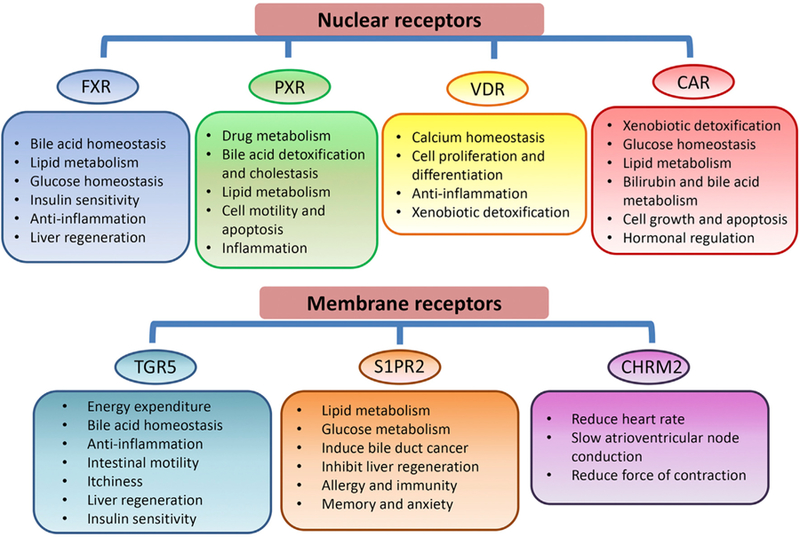
The key functions of bile acid receptors are summarized in the figure. Abbreviations: FXR, farnesoid X receptor; PXR, pregnane X receptor; VDR, vitamin D receptor; CAR, constitutive androstane receptor; TGR5, Takeda G protein receptor 5; S1PR2, sphingosine-1-phosphate receptor 2; CHRM2, cholinergic receptor muscarinic 2.
2.2. Methylation regulates FXR expression and activity
Methylation is a mechanism that affects FXR activity. Silencing the FXR gene through CpG methylation is found in mouse models of colon cancer in adenomatous polyposis coli mutant mice, human colon cancer cells, and human colon cancers.51–53 By direct-sequence analyses of bisulfonated genomic DNA, there are 13 CpG methylation sites located in the region flanking the transcription start site on exon-3 of the FXR.52 In addition, methylation of the FXR is implicated in pregnancy related diseases. The intrahepatic cholestasis of pregnancy is a liver disorder that involves the inter-play between dysregulated BA synthesis, sex hormones, genetic susceptibility, as well as environmental factors. There is a clear relationship between the status of methylation in the FXR promoter and the profile of BAs in intrahepatic cholestasis of pregnancy patients compared with healthy pregnant women.54 Specifically, reduced methylation of the FXR promoter is found in intrahepatic cholestasis of pregnancy cases when compared with healthy pregnancy controls. In addition, increased methylation level at the distal promoter (–1890) is positively correlated with elevated conjugated BAs; whereas methylation level at the proximal promoter (–358) is negatively correlated with serum CA and DCA concentration.54 Methylation of the FXR gene is also implicated in another pregnancy related disease, i.e., preeclampsia. Altered methylation pattern of the FXR as well as liver X receptor (LXR) is found in early onset of preeclampsia based on genome-wide methylation study using cord blood DNAs.55 These findings implicate the potential role of BAs and FXR in immunological disorders.
At the histone level, methylation of H3 and H4 occurs on lysine or arginine and is catalyzed by histone methyltransferases that use S-adenosylmethionine as a methyl donor. It has been shown that methylation by Set7/9, a lysine methyltransferase, increases FXR binding to its target gene leading to increased transcriptional activity.56 In addition, FXR activity incorporates histone methyl-transferase activity within the BSEP gene locus.57 This methyltransferase activity is directed specifically to arginine 17 of H3. By interacting with arginine methyl-transferase type I, the transcriptional activity of FXR is activated, thereby leading to increased expression of SHP and BSEP and decreased CYP7A1.58 Moreover, 5′-deoxy-5′-methylthioadenosine, a methylation inhibitor, reduces the expression of BSEP.58 In consistency, reduced recruitment of H3K4me3 to the BSEP and multidrug resistance-associated protein 2 (Mrp2) promoter of the FXR-binding elements was found in mouse livers after bile duct ligation.59 Thus, histone 3 lysine 4 trimethylation (H3K4me3) is essential to increase the transcription of the BSEP, sodium-taurocholate cotransporting polypeptide (NTCP), and Mrp2 genes that are controlled by FXR.
2.3. Acetylation and FXR activity
The transcriptional activity of FXR can be modulated by sirtuin1 (SIRT1), a protein deacetylase. SIRT1 activity is dependent on nicotinamide adenine dinucleotide (NAD+) levels. Hepatic over-expression of microRNA (miR)-34a, which reduces nicotinamide phosphoribosyltransferase and NAD+ levels, decreases SIRT1 leading to reduced transcriptional activity of FXR.60 It has been shown that the FXR acetylation site targeted by SIRT1 deacetylase and p300 acetylase is at lysine 217.61 Acetylated FXR has increased stability, but reduced capability to dimerize with retinoid X receptor α (RXRα), thereby leading to decreased transcriptional activity. In mouse models of metabolic disease, FXR acetylation level is elevated. Therefore, potentially, inhibiting FXR acetylation by increasing SIRT1 or reducing p300 can be used to treat metabolic disorders.61 Moreover, FXR acetylation increased pro-inflammatory gene expression, macrophage infiltration, and hepatic cytokine and triglyceride levels. Mechanistically, acetylated FXR prevented small ubiquitin-like modifier 2 (SUMO2) modification. SUMOylation of activated FXR increased its interaction with nuclear factor k B (NF-kB), but reduced the dimerization with RXRα.62 Taken together, SIRT1 modulates the FXR signaling pathway by directly deacetylating FXR. Another mechanism by which SIRT1 regulates FXR transcriptional activity is through hepatocyte nuclear factor 1a (HNF1α). Knockout hepatic SIRT1 reduces FXR activity is mainly due to reduced occupancy of HNF1α in the FXR promoter leading to decreased FXR expression.63
The role of SIRT1 in regulating proliferation mediated by FXR is also revealed using a partial hepatectomy model. SIRT1 transgenic mice have increased mortality, impairs hepatocyte proliferation, and BA accumulation after partial hepatectomy. This is in part due to persistent deacetylation and reduced FXR expression. In contrast, 24-nor-ursodeoxycholic acid increases miR-34a and reduces SIRT protein, resulting in increased acetylation of FXR and neighboring histones. Thus, 24-nor-ursodeoxycholic acid is able to establish BA homeostasis and restores liver regeneration capability in SIRT1 transgenic mice.64 Moreover, inversed expression of SIRT1 and FXR is also found in liver cancer; human hepatocellular carcinoma has increased SIRT1 and reduced FXR compared with normal liver.64
At the histone level, the occupancy of FXR and co-activator-associated arginine methyltransferase 1 on the human BSEP locus is associated with increased Arg-17 methylation and Lys-9 acetylation of H3 of the BSEP.57 Moreover, by acetylating histones at the promoter and FXR itself, p300 acetylase is a coactivator of FXR to increase the expression of SHP.65 Taken together, acetylation and deacetylation of FXR should be a dynamic process to maintain FXR activity. Sustained FXR activation and deactivation lead to metabolic imbalance impaired liver regeneration, and potentially carcinogenesis.
2.4. Other mechanisms affecting FXR activity
O-GlcNAc transferase, responsible for O-GlcNAcylation, is a nutrient sensor that links glucose and the hexosamine biosynthetic pathway to the regulation of transcriptional factors that regulate energy homeostasis. By interacting FXR, hepatic carbohydrate response element-binding protein (ChREBP) can regulate glycolytic and lipogenic gene expression. It is interesting to note that FXR as well as ChREBP are both O-GlcNAcylated in response to glucose. High glucose increases FXR O-GlcNAcylation and enhances its stability as well as transcriptional activity. Moreover, in vivo fasting and refeeding experiments show that FXR undergoes O-GlcNAcylation in the fed condition, which is associated with increased expression of FXR target gene.66
MiRNA regulation of protein deacetylases may indirectly affect FXR activity. MiR-34a is one example mentioned above. It is interesting to note that FXR-activation induced miR-22 can also silence SIRT1, which in turn affects FXR stability or transcriptional activation. Such pathway potentially forms a self-regulatory loop.67,68
3. TGR5
3.1. Function of TGR5
In contrast to nuclear receptor FXR, TGR5 is a membrane receptor ubiquitously expressed in adipocytes, endocrine glands, muscles, as well as immune organs.69 It is also known as G protein-coupled bile acid receptor 1 (GPBAR1) or G-protein coupled receptor 19 (GPCR19). TGR5 is also expressed in the gut, liver, and gallbladder, where BAs are produced and stored.70,71 Because TGR5 is expressed in cells of the hematopoietic system, such as monocytes and macrophages, it confers a potent anti-inflammatory property at the systemic level.17,72–76 Our recent publication revealed the potential role of TGR5 in neuro-inflammation as well as neuroplasticity.77 Activation of TGR5 also increases intracellular cyclic adenosine monophosphate (cAMP), thereby activating cAMP response element binding protein. One of the downstream effects of cAMP production is to induce the expression of thyroid hormone deiodinase 2, which generates thyroxine, a key player in basal metabolism.78 In addition to metabolism and inflammation, TGR5 also regulates proliferation, muscle relaxation, and itchiness among many others, which have been reviewed in recent articles (Fig. 1).69,79–82
In the liver, although TGR5 is not expressed in hepatocytes, it is found in Kupffer cells and endothelial and biliary epithelial cells, which are involved in regulating immune, inflammatory signaling, and circulation.83 In the intestine, TGR5 activation induces the expression of the preproglucagon gene (Gcg) and glucagon-like peptide-1 (GLP-1) secretion in the intestinal enteroendocrine L-cells.84,85 GLP-1 is an incretin that potentiates postprandial insulin secretion.86 Activation of TGR5 also releases neuropeptide hormone peptide tyrosine tyrosine (PYY), which regulates immune signaling and intestinal mobility.87
Regarding the ligands, unconjugated BAs such as CDCA, DCA, and ursodeoxycholic acid (UDCA) induce a large cAMP response in neonatal mouse cardiomyocytes.88 Secondary BAs such as LCA and taurine-conjugated LCA are also endogenous ligands for TGR5 (Table 1).16,17 In addition, TGR5 potentially can be activated by many other chemicals. Those include allogregnanolone, betulinic acid, linolenic acid, etc.89–91 We recently showed that supplementation of Western diet-fed mice with epigallocatechin-3-gallate activates TGR5 signaling pathways leading to a lean phenotype.10 Whether the effect is mediated via epigenetic regulation of TGR5 remains to be investigated.
3.2. Regulation of TGR5
FXR induces the expression of the TGR5 gene in mouse intestine. An inverted repeat with one-nucleotide spacing (IR1) that can be occupied by FXR/RXRα has been uncovered in the proximal promoter of the human TGR5 gene.92 FXR and TGR5 are co-expressed in the enteroendocrine L cells, and activation of FXR induces TGR5 to stimulate the secretion of GLP-1.93 Because the expression of the TGR5 gene is transcriptionally regulated by FXR, methylation and acetylation likely influence TGR5 activity, which remains to be proved.
It is interesting to note that INT-777-mediated TGR5 activation induces renal expression of SIRT1 and SIRT3. Increased SIRT3 activity induces acetylation of mitochondrial superoxide dismutase 2 and isocitrate dehydrogenase 2 found in db/db mice.94 Since acetylation has a known role in regulating FXR expression and activity, it is possible TGR5-induced SIRT expression may have an impact on FXR activity as well. The hypothesis that TGR5 and FXR may mutually regulate each other warrants further investigation.
Methylation has a role in TGR5 expression. Methylation status of the TGR5 promoter has been studied in peripheral mononuclear cells in patients with acute-on-chronic hepatitis B liver failure. The frequency of TGR5 promoter methylation is significantly higher in liver failure patients than chronic hepatitis patients. In addition, hyper-methylation is accompanied by reduced TGR5 mRNA level.95 This study concludes that aberrant TGR5 promoter methylation is a potential prognostic marker for acute-on-chronic hepatitis B liver failure. Hyper-methylation of the TGR5 promoter is also found in hepatocellular carcinoma patients by studying their circulating cell-free DNA. Moreover, the methylation rate of TGR5 is age-related, much higher in patients older than 60 than in those younger than 60 years old. It has been suggested that a combination of serum TGR5 promoter methylation level along with the value of a-fetoprotein may increase the sensitivity for hepatocellular carcinoma diagnosis.96
4. S1PR2
S1PR2 or S1P2 is a G protein-coupled receptor for sphingosine-1-phosphate (S1P). S1PR2 was also found to be the receptor for conjugated BAs such as taurocholic acid (TCA) and taurodeoxycholic acid (TDCA) (Table 1).18,97 S1P is a bioactive lipid mediator that regulates proliferation, immunity, cell trafficking, inflammation, etc.98,99 TCA-activated S1PR2 induces the expression and activity of sphingosine kinase (SphK2) to increase the conversion of sphingosine into S1P and leading to increased lipid and sterol metabolism in the liver.18 Thus, conjugated BAs have a pivotal role in S1P singling via SphK2 induction as well as S1PR2. Consistent with these findings, both SphK2 and S1PR2 knockout mice are susceptible to diet-induced fatty liver.97,100
It is interesting to note that nuclear S1P, produced by either induction of SphK2 or inhibition of S1Plyase, binds to histone deacetylases (HDAC) 1 and 2, thereby increasing histone acetylation and up-regulating the expression of metabolic genes.100 Through such HDAC inhibitory mechanism, sphingosine has a role in regulating apoptosis and metabolism. Furthermore, glycochenodeoxycholate (GCDC) via S1PR2 as well as cell entrance have an apoptotic effect in human liver cancer Huh7 cells.101 These results suggested that S1PR2 activation has a pro-apoptotic effect in GCDC-treated liver cancer cells, but the effect is not simply due to just binding between the GCDC and S1PR2.
Conjugated BAs via S1PR2 also activate ERK1/2- and AKT- signaling pathways leading to the growth and invasion of cholangiocarcinoma cells.102 The role of conjugated BAs via S1PR2 to regulate apoptosis or cancer progression remains to be dissected. Nevertheless, there is no doubt that the composition of free and conjugated BAs has an impact on regulating BA receptor activity.
5. Conclusions and perspectives
BA receptors can be found in many types of cells within and outside the digestive tract. By activating G protein-coupled membrane receptors, i.e., TGR5, S1PR2, and muscarinic receptor, BAs exert their effects without crossing the cell membrane. Similarly, those receptors are readily accessible to enzymes that regulate methylation, acetylation, glycosylation, etc. Thus, in addition to transcriptional regulation, it is important to study post–transcriptional modification of those receptors. It is likely that due to transcriptional and post-transcriptional modification, those receptors exert various biological effects ranging across metabolism, energy homeostasis in skeletal muscle and adipose tissue, inflammatory signaling in macrophages, muscle relaxation, hormonal secretion, as well as cell proliferation and apoptosis, etc. The current knowledge limits to acetylation and methylation of a few receptors. More research should be done to understand the mechanism that influences their expression, modification, and biological effects.
Acknowledgments
The authors thank Kyle Mcneil from University of California, Davis, for editing the manuscript. This study was supported by grants funded by the USA National Institutes of Health (NIH) U01CA179582 and R01 CA222490.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Li T, Chiang JY. Bile acids as metabolic regulators. Curr Opin Gastroenterol 2015;31:159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inagaki T, Moschetta A, Lee YK, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A 2006;103:3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev 2009;89:147–191. [DOI] [PubMed] [Google Scholar]
- 4.Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A 2008;105:13580–13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev 2014;66:948–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu HX, Keane R, Sheng L, Wan YJ. Implications of microbiota and bile acid in liver injury and regeneration. J Hepatol 2015;63:1502–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheng L, Jena PK, Liu HX, et al. Gender differences in bile acids and microbiota in relationship with gender dissimilarity in steatosis induced by diet and FXR inactivation. Sci Rep 2017;7:1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jena PK, Sheng L, Liu HX, et al. Western diet-induced dysbiosis in farnesoid X receptor knockout mice causes persistent hepatic inflammation after antibiotic treatment. Am J Pathol 2017;187:1800–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheng L, Jena PK, Hu Y, et al. Hepatic inflammation caused by dysregulated bile acid synthesis is reversible by butyrate supplementation. J Pathol 2017;243:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheng L, Jena PK, Liu HX, et al. Obesity treatment by epigallocatechin-3-gallate-regulated bile acid signaling and its enriched Akkermansia muciniphila. FASEB J 2018. 10.1096/fj.201800370R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makishima M, Okamoto AY, Repa JJ, et al. Identification of a nuclear receptor for bile acids. Science 1999;284:1362–1365. [DOI] [PubMed] [Google Scholar]
- 12.Parks DJ, Blanchard SG, Bledsoe RK, et al. Bile acids: Natural ligands for an orphan nuclear receptor. Science 1999;284:1365–1368. [DOI] [PubMed] [Google Scholar]
- 13.Pellicciari R, Fiorucci S, Camaioni E, et al. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J Med Chem 2002;45:3569–3572. [DOI] [PubMed] [Google Scholar]
- 14.Iguchi Y, Kihira K, Nishimaki-Mogami T, Une M. Structure-activity relationship of bile alcohols as human farnesoid X receptor agonist. Steroids 2010;75: 95–100. [DOI] [PubMed] [Google Scholar]
- 15.Sepe V, Renga B, Festa C, et al. Investigation on bile acid receptor regulators. Discovery of cholanoic acid derivatives with dual G-protein coupled bile acid receptor 1 (GPBAR1) antagonistic and farnesoid X receptor (FXR) modulatory activity. Steroids 2016;105:59–67. [DOI] [PubMed] [Google Scholar]
- 16.Maruyama T, Miyamoto Y, Nakamura T, et al. Identification of membrane-type receptor for bile acids (M-BAR). Biochem Biophys Res Commun 2002;298: 714–719. [DOI] [PubMed] [Google Scholar]
- 17.Kawamata Y, Fujii R, Hosoya M, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem 2003;278:9435–9440. [DOI] [PubMed] [Google Scholar]
- 18.Studer E, Zhou X, Zhao R, et al. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology 2012;55: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staudinger JL, Goodwin B, Jones SA, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A 2001;98:3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie W, Radominska-Pandya A, Shi Y, et al. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci U S A 2001;98:3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodwin B, Gauthier KC, Umetani M, et al. Identification of bile acid precursors as endogenous ligands for the nuclear xenobiotic pregnane X receptor. Proc Natl Acad Sci U S A 2003;100:223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makishima M, Lu TT, Xie W, et al. Vitamin D receptor as an intestinal bile acid sensor. Science 2002;296:1313–1316. [DOI] [PubMed] [Google Scholar]
- 23.Moore LB, Maglich JM, McKee DD, et al. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol Endocrinol 2002;16:977–986. [DOI] [PubMed] [Google Scholar]
- 24.Maglich JM, Stoltz CM, Goodwin B, Hawkins-Brown D, Moore JT, Kliewer SA. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol Pharmacol 2002;62:638–646. [DOI] [PubMed] [Google Scholar]
- 25.Sheikh Abdul Kadir SH, Miragoli M, Abu-Hayyeh S, et al. Bile acid-induced arrhythmia is mediated by muscarinic M2 receptors in neonatal rat cardiomyocytes. PLoS One 2010;5:e9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang JYL, Ferrell JM. Bile acid metabolism in liver pathobiology. Gene Expr 2018;18:71–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang JY. Recent advances in understanding bile acid homeostasis. F1000Res 2017;6:2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiang JYL. Bile acid metabolism and signaling in liver disease and therapy. Liver Res 2017;1:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiang JYL. Linking sex differences in non-alcoholic fatty liver disease to bile acid signaling, gut microbiota, and high fat diet. Am J Pathol 2017;187: 1658–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang XF, Zhao WY, Huang WD. FXR and liver carcinogenesis. Acta Pharmacol Sin 2015;36:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kliewer SA, Mangelsdorf DJ. Bile acids as hormones: The FXR-FGF15/19 pathway. Dig Dis 2015;33:327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gadaleta RM, Cariello M, Sabbà C, Moschetta A. Tissue-specific actions of FXR in metabolism and cancer. Biochim Biophys Acta 2015;1851:30–39. [DOI] [PubMed] [Google Scholar]
- 33.Matsubara T, Li F, Gonzalez FJ. FXR signaling in the enterohepatic system. Mol Cell Endocrinol 2013;368:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Y, Liu H, Zhang M, Guo GL. Fatty liver diseases, bile acids, and FXR. Acta Pharm Sin B 2016;6:409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee FY, Lee H, Hubbert ML, Edwards PA, Zhang Y. FXR, a multipurpose nuclear receptor. Trends Biochem Sci 2006;31:572–580. [DOI] [PubMed] [Google Scholar]
- 36.Wang YD, Chen WD, Moore DD, Huang W. FXR: A metabolic regulator and cell protector. Cell Res 2008;18:1087–1095. [DOI] [PubMed] [Google Scholar]
- 37.Modica S, Gadaleta RM, Moschetta A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl Recept Signal 2010;8:e005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, Li F, Guo GL. Tissue-specific function of farnesoid X receptor in liver and intestine. Pharmacol Res 2011;63:259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Jadhav K, Zhang Y. Bile acid receptors in non-alcoholic fatty liver disease. Biochem Pharmacol 2013;86:1517–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez MJ, Briz O. Bile-acid-induced cell injury and protection. World J Gastroenterol 2009;15:1677–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delzenne NM, Calderon PB, Taper HS, Roberfroid MB. Comparative hepatotoxicity of cholic acid, deoxycholic acid and lithocholic acid in the rat: In vivo and in vitro studies. Toxicol Lett 1992;61:291–304. [DOI] [PubMed] [Google Scholar]
- 42.Neuschwander-Tetri BA, Loomba R, Sanyal AJ, et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): A multicentre, randomised, placebo-controlled trial. Lancet 2015;385:956–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Ge X, Heemstra LA, et al. Loss of FXR protects against diet-induced obesity and accelerates liver carcinogenesis in ob/ob mice. Mol Endocrinol 2012;26:272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang F, Huang X, Yi T, Yen Y, Moore DD, Huang W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Canc Res 2007;67:863–867. [DOI] [PubMed] [Google Scholar]
- 45.Kim I, Morimura K, Shah Y, Yang Q, Ward JM, Gonzalez FJ. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis 2007;28:940–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfe A, Thomas A, Edwards G, Jaseja R, Guo GL, Apte U. Increased activation of the Wnt/b-catenin pathway in spontaneous hepatocellular carcinoma observed in farnesoid X receptor knockout mice. J Pharmacol Exp Ther 2011;338:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li G, Kong B, Zhu Y, et al. Small heterodimer partner overexpression partially protects against liver tumor development in farnesoid X receptor knockout mice. Toxicol Appl Pharmacol 2013;272:299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lax S, Schauer G, Prein K, et al. Expression of the nuclear bile acid receptor/ farnesoid X receptor is reduced in human colon carcinoma compared to nonneoplastic mucosa independent from site and may be associated with adverse prognosis. Int J Canc 2012;130:2232–2239. [DOI] [PubMed] [Google Scholar]
- 49.Liu N, Meng Z, Lou G, et al. Hepatocarcinogenesis in FXR−/−mice mimics human HCC progression that operates through HNF1a regulation of FXR expression. Mol Endocrinol 2012;26:775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su H, Ma C, Liu J, et al. Downregulation of nuclear receptor FXR is associated with multiple malignant clinicopathological characteristics in human hepatocellular carcinoma. Am J Physiol Gastrointest Liver Physiol 2012;303: G1245–G1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torres J, Bao X, Iuga AC, et al. Farnesoid X receptor expression is decreased in colonic mucosa of patients with primary sclerosing cholangitis and colitisassociated neoplasia. Inflamm Bowel Dis 2013;19:275–282. [DOI] [PubMed] [Google Scholar]
- 52.Selmin OI, Fang C, Lyon AM, et al. Inactivation of adenomatous polyposis coli reduces bile acid/farnesoid X receptor expression through Fxr gene CpG methylation in mouse colon tumors and human colon cancer cells. J Nutr 2016;146:236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bailey AM, Zhan L, Maru D, et al. FXR silencing in human colon cancer by DNA methylation and KRAS signaling. Am J Physiol Gastrointest Liver Physiol 2014;306:G48–G58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cabrerizo R, Castanõ GO, Burguenõ AL, et al. Promoter DNA methylation of farnesoid X receptor and pregnane X receptor modulates the intrahepatic cholestasis of pregnancy phenotype. PLoS One 2014;9, e87697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ching T, Ha J, Song MA, et al. Genome-scale hypomethylation in the cord blood DNAs associated with early onset preeclampsia. Clin Epigenet 2015;7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balasubramaniyan N, Ananthanarayanan M, Suchy FJ. Direct methylation of FXR by Set7/9, a lysine methyltransferase, regulates the expression of FXR target genes. Am J Physiol Gastrointest Liver Physiol 2012;302:G937–G947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ananthanarayanan M, Li S, Balasubramaniyan N, Suchy FJ, Walsh MJ. Ligand-dependent activation of the farnesoid X-receptor directs arginine methylation of histone H3 by CARM1. J Biol Chem 2004;279:54348–54357. [DOI] [PubMed] [Google Scholar]
- 58.Rizzo G, Renga B, Antonelli E, Passeri D, Pellicciari R, Fiorucci S. The methyl transferase PRMT1 functions as co-activator of farnesoid X receptor (FXR)/9-cis retinoid X receptor and regulates transcription of FXR responsive genes. Mol Pharmacol 2005;68:551–558. [DOI] [PubMed] [Google Scholar]
- 59.Ananthanarayanan M, Li Y, Surapureddi S, et al. Histone H3K4 trimethylation by MLL3 as part of ASCOM complex is critical for NR activation of bile acid transporter genes and is downregulated in cholestasis. Am J Physiol Gastrointest Liver Physiol 2011;300:G771–G781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi SE, Fu T, Seok S, et al. Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell 2013;12: 1062–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kemper JK, Xiao Z, Ponugoti B, et al. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metabol 2009;10:392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim DH, Xiao Z, Kwon S, et al. A dysregulated acetyl/SUMO switch of FXR promotes hepatic inflammation in obesity. EMBO J 2015;34:184–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purushotham A, Xu Q, Lu J, et al. Hepatic deletion of SIRT1 decreases hepatocyte nuclear factor 1a/farnesoid X receptor signaling and induces formation of cholesterol gallstones in mice. Mol Cell Biol 2012;32:1226–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.García-Rodríguez JL, Barbier-Torres L, Fernández-Álvarez S, et al. SIRT1 controls liver regeneration by regulating bile acid metabolism through farnesoid X receptor and mammalian target of rapamycin signaling. Hepatology 2014;59:1972–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fang S, Tsang S, Jones R, et al. The p300 acetylase is critical for ligand-activated farnesoid X receptor (FXR) induction of SHP. J Biol Chem 2008;283:35086–35095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benhamed F, Filhoulaud G, Caron S, Lefebvre P, Staels B, Postic C. O-GlcNA-cylation links ChREBP and FXR to glucose-sensing. Front Endocrinol (Lausanne) 2015;5:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang F, Hu Y, Liu HX, Wan YJ. MiR-22-silenced cyclin A expression in colon and liver cancer cells is regulated by bile acid receptor. J Biol Chem 2015;290: 6507–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu D, Takeshita F, Hino Y, et al. MiR-22 represses cancer progression by inducing cellular senescence. J Cell Biol 2011;193:409–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duboc H, Taché Y, Hofmann AF. The bile acid TGR5 membrane receptor: From basic research to clinical application. Dig Liver Dis 2014;46:302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maruyama T, Tanaka K, Suzuki J, et al. Targeted disruption of G protein-coupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J Endocrinol 2006;191:197–205. [DOI] [PubMed] [Google Scholar]
- 71.Vassileva G, Golovko A, Markowitz L, et al. Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation. Biochem J 2006;398: 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cipriani S, Mencarelli A, Chini MG, et al. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS One 2011;6:e25637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haselow K, Bode JG, Wammers M, et al. Bile acids PKA-dependently induce a switch of the IL-10/IL-12 ratio and reduce proinflammatory capability of human macrophages. J Leukoc Biol 2013;94:1253–1264. [DOI] [PubMed] [Google Scholar]
- 74.Pols TW, Nomura M, Harach T, et al. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metabol 2011;14:747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor k light-chain enhancer of activated B cells (NF-kB) in mice. Hepatology 2011;54:1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoneno K, Hisamatsu T, Shimamura K, et al. TGR5 signalling inhibits the production of pro-inflammatory cytokines by in vitro differentiated inflammatory and intestinal macrophages in Crohn’s disease. Immunology 2013;139:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jena PK, Sheng L, Di Lucente J, Jin LW, Maezawa I, Wan YY. Dysregulated bile acid synthesis and dysbiosis are implicated in Western diet-induced systemic inflammation, microglial activation, and reduced neuroplasticity. FASEB J 2018;32:2866–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watanabe M, Houten SM, Mataki C, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 2006;439: 484–489. [DOI] [PubMed] [Google Scholar]
- 79.Guo C, Chen WD, Wang YD. TGR5, not only a metabolic regulator. Front Physiol 2016;7:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pols TW, Noriega LG, Nomura M, Auwerx J, Schoonjans K. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J Hepatol 2011;54:1263–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alemi F, Kwon E, Poole DP, et al. The TGR5 receptor mediates bile acidinduced itch and analgesia. J Clin Invest 2013;123:1513–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lieu T, Jayaweera G, Zhao P, et al. The bile acid receptor TGR5 activates the TRPA1 channel to induce itch in mice. Gastroenterology 2014;147: 1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Keitel V, H€aussinger D. Perspective: TGR5 (Gpbar-1) in liver physiology and disease. Clin Res Hepatol Gastroenterol 2012;36:412–419. [DOI] [PubMed] [Google Scholar]
- 84.Thomas C, Gioiello A, Noriega L, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metabol 2009;10:167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Harach T, Pols TW, Nomura M, et al. TGR5 potentiates GLP-1 secretion in response to anionic exchange resins. Sci Rep 2012;2:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hayes MR, Mietlicki-Baase EG, Kanoski SE, De Jonghe BC. Incretins and amylin: neuroendocrine communication between the gut, pancreas, and brain in control of food intake and blood glucose. Annu Rev Nutr 2014;34:237–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De la Fuente M, Bernaez I, Del Rio M, Hernanz A. Stimulation of murine peritoneal macrophage functions by neuropeptide Y and peptide YY. Involvement of protein kinase C. Immunology 1993;80:259–265. [PMC free article] [PubMed] [Google Scholar]
- 88.Ibrahim E, Diakonov I, Arunthavarajah D, et al. Bile acids and their respective conjugates elicit different responses in neonatal cardiomyocytes: Role of Gi protein, muscarinic receptors and TGR5. Sci Rep 2018;8:7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Katsuma S, Hirasawa A, Tsujimoto G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun 2005;329:386–390. [DOI] [PubMed] [Google Scholar]
- 90.Lo SH, Cheng KC, Li YX, Chang CH, Cheng JT, Lee KS. Development of betulinic acid as an agonist of TGR5 receptor using a new in vitro assay. Drug Des Dev Ther 2016;10:2669–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Keitel V, Go€rg B, Bidmon HJ, et al. The bile acid receptor TGR5 (Gpbar-1) acts as a neurosteroid receptor in brain. Glia 2010;58:1794–1805. [DOI] [PubMed] [Google Scholar]
- 92.Pathak P, Liu H, Boehme S, et al. Farnesoid X receptor induces Takeda G-protein receptor 5 cross-talk to regulate bile acid synthesis and hepatic metabolism. J Biol Chem 2017;292:11055–11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pathak P, Xie C, Nichols RG, et al. Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology 2018. 10.1002/hep.29857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang XX, Edelstein MH, Gafter U, et al. G protein-coupled bile acid receptor TGR5 activation inhibits kidney disease in obesity and diabetes. J Am Soc Nephrol 2016;27:1362–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao S, Ji XF, Li F, et al. Aberrant DNA methylation of G-protein-coupled bile acid receptor Gpbar1 predicts prognosis of acute-on-chronic hepatitis B liver failure. J Viral Hepat 2015;22:112–119. [DOI] [PubMed] [Google Scholar]
- 96.Han LY, Fan YC, Mu NN, et al. Aberrant DNA methylation of G-protein-coupled bile acid receptor Gpbar1 (TGR5) is a potential biomarker for hepatitis B virus associated hepatocellular carcinoma. Int J Med Sci 2014;11:164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kwong E, Li Y, Hylemon PB, Zhou H. Bile acids and sphingosine-1-phosphate receptor 2 in hepatic lipid metabolism. Acta Pharm Sin B 2015;5:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nagahashi M, Yuza K, Hirose Y, et al. The roles of bile acids and sphingosine-1-phosphate signaling in the hepatobiliary diseases. J Lipid Res 2016;57: 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aoki M, Aoki H, Ramanathan R, Hait NC, Takabe K. Sphingosine-1-Phosphate signaling in immune cells and inflammation: Roles and therapeutic potential. Mediat Inflamm 2016;2016:8606878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nagahashi M, Takabe K, Liu R, et al. Conjugated bile acid-activated S1P receptor 2 is a key regulator of sphingosine kinase 2 and hepatic gene expression. Hepatology 2015;61:1216–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Webster CR, Anwer MS. Hydrophobic bile acid apoptosis is regulated by sphingosine-1-phosphate receptor 2 in rat hepatocytes and human hepato-cellular carcinoma cells. Am J Physiol Gastrointest Liver Physiol 2016;310: G865–G873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu R, Zhao R, Zhou X, et al. Conjugated bile acids promote cholangiocarcinoma cell invasive growth through activation of sphingosine 1-phosphate receptor 2. Hepatology 2014;60:908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]


