Abstract
Schizophrenia poses an evolutionary-genetic paradox as it exhibits strongly negative fitness effects (early mortality and decreased fecundity), yet it persists at a prevalence of approximately 1% worldwide. Evidence from several studies have suggested that schizophrenia is evolved and maintained in part as a maladaptive byproduct of recent positive selection and adaptive evolution in human beings. However, inconsistent results have been also proposed, challenging the recent positive selection theory to explain the high population frequency of schizophrenia-associated alleles. Here, we used public domain data to locate signatures of positive selection based on genetic diversity, derived allele frequency, differentiation between populations, and long haplotypes at schizophrenia-associated single nucleotide polymorphisms (SNPs) and randomly selected SNPs (as negative controls). We found evidence for positive selection at 10 out of the 105 schizophrenia-associated SNPs, while 5 of these SNPs involved positive selection for the protective allele. Taken together, the absence of widespread positive selection signals at the schizophrenia-associated SNPs, along with the fact that half of the positive selection favored the protective allele, provide little evidence supporting the positive selection theory in schizophrenia.
Keywords: schizophrenia, positive selection, GWAS, evolution
Introduction
Schizophrenia is a severe and debilitating mental disorder, ie, typically characterized by hallucinations and delusions (often involving language), thought disorders, and higher-order cognitive dysfunctions.1,2 Schizophrenia is both highly heritable and polygenic, with an estimated heritability of between 60% and 80%,3 and evidence from recent genome-wide association studies (GWAS) suggested that common alleles may explain between one-third and one-half of the genetic variance in liability.4
Individuals with schizophrenia show decreased reproductive success relative to healthy people and increased mortality due to natural and unnatural causes.5,6 Given that genetic variants associated with reduced fitness are under negative selection pressure, those variants that predispose to schizophrenia should be eliminated from the gene pool. However, schizophrenia is surprisingly prevalent, affecting approximately 1% of the world population, giving rise to the well-known yet still unresolved evolutionary paradox.7–10
As schizophrenia traits appear to be human-specific, several studies have suggested that schizophrenia may have arisen as an unfavorable but inevitable (by-) product of human brain evolution.11,12 According to this view, the high prevalence of schizophrenia is explained by positive selection for genetic variants that are beneficial to higher-order cognitive functions but also predispose risk to schizophrenia. For instance, Crow proposed that positive selection for cerebral flexibility during human evolution allowed for the emergence of language; meanwhile, cerebral flexibility was associated with variation in psychological functioning, which may result in personality disorders observed in schizophrenia.13 In support of this hypothesis, several studies have reported enrichment of genomic markers for positive selection and human-specific evolution in schizophrenia, given advances from recent studies of common genetic variation of schizophrenia.14–17 However, the hypothesis cannot explain why this extreme and maladaptive phenotype persists or from a genetic perspective. Furthermore, as suggested in a more recent study, SNPs under positive selection, as indexed by the composite likelihood ratio (CLR) statistic, are depleted for association with schizophrenia, and no significant relationship between h2SNP and other positive selection or Neanderthal introgression measures (table 1 in the original manuscript) was found after correction for multiple testing, challenging the selective advantages of schizophrenia risk alleles.18 It remains inconclusive whether recent positive selection was enriched within schizophrenia-associated SNPs.
Table 1.
Positive selection signals of each SNP
| No.a | rsID | Chr | Pos.b | P-valc | Tajima’s D | |DDAF| | FST | XPCLR | |UiHS| | |UXPEHH| | Sum | Protective or risk |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
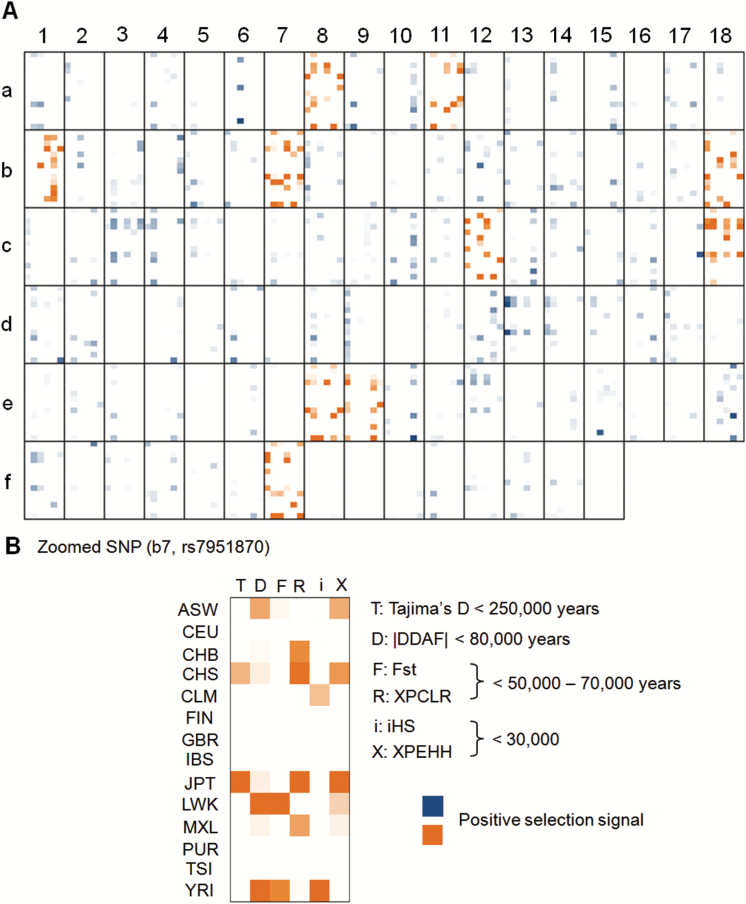
| c7 | rs7951870 | 11 | 46 373 311 | 8.25 E-11 | 2 | 7 | 3 | 5 | 2 | 5 | 24 | Risk |
| b18 | rs11223651 | 11 | 133 841 034 | 2.42 E-11 | 7 | 0 | 3 | 2 | 5 | 1 | 18 | Risk |
| f7 | rs7801375 | 7 | 131 567 263 | 2.26 E-08 | 4 | 4 | 3 | 3 | 1 | 3 | 18 | Protective |
| c18 | rs4766428 | 12 | 110 723 245 | 7.09 E-10 | 3 | 6 | 0 | 5 | 0 | 3 | 17 | Risk |
| e8 | rs9922678 | 16 | 9 946 319 | 6.72 E-09 | 3 | 6 | 1 | 1 | 4 | 1 | 16 | Protective |
| c12 | rs11577346 | 1 | 150 066 530 | 1.47 E-09 | 5 | 0 | 4 | 3 | 1 | 2 | 15 | Protective |
| e9 | rs2068012 | 14 | 30 190 316 | 4.14 E-08 | 6 | 0 | 0 | 0 | 7 | 2 | 15 | Risk |
| a8 | rs4391122 | 5 | 60 598 543 | 1.73 E-13 | 3 | 6 | 0 | 3 | 1 | 2 | 15 | Risk |
| a11 | rs4702 | 15 | 91 426 560 | 2.30 E-12 | 0 | 4 | 0 | 3 | 4 | 4 | 15 | Protective |
| b1 | rs7893279 | 10 | 18 745 105 | 3.56 E-11 | 0 | 0 | 1 | 4 | 9 | 1 | 15 | Protective |
Note: Five SNPs were risk and the other 5 SNPs are protective from schizophrenia. Each number refers to total positive selection signals of all 14 populations for each SNP.
|DDAF|, derived allele frequency; FST, fixation index; XPCLR, cross-population composite likelihood ratio; |UiHS|, integrated haplotype score; |UXPEHH|, cross-population extended haplotype homozygosity.
aTen SNPs exceeded the threshold of 15 positive selection signals to be statistically significant.
bThe UCSC hg19/NCBI build 37 position of the SNP.
cThe P-values of the SNP from the PGC2-SCZ GWAS.
In the present study, we first provide a comprehensive search for signatures of positive selection at schizophrenia-related loci from the Psychiatric Genomics Consortium schizophrenia GWAS (PGC, 36 989 schizophrenia cases and 113 075 controls) (supplementary tables S1 and S2),19 which were then compared with randomly selected SNPs matched for similar recombination rate and allele frequencies (supplementary table S2). Totally, 6 statistical tests of positive selection in 14 populations from the 1000 Genomes Project were employed, including Tajima’s D, difference of derived allele frequency (|DDAF|), fixation index (Wright’s FST), cross-population composite likelihood ratio (XPCLR), integrated haplotype score (|UiHS|) and cross-population extended haplotype homozygosity (|UXPEHH|).20 For replication, we performed similar analyses using data from one more recent schizophrenia GWAS meta-analysis (40 675 cases and 64 643 controls), which identified 50 novel-associated loci and 145 loci in total.18
Methods
Schizophrenia-Associated SNPs, Negative and Positive Control SNPs
Schizophrenia-associated SNPs are from 2 data sets: the PGC schizophrenia GWAS (108 independent loci)19 and one more recent schizophrenia GWAS meta-analysis by Pardiñas et al (145 loci).18 Notably, these 2 GWASs have largely overlapping samples. Meanwhile, randomly selected SNPs matched for their recombination and allele frequencies to the schizophrenia-associated SNPs were selected using the Ensembl database.
To evaluate the reliability of the statistical scores in the dbPSHP database and the statistical methods used in the present study, we employed several positive control SNPs, including the lactase gene (LCT, rs4988235, rs2236783, and rs2322659),21SLC24A5 (rs16891982) and SLC45A2 (rs1426654).22,23
Variant Genotype Properties and Positive Selection Statistical Terms from dbPSHP
The SNP properties included derived allele (DA), ancestral allele (AA), derived allele frequency (DAF), ancestral allele frequency (AAF), Hardy–Weinberg equilibrium (HWE), and heterozygosity (HET) in 14 populations from the 1000 Genomes (supplementary table S2).20 The positive selection statistics were seen in the supplementary materials.
Statistical Methods
For each of the 6 positive selection statistical terms, we counted the total positive selection signals in 2 groups (schizophrenia-associated and negative control SNPs) separately. Because we had 14 populations and 105 SNPs in each group, the maximum number of positive selection signals that could occur for any given test was 105 × 14 = 1470. Individual comparison of frequency for each population was also performed using Pearson Chi-square test. P < .05 was set to be statistically significant.
To determine the SNPs undergoing recent positive selection, we used a Bayesian conjugate beta-binomial analysis to model the distribution of positive signals in the negative control SNPs using the WinBUGS program, as described in detail in previous studies.24 Because we had 14 populations and 6 tests for each SNP, the maximum number of positive selection signals that could occur for any given SNP was 6 × 14 = 84. The actual number of positive signals in the negative control SNPs ranged from 0 to 26 (supplementary figure S1). We set the parameter of a beta distribution (prior), knowing that the mean probability of success across all the tests and populations in the negative control SNPs was .103, and then defined the 95% upper confidence limit (one-tail) for positive selection. The minimum value of the 95% upper confidence limit was 15 positive selection signals across all the tests and populations.
Results
We performed 6 tests to detect potential positive selection throughout our evolutionary history.20,25 The putative positive selection signals were defined using the following criteria: Tajima’D < 0, |DDAF| > 0.20, FST > 0.05, XPCLR > 5, |iHS| > 1.50, and |UXPEHH|> 1, as recommended in a previous study.20
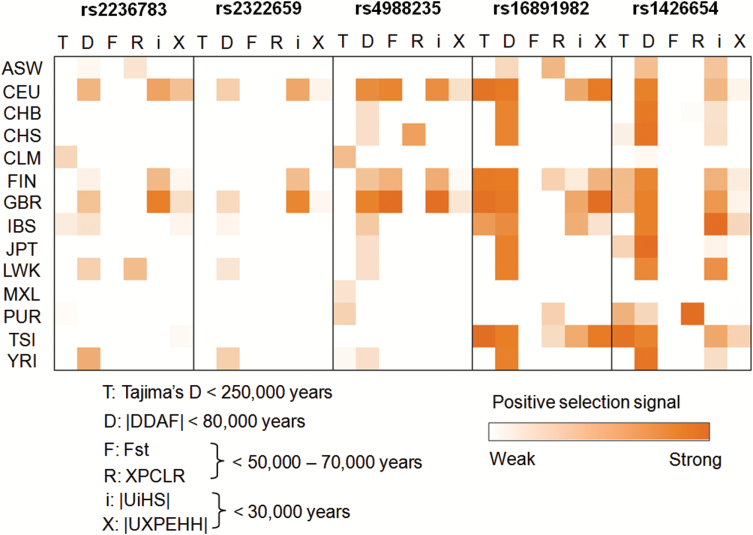
To evaluate the reliability and accuracy of the positive selection signals obtained from the dbPSHP database, we employed 5 well-known SNPs under strong positive selection as positive control SNPs. They were rs4988235, rs2236783, and rs2322659 (from the LCT gene locus) and rs16891982 (SLC24A5) and rs1426654 (SLC45A2) (supplementary table S2).21–23 The statistical scores of 6 tests for each SNP in 14 populations from the 1000 Genomes Phase I (CHB, CHS, JPT, CEU, TSI, GBR, IBS, FIN, ASW, LWK, YRI, MXL, CLM, and PUR) were summarized in supplementary table S3. All these SNPs showed multiple positive selection signals in diverse populations. Especially, 4 out of the 5 SNPs (except rs2322659) had total positive selection signals ≥21 (the maximum positive selection signals that could occur for a given SNP was 6 tests × 14 populations = 84), indicating that these 6 tests can efficiently indicate the positive selection signals (figure 1). We also downloaded the statistical value of the 6 positive selection tests in 14 populations for 105 schizophrenia-associated SNPs and 105 randomly selected SNPs (supplementary table S3).
Fig. 1.
Positive selection heatmap for 5 positive control SNPs. Each single box with border represents a single SNP. Within each box, the horizontal row shows the statistical value of 6 positive selection tests and the column represents 14 different populations. For a given SNP, the maximum positive selection signals that could occur was 6 tests × 14 populations = 84. The red blocks indicated the positive selection signals observed in a specific test-population combination. The detailed statistical results were provided in supplementary table S3. The numbers following each test (ie, <250 000 years) represent the approximate time before the present over which the test can detect positive selection.
Reduction in Genetic Diversity (Tajima’s D, <250 000 years)
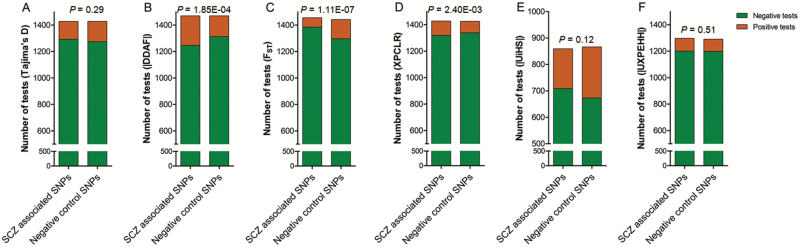
Tajima’s D measures the difference between 2 estimators of the population mutation rate (θw and π), and negative values of Tajima’s D indicate an excess of low frequency alleles, which may result from population expansion or positive selection.26 Totally, 153 positive selection signals were found in 1470 situations (105 loci × 14 populations) for schizophrenia-associated SNPs, while 135 positive signatures were found for the negative controls (χ2 = 1.13, P = .29) (figure 2A and supplementary figure S2).
Fig. 2.
Histogram showing the percentage of positive selection signals in 6 statistical terms in both schizophrenia-associated and negative control SNPs. For each statistical term, there are a maximum of 1470 tests (105 loci × 14 populations) in either the schizophrenia-associated SNPs or negative control SNPs. For some tests, the statistical results were missing from the dbPSHP database. SCZ, schizophrenia.
Considering that the majority of samples from the PGC2 schizophrenia GWAS are of European ancestry, we hypothesized that positive selection signals might be enriched in Europeans but not in other populations. We therefore performed analysis only in European populations, including CEU, FIN, GBR, IBS, and TSI (supplementary table S4). Significantly lower frequency of positive selection signals for Tajima’s D was observed in schizophrenia-associated SNPs (33 positive tests out of 510 situations) compared with the negative control SNPs (71 in 510 tests) (χ2 = 15.46, P = 8.42 E-05), suggesting no widespread positive selection (<250 000 years) at alleles linked to schizophrenia when compared with random SNPs in the genome.
Difference of Derived Allele Frequency (<80 000 years)
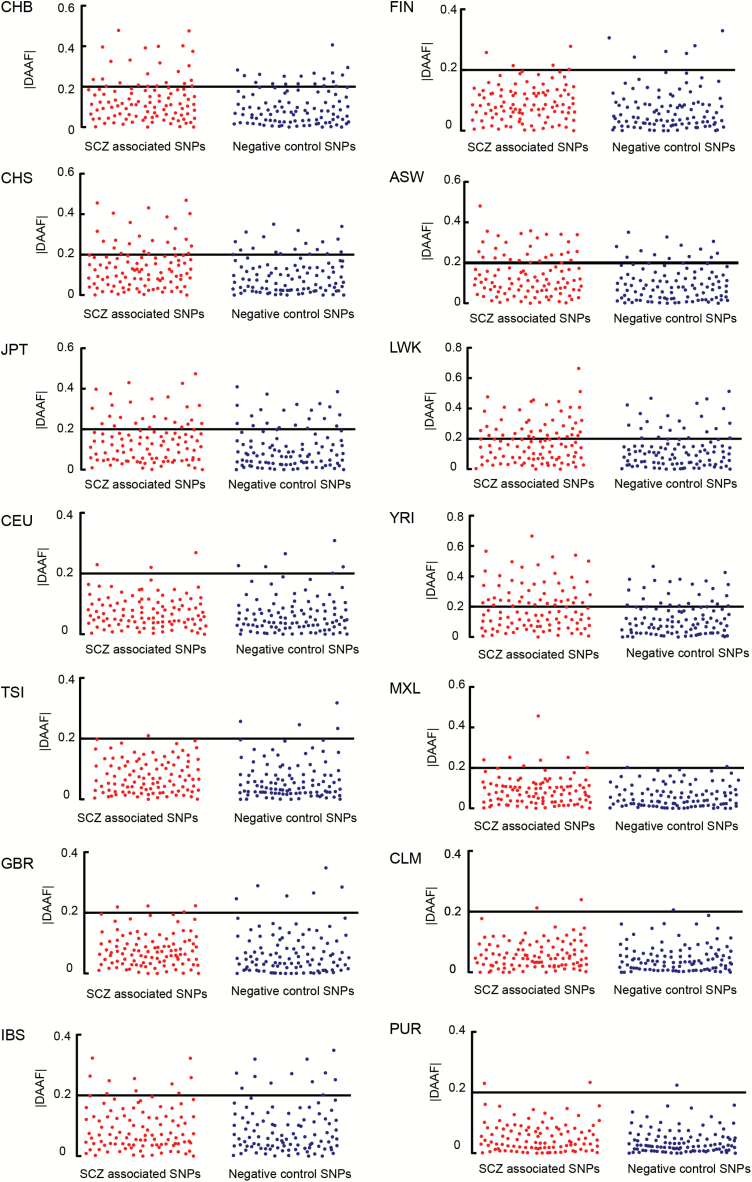
|DDAF| scores were retrieved from the dbPSHP database for all 14 populations. Significant higher frequency of positive selection signals was observed in schizophrenia-associated SNPs when compared with negative control SNPs (χ2 = 13.97, P = 1.85 E-04), which survived for multiple test correction (PCorrected = 1.11 E-03, n = 6 tests) (figures 2B and 3). We then performed individual analysis for each population, and enrichment of positive selection signals was observed in schizophrenia-associated SNPs in 3 populations of African Ancestry, including ASW (χ2 = 7.62, P = 5.77 E-03, PCorrected = .08, n = 14 populations), LWK (χ2 = 6.55, P = .01, PCorrected = .15), and YRI (χ2 = 11.26, P = 7.93 E-04, PCorrected = 0.15). Although a total of 53 schizophrenia-associated SNPs showed |DDAF| > 0.20, in at least 1 of the 3 African populations (supplementary table S3), the derived allele in 25 of these 53 SNPs (47.17%) are of protective rather than risk effect according to the original PGC2-SCZ GWAS (P = .13, two-tailed binomial test). Furthermore, there was no significant difference of positive selection signals for |DDAF| between schizophrenia-associated loci and negative controls in European continental populations (χ2 = 1.89, P = .17) (supplementary table S4).
Fig. 3.
Results of the |DDAF| Test for 14 populations of 1000 Genomes Project. In the left panel, the x-axis is a list of schizophrenia-associated SNPs, while in the right panel, the x-axis refers to the negative control SNPs. The y-axis is the |DDAF| value. The bold x-axis is the threshold for the positive selection (|DDAF| > 0.2). All the statistical tests in 14 populations are detailed in supplementary table S3.
Differentiation between populations (FST and XPCLR, <50 000 to 75 000 years)
Barreiro et al found that global, inter-continental human population differentiation has been largely shaped by natural selection, with negative selection reducing population differentiation and positive selection increasing population differentiation, which primarily affects SNPs in the regulatory regions.27 Significantly lower frequency of positive selection signals for Wright’s FST was observed in schizophrenia-associated SNPs (71 out of 1456 situations) compared with the negative control SNPs (145 in 1442 situations) (χ2 = 28.17, P = 1.11E-07), which survived for multiple test correction (PCorrected = 6.66 E-07, n = 6 tests) (figure 2C and supplementary figure S3). More importantly, no significant difference was observed between schizophrenia-associated SNPs and negative controls in European populations for Wright’s FST (P > .05) (supplementary table S4).
We also employed the XPCLR score to detect the population differentiation, which can model the joint allele frequency spectrum under selection without being affected by population expansion. Limited evidence of positive selection was observed at schizophrenia-associated loci (χ2 = 2.64, P = .10) (figure 2D and supplementary figure S4). Considering that the XPCLR metric largely depends on the reference population used, we performed similar analyses in each continental population separately. No evidence of enrichment of positive selection signals among schizophrenia-associated SNPs was observed when compared with negative controls in African, American, or European populations (all P > .05). Although significant higher frequency of positive selection signals was observed in schizophrenia-associated SNPs in Asian populations (χ2 = 9.24, P = 2.40 E-03), the derived allele in 13 of these 27 SNPs (48.15%) are of protective rather than risk effect according to the original PGC2-SCZ GWAS (P = .33, two-tailed binomial test). Taken together, no preference of recent positive selection (<50 000 years, indicated by population differentiation) was observed in schizophrenia risk loci.
Long-range haplotypes (<30 000 years)
The long-range haplotypes (LRH) test relies on the relationship between the allele’s frequency and the linkage disequilibrium (LD) between that allele and the loci that surround it. Positive selection will result in rapid spread of young alleles, which was thus surrounded by long blocks of LD.25 We applied the UiHS score to measure the haplotype homozygosity extension.28 In addition, the |UXPEHH| statistics (a haplotype-based test that compares 2 population samples) was used, of which the power is increased for alleles that have suffered differential selective pressures since those populations diverged.29 No significant difference of frequency of positive selection signals was observed between schizophrenia-associated SNPs and negative control SNPs for either the |UiHS| (χ2 = 2.486, P = .12; figure 2E and supplementary figure S5) or |UXPEHH| statistics (χ2 = 0.43, P = .51; figure 2F and supplementary figure S6). In European populations, even lower frequency of positive signals regarding |UiHS| was found in schizophrenia-associated SNPs compared with negative controls (supplementary table S4). In consistence with our result, Pardiñas et al used partitioned linkage disequilibrium score regression (LDSR) to test the relationship between schizophrenia-associated alleles and SNP-based signatures of natural selection and found no significant relationship between h2SNP and positive selection measures (iHS, XPEHH, and 2 others).18
In order to increase the statistical power, we combined all the situations from 6 statistical terms together and compared the overall positive selection signals between schizophrenia-associated SNPs and negative control SNPs (the maximum number of tests for each group is 105 SNPs × 14 populations × 6 tests = 8820). Out of the 7924 tests, 804 showed possible positive selection signals for the schizophrenia-associated SNPs (10.2%), which was comparable with the occurrence of positive selection signals (10.3%) among the randomly selected SNPs (χ2 = 0.05, P = .82), further suggesting there was no enrichment of overall wide-spread positive selection at schizophrenia risk loci.
To identify the SNPs that might be under recent positive selection, we built an expected distribution of positive tests for each SNP based on the frequency spectrum of positive selection signals observed in negative control SNPs.24 Assuming that these randomly selected SNPs were not under positive selection, this modeling gave the one-tailed upper 5% confidence limit of 15–17 positive selection signals (the maximum number of positive selection signals that could occur for any given SNP was 6 tests × 14 populations = 84). Using the threshold of ≥15 positive selection signals, 11 out of 105 randomly chosen SNPs might be under positive selection (supplementary table S5 and supplementary figure S7). Among the 105 schizophrenia-associated SNPs, we found 10 SNPs with total positive selection signals ≥15, which might be under positive selection (supplementary table S5 and figure 4).
Fig. 4.
Heatmap of positive selection signals for 105 schizophrenia-associated SNPs. Each single box with border represents a single SNP (detailed information as figure 3). The coordinate of each SNP encoded by the horizontal number and vertical letter can be found in supplementary table S5.
Considering that all the index SNPs from the PGC2 GWAS are causative variants, we then collected all the credible causal SNPs (4212 SNPs) provided online for further analysis (https://www.med.unc.edu/pgc/). Given that the majority of samples used in the original manuscript are of Caucasian ancestry, we restricted our analyses to European populations. Positive selection statistics were available for 1727 out of these 4212 SNPs in European populations (supplementary table S6). Significant enrichment of positive selection signals was observed in schizophrenia-associated SNPs when compared with negative control SNPs for the |DDAF| statistics (χ2 = 78.53, P = 4.73 E-18) (supplementary table S7). However, a total of 217 derived alleles among the 416 SNPs (|DDAF| > 0.2) are protective for schizophrenia susceptibility (P = .38, two-tailed binomial test), suggesting there was no enrichment of widespread positive selection at schizophrenia risk loci.
In order to extend our analyses to even older time frame, we included another positive selection statistics, Extended Lineage Sorting (ELS, <650 000 years), which models explicitly the longer regions produced under selection and includes the fixed differences between archaic and modern human genomes as an additional source of information.30 Comparison was restricted in African populations because archaic introgression in non-African populations could mask events of positive selection by re-introducing ancestral alleles. As reported in the original manuscript, 81 candidate regions of positive selection were detected when requiring a minimum genetic length supported by 2 independent recombination maps.30 However, no individual SNP of either P < 5.0E-08 (totally 12 897 SNPs) or P < 5E-06 (totally 29 598) was located among these 81 candidate regions, compared with 9607 out of the 9 444 230 tested SNPs falling within the candidate regions.19 Taken together, there was no widespread positive selection signatures in common risk alleles associated with schizophrenia.
More recently, Pardiñas et al performed one even larger GWAS and identified 145 independent schizophrenia-related loci.18 For replication of our preliminary results, we performed similar analyses using data from this schizophrenia GWAS. Positive selection statistics were available for 141 out of these 145 loci (supplementary table S8). Compared with randomly selected control SNPs, significant enrichment of positive selection signals was observed in schizophrenia-associated SNPs for the |DDAF| statistics (324 positive signals out of 1935 tests for schizophrenia-associated SNPs and 189 positive signals from 1903 situations for the negative controls, χ2 = 38.45, P = 5.61E-10) (supplementary table S9). Further analysis showed that the derived alleles of 52 out of these 92 SNPs exhibiting positive selection signals in at least one population are protective from schizophrenia susceptibility risk (P = .60, two-tailed binomial test). Besides, no significant increase of positive selection signals were observed for each of the 6 statistics in schizophrenia-related loci when compared with the negative controls in European populations (supplementary table S9).
Discussion
From our analyses, the absence of widespread positive selection signals at the schizophrenia-associated SNPs, along with the fact that part of the positive selection favored the protective allele, provide little evidence supporting the positive selection theory in schizophrenia, which was consistent with several previous studies.16,18 Through employment of partitioned LDSR to test the relationship between schizophrenia-associated alleles and SNP-based signatures of natural selection, Pardiñas et al found no significant relationship between h2SNP and positive selection as well.18 Interestingly, they found a significant depletion of h2SNP in SNPs subject to positive selection as indexed the CLR statistic, which was commonly used for detection of population differentiation.18 Supporting it, significantly lower frequency of positive selection signals was also observed in schizophrenia-associated SNPs when compared to negative control SNPs by using another similar index, Wright’s FST (figure 2C and supplementary figure S3). Similarly, Xu et al reported that genes associated with schizophrenia are under purifying selection, but not positive selection, in spite of the observation that genes near human accelerated regions (HARs) conserved in nonhuman primates (pHARs) are enriched for schizophrenia-associated loci.16
In several other studies, different positive selection metrics have been applied to detect selective sweeps among schizophrenia-associated variants.11,14,15 One study reported nominal positive correlation between schizophrenia associations and the hierarchical boosting score, which, however, didn’t survive multiple testing correction.15 Another study observed a negative correlation between schizophrenia associations and metrics indicative of a Neanderthal selective sweep as evidence for positive selection in schizophrenia.14 It is worth to note that it remains unknown whether the positive selection favored the risk alleles or protective alleles in schizophrenia in these studies. In other words, if positive selection was contributed enormously by the protective alleles, but not the risk alleles, the “positive selection” theory underlying schizophrenia might be unreliable.
Indeed, classic selective sweeps originating from positive selection is not widespread in human beings and it favors nonsynonymous substitutions, especially novel protein structures.31 Instead, polygenic adaptation, the co-occurrence of many subtle allele frequency shifts at many loci, has been proven to be common for complex traits in humans and thus remains an intriguing possibility in schizophrenia.32,33 In addition, other hypotheses also explain, maybe not completely, why schizophrenia risk genes are prevalent in the population, including balancing selection, whereby multiple alleles may be maintained in the gene pool if the genotypes are under different selection pressure34; hitchhiking, whereby risk-associated alleles are maintained by their linkage to positively selected alleles14; epigenetic variation as an evolutionary adaptation to environments35; and theories that attribute these effects to rare variants.9
The main limitation in the current study is that we used the index SNPs of risk loci from the PGC2 schizophrenia GWAS to perform the tests, which, however, may not be the causative variants. The extent of this problem will depend on the extent of linkage disequilibrium between the index and causal SNPs, which for all of these targets is unknown but is presumably high or else the index SNPs would not emerge. An alternative way is to use all the schizophrenia associations with specific threshold of P-value, as suggested in previous studies.14–16,18 We acknowledged that using the overall associations can undoubtedly increase the statistical power; however, it may also inflate the false positive rate due to inclusion of nonfunctional variants (either less significant or in LD with causative variants). The second limitation is that all the 105 SNPs together explain only a small portion of the variance in schizophrenia, and we can’t preclude the possibility that the other risk variants might be under strong positive selection. The third limitation is that the statistics used in the present study are principally able to detect “hard” selection sweeps, while it has been suggested that positive selection may proceed on a background of standing variation or a “soft” sweep. Fourth, although the Bayesian modeling has been widely used to model the distribution of positive signals in another set of negative control SNPs,24 we acknowledged that there may be other and probably better statistical analyses suitable for this case, and comparison between different statistical models are warranted in future studies.
Funding
This work was supported by Sichuan Science and Technology Program (2019YFH0137 to Dr Yao); the Fundamental Research Funds for the Central Universities (No. 2682018CX38 to Dr Rao) and Southwest Jiaotong University (10.13039/501100004536 to the central University colleague).
Supplementary Material
Acknowledgments
The authors have declared that there are no conflicts of interest in relation to the subject of this study. We acknowledge Prof. Stéphane Peyrégne, at Max Planck Institute for Evolutionary Anthropology, for valuable information regarding the ELS statistics.
References
- 1. Brüne M. Schizophrenia-an evolutionary enigma? Neurosci Biobehav Rev. 2004;28(1):41–53. [DOI] [PubMed] [Google Scholar]
- 2. Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60(12):1187–1192. [DOI] [PubMed] [Google Scholar]
- 3. Lichtenstein P, Yip BH, Björk C, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373(9659):234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Purcell SM, Moran JL, Fromer M, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Power RA, Kyaga S, Uher R, et al. Fecundity of patients with schizophrenia, autism, bipolar disorder, depression, anorexia nervosa, or substance abuse vs their unaffected siblings. JAMA Psychiatry. 2013;70(1):22–30. [DOI] [PubMed] [Google Scholar]
- 6. Svensson AC, Lichtenstein P, Sandin S, Hultman CM. Fertility of first-degree relatives of patients with schizophrenia: a three generation perspective. Schizophr Res. 2007;91(1–3):238–245. [DOI] [PubMed] [Google Scholar]
- 7. Burns JK. Psychosis: a costly by-product of social brain evolution in Homo sapiens. Prog Neuropsychopharmacol Biol Psychiatry. 2006;30(5):797–814. [DOI] [PubMed] [Google Scholar]
- 8. Polimeni J, Reiss JP. Evolutionary perspectives on schizophrenia. Can J Psychiatry. 2003;48(1):34–39. [DOI] [PubMed] [Google Scholar]
- 9. Uher R. The role of genetic variation in the causation of mental illness: an evolution-informed framework. Mol Psychiatry. 2009;14(12):1072–1082. [DOI] [PubMed] [Google Scholar]
- 10. Pearlson GD, Folley BS. Schizophrenia, psychiatric genetics, and Darwinian psychiatry: an evolutionary framework. Schizophr Bull. 2008;34(4):722–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crespi B, Summers K, Dorus S. Adaptive evolution of genes underlying schizophrenia. Proc Biol Sci. 2007;274(1627):2801–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burns JK. An evolutionary theory of schizophrenia: cortical connectivity, metarepresentation, and the social brain. Behav Brain Sci. 2004;27(6):831–855; discussion 855. [DOI] [PubMed] [Google Scholar]
- 13. Crow TJ. Schizophrenia as the price that homo sapiens pays for language: a resolution of the central paradox in the origin of the species. Brain Res Brain Res Rev. 2000;31(2–3):118–129. [DOI] [PubMed] [Google Scholar]
- 14. Srinivasan S, Bettella F, Mattingsdal M, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium, The International Headache Genetics Consortium Genetic markers of human evolution are enriched in schizophrenia. Biol Psychiatry. 2016;80(4):284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Polimanti R, Gelernter J. Widespread signatures of positive selection in common risk alleles associated to autism spectrum disorder. PLoS Genet. 2017;13(2):e1006618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu K, Schadt EE, Pollard KS, Roussos P, Dudley JT. Genomic and network patterns of schizophrenia genetic variation in human evolutionary accelerated regions. Mol Biol Evol. 2015;32(5):1148–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Srinivasan S, Bettella F, Hassani S, et al. Probing the association between early evolutionary markers and schizophrenia. PLoS One. 2017;12(1):e0169227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pardiñas AF, Holmans P, Pocklington AJ, et al. ; GERAD1 Consortium:; CRESTAR Consortium:; GERAD1 Consortium; CRESTAR Consortium; GERAD1 Consortium; CRESTAR Consortium Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schizophrenia Working Group of the Psychiatric Genomics. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li MJ, Wang LY, Xia Z, Wong MP, Sham PC, Wang J. dbPSHP: a database of recent positive selection across human populations. Nucleic Acids Res. 2014;42(Database issue):D910–D916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bersaglieri T, Sabeti PC, Patterson N, et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74(6):1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lamason RL, Mohideen MA, Mest JR, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310(5755):1782–1786. [DOI] [PubMed] [Google Scholar]
- 23. Wilde S, Timpson A, Kirsanow K, et al. Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc Natl Acad Sci USA. 2014;111(13):4832–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang G, Speakman JR. Analysis of positive selection at single nucleotide polymorphisms associated with body mass index does not support the “Thrifty Gene” hypothesis. Cell Metab. 2016;24(4):531–541. [DOI] [PubMed] [Google Scholar]
- 25. Sabeti PC, Reich DE, Higgins JM, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–837. [DOI] [PubMed] [Google Scholar]
- 26. Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22(8):437–446. [DOI] [PubMed] [Google Scholar]
- 27. Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40(3):340–345. [DOI] [PubMed] [Google Scholar]
- 28. Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. [DOI] [PubMed] [Google Scholar]
- 30. Peyrégne S, Boyle MJ, Dannemann M, Prüfer K. Detecting ancient positive selection in humans using extended lineage sorting. Genome Res. 2017;27(9):1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vitti JJ, Grossman SR, Sabeti PC. Detecting natural selection in genomic data. Annu Rev Genet. 2013;47:97–120. [DOI] [PubMed] [Google Scholar]
- 32. Fu W, Akey JM. Selection and adaptation in the human genome. Annu Rev Genomics Hum Genet. 2013;14:467–489. [DOI] [PubMed] [Google Scholar]
- 33. Pritchard JK, Di Rienzo A. Adaptation - not by sweeps alone. Nat Rev Genet. 2010;11(10):665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shaner A, Miller G, Mintz J. Schizophrenia as one extreme of a sexually selected fitness indicator. Schizophr Res. 2004;70(1):101–109. [DOI] [PubMed] [Google Scholar]
- 35. Perrin MC, Brown AS, Malaspina D. Aberrant epigenetic regulation could explain the relationship of paternal age to schizophrenia. Schizophr Bull. 2007;33(6):1270–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.