Abstract
We integrate a biocompatible plant virus-based nanotechnology (tobacco mosaic virus, TMV) with S100A9-targeting peptides for its application in imaging and diagnosis of atherosclerosis. S100A9-targeted TMV nanoparticles exhibit remarkable specificity to S100A9 and targeting of atherosclerosis lesions in ApoE−/− mice.
Cardiovascular disease, including myocardial infarction and stroke, is one of the leading causes of death in developed countries (>500 000 deaths annually in the US alone), and it is the most common etiology for adult disability.1 One of the major underlying causes of cardiovascular diseases is atherosclerosis, in which plaques build up inside the arteries.1 Atherosclerosis development often remains asymptomatic until a clinical event such as heart attack or stroke occurs. Blood tests, body weight, and electrocardiograms indicate risk factors for atherosclerosis, but they are not directly indicative of the disease. Imaging approaches, including X-ray angiography,2 optical coherence tomography,3 and intravascular ultrasound4 can locate potential blockages within the arteries. Nevertheless, these techniques only detect luminal narrowing and intimamedia thickening, which do not lay a foundation for predicting atherosclerotic plaques with high risks. Plaque rupture and subsequent fatal cardiovascular events occur more often in vessels with mild-to-moderate luminal stenosis; the risk of a clinical event depends on the molecular composition of the lesion as opposed to plaque size and luminal stenosis.5 Therefore, direct characterization of the molecular composition of the plaques is required for accurate prediction of the likelihood of plaque rupture. Plaques can be differentiated as stable or vulnerable; the latter are characterized by increased infilteration of macrophages and T-cells (i.e., local inflammation) and thrombogenicity,5–8 and are prone to rupture. Only biopsy, followed by histological inspection, gives the molecular information needed for accurate diagnosis of plaques with high risks, but these methods are invasive and associated with significant risks. Therefore, there is a clinical need for non-invasive techniques to aid diagnosis of atherosclerosis.
To enable molecular targeting and high payload delivery of imaging agents, a number of bio/nanotechnologies have been developed; these include micelles,9,10 liposomes,11,12 and inorganic NPs,13,14 as well as protein and virus-based nanoparticles.15–18 These systems have been engineered to target various molecular signatures of plaques, such as vascular cell adhesion molecule 1 (VCAM-1) and intracellular adhesion molecule 1 (ICAM-1).17,19 Since vulnerable plaques have macrophage rich cores, therefore molecular imaging strategies have also explored to target macrophage scavenger receptor 1 (MSR-1) and CD36.20
In this work, we set out to develop a plant virus-based imaging agent targeting S100A9 (a.k.a myeloid-related protein 14). S100A9 is secreted from neutrophils, monocytes, and platelets.21,22 Also, S100A9 was found in atherosclerotic lesions prone to rupture.22 It was identified as one of the strongest predictors of acute myocardial infarction using transcriptional profiling of platelets from patients with acute coronary syndromes.21 Histology and immunoassays of carotid plaques from patients revealed strong association of S100A9 with rupture-prone atherosclerotic lesions compared to stable ones.23 Therefore, S100A9 can be used as a biomarker, indicative of vulnerable (i.e., rupture-prone) plaques, for diagnosis of atherosclerosis with high risk factors.
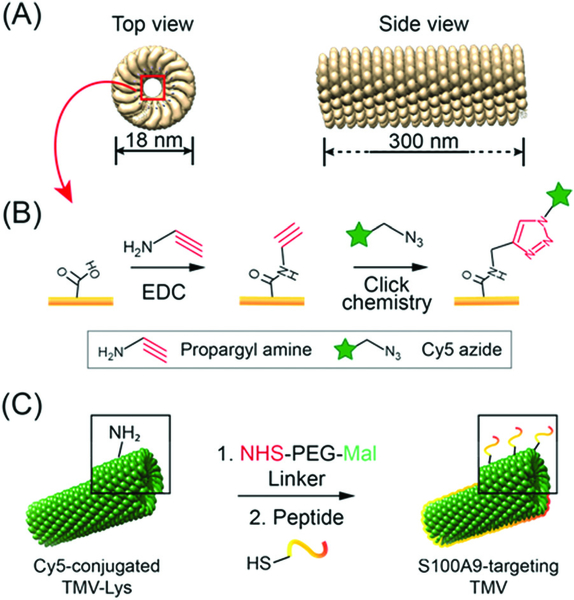
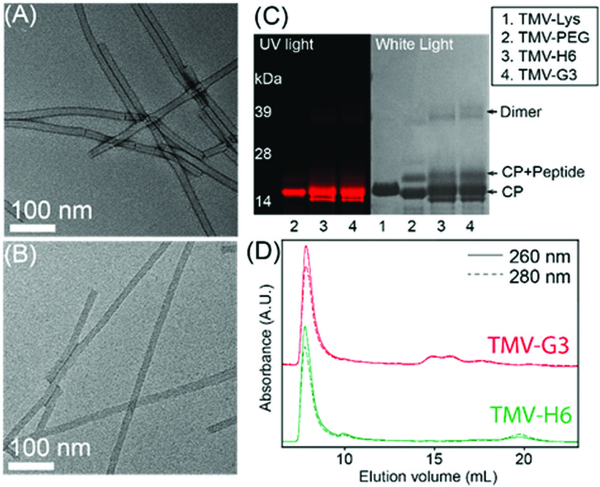
To target S100A9 in the plaques, we chose peptides with demonstrated high affinity toward the protein24 and tobacco mosaic virus (TMV) as a display nanotechnology. The nucleoprotein complex of TMV assembles into 300 × 18 nm hollow nanorods with 4 nm-wide central channel (Scheme 1.). The high aspect ratio shape of TMV provides an advantage for cardiovascular nanomedicine applications; non-spherical materials have increased vascular targeting properties (i.e., enhanced margination to vessel walls), and exhibit reduced clearance by the mononuclear phagocyte system.25–27 The conjugation chemistries on the interior and exterior surfaces are well-established and TMV nanoparticles have been engineered for imaging and drug delivery applications.3,4 TMV-lys (a Lys-added mutant of TMV, in the following denoted as TMV)28 was propagated in and purified from N. benthamiana plants using our established protocols.29 First, amine-reactive Cy5 dyes were conjugated to TMV’s Glu side chains located in the channel using carbodiimide coupling. Then S100A9-targeting, G3 and H6,24 and scrambled, ScG3 and ScH6, were coupled to the engineered Lys side chains at the exterior surface of TMV; Cys-terminated peptides were conjugated via mal-PEG-NHS linkers. The detailed conjugation procedure and the peptide sequence information are described in ESI† Presented in Fig. 1, Fig. S1, and S2 (ESI†) is the collective characterization of S100A9-targeted TMV nanoparticles. The conjugation of Cy5 molecules at the internal surface was confirmed in SDS-PAGE and UV-vis measurement (Fig. 1C and Fig. S2A, ESI†): ~65 Cy5 molecules were attached per TMV. Successful conjugation of the fluorophore was evident by presence of fluorescent coat protein (CP) bands in SDS-PAGE (Fig. 1C). TMV’s CPs have a molecular weight of 17 kDa and peptides (S100A9-specific and scrambled) and PEG molecules are approximately ~2000 Da. The band pattern is consistent with peptide/PEG conjugation (Fig. 1C and Fig. S6, ESI†). Densiometric analysis of the gel images using ImageJ software indicated that 30–35% of total CPs were conjugated with PEG and peptides. In FPLC, modified TMV nanoparticles showed the typical elution profile; intact TMV elutes from the Superose6 column at ~7.5 mL with 260 : 280 nm absorbance ratio of ~1.2 (Fig. 1D). Structural integrity of the TMV nanoparticles was further confirmed by TEM, showing high aspect ratio nanoparticles (Fig. 1A and B), and hydrodynamic size measurement via DLS, revealing that the average hydrodynamic size of TMV remains consistent before and after the peptide conjugation (Fig. S2B, ESI†). The yield of the preparation for S100A9-targeted TMV particles was typically around ~70% of the starting material, and TMV formulations were found to be stable at 4 °C up to 2 months without aggregation.
Scheme 1.
(A) Structure of TMV nanoparticles created by UCSF Chimera and PDB entry (3j06).‡ The side view image shows a partial TMV. (B and C) Illustration of the preparation of fluorescent, S100A9-targeted TMV nanoparticles: (B) internal modification with Cy5 molecules and (C) external modification with S100A9-targeting peptides.
Fig. 1.
(A and B) Transmission electron microscopy (TEM) images of TMV-H6 and TMV-G3. (C) SDS-PAGE of TMV nanoparticles before and after Coomassie staining under (left) UV and (right) white light, respectively. (D) Fast protein liquid chromatography (FPLC) of TMV nanoparticles.
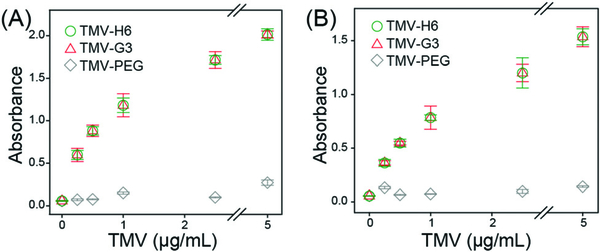
Prior to in vivo studies, we tested the S100A9-specificity of targeted (TMV-H6 and TMV-G3) and non-targeted TMV nanoparticles (TMV-PEG, TMV-ScH6, and TMV-ScG3) using in vitro assays as described in ESI.† Briefly, 96-well plates were coated with S100A9 and then incubated with serial dilutions of TMV formulations for 2 hours. The surface-bound TMV were further captured by using a polyclonal rabbit anti-TMV antibody and secondary horseradish peroxidase-labelled antibody, followed by 30 min colorimetric development using TMB. Fig. 2 shows the dose response plots of S100A9-targeted TMV and non-targeted TMV against human and mouse S100A9 (hS100A9 and mS100A9, respectively). Concentration-dependent binding to S100A9 is shown for the TMV-H6 and TMV-G3 with negligible non-specific binding of the TMV-PEG samples. The binding affinity was determined from the dose response plots (Table S2, ESI†): the estimated KD values of TMV-H6 and TMV-G3 are in the range of tenths of pM. Overall TMV-H6 and TMV-G3 exhibited similar binding affinities and their affinity to mS100A9 is ~2× higher than that to hS100A9. This follows our expectation since the peptides were isolated through a screen against mouse cells.24 However, their cross-reactivity to hS100A9 with high affinity should also be noted. This can be explained by the fact that the protein sequences of mS100A9 and hS100A9 are homologous as evidenced by basic local alignment tool (BLAST)30 analysis (Fig. S3, ESI†). Therefore, the TMV formulations not only provide a tool for preclinical investigation but may also enable transition to human trials without having to change the chemistry of the particles or peptides. Lastly, we compared in vitro specificity of targeted TMV to non-targeted TMV conjugated with scrambled peptides (Fig. S7, ESI†). The scrambled peptide conjugated TMV nanoparticles, TMV-ScH6 and TMV-ScG3, exhibited non-specific binding to S100A9 higher than TMV-PEG, but significantly low binding affinity than TMV-H6 and TMV-G3. Based on these data, we concluded that the peptide sequences are specific to S100A9 proteins and TMV nanoparticles with multivalent presentation of peptides enabled even higher affinities to the target protein.
Fig. 2.
In vitro binding assays of S100A9-targeted TMV nanoparticles for (A) mS100A9 and (B) hS100A9.
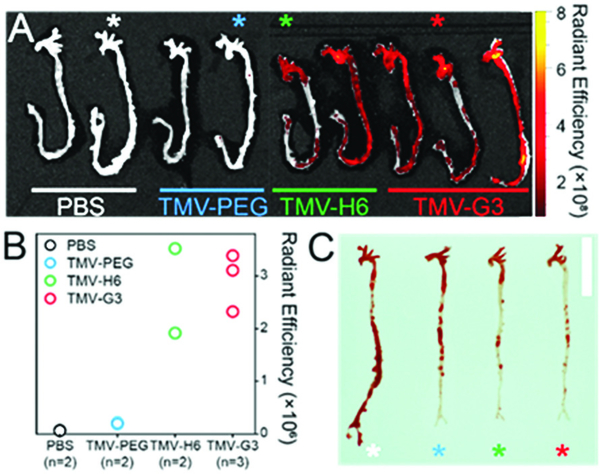
To assess the in vivo targeting capabilities of the engineered TMV probes to S100A9, we studied the well-established atherosclerosis model using apolipoprotein E-deficient (ApoE−/−) mice31 fed on a high fat/high cholesterol diet for at least 16 weeks to ensure the disease establishment.32 All animal studies were carried out under protocols approved by Case Western Reserve University’s IACUC. Mice were randomly assigned and grouped into 4 treatment arms: PBS (n = 2), TMV-PEG (n = 2), TMV-H6 (n = 2), and TMV-G3 (n = 3) at a dosage of 20 mg TMV per kg body weight. Three hours post intravenous administration, mice were sacrificed and aortas were harvested for ex vivo fluorescence imaging and Sudan IV staining establishment. Imaging and quantitative image analysis (Fig. 3A and B) demonstrated excellent binding of the targeted TMV-H6 and TMV-G3 formulation with little to no binding of the TMV-PEG particles – this matches the in vitro results. TMV-H6/G3 exhibited ~40 and ~14-fold increased fluorescence intensities compared to the PBS and TMV-PEG groups, respectively. Lesion development in the aortas was confirmed through staining with Sudan IV (Fig. 3C): significant atherosclerotic lesions (red areas) were established in all aortas. Thus, Fig. 3 supports that S100A9-targeted TMV nanoparticles preferentially accumulated in the atherosclerotic lesions while control particles shows negligible non-specific accumulation in the diseased tissue.
Fig. 3.
(A) Ex vivo fluorescence imaging of the aortas from ApoE−/− mice harvested 3 h post intravenous administration of TMV nanoparticles. The asterisks indicate the aortas used for Sudan IV staining in (C). (B) Quantitative analysis of ex vivo fluorescence images of (A); the total radiant efficiency was normalized to the area of interest. The radiant efficiency units are (p s−1 cm−2 sr−1)/(μW cm−2). (C) Image of aortas after Sudan IV staining. The vertical scale bar is 1 cm.
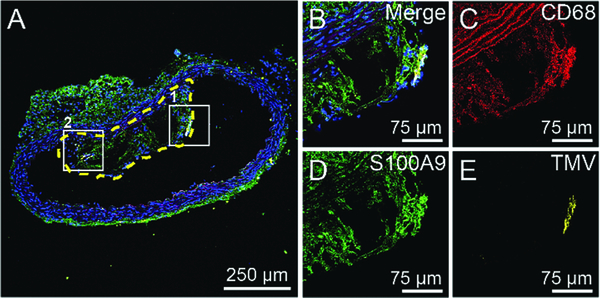
To determine molecular composition of the plaque and where within the S100A9-targeted TMV nanoparticles accumulated, we pursued confocal immunofluorescence imaging of cryosectioned aortas. The major components of atherosclerotic plaques consist of blood-borne inflammatory, immune cells (T cells and macrophages), foam cells (fat-laden macrophage), calcifying areas, etc.33,34 Therefore, 10-micron thin sections of aortas were stained for macrophages (CD68), S100A9 (mS100A9), and nuclei (DAPI). Detailed procedures are described in the ESI† Immunofluorescence images of sectioned and stained aortas from mice from all treatment arms are shown Fig. 4 and Fig. S4 (ESI†). From these images, it is evident that the atherosclerotic lesions formed in ApoE−/− mice are macrophage rich (CD68 positive, in red), which is an indication of vulnerability of the plaques.35 In addition, H&E staining of the sectioned aortas (Fig. S5, ESI†) confirmed the presence of necrotic cores in the atherosclerotic lesions. mS100A9 expression (in green) was confirmed throughout the lesions, attesting to the suitability of the target. mS100A9 expression colocalized with macrophage presence (CD68 staining). This can be attributed to S100A9 being found on several cell types including macrophages, foam cells, and monocytes in the plaque.34,36 Most importantly, our data show that S100A9-targeted TMV nanoparticles (yellow color) were colocalized with S100A9 while TMV-PEG nanoparticles were not detectable (boxed areas in Fig. 4 and Fig. S4, ESI†). TMV particles are located at the intima of the plaques. This is in agreement with a previous study in which VCAM-1-targeted TMV was localized at the surface of the plaques where VCAM-1 receptors are expressed in activated endothelial cells.17 The lack of non-specific accumulation of TMV-PEG and the observation that TMV only accumulates at the intima of the plaque supports molecular specific targeting vs. non-specific accumulation of the nanoparticles. This indicates that the fluorescent signals observed in the ex vivo fluorescence imaging of aortas resulted from specific targeting of S100A9 protein in atherosclerotic lesions by the targeted TMV nanoparticles.
Fig. 4.
Representative confocal fluorescence image of cryosectioned aorta. (A) Aorta section from an ApoE−/− mouse injected with TMV-H6. The yellow-dashed line highlights the plaque in the lumen of the aorta. (B–E) Larger images of box 1 in (A) under different fluorescent channels: merged image with DAPI (blue), CD68 (red), S100A9 (green), and TMV (yellow). The images of box 2 are shown in in Fig. S4A (ESI†).
Understanding of molecular structure of atherosclerosis can lead to new strategies in targeting plaques, and also enable risk stratification for future clinical cardiovascular events. In this paper, we confirmed the abundant expression of S100A9 in macrophage-rich atherosclerotic lesions of ApoE−/− model. The high levels of S100A9 in atherosclerotic plaques is strongly associated with athersoclerotic lesions with necrotic cores (i.e., rupture-prone plaques), which may result in fatal cardiovascular diseases.23 More importantly, S100A9-targeted TMV nanoparticles exhibited exceptional in vitro specificity to S100A9, as well as S100A9 present at the atherosclerotic lesions. Provided that TMV can be loaded with small drugs and protein therapies, as well as MRI contrast agents,37–40 therefore, TMV-based platform for targeting S100A9 may provide an avenue toward theranostic strategies for atherosclerosis. Our technology could be regarded as an alternative approach to the use of spherical nanomaterials specifically designed to be taken up by phagocytes for imaging of macrophage-burden in plaques. Currently, we are working on the development of more clinically relevant diagnostic method (e.g., magnetic resonance imaging, MRI) for atherosclerosis/cardiovascular diseases using the S100A9-targeted TMV nanoparticles.
Supplementary Material
Acknowledgments
This work was funded by the National Institute of Health (NIH R01HL137674). Authors thanks to the Wege Lab for providing the TMV-Lys clones.
Footnotes
Electronic supplementary information (ESI) available: Experimental details and extra figures and tables. See DOI: 10.1039/c8tb02276c
Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco.
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Lloyd-Jones D, Adams R, Brown T, Carnethon M, Dai S, De Simone G, Ferguson T, Ford E, Furie K. and Gillespie C, Circulation, 2010, 121, e46. [Google Scholar]
- 2.Puri R, Tuzcu EM, Nissen SE and Nicholls SJ, Int. J. Cardiol, 2013, 168, 670–679. [DOI] [PubMed] [Google Scholar]
- 3.Rieber J, Meissner O, Babaryka G, Reim S, Oswald M, Koenig A, Schiele TM, Shapiro M, Theisen K. and Reiser MF, Coron. Artery Dis, 2006, 17, 425–430. [DOI] [PubMed] [Google Scholar]
- 4.Hartmann M, Huisman J, Böse D, Jensen LO, Schoenhagen P, Mintz GS, Erbel R. and von Birgelen C, Eur. J. Echocardiogr, 2011, 12, 313–321. [DOI] [PubMed] [Google Scholar]
- 5.Virmani R, Burke AP, Farb A. and Kolodgie FD, J. Am. Coll. Cardiol, 2006, 47, C13–C18. [DOI] [PubMed] [Google Scholar]
- 6.Broisat A, Hernot S, Toczek J, De Vos J, Riou LM, Martin S, Ahmadi M, Thielens N, Wernery U. and Caveliers V, Circ. Res, 2012, 110, 927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Libby P, Ridker PM and Hansson GK, Nature, 2011, 473, 317. [DOI] [PubMed] [Google Scholar]
- 8.Rekhter MD, Cardiovasc. Res, 2002, 54, 36–41. [DOI] [PubMed] [Google Scholar]
- 9.Peters D, Kastantin M, Kotamraju VR, Karmali PP, Gujraty K, Tirrell M. and Ruoslahti E, Proc. Natl. Acad. Sci. U. S. A, 2009, 106, 9815–9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung EJ, Mlinar LB, Nord K, Sugimoto MJ, Wonder E, Alenghat FJ, Fang Y. and Tirrell M, Adv. Healthcare Mater, 2015, 4, 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maiseyeu A, Mihai G, Kampfrath T, Simonetti OP, Sen CK, Roy S, Rajagopalan S. and Parthasarathy S, J. Lipid Res, 2009, 50, 2157–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodside DG, Tanifum EA, Ghaghada KB, Biediger RJ, Caivano AR, Starosolski ZA, Khounlo S, Bhayana S, Abbasi S. and Craft JW, Sci. Rep, 2018, 8, 3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cormode DP, Roessl E, Thran A, Skajaa T, Gordon RE, Schlomka J-P, Fuster V, Fisher EA, Mulder WJ and Proksa R, Radiology, 2010, 256, 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Wang C, Tan H, Cheng L, Liu G, Yang Y, Zhao Y, Zhang Y, Li Y. and Zhang C, Biomaterials, 2016, 108, 71–80. [DOI] [PubMed] [Google Scholar]
- 15.Schwarz B, Uchida M. and Douglas T, Advances in Virus Research, Elsevier, 2017, vol. 97, pp. 1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitagawa T, Kosuge H, Uchida M, Iida Y, Dalman RL, Douglas T. and McConnell MV, J. Magn. Reson. Imaging, 2017, 45, 1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruckman MA, Jiang K, Simpson EJ, Randolph LN, Luyt LG, Yu X. and Steinmetz NF, Nano Lett, 2014, 14, 1551–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun X, Li W, Zhang X, Qi M, Zhang Z, Zhang X-E and Cui Z, Nano Lett, 2016, 16, 6164–6171. [DOI] [PubMed] [Google Scholar]
- 19.Chan CKW, Zhang L, Cheng CK, Yang H, Huang Y, Tian XY and Choi CHJ, Small, 2018, 14, 1702793. [DOI] [PubMed] [Google Scholar]
- 20.Moore KJ and Freeman MW, Arterioscler., Thromb., Vasc. Biol, 2006, 26, 1702–1711. [DOI] [PubMed] [Google Scholar]
- 21.Healy AM, Pickard MD, Pradhan AD, Wang Y, Chen Z, Croce K, Sakuma M, Shi C, Zago AC and Garasic J, Circulation, 2006, 113, 2278–2284. [DOI] [PubMed] [Google Scholar]
- 22.Ionita MG, van den Borne P, Catanzariti LM, Moll FL, de Vries J-PP, Pasterkamp G, Vink A. and de Kleijn DP, Arterioscler., Thromb., Vasc. Biol, 2010, 30, 1842–1848. [DOI] [PubMed] [Google Scholar]
- 23.Ionita MG, Vink A, Dijke IE, Laman JD, Peeters W, van der Kraak PH, Moll FL, de Vries J-PP, Pasterkamp G. and de Kleijn DP, Arterioscler., Thromb., Vasc. Biol, 2009, 29, 1220–1227. [DOI] [PubMed] [Google Scholar]
- 24.Qin H, Lerman B, Sakamaki I, Wei G, Cha SC, Rao SS, Qian J, Hailemichael Y, Nurieva R. and Dwyer KC, Nat. Med, 2014, 20, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T. and Discher DE, Nat. Nanotechnol, 2007, 2, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Champion JA and Mitragotri S, Proc. Natl. Acad. Sci. U. S. A, 2006, 103, 4930–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen AM, Wang Y, Jiang K, Hsu GC, Gao H, Lee KL, Yang AC, Yu X, Simon DI and Steinmetz NF, J. Mater. Chem. B, 2015, 3, 6037–6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geiger FC, Eber FJ, Eiben S, Mueller A, Jeske H, Spatz JP and Wege C, Nanoscale, 2013, 5, 3808–3816. [DOI] [PubMed] [Google Scholar]
- 29.Bruckman MA and Steinmetz NF, Virus Hybrids as Nanomaterials, Springer, 2014, pp. 173–185. [Google Scholar]
- 30.Altschul SF, Gish W, Miller W, Myers EW and Lipman DJ, J. Mol. Biol, 1990, 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 31.Getz GS and Reardon CA, Arterioscler., Thromb., Vasc. Biol, 2012, 32, 1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russell JC and Proctor SD, Cardiovasc. Pathol, 2006, 15, 318–330. [DOI] [PubMed] [Google Scholar]
- 33.Gisterå A. and Hansson GK, Nat. Rev. Nephrol, 2017, 13, 368. [DOI] [PubMed] [Google Scholar]
- 34.McCormick MM, Rahimi F, Bobryshev YV, Gaus K, Zreiqat H, Cai H, Lord RS and Geczy CL, J. Biol. Chem, 2005, 280, 41521–41529. [DOI] [PubMed] [Google Scholar]
- 35.Hansson GK, Libby P. and Tabas I, J. Intern. Med, 2015, 278, 483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eue C. Langer, A. Eckardstein and C. Sorg, Atherosclerosis, 2000, 151, 593–597. [DOI] [PubMed] [Google Scholar]
- 37.Czapar AE, Zheng Y-R, Riddell IA, Shukla S, Awuah SG, Lippard SJ and Steinmetz NF, ACS Nano, 2016, 10, 4119–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pitek A, Hu H, Shukla S. and Steinmetz N, ACS Appl. Mater. Interfaces, 2018, 10, 39468–39477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu H, Zhang Y, Shukla S, Gu Y, Yu X. and Steinmetz NF, ACS Nano, 2017, 11, 9249–9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruckman MA, Hern S, Jiang K, Flask CA, Yu X. and Steinmetz NF, J. Mater. Chem. B, 2013, 1, 1482–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.