Abstract
In mammalian cells, mitochondrial dysfunction triggers the integrated stress response (ISR), in which eIF2α phosphorylation induces the transcription factor ATF41-3. However, how mitochondrial stress is relayed to ATF4 is unknown. We found that HRI is the eIF2α kinase necessary and sufficient for this relay. In a genome-wide CRISPRi screen, we identified factors upstream of HRI: OMA1, a mitochondrial stress-activated protease, and DELE1, a little-characterized protein we found to be associated with the inner mitochondrial membrane. Mitochondrial stress stimulates OMA1-dependent cleavage of DELE1, leading to its accumulation in the cytosol, where it interacts with HRI and activates its eIF2α kinase activity. Additionally, DELE1 is required for ATF4 translation downstream of eIF2α phosphorylation. Blockade of the OMA1-DELE1-HRI pathway triggers an alternative response inducing specific molecular chaperones. Therefore, this pathway is a potential therapeutic target enabling fine-tuning of the ISR for beneficial outcomes in diseases involving mitochondrial dysfunction.
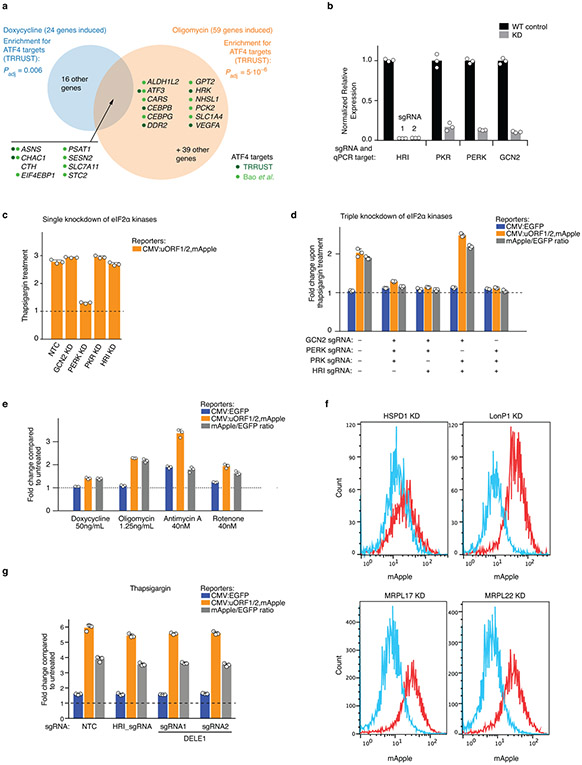
The function of mitochondria, eukaryotic organelles with essential roles in bioenergetics, metabolism and signaling, is challenged in disease and aging4-6. Mitochondrial dysfunction elicits stress responses in mammalian cells, including the mitochondrial unfolded protein response7-11. However, the overwhelming signature in the response of human cells to disruption of mitochondrial functions is the integrated stress response (ISR), as previously published1-3 and confirmed by us (Extended Data Fig. 1a, Supplementary Tables 1, 2). The ISR is mediated through eIF2α phosphorylation under various stress conditions sensed by four eIF2α kinases12,13. eIF2α phosphorylation reduces global protein synthesis but increases translation of specific mRNAs, including ATF4, the master transcriptional regulator of the ISR. How mitochondrial dysfunction triggers the ISR is unknown.
HRI relays mitochondrial stress to ATF4
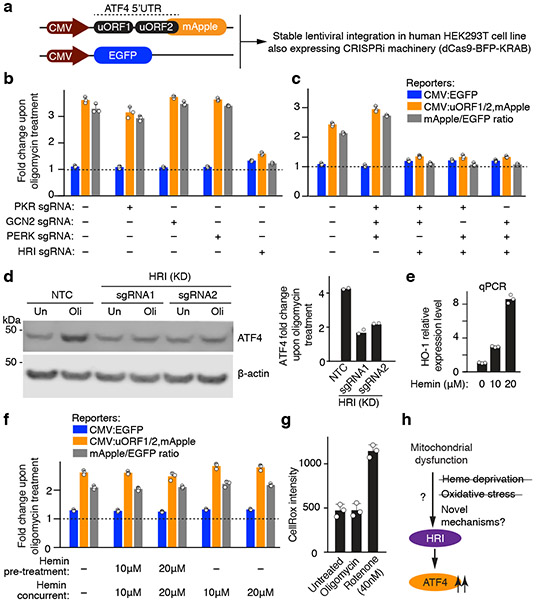
To characterize how mitochondrial dysfunction is signaled to the ISR, we introduced a reporter for ATF4 translation along with the machinery necessary for CRISPR interference (CRISPRi)14,15 into HEK293T cells (Fig. 1a and Methods). The reporter was activated by endoplasmic reticulum stress and a broad range of mitochondrial stresses (Extended Data Fig. 1b-f). We tested the role of the four known eIF2α kinases (GCN2, HRI, PERK, PKR), and found that HRI was necessary and sufficient for the activation of the ATF4 reporter and endogenous ATF4 upon oligomycin-induced mitochondrial stress (Fig. 1b-d). This finding raised the question how dysfunction in the mitochondria is sensed by the cytosolic HRI.
Figure 1. The eIF2α kinase HRI relays mitochondrial stress to ATF4.
(a) ATF4 translational reporter including the upstream open reading frames (uORFs) of the ATF4 5’ untranslated region (5’UTR) followed by mApple replacing the ATF4 coding sequence. Transcription of the reporter and an EGFP transcriptional control is driven by the CMV promoter.
(b,c) Reporter cells expressing either single sgRNAs (b) or triple sgRNAs (c) targeting the indicated eIF2α kinases were exposed to 1.25 ng/mL oligomycin for 16 h before measuring reporter levels by flow cytometry (mean ± s.d., n = 3 culture wells).
(d) Immunoblot of endogenous ATF4. Cells expressing a non-targeting control sgRNA (NTC) or sgRNAs targeting HRI were treated with 1.25 ng/mL of oligomycin for 16 h where indicated. Left, representative blot; Right, quantification of n = 2 blots.
(e) Expression of the heme-induced gene HO-1 after 24-hour incubation with the indicated concentrations of hemin, measured by qPCR (n = 3 technical replicates).
(f) Heme supplementation does not abolish ATF4 induction. Reporter levels in cells were treated with the indicated concentrations of hemin for 24 h before a 16 h treatment with 1.25 ng/mL oligomycin in the presence or absence of hemin (mean ± s.d., n = 3 culture wells).
(g) Oligomycin treatment used in this study does not induce reactive oxygen species (ROS). Cells were treated with 1.25 ng/mL oligomycin or 40 nM rotenone for 16 h and ROS levels were quantified by flow cytometry using the CellROX reagent (mean ± s.d., n = 3 culture wells).
(h) Model.
Since the canonical mechanism of HRI activation is heme depeletion16 and since key steps in heme biosynthesis occur in the mitochondria17, heme depletion could be the signal activating HRI upon mitochondrial dysfunction. However, we rejected this hypothesis because hemin supplementation did not reduce ATF4 induction in response to oligomycin (Fig. 1e,f).
Next, we tested if reactive oxygen species (ROS), which can be produced by dysfunctional mitochondria, are the signal activating HRI through a previously described oxidation mechanism18,19. Unlike rotenone treatment, the oligomycin concentration sufficient to induce ATF4 did not detectably elevate ROS (Fig. 1g), ruling out ROS as the relevant signal. These findings suggested a novel mechanism of mitochondrial stress-induced HRI activation (Fig. 1h).
CRISPRi screen reveals a role for DELE1
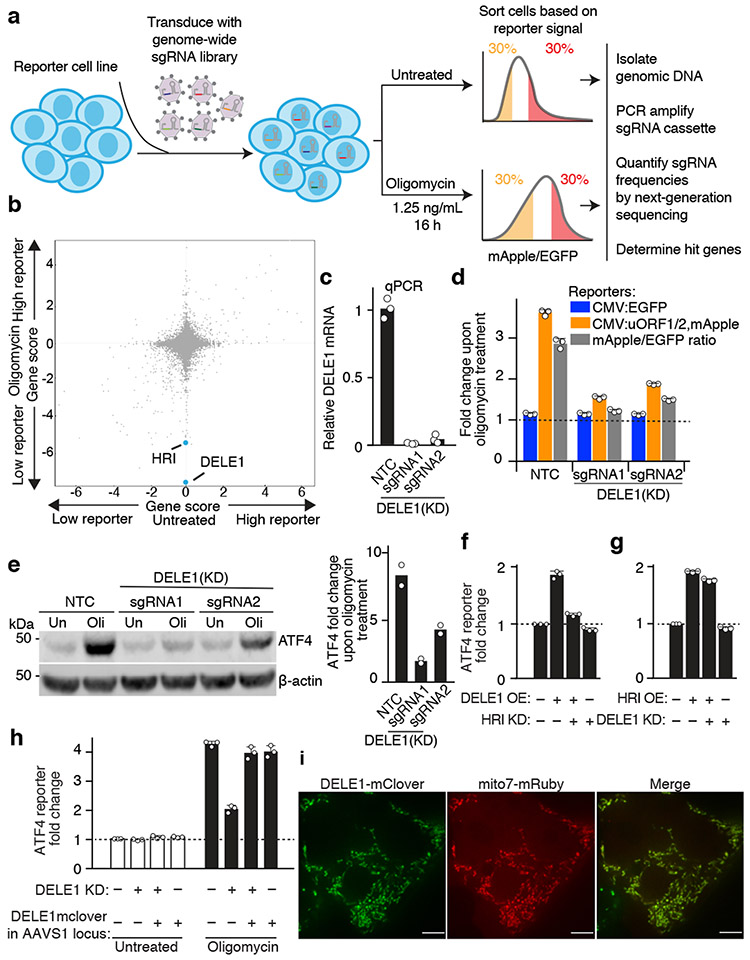
To identify further molecular players mediating ATF4 induction upon mitochondrial stress, we conducted a genome-wide CRISPRi screen (Fig. 2a, Methods and Supplementary Tables 3,4). We focused on hit genes whose knockdown reduced reporter activity in the oligomycin-treated but not untreated cells. HRI was a strong hit, as expected, but the strongest hit in this category was the little-characterized gene DELE1 (Fig. 2b). We validated that DELE1 knockdown inhibited oligomycin-stimulated induction of the ATF4 reporter and endogenous ATF4 (Fig. 2c-e). DELE1 was not required for ATF4 activation in response to ER stress (Extended Data Fig. 1g), suggesting a role specifically in mitochondrial stress.
Fig. 2. A CRISPRi screen identifies a requirement for DELE1 in ATF4 induction.
(a) Schematic of screen.
(b) Comparison of gene scores (defined in Methods) from the screen in untreated and oligomycin-treated conditions.
(c) Knockdown of DELE1 quantified by qPCR (n = 3 technical replicates).
(d) Reporter activation in cells expressing a non-targeting control sgRNA (NTC) or DELE1 sgRNAs (mean ± s.d., n = 3 culture wells).
(e) Immunoblot of endogenous ATF4. Cells were treated with 1.25 ng/mL of oligomycin for 16 hrs (Oli) or untreated (Un). Left, representative blot; Right, quantification of n = 2 blots.
(f) ATF4 reporter activation in cells transiently overexpressing (OE) DELE1-mClover and expressing sgRNA to knockdown (KD) HRI where indicated. (mean ± s.d., n = 3 culture wells).
(g) ATF4 reporter activation in cells transiently overexpressing (OE) HRI and expressing sgRNA to knockdown (KD) DELE1 where indicated. (mean ± s.d., n = 3 culture wells).
(h) DELE1-mClover expression from the AAVS1 safe-harbor locus is not sufficient to induce the ATF4 reporter in the absence of stress, but complements DELE1 knockdown upon oligomycin treatment (mean ± s.d., n = 3 culture wells).
(i) Co-localization of DELE1-mClover with the mitochondrial-targeted mRuby (Mito7-mRuby). Scale bar, 7 μm. Similar results for n = 2 culture wells.
DELE1 was previously identified as a mitochondrial protein with a role in apoptosis20. To further characterize DELE1, for which we were unable to obtain effective antibodies (Extended Data Fig. 2a), we tagged it C-terminally with mClover. Transient transfection with a DELE1-mClover plasmid caused substantial overexpression (Extended Data Fig. 2b) and induced the ATF4 reporter in an HRI-dependent manner (Fig. 2f). Similarly, HRI overexpression induced the ATF4 reporter, but in a DELE1-independent manner, indicating that DELE1 acts upstream of HRI in this pathway (Fig. 2f,g).
To express DELE1-mClover at lower levels enabling its characterization without activating ATF4, we stably integrated it into the AAVS-1 safe harbor locus. This construct was overexpressed to a lesser degree (Extended Data Fig. 2b) and did not induce the ATF4 reporter in the absence of stress (Fig. 2h). Importantly, this construct rescued ATF4 activation in response to oligomycin in DELE1 knockdown cells (Fig. 2h), demonstrating that the mClover tag does not interfere with this function of DELE1. With this transgene, we confirmed mitochondrial localization of DELE1 (Fig. 2i).
OMA1-dependent DELE1 cleavage
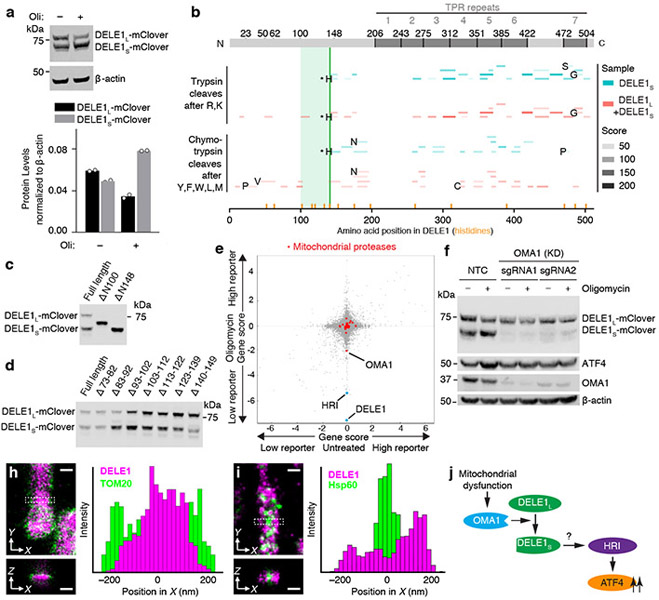
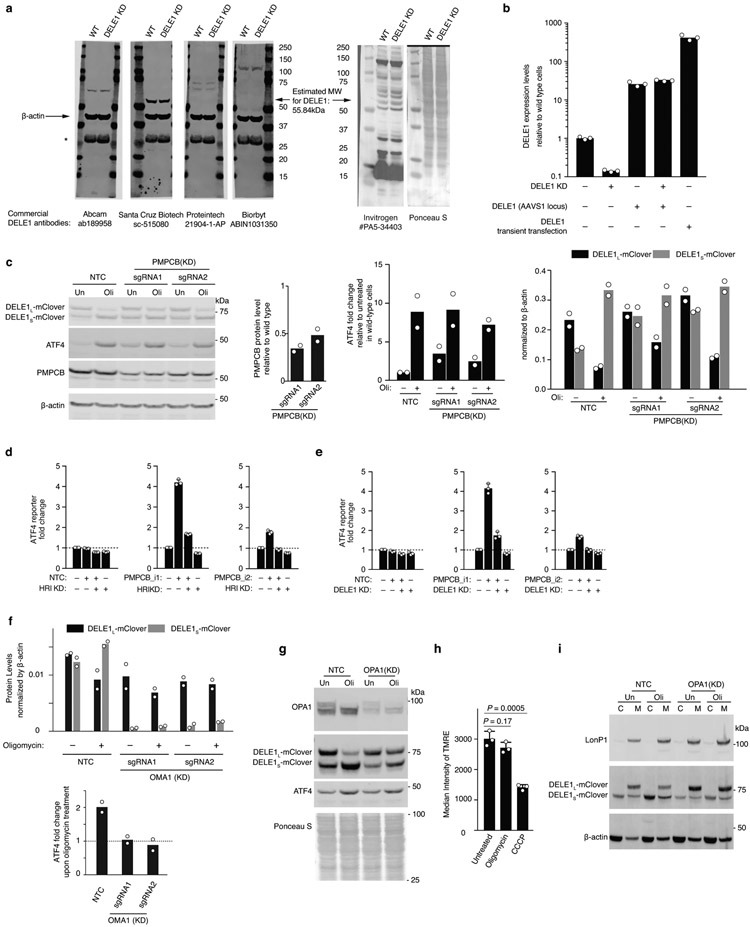
Characterization of DELE1-mClover by immunoblotting revealed two forms, which we named DELE1L and DELE1S. The shorter form, DELE1S, accumulated after oligomycin treatment (Fig. 3a). Because the transgenic construct is cDNA-based, DELE1S cannot represent a splice isoform, but is likely a cleavage product. Proteomics and comparison to defined truncation constructs suggested cleavage at histidine 142 (Fig. 3b,c and Supplementary Discussion). A series of short deletions between amino acids 73 and 149 did not abrogate DELE1 cleavage (Fig. 3d), indicating that cleavage may not require a specific sequence motif. Furthermore, minor bands (Fig. 3d) indicated plausible cleavage events at additional sites. Mitochondrial localization seemed to be required for cleavage, since N-terminal truncation constructs ΔN100 and ΔN148, which no longer localize to mitochondria (Extended Data Fig. 3e) are not cleaved (Fig. 3c). DELE1S was not generated by mitochondrial processing peptidase (MPP), since knockdown of the essential MPP subunit PMPCB did not inhibit DELE1 cleavage (Extended Data Fig. 2c-e).
Fig. 3. A cleaved form of DELE1 accumulates upon mitochondrial stress in an OMA1-dependent manner.
(a) Cells stably expressing DELE1-mClover were treated with 1.25 ng/mL of oligomycin for 16 h. Two DELE1-mClover isoforms (DELE1L and DELE1S) were detected using an anti-GFP antibody. Top, representative blot. Bottom, quantification of n = 2 blots.
(b) Tryptic and chymotrypic peptides of total DELE1 (red) or DELE1S (cyan) detected by mass spectrometry mapped to the amino acid sequence of DELE1. DELE1S lacks peptides derived from the N-terminal 141 amino acids; the light green box indicates the putative cleavage region. Peptides resulting from cleavage at residues that are not canonical targets of trypsin or chymotrypsin, respectively, are labeled with the amino acid N-terminal to the cleavage event. Peptides resulting from cleavage after histidine-142 are labeled with asterisks, and this position is marked with a solid green line. Positions of all histidines in the DELE1 sequence are labeled with orange lines.
(c,d) Immunoblot from cells transiently expressing full-length DELE1-mClover and truncation constructs lacking the indicated amino acids. Similar results in n = 2 independent experiments.
(e) Comparison of gene scores (defined in Methods) from the genome-wide CRISPRi screen in untreated and oligomycin-treated conditions. Highlighted in red are mitochondrial proteases, of which OMA1 knockdown significantly reduces ATF4 activation by oligomycin.
(f) Immunoblot of DELE1-mClover, ATF4 and OMA1 in cells expressing non-targeting control sgRNAs (NTC) or OMA1 sgRNAs, which were treated with 1.25 ng/mL of oligomycin for 16 h where indicated. Similar results in n = 2 technical replicates.
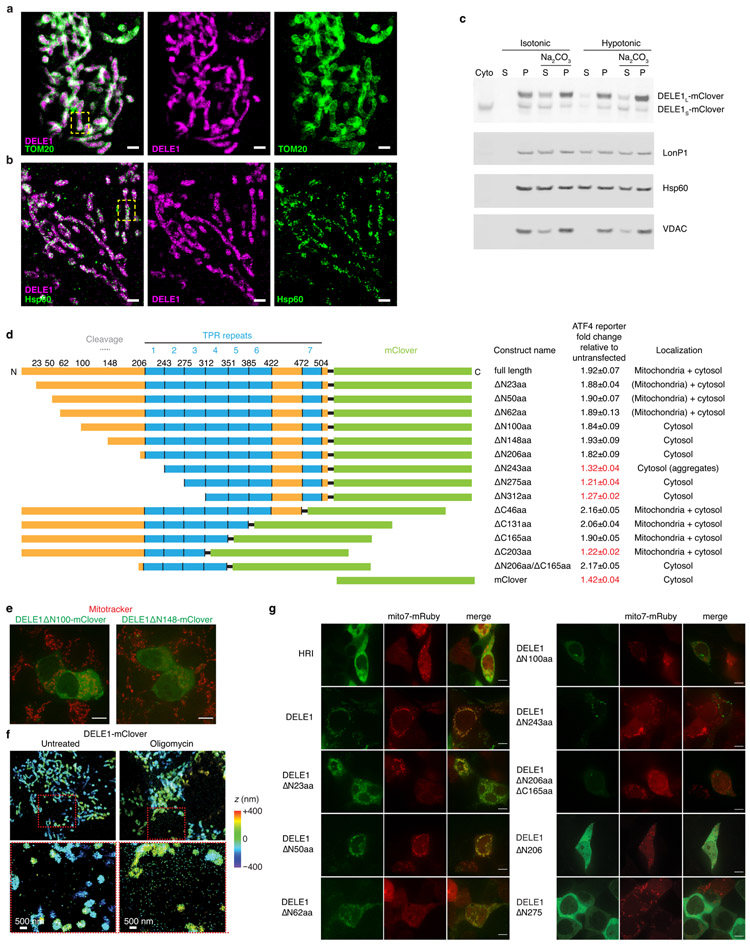
(g-h) Two-color 3D-STORM super-resolution microscopy. Magenta: stably expressed DELE1-mClover, Green: outer mitochondrial membrane protein TOM20 (g) or mitochondrial matrix protein Hsp60 (h). Virtual cross-sections and spatial intensity distributions are shown for the boxed area. Scale bars: 250 nm. Similar results in n = 3 independent experiments.
(i) Model.
Of 28 mitochondrial proteases targeted in our CRISPRi screen21, only knockdown of OMA1, a protease activated by mitochondrial stress22,23, showed substantial abrogation of ATF4 activation (Fig. 3e). OMA1 knockdown dramatically reduced DELE1S levels and blocked ATF4 induction (Fig. 3f, Extended Data Fig. 2f) suggesting that OMA1 cleaves DELE1, which is necessary for ATF4 induction. Oligomycin activated OMA1, as evidenced by cleavage of the canonical OMA1 substrate OPA1, even though the mitochondrial membrane potential was not significantly reduced (Extended Data Fig. 2g,h).
Since OMA1 and OPA1 localize to the inner mitochondrial membrane22,23, we investigated the localization of DELE1. Super-resolution microscopy and biochemical fractionation supported an association of DELE1 with the inner mitochondrial membrane (Fig. 3g,h, Extended Data Fig. 3a-c). (It is challenging to differentiate between peripheral and integral membrane proteins of the inner mitochondrial membrane using classical biochemical approaches24.)
In summary, mitochondrial dysfunction stimulates OMA1-dependent cleavage of DELE1. Given the requirement for both OMA1 and DELE1 for ATF4 induction upon mitochondrial stress, DELE1 cleavage products are likely mediators in this pathway (Fig. 3i).
DELE1 interacts with and activates HRI
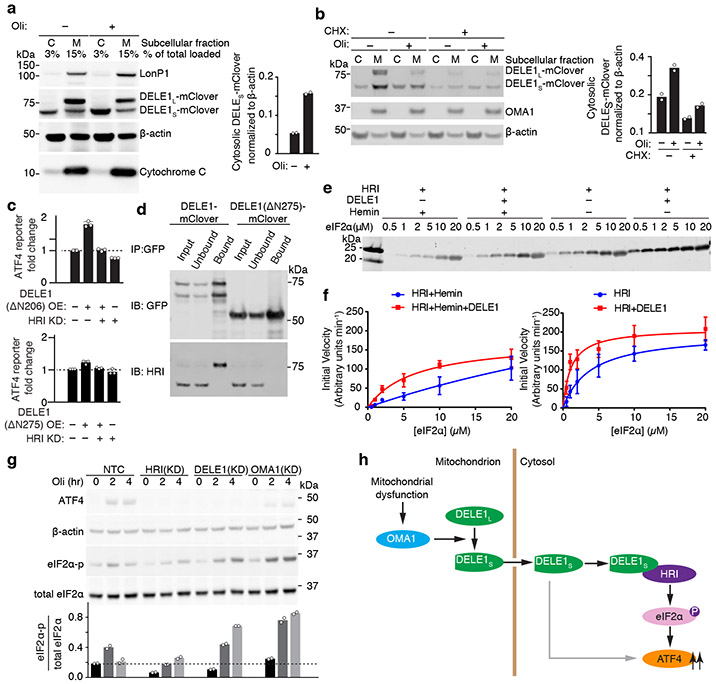
HRI is a cytosolic kinase25,26 (Extended Data Fig. 3g) and we hypothesized that DELE1S might accumulate in the cytosol and interact with HRI. Indeed, whereas DELE1L was exclusively mitochondrially localized, DELE1S accumulated in the cytosol upon mitochondrial stress (Fig. 4a, Extended Data Fig. 3f). This could either indicate that mitochondrially localized DELE1 is cleaved and that the resulting DELE1S exits the mitochondria, or that newly synthesized DELE1 is cleaved during mitochondrial import. Cycloheximide treatment blocking new DELE1 synthesis during mitochondrial stress did not abrogate stress-induced DELE1S accumulation in the cytosol (Fig. 4b), suggesting that cytosolic DELE1S originates from cleavage of pre-existing mitochondrial DELE1L. Next, we asked if OMA1-mediated OPA1 cleavage is required for stress-induced DELE1S cytosolic accumulation. OPA1 knockdown, a condition that mimics OPA1 cleavage and causes mitochondrial fragmentation27, did not increase cytosolic accumulation of DELE1S (Extended Data Fig. 2g), suggesting that OPA1 cleavage is not driving stress-induced cytosolic accumulation of DELE1S.
Fig. 4. Cytosolic DELE1 physically interacts with and activates HRI.
(a) Immunoblot of biochemical fractionations of cells stably expressing DELE1-mClover treated with 1.25 ng/mL oligomycin (Oli) for 16 h where indicated. Left, representative immunoblot of the cytosolic (C) and mitochondrial (M) fractions. LonP1 and cytochrome c were probed as mitochondrial markers. Right, quantification of the cytosolic DELE1S-mClover from n = 2 blots.
(b) Accumulation of DELE1S is protein-synthesis independent. Biochemical fractionation of cells stably expressing DELE1-mClover treated with 1.25 ng/mL oligomycin (Oli) and 20 μg/mL cycloheximide (CHX) for 4 h where indicated. Left, representative immunoblot of the cytosolic (C) and mitochondrial (M) fractions. OMA1 as probed as mitochondrial marker. Right, quantification of the cytosolic DELE1S-mClover from n = 2 blots.
(c) Transient overexpression (OE) of the cytosolically localized DELE1(ΔN206), but not the DELE1(ΔN275) construct induces the ATF4 reporter. Knockdown (KD) of HRI blocks reporter activation (mean ± s.d., n = 3 culture wells).
(d) Co-immunoprecipitation of HRI with transiently expressed full-length DELE1-mClover but not with DELE1(ΔN275)-mClover. Similar results in n = 2 independent experiments.
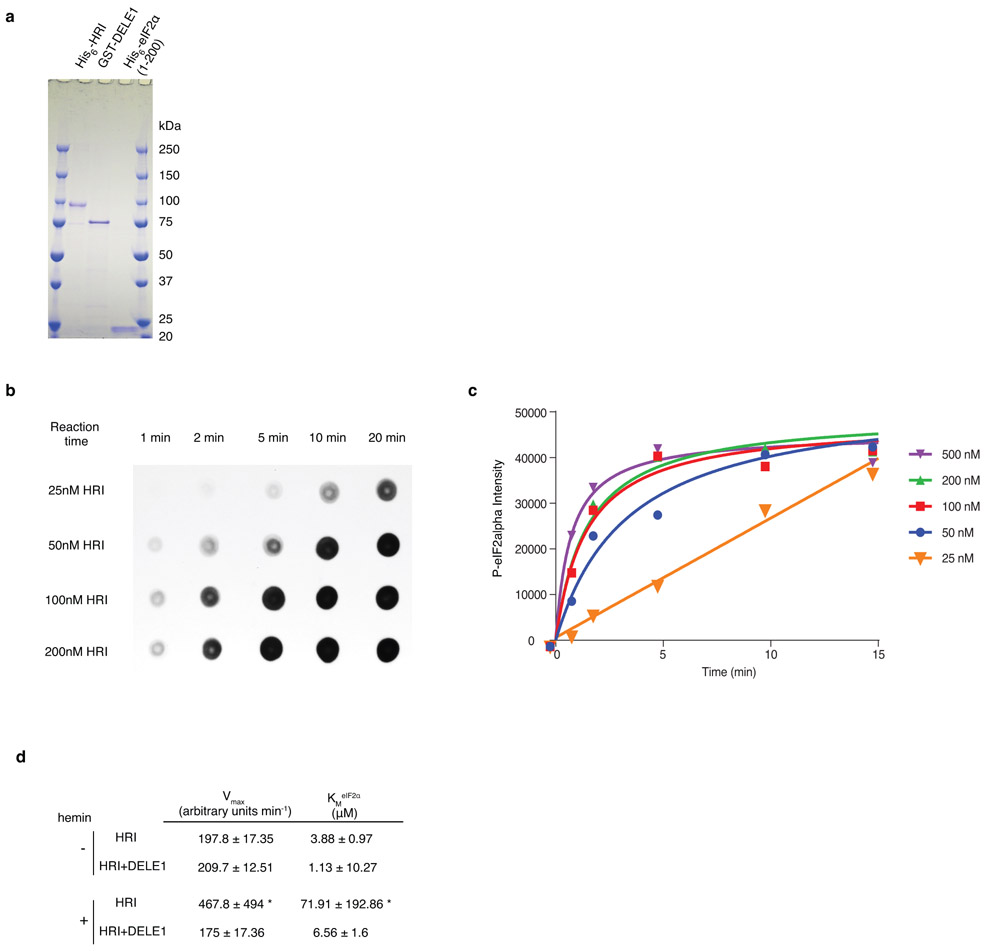
(e,f) HRI enzyme kinetics in the presence or absence of DELE1 with or without 5 μM hemin using different concentrations of the substrate eIF2α. (e) Representative immunoblot. (f) Quantification and fitting to the Michaelis-Menten equation (mean ± s.e.m., n = 3 independent experiments).
(g) Top, immunoblot of ATF4, phospho-eIF2α, total eIF2α in NTC control, HRI KD-, DELE1 KD- and OMA1 KD-cells treated with 1.25ng/mL oligomycin for 0, 2 and 4 hrs. Bottom, quantification of the phospho-eIF2α to total eIF2α ratio from n = 2 blots.
(h) Model.
We dissected the function of different domains of DELE1 in determining its localization and ability to induce ATF4 during transient overexpression (Extended Data Fig. 3d). The C-terminal portion of DELE1 contains 7 predicted TPR repeats. A construct containing all TPR repeats but lacking the N-terminal 206 amino acids no longer localized to the mitochondria (Fig. 3g), but still activated the ATF4 reporter when overexpressed (Fig. 4c). This finding indicated that under overexpression conditions, DELE1 does not need to localize to mitochondria to induce ATF4. However, removal of the most N-terminal TPR repeat (ΔN275) abrogated DELE1 activity (Fig. 4c). A minimal construct consisting of the first four TPR repeats was sufficient to induce the ATF4 reporter (Extended Data Fig. 3d), suggesting that these repeats mediate a protein-protein interaction necessary for ATF4 induction.
We found that HRI and DELE1 interact physically with each other based on co-immunoprecipitation (Fig. 4d). This interaction was dependent on the N-terminal TPR repeats (Fig. 4d), mirroring the requirement for ATF4 activation. Using purified HRI, eIF2α and DELE1, we showed that DELE1 stimulates eIF2α kinase activity of HRI in vitro (Fig. 4e,f, Extended Data Fig. 4, Supplementary Discussion). Taken together, our results suggest that the stress-induced accumulation of DELE1S in the cytosol leads to the activation of HRI through physical interaction.
Surprisingly, knockdown of HRI, but not of DELE1 and OMA1, blocked eIF2α phosphorylation in oligomycin-treated cells (Fig. 4g). This finding suggests that in addition to promoting HRI’s kinase activity under mitochondrial stress, DELE1 has a secondary role in facilitating ATF4 translation, possibly downstream of eIF2α phosphorylation (Fig. 4h) to exempt ATF4 from the general inhibition of protein synthesis. In the context of ER stress, there is a precedent for a factor required in addition to eIF2α phosphorylation to induce ATF4 translation, DDX328. In the absence of DELE1, mitochondrial stress still promoted eIF2α phosphorylation, potentially through HRI itself via a distinct activatory mechanism. Alternatively, the failure to induce ATF4 in response to mitochondrial stress in DELE1 knockdown cells may result in pervasive cellular dysfunction, leading to activation of other eIF2α kinases. While eIF2α phosphorylation in WT cells was transient, DELE1 knockdown prolonged eIF2α phosphorylation (Fig. 4g), consistent with the possibility that in the absence of ATF4 induction, there is no activation of mechanisms downstream of ATF4 that provide a negative-feedback loop (such as transcriptional induction of the GADD34 phosphatase to drive eIF2α dephosphorylation).
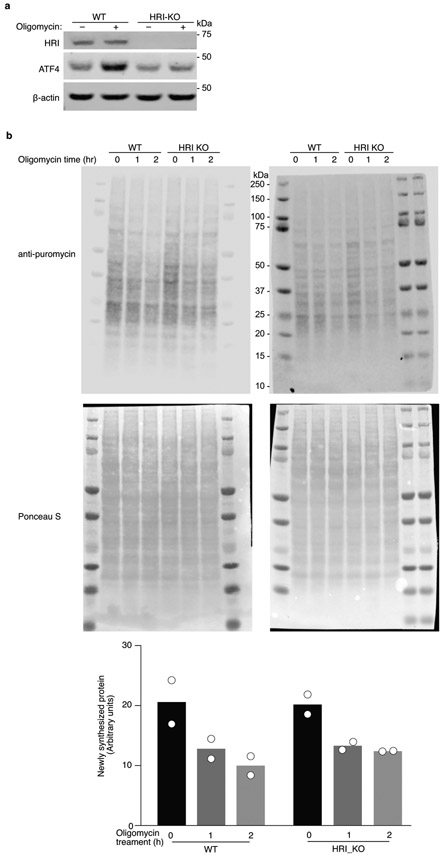
We also investigated rates of protein synthesis, which are known to be reduced downstream of eIF2α phosphorylation. Oligomycin treatment caused a reduction in protein synthesis in WT cells, but also in HRI knockout cells (Extended Data Fig. 5). This suggests that while HRI is required for eIF2α phosphorylation in response to mitochondrial stress (Fig. 4g), a reduction in protein synthesis in response to oligomycin occurs even in the absence of HRI, possibly due to oligomycin-triggered energy depletion interfering with protein synthesis.
Role of the OMA1-DELE1-HRI pathway
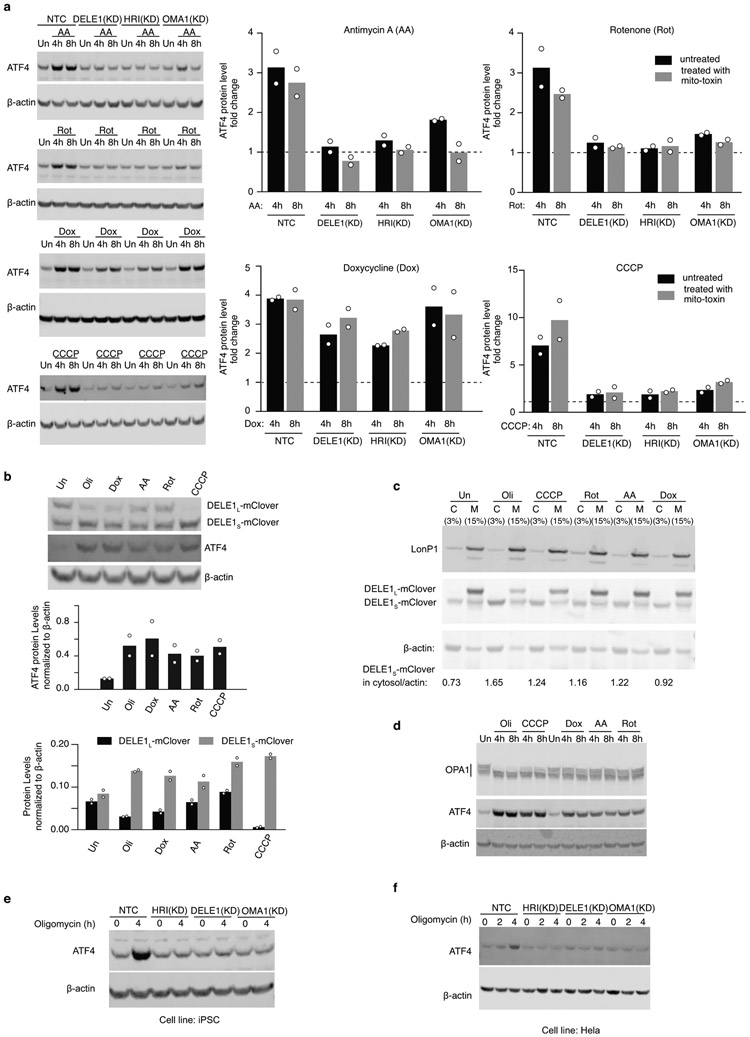
The OMA1-DELE1-HRI pathway was also required for the response to mitochondrial stress in other human cell lines, and mediated the response to other mitochondrial stressors (Extended Data Fig. 6). Doxycycline induction of ATF4 showed less dependence on the pathway (Supplementary Discussion).
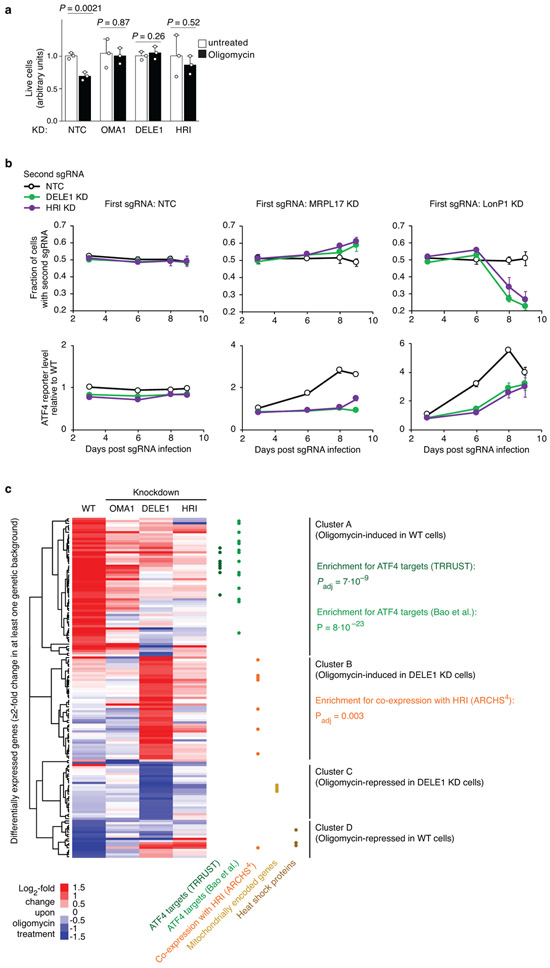
Depending on its context and duration, the ISR can have homeostatic or pro-apoptotic outcomes, and it can be deficient or maladaptive in disease states29,30. We found that blockade of the OMA1-DELE1-HRI pathway was beneficial in response to some mitochondrial stressors, but detrimental in response to others (Extended Data Fig. 7a,b). The fact that DELE1 and HRI knockdown have concordant effects on stress survival suggests that these survival phenotypes are not mediated by eIF2α phosphorylation (which is reduced by knockdown of HRI, but not DELE1).
The transcriptomic response to mitochondrial stress in cells lacking the OMA1-DELE1-HRI pathway (Extended Data Fig. 7c) showed reduced induction of ATF4 targets, as expected. Intriguingly, blockade of ATF4 induction lead to upregulation, instead of downregulation, of cytosolic Hsp70 in the context of mitochondrial stress, possibly contributing to the protective effect of HRI and DELE1 knockdown in some stress contexts. Chaperone induction was previously reported in iron-deficient HRI−/− erythroblasts31.
Discussion
We identified the molecular players that signal mitochondrial stress to the cytosol. Our findings raise new questions for future studies (Supplementary Discussion).
Both inhibition32 and prolongation33 of the ISR are potential therapeutic strategies. However, the context dependence of the ISR, which can be protective or maladaptive, presents a challenge. The OMA1-DELE1-HRI pathway we describe here is a potential therapeutic target to block ATF4 activation in cells experiencing mitochondrial dysfunction, without globally blocking the ISR in all cells. OMA1 ablation was previously found to protect against heart failure in multiple mouse models involving mitochondrial dysfunction34. Our results suggest that this effect may be mediated by attenuation of ATF4 induction. DELE1 could be an even more attractive therapeutic target, since OMA1 performs independent homeostatic functions, and since DELE1 has an additional pro-apoptotic activity20. Furthermore, DELE1 knockdown induced an alternative, likely beneficial, stress response (Extended Data Fig. 7c).
By dissecting the molecular pathways controlling different aspects of the ISR under different stress conditions, additional therapeutic targets may emerge, which could enable fine-tuned manipulation of the cellular response to different stressors to achieve beneficial outcomes tailored to specific disease states.
METHODS
Plasmids
Sequences of oligonucleotides used for cloning are provided in Extended Data Table 1. The ATF4 translational reporter (pXG237) was generated by replacing the venus-IRES-BFP of pMK1163, our previously published integrated stress response vector35, with mApple through Gibson assembly (NEB, E2611). The secondary reporter (pXG260) to control the transcriptional regulation of CMV was generated by replacing ATF4-uORFs1/2-mApple with EGFP through Gibson assembly.
The DELE1 coding sequence was cloned from human cDNA and inserted into pMK125336 through Gibson assembly to generate a DELE1-mClover tagged protein under the EIF1A promoter (pXG286). To obtain a construct that enabled integrate DELE1-mClover at the safe harbor locus (pXG289), the EIF1A promoter-DELE1-mClover cassette was PCR-amplified from pXG286, and inserted between the SpeI and MluI sites of the AAVS-1-targeting vector pMTL337. All truncation constructs were generated through ligation of the corresponding truncated DELE1 into EcoRI- and NotI-digested pXG286. Internal short consecutive deletions covering amino acids 73 to 149 were made by inverse PCR on pXG286.
HRI cDNA was synthesized as a gene block by IDT and cloned into pMK1253 as for pXG286 to obtain pXG272.
To clone individual sgRNAs, top and bottom oligonucleotides (IDT) were annealed and ligated to our optimized lentiviral sgRNA expression vector15. Triple sgRNAs expression constructs were generated as previously published38. Protospacer sequences for sgRNAs are listed in Extended Data Table 2.
Cell lines
HEK293T and HeLa cells were cultured in DMEM (Gibco, 11965-092) with 10% fetal bovine serum (Seradigm # 97068-085, Lot# 076B16), Pen/Strep (Life Technologies # 15140122), and L-glutamine (Life Technologies # 25030081). Human iPSCs (male WTC11 background) were cultured in Stemflex™ Medium (Gibco, A3349401) as previously described37.
The CRISPRi HEK293T cell line (cXG284) was generated by transfecting HEK293T cells with pC13N-dCas9-BFP-KRAB37 and TALENS targeting the human CLYBL intragenic safe harbor locus (between exons 2 and 3) (pZT-C13-R1 and pZT-C13-L1, Addgene #62196, #62197) using DNA-In Stem (VitaScientific). BFP-positive cells were isolated via FACS sorting. The CRISPRi Hela cell line was a gift from Jonathan Weissman. The CRISPRi iPSCs were generated previously37.
The ATF4 translational reporter cell line (cXG289) was generated through lentiviral infection of cXG284 with pXG237 and FACS-based monoclonal selection based on response to mitochondrial stress. The dual reporter cell line (cXG330) was generated via lentiviral infection of the second reporter (pXG260) into cXG289.
CRISPRi knockdown cell lines were generated by lentiviral transduction with plasmids containing individual sgRNAs or triple sgRNAs.
HRI KO cell line was generated via CRISPR/Cas9-mediated gene editing by using two sgRNAs targeting the first and second exon of HRI respectively. The protospacer sequences are: 5’CGGGGTCCGCAAGCGCGAAG3’ and 5’AAACCCACTTCGTTCAAGAC3’.
The cell line expressing a DELE1-mClover transgene line from the AAVS1 locus was generated by transfecting a cXG289 population in which ~ 50% of cells expressed a DELE1-sgRNA and a BFP marker with pXG289 and TALENS targeting the human AAVS1 locus (AAVS1-TALEN_L/R, Addgene #59025 and #59026). Through FACS sorting, cells expressing the DELE1-mClover transgene either in the wild type background (BFP-, GFP+) or in the DELE1 knockdown background (BFP+, GFP+) were isolated.
Drug treatments
HEK293T cells and derived cell lines were seeded at 25% confluency 24 h before drug treatment. An equal volume of DMEM with twice the final drug concentration was added. To trigger ER stress, cells were treated with 75 nM of thapsigargin (Sigma-Aldrich # T9033) for 8 h. For mitochondrial stress, cells were incubated with the following mitochondrial toxins for 16 h unless otherwise stated: 1.25 ng/mL oligomycin (Sigma-Aldrich # 75351), 50 μg/mL doxycycline (Clontech # 631311), 40 nM Antimycin (Sigma-Aldrich # A8674), 40 nM Rotenone (Sigma-Aldrich # R8875) and 5 μM CCCP (Sigma-Aldrich # C2759). For hemin supplementation experiments, 10 μM or 20 μM hemin (Sigma-Aldrich # H9039) was added to the medium when cells were seeded before oligomycin treatment, or hemin was only added during oligomycin treatment. HEK293 cells were treated with 10 μM or 20 μM for 24 h and harvested to quantify mRNA levels of HO-1. For cycloheximide (CHX) (Sigma-Aldrich # A4859) experiments, cells were treated with 20 μg/mL of CHX for 4 h.
Western blot
Cells were lysed using RIPA buffer (Thermo Fisher Scientific #89900). Total protein was quantified via Pierce BCA Protein Assay Kit (Thermo Fisher Scientific #23225). Samples were subjected to SDS-PAGE on NuPage 4-12% Bis-tris gels Bis-tris gels (Thermo Fisher Scientific # NP0336BOX) and transferred to nitrocellulose membrane (Bio-Rad #1704271). The primary antibodies used in this study were: rabbit anti-ATF4 (Cell Signaling Technologies #11815, 1:500) mouse anti-β-actin (Cell Signaling Technologies #3700, 1:5000), mouse anti-GFP (Roche #11814460001, 1:1000), rabbit anti-HRI (Mybiosource #MBS2538144, 1/1000), mouse anti-OMA1 (Santa Cruz Biotech # sc-515788, 1:200), rabbit anti-LonP1 (Cell Signaling Technologies #56266, 1:1000), rabbit anti-Hsp60 (Cell Signaling Technologies #12165, 1:1000), rabbit anti-P-eIF2α (Cell Signaling Technologies #3398, 1:1000), mouse anti-eIF2α (Invitrogen, #AHO0802, 1:1000), rabbit anti-PMPCB (Proteintech #16064-1-AP, 1:1000), mouse anti-OPA1 (BD Transduction Laboratories #612106, 1:1000). Antibodies that failed to detect DELE1 included: Abcam #ab189958, 1:500; Santa Cruz Biotech # sc-515080, 1:100; Proteintech # 21904-1-AP, 1:500, Biorbyt # ABIN1031350, 1:500, Invitrogen, #PA5-34403, 1:500. Blots were incubated with Li-Cor secondary antibodies and imaged via Odyssey Fc Imaging system (Li-Cor #2800). Immunoblot for phospho-eIF2α was detected via HRP method. Digital images were processed and analyzed using Licor ImageStudio™ software. For gel source data, see Supplementary Figure 1.
Flow cytometry
Flow cytometry data were acquired on a BD Celesta. Gating strategies for flow cytometry analysis are shown in Supplementary Figure 2.
RNA sequencing and analyses
RNA was extracted from cells using the Quick-RNA Miniprep Kit (Zymo; Cat. No. R1054) and sent to the DNA Technologies and Expression Analysis Core at the UC Davis Genome Center where they performed 3’ Tag-seq, supported by NIH Shared Instrumentation Grant 1S10OD010786-01.
To obtain transcript abundance counts, sequencing reads from 3’-Tag RNA-seq were mapped to human reference transcriptome (GRCh38, Ensembl Release 97) using Salmon v0.14.139 with the “--noLengthCorrection” option. Gene-level count estimates were obtained using tximport v1.8.040 with default settings. Subsequently, genes with more than 10 counts were retained for differential gene expression analysis, and adjusted P values (Padj) were calculated using DESeq2 v1.20.041. Result files from DESeq2 analysis are provided as Supplementary Tables 1,2,5-7, with the following columns are A: Gene Identifiers, B: mean normalized counts, averaged over all samples from both conditions, C: log2-fold change, D: standard error estimate for the log2-fold change estimate, E: Wald statistic, F: P value for this change, G: P value adjusted for multiple testing using the Benjamini-Hochberg procedure which controls false discovery rate.
Significantly enriched gene sets and their adjusted P values (Padj) were determined using Enrichr37,42. P values for overlap with ATF4 targets from Bao et al. 1 were calculated using Fisher’s exact test. Hierarchical clustering of differentially expressed genes was carried out in Cluster 3.043 and results were visualized using Java Tree View44.
Quantitative RT-PCR
Total RNA was extracted using the Quick-RNA Miniprep Kit (Zymo Research #R1054), and first strand cDNA was synthesized with the SuperScript III First-Strand Synthesis System (Invitrogen #18080-051). qPCR was performed with SensiFAST SYBR Lo-ROX reagents (Bioline #BIO-94005). Expression fold changes were calculated using the ΔΔCt method. qPCR primers used are listed in Extended Data Table 3.
CRISPRi screen
To obtain pooled sgRNA virus, next-generation sgRNA libraries45 were transfected into HEK293T cells together with lentiviral plasmid packaging mix using TransIT®-Lenti Transfection Reagent (Mirus #MIR 6600). The complete set of sgRNA sequences for these libraries was provided in the original publication45 as Supplementary file 3. The dual reporter cell line (cXG330) was then transduced with the pooled sgRNA virus and selected with puromycin (2.5 μg/mL) for 2-3 days until greater than 90% of the cells were BFP-positive. Cells were then cultured for 3 days in the absence of puromycin. Cell populations were cultured such that a representation of at least 1000 cells per sgRNA element was maintained throughout the screen. Cells were seeded at 2.5 million per 10 cm dish (12 dishes total) at a volume of 7.5 mL DMEM on day 0. On day 1, 7.5 mL of additional DMEM with or without 2.5 ng/mL oligomycin was added into each dish. On day 2, after 16 h of oligomycin treatment, both untreated and oligomycin-treated cells were FACS-sorted based on the ratio of mApple to GFP fluorescence intensity, and population corresponding to the top 30% and bottom 30% of cells were collected. This experimental design is optimal for FACS-based screen based on our previous simulations46. The representation in each sorted population was ~500 cells per sgRNA element. Genomic DNA was isolated using a Macherey-Nagel Blood L kit (Machery-Nagel #740954.20). sgRNA-encoding regions were amplified and sequenced as previously published15. Phenotype and P value for each gene were calculated using our recently described bioinformatics pipeline37. Gene scores were defined as the product between the phenotype and -log10(P value). Full screen results are provided as Supplementary Table 3, with the following columns: A: Targeted transcription start site, B: targeted gene, C: Knockdown phenotype for screen in oligomycin-treated cells, D: P value for screen in oligomycin-treated cells, E: Gene score = product of phenotype x −log10(P value) for screen in oligomycin-treated cells, F: Knockdown phenotype for screen in untreated cells, G: P value for screen in untreated cells, H: Gene score = product of phenotype x −log10(P value) for screen in untreated cells. The count information for all sgRNAs is provided as Supplementary Table 4.
Cellular subfractionation
Cytosolic and mitochondrial fractions were separated using the Mitochondrial Isolation Kit for Mammalian Cells (Thermo Fisher Scientific, #89874). The mitochondria were incubated in either isotonic buffer (10 mM Tris HCl, pH 6.7, 0.15 mM MgCl2 0.25mM sucrose, 1 mM DTT, protease inhibitor cocktail (Sigma #5892970001)) or H2O (extreme hypotonic condition) for 5 min, followed by centrifugation (10000 xg for 10min) to separate the supernatant and pellet. The pellet was either resuspended in RIPA buffer or incubated with 0.1M NaCO3 (pH=11.4) for 30 min at 4°C. Supernatant and pellet from NaCO3-treated samples were collected for immunoblotting.
Immunoprecipitation
HEK293T cells were transiently transfected with DELE1-mClover (pXG286) or DELE1(Δ275)-mClover constructs. Cells were collected 24 h after transfection and lysed using a mild lysis buffer (10 mM Tris/Cl pH 7.5; 150 mM NaCl; 0.5 mM EDTA; 0.5% NP-40, 0.09% Na-Azide). The lysates were then incubated with GFP-Trap®_MA beads (Chromtek, #gtma20) for one hour at 4°C. Proteins captured on the magnetic beads were boiled in 2X SDS loading dye for 10 min before subjecting to SDS PAGE and Western blotting.
Confocal microscopy
HEK293T cells and derived cell lines were seeded in the 8-well chamber slide (Ibidi #80824) for 24 h and transfected with DELE1 truncated constructs and/or plasmid mito7-mRuby (a gift from Michael Davidson, Addgene plasmid # 55874) as indicated, using TransIT-Lenti Transfection Reagent (Mirus #MIR 6600). 24 h after transfection, cells were incubated with 100 nM MitoTrackerTM red CMXRos (ThermoFisher Scientific #M7512) for 20 min at 37°C where indicated. Live cell images were then taken using the Yokagawa CSU22 spinning disk confocal and processed using Fiji47.
Super-resolution microscopy
Cell fixation and immunofluorescence
DELE1-mClover cells were seeded on #1.5 glass coverslips and cultured for 24 h, followed by treatment with 1.25 ng/ml oligomycin for 16 h where indicated. Samples were fixed with 3% (w/v) paraformaldehyde and 0.1% (w/v) glutaraldehyde in phosphate-buffered saline (PBS) for 20 min. After reduction with a freshly prepared 0.1% sodium borohydride solution in PBS for 5 min, the samples were permeabilized and blocked in a blocking buffer (3% w/v BSA, 0.1% v/v Triton X-100 in PBS) for 1 h. Afterward, the cells were incubated with primary antibodies (below) in the blocking buffer for 12 h at 4°C. After washing in a washing buffer (0.3% w/v BSA and 0.01% v/v Triton X-100 in PBS) for three times, the cells were incubated with dye-labeled secondary antibodies (below) for 1 h at room temperature. Then, the samples were washed 3 times with the washing buffer and 3 times with PBS. Primary antibodies used: mouse anti-mClover/GFP (Invitrogen #A-11120, 1:350); rabbit anti-Tom20 (Santa Cruz Biotechnology #sc-11415, 1:100); rabbit anti-Hsp60 (Cell Signaling Technology #12165, 1:800). Secondary antibodies used: Alexa Fluor 647-labeled goat anti-mouse (Invitrogen #A21236, 1:400); donkey anti-rabbit (Jackson ImmunoResearch #711-005-152, 1:70) conjugated with CF568 succinimidyl ester (Biotium #92131) or CF680 succinimidyl ester (Biotium #92139).
3D-STORM super-resolution microscopy
3D-STORM super-resolution microscopy48,49 was carried out on a homebuilt setup using a Nikon CFI Plan Apo λ 100x oil immersion objective (NA 1.45), as described previously50. Briefly, the sample was mounted with an imaging buffer consisting of 5% (w/v) glucose, 100 mM cysteamine, 0.8 mg/mL glucose oxidase, and 40 μg/mL catalase in a Tris-HCl buffer (pH 7.5). For two-color imaging of DELE1 and TOM20, the two targets were labeled by Alexa Fluor 647 and CF568, respectively, and were imaged sequentially using 647- and 560-nm excitation lasers. These lasers were passed through an acousto-optic tunable filter and illuminated a few micrometers into the sample at ~2 kW cm−2, thus photoswitching most of the labeled dye molecules in the sample into the dark state while allowing a small, random fraction of molecules to emit across the wield-field over different camera frames. Single-molecule emission was passed through a cylindrical lens of focal length 1 m to introduce astigmatism49, and recorded with an Andor iXon Ultra 897 EM-CCD camera at a framerate of 110 Hz, for a total of ~50,000 frames per image. Data acquisition used publicly available software (https://github.com/ZhuangLab/storm-control). The raw STORM data were analyzed using Insight3 software49 according to previously described methods48,49. For two-color imaging of DELE1 and Hsp60, the two targets were labeled by Alexa Fluor 647 and CF680, respectively, and imaged simultaneously using 647-nm laser excitation. Ratiometric detection was performed using a dichroic mirror (T685lpxr; Chroma) to assign the identity of each detected single molecule, as described previously51,52.
Mass spectrometry
Sample Preparation for Mass Spectrometry
DELE1-mClover was affinity purified from mitochondrial and cytosolic fractions of cells with DELE1-mClover in the AAVS1 locus using GFP-Trap®_MA beads (Chromtek #gtma20).
Beads and affinity-purified proteins were resuspended in a buffer containing 4M guanidine hydrochloride, 0.1M Tris pH 8.0, 5mM tris(2-carboxyethyl)phosphine (Sigma, #C4706), and 10mM 2-chloroacetamide (Sigma #22790). The mixture was incubated at room temperature for 1 hour for protein reduction and alkylation. Subsequently, for trypsin digestion, 0.1M Tris pH 8.0 was used to dilute the concentration of guanidine hydrochloride to 1M while for chymotrypsin digestion, a buffer containing 0.1M Tris pH8.0 and 14mM CaCl2 was used to achieve a final concentration of 1M guanidine hydrochloride and 10mM CaCl2. Next, 5 μg of mass spectrometry grade trypsin (ThermoFisher #90057) and 2 μg of sequencing grade chymotrypsin (Promega #V1061) were added. The trypsin and chymotrypsin digestion reactions were incubated at 37°C and 25°C, respectively, for 24 h on a shaker. Soluble peptides were collected by bead separation on a magnetic stand. Peptides were acidified to a final concentration of 1% trifluoroacetic acid (pH < 3), desalted by SOLA C18 solid phase extraction (SPE) cartridge (ThermoFisher #60109-001), and then dried down in a speed-vac. Dried peptides were stored at −80°C until mass spectrometry analysis.
Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
Desalted peptides were re-constituted in 2% acetonitrile, 0.1% formic acid and diluted to 0.2 μg/μL before mass spectrometry analysis. For each sample, a total of 1 ug of peptides was injected into a Dionex Ultimate 3000 NanoRSLC instrument with a 15-cm Acclaim PEPMAP C18 (Thermo Fisher Scientific #164534) reverse phase column. The samples were separated on a 2-hour non-linear gradient using a mixture of Buffer A (0.1% FA) and B (80% ACN/0.1% FA). The initial flow rate was 0.5 uL/min at 3% B for 13 minutes followed by a drop in flow rate to 0.2 uL/min and a non-linear increase (curve 7) to 40% B for the next 83 minutes. The flow rate was then increased to 0.5 uL/min while Buffer B was linearly ramped up to 99% for the next two minutes. Finally, we maintained the peak flow rate and Buffer B concentration for another five minutes before lowering Buffer B to 3%. Eluted peptides were analyzed with a Thermo Q-Exactive Plus mass spectrometer. The MS survey scan was performed over a mass range of 350-1500 m/z with a resolution of 70,000. The automatic gain control (AGC) was set to 3e6, and the maximum injection time (MIT) was 100 ms. We performed a data-dependent MS2 acquisition at a resolution of 17,500, AGC of 5e4, and MIT of 150 ms. The 15 most intense precursor ions were fragmented in the HCD at a normalized collision energy of 27. Dynamic exclusion was set to 20 seconds to avoid over-sampling of highly abundant species.
MS Data Processing
We analyzed the raw spectral data using MaxQuant version 1.5.1.253 for protein identification by searching against the DELE1 sequence (Q14154 UniProtID). We used the “peptides” output file for downstream analyses.
Recombinant proteins
Full length N-terminal (His)6-tagged rat HRI in construct pET28a-(His)6-HRI54 was co-expressed in Escherichia coli BL21(DE3) with chaperone plasmid pG-KJE8 (Takarabio) at 15°C for 72 h. Cleared cell lysates (lysed in Ni column buffer: 50 mM KPB pH 8.0, 300 mM NaCl, 10% glycerol, 5 mM imidazole, 0.5% CHAPS supplemented with protease inhibitor cocktail) were first applied to a His60 Ni Superflow column (Clontech). Eluates from the His60 Ni column were pooled and dialyzed overnight against 50 mM sodium phosphate pH 7.4 and then loaded onto a DEAE column. (His)6HRI was then step eluted with increasing concentrations of NaCl. Elution fractions with 200 mM, 250 mM and 300 mM NaCl were pooled and concentrated and then further purified through gel filtration on a HiPrep S-300 column and then concentrated and buffer exchanged into PBS with 20% glycerol using an Amicon centrifugal filter column. Purified (His)6-HRI proteins were more than 95% homogeneous as evidenced by SDS-PAGE.
Yeast (His)6-eIF2α (1-200) in the pET-15b vector55 was expressed in Escherichia coli BL21(DE3) at 28°C for 16 h. Protein was purified on a His60 Ni Superflow column (Clontech) according to the manufacturer’s instructions. Briefly, cell pellets were lysed in Ni column buffer and then loaded onto the His60 Ni Superflow column. The column was first washed using lysis buffer supplemented with 20 mM imidazole, and then washed using lysis buffer supplemented with 50 mM imidazole. (His)6-eIF2α was eluted using lysis buffer supplemented with 300 mM imidazole. Different elution fractions were collected and aliquots were checked by SDS-PAGE. Highly pure fractions (greater than 95% homogeneous) were combined and concentrated using an Amicon centrifugal filter column and finally stored in PBS with 20% glycerol.
Full-length recombinant human DELE1 protein fused to GST was purchased from Abcam (#ab160664).
HRI eIF2α kinase assay
25 nM HRI with or without 5 μM hemin were first incubated with 50 nM DELE1 in kinase buffer (20 mM Tris buffer pH 7.4, 40 mM KCl, 3 mM magnesium acetate, 1 mM DTT) at room temperature for 20 min. The reactions were initiated by adding 0.5 μM eIF2α and 50 μM ATP and then incubated at 20°C. The reactions were terminated by the addition of SDS sample buffer. Aliquots were subjected to SDS-PAGE, transferred to nitrocellulose membrane, and then immunoblotted with phospho-eIF2α (Ser52) antibody (Invitrogen, #44-728G, 1:2000). Blots were scanned using the Licor System for quantification. For kinetic analyses of eIF2α phosphorylation, the assay mixture contained 25 nM HRI with or without preincubation with 5 μM hemin, and with or without preincubation with 50 nM DELE1, and 0.5-20 μM of eIF2α. The assay was initiated by the addition of 50 μM ATP and then incubated at 20°C for 5 min (initial velocity conditions). Because HRI kinase reaction rate using 25 nM HRI is linear for the first 15 min (Extended Data Fig. 4b,c), the amount of reaction product (phosphorylated eIF2α) formed per min during the first 5 min was used as the initial velocity. Experiments were performed three separate times with each reaction mixture. Kinetic constants were determined by fitting into the Michaelis-Menten equation using least squares fit using Prism version 6.07.
Measurement of protein synthesis
Protein synthesis was monitored by quantification of the incorporation of puromycin into nascent polypeptide chain as described previously56. Briefly, HEK293T cells were left untreated or treated with 1.25 ng/mL of oligomycin for 1 or 2 hr, followed by addition of 10 ug/mL of puromycin (Sigma #P9620) for 10 min. Cells were lysed for Western blot using anti-puromycin antibody (Sigma #MABE343) to label the newly synthesized proteins.
Data availability
Source data for Immunoblots are provided in Supplementary Figure 1. Gating strategies for flow cytometry experiments are provided in Supplementary Figure 2. RNA sequencing data described in this manuscript (associated with Extended Data Fig. 1 and Extend Data Fig. 7c) are deposited in the NCBI Gene Expression Omnibus (GSE134986). There are no restrictions on data availability.
Code availability
Analysis of CRISPRi screen results was carried out using custom code, MAGeCK-iNC, developed in the Kampmann lab, which was previously described37 and is freely available at
Extended Data
Extended Data Figure 1. Induction of ATF4 target genes under mitochondrial stress conditions and characterization of the ATF4 translational reporter.
(a) HEK293T cells were treated with 50 μg/mL doxycycline or with 1.25 ng/mL of oligomycin for 16 h, and transcript levels were compared to untreated cells using RNA-Seq for n = 2 (doxycyclin) or n = 3 (oligomycin) independent experiments. Differentially expressed genes and P values were determined as described in the Methods (full datasets in Supplementary Tables 1,2). This Figure analyzes significantly induced genes (Padj < 0.05) with at least a 2-fold increase in treated over untreated conditions. Enrichment analysis for targets of transcription factors annotated in the TRRUST database57 (statistical analysis described in the Methods) detected ATF4 as the only significant transcription factor (Padj < 0.05) for both treatments. Genes induced by both treatments are listed, as well as genes annotated as ATF4 targets in TRRUST (dark green dots) or Bao et al.1 (light green dots).
(b) Quantification of the knockdown efficiency of sgRNAs targeting the eIF2α kinases by Quantitative RT-PCR (n = 3 technical replicates). For HRI, two independent sgRNAs, 1 and 2, were characterized.
(c, d) Validation of the reporter cell line using endoplasmic reticulum stressor, tharpsigargin (Tg). Reporter cells expressing either individual or triple sgRNAs targeting the indicated eIF2α kinases were exposed to 75 nM Tg for 8 h before measuring reporter levels by flow cytometry. The induction of ATF4 reporter by Tg is blocked by PERK knockdown. The reporter fold change was quantified as in Fig. 1b (mean ± s.d., n = 3 culture wells).
(e) Pharmacological inhibition of mitochondrial function, using the mitochondrial ribosome inhibitor doxycycline, the electron transport chain inhibitors antimycin A and rotenone, and the ATP synthase inhibitor oligomycin, induces the ATF4 reporter. Reporter cells were exposed to the indicated treatments for 16 h before measuring reporter levels by flow cytometry. The reporter fold change was quantified as in Fig 1b (mean ± s.d., n = 3 culture wells).
(f) CRISPRi knockdown (KD) of factors required for mitochondrial protein homeostasis (HSPD1 and LONP1) and mitochondrial ribosomal proteins (MRPL17 and MRPL22) (red) induce the ATF4 reporter compared to WT cells (blue). Similar results obtained in n > 3 independent experiments.
(g) DELE1 and HRI are not required to trigger the integrated stress response in response to ER stress. Reporter cells expressing non-targeting control sgRNAs (NTC) or sgRNAs targeting HRI or DELE1 were exposed to 75 nM thapsigargin for 8 h before measuring reporter levels by flow cytometry. The reporter fold change (mean ± s.d., n = 3 culture wells) is the ratio of median fluorescence values for thapsigargin over untreated samples.
Extended Data Figure 2. Expression levels of DELE1 constructs and investigation of DELE1 cleavage and export mechanisms.
(a) Commercially available antibodies fail to detect DELE1. Lysates from HEK293T cells that were either WT or expressing a sgRNA knocking down DELE1 were probed with the indicated DELE1 antibodies and an antibody against β-actin. None of the bands detected by the DELE1 antibodies decreases in intensity in DELE1 knockdown cells. *Non-specific band. Similar results obtained in n > 2 independent experiments.
(b) Quantitative RT-PCR quantification of DELE1 mRNA levels in HEK293T cells knocking down (KD) DELE1 by CRISPRi and/or expressing DELE1-mClover stably from the AAVS1 safe-harbor locus or via transient transfection (n = 3 technical replicates).
(c-e) PMPCB knockdown induces the ATF4 translational reporter in the absence of mitochondrial stressors in an HRI-dependent and DELE1-dependent manner. (c) Left, representative immunoblot of ATF4, DELE1-mClover, PMPCB in two PMPCB knockdown cell lines and NTC control cells treated with 1.25 ng/ml oligomycin for 16 hrs where indicated. Right, quantification (mean ± s.d. n = 2 blots). (d,e) HEK293T reporter cells were co-transfected with PMPCB sgRNAs 1 or 2 (with a BFP marker) and HRI (d) or DELE1 (e) sgRNA (with a GFP maker) to generate a cell line with four populations as shown. The intensity of ATF4 reporter was quantified via flow cytometry (mean ± s.d., n = 3 culture wells).
(f) Quantification of immunoblot shown in Fig. 3f. n = 2 blots.
(g) Immunoblot of OPA1, DELE1-mClover, ATF4 in OPA1 knockdown and NTC control cells treated with 1.25ng/mL oligomycin for 16 hrs (Oli) or untreated (Un). Similar results obtained in n > 2 technical replicates.
(h) Measurement of mitochondrial potential by TMRE staining in cells treated with 1.25 ng/mL of oligomycin or 5 μM CCCP for 4 hrs (mean ± s.d., n = 3 culture wells, P values were determined using two-tailed unpaired t test).
(i) Subcellular localization of DELE1L and DELE1S in OPA1 knockdown and NTC control cells treated with 1.25ng/mL oligomycin for 16 hrs (Oli) or untreated (Un). Similar results obtained in n > 2 technical replicates.
For gel source data, see Supplementary Figure 1.
Extended Data Figure 3. Characterization of DELE1 submitochondrial localization.
(a-b) Zoom-out views for two-color 3D-STORM super-resolution images of DELE1-mClover vs. TOM20 and Hsp60. Two-color DELE1-mClover (magenta) vs. (a) TOM20 (green) or (b) Hsp60 (green), followed by the two separated color channels. Scale bars: 1 μm. The boxed regions correspond to Fig. 3g, h. Similar results were obtained in n = 3 independent experiments.
(c) Biochemical fractionation indicates that DELE1 associates with mitochondrial membranes.
Cells stably expressing DELE1-mClover were fractionated into cytosol and mitochondria. Mitochondria were incubated in either isotonic buffer (10 mM Tris HCl, pH 6.7, 0.15 mM MgCl2 0.25mM sucrose,1 mM DTT, protease inhibitor cocktail (Sigma #5892970001)) or H2O (extreme hypotonic condition) for 5 min, followed by centrifugation (10000 xg for 10 min) to separate the supernatant and pellet. The pellet was either dissolved with RIPA buffer or incubated with 0.1M NaCO3 (pH=11.4) for 30 min at 4°C. Supernatant and pellet from NaCO3-treated samples were collected for WB. Unlike soluble matrix protein LonP1 and HSPD1, which can be extracted with H2O incubation, only small proportion of DELE1 is present in the supernatant. NaCO3 can extract the majority of the DELE1S but not DELE1L, which is similar to the pattern of mitochondrial membrane protein VDAC, suggesting that DELE1L is more likely a membrane associated protein. n = 2 independent experiments.
For gel source data, see Supplementary Figure 1.
(d) The indicated DELE1-mClover constructs were transiently overexpressed in reporter cells, and reporter induction was quantified by flow cytometry (mean ± s.d., n = 3 culture wells). Subcellular localization was evaluated by microscopy in cells also expressing mitochondrial-targeted mRuby.
(e) Lack of co-localization of transiently expressed DELE1ΔN100-mClover and DELE1ΔN148-mClover (green) with the mitochondrial stain Mitotracker (red). Scale bar, 7 μm. Similar results obtained in n = 2 culture wells.
(f) Increased detection of DELE1-mClover outside the mitochondria upon oligomycin treatment. 3D-STORM super-resolution images of stably expressed DELE1-mClover (colors indicating depth in the z dimension) in untreated cells (left) and cells treated with 1.25 ng/mL oligomycin for 16 h (right). Areas boxed in red in the top panels are shown in higher magnification in the bottom panels. Similar results were obtained in n = 3 independent experiments.
(g) Co-localization of transiently expressed HRI-mClover and DELE1-mClover with the mitochondrial-targeted mRuby (Mito7-mRuby). Scale bar, 7 μm. n = 1 culture well.
Extended Data Figure 4. The protein kinase assay of purified HRI.
(a) Purified recombinant HRI, DELE1 and eIF2α. 800 ng of each recombinant protein was subjected to SDS-PAGE and stained with Coomassie blue (n = 1 gel).
(b,c) HRI kinase reactions were performed with 1 μM recombinant yeast eIF2α and various amounts of purified recombinant HRI protein. Reactions were stopped at the time points indicated by removing 5-μl aliquots of the kinase reaction mixture and mixing with equal volume of 2×SDS loading buffer. The SDS samples were then dotted on nitrocellulose blots and subjected to immunoblotting analysis with eIF2αP IgGs. (b) Representative dot blot. (c) Densitometric quantification of dot blots expressed as average value from n = 2 individual experiments. To enable a linear range reaction, 25nM HRI and 5 min incubation time were used for all the subsequence experiments.
(d) Enzyme kinetic constants for HRI activity with or without DELE1 in the presence or absence of hemin (mean ± s.e.m., n = 3 individual reactions, fit for data shown in Fig. 4f). Kinetic constants were determined by fitting to the Michaelis-Menten equation using least-squares fit using Prism version 6.07. * The constants calculated from HRI + hemin are not accurate because under the current substrate concentration range, the enzymatic reaction is first order, never reaching Vmax. But higher substrate concentrations cannot be used to obtain Vmax conditions, as purified eIF2α protein will precipitate at higher concentrations.
Extended Data Figure 5. Measurement of protein synthesis under mitochondrial stress.
(a) Immunoblot of HRI and ATF4 in wild type and HRI knockout (KO) cells. Cells were untreated or treated with 1.25 ng/mL oligomycin for 16 h. Similar results obtained in n = 2 independent experiments. For gel source data, see Supplementary Figure 1.
(b) Newly synthesized protein was labeled with puromycin. WT or HRI KO cells were treated with 1.25ng/mL puromycin for 1 or 2 hrs, or left untreated, followed by 10 min puromycin (10ug/mL) incubation. Protein from each sample was quantified by BCA assay (ThermoFisher #23225) and equally loaded. Two immunoblots from separate experiments probed with anti-puromycin (top) and Ponceau S staining (middle). Bottom, quantification of newly synthesized protein (ratio of anti-puromycin signal to Ponceau S signal) for these blots from n = 2 experiments.
Extended Data Figure 6. Examination of mitochondrial stress response with a broad range of mitochondrial toxins and in non-HEK293T cells.
(a) Immunoblot of ATF4 in wild type, DELE1(KD), HRI (KD) and OMA1(KD) HEK293T cell lines under different mitochondrial stress conditions. Cells were left untreated (Un) or treated with 40nM Antimycin A (AA), 40nM Rotenone (Rot), 50 ug/mL Doxycycline (Dox), or 5 μM CCCP for 2 and 4 hrs. Left, representative blots. Right, ATF4 levels were quantified and normalized to β-actin (mean ± s.d., n = 2 blots).
(b) A broad range of mitochondrial toxins stimulates the accumulation of DELE1S. Cells stably expressing DELE1-mClover were untreated or treated with a panel of mitochondrial toxins for 16 h (see Methods for details) and subjected to Western blotting with antibodies detecting DELE1-mClover, ATF4 and actin. Top, representative blot. Middle, bottom, ATF4, DELE1L-mClover and DELE1S-mClover levels were quantified (mean ± s.d., n = 2 blots).
(c) Subcellular localization of DELE1L and DELE1S under a broad range of mitochondrial toxins.
Biochemical fractionation of cells stably expressing DELE1-mClover that were either treated with different mitochondrial toxins as indicated for 16 h or left untreated. β-actin and LonP1 were probed as markers for cytosol and mitochondria, respectively. Similar results obtained in n = 2 independent experiments.
(d) Examination of OPA1 cleavage under a broad range of mitochondrial toxins. Similar results obtained in n = 2 independent experiments.
(e) Immunoblot of ATF4 in wild type, DELE1(KD), HRI (KD) and OMA1(KD) in the WTC11 human iPSC line. Cells were left untreated or treated with 1.25 ng/mL oligomycin for 4 hrs. Similar results obtained in n = 2 independent experiments.
(f) Immunoblot of ATF4 in wild type, DELE1(KD), HRI (KD) and OMA1(KD) in the human Hela cell line. Cells were left untreated or treated with 1.25 ng/mL oligomycin for 2 and 4hrs.
Similar results obtained in n = 2 technical replicates.
For gel source data, see Supplementary Figure 1.
Extended Data Figure 7. The DELE1-HRI pathway can be maladaptive and its blockade induces an alternative program.
(a) OMA1, DELE1, HRI knockdown is protective during oligomycin treatment. HEK293T cells expressing non-targeting control sgRNA (NTC) or an sgRNA knocking down HRI, DELE1 or OMA1 were untreated or treated with 2.5 ng/mL oligomycin for 16 h, and cell numbers were determined by counting (mean ± s.d., n = 3 culture wells, P values were determined by two-tailed unpaired t test).
(b) HRI knockdown is protective for cells with depleted mitochondrial ribosomal protein MRPL17, but sensitizes cells with depleted mitochondrial protease LonP1. HEK293T cells were co-infected with lentiviral construct expressing green fluorescent protein and an sgRNA knocking down HRI, and with a lentiviral construct expressing blue fluorescent protein and a non-targeting control sgRNA (NTC) or an sgRNA knocking down MRPL17, MRPL22, or LONP1. Cells were cultured for 9 days and proportions of cells expressing green and blue fluorescent proteins were quantified on days 3, 6, 8 and 9 post infection by flow cytometry (top). Thus, the effect of HRI knockdown on proliferation in different genetic backgrounds could be evaluated in an internally controlled experiment. In parallel, the ATF4 reporter was quantified (bottom). Mean ± s.d., n = 3 culture wells.
(c) HEK293T cells that were either infected with a non-targeting control sgRNA or in which OMA1, DELE1 or HRI was knocked down were untreated or treated with 1.25 ng/mL oligomycin for 16 h, and transcriptomes were analyzed by RNA sequencing for n = 3 independent experiments. Differentially expressed genes and P values were determined as described in the Methods (full datasets in Supplementary Tables 2,5-7). The heatmap only includes genes expression of which changed significantly upon oligomycin treatment (Padj < 0.05) by at least two-fold in at least one genetic background. Hierarchical clustering reveals four major gene clusters. Gene groups are indicated by dots in different colors: ATF4 targets annotated by the TRRUST database (dark green, Padj value calculated by Enrichr) or Bao et al.1 (light green, P value calculated by Fisher’s exact test), genes co-expressed with HRI in the ARCHS4 database (orange dots, Padj value calculated by Enrichr), mitochondrially encoded genes (golden dots), heat-shock proteins (brown dots).
Extended Data Table 1.
Cloning strategies and oligonucleotide sequences used for plasmids generated
| Plasmid | Insert (5’ to 3’) | Backbone | Cloning Methods |
|---|---|---|---|
| pXG237 | PCR mApple | Inverse PCR on pMK1163 | Gibson Assembly |
| F:cattcctcgattccagcaaagcaccgcaacatggtgtccaagggcgaagagaacaacatg | F:cgcccttggacaccatgttgcggtgctttgctggaatcgagg | ||
| R:agttattaggtccctcgacgaattcgcctaggcttacttgtacagctcgtccattccgcc | R:gacgagctgtacaagtaagcctaggcgaattcgtcgagggacc | ||
| pXG260 | PCR EGFP | Inverse PCR on pMK237 | Gibson Assembly |
| F:ccctcctccccgccctcagggtccacggccaccatggtgagcaagggcgaggagc | F:gtgtccaagggcgaagagaac | ||
| R:attaggtccctcgacgaattcgcctaggcctacttgtacagctcgtccatgc | R:catggtggccgtggaccctg | ||
| pXG286 | PCR DELE1 from human cDNA | Linearize pMK1253 by NheI and BamHI | Gibson Assembly |
| F:agtttttttcttccatttcaggtgtcgtgagctagcgaattcagcgacatgtggcgcctc | |||
| R:tcactgcggttggcgaccggtggatcccgcggccgctgccaaaacctagtcttacaacac | |||
| pXG289 | PCR eIF1A promoter-DELE1-mClover from pXG286 | Digest pMTL3 by SpeI and MluI | Ligation |
| F:tattgactagtggataaccgtattaccgccatgc | |||
| R:gaattcacgcgtttacttgtacagctcgtccatgc | |||
| pXG272 | Gene block containing HRI cDNA from IDT | Digest pMK1253 by EcoRI and XbaI | Ligation |
| pXG294 | PCR DELE1(Δ100aa) from pXG286 | Digest pXG286 with EcoRI and NotI | Ligation |
| pXG336 | PCR DELE1(Δ125aa) from pXG286 | Digest pXG286 with EcoRI and NotI | Ligation |
| F:aattcagcgacatgcgtttcttctcatctcccttgtg | |||
| R:tcactgcggttggcgaccggtggatcccgcggccgctgccaaaacctagtcttacaacac | |||
| pXG305 | PCR DELE1(Δ148aa) from pXG286 | Digest pXG286 with EcoRI and NotI | Ligation |
| F:aattcagcgacatgggcccagctcccaggcac | |||
| R:tcactgcggttggcgaccggtggatcccgcggccgctgccaaaacctagtcttacaacac | |||
| pXG334 (DELE1 Δ140-149) | None | Inverse PCR on pXG286 | Ligation |
| F:ccagctcccaggcacactg | |||
| R:cagtgaggagcatgggtgcc | |||
| pXG313 (DELE1 Δ123-139) | None | Inverse PCR on pXG286 | Ligation |
| F:cgacaacacatcctccccagc | |||
| R:actgtgccaggagcagtgttc | |||
| pXG323 (DELE1 Δ113-122) | None | Inverse PCR on pXG286 | Ligation |
| F:cccctggaccgtttcttctcat | |||
| R:aggtcctgctggcagggatg | |||
| pXG324 (DELE1 Δ103-112) | None | Inverse PCR on pXG286 | Ligation |
| F:cagcgggtagaacactgctcc | |||
| R:gatctgccttgccagctgcag | |||
| pXG325 (DELE1 Δ93-102) | None | Inverse PCR on pXG286 | Ligation |
| F:cacttccaggcatccctgcca | |||
| R:ggccagagtgccccaagatat | |||
| pXG326 (DELE1 Δ83-92) | None | Inverse PCR on pXG286 | Ligation |
| F:gtgctggccctgcagctggc | |||
| R:tagggtgttcggggagacac | |||
| pXG327 (DELE1 Δ73-82) | None | Inverse PCR on pXG286 | Ligation |
| F:tgggatgccatatcttgggg | |||
| R:ccattggaaggcatccttcc |
Extended Data Table 2.
Protospacer sequences of individually cloned sgRNAs
| Target gene | sgRNA | Protospacer (5’ to 3’) |
|---|---|---|
| HRI | sgRNA1 | GTAGCTGCAGCATCGGAGTG |
| sgRNA2 | GACGGCGCTAGCTGCAGCAT | |
| PKR | GGCGGCGGCGCAGGTGAGCA | |
| GCN2 | GCAGCGCTGCGCCCAAGGCA | |
| PERK | GCAGAGGCCGGGCTGAGACG | |
| DELE1 | sgRNA1 | GAGACCAACCCTTGGGACAG |
| sgRNA2 | GGCCCGCCCCCACTCCAGTT | |
| OMA1 | sgRNA1 | GCGAGTAGGATCGTGCCCAG |
| sgRNA2 | GCACTTCCTGCCCGCCATAC | |
| OPA1 | sgRNA1 | GGAGACGGGCCAAGATTGAG |
| PMPCB | sgRNA1 | GCCCTGCCCGACCCACTGTT |
| PMPCB | sgRNA2 | GCGCCACAGATCCGAACAGT |
| HSPD1 | GTTCGGAAAGAAGGACACGT | |
| MRPL17 | GGCCCATACGGCGAAATACG | |
| MRPL22 | GGTAGCGGGAGGGCGAAAGA | |
| LONP1 | GGGGCTATGGCGGCGAGCAC | |
| Non-targeting control (NTC) sgRNAs | 1872 | GTCCACCCTTATCTAGGCTA |
| 2151 | GTGCCAGCTTGTGGTGTCGT | |
| 2152 | GCGACATCGGTTACATGTGG | |
| 2153 | GTAGACGTGCAAAAGCCGCA |
Extended Data Table 3.
qPCR primer sequences
| Genes | Orientation | Sequences (5’ to 3’) |
|---|---|---|
| HO-1 | Forward | GGTCCTTACACTCAGCTTTCT |
| Reverse | CATAGGCTCCTTCCTCCTTTC | |
| HRI | Forward | ACACCAACACATACGTCCAG |
| Reverse | GCTCCATTTCTGTTCCAAACG | |
| PKR | Forward | CGATACATGAGCCCAGAACAG |
| Reverse | AGAATTAGCCCCAAAGCGTAG | |
| PERK | Forward | TGTCTGCACATCTTCCTGC |
| Reverse | ACTAACCCAAAGTCTCCAACC | |
| GCN2 | Forward | TTCCCATTGTGAGTGTGCTAG |
| Reverse | TCTGATGTAAGTTGGCAAGGG | |
| DELE1 | Forward | AGGCTGTGACTTCCATTCAG |
| Reverse | TCGCCACTCTTCATGTTCTC | |
| IMMP2L | Forward | CAATATGATGCTGTGCGAGAA |
| Reverse | AGCCCCATTAAGACATGTGG | |
| β-actin | Forward | ACCTTCTACAATGAGCTGCG |
| Reverse | CCTGGATAGCAACGTACATGG |
Supplementary Material
Acknowledgments:
We thank Greg Mohl, Ben Herken and Danielle Swaney for contributions to preliminary experiments, Jaime Leong for FACS support, Christopher Richards for contributions to data visualization, Morgane Boone, Kevan Shokat and members of the Kampmann lab for discussions, Adam Frost, Isha Jain, Avi Samelson and Emmy Li for comments on the manuscript, Eric Chow (UCSF Center for Advanced Technology) for support with next-generation sequencing, Sarah Elmes (UCSF Laboratory for Cell Analysis) for support with FACS, Delaine Larson (UCSF Nikon Imaging Center) for support with fluorescence microscopy. This work was supported by the National Institutes of Health grants GM119139 (M.K.), DK26506 (M.A.C.), GM44037 (M.A.C.), OD022552 (A.P.W.), the Beckman Young Investigator Program (K.X.) and a Larry L. Hillblom Foundation Postdoctoral Fellowship (X.G.). M.K. and K.X. are Chan Zuckerberg Biohub Investigators.
Footnotes
Competing interests: The authors declare no competing interests.
Supplementary Information is available for this paper.
Main References:
- 1.Bao XR et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife 5, doi: 10.7554/eLife.10575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quiros PM et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol 216, 2027–2045, doi: 10.1083/jcb.201702058 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viader A et al. Aberrant Schwann cell lipid metabolism linked to mitochondrial deficits leads to axon degeneration and neuropathy. Neuron 77, 886–898, doi: 10.1016/j.neuron.2013.01.012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattson MP, Gleichmann M & Cheng A Mitochondria in neuroplasticity and neurological disorders. Neuron 60, 748–766, doi: 10.1016/j.neuron.2008.10.010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun N, Youle RJ & Finkel T The Mitochondrial Basis of Aging. Molecular cell 61, 654–666, doi: 10.1016/j.molcel.2016.01.028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suomalainen A & Battersby BJ Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nature reviews. Molecular cell biology 19, 77–92, doi: 10.1038/nrm.2017.66 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Fiorese CJ et al. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr Biol 26, 2037–2043, doi: 10.1016/j.cub.2016.06.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munch C & Harper JW Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature 534, 710–713, doi: 10.1038/nature18302 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papa L & Germain D Estrogen receptor mediates a distinct mitochondrial unfolded protein response. J Cell Sci 124, 1396–1402, doi: 10.1242/jcs.078220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papa L & Germain D SirT3 regulates the mitochondrial unfolded protein response. Mol Cell Biol 34, 699–710, doi: 10.1128/MCB.01337-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Q et al. A mitochondrial specific stress response in mammalian cells. The EMBO journal 21, 4411–4419, doi: 10.1093/emboj/cdf445 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harding HP et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Molecular cell 11, 619–633 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Wek RC, Jiang HY & Anthony TG Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34, 7–11, doi: 10.1042/BST20060007 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Qi LS et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183, doi: 10.1016/j.cell.2013.02.022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert LA et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661, doi: 10.1016/j.cell.2014.09.029 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JJ & London IM Regulation of protein synthesis by heme-regulated eIF-2 alpha kinase. Trends Biochem Sci 20, 105–108 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Kafina MD & Paw BH Intracellular iron and heme trafficking and metabolism in developing erythroblasts. Metallomics 9, 1193–1203, doi: 10.1039/c7mt00103g (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu L, Han AP & Chen JJ Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol Cell Biol 21, 7971–7980, doi: 10.1128/MCB.21.23.7971-7980.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matts RL, Schatz JR, Hurst R & Kagen R Toxic heavy metal ions activate the heme-regulated eukaryotic initiation factor-2 alpha kinase by inhibiting the capacity of hemin-supplemented reticulocyte lysates to reduce disulfide bonds. The Journal of biological chemistry 266, 12695–12702 (1991). [PubMed] [Google Scholar]
- 20.Harada T, Iwai A & Miyazaki T Identification of DELE, a novel DAP3-binding protein which is crucial for death receptor-mediated apoptosis induction. Apoptosis 15, 1247–1255, doi: 10.1007/s10495-010-0519-3 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Quiros PM, Langer T & Lopez-Otin C New roles for mitochondrial proteases in health, ageing and disease. Nature reviews. Molecular cell biology 16, 345–359, doi: 10.1038/nrm3984 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Baker MJ et al. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. The EMBO journal 33, 578–593, doi: 10.1002/embj.201386474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehses S et al. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol 187, 1023–1036, doi: 10.1083/jcb.200906084 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H, Botelho SC, Park K & Kim H Use of carbonate extraction in analyzing moderately hydrophobic transmembrane proteins in the mitochondrial inner membrane. Protein Sci 24, 2063–2069, doi: 10.1002/pro.2817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berlanga JJ, Herrero S & de Haro C Characterization of the hemin-sensitive eukaryotic initiation factor 2alpha kinase from mouse nonerythroid cells. The Journal of biological chemistry 273, 32340–32346, doi: 10.1074/jbc.273.48.32340 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Delaunay J, Ranu RS, Levin DH, Ernst V & London IM Characterization of a rat liver factor that inhibits initiation of protein synthesis in rabbit reticulocyte lysates. Proceedings of the National Academy of Sciences of the United States of America 74, 2264–2268, doi: 10.1073/pnas.74.6.2264 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H, Smith SB & Yoon Y The short variant of the mitochondrial dynamin OPA1 maintains mitochondrial energetics and cristae structure. The Journal of biological chemistry 292, 7115–7130, doi: 10.1074/jbc.M116.762567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adjibade P et al. DDX3 regulates endoplasmic reticulum stress-induced ATF4 expression. Sci Rep 7, 13832, doi: 10.1038/s41598-017-14262-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter P & Ron D The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086, doi: 10.1126/science.1209038 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Wek RC Role of eIF2alpha Kinases in Translational Control and Adaptation to Cellular Stress. Cold Spring Harb Perspect Biol 10, doi: 10.1101/cshperspect.a032870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S et al. HRI coordinates translation necessary for protein homeostasis and mitochondrial function in erythropoiesis. eLife 8, doi: 10.7554/eLife.46976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou A et al. Inhibition of the integrated stress response reverses cognitive deficits after traumatic brain injury. Proceedings of the National Academy of Sciences of the United States of America 114, E6420–E6426, doi: 10.1073/pnas.1707661114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das I et al. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science 348, 239–242, doi: 10.1126/science.aaa4484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acin-Perez R et al. Ablation of the stress protease OMA1 protects against heart failure in mice. Sci Transl Med 10, doi: 10.1126/scitranslmed.aan4935 (2018). [DOI] [PubMed] [Google Scholar]
Methods References
- 35.Sidrauski C et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife 4, e07314, doi: 10.7554/eLife.07314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen JJ et al. Compromised function of the ESCRT pathway promotes endolysosomal escape of tau seeds and propagation of tau aggregation. The Journal of biological chemistry, doi: 10.1074/jbc.RA119.009432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian R et al. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron 104, 239–255, doi: 10.1016/j.neuron.2019.07.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adamson B et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1882 e1821, doi: 10.1016/j.cell.2016.11.048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patro R, Duggal G, Love MI, Irizarry RA & Kingsford C Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419, doi: 10.1038/nmeth.4197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soneson C, Love MI & Robinson MD Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521, doi: 10.12688/f1000research.7563.2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuleshov MV et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research 44, W90–97, doi: 10.1093/nar/gkw377 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisen MB, Spellman PT, Brown PO & Botstein D Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America 95, 14863–14868, doi: 10.1073/pnas.95.25.14863 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saldanha AJ Java Treeview--extensible visualization of microarray data. Bioinformatics 20, 3246–3248, doi: 10.1093/bioinformatics/bth349 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Horlbeck MA et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 5, doi: 10.7554/eLife.19760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagy T & Kampmann M CRISPulator: a discrete simulation tool for pooled genetic screens. BMC Bioinformatics 18, 347, doi: 10.1186/s12859-017-1759-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682, doi: 10.1038/nmeth.2019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rust MJ, Bates M & Zhuang X Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795, doi: 10.1038/nmeth929 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang B, Wang W, Bates M & Zhuang X Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813, doi: 10.1126/science.1153529 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wojcik M, Hauser M, Li W, Moon S & Xu K Graphene-enabled electron microscopy and correlated super-resolution microscopy of wet cells. Nature Commun. 6, 7384, doi: 10.1038/ncomms8384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bossi M et al. Multicolor far-field fluorescence nanoscopy through isolated detection of distinct molecular species. Nano Lett 8, 2463–2468, doi: 10.1021/nl801471d (2008). [DOI] [PubMed] [Google Scholar]
- 52.Gorur A et al. COPII-coated membranes function as transport carriers of intracellular procollagen I. J Cell Biol 216, 1745–1759, doi: 10.1083/jcb.201702135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372, doi: 10.1038/nbt.1511 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Liao M et al. Impaired dexamethasone-mediated induction of tryptophan 2,3-dioxygenase in heme-deficient rat hepatocytes: translational control by a hepatic eIF2alpha kinase, the heme-regulated inhibitor. J Pharmacol Exp Ther 323, 979–989, doi: 10.1124/jpet.107.124602 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Dey M et al. PKR and GCN2 kinases and guanine nucleotide exchange factor eukaryotic translation initiation factor 2B (eIF2B) recognize overlapping surfaces on eIF2alpha. Molecular and cellular biology 25, 3063–3075, doi: 10.1128/MCB.25.8.3063-3075.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt EK, Clavarino G, Ceppi M & Pierre P SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6, 275–277, doi: 10.1038/nmeth.1314 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Han H et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic acids research 46, D380–D386, doi: 10.1093/nar/gkx1013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Immunoblots are provided in Supplementary Figure 1. Gating strategies for flow cytometry experiments are provided in Supplementary Figure 2. RNA sequencing data described in this manuscript (associated with Extended Data Fig. 1 and Extend Data Fig. 7c) are deposited in the NCBI Gene Expression Omnibus (GSE134986). There are no restrictions on data availability.