Abstract
BACKGROUND:
Microbial invasion of the amniotic cavity resulting in intra-amniotic infection is associated with obstetrical complications such as preterm labor with intact or ruptured membranes, cervical insufficiency, as well as clinical and histological chorioamnionitis. The most widely accepted pathway for intra-amniotic infection is the ascension of microorganisms from the lower genital tract. However, hematogenous dissemination of microorganisms from the oral cavity or intestine, retrograde seeding from the peritoneal cavity through the fallopian tubes, and introduction through invasive medical procedures have also been suggested as potential pathways for intra-amniotic infection. The primary reason that an ascending pathway is viewed as most common is that the microorganisms most often detected in the amniotic fluid are those that are typical inhabitants of the vagina. However, thus far, no studies have shown that microorganisms in the amniotic cavity are simultaneously present in the vagina of the woman from which they were isolated.
OBJECTIVE:
To determine the frequency with which microorganisms isolated from women with intra-amniotic infection are also present in the lower genital tract.
STUDY DESIGN:
This was a cross-sectional study of women with intra-amniotic infection with intact membranes. Intra-amniotic infection was defined as positive culture and elevated concentrations of IL-6 (>2.6ng/mL) in amniotic fluid and/or acute histologic chorioamnionitis and funisitis. Microorganisms isolated from bacterial cultures of amniotic fluid were taxonomically identified through matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) and 16S rRNA gene sequencing. Vaginal swabs were obtained at the time of amniocentesis for the identification of microorganisms in the lower genital tract. The overall bacterial profiles of amniotic fluids and vaginal swabs were characterized through 16S rRNA gene sequencing. The bacterial profiles of vaginal swabs were interrogated for the presence of bacteria cultured from amniotic fluid and for the presence of prominent (> 1% average relative abundance) operational taxonomic units (OTUs) within the overall 16S rRNA gene bacterial profiles of amniotic fluids.
RESULTS:
1) 75% (6/8) of women had bacteria cultured from their amniotic fluid that are typical residents of the vaginal ecosystem. 2) 62.5% (5/8) of women with bacteria cultured from their amniotic fluid also had these bacteria present in their vagina. 3) The microorganisms cultured from amniotic fluid and also detected in the vagina were Ureaplasma urealyticum, Escherichia coli, and Streptococcus agalactiae. 4) 16S rRNA gene sequencing revealed that women with intra-amniotic infection had bacterial profiles dominated by Sneathia, Ureaplasma, Prevotella, Lactobacillus, Escherichia, Gardnerella, Peptostreptococcus, Peptoniphilus, and Streptococcus, many of which had not been cultured from the amniotic fluid samples. 5) 70% (7/10) of the prominent (> 1% average relative abundance) operational taxonomic units found in amniotic fluid were also prominent in the vagina.
CONCLUSIONS:
The majority of women with intra-amniotic infection had bacteria cultured from their amniotic fluid that were typical vaginal commensals and that were detected within the vagina at the time of amniocentesis. Molecular microbiological interrogation of amniotic fluid from women with intra-amniotic infection revealed that the bacterial profiles of amniotic fluid were largely consistent with those of the vagina. These findings indicate that ascension from the lower genital tract is the primary pathway for intra-amniotic infection.
Keywords: bacteria, microbial invasion, amniotic cavity, microbiome, microbiota, pregnancy, chorioamnionitis, preterm birth, funisitis, vaginal flora, Sneathia, Ureaplasma, Gardnerella, amniotic fluid, 16S rRNA sequencing, culture
INTRODUCTION
Microbial invasion of the amniotic cavity resulting in intra-amniotic infection has been associated with obstetrical complications [1–6], including spontaneous preterm labor [3, 7–17], preterm premature rupture of membranes [18, 19], cervical insufficiency [20–26], a sonographic short cervix [27], idiopathic vaginal bleeding [28, 29], and histological [30, 31] and clinical chorioamnionitis [32, 33]. Indeed, intra-amniotic infection has been detected in 6 – 35% of women with preterm labor and intact membranes [3, 8–17, 34], and in 32 – 50% of women with preterm premature rupture of membranes (PPROM) [18, 19]. It has further been detected in 38% of cases of histological chorioamnionitis [30] and in 61% of cases of clinical chorioamnionitis at term [32]. Intra-amniotic infection has thus been associated with labor dysfunction [35–37], maternal morbidity [38–42], and neonatal morbidity and mortality [43–50]. Recent evidence indicates that intra-amniotic infection can be treated [51–56], highlighting the need for further investigation into its etiologies.
Multiple routes of invasion have been proposed for intra-amniotic infection [2, 5, 17, 57, 58], including (1) ascending infection from the lower genital tract through the cervix, (2) hematogenous dissemination from distant sites such as the intestine or the oral cavity through the placenta, (3) retrograde seeding from the peritoneal cavity through the fallopian tube, and (4) accidental introduction of microorganisms at the time of invasive medical procedures. However, ascending infection from the lower genital tract is widely viewed as the primary route by which microbial invasion of the amniotic cavity occurs [2, 5, 17, 57–60]. The principal evidence supporting ascending infection as the primary route for intra-amniotic infection is that the bacterial taxa most often identified in the amniotic cavity are typical members of the human vaginal microbiota [4, 59, 61–71], including Ureaplasma and Mycoplasma spp. [1, 13, 18, 19, 32, 62, 72–77], Gardnerella vaginalis [1, 13, 19, 32, 62, 76, 77], Streptococcus agalactiae [13, 18, 32, 72, 75, 77, 78], Escherichia coli [1, 32], Sneathia and Leptotrichia spp. [13, 19, 32, 72, 73, 75, 76], and Prevotella spp. [62, 77].
Direct evidence of ascending infection as a primary cause of intra-amniotic infection is however lacking. Specifically, demonstration of ascending infection requires showing that bacteria in the amniotic fluid are also present in the vagina of the woman from whom the amniotic fluid was collected. Molecular surveys will be beneficial for this task as they provide greater insight into the diversity of microorganisms inhabiting body sites than can culture [4, 13, 18, 32]. Although molecular surveys have been used to characterize the composition of intra-amniotic [13, 18, 19, 32, 74, 79–82] and vaginal [68–71, 83–93] microbial communities in pregnant women, the concurrent presence of specific microorganisms in the amniotic fluid and the vagina has not been investigated. The objective of this study were therefore to characterize the microorganisms found in amniotic fluid of women with intra-amniotic infection and intact membranes, and to evaluate the frequency of ascending infection by determining whether the microorganisms cultured from, and molecularly identified in, amniotic fluid were also present in the vagina.
MATERIALS AND METHODS
Study Population
This was a cross-sectional study of women who had an amniocentesis performed for the diagnosis of intra-amniotic inflammation and/or infection (see Clinical Definitions below). Patients with intra-amniotic infection were included if they met the following criteria: 1) live intrauterine pregnancy, 2) intact chorioamniotic membranes, 3) the amniotic fluid cultivar was stored and revived, 4) if amniotic fluid was available for molecular microbial characterization, the cultivar was confirmed to be present within the fluid using 16S rRNA sequencing, and 5) vaginal swab samples were collected from the patient within 24 hours of the amniocentesis. Patients were excluded from the study if chromosomal or fetal anomalies were present. The collection of samples and their use for research was approved by the Human Investigation Committee of Wayne State University and the Institutional Review Board of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. All subjects provided written informed consent. Demographic characteristics of the study population are shown in Table 1.
Table 1.
Demographic characteristics of the patients in the study.
| #1 | 26 | Black | 20.1 | 20.7 | Vaginal | 296 |
| #2 | 20 | Black | 31.3 | 31.4 | Vaginal | 1525 |
| #3 | 38 | Black | 22.0 | 22.1 | Vaginal | 471 |
| #4 | 17 | Black | 37.9 | 37.9 | Vaginal | 3060 |
| #5 | 27 | Black | 23.0 | 23.1 | Vaginal | 640 |
| #6 | 31 | Black | 19.6 | 34.9 | Vaginal | 1965 |
| #7 | 24 | Black | 39.9 | 39.9 | Vaginal | 3085 |
| #8 | 21 | White | 22.0 | 22.1 | Vaginal | 494 |
Clinical Definitions
Gestational age was determined by the date of the last menstrual period and confirmed by ultrasound examination. The gestational age derived from sonographic fetal biometry was used if the estimation was inconsistent with menstrual dating. Intra-amniotic inflammation was defined as an amniotic fluid IL-6 concentration ≥ 2.6 ng/mL [12, 33, 94–99] and/or the presence of acute histologic chorioamnionitis and funisitis (see Placental histopathological examination below). Intra-amniotic infection was defined as a positive amniotic fluid culture, including genital mycoplasmas [2, 7, 8, 100, 101] and intra-amniotic inflammation [19, 76, 102–112]. Clinical chorioamnionitis was diagnosed by the presence of maternal fever (temperature >37.8°C) accompanied by two or more of the following criteria: (1) uterine tenderness, (2) foul-smelling amniotic fluid, (3) fetal tachycardia (heart rate >160 beats/min), (4) maternal tachycardia (heart rate >100 beats/min), and (5) maternal leukocytosis (leukocyte count >15,000 cells/mm3) [32, 33, 98, 104–108, 113–119].
Placental histopathological examination
Placentas were examined histologically by perinatal pathologists blinded to clinical diagnoses and obstetrical outcomes according to standardized Perinatology Research Branch protocols [120, 121]. Briefly, three to nine sections of the placenta were examined, and at least one full-thickness section was taken from the center of the placenta; others were taken randomly from the placental disc. Acute inflammatory lesions of the placenta (maternal inflammatory response and fetal inflammatory response) were diagnosed according to established criteria, including staging and grading [120, 122]. The results of placental histopathological examination are presented in Table 2.
Table 2.
Clinical characteristics of the patients in the study.
| #1 | 20 | 126800 | 25 | Ureaplasma urealyticum | Stage 3 | Stage 2 |
| #2 | 206 | 169372 | 1 | Staphylococcus warneri | Stage 3 | Stage 2 |
| #3 | 340 | 171100 | 9 | Streptococcus anginosus | Stage 3 | Stage 1 |
| #4 | 1650 | 29300 | 0.9 | Escherichia coli, Ureaplasma urealyticum | Stage 1 | None |
| #5 | 110 | 148500 | 1 | Escherichia coli | Stage 2 | Stage 2 |
| #6 | 10 | 39200 | 19 | Ureaplasma urealyticum | None | None |
| #7 | 0 | 1485 | 1 | Streptococcus agalactiae | Stage 2 | Stage 2 |
| #8 | 299 | 102800 | 0 | Ureaplasma urealyticum | Stage 3 | Stage 1 |
Acute histologic chorioamnionitis: Stage 1, early, acute subchorionitis/chorionitis; Stage 2, intermediate, acute chorioamnionitis; Stage 3, necrotizing chorioamnionitis.
Funisitis: Stage 1, early umbilical phlebitis/chorionic vasculitis; Stage 2, intermediate, umbilical arteritis.
Amniotic fluid sample collection
Amniotic fluid samples were obtained by transabdominal amniocentesis under antiseptic conditions and monitored by ultrasound in order to detect intra-amniotic inflammation and/or infection in patients with intact membranes. Samples of amniotic fluid were transported to the laboratory in a sterile capped syringe for clinical determinations including amniotic fluid white blood cell count [123], Gram stain examination [124], and glucose concentration [125] (Table 2). The rest of the sample was used for research purposes, which included the determination of IL-6. Determination of IL-6 in amniotic fluid
Amniotic fluid concentrations of IL-6 were determined by using a sensitive and specific enzyme immunoassay obtained from R&D Systems (Minneapolis, MN). The IL-6 concentrations were determined by interpolation from the standard curve. The inter- and intra-assay coefficients of variation for IL-6 were 8.7% and 4.6%, respectively. The detection limit of the IL-6 assay was 0.09 pg/mL. The IL-6 concentrations in amniotic fluid were determined for clinical purposes.
Clinical microbiology
Amniotic fluid was cultured for aerobic and anaerobic bacteria. Specifically, amniotic fluid was inoculated onto chocolate agar, trypticase soy agar with 5% sheep blood, and MacConkey agar culture media. Aerobic plates were incubated at 35°C in an 8% CO2 chamber. Anaerobic plates were incubated at 35°C in 5% CO2, 10% hydrogen and 85% nitrogen. Plates were incubated for 4 days. A Mycofast test kit (MYCOFAST US; Logan, UT) was used for the detection of Mycoplasma and Ureaplasma spp. [126]. Initial taxonomic characterization of cultivars was done via discriminatory biochemical tests and matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) [127].
Vaginal swab collection
Vaginal Dacron swabs (Medical Packaging Swab-Pak, Camarillo, CA) were obtained using a Pederson speculum in the absence of lubrication and turned at a 45° angle to enable the collection of posterior fornix fluid samples. All vaginal samples were collected within 24 hours of amniocentesis, and prior to any rupture of membranes. All swabs were stored at ≤ −70 °C until analysis.
Isolation of DNA from amniotic fluid and vaginal samples
Prior to DNA extraction, the order of all samples was randomized to avoid cross-contamination of DNA between samples based on body site or patient identity. Total DNA was extracted from vaginal swabs, 200 μl of amniotic fluid supernatant, and 200 μl of amniotic fluid pellet (all pellets were resuspended in 450 μl sterile PBS) using the DNeasy PowerLyzer PowerSoil Kit (QIAGEN, Valencia, CA) with four modifications to the manufacturer’s protocol: 1) vaginal swabs were immersed in 500 μl of the supplied bead solution and 200 μl of phenol:chloroform:isoamyl alcohol pH 7–8 solution for 10 minutes prior to cell lysis by mechanical disruption rather than being suspended in 700 μl of supplied bead solution alone; 2) 100 μl of solution C2, 100 μl of solution C3, and 1 μl of RNase A were added and samples were incubated at 4°C for 5 minutes prior to centrifugation rather than being added to samples over two steps; 3) lysates were combined with 650 μl of solution C4 and 650 μl of 100% ethanol prior to loading samples on the supplied spin column rather than being combined with 1200 μl of solution C4 alone; 4) DNA was eluted in 60 μl solution C6 rather than 100 μl solution C6. To assess potential background DNA contamination within the DNA extraction kit or reagents, three blank DNA extraction kit control samples were processed alongside biological samples and sequenced. Purified DNA was stored at −20 ° C.
Amplification and sequencing of bacterial 16S rRNA genes in amniotic fluid and vaginal swabs
The 16S rRNA gene is widely used as a phylogenetic marker gene for characterizing bacterial communities in environmental and clinical samples. To characterize and compare the bacterial communities in paired amniotic fluid and vaginal samples, the V4 region of the 16S rRNA gene was amplified (515F/806R primers) from sample DNA extracts and sequenced at the University of Michigan’s Microbial Systems Molecular Biology Laboratory [https://microbe.med.umich.edu/services] using the dual indexing strategy developed by Kozich et al [128]. Sequencing was done on the Illumina MiSeq platform, using a MiSeq V2 500 cycle Reagent Kit (MS102–2003), according to the manufacturer’s instructions with modifications found in Kozich et al [128]. AccuPrime High Fidelity Taq (Life Technologies, 12346094) was used instead of AccuPrime Pfx SuperMix. Each PCR reaction contained 1.0 μM of each primer, 3 μl template DNA, and 0.15 μl AccuPrime HiFi Polymerase and DNase-free water to produce a final volume of 20 μl. PCR was performed using the following conditions: 95 °C for 2 min, followed by 30 cycles of 95 °C for 20 sec, 55 °C for 30 sec and 72 °C for 5 min, with an additional elongation at 72 °C for 10 min. To determine if bacteria cultured from amniotic fluid were also present in the vagina of respective patients (i.e. the 16S rRNA gene of the cultured bacterium was also detected in the vagina), the V4 region of the 16S rRNA gene was amplified (515F/806R primers) from vaginal DNA extracts and sequenced at Michigan State University’s Research Technology Support Facility [https://rtsf.natsci.msu.edu/] using protocols established by Caporaso et al [129].
16S rRNA gene sequence processing and bacterial community characterization
Mothur software (v 1.39.5) was used to assemble paired-read contigs from FASTQ files, to trim, filter, and align sequences, to identify chimeras, to assign sequences to taxonomies, and to assign sequences to operational taxonomic units (OTUs) based on their percent nucleotide similarity [130]. Briefly, quality-filtered sequences (maximum length 300 bp, with no ambiguous base calls and homopolymers of no more than 8 bp) were aligned to the SILVA 16S rRNA gene reference database (release 102) [131]. Chimeras were identified using the method of Edgar et al [132] as implemented in mothur, and these sequences were subsequently removed. A preclustering step (diffs = 2) was performed to reduce the impact of sequencing errors. OTUs were defined by clustering 16S rRNA gene sequences at a nucleotide similarity level of 99%. The remaining sequences were classified against the SILVA 16S rRNA gene reference database using a k-nearest neighbor approach with a confidence threshold of 80%. Any reads derived from an unknown domain, Eukaryota, chloroplasts, mitochondria, or Archaea, were removed.
Good’s coverage values (an indicator of sample coverage) for all amniotic fluid supernatant and pellet samples, vaginal swabs, and the three blank DNA extraction kit controls exceeded 98.0%. Raw OTU count data were converted to percentages within each subject’s dataset. There was no difference between the structure of the bacterial profiles of the supernatant and pellet portions of amniotic fluid samples (N = 6; NPMANOVA; Bray-Curtis: F = 0.698, p = 0.590), yet they were highly patient-specific (F = 3.337, p = 0.004). Therefore, bacterial community data for amniotic fluid supernatants and pellets for individual patients were combined bioinformatically to generate a single amniotic fluid bacterial community profile per patient. The profiles of amniotic fluid and blank DNA extraction kits differed in both composition (Jaccard; F = 1.30 p = 0.014) and structure (Bray-Curtis; F = 2.11 p = 0.045). Forty OTUs were identified in the three blank DNA extraction kit samples. The two OTUs with the highest mean relative abundance in extraction kit samples (OTU 87: Bacteroides and OTU 8: Escherichia) were found in two of the three kit controls. Only two OTUs (OTU 5: Lactobacillus and OTU 38: Streptococcus) were present in all three kit controls, and they were present at lower mean relative abundances in kit controls than in amniotic fluid samples. Therefore, we did not remove any OTUs from the dataset.
16S rRNA gene sequencing and taxonomic identification of bacteria cultured from amniotic fluid
Amniotic fluid bacterial cultivars were recovered from frozen stocks on chocolate agar, Columbia CNA with sheep blood agar, MacConkey agar, or SP4 broth with urea media. Genomic DNA was extracted from pure cultures of the bacteria using an UltraClean Microbial DNA Isolation kit (MoBio) following the manufacturer’s protocol. The 16S rRNA gene was first amplified from purified DNA with the 8F/1492R primer set and then bidirectionally sequenced using the Sanger chain termination method with the 515F and 806R primers targeting the V4 hypervariable region of the gene. Forward and reverse reads were trimmed using DNA Baser software (http://www.dnabaser.com/) with default settings, and assembled using the CAP (contig assembly program) of BioEdit software (v7.2.5) with default settings. The initial taxonomic identity of each bacterial cultivar as determined by MALDI-TOF was then confirmed by searching for similarity by BLAST against cultured bacterial type strain 16S rRNA gene sequences ≥1200 bp (12,736 sequences) contained within the RDP (Ribosomal Database Project) database (Release 11, Update 5) [133, 134]. BLAST results were consistent with those of MALDI-TOF for each of the bacterial cultivars.
Determining if bacteria cultured from amniotic fluid are also present in the vagina
To determine if bacterial cultivars from amniotic fluid were also present in the patient’s vagina, the 16S rRNA gene sequences obtained from each patient’s vaginal sample were screened for the presence of the 16S rRNA gene sequence of that patient’s respective cultivar(s). Specifically, the V4 region of the 16S rRNA gene sequence for each cultivar was trimmed to exclude the 515F and 806R primer regions, and a matching sequence was identified by searching for similarity using BLAST [133] against individual databases containing each woman’s vaginal 16S rRNA gene sequence library. Sequence libraries used to construct BLAST databases were generated prior to the preclustering step in the mothur protocol described above.
Statistical analysis
Non-parametric multivariate analysis of variance (NPMANOVA) with 10,000 permutations was conducted to evaluate variation in the composition and structure of the bacterial profiles of amniotic fluid and vaginal samples. Bacterial community composition and structure were characterized using the Jaccard and Bray Curtis similarity indices, respectively [135]. All analyses were conducted using PAST software (v2.17) [136].
RESULTS
Clinical characteristics of the study population
Table 1 describes the demographic characteristics of the patients in this study. Seven of the eight patients (87.5%) delivered within one week of amniocentesis and their amniotic fluid testing positive for bacterial cultures. Most of the patients included in this study underwent preterm labor (87.5%) and/or were diagnosed with clinical chorioamnionitis (50%). Table 2 describes the clinical laboratory determinations and the microorganisms cultured from amniotic fluid, as well as the results of placental histopathological examination.
Are the bacteria cultured from amniotic fluid also present in the vagina?
Six of the eight (75%) patients had bacteria cultured from their amniotic fluid that are typical vaginal commensals (Figure 1a), specifically Ureaplasma urealyticum, Escherichia coli, and Streptococcus agalactiae. The other two patients (25%) had bacteria in their amniotic fluid that are not commonly associated with the vagina (Figure 1a), namely Staphylococcus warneri and Streptococcus anginosus. When vaginal swabs were interrogated for the presence of amniotic fluid isolates, matches were found in five of the eight (62.5%) patients. Precisely, in these five cases, bacteria cultured from amniotic fluid were detected in paired vaginal swabs based on matching 16S rRNA gene sequences (≥ 99.5% shared nucleotide identity) (Figure 2). The bacterial cultivars with matching 16S rRNA gene sequences in the vagina were identified as Ureaplasma urealyticum (3 cultivars), Escherichia coli (2 cultivars), and Streptococcus agalactiae (1 cultivar) (Table 3). These bacteria were typically present at low relative abundances within their respective vaginal bacterial communities (Figure 2). These results show that most women with intra-amniotic infection are colonized by vaginal microorganisms.
Figure 1.
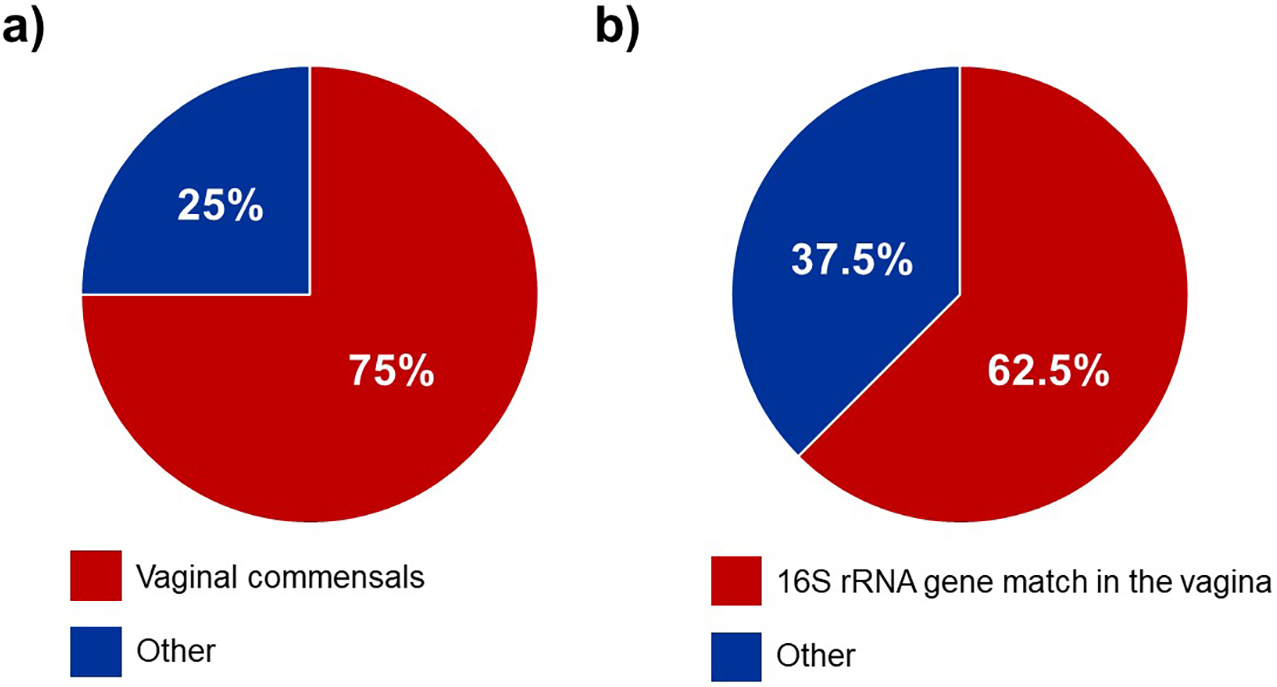
Pie charts illustrating the relationships between bacteria isolated from the amniotic fluid and vaginal microbiota. The percentage of women for whom the bacteria isolated from their amniotic fluid samples a) were typical commensals of the human vaginal ecosystem, and b) had exact matches of their 16S rRNA genes also detected among the vaginal microbiota.
Figure 2.
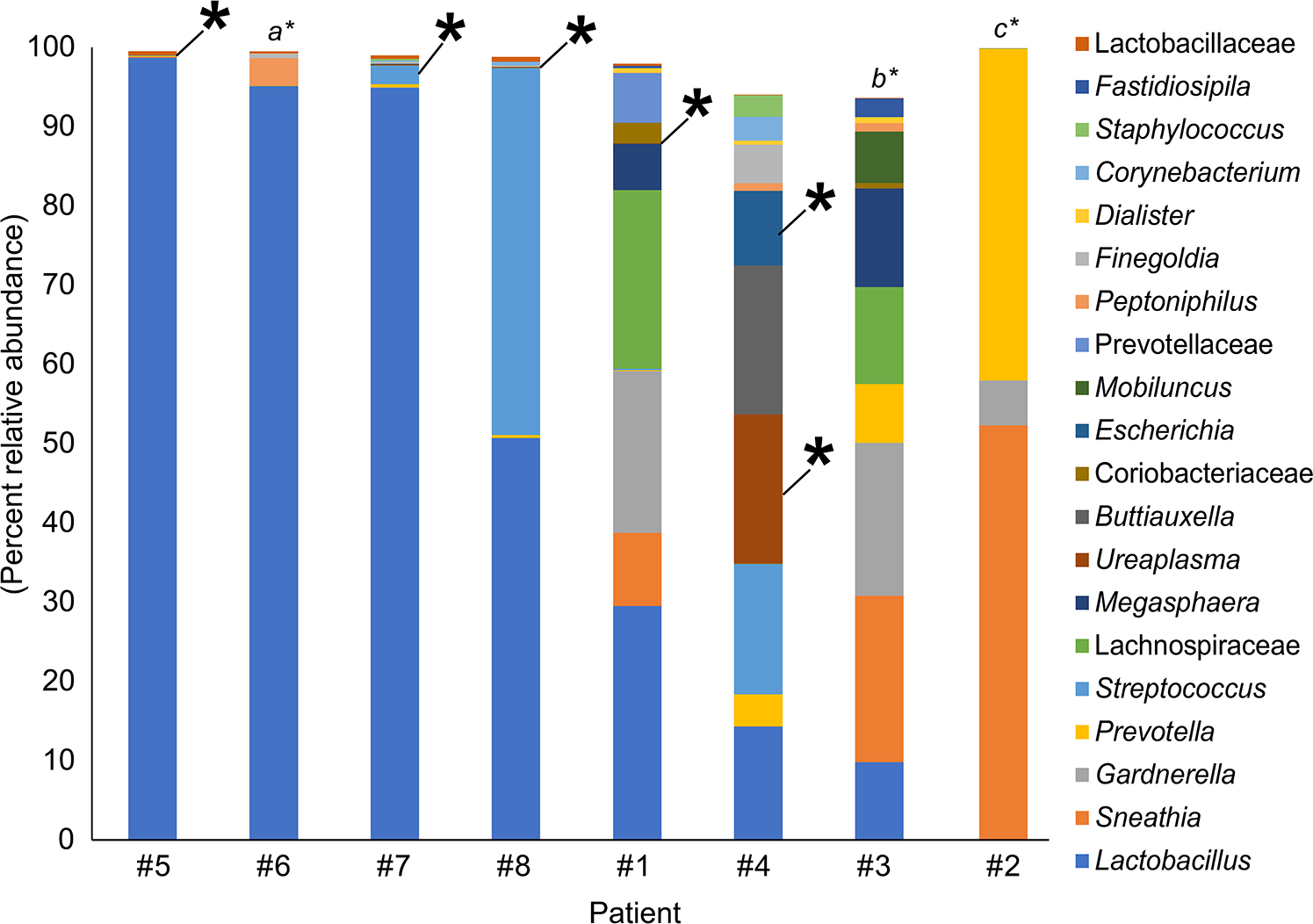
Percent relative abundance of bacterial taxa in the vaginal samples of eight patients with intra-amniotic infection confirmed through culture. The top 20 bacterial taxa, based on average percent relative abundance among the vaginal samples, are displayed. Asterisks indicate a match between the 16S rRNA gene sequence of the bacterial cultivar obtained from amniotic fluid and 16S rRNA gene sequences in the same subject’s vaginal sample. The taxonomic identities of intra-amniotic bacterial cultures without matching sequences in their subject’s vaginal sample were a) Ureaplasma urealyticum, b) Streptococcus anginosus, and c) Staphylococcus warneri.
Table 3.
The frequency of detection of bacteria cultured from amniotic fluid within the microbiota of paired vaginal samples using 16S rRNA gene sequencing.
| Ureaplasma urealtyticum | 4 | 3 |
| Escherichia coli | 2 | 2 |
| Streptococcus agalactiae | 1 | 1 |
| Staphylococcus warneri | 1 | 0 |
| Streptococcus anginosus | 1 | 0 |
Comparison of the molecular bacterial profiles of paired amniotic fluid and vaginal samples
Six of the eight women with intra-amniotic infection had a sufficient volume of amniotic fluid to generate 16S rRNA gene profiles of the fluid. Among these six women, there were 10 prominent (≥ 1% average relative abundance) operational taxonomic units (OTUs) among the amniotic fluid samples (Figure 3, taxa in red font). Their taxonomic identities at the genus level, in order of most to least relative abundance in the amniotic fluid, were Sneathia, Ureaplasma, Prevotella, Lactobacillus, Escherichia, Gardnerella, Peptostreptococcus, Peptoniphilus, and Streptococcus (Figure 3). Seven of the 10 (70.0%) prominent OTUs in amniotic fluid were also prominent in vaginal bacterial communities (Figure 3, taxa in bold red font). At the genus level, the seven prominent OTUs shared between amniotic fluid and the vagina were Lactobacillus, Sneathia, Gardnerella, Prevotella, Ureaplasma, and Escherichia (Figure 3, taxa in bold red font). In four of six (66.7%) patients with paired amniotic fluid and vaginal samples, at least 50% of the prominent OTUs in the amniotic fluid were also present in the vagina (Figure 3, case #1, 2, 3 & 4). In 50.0% (3/6) of patients with paired amniotic fluid and vaginal samples, at least 88.9% of the prominent OTUs in the amniotic fluid were also present in the vagina (Figure 3, case #1, 2 & 3). The prominent OTUs most commonly shared between paired amniotic fluid and vaginal samples were Sneathia (OTUs 3 and 6), Lactobacillus (OTUs 5 and 14), Gardnerella (OTU 9), and Prevotella (OTU 10). In general, among women with intra-amniotic infection, there was overlap between the bacterial profiles of amniotic fluid and the vagina.
Figure 3.
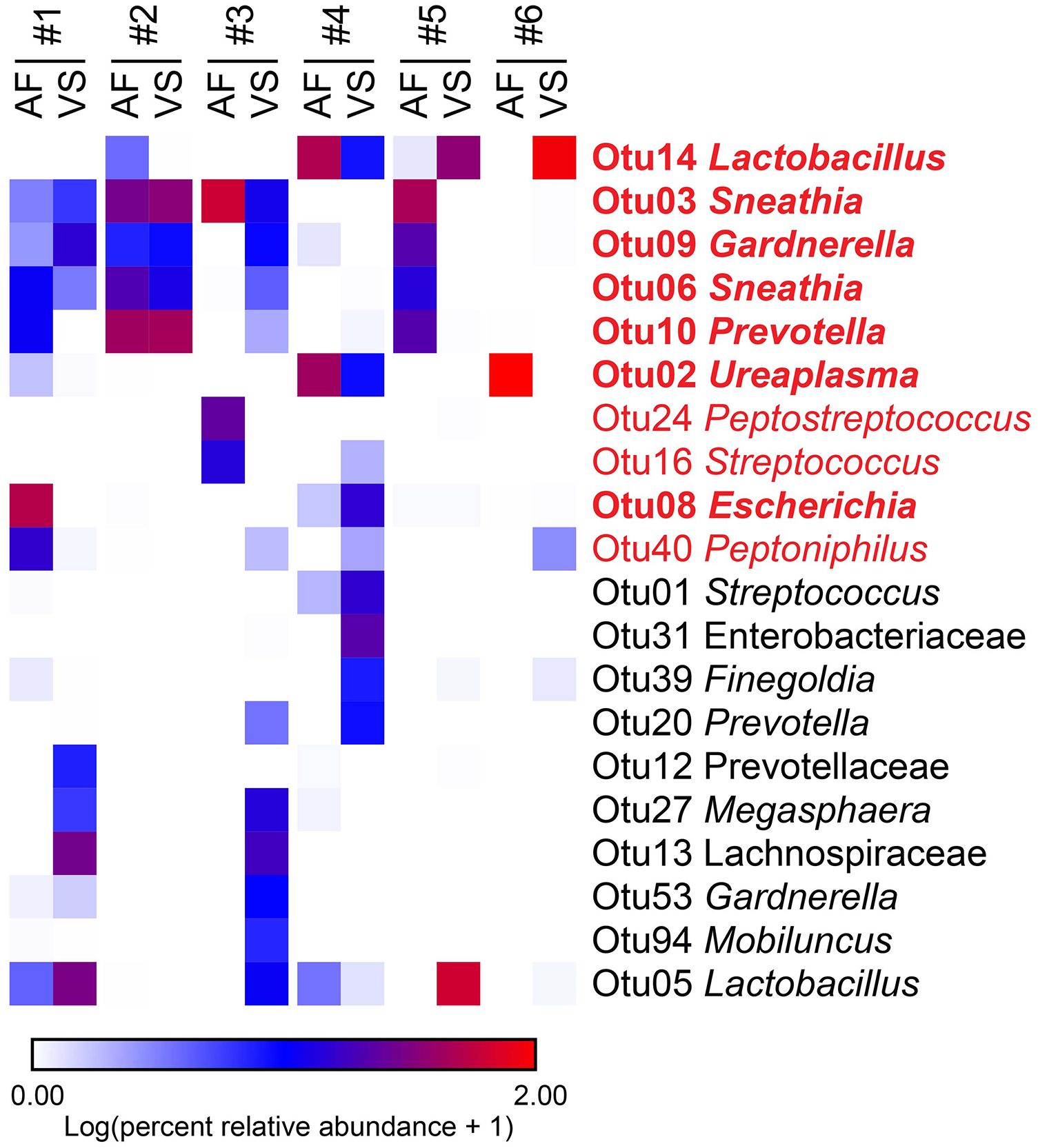
Heat map illustrating similarity in percent relative abundances of prominent (≥ 1% average relative abundance) operational taxonomic units (OTUs) among paired amniotic fluid (AF) and vaginal swab (VS) samples of six women with intra-amniotic infection confirmed through culture. OTUs that were prominent among amniotic fluid samples are highlighted in red. OTUs that were prominent only among vaginal swab samples are in black. OTUs that were prominent in both amniotic fluid and vaginal swab samples are in bold red font.
DISCUSSION
Principal findings of the study
1) Seventy five percent (6/8) of women had bacteria cultured from their amniotic fluid that are typical residents of the vaginal ecosystem; 2) Sixty two and one-half percent (5/8) of women had the bacteria cultured from their amniotic fluid also present in the bacterial communities of their paired vaginal sample; 3) the bacterial cultivars associated with ascending infection were Ureaplasma urealyticum, Escherichia coli, and Streptococcus agalactiae; 4) as assessed by 16S rRNA gene sequencing, the bacterial communities of amniotic fluids with positive bacterial cultures were dominated by Sneathia, Ureaplasma, Prevotella, Lactobacillus, Escherichia, Gardnerella, Peptostreptococcus, Peptoniphilus, and Streptococcus, indicating that microbial cultures do not detect many bacteria present in amniotic fluid; and 5) seventy percent (7/10) of prominent (> 1% average relative abundance) operational taxonomic units (OTUs) in amniotic fluid bacterial communities were also prominent in bacterial communities of the vagina.
Evidence that ascending infection from the lower genital tract is responsible for intra-amniotic infection
Although several routes of invasion have been proposed for intra-amniotic infection, ascension of microorganisms from the vagina has been considered the most common [2, 5, 17, 57–60]. This hypothesis is based on indirect evidence [17]. First, microorganisms detected in amniotic fluid are often those associated with the human vagina [4, 59, 61–63, 67]. Second, in twin pregnancies, when there is intra-amniotic infection, the microorganisms are found in the presenting (rather than non-presenting) sac [100, 137]. Third, women with bacterial vaginosis, a condition in which there is a change in the microbial ecosystem of the vagina, are more likely to have intra-amniotic infection [1, 138, 139]. Fourth, in some cases of early neonatal sepsis or neonatal pneumonia, the microorganisms involved are similar to those found in the vagina [137, 140–142]. Lastly, in an experimental study of non-pregnant women, carbon nanoparticles were placed within the vagina prior to hysterectomy, and these particles were recovered from the Fallopian tubes within 28–34 minutes [143]. The authors concluded that uterine contractions, induced by oxytocin administered prior to surgery, likely played a role in ascension of carbon particles from the vagina to Fallopian tubes [143]. These data suggest that uterine contractions, particularly during prolonged labor, could promote ascending infection.
Mechanisms responsible for ascending infection
A fundamental question which remains unresolved is why some women develop intra-amniotic infection and others do not [5]. The uterine cervix, chorioamniotic membranes, and amniotic fluid, as well as microbial pathogenic factors, may play a role in the likelihood of ascending infection. During pregnancy, the uterine cervix produces a mucous plug, which has been shown to have antimicrobial properties [144–152]. Therefore, some women with cervical shortening and the loss of the mucous plug can develop intra-amniotic infection, and, even if the mucous plug is present, its anti-microbial properties may be inadequate, which can also result in ascending infection [149, 152–154]. The chorioamniotic membranes represent a physical and biochemical barrier to microorganisms, given that they produce antimicrobial peptides [155–157] and contain cells of the innate immune system capable of protecting the host against bacteria [158–163]. Therefore, when the membranes rupture, ascension of bacteria into the amniotic cavity can occur. This is supported by the high frequency of intra-amniotic infection in patients with preterm PROM [18, 34, 164, 165]. Indeed, patients with preterm PROM have higher frequencies of intra-amniotic infection than those with intact membranes [19, 102]. Amniotic fluid also contains multiple antimicrobial factors which could control proliferation of bacteria gaining access to the amniotic cavity [97, 99, 166–174]. Similarly, amniotic fluid contains cells of the innate immune system, which represent another means of host defense [117, 175–185].
All women have microorganisms in the lower genital tract; however, changes in the microbial ecosystem, such as those observed in bacterial vaginosis, are associated with intra-amniotic infection [1, 138, 139, 186]. It is possible that the dysbiotic shift of the vaginal microbiota from communities dominated by Lactobacillus to those comprised primarily of Gardnerella, Prevotella, Porphyromonas, Bacteroides, Peptostreptocccus, Megasphaera, and Sneathia, or related changes in bacterial load [187–189], predispose to ascending infection. Virulence factors of microorganisms may also explain why some gain access to the amniotic cavity while others do not – for example, genital mycoplasmas are the most common organisms found in amniotic fluid in cases of intra-amniotic infection [26, 75, 76, 190, 191]. Although these microorganisms are considered to have less pathogenic potential than others (e.g. Streptococcus agalactiae, or GBS [192–195]), and are present in the vaginal ecosystems of many normal pregnant women [69, 70, 196], a consistent observation is that these organisms are the most frequent microorganisms responsible for intra-amniotic infection [26, 75, 76, 190, 191]. Virulence factors have been identified in Ureaplasma spp. [197, 198] and Mycoplasma spp. [199], which may explain their invasive potential.
What is the origin of intra-amniotic infections in which bacteria in amniotic fluid could not be detected in the lower genital tract?
Three patients in our study had microorganisms in the amniotic cavity which were not detected in vaginal swabs, suggesting that alternative sources for microbial invasion of the amniotic cavity should exist. Previous reports have identified microorganisms in amniotic fluid which had also been identified in the oral cavity [200–202]. Indeed, for animal models of periodontal disease, in which the microorganisms involved are found in the peripheral blood, hematogenous dissemination has been proposed as a route for intra-amniotic and fetal infection [203–209]. Other sources of microorganisms could be the gastrointestinal tract, as a leaky gut can result in bacteremia [210–213]. For example, in the current study Staphylococcus warneri and Streptococcus anginosus were cultured from amniotic fluid but were not present in the matching vaginal swabs. Strains of Streptococcus anginosus have been isolated from the sinus, mouth, throat, and feces, and they are associated with a number of infections [214]. In addition, Staphylococcus warneri, a normal inhabitant of human epithelia and mucosal membranes, has also been associated with orthopedic infections [215, 216], meningitis [217], and endocarditis [218]. Notably, each has been previously reported as etiological agents of bacteremia [219, 220]. Therefore, the absence of each of these bacteria in paired vaginal samples may be due to the hematogenous spread of the microbe originating from a distant body site to the amniotic cavity, as opposed to ascending from the vagina. Alternatively, it is possible that the molecular microbiological methods used in this study have not identified all potential microorganisms present in the vagina; further studies with a large sample size and sampling of other body sites are required to address this question.
Clinical implications of ascending infection
Establishing that microorganisms present in the vagina are responsible for most cases of intra-amniotic infection increases the value of studying the vaginal microbiota. Although the issue is controversial, there is an increasing body of evidence that changes in the vaginal microbiota precede spontaneous preterm birth [84–93]. How such alterations translate to increased frequency of ascending intra-amniotic infection remains to be determined.
Research implications
Additional studies are required to determine why some microorganisms ascend and others do not. For example, why are Lactobacillus so frequent in the vagina and not found in amniotic cavity? Why are Sneathia and Ureaplasmas, which are rare in the vaginal ecosystem, so frequently found in the amniotic cavity? A possible explanation is that these microorganisms possess pathogenic factors that allow them to ascend and invade the intra-amniotic space [71, 199].
Strengths and limitations
This is the first sequencing-based study to compare the amniotic fluid and vaginal microbiota in patients with intra-amniotic infection confirmed through bacterial culture. It provides evidence of ascending infection as the primary cause of intra-amniotic infection. Nevertheless, the observed associations between the vaginal microbiota and amniotic fluid infection are implied and not causally demonstrated. Future studies are required to comprehensively characterize the cervical, vaginal, oral, and intestinal microbiota in women susceptible to microbial invasion of the amniotic cavity in order to confirm the origins of intra-amniotic infection. This will be fundamental to furthering prevention and treatment of intra-amniotic infection.
Conclusions
In most women, the microorganisms causing intra-amniotic infection are members of the vaginal ecosystem, supporting the hypothesis that an ascending pathway is the most common route for microbial invasion of the amniotic cavity.
ACKNOWLEDGEMENTS
This research was supported, in part, by the Perinatology Research Branch (PRB), Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U. S. Department of Health and Human Services (NICHD/NIH/DHHS), and, in part, with federal funds from the NICHD/NIH/DHHS under Contract No. HHSN275201300006C. This research was also supported by the Wayne State University Perinatal Initiative in Maternal, Perinatal and Child Health. We gratefully acknowledge the PRB Translational Research Laboratory for their contributions to the execution of this study. We thank the physicians and nurses from the Center for Advanced Obstetrical Care and Research and the Intrapartum Unit for their help in collecting human samples. The authors also thank the staff members of the PRB Clinical Laboratory and the PRB Histology/Pathology Unit for the processing and examination of the pathological sections.
Footnotes
Declaration of Interests Statement
We have no conflicts of interest to declare.
REFERENCES
- 1.Gravett MG, Eschenbach DA. Possible role of Ureaplasma urealyticum in preterm premature rupture of the fetal membranes. Pediatr Infect Dis. 1986;5(6 Suppl):S253–7. [DOI] [PubMed] [Google Scholar]
- 2.Romero R, Mazor M. Infection and preterm labor. Clin Obstet Gynecol. 1988;31(3):553–84. [DOI] [PubMed] [Google Scholar]
- 3.Watts DH, Krohn MA, Hillier SL, Eschenbach DA. The association of occult amniotic fluid infection with gestational age and neonatal outcome among women in preterm labor. Obstet Gynecol. 1992;79(3):351–7. [DOI] [PubMed] [Google Scholar]
- 4.DiGiulio DB. Diversity of microbes in amniotic fluid. Semin Fetal Neonatal Med. 2012;17(1):2–11. [DOI] [PubMed] [Google Scholar]
- 5.Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science. 2014;345(6198):760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strauss JF 3rd, Romero R, Gomez-Lopez N, Haymond-Thornburg H, Modi BP, Teves ME, et al. Spontaneous preterm birth: advances toward the discovery of genetic predisposition. Am J Obstet Gynecol. 2018;218(3):294–314 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romero R, Mazor M, Wu YK, Sirtori M, Oyarzun E, Mitchell MD, et al. Infection in the pathogenesis of preterm labor. Semin Perinatol. 1988;12(4):262–79. [PubMed] [Google Scholar]
- 8.Romero R, Sirtori M, Oyarzun E, Avila C, Mazor M, Callahan R, et al. Infection and labor. V. Prevalence, microbiology, and clinical significance of intraamniotic infection in women with preterm labor and intact membranes. Am J Obstet Gynecol. 1989;161(3):817–24. [DOI] [PubMed] [Google Scholar]
- 9.Skoll MA, Moretti ML, Sibai BM. The incidence of positive amniotic fluid cultures in patients preterm labor with intact membranes. Am J Obstet Gynecol. 1989;161(3):813–6. [DOI] [PubMed] [Google Scholar]
- 10.Hillier SL, Witkin SS, Krohn MA, Watts DH, Kiviat NB, Eschenbach DA. The relationship of amniotic fluid cytokines and preterm delivery, amniotic fluid infection, histologic chorioamnionitis, and chorioamnion infection. Obstet Gynecol. 1993;81(6):941–8. [PubMed] [Google Scholar]
- 11.Gonzalez-Bosquet E, Cerqueira MJ, Dominguez C, Gasser I, Bermejo B, Cabero L. Amniotic fluid glucose and cytokines values in the early diagnosis of amniotic infection in patients with preterm labor and intact membranes. J Matern Fetal Med. 1999;8(4):155–8. [DOI] [PubMed] [Google Scholar]
- 12.Yoon BH, Romero R, Moon JB, Shim SS, Kim M, Kim G, et al. Clinical significance of intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Obstet Gynecol. 2001;185(5):1130–6. [DOI] [PubMed] [Google Scholar]
- 13.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. 2008;3(8):e3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Combs CA, Gravett M, Garite TJ, Hickok DE, Lapidus J, Porreco R, et al. Amniotic fluid infection, inflammation, and colonization in preterm labor with intact membranes. Am J Obstet Gynecol. 2014;210(2):125.e1–.e15. [DOI] [PubMed] [Google Scholar]
- 15.Combs CA, Garite TJ, Lapidus JA, Lapointe JP, Gravett M, Rael J, et al. Detection of microbial invasion of the amniotic cavity by analysis of cervicovaginal proteins in women with preterm labor and intact membranes. Am J Obstet Gynecol. 2015;212(4):482.e1–.e12. [DOI] [PubMed] [Google Scholar]
- 16.Cobo T, Vives I, Rodriguez-Trujillo A, Murillo C, Angeles MA, Bosch J, et al. Impact of microbial invasion of amniotic cavity and the type of microorganisms on short-term neonatal outcome in women with preterm labor and intact membranes. Acta Obstet Gynecol Scand. 2017;96(5):570–9. [DOI] [PubMed] [Google Scholar]
- 17.Romero R, Gomez R, Chaiworapongsa T, Conoscenti G, Kim JC, Kim YM. The role of infection in preterm labour and delivery. Paediatr Perinat Epidemiol. 2001;15 Suppl 2:41–56. [DOI] [PubMed] [Google Scholar]
- 18.DiGiulio DB, Romero R, Kusanovic JP, Gomez R, Kim CJ, Seok KS, et al. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am J Reprod Immunol. 2010;64(1):38–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romero R, Miranda J, Chaemsaithong P, Chaiworapongsa T, Kusanovic JP, Dong Z, et al. Sterile and microbial-associated intra-amniotic inflammation in preterm prelabor rupture of membranes. J Matern Fetal Neonatal Med. 2015;28(12):1394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodlin RC. Cervical incompetence, hourglass membranes, and amniocentesis. Obstet Gynecol. 1979;54(6):748–50. [PubMed] [Google Scholar]
- 21.Romero R, Gonzalez R, Sepulveda W, Brandt F, Ramirez M, Sorokin Y, et al. Infection and labor. VIII. Microbial invasion of the amniotic cavity in patients with suspected cervical incompetence: prevalence and clinical significance. Am J Obstet Gynecol. 1992;167(4 Pt 1):1086–91. [DOI] [PubMed] [Google Scholar]
- 22.Mays JK, Figueroa R, Shah J, Khakoo H, Kaminsky S, Tejani N. Amniocentesis for selection before rescue cerclage. Obstet Gynecol. 2000;95(5):652–5. [DOI] [PubMed] [Google Scholar]
- 23.Bujold E, Morency AM, Rallu F, Ferland S, Tetu A, Duperron L, et al. Bacteriology of amniotic fluid in women with suspected cervical insufficiency. J Obstet Gynaecol Can. 2008;30(10):882–7. [DOI] [PubMed] [Google Scholar]
- 24.Lee SE, Romero R, Park CW, Jun JK, Yoon BH. The frequency and significance of intraamniotic inflammation in patients with cervical insufficiency. Am J Obstet Gynecol. 2008;198(6):633.e1–8. [DOI] [PubMed] [Google Scholar]
- 25.Airoldi J, Pereira L, Cotter A, Gomez R, Berghella V, Prasertcharoensuk W, et al. Amniocentesis prior to physical exam-indicated cerclage in women with midtrimester cervical dilation: results from the expectant management compared to Physical Exam-indicated Cerclage international cohort study. Am J Perinatol. 2009;26(1):63–8. [DOI] [PubMed] [Google Scholar]
- 26.Oh KJ, Lee SE, Jung H, Kim G, Romero R, Yoon BH. Detection of ureaplasmas by the polymerase chain reaction in the amniotic fluid of patients with cervical insufficiency. J Perinat Med. 2010;38(3):261–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassan S, Romero R, Hendler I, Gomez R, Khalek N, Espinoza J, et al. A sonographic short cervix as the only clinical manifestation of intra-amniotic infection. J Perinat Med. 2006;34(1):13–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez R, Romero R, Nien JK, Medina L, Carstens M, Kim YM, et al. Idiopathic vaginal bleeding during pregnancy as the only clinical manifestation of intrauterine infection. J Matern Fetal Neonatal Med. 2005;18(1):31–7. [DOI] [PubMed] [Google Scholar]
- 29.Madan I, Romero R, Kusanovic JP, Mittal P, Chaiworapongsa T, Dong Z, et al. The frequency and clinical significance of intra-amniotic infection and/or inflammation in women with placenta previa and vaginal bleeding: an unexpected observation. J Perinat Med. 2010;38(3):275–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romero R, Salafia CM, Athanassiadis AP, Hanaoka S, Mazor M, Sepulveda W, et al. The relationship between acute inflammatory lesions of the preterm placenta and amniotic fluid microbiology. Am J Obstet Gynecol. 1992;166(5):1382–8. [DOI] [PubMed] [Google Scholar]
- 31.Latino MA, Botta G, Badino C, Maria D, Petrozziello A, Sensini A, et al. Association between genital mycoplasmas, acute chorioamnionitis and fetal pneumonia in spontaneous abortions. J Perinat Med. 2018;46(5):503–8. [DOI] [PubMed] [Google Scholar]
- 32.Romero R, Miranda J, Kusanovic JP, Chaiworapongsa T, Chaemsaithong P, Martinez A, et al. Clinical chorioamnionitis at term I: microbiology of the amniotic cavity using cultivation and molecular techniques. J Perinat Med. 2015;43(1):19–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaiyasit N, Romero R, Chaemsaithong P, Docheva N, Bhatti G, Kusanovic JP, et al. Clinical chorioamnionitis at term VIII: a rapid MMP-8 test for the identification of intra-amniotic inflammation. J Perinat Med. 2017;45(5):539–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero R, Quintero R, Oyarzun E, Wu YK, Sabo V, Mazor M, et al. Intraamniotic infection and the onset of labor in preterm premature rupture of the membranes. Am J Obstet Gynecol. 1988;159(3):661–6. [DOI] [PubMed] [Google Scholar]
- 35.Duff P, Sanders R, Gibbs RS. The course of labor in term patients with chorioamnionitis. Am J Obstet Gynecol. 1983;147(4):391–5. [DOI] [PubMed] [Google Scholar]
- 36.Mark SP, Croughan-Minihane MS, Kilpatrick SJ. Chorioamnionitis and uterine function. Obstetrics & Gynecology. 2000;95(6):909–12. [PubMed] [Google Scholar]
- 37.Edwards RK. Chorioamnionitis and labor. Obstet Gynecol Clin North Am. 2005;32(2):287–96, x. [DOI] [PubMed] [Google Scholar]
- 38.Oleen MA, Mariano JP. Controlling refractory atonic postpartum hemorrhage with Hemabate sterile solution. Am J Obstet Gynecol. 1990;162(1):205–8. [DOI] [PubMed] [Google Scholar]
- 39.Munn MB, Owen J, Vincent R, Wakefield M, Chestnut DH, Hauth JC. Comparison of two oxytocin regimens to prevent uterine atony at cesarean delivery: a randomized controlled trial. Obstet Gynecol. 2001;98(3):386–90. [DOI] [PubMed] [Google Scholar]
- 40.Rouse DJ, Landon M, Leveno KJ, Leindecker S, Varner MW, Caritis SN, et al. The Maternal-Fetal Medicine Units cesarean registry: Chorioamnionitis at term and its duration-relationship to outcomes. Am J Obstet Gynecol. 2004;191(1):211–6. [DOI] [PubMed] [Google Scholar]
- 41.Tran TS, Jamulitrat S, Chongsuvivatwong V, Geater A. Risk factors for postcesarean surgical site infection. Obstet Gynecol. 2000;95(3):367–71. [DOI] [PubMed] [Google Scholar]
- 42.Incerpi MH. Chorio-amnionitis and postpartum endometritis In: Goodwin TM, editor. Management of Common Problems in Obstetrics and Gynecology (Goodwin TM, et al, eds) Oxford, UK: Wiley-Blackwell; 2010. p. 48–51. [Google Scholar]
- 43.Strunk T, Doherty D, Jacques MBiostat A, Simmer K, Richmond P, Kohan R, et al. Histologic chorioamnionitis is associated with reduced risk of late-onset sepsis in preterm infants. Pediatrics-English Edition. 2012;129(1):e134. [DOI] [PubMed] [Google Scholar]
- 44.Alexander JM, McIntire DM, Leveno KJ. Chorioamnionitis and the prognosis for term infants. Obstetrics & Gynecology. 1999;94(2):274–8. [DOI] [PubMed] [Google Scholar]
- 45.De Felice C, Toti P, Laurini RN, Stumpo M, Picciolini E, Todros T, et al. Early neonatal brain injury in histologic chorioamnionitis. The Journal of pediatrics. 2001;138(1):101–4. [DOI] [PubMed] [Google Scholar]
- 46.Wu YW, Escobar GJ, Grether JK, Croen LA, Greene JD, Newman TB. Chorioamnionitis and cerebral palsy in term and near-term infants. Jama. 2003;290(20):2677–84. [DOI] [PubMed] [Google Scholar]
- 47.Johnson CT, Farzin A, Burd I. Current management and long-term outcomes following chorioamnionitis. Obstet Gynecol Clin North Am. 2014;41(4):649–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willoughby RE, Jr., Nelson KB. Chorioamnionitis and brain injury. Clin Perinatol. 2002;29(4):603–21. [DOI] [PubMed] [Google Scholar]
- 49.Randis TM, Rice MM, Myatt L, Tita ATN, Leveno KJ, Reddy UM, et al. Incidence of early-onset sepsis in infants born to women with clinical chorioamnionitis. J Perinat Med. 2018;46(8):926–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh KJ, Park JY, Lee J, Hong JS, Romero R, Yoon BH. The combined exposure to intra-amniotic inflammation and neonatal respiratory distress syndrome increases the risk of intraventricular hemorrhage in preterm neonates. J Perinat Med. 2018;46(1):9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fidel P, Ghezzi F, Romero R, Chaiworapongsa T, Espinoza J, Cutright J, et al. The effect of antibiotic therapy on intrauterine infection-induced preterm parturition in rabbits. J Matern Fetal Neonatal Med. 2003;14(1):57–64. [DOI] [PubMed] [Google Scholar]
- 52.Lee J, Romero R, Kim SM, Chaemsaithong P, Park CW, Park JS, et al. A new anti-microbial combination prolongs the latency period, reduces acute histologic chorioamnionitis as well as funisitis, and improves neonatal outcomes in preterm PROM. J Matern Fetal Neonatal Med. 2016;29(5):707–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee J, Romero R, Kim SM, Chaemsaithong P, Yoon BH. A new antibiotic regimen treats and prevents intra-amniotic inflammation/infection in patients with preterm PROM. J Matern Fetal Neonatal Med. 2016;29(17):2727–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoneda S, Shiozaki A, Yoneda N, Ito M, Shima T, Fukuda K, et al. Antibiotic therapy increases the risk of preterm birth in preterm labor without intra-amniotic microbes, but may prolong the gestation period in preterm labor with microbes, evaluated by rapid and high-sensitive pcr system. Am J Reprod Immunol. 2016;75(4):440–50. [DOI] [PubMed] [Google Scholar]
- 55.Oh KJ, Romero R, Park JY, Lee J, Conde-Agudelo A, Hong JS, et al. Evidence that antibiotic administration is effective in the treatment of a subset of patients with intra-amniotic infection/inflammation presenting with cervical insufficiency. Am J Obstet Gynecol. 2019;221(2):140.e1–.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoon BH, Romero R, Park JY, Oh KJ, Lee J, Conde-Agudelo A, et al. Antibiotic administration can eradicate intra-amniotic infection or intra-amniotic inflammation in a subset of patients with preterm labor and intact membranes. Am J Obstet Gynecol. 2019;221(2):142.e1–.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goldenberg RL, Hauth JC, Andrews WW. Intrauterine infection and preterm delivery. N Engl J Med. 2000;342(20):1500–7. [DOI] [PubMed] [Google Scholar]
- 58.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371(9606):75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Romero R, Espinoza J, Kusanovic JP, Gotsch F, Hassan S, Erez O, et al. The preterm parturition syndrome. BJOG. 2006;113 Suppl 3:17–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Payne MS, Bayatibojakhi S. Exploring preterm birth as a polymicrobial disease: an overview of the uterine microbiome. Front Immunol. 2014;5:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martius J, Eschenbach DA. The role of bacterial vaginosis as a cause of amniotic fluid infection, chorioamnionitis and prematurity--a review. Arch Gynecol Obstet. 1990;247(1):1–13. [DOI] [PubMed] [Google Scholar]
- 62.Hillier SL, Krohn MA, Cassen E, Easterling TR, Rabe LK, Eschenbach DA. The role of bacterial vaginosis and vaginal bacteria in amniotic fluid infection in women in preterm labor with intact fetal membranes. Clin Infect Dis. 1995;20 Suppl 2:S276–8. [DOI] [PubMed] [Google Scholar]
- 63.Krohn MA, Hillier SL, Nugent RP, Cotch MF, Carey JC, Gibbs RS, et al. The genital flora of women with intraamniotic infection. Vaginal Infection and Prematurity Study Group. J Infect Dis. 1995;171(6):1475–80. [DOI] [PubMed] [Google Scholar]
- 64.Zhou X, Brotman RM, Gajer P, Abdo Z, Schuette U, Ma S, et al. Recent advances in understanding the microbiology of the female reproductive tract and the causes of premature birth. Infect Dis Obstet Gynecol. 2010;2010:737425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lamont RF, Sobel JD, Akins RA, Hassan SS, Chaiworapongsa T, Kusanovic JP, et al. The vaginal microbiome: new information about genital tract flora using molecular based techniques. BJOG. 2011;118(5):533–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4(132):132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mendz GL, Kaakoush NO, Quinlivan JA. Bacterial aetiological agents of intra-amniotic infections and preterm birth in pregnant women. Front Cell Infect Microbiol. 2013;3:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeoman CJ, Thomas SM, Miller MEB, Ulanov AV, Torralba M, Lucas S, et al. A multi-omic systems-based approach reveals metabolic markers of bacterial vaginosis and insight into the disease. PLoS One. 2013;8(2):e56111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Bieda J, et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014;2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. 2014;2(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lannon SMR, Adams Waldorf KM, Fiedler T, Kapur RP, Agnew K, Rajagopal L, et al. Parallel detection of lactobacillus and bacterial vaginosis-associated bacterial DNA in the chorioamnion and vagina of pregnant women at term. J Matern Fetal Neonatal Med. 2018:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Han YW, Shen T, Chung P, Buhimschi IA, Buhimschi CS. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J Clin Microbiol. 2009;47(1):38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim MJ, Romero R, Gervasi MT, Kim JS, Yoo W, Lee DC, et al. Widespread microbial invasion of the chorioamniotic membranes is a consequence and not a cause of intra-amniotic infection. Lab Invest. 2009;89(8):924–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.DiGiulio DB, Gervasi M, Romero R, Mazaki-Tovi S, Vaisbuch E, Kusanovic JP, et al. Microbial invasion of the amniotic cavity in preeclampsia as assessed by cultivation and sequence-based methods. J Perinat Med. 2010;38(5):503–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oh KJ, Lee KA, Sohn YK, Park CW, Hong JS, Romero R, et al. Intraamniotic infection with genital mycoplasmas exhibits a more intense inflammatory response than intraamniotic infection with other microorganisms in patients with preterm premature rupture of membranes. Am J Obstet Gynecol. 2010;203(3):211.e1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Romero R, Miranda J, Chaiworapongsa T, Chaemsaithong P, Gotsch F, Dong Z, et al. A novel molecular microbiologic technique for the rapid diagnosis of microbial invasion of the amniotic cavity and intra-amniotic infection in preterm labor with intact membranes. Am J Reprod Immunol. 2014;71(4):330–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodríguez‐Trujillo A, Cobo T, Vives I, Bosch J, Kacerovsky M, Posadas DE, et al. Gestational age is more important for short‐term neonatal outcome than microbial invasion of the amniotic cavity or intra‐amniotic inflammation in preterm prelabor rupture of membranes. Acta Obstet Gynecol Scand. 2016;95(8):926–33. [DOI] [PubMed] [Google Scholar]
- 78.Moster D, Lie RT, Markestad T. Long-term medical and social consequences of preterm birth. N Engl J Med. 2008;359(3):262–73. [DOI] [PubMed] [Google Scholar]
- 79.DiGiulio DB, Gervasi MT, Romero R, Vaisbuch E, Mazaki-Tovi S, Kusanovic JP, et al. Microbial invasion of the amniotic cavity in pregnancies with small-for-gestational-age fetuses. J Perinat Med. 2010;38(5):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016;6:23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rowlands S, Danielewski JA, Tabrizi SN, Walker SP, Garland SM. Microbial invasion of the amniotic cavity in midtrimester pregnancies using molecular microbiology. Am J Obstet Gynecol. 2017;217(1):71.e1–.e5. [DOI] [PubMed] [Google Scholar]
- 82.Rehbinder EM, Lodrup Carlsen KC, Staff AC, Angell IL, Landro L, Hilde K, et al. Is amniotic fluid of women with uncomplicated term pregnancies free of bacteria? Am J Obstet Gynecol. 2018;219(3):289 e1–e12. [DOI] [PubMed] [Google Scholar]
- 83.Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7(6):e36466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hyman RW, Fukushima M, Jiang H, Fung E, Rand L, Johnson B, et al. Diversity of the vaginal microbiome correlates with preterm birth. Reprod Sci. 2014;21(1):32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A. 2015;112(35):11060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Callahan BJ, DiGiulio DB, Goltsman DSA, Sun CL, Costello EK, Jeganathan P, et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc Natl Acad Sci U S A. 2017;114(37):9966–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kindinger LM, Bennett PR, Lee YS, Marchesi JR, Smith A, Cacciatore S, et al. The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk. Microbiome. 2017;5(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haque MM, Merchant M, Kumar PN, Dutta A, Mande SS. First-trimester vaginal microbiome diversity: A potential indicator of preterm delivery risk. Sci Rep. 2017;7(1):16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stout MJ, Zhou Y, Wylie KM, Tarr PI, Macones GA, Tuuli MG. Early pregnancy vaginal microbiome trends and preterm birth. Am J Obstet Gynecol. 2017;217(3):356.e1–.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Freitas AC, Bocking A, Hill JE, Money DM. Increased richness and diversity of the vaginal microbiota and spontaneous preterm birth. Microbiome. 2018;6(1):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Elovitz MA, Gajer P, Riis V, Brown AG, Humphrys MS, Holm JB, et al. Cervicovaginal microbiota and local immune response modulate the risk of spontaneous preterm delivery. Nat Commun. 2019;10(1):1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, et al. The vaginal microbiome and preterm birth. Nat Med. 2019;25(6):1012–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tabatabaei N, Eren AM, Barreiro LB, Yotova V, Dumaine A, Allard C, et al. Vaginal microbiome in early pregnancy and subsequent risk of spontaneous preterm birth: a case-control study. BJOG. 2019;126(3):349–58. [DOI] [PubMed] [Google Scholar]
- 94.Chaemsaithong P, Romero R, Korzeniewski SJ, Martinez-Varea A, Dong Z, Yoon BH, et al. A rapid interleukin-6 bedside test for the identification of intra-amniotic inflammation in preterm labor with intact membranes. J Matern Fetal Neonatal Med. 2016;29(3):349–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chaemsaithong P, Romero R, Korzeniewski SJ, Martinez-Varea A, Dong Z, Yoon BH, et al. A point of care test for interleukin-6 in amniotic fluid in preterm prelabor rupture of membranes: a step toward the early treatment of acute intra-amniotic inflammation/infection. J Matern Fetal Neonatal Med. 2016;29(3):360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Romero R, Chaemsaithong P, Chaiyasit N, Docheva N, Dong Z, Kim CJ, et al. CXCL10 and IL-6: Markers of two different forms of intra-amniotic inflammation in preterm labor. Am J Reprod Immunol. 2017;78(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Varrey A, Romero R, Panaitescu B, Miller D, Chaiworapongsa T, Patwardhan M, et al. Human beta-defensin-1: A natural antimicrobial peptide present in amniotic fluid that is increased in spontaneous preterm labor with intra-amniotic infection. Am J Reprod Immunol. 2018;80(4):e13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gomez-Lopez N, Romero R, Maymon E, Kusanovic JP, Panaitescu B, Miller D, et al. Clinical chorioamnionitis at term IX: in vivo evidence of intra-amniotic inflammasome activation. J Perinat Med. 2019;47(3):276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Para R, Romero R, Miller D, Panaitescu B, Varrey A, Chaiworapongsa T, et al. Human beta-defensin-3 participates in intra-amniotic host defense in women with labor at term, spontaneous preterm labor and intact membranes, and preterm prelabor rupture of membranes. J Matern Fetal Neonatal Med. 2019:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Romero R, Shamma F, Avila C, Jimenez C, Callahan R, Nores J, et al. Infection and labor. VI. Prevalence, microbiology, and clinical significance of intraamniotic infection in twin gestations with preterm labor. Am J Obstet Gynecol. 1990;163(3):757–61. [DOI] [PubMed] [Google Scholar]
- 101.Romero R, Ghidini A, Mazor M, Behnke E. Microbial invasion of the amniotic cavity in premature rupture of membranes. Clin Obstet Gynecol. 1991;34(4):769–78. [DOI] [PubMed] [Google Scholar]
- 102.Romero R, Miranda J, Chaiworapongsa T, Korzeniewski SJ, Chaemsaithong P, Gotsch F, et al. Prevalence and clinical significance of sterile intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Reprod Immunol. 2014;72(5):458–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Romero R, Miranda J, Chaiworapongsa T, Chaemsaithong P, Gotsch F, Dong Z, et al. Sterile intra-amniotic inflammation in asymptomatic patients with a sonographic short cervix: prevalence and clinical significance. J Matern Fetal Neonatal Med. 2014:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Romero R, Chaemsaithong P, Docheva N, Korzeniewski SJ, Kusanovic JP, Yoon BH, et al. Clinical chorioamnionitis at term VI: acute chorioamnionitis and funisitis according to the presence or absence of microorganisms and inflammation in the amniotic cavity. J Perinat Med. 2016;44(1):33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Romero R, Chaemsaithong P, Docheva N, Korzeniewski SJ, Tarca AL, Bhatti G, et al. Clinical chorioamnionitis at term V: umbilical cord plasma cytokine profile in the context of a systemic maternal inflammatory response. J Perinat Med. 2016;44(1):53–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Romero R, Chaemsaithong P, Docheva N, Korzeniewski SJ, Tarca AL, Bhatti G, et al. Clinical chorioamnionitis at term IV: the maternal plasma cytokine profile. J Perinat Med. 2016;44(1):77–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Romero R, Chaemsaithong P, Korzeniewski SJ, Kusanovic JP, Docheva N, Martinez-Varea A, et al. Clinical chorioamnionitis at term III: how well do clinical criteria perform in the identification of proven intra-amniotic infection? J Perinat Med. 2016;44(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Romero R, Chaemsaithong P, Korzeniewski SJ, Tarca AL, Bhatti G, Xu Z, et al. Clinical chorioamnionitis at term II: the intra-amniotic inflammatory response. J Perinat Med. 2016;44(1):5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chaemsaithong P, Romero R, Docheva N, Chaiyasit N, Bhatti G, Pacora P, et al. Comparison of rapid MMP-8 and interleukin-6 point-of-care tests to identify intra-amniotic inflammation/infection and impending preterm delivery in patients with preterm labor and intact membranes. J Matern Fetal Neonatal Med. 2018;31(2):228–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kusanovic JP, Romero R, Martinovic C, Silva K, Erez O, Maymon E, et al. Transabdominal collection of amniotic fluid “sludge” and identification of Candida albicans intra-amniotic infection. J Matern Fetal Neonatal Med. 2018;31(10):1279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oh KJ, Romero R, Park JY, Kang J, Hong JS, Yoon BH. A high concentration of fetal fibronectin in cervical secretions increases the risk of intra-amniotic infection and inflammation in patients with preterm labor and intact membranes. J Perinat Med. 2019;47(3):288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pacora P, Romero R, Erez O, Maymon E, Panaitescu B, Kusanovic JP, et al. The diagnostic performance of the beta-glucan assay in the detection of intra-amniotic infection with Candida species. J Matern Fetal Neonatal Med. 2019;32(10):1703–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gibbs RS, Blanco JD, St Clair PJ, Castaneda YS. Quantitative bacteriology of amniotic fluid from women with clinical intraamniotic infection at term. J Infect Dis. 1982;145(1):1–8. [DOI] [PubMed] [Google Scholar]
- 114.Gibbs RS, Dinsmoor MJ, Newton ER, Ramamurthy RS. A randomized trial of intrapartum versus immediate postpartum treatment of women with intra-amniotic infection. Obstet Gynecol. 1988;72(6):823–8. [DOI] [PubMed] [Google Scholar]
- 115.Gilstrap LC 3rd, Cox SM. Acute chorioamnionitis. Obstet Gynecol Clin North Am. 1989;16(2):373–9. [PubMed] [Google Scholar]
- 116.Gibbs RS, Duff P. Progress in pathogenesis and management of clinical intraamniotic infection. Am J Obstet Gynecol. 1991;164(5 Pt 1):1317–26. [DOI] [PubMed] [Google Scholar]
- 117.Martinez-Varea A, Romero R, Xu Y, Miller D, Ahmed AI, Chaemsaithong P, et al. Clinical chorioamnionitis at term VII: the amniotic fluid cellular immune response. J Perinat Med. 2017;45(5):523–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Oh KJ, Kim SM, Hong JS, Maymon E, Erez O, Panaitescu B, et al. Twenty-four percent of patients with clinical chorioamnionitis in preterm gestations have no evidence of either culture-proven intraamniotic infection or intraamniotic inflammation. Am J Obstet Gynecol. 2017;216(6):604.e1–.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Romero R, Gomez-Lopez N, Kusanovic JP, Pacora P, Panaitescu B, Erez O, et al. Clinical chorioamnionitis at term: new insights into the etiology, microbiology, and the fetal, maternal and amniotic cavity inflammatory responses. Nogyogyaszati Szuleszeti Tovabbkepzo Szemle. 2018;20(3):103–12. [PMC free article] [PubMed] [Google Scholar]
- 120.Kim CJ, Romero R, Chaemsaithong P, Chaiyasit N, Yoon BH, Kim YM. Acute chorioamnionitis and funisitis: definition, pathologic features, and clinical significance. Am J Obstet Gynecol. 2015;213(4 Suppl):S29–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Romero R, Kim YM, Pacora P, Kim CJ, Benshalom-Tirosh N, Jaiman S, et al. The frequency and type of placental histologic lesions in term pregnancies with normal outcome. J Perinat Med. 2018;46(6):613–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Redline RW. Classification of placental lesions. Am J Obstet Gynecol. 2015;213(4 Suppl):S21–8. [DOI] [PubMed] [Google Scholar]
- 123.Romero R, Quintero R, Nores J, Avila C, Mazor M, Hanaoka S, et al. Amniotic fluid white blood cell count: a rapid and simple test to diagnose microbial invasion of the amniotic cavity and predict preterm delivery. Am J Obstet Gynecol. 1991;165(4 Pt 1):821–30. [DOI] [PubMed] [Google Scholar]
- 124.Romero R, Emamian M, Quintero R, Wan M, Hobbins JC, Mazor M, et al. The value and limitations of the Gram stain examination in the diagnosis of intraamniotic infection. Am J Obstet Gynecol. 1988;159(1):114–9. [DOI] [PubMed] [Google Scholar]
- 125.Romero R, Jimenez C, Lohda AK, Nores J, Hanaoka S, Avila C, et al. Amniotic fluid glucose concentration: a rapid and simple method for the detection of intraamniotic infection in preterm labor. Am J Obstet Gynecol. 1990;163(3):968–74. [DOI] [PubMed] [Google Scholar]
- 126.Redelinghuys MJ, Ehlers MM, Dreyer AW, Lombaard HA, Kock MM. Comparison of the new Mycofast Revolution assay with a molecular assay for the detection of genital mycoplasmas from clinical specimens. BMC Infect Dis. 2013;13:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015;6:791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME journal. 2012;6(8):1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42(Database issue):D633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Magurran A, McGill BJ. Biological diversity: frontiers in measurement and assessment. Oxford: Oxford University Press; 2010. [Google Scholar]
- 136.Hammer O, Harper D. PAleontological STatistics software package for education and data analysis. Palaeontologia Electronica. 2001;4:1–9. [Google Scholar]
- 137.Benirschke K. and Routes and types of infection in the fetus and the newborn. AMA J Dis Child. 1960;99:714–21. [DOI] [PubMed] [Google Scholar]
- 138.Eschenbach DA, Gravett MG, Chen KC, Hoyme UB, Holmes KK. Bacterial vaginosis during pregnancy. An association with prematurity and postpartum complications. Scand J Urol Nephrol Suppl. 1984;86:213–22. [PubMed] [Google Scholar]
- 139.Gravett MG, Hummel D, Eschenbach DA, Holmes KK. Preterm labor associated with subclinical amniotic fluid infection and with bacterial vaginosis. Obstet Gynecol. 1986;67(2):229–37. [DOI] [PubMed] [Google Scholar]
- 140.Blanc WA. Amniotic infection syndrome; pathogenesis, morphology, and significance in circumnatal mortality. Clin Obstet Gynecol. 1959;2:705–34. [PubMed] [Google Scholar]
- 141.Blanc WA. Pathways of fetal and early neonatal infection. Viral placentitis, bacterial and fungal chorioamnionitis. J Pediatr. 1961;59:473–96. [DOI] [PubMed] [Google Scholar]
- 142.Naeye RL, Dellinger WS, Blanc WA. Fetal and maternal features of antenatal bacterial infections. J Pediatr. 1971;79(5):733–9. [DOI] [PubMed] [Google Scholar]
- 143.Egli GE, Newton M. The transport of carbon particles in the human female reproductive tract. Fertil Steril. 1961;12:151–5. [DOI] [PubMed] [Google Scholar]
- 144.Svinarich DM, Wolf NA, Gomez R, Gonik B, Romero R. Detection of human defensin 5 in reproductive tissues. Am J Obstet Gynecol. 1997;176(2):470–5. [DOI] [PubMed] [Google Scholar]
- 145.Hein M, Helmig RB, Schonheyder HC, Ganz T, Uldbjerg N. An in vitro study of antibacterial properties of the cervical mucus plug in pregnancy. Am J Obstet Gynecol. 2001;185(3):586–92. [DOI] [PubMed] [Google Scholar]
- 146.Hein M, Valore EV, Helmig RB, Uldbjerg N, Ganz T. Antimicrobial factors in the cervical mucus plug. Am J Obstet Gynecol. 2002;187(1):137–44. [DOI] [PubMed] [Google Scholar]
- 147.Cole AM. Innate host defense of human vaginal and cervical mucosae. Curr Top Microbiol Immunol. 2006;306:199–230. [PubMed] [Google Scholar]
- 148.Sawada M, Otsuki K, Mitsukawa K, Yakuwa K, Nagatsuka M, Okai T. Cervical inflammatory cytokines and other markers in the cervical mucus of pregnant women with lower genital tract infection. Int J Gynaecol Obstet. 2006;92(2):117–21. [DOI] [PubMed] [Google Scholar]
- 149.Becher N, Adams Waldorf K, Hein M, Uldbjerg N. The cervical mucus plug: structured review of the literature. Acta Obstet Gynecol Scand. 2009;88(5):502–13. [DOI] [PubMed] [Google Scholar]
- 150.Lee DC, Hassan SS, Romero R, Tarca AL, Bhatti G, Gervasi MT, et al. Protein profiling underscores immunological functions of uterine cervical mucus plug in human pregnancy. J Proteomics. 2011;74(6):817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Radtke AL, Quayle AJ, Herbst-Kralovetz MM. Microbial products alter the expression of membrane-associated mucin and antimicrobial peptides in a three-dimensional human endocervical epithelial cell model. Biol Reprod. 2012;87(6):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hansen LK, Becher N, Bastholm S, Glavind J, Ramsing M, Kim CJ, et al. The cervical mucus plug inhibits, but does not block, the passage of ascending bacteria from the vagina during pregnancy. Acta Obstet Gynecol Scand. 2014;93(1):102–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gomez R, Romero R, Nien JK, Chaiworapongsa T, Medina L, Kim YM, et al. A short cervix in women with preterm labor and intact membranes: a risk factor for microbial invasion of the amniotic cavity. Am J Obstet Gynecol. 2005;192(3):678–89. [DOI] [PubMed] [Google Scholar]
- 154.Critchfield AS, Yao G, Jaishankar A, Friedlander RS, Lieleg O, Doyle PS, et al. Cervical mucus properties stratify risk for preterm birth. PLoS One. 2013;8(8):e69528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Erez O, Romero R, Tarca AL, Chaiworapongsa T, Kim YM, Than NG, et al. Differential expression pattern of genes encoding for anti-microbial peptides in the fetal membranes of patients with spontaneous preterm labor and intact membranes and those with preterm prelabor rupture of the membranes. J Matern Fetal Neonatal Med. 2009;22(12):1103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Boldenow E, Jones S, Lieberman RW, Chames MC, Aronoff DM, Xi C, et al. Antimicrobial peptide response to group B Streptococcus in human extraplacental membranes in culture. Placenta. 2013;34(6):480–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zare-Bidaki M, Sadrinia S, Erfani S, Afkar E, Ghanbarzade N. Antimicrobial properties of amniotic and chorionic membranes: a comparative study of two human fetal sacs. J Reprod Infertil. 2017;18(2):218–24. [PMC free article] [PubMed] [Google Scholar]
- 158.Whidbey C, Harrell MI, Burnside K, Ngo L, Becraft AK, Iyer LM, et al. A hemolytic pigment of Group B Streptococcus allows bacterial penetration of human placenta. J Exp Med. 2013;210(6):1265–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Boldenow E, Gendrin C, Ngo L, Bierle C, Vornhagen J, Coleman M, et al. Group B Streptococcus circumvents neutrophils and neutrophil extracellular traps during amniotic cavity invasion and preterm labor. Sci Immunol. 2016;1(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gomez-Lopez N, Romero R, Leng Y, Garcia-Flores V, Xu Y, Miller D, et al. Neutrophil extracellular traps in acute chorioamnionitis: A mechanism of host defense. Am J Reprod Immunol. 2017;77(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Doster RS, Kirk LA, Tetz LM, Rogers LM, Aronoff DM, Gaddy JA. Staphylococcus aureus infection of human gestational membranes induces bacterial biofilm formation and host production of cytokines. J Infect Dis. 2017;215(4):653–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kothary V, Doster RS, Rogers LM, Kirk LA, Boyd KL, Romano-Keeler J, et al. Group B Streptococcus induces neutrophil recruitment to gestational tissues and elaboration of extracellular traps and nutritional immunity. Front Cell Infect Microbiol. 2017;7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Doster RS, Sutton JA, Rogers LM, Aronoff DM, Gaddy JA. Streptococcus agalactiae induces placental macrophages to release extracellular traps loaded with tissue remodeling enzymes via an oxidative burst-dependent mechanism. MBio. 2018;9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim KW, Romero R, Park HS, Park CW, Shim SS, Jun JK, et al. A rapid matrix metalloproteinase-8 bedside test for the detection of intraamniotic inflammation in women with preterm premature rupture of membranes. Am J Obstet Gynecol. 2007;197(3):292.e1–5. [DOI] [PubMed] [Google Scholar]
- 165.Tchirikov M, Schlabritz-Loutsevitch N, Maher J, Buchmann J, Naberezhnev Y, Winarno AS, et al. Mid-trimester preterm premature rupture of membranes (PPROM): etiology, diagnosis, classification, international recommendations of treatment options and outcome. J Perinat Med. 2018;46(5):465–88. [DOI] [PubMed] [Google Scholar]
- 166.Galask RP, Snyder IS. Antimicrobial factors in amniotic fluid. Am J Obstet Gynecol. 1970;106(1):59–65. [DOI] [PubMed] [Google Scholar]
- 167.Larsen B, Snyder IS, Galask RP. Bacterial growth inhibition by amniotic fluid. I. In vitro evidence for bacterial growth-inhibiting activity. Am J Obstet Gynecol. 1974;119(4):492–6. [DOI] [PubMed] [Google Scholar]
- 168.Schlievert P, Johnson W, Galask RP. Isolation of a low-molecular-weight antibacterial system from human amniotic fluid. Infect Immun. 1976;14(5):1156–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tafari N, Ross SM, Naeye RL, Galask RP, Zaar B. Failure of bacterial growth inhibition by amniotic fluid. Am J Obstet Gynecol. 1977;128(2):187–9. [DOI] [PubMed] [Google Scholar]
- 170.Niemela A, Kulomaa M, Vija P, Tuohimaa P, Saarikoski S. Lactoferrin in human amniotic fluid. Hum Reprod. 1989;4(1):99–101. [DOI] [PubMed] [Google Scholar]
- 171.Pacora P, Maymon E, Gervasi MT, Gomez R, Edwin SS, Yoon BH, et al. Lactoferrin in intrauterine infection, human parturition, and rupture of fetal membranes. Am J Obstet Gynecol. 2000;183(4):904–10. [DOI] [PubMed] [Google Scholar]
- 172.Espinoza J, Chaiworapongsa T, Romero R, Edwin S, Rathnasabapathy C, Gomez R, et al. Antimicrobial peptides in amniotic fluid: defensins, calprotectin and bacterial/permeability-increasing protein in patients with microbial invasion of the amniotic cavity, intra-amniotic inflammation, preterm labor and premature rupture of membranes. J Matern Fetal Neonatal Med. 2003;13(1):2–21. [DOI] [PubMed] [Google Scholar]
- 173.Soto E, Espinoza J, Nien JK, Kusanovic JP, Erez O, Richani K, et al. Human beta-defensin-2: a natural antimicrobial peptide present in amniotic fluid participates in the host response to microbial invasion of the amniotic cavity. J Matern Fetal Neonatal Med. 2007;20(1):15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Romero R, Kusanovic JP, Gotsch F, Erez O, Vaisbuch E, Mazaki-Tovi S, et al. Isobaric labeling and tandem mass spectrometry: a novel approach for profiling and quantifying proteins differentially expressed in amniotic fluid in preterm labor with and without intra-amniotic infection/inflammation. J Matern Fetal Neonatal Med. 2010;23(4):261–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Romero R, Emamian M, Wan M, Yarkoni S, McCormack W, Mazor M, et al. The value of the leukocyte esterase test in diagnosing intra-amniotic infection. Am J Perinatol. 1988;5(1):64–9. [DOI] [PubMed] [Google Scholar]
- 176.Sampson JE, Theve RP, Blatman RN, Shipp TD, Bianchi DW, Ward BE, et al. Fetal origin of amniotic fluid polymorphonuclear leukocytes. Am J Obstet Gynecol. 1997;176(1 Pt 1):77–81. [DOI] [PubMed] [Google Scholar]
- 177.Macias AE, Wong SW, Sadowsky DW, Luetjens CM, Axthelm MK, Gravett MG, et al. Maternal or fetal origin of rhesus monkey (Macaca mulatta) amniotic fluid leukocytes can be identified by polymerase chain reaction using the zinc finger Y gene. Am J Primatol. 2001;55(3):159–70. [DOI] [PubMed] [Google Scholar]
- 178.Romero R, Kusanovic JP, Gomez R, Lamont R, Bytautiene E, Garfield RE, et al. The clinical significance of eosinophils in the amniotic fluid in preterm labor. J Matern Fetal Neonatal Med. 2010;23(4):320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Marquardt N, Ivarsson MA, Sundstrom E, Akesson E, Martini E, Eidsmo L, et al. Fetal CD103+ IL-17-producing Group 3 innate lymphoid cells represent the dominant lymphocyte subset in human amniotic fluid. J Immunol. 2016;197(8):3069–75. [DOI] [PubMed] [Google Scholar]
- 180.Gomez-Lopez N, Romero R, Garcia-Flores V, Xu Y, Leng Y, Alhousseini A, et al. Amniotic fluid neutrophils can phagocytize bacteria: A mechanism for microbial killing in the amniotic cavity. Am J Reprod Immunol. 2017;78(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gomez-Lopez N, Romero R, Xu Y, Leng Y, Garcia-Flores V, Miller D, et al. Are amniotic fluid neutrophils in women with intraamniotic infection and/or inflammation of fetal or maternal origin? Am J Obstet Gynecol. 2017;217(6):693.e1–.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gomez-Lopez N, Romero R, Xu Y, Miller D, Unkel R, Shaman M, et al. Neutrophil extracellular traps in the amniotic cavity of women with intra-amniotic infection: a new mechanism of host defense. Reprod Sci. 2017;24(8):1139–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gomez-Lopez N, Romero R, Xu Y, Miller D, Leng Y, Panaitescu B, et al. The immunophenotype of amniotic fluid leukocytes in normal and complicated pregnancies. Am J Reprod Immunol. 2018;79(4):e12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Miller D, Motomura K, Garcia-Flores V, Romero R, Gomez-Lopez N. Innate lymphoid cells in the maternal and fetal compartments. Front Immunol. 2018;9:2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gomez-Lopez N, Roberto R, Galaz J, Xu Y, Panaitescu B, Slutsky R, et al. Cellular immune responses in amniotic fluid of women with preterm labor and intra-amniotic infection or intra-amniotic inflammation. Am J Reprod Immunol. 2019:e13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Romero R, Chaiworapongsa T, Kuivaniemi H, Tromp G. Bacterial vaginosis, the inflammatory response and the risk of preterm birth: a role for genetic epidemiology in the prevention of preterm birth. Am J Obstet Gynecol. 2004;190(6):1509–19. [DOI] [PubMed] [Google Scholar]
- 187.Zozaya-Hinchliffe M, Lillis R, Martin DH, Ferris MJ. Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J Clin Microbiol. 2010;48(5):1812–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hilbert DW, Smith WL, Chadwick SG, Toner G, Mordechai E, Adelson ME, et al. Development and validation of a highly accurate quantitative real-time PCR assay for diagnosis of bacterial vaginosis. J Clin Microbiol. 2016;54(4):1017–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Plummer EL, Garland SM, Bradshaw CS, Law MG, Vodstrcil LA, Hocking JS, et al. Molecular diagnosis of bacterial vaginosis: Does adjustment for total bacterial load or human cellular content improve diagnostic performance? J Microbiol Methods. 2017;133:66–8. [DOI] [PubMed] [Google Scholar]
- 190.Yoon BH, Romero R, Kim M, Kim EC, Kim T, Park JS, et al. Clinical implications of detection of Ureaplasma urealyticum in the amniotic cavity with the polymerase chain reaction. Am J Obstet Gynecol. 2000;183(5):1130–7. [DOI] [PubMed] [Google Scholar]
- 191.Yoon BH, Romero R, Lim JH, Shim SS, Hong JS, Shim JY, et al. The clinical significance of detecting Ureaplasma urealyticum by the polymerase chain reaction in the amniotic fluid of patients with preterm labor. Am J Obstet Gynecol. 2003;189(4):919–24. [DOI] [PubMed] [Google Scholar]
- 192.Sitkiewicz I, Green NM, Guo N, Bongiovanni AM, Witkin SS, Musser JM. Transcriptome adaptation of group B Streptococcus to growth in human amniotic fluid. PLoS One. 2009;4(7):e6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gendrin C, Vornhagen J, Armistead B, Singh P, Whidbey C, Merillat S, et al. A nonhemolytic Group B Streptococcus strain exhibits hypervirulence. J Infect Dis. 2018;217(6):983–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Korir ML, Flaherty RA, Rogers LM, Gaddy JA, Aronoff DM, Manning SD. Investigation of the role that NADH peroxidase plays in oxidative stress survival in Group B Streptococcus. Front Microbiol. 2018;9:2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Vornhagen J, Armistead B, Santana-Ufret V, Gendrin C, Merillat S, Coleman M, et al. Group B streptococcus exploits vaginal epithelial exfoliation for ascending infection. J Clin Invest. 2018;128(5):1985–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Donders GGG, Ruban K, Bellen G, Petricevic L. Mycoplasma/Ureaplasma infection in pregnancy: to screen or not to screen. J Perinat Med. 2017;45(5):505–15. [DOI] [PubMed] [Google Scholar]
- 197.Knox CL, Dando SJ, Nitsos I, Kallapur SG, Jobe AH, Payton D, et al. The severity of chorioamnionitis in pregnant sheep is associated with in vivo variation of the surface-exposed multiple-banded antigen/gene of Ureaplasma parvum. Biol Reprod. 2010;83(3):415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sweeney EL, Kallapur SG, Meawad S, Gisslen T, Stephenson S-A, Jobe AH, et al. Ureaplasma species Multiple Banded Antigen (MBA) variation is associated with the severity of inflammation in vivo and in vitro in human placentae. Front Cell Infect Mi. 2017;7(123). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Allen-Daniels MJ, Serrano MG, Pflugner LP, Fettweis JM, Prestosa MA, Koparde VN, et al. Identification of a gene in Mycoplasma hominis associated with preterm birth and microbial burden in intraamniotic infection. Am J Obstet Gynecol. 2015;212(6):779.e1–.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hill GB. Investigating the source of amniotic fluid isolates of fusobacteria. Clin Infect Dis. 1993;16 Suppl 4:S423–4. [DOI] [PubMed] [Google Scholar]
- 201.Leon R, Silva N, Ovalle A, Chaparro A, Ahumada A, Gajardo M, et al. Detection of Porphyromonas gingivalis in the amniotic fluid in pregnant women with a diagnosis of threatened premature labor. J Periodontol. 2007;78(7):1249–55. [DOI] [PubMed] [Google Scholar]
- 202.Radochova V, Kacerovska Musilova I, Stepan M, Vescicik P, Slezak R, Jacobsson B, et al. Periodontal disease and intra-amniotic complications in women with preterm prelabor rupture of membranes. J Matern Fetal Neonatal Med. 2018;31(21):2852–61. [DOI] [PubMed] [Google Scholar]
- 203.Lin D, Smith MA, Elter J, Champagne C, Downey CL, Beck J, et al. Porphyromonas gingivalis infection in pregnant mice is associated with placental dissemination, an increase in the placental Th1/Th2 cytokine ratio, and fetal growth restriction. Infect Immun. 2003;71(9):5163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Boggess KA, Price WA, Preisser JS, Moise KJ Jr., Offenbacher S. Insulin-like growth factor and interleukin-1beta levels and subsequent fetal size in response to chronic Porphyromonas gingivalis exposure in the pregnant rabbit. Am J Obstet Gynecol. 2005;193(3 Pt 2):1219–23. [DOI] [PubMed] [Google Scholar]
- 205.Arce RM, Barros SP, Wacker B, Peters B, Moss K, Offenbacher S. Increased TLR4 expression in murine placentas after oral infection with periodontal pathogens. Placenta. 2009;30(2):156–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fardini Y, Chung P, Dumm R, Joshi N, Han YW. Transmission of diverse oral bacteria to murine placenta: evidence for the oral microbiome as a potential source of intrauterine infection. Infect Immun. 2010;78(4):1789–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ao M, Miyauchi M, Furusho H, Inubushi T, Kitagawa M, Nagasaki A, et al. Dental infection of Porphyromonas gingivalis induces preterm birth in mice. PLoS One. 2015;10(8):e0137249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Stockham S, Stamford JE, Roberts CT, Fitzsimmons TR, Marchant C, Bartold PM, et al. Abnormal pregnancy outcomes in mice using an induced periodontitis model and the haematogenous migration of Fusobacterium nucleatum sub-species to the murine placenta. PLoS One. 2015;10(3):e0120050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Liang S, Ren H, Guo H, Xing W, Liu C, Ji Y, et al. Periodontal infection with Porphyromonas gingivalis induces preterm birth and lower birth weight in rats. Mol Oral Microbiol. 2018;33(4):312–21. [DOI] [PubMed] [Google Scholar]
- 210.Fukui H How leaky gut and endotoxemia induce bacterial infection in cirrhosis and gastrointestinal hemorrhage? J Gastroenterol Hepatol. 2011;26(3):423–5. [DOI] [PubMed] [Google Scholar]
- 211.Kim BI, Kim HJ, Park JH, Park DI, Cho YK, Sohn CI, et al. Increased intestinal permeability as a predictor of bacterial infections in patients with decompensated liver cirrhosis and hemorrhage. J Gastroenterol Hepatol. 2011;26(3):550–7. [DOI] [PubMed] [Google Scholar]
- 212.Panpetch W, Chancharoenthana W, Bootdee K, Nilgate S, Finkelman M, Tumwasorn S, et al. Lactobacillus rhamnosus L34 attenuates gut translocation-induced bacterial sepsis in murine models of leaky gut. Infect Immun. 2018;86(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tamburini FB, Andermann TM, Tkachenko E, Senchyna F, Banaei N, Bhatt AS. Precision identification of diverse bloodstream pathogens in the gut microbiome. Nat Med. 2018;24(12):1809–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ruoff KL. Streptococcus anginosus (“Streptococcus milleri”): the unrecognized pathogen. Clin Microbiol Rev. 1988;1(1):102–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Announ N, Mattei JP, Jaoua S, Fenollar F, Sati H, Chagnaud C, et al. Multifocal discitis caused by Staphylococcus warneri. Joint Bone Spine. 2004;71(3):240–2. [DOI] [PubMed] [Google Scholar]
- 216.Campoccia D, Montanaro L, Visai L, Corazzari T, Poggio C, Pegreffi F, et al. Characterization of 26 Staphylococcus warneri isolates from orthopedic infections. Int J Artif Organs. 2010;33(9):575–81. [DOI] [PubMed] [Google Scholar]
- 217.Incani RN, Hernandez M, Cortez J, Gonzalez ME, Salazar YD. Staphylococcus warneri meningitis in a patient with Strongyloides stercoralis hyperinfection and lymphoma: first report of a case. Rev Inst Med Trop Sao Paulo. 2010;52(3):169–70. [DOI] [PubMed] [Google Scholar]
- 218.Stollberger C, Wechsler-Fordos A, Geppert F, Gulz W, Brownstone E, Nicolakis M, et al. Staphylococcus warneri endocarditis after implantation of a lumbar disc prosthesis in an immunocompetent patient. J Infect. 2006;52(1):e15–8. [DOI] [PubMed] [Google Scholar]
- 219.Kamath U, Singer C, Isenberg HD. Clinical significance of Staphylococcus warneri bacteremia. J Clin Microbiol. 1992;30(2):261–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Bert F, Bariou-Lancelin M, Lambert-Zechovsky N. Clinical significance of bacteremia involving the “Streptococcus milleri” group: 51 cases and review. Clin Infect Dis. 1998;27(2):385–7. [DOI] [PubMed] [Google Scholar]


