Abstract
Recent years have witnessed significant progress in understanding how memories are encoded, from the molecular to the cellular and the circuit/systems levels. With a good compromise between brain complexity and behavioral sophistication, the fruit fly Drosophila melanogaster is one of the preeminent animal models of learning and memory. Here we review how memories are encoded in Drosophila, with a focus on short-term memory and an eye toward future directions. Forward genetic screens have revealed a large number of genes and transcripts necessary for learning and memory, some acting cell-autonomously. Further, the relative numerical simplicity of the fly brain has enabled the reverse engineering of learning circuits with remarkable precision, in some cases ascribing behavioral phenotypes to single neurons. Functional imaging and physiological studies have localized and parsed the plasticity that occurs during learning at some of the major loci. Connectomics projects are significantly expanding anatomical knowledge of the nervous system, filling out the roadmap for ongoing functional/physiological and behavioral studies, which are being accelerated by simultaneous tool development. These developments have provided unprecedented insight into the fundamental neural principles of learning, and lay the groundwork for deep understanding in the near future.
If the human brain were so simple, That we could understand it, We would be so simple, That we couldn’t.
Emerson M. Pugh
Introduction
The human brain is among the most complex organs in the body. Understanding its functionality is, as Emerson Pugh’s famous historical quip highlights, a longstanding challenge for biological inquiry (Pugh, 1977). Within the field of neuroscience, deciphering how memories are formed and maintained is a major area of focus. Memories of our past experiences interact with present sensory perceptions to influence our behaviors, but what is the neural substrate of these interactions? What molecular pathways drive the modifications of neural activity supporting the formation of memories, and how are these manifested at the circuit and systems levels? Our ability to distill these broad questions into meaningful, experimentally-tractable derivatives will be the one of the greatest determinants of the success of neuroscience research. Given the immense challenge of reverse engineering the human brain, with its 86 billion neurons and estimated ~100 trillion synapses, neuroscience research heavily leverages model organisms with numerically-reduced nervous systems. Offering a good compromise between relative brain simplicity and behavioral sophistication, the fruit fly Drosophila melanogaster has been a highly informative model organism for the study of behaviors ranging from circadian rhythms to learning and memory (Hardin et al., 1992; Heisenberg et al., 1985; McBride et al., 1999; Nitabach and Taghert, 2008; Zars, 2010; Zars et al., 2000a).
Behavior and genetics
Here we review the mechanisms of learning and memory in Drosophila, with a focus on the encoding of short-term memories, and an eye toward potential future directions for the field. Learning has been studied with a variety of assays in flies, the most common being olfactory classical conditioning (Tully and Quinn, 1985). In this assay, flies are presented with an odor, the conditioned stimulus (CS+), which is paired with a second stimulus, the unconditioned stimulus (US). The US can be aversive, such as electric shock, or appetitive, such as sucrose (Busto et al., 2010). A second unpaired odor (the CS−) is also presented, either before or (usually) after pairing the CS+ and US. After conditioning, memory is tested in a T-maze, providing arms containing each of the odors, and allowing the flies to distribute between them. A performance index is calculated as the proportion of flies that choose the CS+ over the CS−.
Forward genetic screens have identified many genes necessary for olfactory learning and memory. Previous reviews have covered these in detail, e.g., (Davis, 2005; Dudai, 1988; Tomchik and Davis, 2013; Waddell and Quinn, 2001), so we will focus here on the major themes, with an emphasis on the learning (acquisition) phase. While multiple signaling cascades are involved in learning, both unbiased genetic screens and hypothesis-driven studies based on data from other systems have converged on genes involved in cAMP signaling cascades. In particular, the groundbreaking research of Troy Zars provided some of the earliest support for the role of one cAMP signaling molecule, Rutabaga, describing its tissue-specific function in learning and memory (Zars et al., 2000a; Zars et al., 2000b). Some of the more well-studied examples of cAMP signaling molecules include Rutabaga, Dunce, and DC0. Rutabaga (rut) encodes a type I Ca2+/calmodulin-dependent adenylyl cyclase, Dunce (dnc) encodes a cAMP-specific phosphodiesterase, and dc0 the catalytic subunit of the cAMP-dependent protein kinase A (PKA). These proteins regulate cAMP signaling antagonistically: Rut synthesizes cAMP, while Dnc metabolizes it, regulating downstream DC0 signaling antagonistically (Davis et al., 1995; Levin et al., 1992; Skoulakis et al., 1993). Alterations to the activity of other members of the cAMP signaling pathway, including Gαs (Connolly et al., 1996) as well as the regulatory (RI) subunits of PKA, are also required for learning (Goodwin et al., 1997). Mutations in dopa decarboxylase were shown to impair learning, which provided the first insight into the neurotransmitter systems (dopamine and/or serotonin) that potentially activate cAMP signaling (Tempel et al., 1984).
Dopaminergic circuits play particularly important roles in the US processing, and we will describe their roles in detail below. Nonetheless, it should be noted that other neuromodulators, signaling proteins, and cells/circuits that express and release them, are involved in learning and memory. Along with dopamine, the biogenic amines octopamine and serotonin contribute to the neuromodulation underlying memory formation. Octopamine plays a prominent role in appetitive learning (Burke et al., 2012; Han et al., 1996; Huetteroth et al., 2015; Kim et al., 2013; Schwaerzel et al., 2003). It is also involved (to a lesser extent) in aversive learning, as tβh mutants that cannot produce octopamine show aversive memory deficits (Iliadi et al., 2017). In addition, serotonin has been implicated in learning, and in some cases exerts effects indirectly via actions on dopaminergic neurons (Johnson et al., 2011; Scheunemann et al., 2018). Both serotonin and octopamine contribute in parallel to the formation of a specific form of consolidated memory (Wu et al., 2013). Octopaminergic and serotonergic mechanisms have received less attention than dopaminergic mechanisms overall, and represent additional layers of putative regulation of learning and memory. In addition to these neuromodulatory pathways, various signal transduction cascades are involved in memory formation as well. The scaffolding protein Leonardo is highly expressed in relevant brain regions and necessary for olfactory learning (Broadie et al., 1997; Skoulakis and Davis, 1996). In addition to PKA, other protein kinases, such as S6KII, CaMKII, PKC, and PKG, also modulate memory (Brembs and Plendl, 2008; Griffith et al., 1993; Kane et al., 1997; Kaun et al., 2007; Mery et al., 2007 Neuser et al., 2008; Putz et al., 2004). GABAergic circuits are involved in sparsening the sensory representation in the olfactory pathway, enhancing memory encoding (Honegger et al., 2011; Lin et al., 2014; Liu et al., 2009; Liu and Davis, 2009) as well as playing a role in memory consolidation (Haynes et al., 2015). Muscarinic acetylcholine receptors are necessary on MB neurons for aversive memory formation (Bielopolski et al., 2019; Silva et al., 2015), and shape odor coding in the antennal lobe (Rozenfeld et al., 2019). A major open question is how (specifically) all of these neuromodulators, receptors, and intracellular signaling cascades alter cellular physiology to generate the plasticity in the olfactory pathway neurons that underlies learning.
Fundamental anatomy of the olfactory pathway
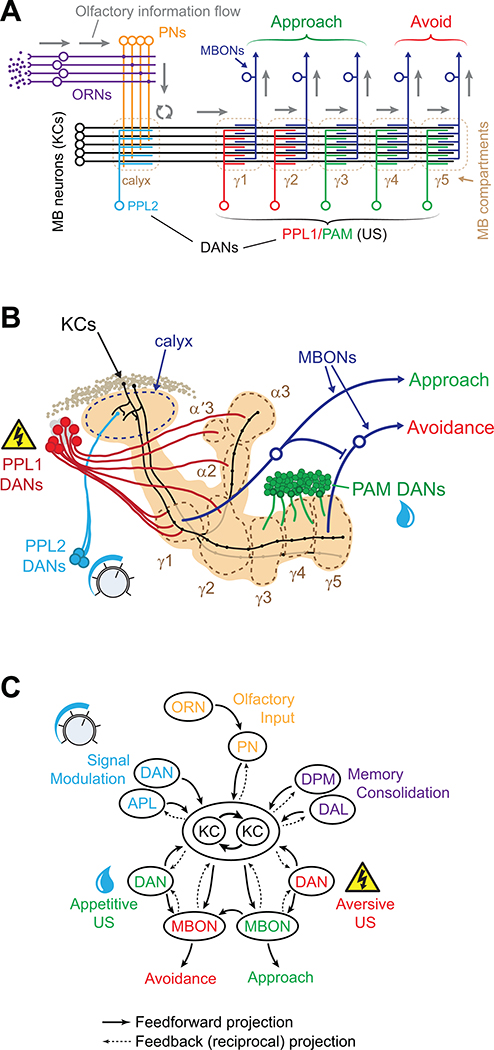
The mushroom body (MB) is a critical anatomical structure involved in olfactory memory formation, as well as some types of visual and courtship memory (McBride et al., 1999; Vogt et al., 2014). The MB is situated in the olfactory pathway, as the tertiary structure, hierarchically similar to the mammalian amygdala or piriform cortex (Su et al., 2009). Olfactory stimuli are initially detected by olfactory receptor neurons (ORNs) in the periphery, which transmit information to projection neurons (PNs), and subsequently the MB and another structure, the lateral horn (Davis, 2005; Fiala, 2007). The intrinsic MB neurons, also called Kenyon Cells (KCs), relay information to mushroom body output neurons (MBONs) (Aso et al., 2014a; Tanaka et al., 2008) via cholinergic synapses (Barnstedt et al., 2016). The MB is innervated by modulatory neurons, such as dopaminergic neurons (DANs), which are critical for learning and memory (Tanaka et al., 2008). This description includes the basic circuit elements (ORN→PN→KC→MBON; w/ modulatory DANs) (Fig. 1A,B), and will be further elaborated below. Note that while the general flow of information is most easily conceptualized as unidirectional, some of these connections exhibit both pre- and post-synaptic zones indicative of bidirectional communication (Christiansen et al., 2011; Pauls et al., 2010; Rolls et al., 2007) (Fig. 1C). This bidirectional communication adds a layer of complexity with behavioral and computational implications that are largely unknown currently.
Figure 1.
Simplified diagram of the anatomical connectivity and information processing that underlies olfactory learning in Drosophila. A. Simplified diagram of the olfactory pathway through the mushroom body (MB) and mushroom body output neurons (MBONs), highlighting the major anatomical structures involved in learning. Connections to the lateral horn are not shown. Gray arrows indicate the inward information flow from peripheral olfactory sensory receptors. ORNs: olfactory receptor neurons, PNs: projection neurons, KCs: Kenyon Cells, DAN: dopaminergic neuron(s), MBON: mushroom body output neuron, PPL1/2: paired posterior lateral (dopaminergic) neurons, PAM: protocerebral anterior medial (dopaminergic) neurons. B. Drawing of the mushroom body, showing the major anatomical subdivisions and classes of neurons relevant for learning. Anatomical compartments (γ1-γ5, α2, α3, α′3) are outlined with dashed lines. For clarity, only one γ KC and one α/β KC are drawn (out of ~2500 total); likewise, two MBONs are drawn (out of 34 total). C. Flowchart highlighting major connections in the olfactory pathway that are critical for olfactory learning and memory. DPM: dorsal paired medial neuron, DAL: dorsal anterior lateral, APL: anterior paired lateral neuron.
The MBs receive olfactory input from the antennal lobe via the PNs (Su et al., 2009). They are composed of approximately 2500 pseudounipolar KCs (Fig. 1B). Their somata are located in the posterior dorsal region of the brain. The KC dendrites form a calyx structure in each hemisphere, and their axons fasciculate together, projecting to the anterior face of the brain. Once they reach the anterior face, they divide into vertical (α, α′) and horizontal (β, β′ and γ) lobes, forming a distinctive lobular anatomical structure (Fig. 1B; Table 1) (Crittenden et al., 1998; Guven-Ozkan and Davis, 2014). The MBs function as a critical site of plasticity during learning, integrating information about olfactory stimuli with US signals conveyed by modulatory interneurons (Burke et al., 2012; Claridge-Chang et al., 2009; Gervasi et al., 2010; Schroll et al., 2006; Tomchik and Davis, 2009; Ueno et al., 2017). Flies with abnormal MB morphology show memory defects, as do flies in which the MBs are chemically ablated (de Belle and Heisenberg, 1994). The MBs are specialized for higher-order processes. Insects without mushroom bodies are generally healthy, and can move and perceive multiple sensory modalities (Wolf et al., 1998). Other forms of memory, such as place conditioning, are unaffected by loss of mushroom bodies (Zars et al., 2000b). Many proteins required for olfactory learning and memory exhibit preferential expression in the MBs (e.g., Dnc, Rut, DC0) (Nighorn et al., 1991; Skoulakis et al., 1993).
Table 1.
Nomenclature for neurons discussed in this review. While MB subsets have consistent nomenclature, several systems have been used to label DANs and MBONs. We use the system of Aso et al. in this review.
| Subset | Nomenclature1 | Nomenclature2 | Nomenclature3 | |
|---|---|---|---|---|
| Mushroom body (MB) neurons; = Kenyon Cells (KCs) | α/β | |||
| α′/β′ | ||||
| γ | ||||
| Dopaminergic neurons (DANs) | PPL1 neurons | PPL1-γ1pedc | MB-MP1 | |
| PPL1-α2α′2 | MB-V2 | |||
| PAM neurons | PAM-γ3 | MB-M2 | ||
| PAM-γ5 | asp13 | |||
| PAM-β2β′2a | PAM-M3 | |||
| Mushroom body output neurons (MBONs) | β1>α | MB-MV2 | ||
| γ1ped>α/β | MB-MVP2 | |||
| γ4>γ1γ2 | ||||
| γ5β′2a | MB-M6 | MB-M6 |
Nomenclature used in Aso et al. (2014)
Nomenclature used in Tanaka et al. (2008)
Nomenclature used in Zhao et al. (2018)
Molecular biology and circuits underlying learning
The MBs are believed to encode olfactory memories by associating olfactory cues (odors) with US information, altering the flow of olfactory information following learning. KCs initially encode odors in a sparse representation (Turner et al., 2008). At the circuit level, they are innervated by multiple sets of extrinsic neurons (Tanaka et al., 2008), including DANs (Fig. 1), which are particularly important for US processing during learning (Qin et al., 2012; Schroll et al., 2006; Schwaerzel et al., 2003). The DANs respond strongly to sensory stimuli, particularly stimuli with positive and negative valence, such as electric shock and sucrose (Liu et al., 2012; Louis et al., 2018; Mao and Davis, 2009). During conditioning, a subset of KCs responds to odors with depolarization and receives a strong dopaminergic signal during US presentation. This synergistically elevates cAMP and PKA in KCs in a Rut-dependent manner (Gervasi et al., 2010; Tomchik and Davis, 2009). These observations experimentally reinforced a key prediction of the coincidence detection model, in which the Ca2+-sensitive adenylyl cyclase Rut, expressed in the MB, functions to integrate activity state of the KCs with signaling from GPCRs to elevate cAMP (Heisenberg, 2003; Livingstone et al., 1984; Zars et al., 2000a; Zars et al., 2000b). Upstream of cAMP signaling, the D1-like dopamine receptor DopR is necessary for learning (Kim et al., 2007), and the circuit roles of dopamine will be discussed in detail below. Downstream effectors generate plasticity in a cAMP/PKA-dependent manner, presumably through modulation of local excitability, compartmentalized signaling, and/or modulation of synaptic vesicle cycling and release. cAMP/PKA activation can alter the properties of KCs over short time scales via mechanisms such as K+ channel phosphorylation (Drain et al., 1994). It also potentially modifies synaptic output through effectors such as synapsin, which modulates vesicular pool dynamics (Knapek et al., 2010; Michels et al., 2011; Michels et al., 2005; Niewalda et al., 2015). cAMP exerts cellular effects in a PKA-independent manner through cyclic-nucleotide-gated channels, with potential effects on MB physiology and memory (Pavot et al., 2015). Finally, over longer time scales, cAMP/PKA signaling alters gene transcription via CREB (Chen et al., 2012; Miyashita et al., 2018; Widmer et al., 2018; Yin et al., 1995; Zhang et al., 2015). It should be noted that while mutants for cAMP signaling molecules are deficient in olfactory memory, there is residual memory. For instance, rut mutants exhibit memory scores approximately half of that of controls, demonstrating that there are rut-independent memory pathways (Tan et al., 2010; Tully and Quinn, 1985; Zars et al., 2000a).
While the MB is a critical locus for learning-induced plasticity, such plasticity is not restricted to the MB. Experiments using in vivo calcium imaging – examining odor-evoked responses before and after conditioning while looking for changes resulting from learning – have revealed plasticity across multiple loci in the olfactory pathway. These regions include antennal lobe local neurons (Scheunemann et al., 2012), PNs (Yu et al., 2004), KCs (Akalal et al., 2010; Louis et al., 2018; Wang et al., 2008; Yu et al., 2006; Yu et al., 2004; Zhang and Roman, 2013), dorsal paired medial (DPM) neurons (Cervantes-Sandoval and Davis, 2012; Yu et al., 2005), GABAergic anterior paired lateral (APL) neurons (Liu and Davis, 2009), dorsal anterior lateral (DAL) neurons (Chen et al., 2012), and MB output neurons (MBONs) (Berry et al., 2018; Owald et al., 2015; Sejourne et al., 2011) (Fig. 1). These loci exhibit changes at various time points following conditioning. Despite the somewhat distributed spatiotemporal pattern of plasticity, it is clear that the KCs represent a central node in the circuit where cAMP-dependent plasticity is necessary for memory formation. Cell-targeted genetic rescue experiments of cAMP signaling molecules almost invariably require MB expression for behavioral rescue, and expression in KCs is sufficient for rescue in many cases (Kim et al., 2007; Michels et al., 2011; Scheunemann et al., 2012; Zars et al., 2000a). Indeed, the MB appears to be where the CS and US are integrated, where the olfactory information is imparted with a valence (positive or negative association) following classical conditioning. At the circuit level, this process critically depends on the DANs that innervate the MB.
Dopaminergic neurons: key players in US processing
Several major anatomical classes of DANs innervate the MB, and they play distinct roles in memory formation (Fig. 1; Table 1). PPL1 neurons innervate the MB vertical lobes, and are necessary for aversive olfactory learning (Galili et al., 2014). The neurons respond strongly to the electric shock that is typically used as the US in aversive conditioning studies (Cervantes-Sandoval et al., 2017; Mao and Davis, 2009; Riemensperger et al., 2005). Finally, pairing an odor with stimulation of PPL1 neurons is sufficient to induce the formation of aversive memory (Aso et al., 2010; Aso et al., 2012; Claridge-Chang et al., 2009; Schroll et al., 2006). This demonstrates that PPL1 neurons are intimately involved in processing of the aversive US during learning, and may convey a portion of the US signal to the MB. In comparison, PAM DANs innervate the MB horizontal lobes, and are critical for US processing during appetitive olfactory classical conditioning (Fig. 1B). A subset of these neurons respond strongly to the sucrose stimulus that is provided as the US in appetitive classical conditioning (Yamagata et al., 2016; Yamagata et al., 2015). They function in an analogous manner to PPL1 neurons, except in appetitive conditioning rather than aversive conditioning: their activity is necessary and sufficient as an appetitive US (Liu et al., 2012). Thus, the PPL1 and PAM neurons perform similar functions in US processing, though with opposite valence (negative vs. positive, respectively). These classes of neurons can be further subdivided functionally. For instance, the γ1-pedc subset of PPL1 neurons innervates the heel region of the mushroom body (the γ1 compartment) and is necessary for aversive learning in olfactory classical conditioning (Aso et al., 2012). Another subset innervates the MB vertical lobes dorsal to that region, and is particularly important for taste learning (Masek et al., 2015). Some neurons within these subsets perform roles that are antagonistic to the overall, net effect of the subset as a whole. For example, while the PAM DANs have a net appetitive role, the PAM-β2β′2a neurons modulate aversive memory (Aso et al., 2012). Bidirectional modulation of the activity of at least some DANs can produce opposing effects on behavior. For instance, PAM DANs innervating the γ3 compartment of the MB function bidirectionally, with activation driving aversive memory and blockade driving appetitive memory (Yamagata et al., 2016).
In addition to the role that DANs play in valence/US processing, one subset plays a unique role in memory, driving plasticity in the MB and modulating memory strength without encoding valence. PPL2ab DANs innervate the dendritic MB region in the calyx (among other brain regions) (Kuo et al., 2015; Mao and Davis, 2009) (Fig. 1B). Pairing stimulation of the PPL2ab neurons with odor presentation enhances odor-evoked activity in MB γ neurons; this effect is associated with increased aversive memory performance (Boto et al., 2019). These results imply that dopamine acting on MB neurons can either drive a valence signal (US) or act as a gain control mechanism, modulating the strength of the memory. The behavioral outcome appears to depend on which dopaminergic pathway is activated and which spatial MB compartment the neurons innervate. This compartmental topography and division of labor represents a fundamental organizing principle on which the MB operates (Fig. 1 A,B).
Circuit biology: how is olfactory information rerouted by learning?
A longstanding model postulates that the MB modulates conditioned behaviors by altering the flow of olfactory information through downstream circuits that mediate approach and avoidance behavior (Heisenberg, 2003). This may occur in part via dopaminergic modulation of distinct spatial compartments of the MB (Fig. 1 A,B). The lobes of the MB are divided into compartmental zones that each receive dopaminergic input from distinct DANs, and send outputs to distinct downstream MBONs (Aso et al., 2014a; Aso et al., 2014b; Mao and Davis, 2009; Tanaka et al., 2008) (Fig. 1A). In naïve situations, DAN activity modulates Ca2+ responses in a compartmentalized fashion in the MB γ lobe. Odor-evoked Ca2+ responses show non-uniform distribution along the axon (Cohn et al., 2015). Under normal circumstances, larger responses were observed in the γ2/γ3 (proximal) compartments than the γ4/γ5 (distal) compartments. Upon activation of DAN neurons innervating the γ lobe, the profile of odor-evoked responses inverted, increasing the responses in distal compartments relative to the proximal ones, resembling the effect of sucrose ingestion. DANs also produce plasticity in odor-evoked Ca2+ responses in the MB following olfactory classical conditioning (Louis et al., 2018). In this context, conditioning produced spatially-broad facilitation of odor-evoked Ca2+ responses that were specific to appetitive conditioning (no net changes were observed with aversive conditioning). This potentiation was recapitulated when comparing odor-evoked Ca2+ responses before and after appetitive conditioning, and it was dependent on Rutabaga. Stimulation of “appetitive” PAM DANs paired with odor presentation also induces elevation of cAMP levels (Boto et al., 2014; Handler et al., 2019) and a facilitation in the odor-evoked Ca2+ responses across the MB lobes (Boto et al., 2014). This suggests that appetitive conditioning exerts strong influence over odor-evoked Ca2+ in the MB. How this Ca2+ signal correlates with synaptic plasticity is an open question, as there are multiple sources of intracellular Ca2+; for instance, the endoplasmic reticulum in KCs exhibits Ca2+ flux under learning-relevant conditions (Handler et al., 2019).
Several examples of learning-induced plasticity in KCs have been documented. Changes in synaptic output from KCs have been observed with synaptophluorin following conditioning. Aversive conditioning decreases synaptic CS− responses, and this effect is dependent on the activity of the heterotrimeric G protein subunit Gαo (Zhang and Roman, 2013). Synaptic content of the MB and DANs has been associated with increased memory strength in a developmental context (Phan et al., 2019). In vitro experiments using synaptophluorin have observed integration of signals in the MB from the antennal nerves and ascending fibers of the ventral nerve cord, which putatively carry somatosensory US information, and plasticity when these pathways are stimulated in tandem (Ueno et al., 2013). Dopamine application can replace stimulation of the ventral nerve fibers, and the plasticity is dependent on Rut expression in the MB (Ueno et al., 2017).
The studies reviewed above suggest that the MB encodes odors, is innervated by DANs and other modulatory neurons that carry positive and negative valence signals, and conveys processed olfactory information to downstream neurons that can bias behavioral decisions. This provides a substrate through which learning-induced plasticity could alter the flow of information to generate conditioned responses following learning. At the synapse between the KCs and MBONs, heterosynaptic depression has been observed postsynaptically following pairing of odor with stimulation of DANs (Hige et al., 2015). This drives alterations in the responses of MBONs, both via direct effects at local synapses and via downstream network effects (e.g., feedforward inhibition) (Berry et al., 2018; Felsenberg et al., 2018; Hige et al., 2015; Owald et al., 2015; Perisse et al., 2016; Sejourne et al., 2011). Notwithstanding the lack of compartmentalized plasticity at the level of Ca2+ in some paradigms, other evidence suggests that presynaptic, compartmentalized plasticity in KCs could contribute to coherent learning-induced plasticity across the MBONs. Stimulation of DANs produces compartmentalized cAMP transients in KCs (Boto et al., 2014). Since many of the MBONs have an innate valence, and bias an animal toward or away from an odor, compartmentalized neuromodulatory effects in KCs could contribute to differential activation of some MBONs relative to others following learning (Aso et al., 2014a; Cohn et al., 2015; Handler et al., 2019). MBON display also biased innervation towards specific KC depending on their role in avoidance or attraction, as it has been reported that non-overlapping populations of KC can be functionally subdivided depending on the valence they encode. (Perisse et al., 2013; Yamazaki et al., 2018).
Circuit complexity: interconnections in the olfactory nervous system
As more anatomical detail is mapped in the Drosophila olfactory pathway, it is becoming clear that there is substantial reciprocal connectivity, recurrent feedback, and lateral connectivity among the neuronal elements. These circuit motifs are relevant for emerging circuit models of learning and memory, though the behavioral implications are incompletely understood. At the input to the MB, antennal lobe PNs form bidirectional connections with the KC dendrites in the calyx (Christiansen et al., 2011; Pech et al., 2015). Between KCs and DANs, the simplest circuit model of connectivity would be unidirectional, with the DANs presynaptic to the KCs. DANs release dopamine that acts on MB neurons to elevate cAMP via D1-like receptors, demonstrating functional connectivity in this direction (Boto et al., 2014). However, recent studies have also documented reciprocal connectivity between these cell types (Fig. 1 B,C). Multiple examples have been documented, including PAM-γ4<γ1γ2, PPL1 α2α′2 DANs, PAM-γ5 (via an MBON loop), and larval protocerebral anterior PAM neurons (Aso et al., 2014a; Cervantes-Sandoval et al., 2017; Handler et al., 2019; Lyutova et al., 2019; Zhao et al., 2018). In all of these instances, KC activity drives activation of the DANs, which ultimately influences learning and memory by modulating the release of dopamine onto the KCs. Reciprocal connectivity of KCs and DANs may function, along with inputs from somatosensory circuits that convey US information from the body, to drive plasticity via nicotinic acetylcholine receptor-dependent mechanisms (Ueno et al., 2017). Recurrent connections between KCs and MBONs have also been shown to play key roles in modulating learned behaviors. For instance, one cell type connects the peduncle of the MB γ neurons back to the MB calyx, providing recurrent feedback from the MB output to its input layer (Zheng et al., 2018). MBON recurrent connections have been shown to play a role in long-term memory (Ichinose et al., 2015; Pavlowsky et al., 2018). Thus, recurrent connectivity among layers in the olfactory pathway is a fundamental circuit motif that presumably plays a role in shaping the plasticity driving learned behaviors (Aso et al., 2014a; Eichler et al., 2017; Takemura et al., 2017).
At the circuit level, the complex reciprocal interconnectivity between KCs & KCs, PNs & KCs, KC & DANs, KCs & MBONs, DANs & MBONs has led to increasingly complex neuronal circuitry models and questions (Fig. 1C). For instance, lateral connectivity between KCs, involving both gap junctions and chemical synapses, has also been recently discovered (Liu et al., 2016; Takemura et al., 2017). The functional significance of this intriguing circuit feature is a mystery. While electrical synapses between KC have been implicated in the retrieval of a specific form of consolidated memory (Shyu et al., 2019), connectomics studies have recently reported rosette-like synapses, interpreted as KC>KC reciprocal contacts. Most of these KC>KC connections have one additional postsynaptic partner, most frequently an MBON. KC>KC synapses could enable the excitation of one KC to spread to adjacent KCs (Takemura et al., 2017). In contrast, reciprocal synapses between KCs and DANs have received more attention, and the function of these connections may be to amplify the signals from the KCs, generating synaptic plasticity under biologically-relevant conditions (Zhao et al., 2018). Finally, in addition to innervating KCs, the DANs also synapse with the MBONs one synapse downstream (Eichler et al., 2017; Takemura et al., 2017). The behavioral roles of most of these types of connections are not yet well understood.
After the MBs integrate and process information, it is fed to MBONs. Each type of MBON receives dendritic input from at least one of the MB compartments. In turn, most MBONs project to different neuropil regions in the protocerebrum, though three MBONs provide recurrent feedback to the MBs (Aso et al., 2014a). In marked contrast to the sparse odor responsivity among KCs, MBONs respond broadly to odors, and the odor tuning varies between individual flies (Hige et al., 2015). Activation of different output neurons generates approach/avoidance behavior, leading to the idea that MBONs encode valence (value associated with a stimulus) rather than stimulus identity (Aso et al., 2014b). Additional complexity will likely be incorporated into our view of MB function as additional anatomical, behavioral, and physiological data are collected and integrated. For instance, MBONs exhibit significant anatomical complexity, with at least 3 recurrent connections between MBONs and KCs (Aso et al., 2014a), feedforward inhibition (Perisse et al., 2016), and intricate dendritic and axonal fields (Felsenberg et al., 2018). The γ1pedc>α/β MBON plays a key role in state-dependent bidirectional signaling associated with olfactory classical conditioning (Perisse et al., 2016; Tsao et al., 2018). The glutamatergic γ4>γ1γ2 is activated over time to produce a forgetting phenotype that is not acute but cumulative (Shuai et al., 2015). The β1>α MBON is another glutamatergic output neuron that currently has not been associated with learning and memory, but when activated produces an avoidance response (Aso et al., 2014a).
The MBs and MBONs regulate learned behaviors in complex, combinatorial ways. Information about prior experience (learning) and internal state converges in the MB and is transmitted to the MBONs. The MBONs are responsible for valence assignment in a combinatorial way. That is, the final value of a stimulus is believed to be computed as some weighted combination of activity across the “positive” and “negative” MBONs (Owald et al., 2015). The activity of several MBONs is necessary for memory retrieval in the olfactory classical conditioning paradigm (Bouzaiane et al., 2015; Owald et al., 2015; Sejourne et al., 2011). Relatedly, physiological changes in several MBONs have been reported as a result of memory formation (Berry et al., 2018; Bouzaiane et al., 2015; Hige et al., 2015; Owald et al., 2015; Sejourne et al., 2011).
Behavioral complexity: shared circuits regulate diverse behaviors
While the MB is critical for olfactory learning, it is also, more generally, a multimodal sensory integration center (Kirkhart and Scott, 2015; Ren et al., 2012; Vogt et al., 2014; Yagi et al., 2016). The circuits that drive approach and avoidance are involved in modulating other behaviors as well. For instance, male mating behavior relies upon the γ5β′2a output neuron circuit (Keleman et al., 2007; Manoli et al., 2005; Yu et al., 2010; Zhao et al., 2018). This circuit is necessary for inhibition of courtship to unreceptive females. γ KCs synapse onto γ5β′2a MBONs, which subsequently activate the PAM-γ5 DANs (Zhao et al., 2018). If this reciprocal feedback circuit is altered, male courtship conditioning is impaired. In addition, MBONs and DANs also modulate sleep, visual memory, and food-seeking behavior (Aso et al., 2014b; Joiner et al., 2006; Landayan et al., 2018; Pitman et al., 2006; Tsao et al., 2018). Recurrent circuits are utilized for other learned behaviors that are not necessarily directly reliant upon olfaction. For instance, male aggression state is modulated by the γ1pedc>α/β MBON and PPL1-γ1pedc DAN (Kim et al., 2018). This demonstrates that these networks can be co-opted for the alteration of complex behavioral dynamics involving multisensory input.
Conclusions and future outlook
Looking forward, several major themes emerge as likely avenues of intensive research in pursuit of a complete understanding of the fly brain. First, we will need a more complete understanding of the genetics, and synaptic/cellular signaling underlying learning and memory. For instance, single cell transcriptomic studies are leading to the identification of a plethora of novel candidate genes underlying learning and memory (Crocker et al., 2016; Croset et al., 2018; Li et al., 2017; Shih et al., 2019). Second, further anatomical detail will be necessary to understand the connectivity of the nervous system as a whole. Large-scale projects are underway to map the connectome of Drosophila with varying scope and resolution (Eichler et al., 2017; Zheng et al., 2018). This will provide a ground plan necessary to interrogate the nervous system of this key model system with unprecedented specificity. The experience with organisms such as C. elegans and the stomatogastric ganglion of C. borealis has shown that a connectome is a highly valuable contribution to accelerate discovery from functional studies. While general principles of neuronal circuit function can be gleaned from detailed cell-by-cell analysis, it is hard to imagine completely understanding the nervous system without elucidating at least the majority of its “road map”. This points the way toward the third major direction for future research, the functional analysis of neuronal circuits in parallel with mapping projects. This will include the refinement of tools for observing (imaging) and manipulating neuronal circuits to parse their function. Finally, we expect that new conceptual and computational insights will be necessary in order to fully understand the function of the nervous system. Future directions in this realm are more difficult to anticipate, and the punctuated equilibrium of progress is more apparent. In all of these areas, given the conservation of genetics, cell biology, and fundamental neuronal circuit function, developing a thorough understanding of how learning and memory works in the Drosophila brain will provide key insight into the fundamental principles at play across taxa, including humans.
References
- Akalal DB, Yu D, and Davis RL (2010). A late-phase, long-term memory trace forms in the gamma neurons of Drosophila mushroom bodies after olfactory classical conditioning. J Neurosci 30, 16699–16708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, et al. (2014a). The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3, e04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Herb A, Ogueta M, Siwanowicz I, Templier T, Friedrich AB, Ito K, Scholz H, and Tanimoto H (2012). Three dopamine pathways induce aversive odor memories with different stability. PLoS Genet 8, e1002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Siwanowicz I, Bräcker L, Ito K, Kitamoto T, Tanimoto H (2010). Specific dopaminergic neurons for the formation of labile aversive memory. Curr Biol 20: 1445–1451. 10.1016/j.cub.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Sitaraman D, Ichinose T, Kaun KR, Vogt K, Belliart-Guerin G, Placais PY, Robie AA, Yamagata N, Schnaitmann C, et al. (2014b). Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnstedt O, Owald D, Felsenberg J, Brain R, Moszynski JP, Talbot CB, Perrat PN, and Waddell S (2016). Memory-Relevant Mushroom Body Output Synapses Are Cholinergic. Neuron 89, 1237–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JA, Phan A, and Davis RL (2018). Dopamine Neurons Mediate Learning and Forgetting through Bidirectional Modulation of a Memory Trace. Cell Rep 25, 651–662 e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielopolski N, Amin H, Apostolopoulou AA, Rozenfeld E, Lerner H, Huetteroth W, Lin AC, and Parnas M (2019). Inhibitory muscarinic acetylcholine receptors enhance aversive olfactory learning in adult Drosophila. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boto T, Louis T, Jindachomthong K, Jalink K, and Tomchik SM (2014). Dopaminergic Modulation of cAMP Drives Nonlinear Plasticity across the Drosophila Mushroom Body Lobes. Current Biology 24, 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boto T, Stahl A, Zhang XF, Louis T, and Tomchik SM (2019). Independent Contributions of Discrete Dopaminergic Circuits to Cellular Plasticity, Memory Strength, and Valence in Drosophila. Cell Reports 27, 2014–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzaiane E, Trannoy S, Scheunemann L, Placais PY, and Preat T (2015). Two Independent Mushroom Body Output Circuits Retrieve the Six Discrete Components of Drosophila Aversive Memory. Cell Reports 11, 1280–1292. [DOI] [PubMed] [Google Scholar]
- Brembs B, and Plendl W (2008). Double dissociation of PKC and AC manipulations on operant and classical learning in Drosophila. Curr Biol 18, 1168–1171. [DOI] [PubMed] [Google Scholar]
- Broadie K, Rushton E, Skoulakis EM, and Davis RL (1997). Leonardo, a Drosophila 14–3-3 protein involved in learning, regulates presynaptic function. Neuron 19, 391–402. [DOI] [PubMed] [Google Scholar]
- Burke CJ, Huetteroth W, Owald D, Perisse E, Krashes MJ, Das G, Gohl D, Silies M, Certel S, and Waddell S (2012). Layered reward signalling through octopamine and dopamine in Drosophila. Nature 492, 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busto GU, Cervantes-Sandoval I, and Davis RL (2010). Olfactory learning in Drosophila. Physiology (Bethesda, Md) 25, 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Sandoval I, and Davis RL (2012). Distinct traces for appetitive versus aversive olfactory memories in DPM neurons of Drosophila. Curr Biol 22, 1247–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Sandoval I, Phan A, Chakraborty M, and Davis RL (2017). Reciprocal synapses between mushroom body and dopamine neurons form a positive feedback loop required for learning. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Wu JK, Lin HW, Pai TP, Fu TF, Wu CL, Tully T, and Chiang AS (2012). Visualizing long-term memory formation in two neurons of the Drosophila brain. Science 335, 678–685. [DOI] [PubMed] [Google Scholar]
- Christiansen F, Zube C, Andlauer TF, Wichmann C, Fouquet W, Owald D, Mertel S, Leiss F, Tavosanis G, Luna AJ, et al. (2011). Presynapses in Kenyon cell dendrites in the mushroom body calyx of Drosophila. J Neurosci 31, 9696–9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Roorda RD, Vrontou E, Sjulson L, Li H, Hirsh J, and Miesenbock G (2009). Writing memories with light-addressable reinforcement circuitry. Cell 139, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn R, Morantte I, and Ruta V (2015). Coordinated and Compartmentalized Neuromodulation Shapes Sensory Processing in Drosophila. Cell 163, 1742–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly JB, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, and O’Kane CJ (1996). Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science (New York, NY) 274, 2104–2107. [DOI] [PubMed] [Google Scholar]
- Crittenden JR, Skoulakis EM, Han KA, Kalderon D, and Davis RL (1998). Tripartite mushroom body architecture revealed by antigenic markers. Learn Mem 5, 38–51. [PMC free article] [PubMed] [Google Scholar]
- Crocker A, Guan XJ, Murphy CT, and Murthy M (2016). Cell-Type-Specific Transcriptome Analysis in the Drosophila Mushroom Body Reveals Memory-Related Changes in Gene Expression. Cell Rep 15, 1580–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V, Treiber CD, and Waddell S (2018). Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL (2005). Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci 28, 275–302. [DOI] [PubMed] [Google Scholar]
- Davis RL, Cherry J, Dauwalder B, Han PL, and Skoulakis E (1995). The cyclic AMP system and Drosophila learning. Molecular and cellular biochemistry 149–150, 271–278. [DOI] [PubMed] [Google Scholar]
- de Belle JS, and Heisenberg M (1994). Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science (New York, NY) 263, 692–695. [DOI] [PubMed] [Google Scholar]
- Drain P, Dubin AE, and Aldrich RW (1994). Regulation of Shaker K+ channel inactivation gating by the cAMP-dependent protein kinase. Neuron 12, 1097–1109. [DOI] [PubMed] [Google Scholar]
- Dudai Y (1988). Neurogenetic dissection of learning and short-term memory in Drosophila. Annu Rev Neurosci 11, 537–563. [DOI] [PubMed] [Google Scholar]
- Eichler K, Li F, Litwin-Kumar A, Park Y, Andrade I, Schneider-Mizell CM, Saumweber T, Huser A, Eschbach C, Gerber B, et al. (2017). The complete connectome of a learning and memory centre in an insect brain. Nature 548, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenberg J, Jacob PF, Walker T, Barnstedt O, Edmondson-Stait AJ, Pleijzier MW, Otto N, Schlegel P, Sharifi N, Perisse E, et al. (2018). Integration of Parallel Opposing Memories Underlies Memory Extinction. Cell 175, 709–722 715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala A (2007). Olfaction and olfactory learning in Drosophila: recent progress. Curr Opin Neurobiol 17, 720–726. [DOI] [PubMed] [Google Scholar]
- Galili DS, Dylla KV, Ludke A, Friedrich AB, Yamagata N, Wong JY, Ho CH, Szyszka P, and Tanimoto H (2014). Converging circuits mediate temperature and shock aversive olfactory conditioning in Drosophila. Curr Biol 24, 1712–1722. [DOI] [PubMed] [Google Scholar]
- Gervasi N, Tchenio P, and Preat T (2010). PKA dynamics in a Drosophila learning center: coincidence detection by rutabaga adenylyl cyclase and spatial regulation by dunce phosphodiesterase. Neuron 65, 516–529. [DOI] [PubMed] [Google Scholar]
- Goodwin SF, Del Vecchio M, Velinzon K, Hogel C, Russell SR, Tully T, and Kaiser K (1997). Defective learning in mutants of the Drosophila gene for a regulatory subunit of cAMP-dependent protein kinase. The Journal of neuroscience : the official journal of the Society for Neuroscience 17, 8817–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith LC, Verselis LM, Aitken KM, Kyriacou CP, Danho W, and Greenspan RJ (1993). Inhibition of calcium/calmodulin-dependent protein kinase in Drosophila disrupts behavioral plasticity. Neuron 10, 501–509. [DOI] [PubMed] [Google Scholar]
- Guven-Ozkan T, and Davis RL (2014). Functional neuroanatomy of Drosophila olfactory memory formation. Learn Mem 21, 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KA, Millar NS, Grotewiel MS, and Davis RL (1996). DAMB, a novel dopamine receptor expressed specifically in Drosophila mushroom bodies. Neuron 16, 1127–1135. [DOI] [PubMed] [Google Scholar]
- Handler A, Graham TGW, Cohn R, Morantte I, Siliciano AF, Zeng J, Li Y, and Ruta V (2019). Distinct Dopamine Receptor Pathways Underlie the Temporal Sensitivity of Associative Learning. Cell 178, 60–75.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, and Rosbash M (1992). Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci U S A 89, 11711–11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes PR, Christmann BL, and Griffith LC (2015). A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M (2003). Mushroom body memoir: from maps to models. Nat Rev Neurosci 4, 266–275. [DOI] [PubMed] [Google Scholar]
- Heisenberg M, Borst A, Wagner S, and Byers D (1985). Drosophila mushroom body mutants are deficient in olfactory learning. Journal of neurogenetics 2, 1–30. [DOI] [PubMed] [Google Scholar]
- Hige T, Aso Y, Modi MN, Rubin GM, and Turner GC (2015). Heterosynaptic Plasticity Underlies Aversive Olfactory Learning in Drosophila. Neuron 88, 985–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger KS, Campbell RA, and Turner GC (2011). Cellular-resolution population imaging reveals robust sparse coding in the Drosophila mushroom body. J Neurosci 31, 11772–11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huetteroth W, Perisse E, Lin S, Klappenbach M, Burke C, and Waddell S (2015). Sweet taste and nutrient value subdivide rewarding dopaminergic neurons in Drosophila. Curr Biol 25, 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose T, Aso Y, Yamagata N, Abe A, Rubin GM, and Tanimoto H (2015). Reward signal in a recurrent circuit drives appetitive long-term memory formation. Elife 4, e10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliadi KG, Iliadi N, and Boulianne GL (2017). Drosophila mutants lacking octopamine exhibit impairment in aversive olfactory associative learning. Eur J Neurosci 46, 2080–2087. [DOI] [PubMed] [Google Scholar]
- Johnson O, Becnel J, and Nichols CD (2011). Serotonin receptor activity is necessary for olfactory learning and memory in Drosophila melanogaster. Neuroscience 192, 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, and Sehgal A (2006). Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441, 757–760. [DOI] [PubMed] [Google Scholar]
- Kane NS, Robichon A, Dickinson JA, and Greenspan RJ (1997). Learning without performance in PKC-deficient Drosophila. Neuron 18, 307–314. [DOI] [PubMed] [Google Scholar]
- Kaun KR, Hendel T, Gerber B, Sokolowski MB (2007) Natural variation in Drosophila larval reward learning and memory due to a cGMP-dependent protein kinase. Learn Mem 14: 342–349. 10.1101/lm.505807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleman K, Kruttner S, Alenius M, and Dickson BJ (2007). Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci 10, 1587–1593. [DOI] [PubMed] [Google Scholar]
- Kim YC, Lee HG, and Han KA (2007). D1 dopamine receptor dDA1 is required in the mushroom body neurons for aversive and appetitive learning in Drosophila. J Neurosci 27, 7640–7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Lee HG, Lim J, and Han KA (2013). Appetitive learning requires the alpha1-like octopamine receptor OAMB in the Drosophila mushroom body neurons. J Neurosci 33, 1672–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Saver M, Simon J, Kent CF, Shao LS, Eddison M, Agrawal P, Texada M, Truman JW, and Heberlein U (2018). Repetitive aggressive encounters generate a long-lasting internal state in Drosophila melanogaster males. P Natl Acad Sci USA 115, 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkhart C, and Scott K (2015). Gustatory learning and processing in the Drosophila mushroom bodies. J Neurosci 35, 5950–5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapek S, Gerber B, and Tanimoto H (2010). Synapsin is selectively required for anesthesia-sensitive memory. Learn Mem 17, 76–79. [DOI] [PubMed] [Google Scholar]
- Kuo SY, Wu CL, Hsieh MY, Lin CT, Wen RK, Chen LC, Chen YH, Yu YW, Wang HD, Su YJ, et al. (2015). PPL2ab neurons restore sexual responses in aged Drosophila males through dopamine. Nat Commun 6, 7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landayan D, Feldman DS, and Wolf FW (2018). Satiation state-dependent dopaminergic control of foraging in Drosophila. Sci Rep 8, 5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin LR, Han PL, Hwang PM, Feinstein PG, Davis RL, and Reed RR (1992). The Drosophila learning and memory gene rutabaga encodes a Ca2+/Calmodulin-responsive adenylyl cyclase. Cell 68, 479–489. [DOI] [PubMed] [Google Scholar]
- Li HJ, Horns F, Wu B, Xie QJ, Li JF, Li TC, Luginbuhl DJ, Quake SR, and Luo LQ (2017). Classifying Drosophila Olfactory Projection Neuron Subtypes by Single-Cell RNA Sequencing. Cell 171, 1206–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Bygrave AM, de Calignon A, Lee T, and Miesenbock G (2014). Sparse, decorrelated odor coding in the mushroom body enhances learned odor discrimination. Nat Neurosci 17, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Placais PY, Yamagata N, Pfeiffer BD, Aso Y, Friedrich AB, Siwanowicz I, Rubin GM, Preat T, and Tanimoto H (2012). A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature 488, 512–516. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yang X, Tian J, Gao Z, Wang M, Li Y, Guo A (2016). Gap junction networks in mushroom bodies participate in visual learning and memory in Drosophila. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Buchanan ME, Han KA, and Davis RL (2009). The GABAA receptor RDL suppresses the conditioned stimulus pathway for olfactory learning. J Neurosci 29, 1573–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, and Davis RL (2009). The GABAergic anterior paired lateral neuron suppresses and is suppressed by olfactory learning. Nat Neurosci 12, 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone MS, Sziber PP, and Quinn WG (1984). Loss of calcium/calmodulin responsiveness in adenylate cyclase of rutabaga, a Drosophila learning mutant. Cell 37, 205–215. [DOI] [PubMed] [Google Scholar]
- Louis T, Stahl A, Boto T, and Tomchik SM (2018). Cyclic AMP-dependent plasticity underlies rapid changes in odor coding associated with reward learning. Proc Natl Acad Sci U S A 115, E448–E457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyutova R, Selcho M, Pfeuffer M, Segebarth D, Habenstein J, Rohwedder A, Frantzmann F, Wegener C, Thum AS, and Pauls D (2019). Reward signaling in a recurrent circuit of dopaminergic neurons and peptidergic Kenyon cells. Nat Commun 10, 3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli DS, Foss M, Villella A, Taylor BJ, Hall JC, and Baker BS (2005). Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature 436, 395–400. [DOI] [PubMed] [Google Scholar]
- Mao Z, and Davis RL (2009). Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front Neural Circuits 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek P, Worden K, Aso Y, Rubin GM, and Keene AC (2015). A dopamine-modulated neural circuit regulating aversive taste memory in Drosophila. Curr Biol 25, 1535–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, Giuliani G, Choi C, Krause P, Correale D, Watson K, Baker G, and Siwicki KK (1999). Mushroom body ablation impairs short-term memory and long-term memory of courtship conditioning in Drosophila melanogaster. Neuron 24, 967–977. [DOI] [PubMed] [Google Scholar]
- Mery F, Belay AT, So AK, Sokolowski MB, Kawecki TJ (2007) Natural polymorphism affecting learning and memory in Drosophila. Proceedings of the National Academy of Sciences, 104, 13051–13055. 10.1073/pnas.0702923104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels B, Chen YC, Saumweber T, Mishra D, Tanimoto H, Schmid B, Engmann O, and Gerber B (2011). Cellular site and molecular mode of synapsin action in associative learning. Learn Mem 18, 332–344. [DOI] [PubMed] [Google Scholar]
- Michels B, Diegelmann S, Tanimoto H, Schwenkert I, Buchner E, and Gerber B (2005). A role for Synapsin in associative learning: the Drosophila larva as a study case. Learning & memory (Cold Spring Harbor, NY) 12, 224–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T, Kikuchi E, Horiuchi J, and Saitoe M (2018). Long-Term Memory Engram Cells Are Established by c-Fos/CREB Transcriptional Cycling. Cell Rep 25, 2716–2728 e2713. [DOI] [PubMed] [Google Scholar]
- Neuser K, Triphan T, Mronz M, Poeck B, and Strauss R (2008). Analysis of a spatial orientation memory in Drosophila. Nature 453, 1244–1247. [DOI] [PubMed] [Google Scholar]
- Niewalda T, Michels B, Jungnickel R, Diegelmann S, Kleber J, Kahne T, and Gerber B (2015). Synapsin determines memory strength after punishment- and relief-learning. J Neurosci 35, 7487–7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nighorn A, Healy MJ, and Davis RL (1991). The cyclic AMP phosphodiesterase encoded by the Drosophila dunce gene is concentrated in the mushroom body neuropil. Neuron 6, 455–467. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, and Taghert PH (2008). Organization of the Drosophila circadian control circuit. Current biology : CB 18, R84–93. [DOI] [PubMed] [Google Scholar]
- Owald D, Felsenberg J, Talbot CB, Das G, Perisse E, Huetteroth W, and Waddell S (2015). Activity of defined mushroom body output neurons underlies learned olfactory behavior in Drosophila. Neuron 86, 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls D, Selcho M, Gendre N, Stocker RF, and Thum AS (2010). Drosophila larvae establish appetitive olfactory memories via mushroom body neurons of embryonic origin. J Neurosci 30, 10655–10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlowsky A, Schor J, Placais PY, and Preat T (2018). A GABAergic Feedback Shapes Dopaminergic Input on the Drosophila Mushroom Body to Promote Appetitive Long-Term Memory. Curr Biol 28, 1783–1793 e1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavot P, Carbognin E, and Martin JR (2015). PKA and cAMP/CNG Channels Independently Regulate the Cholinergic Ca(2+)-Response of Drosophila Mushroom Body Neurons. eNeuro 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech U, Revelo NH, Seitz KJ, Rizzoli SO, and Fiala A (2015). Optical dissection of experience-dependent pre- and postsynaptic plasticity in the Drosophila brain. Cell Rep 10, 2083–2095. [DOI] [PubMed] [Google Scholar]
- Perisse E, Owald D, Barnstedt O, Talbot CB, Huetteroth W, and Waddell S (2016). Aversive Learning and Appetitive Motivation Toggle Feed-Forward Inhibition in the Drosophila Mushroom Body. Neuron 90, 1086–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisse E, Yin Y, Lin AC, Lin S, Huetteroth W, and Waddell S (2013). Different kenyon cell populations drive learned approach and avoidance in Drosophila. Neuron 79, 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A, Thomas CI, Chakraborty M, Berry JA, Kamasawa N, and Davis RL (2019). Stromalin Constrains Memory Acquisition by Developmentally Limiting Synaptic Vesicle Pool Size. Neuron 101, 103–118.e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, and Allada R (2006). A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441, 753–756. [DOI] [PubMed] [Google Scholar]
- Pugh GE (1977). The Biological Origin of Human Values (New York: Basic Books, Inc.). [Google Scholar]
- Putz G, Bertolucci F, Raabe T, Zars T, and Heisenberg M (2004). The S6KII (rsk) gene of Drosophila melanogaster differentially affects an operant and a classical learning task. J Neurosci 24, 9745–9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Cressy M, Li W, Coravos JS, Izzi SA, and Dubnau J (2012). Gamma neurons mediate dopaminergic input during aversive olfactory memory formation in Drosophila. Curr Biol 22, 608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Li H, Wu Y, Ren J, and Guo A (2012). A GABAergic inhibitory neural circuit regulates visual reversal learning in Drosophila. J Neurosci 32, 11524–11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemensperger T, Voller T, Stock P, Buchner E, and Fiala A (2005). Punishment prediction by dopaminergic neurons in Drosophila. Curr Biol 15, 1953–1960. [DOI] [PubMed] [Google Scholar]
- Rolls MM, Satoh D, Clyne PJ, Henner AL, Uemura T, and Doe CQ (2007). Polarity and intracellular compartmentalization of Drosophila neurons. Neural Dev 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenfeld E, Lerner H, and Parnas M (2019). Muscarinic Modulation of Antennal Lobe GABAergic Local Neurons Shapes Odor Coding and Behavior. Cell Rep 29, 3253–3265 e3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheunemann L, Jost E, Richlitzki A, Day JP, Sebastian S, Thum AS, Efetova M, Davies SA, and Schwarzel M (2012). Consolidated and labile odor memory are separately encoded within the Drosophila brain. The Journal of neuroscience : the official journal of the Society for Neuroscience 32, 17163–17171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheunemann L, Placais PY, Dromard Y, Schwarzel M, and Preat T (2018). Dunce Phosphodiesterase Acts as a Checkpoint for Drosophila Long-Term Memory in a Pair of Serotonergic Neurons. Neuron 98, 350–365.e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroll C, Riemensperger T, Bucher D, Ehmer J, Voller T, Erbguth K, Gerber B, Hendel T, Nagel G, Buchner E, et al. (2006). Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol 16, 1741–1747. [DOI] [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, and Heisenberg M (2003). Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci 23, 10495–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sejourne J, Placais PY, Aso Y, Siwanowicz I, Trannoy S, Thoma V, Tedjakumala SR, Rubin GM, Tchenio P, Ito K, et al. (2011). Mushroom body efferent neurons responsible for aversive olfactory memory retrieval in Drosophila. Nat Neurosci 14, 903–U129. [DOI] [PubMed] [Google Scholar]
- Shih MFM, Davis FP, Henry GL, and Dubnau J (2019). Nuclear Transcriptomes of the Seven Neuronal Cell Types That Constitute the Drosophila Mushroom Bodies. G3-Genes Genom Genet 9, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai YC, Hirokawa A, Ai YL, Zhang M, Li WH, and Zhong Y (2015). Dissecting neural pathways for forgetting in Drosophila olfactory aversive memory. P Natl Acad Sci USA 112, E6663–E6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu WH, Lee WP, Chiang MH, Chang CC, Fu TF, Chiang HC, Wu T, and Wu CL (2019). Electrical synapses between mushroom body neurons are critical for consolidated memory retrieval in Drosophila. PLoS genetics 15, e1008153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva B, Molina-Fernandez C, Ugalde MB, Tognarelli EI, Angel C, and Campusano JM (2015). Muscarinic ACh Receptors Contribute to Aversive Olfactory Learning in Drosophila. Neural Plast 2015, 658918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulakis EM, and Davis RL (1996). Olfactory learning deficits in mutants for leonardo, a Drosophila gene encoding a 14–3-3 protein. Neuron 17, 931–944. [DOI] [PubMed] [Google Scholar]
- Skoulakis EM, Kalderon D, and Davis RL (1993). Preferential expression in mushroom bodies of the catalytic subunit of protein kinase A and its role in learning and memory. Neuron 11, 197–208. [DOI] [PubMed] [Google Scholar]
- Su CY, Menuz K, and Carlson JR (2009). Olfactory perception: receptors, cells, and circuits. Cell 139, 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Aso Y, Hige T, Wong A, Lu Z, Xu CS, Rivlin PK, Hess H, Zhao T, Parag T, et al. (2017). A connectome of a learning and memory center in the adult Drosophila brain. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Yu D, Pletting J, and Davis RL (2010). Gilgamesh is required for rutabaga-independent olfactory learning in Drosophila. Neuron 67, 810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka NK, Tanimoto H, and Ito K (2008). Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol 508, 711–755. [DOI] [PubMed] [Google Scholar]
- Tempel BL, Livingstone MS, and Quinn WG (1984). Mutations in the dopa decarboxylase gene affect learning in Drosophila. Proc Natl Acad Sci U S A 81, 3577–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomchik SM, and Davis RL (2009). Dynamics of learning-related cAMP signaling and stimulus integration in the Drosophila olfactory pathway. Neuron 64, 510–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomchik SM, and Davis RL (2013). Memory Research Through Four Eras: Genetic, Molecular Biology, Neuroanatomy, and Systems Neuroscience In Invertebrate Learning and Memory, Menzel R, and Benjamin P, eds. (London: Elsevier; ), pp. 359–377. [Google Scholar]
- Tsao CH, Chen CC, Lin CH, Yang HY, and Lin S (2018). Drosophila mushroom bodies integrate hunger and satiety signals to control innate food-seeking behavior. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, and Quinn WG (1985). Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A 157, 263–277. [DOI] [PubMed] [Google Scholar]
- Turner GC, Bazhenov M, and Laurent G (2008). Olfactory representations by Drosophila mushroom body neurons. J Neurophysiol 99, 734–746. [DOI] [PubMed] [Google Scholar]
- Ueno K, Naganos S, Hirano Y, Horiuchi J, and Saitoe M (2013). Long-term enhancement of synaptic transmission between antennal lobe and mushroom body in cultured Drosophila brain. The Journal of physiology 591, 287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K, Suzuki E, Naganos S, Ofusa K, Horiuchi J, and Saitoe M (2017). Coincident postsynaptic activity gates presynaptic dopamine release to induce plasticity in Drosophila mushroom bodies. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt K, Schnaitmann C, Dylla KV, Knapek S, Aso Y, Rubin GM, and Tanimoto H (2014). Shared mushroom body circuits underlie visual and olfactory memories in Drosophila. Elife 3, e02395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell S, and Quinn WG (2001). Flies, genes, and learning. Annu Rev Neurosci 24, 1283–1309. [DOI] [PubMed] [Google Scholar]
- Wang Y, Mamiya A, Chiang AS, and Zhong Y (2008). Imaging of an early memory trace in the Drosophila mushroom body. J Neurosci 28, 4368–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer YF, Fritsch C, Jungo MM, Almeida S, Egger B, and Sprecher SG (2018). Multiple neurons encode CrebB dependent appetitive long-term memory in the mushroom body circuit. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf R, Wittig T, Liu L, Wustmann G, Eyding D, and Heisenberg M (1998). Drosophila mushroom bodies are dispensable for visual, tactile, and motor learning. Learning & memory (Cold Spring Harbor, NY) 5, 166–178. [PMC free article] [PubMed] [Google Scholar]
- Wu CL, Shih MF, Lee PT, and Chiang AS (2013). An octopamine-mushroom body circuit modulates the formation of anesthesia-resistant memory in Drosophila. Curr Biol 23, 2346–2354. [DOI] [PubMed] [Google Scholar]
- Yagi R, Mabuchi Y, Mizunami M, and Tanaka NK (2016). Convergence of multimodal sensory pathways to the mushroom body calyx in Drosophila melanogaster. Sci Rep 6, 29481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata N, Hiroi M, Kondo S, Abe A, and Tanimoto H (2016). Suppression of Dopamine Neurons Mediates Reward. PLoS Biol 14, e1002586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata N, Ichinose T, Aso Y, Placais PY, Friedrich AB, Sima RJ, Preat T, Rubin GM, and Tanimoto H (2015). Distinct dopamine neurons mediate reward signals for short- and long-term memories. Proc Natl Acad Sci U S A 112, 578–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Hiroi M, Abe T, Shimizu K, Minami-Ohtsubo M, Maeyama Y, Horiuchi J, and Tabata T (2018). Two Parallel Pathways Assign Opposing Odor Valences during Drosophila Memory Formation. Cell Rep 22, 2346–2358. [DOI] [PubMed] [Google Scholar]
- Yin JC, Wallach JS, Wilder EL, Klingensmith J, Dang D, Perrimon N, Zhou H, Tully T, and Quinn WG (1995). A Drosophila CREB/CREM homolog encodes multiple isoforms, including a cyclic AMP-dependent protein kinase-responsive transcriptional activator and antagonist. Molecular and cellular biology 15, 5123–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Akalal DB, and Davis RL (2006). Drosophila alpha/beta mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron 52, 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Keene AC, Srivatsan A, Waddell S, and Davis RL (2005). Drosophila DPM neurons form a delayed and branch-specific memory trace after olfactory classical conditioning. Cell 123, 945–957. [DOI] [PubMed] [Google Scholar]
- Yu D, Ponomarev A, and Davis RL (2004). Altered representation of the spatial code for odors after olfactory classical conditioning; memory trace formation by synaptic recruitment. Neuron 42, 437–449. [DOI] [PubMed] [Google Scholar]
- Yu JY, Kanai MI, Demir E, Jefferis GSXE, and Dickson BJ (2010). Cellular Organization of the Neural Circuit that Drives Drosophila Courtship Behavior. Current Biology 20, 1602–1614. [DOI] [PubMed] [Google Scholar]
- Zars T (2010). Short-term memories in Drosophila are governed by general and specific genetic systems. Learning & memory (Cold Spring Harbor, NY) 17, 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zars T, Fischer M, Schulz R, and Heisenberg M (2000a). Localization of a short-term memory in Drosophila. Science 288, 672–675. [DOI] [PubMed] [Google Scholar]
- Zars T, Wolf R, Davis R, and Heisenberg M (2000b). Tissue-specific expression of a type I adenylyl cyclase rescues the rutabaga mutant memory defect: in search of the engram. Learn Mem 7, 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tanenhaus AK, Davis JC, Hanlon BM, and Yin JC (2015). Spatio-temporal in vivo recording of dCREB2 dynamics in Drosophila long-term memory processing. Neurobiol Learn Mem 118, 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, and Roman G (2013). Presynaptic inhibition of gamma lobe neurons is required for olfactory learning in Drosophila. Curr Biol 23, 2519–2527. [DOI] [PubMed] [Google Scholar]
- Zhao XL, Lenek D, Dag U, Dickson BJ, and Keleman K (2018). Persistent activity in a recurrent circuit underlies courtship memory in Drosophila. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Lauritzen JS, Perlman E, Robinson CG, Nichols M, Milkie D, Torrens O, Price J, Fisher CB, Sharifi N, et al. (2018). A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster. Cell 174, 730–743 e722. [DOI] [PMC free article] [PubMed] [Google Scholar]