Abstract
Background:
Isolated pulmonary involvement is uncommon in metastatic hormone-sensitive prostate cancer (mHSPC). To characterize outcomes and molecular alterations of this unique patient subset, we conducted a retrospective review of patients with hormone-naïve prostate cancer presenting with lung-only metastases.
Methods:
This was a retrospective single-institution study. Medical records of 25 patients presenting with pulmonary-only metastases before receiving androgen deprivation therapy (ADT) were analyzed. Germline and/or somatic genomic results, where available (n = 16), were documented. Tumor tissue was analyzed using clinical-grade next-generation DNA sequencing assays. Clinical endpoints included complete prostate-specific antigen (PSA) response to ADT (<0.1 ng/mL), median overall survival (OS) from time of ADT initiation, median PSA progression-free survival (PSA-PFS), and failure-free survival (FFS) at 4 years.
Results:
Baseline characteristics were notable for 48% of men (12 of 25) having first or second-degree relatives with prostate cancer, compared with 20% expected. Complete PSA responses to ADT were noted in 52% of men, with a median PSA-PFS of 66 months, a 4-year FFS rate of 72%, and a median OS that was not reached after 190 months. In evaluable patients, molecular drivers were enriched for mismatch repair mutations (4 of 16, 25%) and homologous-recombination deficiency mutations (4 of 16, 25%). These results are limited by the small sample size and retrospective nature of this analysis.
Conclusions:
This exploratory study represents one of the largest cohorts of lung-only mHSPC patients to-date. The prevalence of actionable DNA-repair gene alterations was higher than anticipated (any DNA-repair mutation: 8 of 16, 50%). Compared to historical data, these patients appear to have exceptional and durable responses to first-line ADT. This study suggests that pulmonary-tropic mHSPC biology may be fundamentally different from nonpulmonary mHSPC.
Keywords: DNA repair, homologous recombination, mismatch repair, prostate cancer, pulmonary metastases
1 ∣. INTRODUCTION
Lung metastases occur in over 40% of men with prostate cancer on postmortem analyses, though only 1% to 4% of cases demonstrate pulmonary-only metastases without bone or visceral involvement.1,2 Only a handful of pulmonary-only prostate cancer cases have been reported.3-6 In newly diagnosed metastatic hormone-sensitive prostate cancer (mHSPC) patients, only about 4% to 5% of cases have lung involvement as part of their initial metastatic spectrum,7 and even fewer (about 0.2%) have lung-only metastases.8 Presently, pulmonary metastases are considered visceral disease and are considered high-risk to patients.9
While preliminary data in small groups suggest favorable disease course and outcomes to androgen deprivation therapy (ADT) in these patients, the experience in the lung-only mHSPC population remains quite limited with no large cohort studies available.
Given the rarity of lung-only mHSPC, and the lack of information regarding the associated molecular features, we compiled a single-institution cohort of 25 cases spanning 15 years to evaluate clinical and genomic characteristics of these patients. We hypothesized that this may represent a distinct molecular and clinical subtype of the disease with favorable outcomes.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Patients
Our study included 25 mHSPC patients presenting with lung-only metastases who were managed at the Johns Hopkins Hospital between 2003 and 2018. These patients were retrospectively identified based on having isolated pulmonary metastases at the time of initial diagnosis of metastatic disease in the hormone-sensitive state, without prior knowledge of other clinical or molecular factors. All patients underwent a staging workup including a complete history and physical, baseline bloods, bone scintigraphy and computerized tomography of the chest, abdomen, and pelvis. Patients had to have histologically confirmed prostatic adenocarcinoma from a prostate or lung biopsy. Eight patients received intermittent ADT, 16 had continuous ADT, and one had ADT plus abiraterone. The Johns Hopkins University Institutional Review Board approved this retrospective study.
2.2 ∣. Study design and molecular analyses
Demographic, clinical, and histopathologic characteristics were collected from electronic charts for all patients. Patients had prostate-specific antigen (PSA) levels measured at regular intervals of every 3 to 6 months. Sixteen of the 25 men in the study had been offered germline (Color Genomics, Invitae) and/or somatic (Foundation Medicine, Personal Genome Diagnostics) genomic testing for clinical purposes at different stages of treatment utilizing various commercial next-generation DNA sequencing assays. Only protein-truncating alterations (frameshift insertions or deletions, nonsense mutations, and canonical splice site mutations) were coded as pathogenic or likely pathogenic in the current analysis. Otherwise, unless specifically designated as pathogenic in the ClinVar database,10 missense changes and other variants of undetermined significance were excluded. In instances of multiple pathogenic alterations, “driver” mutations were defined as those most likely to account for the overall pattern of mutations observed.11
2.3 ∣. Outcome measures
Clinical outcome measures were defined using conventional methodology, generally following the PCWG3 criteria. Complete PSA response was defined as an undetectable PSA level (<0.1 ng/mL) at any time following initiation of primary ADT; overall survival (OS) was defined as the interval of time from ADT initiation to death from any cause; and PSA progression-free survival (PSA-PFS) was defined as the time interval to developing a greater than or equal to 25% increase in the PSA level from baseline or nadir (and by ≥2 ng/mL) that required confirmation more than or equal to 3 weeks later (PCWG3 criteria).12 Failure-free survival was defined as symptomatic progression (new cancer-related complications or worsening of existing disease-related symptoms), or radiologic progression (≥2 new bone lesions not due to flare or ≥20% enlargement on computed tomography (CT) scans in sum diameter of target lesions according to RECIST criteria or any new lesions), or PSA progression as per above PCWG3 criteria, or death; whichever occurred first.12,13
2.4 ∣. Statistical analyses
The sample size for this observational study was not prospectively defined and was based on available cases in our clinical and pathology databases that met study criteria. Kaplan-Meier curves were used to visualize time-to-event data. Hazard ratios (HR), associated 95% confidence intervals (CIs), and differences between groups were evaluated with the use of a Cox proportional-hazards model. GraphPad Prism version 8.0.1 and MEDCALC version 18 statistical software was utilized for statistical analyses. Fisher’s exact test was used to compare the mutation frequency between our samples and The Cancer Genome Atlas (TCGA)/Stand Up To Cancer (SU2C) data, and P < .01 were considered of significance to account for multiple comparisons.
3 ∣. RESULTS
3.1 ∣. Clinical characteristics
A total of 25 mHSPC patients, treated from 2003-2018, with lung-only metastatic sites were identified using the Johns Hopkins EPIC EMR and the Johns Hopkins Pathology database. Pulmonary-metastatic adenocarcinoma of the prostate was confirmed either by radiographic response to ADT or in all cases in which a lung biopsy had been performed (13 of 13, Table S1). Lung-only mHSPC patients had either no detectable lymphadenopathy (N = 21/25), or lymph nodes less than 1.5 cm in the short-axis diameter on conventional imaging (N = 4/26), as suggested by PCWG3 criteria.12 Baseline clinical and pathologic characteristics are summarized in Table 1. Median age at initial diagnosis was 60 years, median follow-up from diagnosis was 16 years (190 months), 76% of patients were Caucasian, the majority of patients had Gleason sum greater than or equal to 8%, and 32% of men presented with metastatic disease at first diagnosis. Intraductal/ductal histology was found in 28%, lymphovascular invasion in 16%, and perineural invasion in 48% of cases. Family history of cancer was notable for 48% of men having a first or second-degree relative with prostate cancer, and 36% having a first or second-degree relative with any cancer. Overall, 68% of patients were nonsmokers.
TABLE 1.
Baseline demographic, clinical, and pathological characteristics
| Patients with lung-only metastases (N = 25) |
|
|---|---|
| Median age (y), range | 60 (45–78) |
| Race, % (N) | |
| White | 76% (19) |
| Black | 16% (4) |
| Asian | 4% (1) |
| Other | 4% (1) |
| Baseline PSA at diagnosis | 5 ng/mL |
| Median, range | 0-134 |
| Follow-up time (mo), median, range | 190 (13-244) |
| Type of tissue used for histological analysis, % (N) | |
| Prostatectomy | 28% (7) |
| Prostate biopsy | 72% (18) |
| Grade group/Gleason sum at diagnosis, % (N) | |
| Grade group 1: 3 + 3 | 8% (2) |
| Grade group 2: 3 + 4 | 8% (2) |
| Grade group 3: 4 + 3 | 16% (4) |
| Grade group 4: 4 + 4 | 36% (9) |
| Grade group 5: 4 + 5, 5 + 4, or 5 + 5 | 32% (8) |
| Gleason sum, % (N) | |
| ≤7 | 32% (8) |
| ≥8 | 68% (17) |
| Presence of Intraductal or ductal histology, % (N) | |
| Yes | 28% (7) |
| No | 68% (17) |
| N/A | 4% (1) |
| Presence of lymphovascular invasion, % (N) | |
| Yes | 16% (4) |
| No | 80% (20) |
| N/A | 4% (1) |
| Presence of perineural invasion, % (N) | |
| Yes | 48% (12) |
| No | 48% (12) |
| N/A | 4% (1) |
| M1 disease at diagnosis, % (N) | 32% (8) |
| Biopsy proven lung metastases, % (N) | |
| Yes/no | 52% (13)/48% (12) |
| First or second-degree relative with prostate cancer? | 48% (12) |
| First or second-degree relative with breast, ovarian, uterine, colon, gastric, or pancreatic cancer? | 36% (9) |
| First or second-degree relative with any cancer? | 36% (9) |
| Smoker or former smoker? | |
| Yes/no | 32% (8)/68% (17) |
| Complete PSA response on ADT? | |
| Yes | 52% (13) |
| No | 48% (12) |
| NA | 4% (1) |
| 4-y failure-free rate | |
| Progressed under 4 y (clinical/PSA/radiographic) | 28% (5) |
| Free of progression | 72% (13) |
| Follow-up of <4 y | 7 |
Abbreviations: ADT, androgen deprivation therapy; PSA, prostate-specific antigen.
3.2 ∣. Genomic analyses
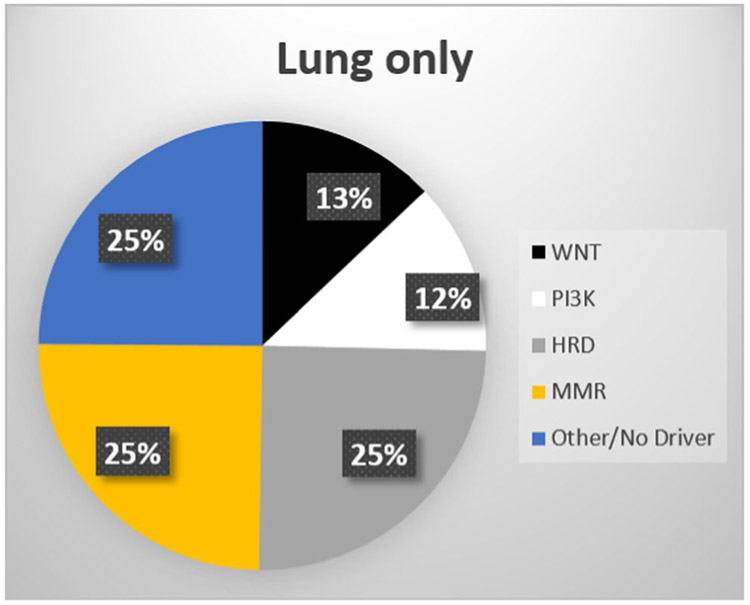
Sixteen (16) of the 25 patients had previously undergone commercial-grade somatic and/or germline genetic testing as part of routine clinical care. Germline testing was always performed from saliva, and somatic testing was performed either from primary tumor tissue (N = 5), plasma DNA (N = 3) or from metastatic lung biopsies (N = 8). Overall, genomic alterations were relatively common and are listed in Table S2. These included alterations in mismatch repair (MMR) genes (4 of 16, 25%), homologous-recombination deficiency (HRD) genes (8 of 16, 50%), PI3K-pathway genes (8 of 16, 50%), Wnt-signaling pathway genes (6 of 16, 38%), as well as TP53 mutations (2 of 13, 16%). The relative breakdown of putative “driver” mutations is summarized in Table 2 and Figure 1. Notably, drivers were enriched for actionable MMR mutations (4 of 16, 25%) and HRD mutations (4 of 16, 25%).
TABLE 2.
List of putative “driver” mutations, by patient
| Patient No. | Germline | MMR | HRD | PI3K/Akt | Wnt | TP53 |
|---|---|---|---|---|---|---|
| 1 | – | Yes (MSH6) | – | – | – | Yes |
| 2 | Yes (BRCA2) | – | Yes (BRCA2) | – | – | – |
| 3 | – | – | – | – | Yes (CTNNB1) | – |
| 4 | – | – | Yes (BRCA2) | – | Yes (APC) | – |
| 5 | – | – | Yes (ATM) | – | – | – |
| 6 | – | – | – | Yes (PTEN) | – | – |
| 7 | – | Yes (MSH2) | – | – | – | – |
| 8 | – | Yes (MSH2) | – | – | – | – |
| 11 | – | – | – | – | Yes (APC) | – |
| 15 | – | – | – | – | – | – |
| 16 | – | – | – | – | – | – |
| 17 | – | – | – | Yes (PIK3CA) | – | – |
| 18 | – | – | – | – | – | Yes |
| 19 | – | – | – | – | – | – |
| 20 | – | Yes (MLH1) | – | – | – | – |
| 25 | Yes (BRCA2) | – | Yes (BRCA2) | – | – | – |
Note: Those mutations that were confirmed to arise in the germline are shown in the first column. Mutations depicted in bold are considered to be the “driver” mutations in each case.
Abbreviations: HRD, homologous-recombination deficiency; MMR, mismatch repair.
FIGURE 1.
Distribution of putative “driver” mutations in lung-only mHSPC patients. HRD, homologous-recombination deficiency; mHSPC, metastatic hormone-sensitive prostate cancer; MMR, mismatch repair [Color figure can be viewed at wileyonlinelibrary.com]
This cohort appears to be significantly enriched for DNA-repair gene alterations compared with the overall advanced prostate cancer population included in our prostate cancer genomics database (published elsewhere14,15), where 22% of men with metastatic disease have an HRD mutation and only 4% have an MMR mutation.14,15 The relatively frequent molecular enrichment of MMR gene and HRD mutations is notable, with prevalence being higher than anticipated even for advanced prostate cancer populations. Statistical comparisons between the genomics of our lung-only mHSPC patients and those extracted from the published genomic datasets (TCGA16 and SU2C-Prostate Cancer Foundation [PCF]17), including alterations beyond the MMR and HRD pathways, are depicted in Table 3.
TABLE 3.
Recurrent genomic alterations in lung-only mHSPC cases compared to men with sporadic localized and castration-resistant prostate cancer
| Lung-only |
TCGAa |
SU2Cb |
|||||
|---|---|---|---|---|---|---|---|
| N = 16 |
N = 333 |
N = 150 |
Lung-only vs TCGA |
Lung-only vs SU2C |
|||
| Gene/pathway | No. of mutations (% of men) | RR (95% CI) | P value | RR (95% CI) | P value | ||
| Any DDR alteration | 10 (63) | 62 (19) | 34 (23) | 3.36 (2.16-5.22) | <.001 | 2.76 (1.70-4.46) | <.001 |
| Any MMR alteration | 4 (25) | 11 (3) | 3 (2) | 8.07 (2.91-22.40) | <.001 | 13.33 (3.29-54.05) | <.001 |
| MSH2 | 2 (13) | 5 (2) | 3 (2) | 8.33 (1.75-39.67) | .008 | 6.25 (1.13-34.67) | .036 |
| MLH1 | 1 (6) | 1 (0.3) | 1 (0.7) | 20.81 (1.36-317.87) | .029 | 9.38 (0.62-142.82) | .107 |
| MSH6 | 1 (6) | 6 (2) | 0 | 3.47 (0.44-27.13) | .236 | – | – |
| PMS2 | 0 | 4 (1) | 0 | 2.18 (0.12-38.92) | .595 | – | – |
| Any HRD alterations | 8 (50) | 44 (13) | 29 (19) | 3.78 (2.16-6.64) | <.0001 | 2.59 (1.44-4.66) | .002 |
| BRCA1 | 1 (6) | 4 (1) | 1 (0.7) | 5.20 (0.62-43.92) | .130 | 9.38 (0.62-142.82) | .107 |
| BRCA2 | 4 (25) | 11 (3) | 19 (13) | 7.57 (2.71-21.17) | <.001 | 1.95 (0.76-5.02) | .168 |
| ATM | 3 (19) | 24 (7) | 7 (7.3) | 2.60 (0.87-7.74) | .086 | 4.02 (1.15-14.03) | .029 |
| PALB2 | 0 | 5 (2) | 0 | 1.79 (0.10-30.99) | .690 | – | – |
| CDK12 | 0 | 5 (2) | 7 (5) | 1.25 (0.07-21.70) | .878 | 0.592 (0.04-9.92) | .716 |
| Wnt-pathway | 6 (38) | 27 (8) | 19 (13) | 4.63 (2.23-9.58) | <.0001 | 2.96 (1.39-6.33) | .005 |
| CTNNB1 | 3 (19) | 9 (3) | 6 (4) | 6.94 (2.08-23.18) | .002 | 4.69 (1.29-16.97) | .019 |
| APC | 3 (19) | 18 (5) | 13 (9) | 3.47 (1.14-10.57) | .029 | 2.16 (0.69-6.80) | .186 |
| PI3K-mTOR pathway | 8 (50) | 103 (31) | 73 (49) | 1.62 (0.97-2.71) | .068 | 1.03 (0.61-1.72) | .918 |
| PTEN | 3 (19) | 58 (17) | 61 (41) | 1.08 (0.38-3.07) | .890 | 0.46 (0.16-1.30) | .144 |
| PIK3CA | 3 (19) | 16 (5) | 8 (5) | 3.90 (1.27-12.04) | .018 | 3.52 (1.04-11.94) | .044 |
| AKT1 | 2 (13) | 7 (2) | 2 (1) | 5.95 (1.34-26.37) | .019 | 9.38 (1.41-62.12) | .020 |
| TSC2 | 0 | 5 (2) | 6 (4) | 1.79 (0.10-31.00) | .690 | 0.68 (0.04-11.61) | .792 |
| Other ETS fusions |
3 (19) | 199 (60) | 84 (56) | 0.31 (0.11-0.87) | .027 | 0.33 (0.12-0.94) | .037 |
| TP53 | 2 (13) | 25 (8) | 80 (54) | 1.67 (0.43-6.42) | .459 | 0.23 (0.06-0.86) | .029 |
Note: P values and RR are in comparison to the lung-only patients. Mutational frequencies for comparator datasets were derived from either the primary publications or extracted from cBioPortal.16-19 P < .01 were considered of significance to account for multiple comparisons and are bolded for emphasis. Abbreviations: CI, confidence interval; HRD, homologous-recombination deficiency; mHSPC, metastatic hormone-sensitive prostate cancer; MMR, mismatch repair; RR, relative risks; SU2C-PCF, Stand Up To Cancer-Prostate Cancer Foundation; TCGA, The Cancer Genome Atlas.
TCGA data set is comprised of 333 men with localized prostate cancer.16
SU2C-PCF International Prostate Cancer Dream Team discovery set is comprised of 150 men with metastatic castration-resistant prostate cancer. Genomic landscape of this population has been previously published.17
3.3 ∣. Efficacy of first-line ADT
Complete PSA responses to first-line ADT were seen in 52% of cases. Median PSA-PFS to first-line ADT was 66 months (Figure 2A), which is longer than anticipated. Failure-free survival (radiographic, clinical, or PSA progression, or death) at 4 years was 72% (13 of 18 men with >4 years follow-up), with seven additional patients demonstrating ongoing response with shorter follow-up times. Presence of DNA-repair gene mutations—in aggregate—did not appear to account for the observed difference in ADT sensitivity (Figure 2C), and insufficient sample size was available to assess HRD and MMR contributions separately. All deaths in this population were due to prostate cancer (N = 2; one at 50 months of follow-up, and one at 66 months of follow-up), with median OS after first-line ADT not being reached for the overall cohort after more than 160 months (Figure 2B). Of note, one patient received first-line ADT plus abiraterone. In addition, one patient was not analyzed due to receiving first-line degarelix and pembrolizumab on a clinical trial.
FIGURE 2.
Kaplan-Meier curves showing (A) PSA-PFS, (B) OS from ADT initiation, and (C) PSA-PFS in DNA-repair mutated or intact patients. One patient was excluded from analysis due to starting ADT+pembrolizumab as first-line systemic treatment rather than ADT alone. ADT, androgen deprivation therapy; OS, overall survival; PFS, progression-free survival; PSA, prostate-specific antigen [Color figure can be viewed at wileyonlinelibrary.com]
Three patients had complete radiographic responses in lung lesions by CT scan, in addition to complete PSA responses. One of these patients was a 69-year-old Caucasian male, nonsmoker, with Gleason 4 + 4 = 8 prostate adenocarcinoma and ductal features. He had undergone a radical prostatectomy in 2011 (pT3a N0 R0) and achieved an undetectable postoperative PSA level. His PSA began to rise in 2012 (1 year after surgery) with a PSA doubling time of 7 months for which he received salvage radiotherapy leading to a PSA nadir of 0.2 ng/mL, but increasing again in April 2013. CT scanning at that time showed innumerable pulmonary metastases (Figure 3A). He was started on ADT in July 2015, which he received for 18 months, and achieved a complete biochemical (PSA <0.1 ng/mL) and radiographic response. The ADT was then discontinued in February 2017, and he was radiographically free of disease recurrence for more than 2 years (Figure 3B). Genomic analysis from this patient did not yield adequate DNA for somatic sequencing.
FIGURE 3.
Effect of androgen deprivation therapy (ADT) in a lung-only metastatic prostate cancer patient, (A) before ADT therapy, and (B) 12 months post-ADT therapy [Color figure can be viewed at wileyonlinelibrary.com]
4 ∣. DISCUSSION
Isolated lung metastases in hormone-sensitive prostate cancer are extremely rare, with a possible estimate being 0.2%.8 Preliminary data in small groups suggest favorable disease course and outcomes to ADT, prompting us to hypothesize that these patients constitute a unique molecular and clinical subgroup. To our knowledge, this is the largest published cohort of mHSPC patients with lung-only metastases and the first to genomically characterize these tumors.
The results of our hypothesis-generating study show that lung-only mHSPC patients are enriched for actionable driver mutations: 25% have loss-of-function mutations in MMR genes, 25% have HRD alterations, and 50% overall have any DNA-repair gene alteration. The 25% prevalence of MMR alterations (4 of 16) is significantly higher than that previously reported by the TCGA group for primary prostate cancers16 (25% vs 2%, P < .001), and is also significantly higher than in unselected metastatic prostate cancers as reported by the SU2C-PCF set of metastatic castration-resistant prostate cancer (mCRPC) individuals17 (25% vs 3%, P < .001). The rarity of MMR gene mutations seen in the overall SU2C-PCF cohort has been confirmed by other studies and is estimated to be in the 2% to 5% range.20,21 The 50% prevalence of HRD alterations (8 of 16, half of which were a consequence of a primary MMR mutation) is significantly higher than that reported by the TCGA (50% vs 13%, P < .0001), and is also higher than that reported by the SU2C-PCF (50% vs 19%, P = .002).
Furthermore, although Wnt-pathway alterations accounted for about 13% of driver mutations, they constituted 38% (6 of 16) of the overall pathogenic recurrent mutations, which again is significantly higher than that reported by the TCGA (38% vs 8%, P < .0001), and is also higher than that reported by the SU2C-PCF (38% vs 13%, P = .005), but the significance of this is unclear. Notably, PI3K-signaling pathway mutations, TP53 mutations, and ETS gene rearrangements were not significantly different compared to both primary (TCGA) and metastatic (SU2C-PCF) prostate cancer datasets.16,17 Interestingly, consensus in the literature is building that aggressive histological subtypes of localized prostate cancer (including ductal prostate cancer22 and primary Gleason pattern 5 adenocarcinomas23) may also be enriched for MMR gene defects.
Clinically, lung-only mHSPC patients demonstrated enhanced sensitivity to standard ADT that greatly exceeded historical estimates (median PSA-PFS of 66 months in our study; vs 12 months in the CHAARTED9 control arm, and 7 months in the LATITUDE24 control arm). Likewise, 4-year failure-free survival in our cohort was 67%; compared to 20% in the LATITUDE control arm, and 15% in the STAMPEDE control arm. Remarkably, these patients also exhibited longevity well beyond historical mHSPC cohorts, with median follow-up of 16 years (only 2 of 16 deaths in this period) and an OS estimate from ADT initiation of more than 160 months, which greatly exceeds the median OS for high-volume disease patients (defined in CHAARTED as any patients with visceral disease, as in this study) in both the ADT-alone arm (median, 34 months) and even the ADT-docetaxel arm (median, 49 months). Indeed, at 3 years all of the patients in this study were alive, and at 5 years 88% remained alive; compared to 60% OS at 3 years for STAMPEDE,25 50% OS at 3 years for LATITUDE, and 50% OS at 3 years (and only 10% OS at 5 years) for CHAARTED high-volume mHSPC patients.
If prospectively confirmed, our findings may impact clinical decisions and aid therapy selection for mHSPC patients with lung-only metastatic disease, especially as they appear to be more accurately classified as “low-burden, good-risk patients” rather than “high-burden, high-risk” disease as defined by the pivotal CHAARTED study. Furthermore, this classification would be akin to pulmonary-only visceral disease in metastatic testicular cancer, that is considered to be a good prognostic finding compared to non–lung visceral disease which is a poor prognostic marker—a previously unappreciated distinction in prostate cancer.26 In addition, the National Comprehensive Cancer Network (NCCN) 2019 Prostate Cancer guidelines27 now recommend germline DNA testing for all men diagnosed with mCRPC, but our data suggest that germline and somatic clinical-grade next-generation DNA sequencing can provide additional information in specific mHSPC populations.
This study has several limitations. First, causal inferences cannot be made despite the strong associations seen, as this was a retrospective study for which the sample size was not determined a prior using hypothesis testing. Second, despite our best efforts to select all available patients with lung metastases from our EMR and the pathology database, this retrospective study design inherently suffers from information and selection bias. Third, the interpretation of Kaplan-Meier curves should be done with caution due to the small sample sizes (25 patients), resulting in wide CIs. Fourth, we were unable to adjust or control for potential baseline clinical discrepancies arising from the small sample size and the hypothesis-generating nature of this study. Last, the enrichment of HRD and MMR mutations did not appear to drive the observed PSA-PFS or OS improvements, indicating that unaccounted alternative genetic/epigenetic or clinical drivers may underlie pulmonary-tropic prostate cancer. Furthermore, our study is limited by only having genomic information on 16 of 25 cases and not utilizing whole-exome sequencing (meaning that we may have a lower sensitivity for detecting complex genomic rearrangements such as in the MMR genes that could be missed by panels that only sequence exonic regions like the clinical assays utilized herein28,29). Consequently, we may be under-calling the true prevalence of pathogenic MMR or HRD mutations in our study. These results indicate that whole-exome sequencing and RNA sequencing/expression analyses are warranted to compare the lung-only and nonpulmonary mHSPC cohorts, but are currently beyond the scope of this study.
5 ∣. CONCLUSIONS
Our preliminary findings suggest that mHSPC patients with lung-only metastases may be a unique molecular and clinical subgroup with significant enrichment for MMR and HDR mutations, and favorable clinical outcomes to first-line ADT treatment (which does not appear to be driven by enrichment in HRD and MMR alterations alone), and may imply additional unique biologic features. These results also indicate that patients with lung-only metastatic disease should be offered germline and somatic DNA sequencing as they may have a high prevalence of actionable DNA-repair gene mutations. Specifically, more than half (collectively) of these men would potentially be eligible for FDA approved on-label second-line pembrolizumab treatment (for MMR alterations) or investigational PARP inhibitor or platinum-based chemotherapy (for HRD mutations).30 These intriguing findings, interpreted in the context of the limitations inherent to the design of our retrospective study, should be considered hypothesis-generating and will require prospective validation before alteration in clinical management can be recommended.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Bloomberg-Kimmel Institute for Cancer Immunotherapy and the NCI NIH SKCCC core grant P30CA006973. ES is partially supported by an ASCO Conquer Cancer Foundation Young Investigator Award. ESA is partially supported by National Institutes of Health grants R01CA185297 and P30CA006973, and Department of Defense Prostate Cancer Research Program grants W81XWH-15-2-0050.
Funding information
Conquer Cancer Foundation, Grant/Award Number: Young Investigator Award to ES; National Cancer Institute of the National Institute of Health, Grant/Award Number: R01CA185297; National Cancer Institute of the National Insitute of Health Sidney Kimmel Cancer Center Core Grant, Grant/Award Number: P30CA006973; The Bloomberg-Kimmel Institute for Cancer Immunotherapy; Congressionally Directed Medical Research Programs, Grant/Award Number: W81XWH-15-2-0050
Footnotes
DISCLOSURES
ES is a founder of LifeImmune, an allergy diagnostics company, and has an allergy-related patent pending. ESA is a paid consultant/advisor to Janssen, Astellas, Pfizer, Sanofi, Dendreon, AstraZeneca, Clovis, and Merck; he has received research funding to his institution from Janssen, Johnson & Johnson, Pfizer, Sanofi, Dendreon, Genentech, Novartis, Bristol-Myers Squibb, AstraZeneca, Clovis, and Merck; and he is the coinventor of a biomarker technology that has been licensed to Qiagen. MAE is an independent board of directors member and a stockholder of VERU, Inc. DP is a consultant for Aduro Biotech, Amgen, Bayer, Dynavax, Ervaxx, Five Prime, FLX Bio, Immunomic Therapeutics, Janssen, Merck, Rock Springs Capitol, Tizona, and Trieza. DP is on the Board of Directors of DNAtrix and on the Scientific Advisory Boards of WindMil, Camden Partners, Potenza. DP has research support from AstraZeneca, Bristol-Myers Squibb, and Compugen. All other authors have no disclosures related to this work.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- 1.Saitoh H, Hida M, Shimbo T, Nakamura K, Yamagata J, Satoh T. Metastatic patterns of prostatic cancer. Correlation between sites and number of organs involved. Cancer. 1984;54(12):3078–3084. [DOI] [PubMed] [Google Scholar]
- 2.Bubendorf L, Schopfer A, Wagner U, et al. Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Hum Pathol. 2000;31(5):578–583. [DOI] [PubMed] [Google Scholar]
- 3.Gago JP, Camara G, Dionisio J, Opiniao A. Pulmonary metastasis as sole manifestation of relapse in previously treated localised prostate cancer: three exceptional case reports. Ecancermedicalscience. 2016; 10:645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kume H, Takai K, Kameyama S, Kawabe K. Multiple pulmonary metastasis of prostatic carcinoma with little or no bone or lymph node metastasis. Report of two cases and review of the literature. Urol Int. 1999;62(1):44–47. [DOI] [PubMed] [Google Scholar]
- 5.Smith CP, Sharma A, Ayala G, Cagle P, Kadmon D. Solitary pulmonary metastasis from prostate cancer. J Urol. 1999;162(6):2102–2102. [DOI] [PubMed] [Google Scholar]
- 6.Hofland CA, Bagg MD. An isolated pulmonary metastasis in prostate cancer. Mil Med. 2000;165(12):973–974. [PubMed] [Google Scholar]
- 7.Parker CC, James ND, Brawley CD, et al. Radiotherapy to the primary tumour for newly diagnosed, metastatic prostate cancer (STAMPEDE): a randomised controlled phase 3 trial. Lancet. 2018;392(10162): 2353–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabozzi SJ, Schellhammer PF, el-Mahdi AM. Pulmonary metastases from prostate cancer. Cancer. 1995;75(11):2706–2709. [DOI] [PubMed] [Google Scholar]
- 9.Sweeney CJ, Chen Y-H, Carducci M, et al. Chemohormonal therapy in metastatic hormone-sensitive prostate cancer. N Engl J Med. 2015; 373(8):737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016; 44(D1):D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61(8): 3230–3239. [PubMed] [Google Scholar]
- 12.Scher HI, Morris MJ, Stadler WM, et al. Trial design and objectives for castration-resistant prostate cancer: Updated recommendations from the Prostate Cancer Clinical Trials Working Group 3. J Clin Oncol. 2016;34(12):1402–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. [DOI] [PubMed] [Google Scholar]
- 14.Antonarakis ES, Shaukat F, Isaacsson Velho P, et al. Clinical features and therapeutic outcomes in men with advanced prostate cancer and dna mismatch repair gene mutations. Eur Urol. 2018;74:218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antonarakis ES, Lu C, Luber B, et al. Germline DNA-repair gene mutations and outcomes in men with metastatic castration-resistant prostate cancer receiving first-line abiraterone and enzalutamide. Eur Urol. 2018;74(2):218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abeshouse A, Ahn J, Akbani R, et al. The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson D, Van Allen Eliezer M, Wu Y-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le DT, Durham JN, Smith KN, et al. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schweizer MT, Antonarakis ES, Bismar TA, et al. Genomic characterization of prostatic ductal adenocarcinoma identifies a high prevalence of DNA repair gene mutations. JCO Precis Oncol. 2019;(3):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guedes LB, Antonarakis ES, Schweizer MT, et al. MSH2 loss in primary prostate cancer. Clin Cancer Res. 2017;23(22):6863–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fizazi K, Tran N, Fein L, et al. Abiraterone plus prednisone in metastatic, castration-sensitive prostate cancer. N Engl J Med. 2017; 377(4):352–360. [DOI] [PubMed] [Google Scholar]
- 25.James ND, de Bono JS, Spears MR, et al. Abiraterone for prostate cancer not previously treated with hormone therapy. N Engl J Med. 2017;377(4):338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Germ Cell Cancer Collaborative Group. International Germ Cell Consensus Classification: a prognostic factor-based staging system for metastatic germ cell cancers. J Clin Oncol. 1997;15(2):594–603. [DOI] [PubMed] [Google Scholar]
- 27.National Comprehensive Cancer Network. Genetic/familial high-risk assessment: breast and ovarian. Plymouth Meeting, PA: National Comprehensive Cancer Network; 2019. https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf. Accessed May 1, 2019. [Google Scholar]
- 28.Pritchard CC, Salipante SJ, Koehler K, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn. 2014;16(1):56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pritchard CC, Morrissey C, Kumar A, et al. Complex MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nat Commun. 2014;5:4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.United States Food and Drug Administration. FDA grants accelerated approval to pembrolizumab for first tissue/site agnostic indication. Silver Spring, MD: USFDA; 2017. 37 https://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm560040.htm. Accessed May 2, 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.