Abstract
Objective
This study was designed to evaluate the roles of microRNAs (miRNAs) in slow transit constipation (STC).
Design
All human tissue samples were from the muscularis externa of the colon. Expression of 372 miRNAs was examined in a discovery cohort of four patients with STC versus three age/sex-matched controls by a quantitative PCR array. Upregulated miRNAs were examined by quantitative reverse transcription qPCR (RT-qPCR) in a validation cohort of seven patients with STC and age/sex-matched controls. The effect of a highly differentially expressed miRNA on a custom human smooth muscle cell line was examined in vitro by RT-qPCR, electrophysiology, traction force microscopy, and ex vivo by lentiviral transduction in rat muscularis externa organotypic cultures.
Results
The expression of 13 miRNAs was increased in STC samples. Of those miRNAs, four were predicted to target SCN5A, the gene that encodes the Na+ channel NaV1.5. The expression of SCN5A mRNA was decreased in STC samples. Let-7f significantly decreased Na+ current density in vitro in human smooth muscle cells. In rat muscularis externa organotypic cultures, overexpression of let-7f resulted in reduced frequency and amplitude of contraction.
Conclusions
A small group of miRNAs is upregulated in STC, and many of these miRNAs target the SCN5A-encoded Na+ channel NaV1.5. Within this set, a novel NaV1.5 regulator, let-7f, resulted in decreased NaV1.5 expression, current density and reduced motility of GI smooth muscle. These results suggest NaV1.5 and miRNAs as novel diagnostic and potential therapeutic targets in STC.
Keywords: motility disorders, intestinal motility, intestinal ion transport, genetics, constipation
Significance of this study.
What is already known on this subject?
Slow transit constipation is defined as prolonged transit time through the colon in the absence of obstruction or structural abnormalities. Slow transit constipation pathophysiology is poorly understood.
The voltage-gated mechanosensitive Na+ channel NaV1.5 is expressed in GI human and rat smooth muscle cells and regulates smooth muscle electrical and mechanical function.
Dysregulation of NaV1.5 due to altered expression of microRNAs (miRNAs) has pathological consequences in the heart.
What are the new findings?
In slow transit constipation, SCN5A-encoded NaV1.5 expression is decreased, a subset of 13 miRNAs is overexpressed and 4 of these are predicted to target SCN5A.
Let-7f is a novel miRNA regulator of NaV1.5. It was one of the overexpressed miRNAs in slow transit constipation; it decreased functional NaV1.5 channel expression, NaV1.5 currents in human smooth muscle cells and rat GI smooth muscle contractility.
Let-7f as a regulator of NaV1.5 function is a novel finding and has broad interest given the physiological roles of NaV1.5 in heart and gut.
How might it impact on clinical practice in the foreseeable future?
miRNAs are differentially regulated in slow transit constipation, which may be of diagnostic significance.
miRNA regulation of NaV1.5 current density may be important in slow transit constipation pathophysiology.
Therefore, miRNAs and NaV1.5 may be novel therapeutic targets in slow transit constipation.
Introduction
GI motility is critical for the normal function of the human GI tract. Disruptions in GI motility are associated with several diseases or disorders with onsets that vary from acute, for example, postoperative ileus, to insidious, such as pseudo-obstruction. Slow transit constipation (STC), a chronic disabling disease hallmarked by delays in colonic transit in the absence of outlet obstruction, may be refractory to medication, resulting in a colectomy.1 2 The pathophysiology of STC is not well defined. Previous studies show a loss of enteric neurons3 and interstitial cells of Cajal (ICC)—the pacemakers of the gut.4 5 However, in the majority of cases, a histological analysis is not diagnostic,6 which may suggest functional pathology at the molecular level.
Ion channels are indispensable for electrical excitability and electromechanical coupling in GI smooth muscle.7 Therefore, tight regulation of ion channel expression and function are paramount for normal GI motility. Voltage-gated Na+ channels, in particular, are important for the generation and propagation of signals in electrically excitable cells, such as myocytes, cardiomyocytes and neurons.8 Even in GI smooth muscle, where Ca2+ signaling is fundamental for excitability and motility, Na+ channels contribute in a species-dependent fashion to the regulation of electrical and contractile function.9–13 A voltage-gated Na+ channel NaV1.5, encoded by the SCN5A gene, is traditionally considered to be a ‘cardiac’ Na+ channel,14 but NaV1.5 is expressed in human GI smooth muscle and ICC in jejunum and colon,10 15 where it is important for electrophysiological function11 16 and contractility.13 It is also present and functionally relevant in the intestinal smooth muscle of rat11 and myenteric neurons but not smooth muscle of mouse.12 17
SCN5A mutations cause cardiac conduction disorders, ‘channelopathies’, such as long QT syndrome and Brugada syndrome,18 and SCN5A is the only non-structural gene implicated in dilated cardiomyopathy, a cardiac contractility disorder.19–21 Patients with SCN5A channelopathies and known cardiac arrhythmias have an increased prevalence of functional GI diseases,22 and conversely patients with IBS are more likely to have SCN5A channelopathies, with a greater percentage of patients with SCN5A channelopathies having constipation-predominant IBS.23 Furthermore, NaV1.5 block by ranolazine, a Food and Drug Administration-approved medication, is strongly associated with constipation24 due to the inhibition of NaV1.5 mechanosensitivity and reduced contractility of human colon smooth muscle.13
However, channelopathies are relatively rare. More frequently, diseases are caused by altered ion channel expression,25 especially because changes in ionic current as little as 1% can lead to fatal diseases.26 Therefore, ion channel expression is tightly regulated by multiple mechanisms, including at the epigenetic, mRNA and protein levels. microRNAs (miRNAs) are a class of small non-coding RNAs that modulate gene expression at the post-transcriptional level and are involved in the pathogenesis of many diseases.27 Ion channel densities, including NaV1.5, are frequently regulated by miRNAs, and this mechanism is associated with cardiac conduction disorders.28 In the GI tract, miRNAs regulate ion channel densities and contribute to functional GI disorders.29 30 Since NaV1.5 current density is involved in GI smooth muscle function, where NaV1.5 inhibition leads to constipation and delayed transit, we hypothesised in this study that miRNAs might regulate NaV1.5 density in STC.
Methods
Human colon samples and RNA/miRNA extraction
The Mayo Clinic Institutional Review Board approved the use of human colonic tissue obtained as surgical waste. Colon muscularis externa was dissected from human colon tissue from patients with STC undergoing subtotal colectomy or control patients undergoing colon resections for colon cancer (table 1). Tissue was flash-frozen in liquid nitrogen and stored at −80°C until use. Total RNA, including the miRNA fraction, was extracted using miRNeasy mini kit (Qiagen) according to manufacturer instructions.
Table 1.
Demographic information of patients
| Patient | Sex | Age (years) |
| CTRL 1 | Female | 58 |
| CTRL 2 | Female | 48 |
| CTRL 3 | Female | 64 |
| CTRL 4 | Female | 49 |
| CTRL 5 | Female | 69 |
| CTRL 6 | Female | 43 |
| CTRL 7 | Female | 52 |
| CTRL 8 | Female | 54 |
| CTRL 9 | Female | 58 |
| CTRL 10 | Female | 62 |
| STC 1 | Female | 54 |
| STC 2 | Female | 51 |
| STC 3 | Female | 54 |
| STC 4 | Female | 61 |
| STC 5 | Female | 54 |
| STC 6 | Female | 50 |
| STC 7 | Female | 55 |
| STC 8 | Female | 60 |
| STC 9 | Female | 61 |
| STC 10 | Female | 54 |
| STC 11 | Female | 60 |
CTRL, control; STC, slow transit constipation.
Quantitative PCR (qPCR) arrays
Reverse transcription to prepare cDNA for mature miRNA profiling was carried out using the miScript II RT Kit (Qiagen) and 400 ng of total RNA according to the manufacturer instructions. The qPCR arrays used were the miScript miRNA PCR Array Human miFinder 384HC (MIHS-3001Z, Qiagen). These continuous data were analysed using Qiagen software and significance was assigned if p<0.05.
qRT-PCR
Reverse transcription reactions for mRNA of interest were conducted using the SuperScript VILO cDNA Synthesis Kit (Life Technologies). Reverse transcription reactions for miRNA of interest were done using the MicroRNA RT Kit and specific MicroRNA primers (TaqMan, ThermoFisher), according to specifications. qPCR was done on LightCycler 480 system with SYBRGreen I Master Mix (Roche Applied Science). Results were calculated as expression relative to the housekeeper (hypoxanthine phosphoribosyltransferase 1 (HPRT1)) using 2ˆ(-ΔCT), and statistics on these categorical data were done using Mann-Whitney with significance assigned if p<0.05.
Cell lines
Primary human smooth muscle cells (HuSMCs) were dissociated from the jejunum of a patient (female, 52) undergoing conversion to Roux-en-y gastric bypass surgery, and were immortalised by transformation with the SV40 large T antigen.31 HuSMCs were cultured in complete Clonetics SmGM−2 Smooth Muscle Growth Medium-2 (Lonza) containing 1% antimycotic-antibiotic (Gibco).
miRNA transfection
HuSMCs were transfected using LIPOFECTAMINE RNAiMax reagent (Invitrogen) and 50 nM of miRNA mimics (Thermo Fisher) in Opti-MEM according to the manufacturer instructions. The cells were incubated for 48 hours before RNA/miRNA extraction and electrophysiology experiments.
miRNA modulation of SCN5A expression
HEK293 cells were plated in six-well plates for 72 hours in minimum essential medium (MEM), supplemented with 10% fetal bovine serum and penicillin/streptomycin), then transfected by Lipofectamine 3000 (Invitrogen) with SCN5A full-length 3′ UTR (2259 bp) reporter vector (HmiT016601-MT05, Genecopoeia) and/or 50 nM of let-7f miRNA mimic (hsa-let7f-5p, Life Technologies) in a total volume of 2.4 mL per well. After 24 hours, Opti-MEM was replaced with fresh MEM. At 48 hours, conditioned MEM media was collected into 1.5 mL tubes, and Gaussia luciferase (GLuc) and secreted alkaline phosphatase (SEAP) activities were measured by Secrete-Pair Dual Luminescence Assay (Genecopoeia). Luminescence was read on a plate reader within 10 min at 1 s integration time. Normalised luminescence units (NLUs) were calculated by the equation: NLU = (GLucX – GLuc–UTR)/(SEAPX – SEAP–UTR), in which GLuc or SEAP are activities (arbitrary units) of luciferase or SEAP in conditioned media, –UTR is conditioned media from cells transfected without the SCN5A 3′ UTR reporter vector and X was conditioned media from cells transfected without (control) or with miRNA mimic (let-7f).
Protein extraction and western blotting
HuSMC were gently washed twice with ice-cold phosphate-buffered saline and collected by scraping the plates. Cells were pelleted at 4°C for 5 min and used for protein extraction and immunoblotting as previously described32 (see online supplementary methods).
gutjnl-2019-318747supp001.pdf (568.1KB, pdf)
Electrophysiology
Data were recorded on an Axopatch 200B amplifier with pClampfit 10 software. Whole-cell Na+ currents were elicited by stepping from the holding potential at −120 mV to 30 ms test pulses at −80 through +20 mV. Peak Na+ currents at the −20 mV step were normalised to whole cell capacitance. Solutions used are described in online supplementary methods.
Traction force microscopy
Traction analysis was conducted as previously described33 (see online supplementary methods).
Lentiviral transduction
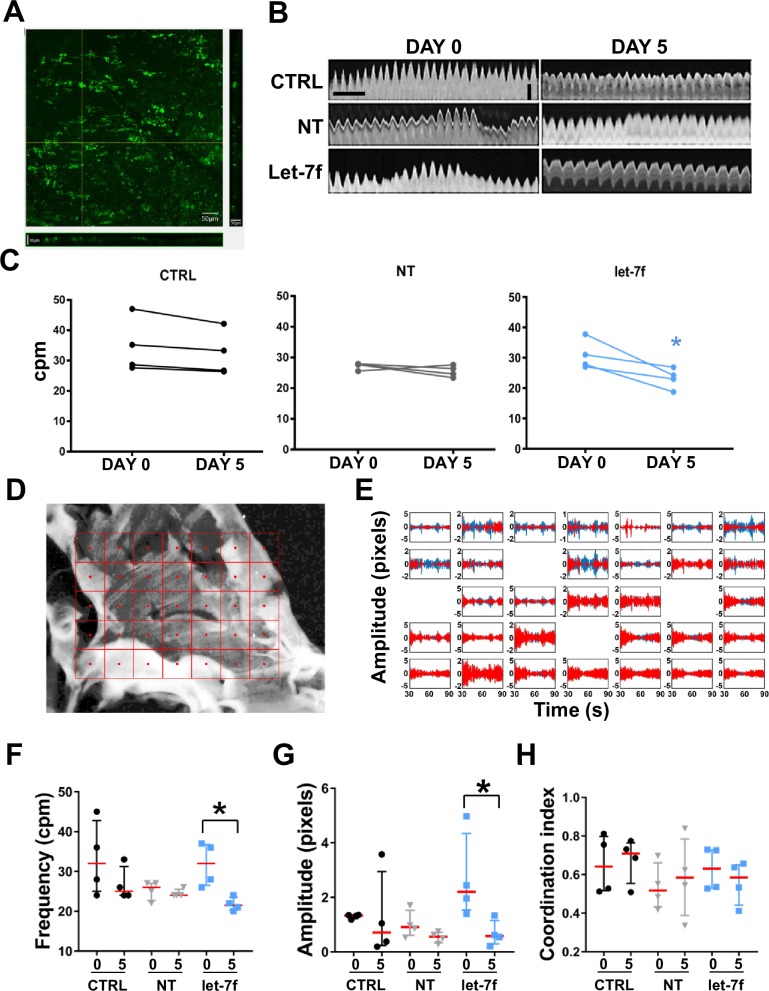
Muscularis externa was dissected from the jejunum of 6–8 weeks old Sprague Dawley rat as previously described.11 Tissues were transduced with non-targeting or let-7f lentiviral particles (Dharmacon) with a multiplicity of infection of 5. At day 0 and day 5, about 2 min long movies were recorded at a rate of 14 frames/second using a DP22 camera (Olympus) connected to a SZ61 stereomicroscope (Olympus) and used for contractility measurement and spatial–temporal mapping.
Contractility measurements
ImageJ software was used to import and convert images for contractility measurements as described in online supplementary methods.
Spatial–temporal mapping
Individual frames over 90 s were extracted as grey-scale images from each video and enhanced in MATLAB (2015a, MathWorks). Displacements, frequency and coordination were measured as detailed in online supplementary methods.
Results
Multiple miRNAs predicted to target SCN5A are overexpressed in STC
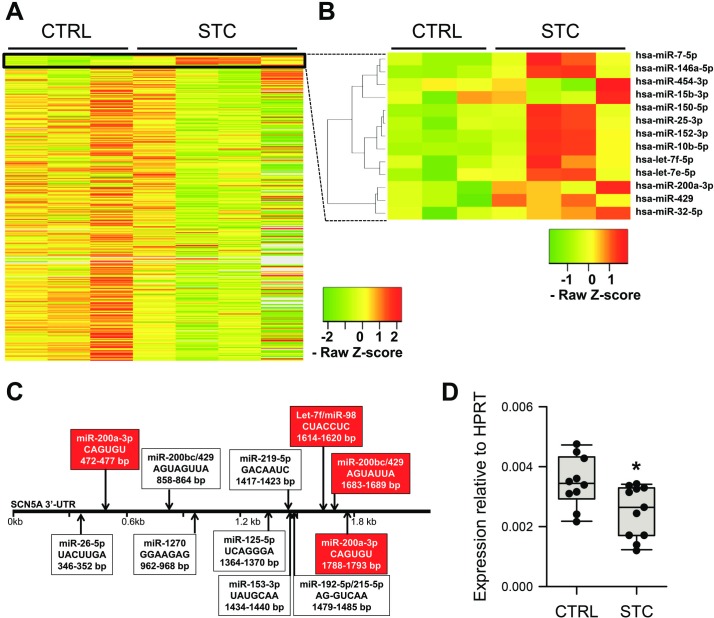
We examined 372 of the most common miRNAs in the miRBase database by a qPCR miRNA array to identify miRNAs for further investigation from four female patients and three age and sex-matched controls. We found that 13 of 372 miRNAs (3.5%) were threefold to eightfold upregulated in STC tissues (figure 1A; table 2). None of the examined miRNAs were significantly downregulated in STC. An unsupervised hierarchical cluster analysis based on the expression patterns of the 13 identified miRNAs using average linkage separated them into two major clusters (figure 1B). Interestingly, both clusters contained miRNAs that are known regulators of SCN5A, 34 the gene encoding for the mechanosensitive ion channel NaV1.5, a well-known player in multiple GI motility disorders.13 22 23 35 36 The first cluster, with 10 of the 13 identified miRNAs, included two members of the miR-98 family (let-7e and let-7f), and the second included two members of the miR-200 family (miR-200a-3p and miR-429). Those miRNAs are predicted or previously shown to modulate NaV1.5 expression by binding to the 3′UTR of the SCN5A mRNA (figure 1C). These results suggest that a specific miRNA set is enriched in STC, and >30% of these may target SCN5A.
Figure 1.
Thirteen of 372 miRNA examined are differentially expressed in slow transit constipation. (A) Heatmap plotting negative Ct values from the miRNA qPCR array organised by increasing p values. The box indicates the ones with p<0.05. The expression level of miRNAs is colour-coded as indicated in the legend. (B) Blowup of highlighted group of miRNAs, with hierarchical clustering analysis on the left. (C) Map of SCN5A mRNA 3′-UTR with binding sites for miRNAs obtained from multiple prediction algorithms (online supplementary table 1) and literature. In the boxes are position and sequence of target region complementary to each miRNA seed. In red are miRNAs identified by qPCR array screening. (D) SCN5A mRNA expression is reduced in STC smooth muscle. Data are median±IQR. n=10–11. *p<0.05, Mann-Whitney test. CTRL, control; miRNA, microRNA; qPCR, quantitative PCR; STC, slow transit constipation.
Table 2.
List of miRNA differentially expressed in slow transit constipation compared with controls
| miRNA | Fold change |
P value (t-test) |
| hsa-miR-429 | 8.28 | 0.005983 |
| hsa-miR-32-5 p | 6.79 | 0.040771 |
| hsa-miR-10b-5p | 6.71 | 0.040412 |
| hsa-let-7e-5p | 6.56 | 0.042515 |
| hsa-let-7f-5p | 6.46 | 0.046753 |
| hsa-miR-150-5 p | 6.34 | 0.046574 |
| hsa-miR-15b-3p | 6.26 | 0.048789 |
| hsa-miR-25-3 p | 6.23 | 0.042496 |
| hsa-miR-454-3 p | 4.97 | 0.047989 |
| hsa-miR-7-5 p | 4.76 | 0.03512 |
| hsa-miR-152-3 p | 4.62 | 0.044643 |
| hsa-miR-146a-5p | 4.57 | 0.026612 |
| hsa-miR-200a-3p | 3.25 | 0.020789 |
Results of the statistical analysis carried out on the data obtained by the miRNA qPCR array screening using the SABioscience analysis software (http://pcrdataanalysis.sabiosciences.com/mirna). miRNA predicted to bind to the 3′UTR of SCN5A is given in bold.
miRNA, microRNA; qPCR, quantitative PCR.
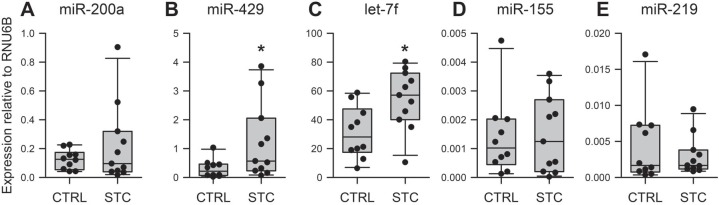
These results prompted us to examine if SCN5A expression is altered in STC. We expanded our cohort and collected tissue from seven additional patients with STC and seven age-matched controls. We found a robust decrease of SCN5A mRNA expression by about 35% in STC (figure 1D). We used this expanded cohort to validate the array results by quantitative reverse transcription PCR of individual miRNAs. We selected differentially expressed miRNAs that were predicted to target SCN5A: miR-429 and miR-200a-3p from one cluster and let-7f from the other. We found no difference in expression between the STC and control groups for miR-200a-3p, likely due to low expression of this miRNA in our samples (figure 2A), but we confirmed significant differential expression for let-7f and miR-429 (figure 2B, C). We also tested the expression of two negative controls. The first is miR-155, a miRNA that was not predicted to target the 3′-UTR of SCN5A and was not differentially expressed in our array screening but reported to be affected in some pathologies of mechanosensitivity.37 38 The second control was miR-219, a miRNA that targets SCN5A,34 but was not differentially expressed in the array screening. Neither miRNA was differentially expressed between the control and STC samples (figure 2D, E).
Figure 2.
The expression of let-7f miRNA is significantly increased in STC. (A) miR-200a is not differentially expressed between CTRL and STC. Expression of miR-429 (B) and Let-7f (C) is increased in STC. Expression of miR-155 (D) and miR-219 (E) is not altered in STC. Data are median±IQR. n=10–12, *p<0.05, Mann-Whitney test. CTRL, control; miRNA, microRNA; STC, slow transit constipation.
HuSMC line retains electrical excitability and a contractile phenotype
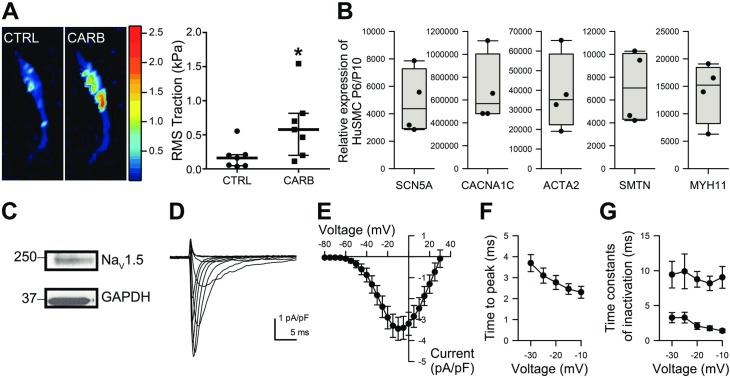
Smooth muscle cells (SMCs) frequently and rapidly lose their phenotype in primary cultures,39 and they are difficult to transfect. Therefore, we immortalised HuSMCs by using the SV40 large T antigen. We used several techniques to verify that HuSMC maintained a smooth muscle phenotype. First, by traction force microscopy, we quantified stimulus-driven tensile forces exerted by those cells in the presence of contractile stimuli. The traction forces generated by HuSMCs were significantly higher in the presence of carbachol, a typical SMC stimulant (figure 3A). We determined by PCR that HuSMCs were highly enriched in mRNA for relevant smooth muscle ion channels—NaV1.5-coding SCN5A and the L-type calcium channel α1C (CACNA1C), as well as the typical SMC markers such as smooth muscle actin (ACTA2) and myosin-11 (MYH11), and smoothelin (SMTN), compared with later passages (>11), which lose the smooth muscle phenotype and NaV1.5 currents (figure 3B). Western blot experiments showed that HuSMCs also express NaV1.5 protein (figure 3C). We used whole-cell voltage clamp to test HuSMC for NaV1.5 function directly and found robust NaV1.5 currents with density and kinetic properties comparable to freshly dissociated HuSMCs10–12 15 (figure 3D–G). These data suggested that HuSMC is a suitable SMC model to examine the effects of miRNAs’ regulation on NaV1.5 and SMC function.
Figure 3.
HuSMCs have characteristics similar to GI smooth muscle cells. (A) Representative images of cell tractions’ colour maps generated by the same HuSMC in basal conditions (left) and response to carbachol (right). The colours correspond to magnitudes of traction forces generated as indicated in the colour bar. The RMS traction forces generated are quantified in the graph. Data are median±IQR, n=7, *p<0.05, Mann-Whitney test. (B) HuSMCs (passage 6, P6) are highly enriched for mRNA for smooth muscle cell marker genes, including SCN5A and CACNA1C, compared with later passages (>10, P10), which lose smooth muscle phenotype (n=4). (C) HuSMCs express NaV1.5 protein by western blot. (D) Representative traces of Na+ currents recorded from HuSMCs. (E) I/V plot with Na+ currents recorded from HuSMCs. Time to peak (F) and time constants of inactivation (G) calculated from currents recorded at different voltages. ACTA2, smooth muscle actin; CACNA1C, L-type calcium channel α1C; CARB, carbachol; CTRL, control; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HuSMC, human smooth muscle cells; MYH11, myosin-11; pA, peak Na+ currents; pF, whole cell capacitance; RMS, root mean square; SCN5A, sodium channel NaV1.5; SMTN, smoothelin.
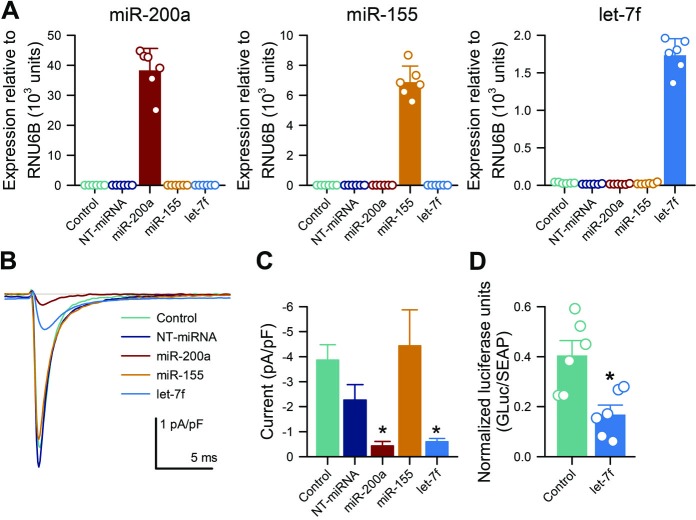
Let-7f overexpression in HuSMCs significantly reduced SCN5A expression, NaV1.5 current density and cellular contractility
We first used a luciferase reporter driven by SCN5A 3′UTR to determine if let-7f suppresses SCN5A expression. We found that 48 hours after transfection, let-7f miRNA mimics decreased normalised luciferase units by 55.9%±8.2% (0.169±0.037 NLU) in HEK293 cells cotransfected with let-7f miRNA mimic and SCN5A 3′ UTR, compared with cells transfected with the 3′ UTR vector alone (0.406±0.058 NLU; n=6; p=0.013, let-7f mimic to no-mimic controls by a two-tailed paired t-test; figure 4D). Since let-7f targets SCN5A and was enriched in STC, we next examined whether it had functional effects on HuSMCs. We transfected HuSMC with let-7f mimics. For positive control, we transfected miR-200a, and for negative control we used miR-155. After transient transfection with mimics, we detected a substantial increase in miRNA of interest (figure 4A). We then evaluated the effect of transfection of miRNAs on the function of NaV1.5 protein using electrophysiology. Whole-cell voltage-clamp showed that 48 hours after transfection of let-7f mimics HuSMC NaV1.5 currents were significantly decreased compared with untransfected HuSMC, while transfection with non-targeted (NT) miRNA had no effect (figure 4C, D). Similar effects were obtained with the transfection of HuSMC with the positive control miR-200a, while miR-155 did not affect HuSMC NaV1.5 currents (figure 4C, D). These data indicate that overexpression of let-7f led to decreased NaV1.5 current density.
Figure 4.
Overexpression of Let-7f significantly reduced Na+ current density in HuSMCs and resulted in changes in the cells’ properties. (A) RT-qPCR show efficient delivery of miRNA after transfection of mimics in HuSMCs (means±SDs, n=6). (B) Representative patch-clamp traces for HuSMC±miRNA mimics. (C) Peak Na+ current densities were reduced after transfection with let-7f (means±SEMs, n=5–17 cells, *p<0.05 by a one-way analysis of variance with Dunnett’s post-test). (D) NLUs of HEK-293 cells transfected with SCN5A 3′UTR vector alone (control) or cotransfected with the 3′UTR vector and miRNA mimic (let-7f) (means±SEMs, n=6 transfections, *p<0.05 to same-plate controls by a two-tailed paired t-test). GLuc, Gaussia luciferase; HuSMC, human smooth muscle cells; miRNA, microRNA; NLU, normalised luciferase unit; NT, non-targeted; pA, peak Na+ currents; pF, whole cell capacitance; SEAP, secreted alkaline phosphatase.
Overexpression of let-7f in rat intestinal organotypic cultures results in altered smooth muscle contractility
We previously established that the rat,11 but not mouse,12 GI smooth muscle expresses SCN5A and has functional NaV1.5, which makes it a relevant model to study of the role NaV1.5 in GI physiology and pathophysiology.11 Sequence pile-up using the Clustal Omega software showed that the sequence of let-7f miRNA is 98.9% conserved between rats and humans (online supplementary figure 1A). TargetScan analysis also showed that the predicted target site for let-7f is conserved in the rat SCN5A 3′UTR (online supplementary figure 1B). Therefore, we chose to use rat intestinal organotypic cultures we previously developed.11 To study the effects of let-7f overexpression on GI smooth muscle contractility in this system, we optimised conditions for lentiviral transduction and long-term contractility measurements. Five days after transduction, the lentiviral-mediated overexpression of let-7f resulted in abundant expression of green fluorescent protein (GFP)-tagged let-7f throughout the smooth muscle layer (figure 5A), while no signal was observed in control tissues or tissues treated with non-targeting particles. We also measured rhythmic contractions of rat intestinal smooth muscle strips in the presence or absence of lentiviral-mediated overexpression of let-7f by measuring the frequency of contraction of smooth muscle strips at day 0 and day 5 in culture in the same tissue (figure 5B). Control tissues showed no significant differences in frequency of contractility between the two time points, as did the tissue treated with non-target lentiviral particles. However, tissues treated with let-7f lentiviral particles exhibited a reduction in contractility of about 30% after 5 days (figure 5C). We used spatial–temporal mapping analysis of the recorded movies to detect the displacements of tissue features across successive frames based on an overlaid grid (figure 5D). The frequency and displacement of contractions were detected from all the fields of view containing regions of interest (figure 5E). The contraction patterns were reduced by about 35% over time in tissues transduced with let-7f lentiviral particles, while no differences were observed in controls and tissues treated with non-targeting particles (figure 5F; table 3). These findings are consistent with our previous analysis, and the frequencies measured using the two methods are not significantly different between the methods (using two-way analysis of variance with Sidak’s multiple comparison test). We also analysed the amplitude of contraction, measured as pixel displacement. The amplitudes were not significantly different in controls and in the NT group, while they decreased by 78% between day 0 and day 5 for the let-7f group (figure 5G; table 3). We also measured a coordination index of the tissues, as a measure of the ratio of vectors pointing in the same direction. Interestingly, coordination showed no difference in any of the group examined (figure 5H; table 3), suggesting that the overall direction of contraction was unchanged, even if the frequency and amplitude were affected by the overexpression of let-7f. These data suggest that the function of ICC was not altered to the extent that coordination was affected.40 41 Our results suggest that let-7f has a direct negative effect on regulating smooth muscle contractility.
Figure 5.
Overexpression of Let-7f in rat small intestinal organotypic cultures results in altered smooth muscle contractility. (A) Representative images from transduced tissue showing expression of let-7f-GFP throughout its full depth. Scale bar sizes are indicated in the figure. (B) Representative contraction patterns using ImageJ. Horizontal scale bar=2.5 s, vertical scale bar=35 pixels. (C) Frequency of contraction from ImageJ analyses reported as cpm, *p<0.05, n=4, paired t-test. (D) Representative grid overlay with tracked ROIs and the centroid used for calculations. (E) Displacements in the horizontal (x, blue) and vertical (y, red) directions for each ROI in the representative tissue. (F) Frequency of contraction from spatial–temporal mapping reported as cpm. (G) Amplitude of contraction calculated as displacement of pixels. (H) Coordination metric measured as ratio of vectors pointing in the same direction. For F, G and H, data are median±IQR, *p<0.05, n=4, one-way analysis of variance with Dunn’s multiple comparison test. cpm, contraction per minute; CTRL, control; ROI, region of interest; NT, non-targeted.
Table 3.
Summary of the spatial–temporal mapping analysis
| Day 0 CTRL | Day 5 CTRL | Day 0 NT | Day 5 NT | Day 0 let-7f | Day 5 let-7f | |
| Frequency (cpm) | 32±25, 42 | 25±24, 31 | 26±22, 25 | 24±24, 25 | 32±26, 36 | 21±20, 23* |
| Amplitude (pixel) | 1.3±1.2,1.3 | 0.7±0.2, 2.9 | 0.9±0.6,1.5 | 0.6±0.3, 0.7 | 2.2±1.5, 4.3 | 0.5±0.3, 1.1* |
| Coordination | 0.64±0.5, 0.7 | 0.70±0.5, 0.7 | 0.51±0.4, 0.6 | 0.58±0.3, 0.7 | 0.63±0.5, 0.7 | 0.58±0.4, 0.6 |
*p<0.05, n=4, one-way analysis of variance with Dunn’s multiple comparisons test.
cpm, contraction per minute; CTRL, control; NT, non-targeted.
Discussion
The goal of this study was to improve our understanding of STC. We focused on miRNAs because they control GI and vascular smooth muscle phenotype and function.42–44 Previous studies in mice that entirely removed the ability of SMCs to generate miRNAs showed a dramatic smooth muscle dysfunction, hallmarked by decreases in vascular smooth muscle contractility44 and bowel hypomotility and dilation.42 However, complete miRNA knockout made the clinical relevance of these studies unclear. In this study, we found in human STC colonic smooth muscle a surprisingly small subset of dysregulated miRNAs, suggesting functional roles of miRNAs in human GI smooth muscle contractility and dysmotility.
We pursued the mechanism of miRNA regulation of smooth muscle function. We focused on miRNA roles in regulating the availability of mRNA for ion channels, since ion channels are crucial for SMC function, and miRNAs are highly effective regulators of electrical excitability and excitation–contraction coupling.45 46 For example, miRNAs regulated visceral sensation47 and cardiac motility48 by controlling ion channel mRNA density. In our miRNA set, we found that 30% of the miRNAs we identified were predicted to target SCN5A, which codes for a mechanosensitive voltage-gated Na+ channel NaV1.5 that is associated with motility disorders characterised by compromised contractile properties of smooth muscle.13 22 23 35 36 Since miRNAs have hundreds of targets,49 genomic analyses are intrinsically limited,50 and functional assessment was required. We focused our investigation on let-7f for three reasons. First, it is a part of the miR-98 family, which was recently shown to be upregulated in STC in a separate study.51 Second, it was abundantly expressed in our samples. Third, it has been suggested that other members of the miR-98 family regulate SCN5A.34 We found that let-7f overexpression led to a decrease in SCN5A expression, NaV1.5 current density and electrical excitability in human SMC.
Given that decreased NaV1.5 is associated with a decrease in excitability and contractility in the human colon13 and rat model,11 we used rat organotypic cultures with lentiviral transduction of let-7f. We found that let-7f decreased smooth muscle contractility through a decrease in the amplitude and frequency of contractions, leaving the coordination index unchanged, similar to a myopathic process on manometry. While let-7f may have several cellular targets, our results suggest that let-7f may regulate smooth muscle function potentially via regulation of NaV1.5 in SMCs. In addition to let-7f, we found three miRNAs that target SCN5A mRNA by seed sequences that are spread through the 3′UTR, suggesting that their effects may be additive. In all, our novel findings have broad implications in other excitable systems where NaV1.5 plays vital roles, such as cardiac,52 vascular smooth muscle44 and sensory neurons,17 53 but also non-excitable cells54 and even cancer.55 56
There are intriguing mechanistic possibilities regarding how lower NaV1.5 density decreases GI smooth muscle contractility, including via NaV1.5 function as an ion channel or via NaV1.5 role as a central component of the macromolecular complex that participates in myocyte cellular structure.57 Interestingly, SCN5A mutations are not only linked to dilated cardiomyopathy, a disease characterised by impaired cardiac contractility, but also SCN5A is one of only few ion channel genes involved.19 58 SCN5A channelopathies may contribute to hypomotility in both smooth and cardiac muscles via decreased myocyte excitability.59 60 In GI smooth muscle, NaV1.5 contributes to the initial, fast-rising depolarisation of the slow wave in SMC, and it is important for the regulation of amplitude and duration of the slow wave,10 11 key parameters for the activation of SMC L-type Ca2+ channels. It is possible that the let-7f-driven decrease in NaV1.5 current density may cause decreased SMC and cardiomyocyte contractility via an imbalance in the function of Na+/K+ pump or Na+/Ca2+ exchanger, like in vascular smooth muscle,61 ultimately leading to decreased [Ca2+]i. Another possibility is via NaV1.5 role as the central component of a macromolecular complex that contributes to myocyte structure by interaction with structural genes, like syntrophin and telethonin, both in the heart62 and GI smooth muscle.59 60 Consequently, reduced expression of NaV1.5 may remove a critical component that supports the cytoskeleton and intercalated discs in the cardiac cell63 or the cytoskeleton and the plasma membrane in SMC,16 leading to cell structure alterations and impaired contractility. In all, while NaV1.5 role in dysmotility is established, future studies need to determine its roles in disease.
This study also provides important clues regarding GI SMC biology. Let-7f and other STC-associated miRNAs target GI SMC genes in addition to SCN5A. Even when mature and fully differentiated, SMCs retain remarkable plasticity. SMCs can lose the contractile function in favour of proliferating capability in response to environmental signals.64 An imbalance between the two states can lead to disease.64 Indeed, altered expression of let-7g, another member of the mir-98 family, plays a key role in the switch from contractile to proliferative phenotypes in vascular SMC.65 In GI smooth muscle, miRNAs have been shown to work in conjunction with serum response factor, to drive a switch in smooth muscle phenotype in the opposite direction from proliferative to contractile.66 In a subset of myopathic chronic intestinal pseudo-obstruction, impaired GI motility may be a consequence of loss of contractile SMCs and reversion to a more immature and proliferative state.67 Our STC patient cohort was carefully selected, but we were not able to precisely control the location of the tissue we received for analysis. Therefore, our cohort is likely a subset of STC patients and tissues. Further investigation is required to generalise our findings and to establish the GI SMC relationships between proliferation and differentiation pathways in health and STC. These questions may be illuminated by further work in humans, especially as full-thickness biopsies for GI motility disorders come of age.68
The causes of miRNA dysregulation in STC are not known, and we can only speculate. An intriguing possibility is through diet, which plays a crucial role in altering the expression of miRNAs in the GI tract, with physiological consequences on intestinal transit.69 The mechanism may involve microbial-derived metabolites such as short-chain fatty acid and bile acids that modulate gut motility70 and indirectly alter miRNA expression. It is also possible that microbiome either dependently or independently of diet71 may directly alter miRNAs expression as recently proposed for some GI diseases.72
In summary, we found in the colonic smooth muscle of patients with STC an increased expression of a small subset of miRNAs. A subset of these target NaV1.5, leading to decreased HuSMC NaV1.5 current density, and GI smooth muscle contractility. Thus, our observations suggest a role for miRNAs in regulation of SMC function and the pathogenesis and pathophysiology of STC.
gutjnl-2019-318747supp002.xlsx (107.4KB, xlsx)
Acknowledgments
The authors thank Kristy Zodrow for administrative assistance and Denika S Kerska for the help with the analyses on the frequency of contraction. The authors also thank the funding bodies for their support in completing this study.
Footnotes
Twitter: @beyderlab
Contributors: AM designed and performed experiments, analysed data and wrote the manuscript. PRS, VJ, YH, MER, CA and AJH performed experiments and analysed data; TO, PJM, SC, DT, RG, SJG and PD designed experiments and analysed data; CEB coordinated identification and retrieval of surgical tissue; RRC, DWL and HKC performed surgeries and provided tissues; AB and GF designed the research, analysed data and wrote the manuscript. All authors critically reviewed and approved the manuscript.
Funding: This study was funded by National Institute of Diabetes and Digestive and Kidney Diseases NIH R01 (DK52766) to GF and by NIH K08 (DK106456), American Gastroenterological Association Research Scholar Award (AGA RSA) to AB and The Rutherford Discovery Fellowship by The Rutherford Foundation, Royal of Society of New Zealand to PD.
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as supplementary information.
References
- 1. De Marco P, Militello G, Tutino R, et al. The management of the slow transit constipation in the laparoscopic era. G Chir 2018;34:297–302. [PubMed] [Google Scholar]
- 2. Knowles CH, Grossi U, Horrocks EJ, et al. Surgery for constipation: systematic review and clinical guidance: Paper 1: Introduction & Methods. Colorectal Dis 2017;19:5–16. 10.1111/codi.13774 [DOI] [PubMed] [Google Scholar]
- 3. Bassotti G, Villanacci V. Slow transit constipation: a functional disorder becomes an enteric neuropathy. World J Gastroenterol 2006;12:4609–13. 10.3748/wjg.v12.i29.4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. He CL, Burgart L, Wang L, et al. Decreased interstitial cell of Cajal volume in patients with slow-transit constipation. Gastroenterology 2000;118:14–21. 10.1016/s0016-5085(00)70409-4 [DOI] [PubMed] [Google Scholar]
- 5. Lyford GL, He C-L, Soffer E, et al. Pan-colonic decrease in interstitial cells of Cajal in patients with slow transit constipation. Gut 2002;51:496–501. 10.1136/gut.51.4.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tillou J, Poylin V. Functional disorders: slow-transit constipation. Clin Colon Rectal Surg 2017;30:076–86. 10.1055/s-0036-1593436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beyder A, Farrugia G. Targeting ion channels for the treatment of gastrointestinal motility disorders. Therap Adv Gastroenterol 2012;5:5–21. 10.1177/1756283X11415892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kass RS. The channelopathies: novel insights into molecular and genetic mechanisms of human disease. J Clin Invest 2005;115:1986–9. 10.1172/JCI26011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Strege PR, Ou Y, Sha L, et al. Sodium current in human intestinal interstitial cells of Cajal. Am J Physiol Gastrointest Liver Physiol 2003;285:G1111–21. 10.1152/ajpgi.00152.2003 [DOI] [PubMed] [Google Scholar]
- 10. Holm AN, Rich A, Miller SM, et al. Sodium current in human jejunal circular smooth muscle cells. Gastroenterology 2002;122:178–87. 10.1053/gast.2002.30346 [DOI] [PubMed] [Google Scholar]
- 11. Beyder A, Gibbons SJ, Mazzone A, et al. Expression and function of the Scn5a-encoded voltage-gated sodium channel Nav 1.5 in the rat jejunum. Neurogastroenterol Motil 2016;28:64–73. 10.1111/nmo.12697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Strege PR, Mazzone A, Kraichely RE, et al. Species dependent expression of intestinal smooth muscle mechanosensitive sodium channels. Neurogastroenterol Motil 2007;19:135–43. 10.1111/j.1365-2982.2006.00844.x [DOI] [PubMed] [Google Scholar]
- 13. Neshatian L, Strege PR, Rhee P-L, et al. Ranolazine inhibits voltage-gated mechanosensitive sodium channels in human colon circular smooth muscle cells. Am J Physiol Gastrointest Liver Physiol 2015;309:G506–12. 10.1152/ajpgi.00051.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gellens ME, George AL, Chen LQ, et al. Primary structure and functional expression of the human cardiac tetrodotoxin-insensitive voltage-dependent sodium channel. Proc Natl Acad Sci U S A 1992;89:554–8. 10.1073/pnas.89.2.554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ou Y, Gibbons SJ, Miller SM, et al. Scn5A is expressed in human jejunal circular smooth muscle cells. Neurogastroenterol Motil 2002;14:477–86. 10.1046/j.1365-2982.2002.00348.x [DOI] [PubMed] [Google Scholar]
- 16. Strege PR, Holm AN, Rich A, et al. Cytoskeletal modulation of sodium current in human jejunal circular smooth muscle cells. Am J Physiol Cell Physiol 2003;284:C60–6. 10.1152/ajpcell.00532.2001 [DOI] [PubMed] [Google Scholar]
- 17. Osorio N, Korogod S, Delmas P. Specialized functions of Nav1.5 and Nav1.9 channels in electrogenesis of myenteric neurons in intact mouse ganglia. J Neurosci 2014;34:5233–44. 10.1523/JNEUROSCI.0057-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ackerman MJ. The long QT syndrome: ion channel diseases of the heart. Mayo Clin Proc 1998;73:250–69. 10.4065/73.3.250 [DOI] [PubMed] [Google Scholar]
- 19. McNair WP, Sinagra G, Taylor MRG, et al. Scn5A mutations associate with arrhythmic dilated cardiomyopathy and commonly localize to the voltage-sensing mechanism. J Am Coll Cardiol 2011;57:2160–8. 10.1016/j.jacc.2010.09.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koenig X, Dysek S, Kimbacher S, et al. Voltage-Gated ion channel dysfunction precedes cardiomyopathy development in the dystrophic heart. PLoS One 2011;6:e20300 10.1371/journal.pone.0020300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hesse M, Kondo CS, Clark RB, et al. Dilated cardiomyopathy is associated with reduced expression of the cardiac sodium channel SCN5A. Cardiovasc Res 2007;75:498–509. 10.1016/j.cardiores.2007.04.009 [DOI] [PubMed] [Google Scholar]
- 22. Locke GR, Ackerman MJ, Zinsmeister AR, et al. Gastrointestinal symptoms in families of patients with an SCN5A-encoded cardiac channelopathy: evidence of an intestinal channelopathy. Am J Gastroenterol 2006;101:1299–304. 10.1111/j.1572-0241.2006.00507.x [DOI] [PubMed] [Google Scholar]
- 23. Beyder A, Mazzone A, Strege PR, et al. Loss-Of-Function of the voltage-gated sodium channel Nav1.5 (channelopathies) in patients with irritable bowel syndrome. Gastroenterology 2014;146:1659–68. 10.1053/j.gastro.2014.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nash DT, Nash SD. Ranolazine for chronic stable angina. Lancet 2008;372:1335–41. 10.1016/S0140-6736(08)61554-8 [DOI] [PubMed] [Google Scholar]
- 25. Ashcroft FM. Ion channels and disease. Elsevier, 2000. [Google Scholar]
- 26. Bennett PB, Yazawa K, Makita N, et al. Molecular mechanism for an inherited cardiac arrhythmia. Nature 1995;376:683–5. 10.1038/376683a0 [DOI] [PubMed] [Google Scholar]
- 27. Bartel DP. Micrornas: target recognition and regulatory functions. Cell 2009;136:215–33. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim GH. Microrna regulation of cardiac conduction and arrhythmias. Transl Res 2013;161:381–92. 10.1016/j.trsl.2012.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Q, Costinean S, Croce CM, et al. Microrna 29 targets nuclear factor-κB-repressing factor and claudin 1 to increase intestinal permeability. Gastroenterology 2015;148:158–69. 10.1053/j.gastro.2014.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou Q, Yang L, Larson S, et al. Decreased miR-199 augments visceral pain in patients with IBS through translational upregulation of TRPV1. Gut 2016;65:797–805. 10.1136/gutjnl-2013-306464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ahuja D, Sáenz-Robles MT, Pipas JM. Sv40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 2005;24:7729–45. 10.1038/sj.onc.1209046 [DOI] [PubMed] [Google Scholar]
- 32. Koval OM, Snyder JS, Wolf RM, et al. Ca2+/Calmodulin-Dependent protein kinase II-based regulation of voltage-gated Na+ channel in cardiac disease. Circulation 2012;126:2084–94. 10.1161/CIRCULATIONAHA.112.105320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marinković A, Mih JD, Park J-A, et al. Improved throughput traction microscopy reveals pivotal role for matrix stiffness in fibroblast contractility and TGF-β responsiveness. Am J Physiol Lung Cell Mol Physiol 2012;303:L169–80. 10.1152/ajplung.00108.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Daimi H, Lozano-Velasco E, Haj Khelil A, et al. Regulation of SCN5A by microRNAs: miR-219 modulates SCN5A transcript expression and the effects of flecainide intoxication in mice. Heart Rhythm 2015;12:1333–42. 10.1016/j.hrthm.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 35. Saito YA, Strege PR, Tester DJ, et al. Sodium channel mutation in irritable bowel syndrome: evidence for an ion channelopathy. Am J Physiol Gastrointest Liver Physiol 2009;296:G211–8. 10.1152/ajpgi.90571.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Strege PR, Mazzone A, Bernard CE, et al. Irritable bowel syndrome patients have SCN5A channelopathies that lead to decreased NaV1.5 current and mechanosensitivity. Am J Physiol Gastrointest Liver Physiol 2018;314:G494–503. 10.1152/ajpgi.00016.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weber M, Kim S, Patterson N, et al. MiRNA-155 targets myosin light chain kinase and modulates actin cytoskeleton organization in endothelial cells. Am J Physiol Heart Circ Physiol 2014;306:H1192–203. 10.1152/ajpheart.00521.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seok HY, Chen J, Kataoka M, et al. Loss of microRNA-155 protects the heart from pathological cardiac hypertrophy. Circ Res 2014;114:1585–95. 10.1161/CIRCRESAHA.114.303784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Campbell JH, Campbell GR. Smooth muscle phenotypic modulation--a personal experience. Arterioscler Thromb Vasc Biol 2012;32:1784–9. 10.1161/ATVBAHA.111.243212 [DOI] [PubMed] [Google Scholar]
- 40. Singh RD, Gibbons SJ, Saravanaperumal SA, et al. Ano1, a Ca2+-activated Cl- channel, coordinates contractility in mouse intestine by Ca2+ transient coordination between interstitial cells of Cajal. J Physiol 2014;592:4051–68. 10.1113/jphysiol.2014.277152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Malysz J, Gibbons SJ, Saravanaperumal SA, et al. Conditional genetic deletion of Ano1 in interstitial cells of Cajal impairs Ca2+ transients and slow waves in adult mouse small intestine. Am J Physiol Gastrointest Liver Physiol 2017;312:G228–45. 10.1152/ajpgi.00363.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park C, Yan W, Ward SM, et al. Micrornas dynamically remodel gastrointestinal smooth muscle cells. PLoS One 2011;6:e18628 10.1371/journal.pone.0018628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krishna CV, Singh J, Thangavel C, et al. Role of microRNAs in gastrointestinal smooth muscle fibrosis and dysfunction: novel molecular perspectives on the pathophysiology and therapeutic targeting. Am J Physiol Gastrointest Liver Physiol 2016;310:G449–59. 10.1152/ajpgi.00445.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Albinsson S, Skoura A, Yu J, et al. Smooth muscle miRNAs are critical for post-natal regulation of blood pressure and vascular function. PLoS One 2011;6:e18869 10.1371/journal.pone.0018869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luo X, Zhang H, Xiao J, et al. Regulation of human cardiac ion channel genes by microRNAs: theoretical perspective and pathophysiological implications. Cell Physiol Biochem 2010;25:571–86. 10.1159/000315076 [DOI] [PubMed] [Google Scholar]
- 46. Wang Z. The role of microRNA in cardiac excitability. J Cardiovasc Pharmacol 2010;56:460–70. 10.1097/FJC.0b013e3181edb22c [DOI] [PubMed] [Google Scholar]
- 47. Zhou Q, Verne GN. Role of microRNA in chronic visceral nociception. Pain 2013;154:9–10. 10.1016/j.pain.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang K-C, Nerbonne JM. Mechanisms contributing to myocardial potassium channel diversity, regulation and remodeling. Trends Cardiovasc Med 2016;26:209–18. 10.1016/j.tcm.2015.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 50. Seitz H. Redefining microRNA targets. Curr Biol 2009;19:870–3. 10.1016/j.cub.2009.03.059 [DOI] [PubMed] [Google Scholar]
- 51. Zhao S, Chen Q, Kang X, et al. Aberrantly expressed genes and miRNAs in slow transit constipation based on RNA-seq analysis. Biomed Res Int 2018;2018:1–14. 10.1155/2018/2617432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Veerman CC, Wilde AAM, Lodder EM. The cardiac sodium channel gene SCN5A and its gene product Nav1.5: role in physiology and pathophysiology. Gene 2015;573:177–87. 10.1016/j.gene.2015.08.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Frenz CT, Hansen A, Dupuis ND, et al. Nav1.5 sodium channel window currents contribute to spontaneous firing in olfactory sensory neurons. J Neurophysiol 2014;112:1091–104. 10.1152/jn.00154.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Black JA, Waxman SG. Noncanonical roles of voltage-gated sodium channels. Neuron 2013;80:280–91. 10.1016/j.neuron.2013.09.012 [DOI] [PubMed] [Google Scholar]
- 55. House CD, Vaske CJ, Schwartz AM, et al. Voltage-Gated Na+ channel SCN5A is a key regulator of a gene transcriptional network that controls colon cancer invasion. Cancer Res 2010;70:6957–67. 10.1158/0008-5472.CAN-10-1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Baptista-Hon DT, Robertson FM, Robertson GB, et al. Potent inhibition by ropivacaine of metastatic colon cancer SW620 cell invasion and Nav1.5 channel function. Br J Anaesth 2014;113:i39–48. 10.1093/bja/aeu104 [DOI] [PubMed] [Google Scholar]
- 57. Abriel H. Cardiac sodium channel Na(v)1.5 and interacting proteins: Physiology and pathophysiology. J Mol Cell Cardiol 2010;48:2–11. 10.1016/j.yjmcc.2009.08.025 [DOI] [PubMed] [Google Scholar]
- 58. Olson TM, Michels VV, Ballew JD, et al. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA 2005;293:447–54. 10.1001/jama.293.4.447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ou Y, Strege P, Miller SM, et al. Syntrophin gamma 2 regulates SCN5A gating by a PDZ domain-mediated interaction. J Biol Chem 2003;278:1915–23. 10.1074/jbc.M209938200 [DOI] [PubMed] [Google Scholar]
- 60. Mazzone A, Strege PR, Tester DJ, et al. A mutation in telethonin alters Nav1.5 function. J Biol Chem 2008;283:16537–44. 10.1074/jbc.M801744200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nakamura Y, Ohya Y, Abe I, et al. Sodium-Potassium pump current in smooth muscle cells from mesenteric resistance arteries of the guinea-pig. J Physiol 1999;519 Pt 1:203–12. 10.1111/j.1469-7793.1999.0203o.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rook MB, Evers MM, Vos MA, et al. Biology of cardiac sodium channel Nav1.5 expression. Cardiovasc Res 2012;93:12–23. 10.1093/cvr/cvr252 [DOI] [PubMed] [Google Scholar]
- 63. Abriel H, Rougier J-S, Jalife J. Ion channel macromolecular complexes in cardiomyocytes: roles in sudden cardiac death. Circ Res 2015;116:1971–88. 10.1161/CIRCRESAHA.116.305017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev 2004;84:767–801. 10.1152/physrev.00041.2003 [DOI] [PubMed] [Google Scholar]
- 65. Wang T-M, Chen K-C, Hsu P-Y, et al. microRNA let-7g suppresses PDGF-induced conversion of vascular smooth muscle cell into the synthetic phenotype. J Cell Mol Med 2017;21:3592–601. 10.1111/jcmm.13269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Park C, Hennig GW, Sanders KM, et al. Serum response factor-dependent microRNAs regulate gastrointestinal smooth muscle cell phenotypes. Gastroenterology 2011;141:164–75. 10.1053/j.gastro.2011.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yamaji M, Mahmoud M, Evans IM, et al. Neuropilin 1 is essential for gastrointestinal smooth muscle contractility and motility in aged mice. PLoS One 2015;10:e0115563 10.1371/journal.pone.0115563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rajan E, Al-Bawardy B, Gostout CJ, et al. Endoscopic muscle biopsy sampling of the duodenum and rectum: a pilot survival study in a porcine model to detect myenteric neurons. Gastrointest Endosc 2018;87:600–6. 10.1016/j.gie.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nezami BG, Mwangi SM, Lee JE, et al. Microrna 375 mediates palmitate-induced enteric neuronal damage and high-fat diet-induced delayed intestinal transit in mice. Gastroenterology 2014;146:473–83. 10.1053/j.gastro.2013.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ge X, Zhao W, Ding C, et al. Potential role of fecal microbiota from patients with slow transit constipation in the regulation of gastrointestinal motility. Sci Rep 2017;7:441 10.1038/s41598-017-00612-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kashyap PC, Marcobal A, Ursell LK, et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology 2013;144:967–77. 10.1053/j.gastro.2013.01.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Feng Q, Chen W-D, Wang Y-D. Gut microbiota: an integral Moderator in health and disease. Front Microbiol 2018;9:151 10.3389/fmicb.2018.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2019-318747supp001.pdf (568.1KB, pdf)
gutjnl-2019-318747supp002.xlsx (107.4KB, xlsx)