Abstract
Medico-scientific concepts are not easily understood by laypeople that frequently use lay synonyms. For this reason, strategies that help users formulate health queries are essential. Health Suggestions is an existing extension for Google Chrome that provides suggestions in lay and medico-scientific terminologies, both in English and Portuguese. This work proposes, evaluates, and compares further strategies for generating suggestions based on the initial consumer query, using multi-concept recognition and the Unified Medical Language System (UMLS). The evaluation was done with an English and a Portuguese test collection, considering as baseline the suggestions initially provided by Health Suggestions. Given the importance of understandability, we used measures that combine relevance and understandability, namely, uRBP and uRBPgr. Our best method merges the Consumer Health Vocabulary (CHV)-preferred expression for each concept identified in the initial query for lay suggestions and the UMLS-preferred expressions for medico-scientific suggestions. Multi-concept recognition was critical for this improvement.
Keywords: Health information retrieval, Cross-language information retrieval, Cross-terminology information retrieval, Query suggestion
Introduction
Search engines are commonly used to seek health information, an activity that is considered the third most popular activity on the Internet [1]. Despite the increasing use of the Web to search for health-related information, there may exist inequalities in access to health information [6]. Users with low levels of health literacy can struggle to satisfy their information needs because health-related information usually contains medico-scientific expressions that are not easily understandable [13]. The gap between lay and medico-scientific terminologies limits this access and can be assisted through query modification techniques [10]. There is evidence that multilingual query suggestions in lay and medico-scientific terminologies improve health information retrieval by laypeople [9].
Taking this into account, Health Suggestions was developed as an extension for Google Chrome, suggesting queries in lay and medico-scientific terminologies, both in English and Portuguese, based on the Consumer Health Vocabulary (CHV) [8]. To improve the system, we propose and evaluate strategies for query suggestion that involve multi-concept recognition and information from the Unified Medical Language System (UMLS). For evaluation, the new generated query is used to retrieve documents from an English and a Portuguese test collection. The strategies are evaluated, taking into account the relevance of the documents and its understandability by lay users, comparing them with the results of queries initially suggested by Health Suggestions.
Related Work
When users are trying to express their information need, they might use keywords that are too general or different from the ones included in documents, as well as an insufficient number of terms, making the query difficult to “be understood” by the system [5]. Techniques such as query expansion, query refinement, and query suggestion have been proposed to solve this problem, improving the relevance and comprehension of the retrieved documents.
Zeng et al. [12] developed a system that suggests alternative or additional terms to the query using logs and the co-occurrence of concepts in medical documents, as well as the semantic relationships existing in medical vocabularies. Liu and Wesley [7] proposed a query expansion method that exploited the UMLS, appending additional relevant terms to the original query.
A query suggestion system was developed by Lopes and Ribeiro [9], combining multilingual alternatives (in Portuguese and English) with the use of lay and medico-scientific terminology. Authors used the CHV that maps technical terms to consumer-friendly language. For each query, they identify the associated concept and then return its CHV and UMLS-preferred names in English and Portuguese. Lopes and Fernandes [8] created HealthSuggestions, an extension for Google Chrome to assist users in obtaining high-quality search results in the health domain using the CHV.
Proposed Methods for Suggesting Queries
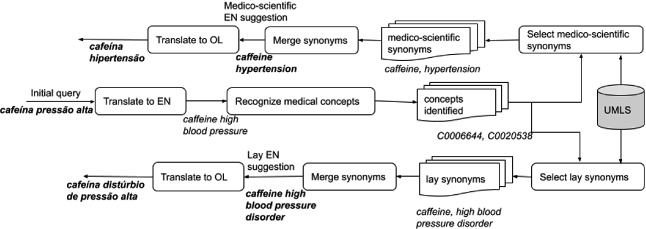
To generate the query suggestions, we implemented several methods that use multi-concept recognition to detect the medical concepts included in the initial query and use the information from UMLS as a knowledge source. All methods follow the approach described in Fig. 1. Briefly, the initial query is translated into English, and its medical concepts are identified. For each of these concepts, we select lay and medico-scientific expressions, concatenate them to compose the corresponding suggestions in English and, in the end, we translate them to the original language. All translations are done with Google Translator.
Fig. 1.
Process followed for the generation of query suggestions.
Several strategies were analyzed for multi-concept recognition, and we decided to use MetaMap, a rule-based system of concept recognition, to discover UMLS concepts referred to in free text [2], which is interesting because we use UMLS as our knowledge source. MetaMap provides a list of mappings for each identified concept. In each query suggestion method, we used two approaches to select the best mapping. In the first approach, we choose the first mapping, that is, the one with the highest score. In the second, we used the Word-Sense Disambiguation (WSD) feature that favors those that are semantically consistent with the surrounding text [3]. For each approach, we used the UMLS Concept Unique Identifier (CUI) and the name of the concepts as input.
The selection of lay and medico-scientific synonyms is what differentiates the suggestion methods. All the methods use the UMLS, a knowledge base that aggregates multiple thesauri of the medical domain [4], each composed of concepts related to health, their various names, and the relationships that exist between them. One of the UMLS vocabularies is the CHV1, a vocabulary that connects simple, everyday health words to technical terms used by health care professionals. For each concept, it stores the best way to express it for a lay audience (CHV-preferred) and the same for a professional audience (UMLS-preferred).
Differences between the methods are summarized in Table 1. In the CHV-preferred/UMLS-preferred method, the selected synonyms correspond to the CHV-preferred and UMLS-preferred expressions for each concept. This is the only method using exclusively one vocabulary.
Table 1.
Proposed methods.
| Name | Vocabulary | Atoms | Relationships | Type of relationships |
|---|---|---|---|---|
| CHV-preferred/UMLS-preferred | Only CHV | All | – | – |
| Preferred Atoms | UMLS ones | Preferred atoms | – | – |
| All preferred/synonym atoms | UMLS ones |
All English preferred/ synonym atoms |
– | – |
| All Atoms | UMLS ones | All English atoms | – | – |
|
All Atoms + Child/ Parent/Same Relations |
UMLS ones | All English atoms | With atoms | Child/Parent/Same |
| Broader/Narrower Concepts | UMLS ones | All English atoms | With concepts | Broader/Narrower |
The other methods use the overall UMLS to obtain an expression or a subset of expressions, from which we select the lay and medico-scientific synonymous. The lay synonymous is the expression with the highest value of similarity with the lay terminology, and the medico-scientific one is the expression closest to the medico-scientific terminology. To determine the closeness of the expressions to these terminologies, we used a previously created algorithm [11].
The Preferred Atoms method uses the default preferred atom associated with the CUI. The All preferred/synonym atoms method retrieves a list of all English atoms that are the preferred names or a synonym in the various vocabularies of the UMLS. The All Atoms method retrieves all the English atoms, instead of extracting only the preferred and synonym ones. To explore other atoms associated with a concept, the method All Atoms + Child/Parent/Same Relations identifies all English atoms associated with a concept and then retrieves atoms related to the first one through parent/child/same relationships. Finally, the Broader/Narrower Concepts recovers broader and narrower atoms that are directly connected with the initial identified concept, instead of looking for atoms associated with the concept.
Evaluation
To assess and compare the effectiveness of the developed methods, we used two test collections, one in English and the other in Portuguese. The English collection is provided by the Consumer Health Search Task in the 2018 edition of the CLEF eHealth Lab2. This task uses a set of 50 English queries and a document corpus with 5,535,120 web pages acquired from a CommonCrawl dump. It also provides 26,025 judgments of relevance and understandability.
The Portuguese collection was explicitly built for this work. We used the English queries provided by the User-Centred Health Information Retrieval3 and Patient-Centred Information Retrieval4 Tasks of the 2015 and 2016 editions of the CLEF eHealth Lab. We translated the 208 queries to the Portuguese language with the collaboration of a medical doctor. Although the dataset of the 2015 edition had Portuguese translations of the queries, they were in some cases in PT-BR, and for this reason, we decided to translate them to PT-PT manually.
The queries were used in a user study with 104 participants. These participants were students, and as part of one work assignment, they were assigned two tasks regarding two different queries. In each task, they were asked to judge the relevance and understandability of the 30-top documents retrieved by four search engines: Google, Bing, Yahoo!, and HONSearch. The 16,505 assessed documents and the judgments of the participants complete this collection5. The number of documents is different from 24,960 (208*4*30) because there was an overlap between documents retrieved by the four search engines and because the number of retrieved results may be inferior to 30.
We have indexed the document corpora in Elastic Search. For each query, we compute four types of suggestions, in lay and medico-scientific suggestions, both in English and Portuguese. Using the judgments of each test collection as ground truth, we assessed the performance of each suggestion through the top-10 documents retrieved by Elastic Search for that query. For this evaluation, our baseline is the performance of the suggestions provided by Health Suggestions.
The performance was assessed through the Understandability-based RBP (uRBP) and uRBP graded (uRBPgr). uRBP is a measure that increases when the user chooses a document that is considered both relevant and understandable, based on binary assessments. The uRBPgr allows graded assessment values [14]. For each method, we conduct one evaluation considering word-sense disambiguation and one without it.
Results
The best methods select the CHV-preferred expressions for lay suggestions and the UMLS-preferred expression for the medico-scientific suggestions (Table 2). Both methods outperform the baseline.
Table 2.
Evaluation of the methods using the English and Portuguese test collections.
| Terminology | Method | English | Portuguese | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Without WSD | With WSD | Without WSD | With WSD | ||||||
| uRBP | uRBPgr | uRBP | uRBPgr | uRBP | uRBPgr | uRBP | uRBPgr | ||
| Lay | HealthSuggestions (Baseline) | 0.2869 | 0.1257 | 0.2869 | 0.1257 | 0.0404 | 0.0567 | 0.0404 | 0.0567 |
| CHV-preferred | 0.4961 | 0.2372 | 0.4846 | 0.2298 | 0.1237 | 0.1496 | 0.1258 | 0.1500 | |
| All preferred/synonym atoms | 0.3618 | 0.1750 | 0.3221 | 0.1558 | 0.0878 | 0.1070 | 0.0879 | 0.1102 | |
| All Atoms | 0.3189 | 0.1634 | 0.3530 | 0.1763 | 0.0813 | 0.1006 | 0.0888 | 0.1091 | |
| All Atoms + Child Relations | 0.1460 | 0.0665 | 0.2341 | 0.1001 | 0.0400 | 0.0507 | 0.0466 | 0.0578 | |
| All Atoms + Parent Relations | 0.1972 | 0.0948 | 0.2685 | 0.1226 | 0.0705 | 0.0918 | 0.0675 | 0.0870 | |
| All Atoms + Same Relations | 0.3477 | 0.1645 | 0.3693 | 0.1731 | 0.0919 | 0.1126 | 0.0910 | 0.1096 | |
| Broader Concepts | 0.2852 | 0.1307 | 0.3321 | 0.1525 | 0.0839 | 0.1056 | 0.0928 | 0.1147 | |
| Narrower Concepts | 0.1617 | 0.0775 | 0.2397 | 0.1121 | 0.0590 | 0.0772 | 0.0801 | 0.0999 | |
| Medico-scientific | HealthSuggestions (Baseline) | 0.2610 | 0.1167 | 0.2610 | 0.1167 | 0.0385 | 0.0537 | 0.0385 | 0.0537 |
| UMLS-preferred | 0.4155 | 0.2073 | 0.4280 | 0.2122 | 0.0969 | 0.1214 | 0.1081 | 0.1334 | |
| All preferred/synonym atoms | 0.3164 | 0.1610 | 0.2510 | 0.1279 | 0.0821 | 0.1022 | 0.0823 | 0.1020 | |
| All Atoms | 0.3381 | 0.1690 | 0.3269 | 0.1634 | 0.0628 | 0.0805 | 0.0716 | 0.0894 | |
| All Atoms + Child Relations | 0.1192 | 0.0531 | 0.1772 | 0.0716 | 0.0496 | 0.0632 | 0.0573 | 0.0744 | |
| All Atoms + Parent Relations | 0.2031 | 0.0999 | 0.2655 | 0.1231 | 0.0715 | 0.0933 | 0.0715 | 0.0922 | |
| All Atoms + Same Relations | 0.3480 | 0.1753 | 0.3719 | 0.1873 | 0.0721 | 0.0932 | 0.0673 | 0.0860 | |
| Broader Concepts | 0.2273 | 0.1042 | 0.2798 | 0.1243 | 0.0848 | 0.1031 | 0.1017 | 0.1228 | |
| Narrower Concepts | 0.1774 | 0.0828 | 0.2547 | 0.1188 | 0.0597 | 0.0781 | 0.0803 | 0.1012 | |
| Both | Preferred Atoms | 0.3662 | 0.1905 | 0.3469 | 0.1754 | 0.1011 | 0.1295 | 0.1084 | 0.1369 |
Globally, the methods with better performance are the ones that consider the preferred atoms of the different vocabularies from the UMLS, mainly the CHV. Using child relations does not help, probably due to the specificity of the suggestion. Using broader terms (parent and broader relations) proved to be more useful since other designations for the same concept are being explored.
In the English test collection, the use of WSD does not improve the performance of the methods that use UMLS-preferred terms but is useful when exploring relations. In the Portuguese collection, in general, there are slightly better results when using WSD. Nevertheless, this difference is so small that we conclude that it is better to disambiguate in methods that explore relations and the other way around in methods that pick the preferred terms. Note that context is essential in methods that use relations that may justify the importance of disambiguation.
The average number of seconds to formulate a suggestion is presented, for each method, in Table 3. As can be seen, methods that consider the relationships of atoms take a longer time compared to the others. The use of the relations from concepts should be preferred since it takes less time to process them, and the performance is similar. In English, the use of WSD helps to reduce the processing time because fewer atoms are retrieved and, therefore, less processing is needed afterward. The CHV/UMLS-preferred are the fastest methods since they only need to identify the concept and retrieve the corresponding CHV/UMLS-preferred expression.
Table 3.
Average number of seconds to generate a suggestion.
| Method | English | Portuguese | ||
|---|---|---|---|---|
| Without WSD | With WSD | Without WSD | With WSD | |
| CHV/UMLS-preferred | 0.42 | 0.34 | 0.22 | 0.26 |
| Preferred Atoms | 1.34 | 1.08 | 1.52 | 1.45 |
| All preferred/synonym atoms | 30.54 | 25.72 | 38.59 | 32.55 |
| All Atoms | 50.02 | 41.60 | 64.85 | 46.98 |
| All Atoms + Child Relations | 111.5 | 99.30 | 200.98 | 207.29 |
| All Atoms + Parent Relations | 175.44 | 134.72 | 321.52 | 250.51 |
| All Atoms + Same Relations | 127.34 | 100.72 | 257.44 | 205.61 |
| Broader Concepts | 17.42 | 10.80 | 18.29 | 21.04 |
| Narrower Concepts | 3.98 | 3.66 | 5.82 | 5.42 |
Conclusions
The majority of the developed methods proved to be better than the baseline, helping the user to retrieve more relevant and understandable documents. Using UMLS-preferred terms resulted in a better performance. Others explored broader terms, more specific terms, and similar terms but did not retrieve as good results. The best method to suggest lay queries is the one that uses the CHV-preferred expressions (the most familiar ones) to substitute the identified concepts. The best method to suggest medico-scientific suggestions uses UMLS-preferred expressions. These methods are better in the relevance and understandability but are also better in generation time. Since the word-sense disambiguation reduces the time that is necessary to generate new suggestions, and slightly improves or does not affect the overall performance, we conclude it should be used.
Acknowledgments
This work was financed by the Portuguese funding agency, FCT - Fundação para a Ciência e a Tecnologia, through national funds, and co-funded by the FEDER, where applicable.
Footnotes
Available at https://rdm.inesctec.pt/dataset/cs-2020-004.
Contributor Information
Joemon M. Jose, Email: joemon.jose@glasgow.ac.uk
Emine Yilmaz, Email: emine.yilmaz@ucl.ac.uk.
João Magalhães, Email: jm.magalhaes@fct.unl.pt.
Pablo Castells, Email: pablo.castells@uam.es.
Nicola Ferro, Email: ferro@dei.unipd.it.
Mário J. Silva, Email: mjs@inesc-id.pt
Flávio Martins, Email: flaviomartins@acm.org.
Paulo Miguel Santos, Email: up201403745@fe.up.pt.
Carla Teixeira Lopes, Email: ctl@fe.up.pt.
References
- 1.Akerkar S, Bichile L. Health information on the internet: patient empowerment or patient deceit? Indian J. Med. Sci. 2004;58(8):321–6. [PubMed] [Google Scholar]
- 2.Aronson, A.R.: Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. In: Proceedings of AMIA Symposium, pp. 17–21 (2001). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2243666/ [PMC free article] [PubMed]
- 3.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J. Am. Med. Inform. Assoc. 2010;17(3):229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database Issue):267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ermakova, L., Mothe, J., Nikitina, E.: Proximity relevance model for query expansion. In: Proceedings of the 31st Annual ACM Symposium on Applied Computing, SAC 2016, pp. 1054–1059. ACM, New York (2016). 10.1145/2851613.2851696
- 6.Jacobs W, Amuta AO, Jeon KC. Health information seeking in the digital age: an analysis of health information seeking behavior among US adults. Cogent Soc. Sci. 2017;3(1):1–11. doi: 10.1080/23311886.2017.1302785. [DOI] [Google Scholar]
- 7.Liu Z, Chu WW. Knowledge-based query expansion to support scenario-specific retrieval of medical free text. Inf. Retrieval. 2007;10(2):173–202. doi: 10.1007/s10791-006-9020-6. [DOI] [Google Scholar]
- 8.Lopes CT, Fernandes TA, et al. Health suggestions: a chrome extension to help laypersons search for health information. In: Fuhr N, et al., editors. Experimental IR Meets Multilinguality, Multimodality, and Interaction; Cham: Springer; 2016. pp. 241–246. [Google Scholar]
- 9.Lopes CT, Ribeiro C, et al. Effects of language and terminology on the usage of health query suggestions. In: Fuhr N, et al., editors. Experimental IR Meets Multilinguality, Multimodality, and Interaction; Cham: Springer; 2016. pp. 83–95. [Google Scholar]
- 10.Ooi, J., Ma, X., Qin, H., Liew, S.C.: A survey of query expansion, query suggestion and query refinement techniques. In: 2015 4th International Conference on Software Engineering and Computer Systems (ICSECS), pp. 112–117 (2015). 10.1109/ICSECS.2015.7333094
- 11.Santos, P., Lopes, C.T.: Is it a lay or medico-scientific concept? Automatic classification in two languages. In: 2019 14th Iberian Conference on Information Systems and Technologies (CISTI), pp. 1–4 (2019). 10.23919/CISTI.2019.8760745
- 12.Zeng QT, Crowell J, Plovnick RM, Kim E, Ngo L, Dibble E. Assisting consumer health information retrieval with query recommendations. J. Am. Med. Inform. Assoc. 2006;13(1):80–90. doi: 10.1197/jamia.M1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng QT, Kogan S, Plovnick RM, Crowell J, Lacroix EM, Greenes RA. positive attitudes and failed queries: an exploration of the conundrums of consumer health information retrieval. Int. J. Med. Inform. 2004;73(1):45–55. doi: 10.1016/j.ijmedinf.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 14.Zuccon G, et al. Understandability biased evaluation for information retrieval. In: Ferro N, et al., editors. Advances in Information Retrieval; Cham: Springer; 2016. pp. 280–292. [Google Scholar]