Schizorhodopsins, recently discovered rhodopsins in Asgard archaea, function as light-driven inward H+ pumps.
Abstract
Schizorhodopsins (SzRs), a rhodopsin family first identified in Asgard archaea, the archaeal group closest to eukaryotes, are present at a phylogenetically intermediate position between typical microbial rhodopsins and heliorhodopsins. However, the biological function and molecular properties of SzRs have not been reported. Here, SzRs from Asgardarchaeota and from a yet unknown microorganism are expressed in Escherichia coli and mammalian cells, and ion transport assays and patch clamp analyses are used to demonstrate SzR as a novel type of light-driven inward H+ pump. The mutation of a cytoplasmic glutamate inhibited inward H+ transport, suggesting that it functions as a cytoplasmic H+ acceptor. The function, trimeric structure, and H+ transport mechanism of SzR are similar to that of xenorhodopsin (XeR), a light-driven inward H+ pumping microbial rhodopsins, implying that they evolved convergently. The inward H+ pump function of SzR provides new insight into the photobiological life cycle of the Asgardarchaeota.
INTRODUCTION
Microbial rhodopsins are photoreceptive membrane proteins with all-trans retinylidene chromophore (all-trans retinal) widely identified in microorganisms (e.g., bacteria, archaea, algae, and fungi) and giant viruses (1, 2). The functions of microbial rhodopsins are very diverse: light-driven ion pumps that actively transport various ions (H+, Na+, Cl−, etc.) against electrochemical potential, passively transporting light-gated ion channels, (photo)sensory rhodopsins, light-dependent enzymes, and so on (1, 3, 4). Despite the diversity of their function, all microbial rhodopsins share a heptahelical-transmembrane structural scaffold in which the all-trans retinal chromophore is bound to a conserved lysine residue in the seventh transmembrane helix (helix G) (1). A recently discovered rhodopsin family called heliorhodopsins (HeRs), distinct from typical microbial rhodopsins, was found in diverse microorganisms and giant viruses (5). HeRs are phylogenetically distinct from typical microbial rhodopsins and show little sequence homology (the identity between HeRs and typical microbial rhodopsins is <15%). Although the biological function of HeRs has not yet been revealed, it further reaffirms the diverse photobiological role of rhodopsins in microbial species.
Phylogenomic analysis has suggested that the Asgardarchaeota are the closest prokaryote relatives of eukaryotes (6–8). In addition to many typical microbial rhodopsins and HeR, a previously unknown rhodopsin family, schizorhodopsin (SzR), has recently been identified in Asgardarchaeota genomes (8). Phylogenetic analysis indicates that SzR is distinct from both typical microbial rhodopsin and HeR and is found in an intermediate position between them (Fig. 1A). Although SzR is expected to provide insights into the molecular evolution of rhodopsins in microorganisms, its function remains unknown. The aim of this study was to provide insight into the photobiological lifecycle of Asgardarchaeota.
Fig. 1. Phylogenetic and sequence analyses of SzRs with microbial rhodopsins and HeRs.
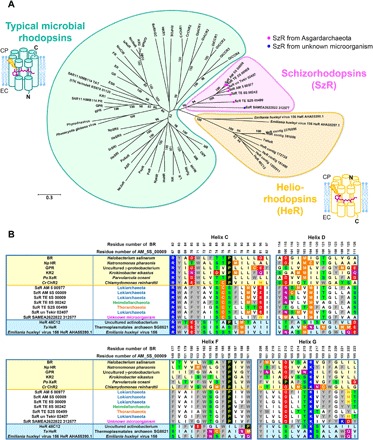
(A) Phylogenetic tree of the representative typical microbial rhodopsins (green), HeRs (yellow), and SzRs (pink). The SzRs originated from Asgardarchaeota and from unknown microorganism are indicated with magenta and blue circles, respectively. The molecular orientations of the typical microbial rhodopsins and HeRs are schematically shown. N, N-terminus; C, C-terminus; EC, extracellular side; CP, cytoplasmic side. (B) Multiple amino acid alignment of SzRs with typical microbial rhodopsins and HeRs for the helices C, D, F, and G. Their full sequences are shown in fig. S2.
Here, the biological function of SzRs was explored by expressing SzR genes in Escherichia coli and mammalian cells. Ion transport assays of E. coli cells with a pH electrode and electrophysiological measurements in mammalian cells revealed that SzR is a formerly unknown type of light-driven inward H+ pump, which actively transports H+ into the cytoplasm against the electrochemical potential. The purified SzR protein was shown to have a trimeric structure by circular dichroic (CD) spectroscopy and high-speed atomic force microscopy (HS-AFM), as well as a photocycle involving a large accumulation of blue-shifted M intermediate, representing the deprotonation of retinal Schiff base (RSB) after photoisomerization of the all-trans retinal chromophore to 13-cis form upon light illumination by laser flash photolysis. Furthermore, Fourier transform infrared (FTIR) spectroscopy indicated the presence of several water molecules around the chromophore, whose hydrogen bonding structure was altered upon retinal isomerization. Mutational experiments suggested that a cytoplasmic glutamate works as an H+ acceptor, receiving an H+ from RSB upon M formation, which is indispensable for the inward H+ transport. A cysteine in helix C (the third transmembrane helix) conserved in the SzR family at the same position as in channelrhodopsins (ChRs) and enzymatic rhodopsins was also shown to be critical for its function. The function, trimeric structure, and H+ transport mechanism of SzR show many common aspects with xenorhodopsin (XeR), another inward H+ pump found in typical microbial rhodopsins (9–11). Our results suggest that convergent molecular evolution occurred between these two unrelated rhodopsin families.
RESULTS
Comparison of SzR sequence with type 1 rhodopsin and HeR
Although SzR was first identified in Asgardarchaeota (8), we also identified a homologous gene of unknown microbial origin (SAMEA 2622822_312577 amino acid sequence is shown in the Supplementary Materials) in a database of contigs assembled from the Tara Oceans metagenomic datasets of bacterial (12), viral (13), and girus (14) samples [the assemblies were generated as described elsewhere (15)], suggesting SzRs are also present in species other than Asgardarchaeota. In the assemblies, a total of 63 SzRs were identified (fig. S1). The amino acid sequences of six previously reported SzRs and the SzR SAMEA 2622822_312577 were aligned with representative type 1 rhodopsins and HeRs in Fig. 1B and fig. S2. This alignment suggests that helix C in SzR is more similar to that of HeR than type 1 rhodopsin [e.g., Trp68, Ser74, and Ser76 in SzR AM_5S_00009 (hereafter SzR AM_5S_00009 and SAMEA 2622822_312577 are referred as SzR1 and SzR2, respectively) are conserved in most HeRs but not in type 1 rhodopsins] (Fig. 1B). In contrast, helices F and G show a higher similarity to type 1 rhodopsin than HeR {e.g., Trp154, Pro158, Trp161, Asp184, and Phe191 in SzR1 are conserved in most type 1 rhodopsins [corresponding to Trp182, Pro186, Trp189, Asp212, and Phe219 in bacteriorhodopsin (BR)] but not in HeRs} (Fig. 1B). The previously reported phylogenetic analysis of rhodopsins suggested that SzRs at the intermediate position of type 1 rhodopsins and HeRs (Fig. 1A) (8), and the similarity to type 1 rhodopsin and HeR are heterogeneously different in each helix. Another characteristic residue in SzRs is a phenylalanine in helix C at the 85th position in BR. Generally, this is a position of RSB counterion, which is an Asp and a Glu in most type 1 rhodopsins and HeRs, respectively. SzR is the first microbial rhodopsin with a phenylalanine at this position. Furthermore, SzR has a cysteine in helix C at the 90th position in BR, which is homologous to a cysteine in ChRs, forming a DC (aspartic acid-cystein) gate critical for ion transport (16), and rhodopsin guanylyl cyclase (Rh-GC) and rhodopsin phosphodiesterase (Rh-PDE) also have a homologous cysteine (17, 18).
Light-driven inward proton transport by SzRs
To investigate the ion transport function of SzRs, all proteins were expressed in E. coli cells. All the cells, except for those transformed with SzR un_Tekir_02407, showed pink or purple colors (Fig. 2A), indicating the formation of functional rhodopsins. The ion transport activity of the E. coli cells was assayed by observing the light-induced changes in the pH of the external solvent, which was used to characterize the function of outward and inward H+, inward Cl−, and outward Na+ pumps (9, 19). In the case of outward H+-pumping green absorbing proteorhodopsin (GPR), a pH drop induced by an H+ release from the cytoplasm was observed (Fig. 2A). We observed large pH increases for all six SzR expressed in the cells, indicating inward H+ transport (Fig. 2A). A similar pH increase was observed for Parvularcula oceani XeR (PoXeR), the first reported natural inward H+ pump (9). The pH increases observed for SzR were substantially larger than that of PoXeR (Fig. 2A). Because H+ gradient and the membrane potential across the cell membrane cannot be regulated in E. coli cells, we need to carefully consider two possibilities to explain the pH increases observed for the SzRs, i.e., active inward H+ pump as PoXeR and passive H+ channel, as reported previously using artificial rhodopsin mutants (20, 21). To clarify this, we conducted electrophysiology experiments, in which photocurrents could be measured under clamped membrane voltage. The results for SzRs expressed in mammalian (ND7/23) cells are shown in Fig. 2B and fig. S3. All seven SzR were expressed in ND7/23 cells and showed inward currents consistent with inward H+ transport in E. coli cells. Note that, despite the membrane potential being scanned from −80 to +100 mV, a full inward current was observed. The photocurrent amplitudes from SzR1 and SzR2 were rather small (20 to 60 pA), probably due to the poor expression level. Furthermore, we investigated the effect of the H+ gradient on the photocurrent of SzR TE_S2S_00499 (hereafter SzR3), which showed the largest photocurrent in ND7/23 cells compared with other SzRs, and no dependence upon outer pH (pHo) was observed (Fig. 2C). Hence, the SzRs actively and inwardly transport H+, indicating that it is a formerly unknown light-driven inward H+ pump rhodopsin. Compared with PoXeR, whose photocurrent was reported to be ~−10 pA, the photocurrent of SzRs (between −80 and −300 pA) is substantially larger and comparable to that of Nanosalina XeR (NsXeR), which has been suggested to be a highly functional XeR (Fig. 2B and fig. S3). We observed that the off-phase kinetics of the photocurrents differ in each SzR and that the photocurrent of SzR3 was the most rapidly shut off (3 to 10 ms; Fig. 2D) once light was removed (Fig. 2B). The action spectrum of SzR3 photocurrent showed a broad peak, which was centered at ~525 nm (Fig. 2E).
Fig. 2. Light-driven active inward H+ transport by SzR.
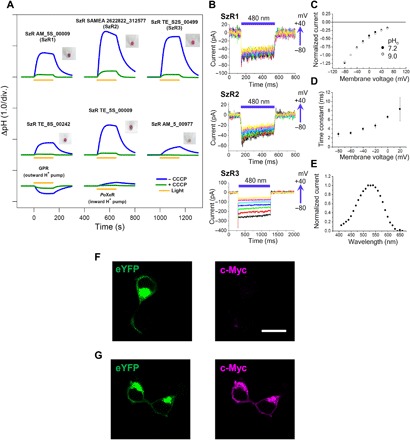
(A) Ion transport activity assay of SzRs in E. coli cells. The cells were illuminated with light (λ > 500 nm) for 150 s (yellow line). The pictures of the pellets of E. coli cells expressing each SzR are shown next to the corresponding results. (B) Electrophysiological measurements of SzR-driven photocurrent in ND7/23 cells. The cells were illuminated with light (λ = 480 nm, 12.3 mW/mm2) during the time region shown by blue bars. The membrane voltage was clamped from −80 to +100 mV for every 20-mV step. (C to E) I-V plot at pHo 7.2 and 9.0 (C), membrane-voltage dependence of the off-kinetics time constant (D), and action spectrum (E) of the current of SzR3. For action spectrum measurement, the light intensity of each wavelength was adjusted to 0.2 mW/mm2. (F and G) eYFP fluorescence (left, green) and immunofluorescence staining observation of SzR1 with a c-Myc epitope tag at the C terminus in cultured ND7/23 cells (right, magenta) in unpermeabilized (F) and permeabilized conditions with detergent (Triton X-100) (G). Scale bar, 20 μm.
The orientation of SzR in the plasma membrane
Next, we investigated the molecular mechanism of the inward H+ pump function in SzRs. To determine the direction of H+ transport, the molecular orientation in the plasma membrane was investigated. Whereas the N and C termini of typical type 1 rhodopsins are oriented to the extracellular and cytoplasmic spaces, respectively, HeRs show an inverted orientation (Fig. 1A) (5). SzRs are present at a phylogenetically intermediate position between type 1 rhodopsins and HeRs, and the molecular orientation of SzRs is insightful to investigate the molecular mechanism of inward H+ pump function in SzRs. To investigate the molecular orientation of SzR in the plasma membrane, we expressed SzR1 with an enhanced yellow fluorescent protein (eYFP) and a c-Myc epitope tag at the C terminus in cultured ND7/23 cells. While no immunofluorescence was observed when the cells were treated with goat anti-rabbit immunoglobulin G (IgG) secondary antibody conjugated to Alexa Fluor 594 (Fig. 2F), SzR1 was immunostained in the cells whose plasma membrane was permeabilized with detergent (Triton X-100) (Fig. 2G). This immunostaining pattern suggests that the C terminus of SzR is oriented toward the cytoplasmic side of the cells, as in type 1 rhodopsins. Hence, the inward H+ pumping in SzR transports H+ from the N terminus facing the extracellular side to the C terminus facing the cytoplasmic side.
Molecular properties and photocycles of SzRs
To determine the photoreaction of SzR, we purified Asgardarchaeota SzR1 and SzR2 from a non-Asgardarchaeota species. SzR1 and SzR2 showed maximum absorption wavelengths (λmax) at 557 and 542 nm, respectively, in n-dodecyl-β-d-maltoside (DDM) (Fig. 3A). The CD spectra of both SzR showed negative and positive peaks at the short and long wavelength sides of their λmax, respectively, which is characteristic of rhodopsins in trimeric form (Fig. 3B) (22). The oligomeric structure of SzR1 was determined using high-speed AFM imaging, which indicated its trimeric structure (Fig. 3C). Hence, the correlation between the CD spectrum and the oligomeric structure is also conserved in phylogenetically distant SzRs.
Fig. 3. Absorption and CD spectra and high-speed AFM imaging of purified SzR proteins.
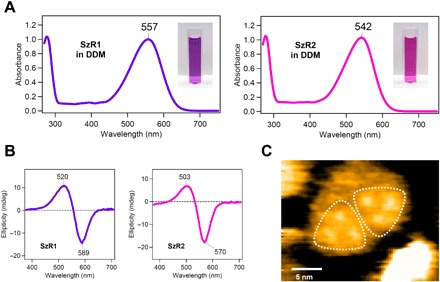
(A and B) UV-vis absorption (A) and CD (B) spectra of SzR1 (left) and SzR2 (right) in 100 mM NaCl, 20 mM tris-HCl (pH 8.0), and 0.05% DDM. (C) High-speed AFM image of SzR1 trimers in lipid bilayers.
The absorption peak of SzR1 was highly blue shifted at alkaline pH (fig. S4A), indicating the deprotonation of RSB. By fitting the absorption change with the Henderson-Hasselbalch equation (fig. S4B), the pKa of RSB in SzR1 was estimated to be ~13.5, which is similar to the cases of most microbial rhodopsins [e.g., the pKa of RSB in BR is 13.3 ± 0.3 (23)] and higher than that of HeR (5). In contrast, no significant change in the absorption was observed on the acidic side (pH 7→2.3) (fig. S4A), indicating the putative counterion SzR1 Asp184 that is highly conserved and deprotonated to stabilize the protonated RSB in most microbial rhodopsins does not change its protonation state up to pH 2.3 and its pKa <2. The pH dependence of SzR2 was also investigated, and it showed lower deprotonation of RSB than SzR1 at alkaline pH, suggesting that the pKa of RSB in SzR2 is >13.5 (fig. S4C).
The retinal configuration in SzR1 was investigated by high-performance liquid chromatography (HPLC) analysis on retinal oximes produced by the hydrolysis reaction of RSB in SzR with hydroxylamine and extracted using hexane. Most of the chromophore extracted from the dark-adapted protein showed an all-trans configuration with a trace amount of 11-cis form (fig. S5 and table S1). After a 1-min illumination at λ > 500 nm, the 13-cis form was accumulated with increased 11-cis fractions in the light-adapted state, where >70% chromophore remained in all-trans configuration (fig. S5 and table S1). As such, the all-trans form represents the functional state of SzR, as in typical microbial rhodopsins and HeRs (1, 5).
Next, the transient absorption changes in SzR1 and SzR2 were measured using the laser flash photolysis method (Fig. 4). At the fastest time point (t = 3 μs), only a negative (bleach) signal was observed (Fig. 4, A and B). Then, a blue-shifted M intermediate was observed at 390 to 400 nm, along with an increase in the bleaching signal. The increased bleaching signal suggests that the transient absorption of the K and L intermediates overlapped with the bleaching at the faster time points. The bleaching signal was then recovered with a double-exponential decay of the M intermediate at 10 to 200 ms. Hence, the initial state is directly recovered from the M, which varies from the recovery process of PoXeR, in which 57 and 43% of the M decay to the initial and PoXeR13C state with 13-cis-15-syn retinal, respectively (24). The direct recovery from the M intermediate results in a turnover rate in SzR that is faster than that of PoXeR, which would result in a stronger inward H+ pump efficiency, and the models of photocycles of SzR1 and SzR2 were suggested from the multiexponential kinetic analysis of the transient absorption change (Fig. 4, A and B).
Fig. 4. Transient absorption changes and the photocycles of SzRs.
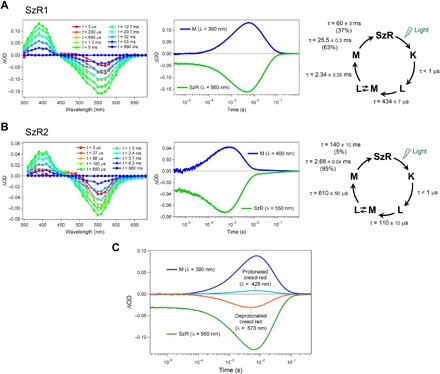
(A and B) Transient absorption spectra (left), time evolutions of transient absorption changes at specific wavelengths (middle), and the photocycles determined by analyzing the time evolution with multiexponential functions (right) of SzR1 (A) and SzR2 (B) in POPE/POPG (molar ratio, 3:1) membrane. (C) Transient absorption change of cresol red accompanying the photoreaction of SzR1 in 0.1% DDM. Positive and negative absorption changes at λ = 429 and 573 nm, respectively, indicate the protonation of cresol red.
The H+ release and uptake processes between SzR1 and the solvent phase were investigated by monitoring the transient absorption change accompanying the changes in protonation state of a pH-indicating dye, cresol red. Cresol red shows absorption peaks at 429 and 573 nm in protonated and deprotonated forms, respectively (5). Along the M accumulation at t = ~100 to ~7 ms, we observed a decrease in the absorption of cresol red at 573 nm and an increase at 429 nm (Fig. 4C). This indicates that an H+ was released from the protein to the solvent. Subsequently, the changes in the absorption of cresol red at 429 and 573 nm were associated with M decay and the recovery of initial state (t = ~7 to ~20 ms). In the case of the photocycle of PoXeR, a significant delay was observed between the M rise and the H+ release to the solvent, where the H+ is transiently trapped in the protein in the L/M intermediate (24). In contrast, the protonation of cresol red occurred without any delay from the M rise of SzR1 (Fig. 4C). The amount of H+ released was proportional to the M accumulation. Hence, the H+ is not trapped in the protein in the L/M intermediate of SzR1, unlike PoXeR.
FTIR spectroscopy of photointermediates of SzR
To obtain structural insights of SzR, light-induced low-temperature FTIR spectroscopy was carried out on SzR1 (Fig. 5). The red-shifted K intermediate was trapped at T = 110 K, which was indicated by low-temperature ultraviolet-visible (UV-vis) spectroscopy under the same temperature and light illumination condition (fig. S6). The K-minus-dark spectrum shows a 1200(−)/1192(+)-cm−1 peak pair, indicating that all-trans to 13-cis retinal isomerization occurs upon photoexcitation, as in most type 1 rhodopsins and HeRs. The L, L/M, and M intermediates were trapped by raising the temperature to 190, 210, and 230 K, respectively. The negative band at 1526 cm−1 originated from the ethylenic stretching C=C vibration of the all-trans chromophore in the dark state. The wavenumber of this band is known to show a correlation with λmax, and a downshift is expected to be observed for the red-shifted intermediate (25). Although no significant downshifted positive peak was observed upon the K formation, the band observed at 1519 cm−1 was tentatively assigned to the ethylenic stretching C=C vibration of the 13-cis chromophore in the K. Hydrogen out-of-plane (HOOP), N-D in-plane bending, and methyl rocking vibrations appear in the 1110 to 900 cm−1 region. While the large positive peaks observed in this region upon K formation represent strongly distorted 13-cis chromophore in many microbial rhodopsins and HeRs (5, 26), the K-intermediate of SzR1 shows relatively small positive peaks at 1010, 951, and 937 cm−1 (Fig. 5A). This weak intensity indicates that 13-cis chromophore in the K intermediate of SzR1 appears in a more relaxed form, compared with other microbial rhodopsins and HeRs. In all light-induced FTIR spectra of SzR1, a sharp negative peak was observed at 1444 cm−1. Previously, ASR and PoXeR were reported to show a doublet peak at the same region and were considered to originate from a proline conserved in XeRs, at the position corresponding to BR Asp212. However, this proline is not conserved in SzRs, and no other SzR-specific proline is present in the transmembrane (TM) region of SzR. Furthermore, since the 1444(−) cm−1 peak disappeared in D2O, which is different from the case of ASR and PoXeR, this SzR-specific band must have originated from an amino acid other than proline. The assignment of this band lies outside the focus of this study and will be studied elsewhere in the future.
Fig. 5. Light-induced low-temperature difference FTIR spectroscopy of SzR1.
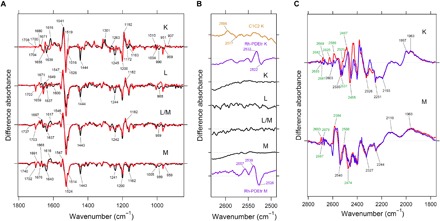
(A and B) Light-induced low-temperature K-minus-dark, L-minus-dark, L/M-minus-dark, and M-minus-dark difference FTIR spectra of SzR1 obtained at T = 110, 190, 210, and 230 K, respectively, in the 1800 to 850 (A) and 2630 to 2490 (B) cm−1 regions. In (B), the light-induced low-temperature K-minus-dark FTIR spectra of C1C2 and Rh-PDEtr and low-temperature M-minus-dark FTIR spectrum of Rh-PDEtr were reproduced from (18) and (28). (C) Light-induced low-temperature K-minus-dark and M-minus-dark difference FTIR spectra of SzR1 obtained at T = 110 and 230 K, respectively, in the 2800 to 1760 cm−1 region. The normalization factors multiplied to each spectrum are listed in tables S2 and S3.
In PoXeR, an hydrogen/deuterium (H/D)–sensitive positive peak from protonated Asp216 in helix G was observed at 1736 cm−1 on the M intermediate. In contrast, no H/D-sensitive band was observed in the 1700 to 1800 cm−1 region of any of the photointermediates of SzR (Fig. 5A). This difference could be due to the different H+ release processes between SzR and PoXeR: While an H+ is metastably trapped at Asp216 in PoXeR, it is directly released into the cytoplasmic milieu in SzR, which is consistent with the H+ release process without trapping in the protein on the L/M state (see above).
As mentioned above, SzR has a characteristic cysteine (SzR1 Cys75) in helix C. In the case of the chimeric protein between Chlamydomonas reinhardtii ChR1 and ChR2 (C1C2) and the truncated rhodopsin domain of Rh-PDE, which also has homologous cysteines, an upshift of the peaks in the 2630 to 2490 cm−1 region was observed, indicating that the hydrogen bonds of the S-H group of cysteines became weaker upon the photoactivation of these rhodopsins (Fig. 5B) (18, 27, 28). In contrast, no apparent peak was observed in this region for any of the photointermediates of SzR1. This suggests that the hydrogen bonding strength of the conserved cysteine (SzR1 Cys75) is not altered in the photocycle of SzR.
In Fig. 5C, X-D stretching vibrations are observed in the 2800 to 1760 cm−1 region, where O-D stretching vibrations derived from water molecules showed a downshift upon the exchange of D2O with D218O. In the K-minus-dark spectrum, six and four O-D vibrations of water molecules were observed for the K and dark states, respectively, suggesting that several water molecules are found near RSB, which show hydrogen bonding alterations upon retinal isomerization. In the M-minus-dark spectrum, the number of peaks of O-D stretching vibrations of water molecules was decreased compared with the K-minus-dark spectrum. This suggests that the altered hydrogen bonding strength of certain water molecules in the K intermediate relaxed during the K-to-M process. The most downshifted O-D stretching vibrations of the outward H+-pumping rhodopsins appear in the region of <2400 cm−1, suggesting that a strong hydrogen-bonded water molecule is a prerequisite for outward H+ pumps (29). This is also the case for PoXeR (30), but not for SzR. Therefore, the presence of a strongly hydrogen-bonded water molecule is not common for inward H+ pumps, in contrast to outward H+ pumps.
Ion transport activity of SzR mutants
The H+ transport pathway in SzR was studied by investigating the inward H+ transport activity of site-directed SzR mutants. Acidic amino acid residues play a critical role in H+ transport in both outward and inward H+ pumps. Five acidic residues (Glu2, Glu3, Glu81, Glu95, and Asp184) are conserved in the transmembrane helices in SzRs. As such, we performed D-to-N or E-to-Q mutations in these residues (Fig. 6A). As a result, SzR1 E81Q completely lost the function, and, since SzR1 D184N did not show any pigment formation in E. coli cells, we were unable to measure its transport activity. Glu81 and Glu95 are suggested to be present on the cytoplasmic side of the RSB on the basis of sequential alignments (Fig. 1B), and the loss-of-the-function mutation of the former into a glutamine suggests that Glu81 works as a cytoplasmic H+ acceptor in helix C, similar to PoXeR Asp216 in helix G (9). SzR1 Glu81 is replaced by a histidine in SzR2 (His82). Although the histidine is able to accept H+, SzR2 H82F did not show any significant reduction in H+ transport activity (Fig. 6B), and the histidine is not related to the function of transport. As such, the residue that acts as an H+ acceptor in SzR2 needed to be identified. A unique aspartate (Asp197) is present in the cytoplasmic half of helix G in SzR2. The mutation of this aspartate to glutamate or asparagine, however, did not alter the transport activity (Fig. 6B). Hence, no residue seems act as an H+ acceptor in SzR2. Although it is possible that an unidentified H+-accepting residue exists, our results suggest that H+ is directly released to cytoplasmic milieu from RSB.
Fig. 6. H+ transport activity of SzR mutants and inward H+ transport model of SzR.
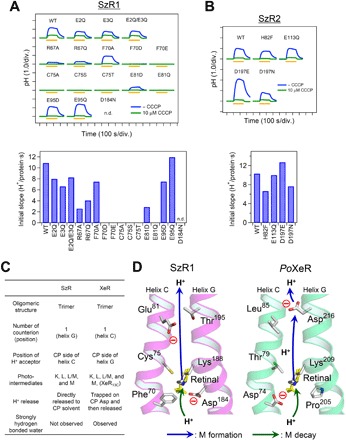
(A and B) Ion transport activity assay in E. coli cells (upper) and initial slopes normalized by the amount of expressed proteins (lower) of the mutants of SzR1 (A) and SzR2 (B). n.d., not determined. (C) Different and similar molecular characters between SzRs and XeRs. (D) H+ transport pathway in SzR1 (left) suggested in this study and in PoXeR (right) (9, 24). The structure of both proteins was drawn on the basis of the x-ray crystallographic structure of NsXeR (PDB code: 6EYU) (10). The proton release and uptake pathways on the M formation and M decay are indicated by blue and green arrows, respectively.
Proteorhodopsin is a light-driven outward H+ pump with a DTE (aspartic acid-threonine-glutamic acid) motif in helix C (19). Since SzR1 has an FTE (phenylalanine-threonine-glucamic acid) motif, we attempted to convert it to an outward H+ pump by altering the motif to DTE.SzR1 F70D, however, did not show any ion transport, and SzR1 F70E with ETE lost this function (Fig. 6A). As such, motif alteration alone is not sufficient to invert the direction of the H+ pump of SzR. Even if we mutate Phe70 to alanine (SzR1 F70A), no significant transport inhibition was observed (Fig. 6A), suggesting that the size of this hydrophobic residue is not critical for the inward H+ pump function. In most microbial rhodopsins and HeRs, a conserved arginine residue is found at BR Arg82. The mutation of this residue in SzR1, i.e., Arg67, to alanine and glutamine highly reduced the H+ transport activity (SzR1 R67A and R67Q) (Fig. 6A).
DISCUSSION
SzR was first identified in Asgardarchaeota and is phylogenetically positioned between typical microbial rhodopsins and HeRs (Fig. 1). In this study, we showed that SzR is a new type of light-driven inward H+ pump (Fig. 2). XeR was previously reported as an inward H+ pump in the typical microbial rhodopsin family (9–11). Although the sequential homology between the two subfamilies is low (SzR1 and PoXeR show 15.7% identity and 42.6% similarity), the trimeric structure and the photocycle with a large M accumulation not accompanied by N and O intermediates of SzR are similar to those reported for XeR (9–11), despite a large phylogenetic distance between them. This suggests that XeR and SzR underwent convergent evolution at the molecular level to achieve the same biological function. The differences and similarities between SzR and XeR are listed in Fig. 6C. Asgardarchaeota contain not only SzRs but also typical microbial rhodopsins with a DTK motif in helix C and HeRs (8). Although the function of this DTK-type rhodopsin has not yet been reported, it is considered to be an outward H+ pump, because Exiguobacterium sibiricum rhodopsin, which also has a DTK motif but is phylogenetically isolated from the DTK-type rhodopsins of Asgardarchaeota, functions as a light-driven outward H+ pump (31). The ion transport assay of DTK rhodopsin from Heimdallarchaeota (a member of Asgardarchaeota) showed outward H+ transport activity in E. coli cells (fig. S7). Hence, Asgardarchaeota have two types of light-driven H+ pumps, transporting protons in opposite directions. These pumps may be used in different situations; otherwise, their transports may cancel each other.
Our immunostaining results suggest that the orientation of SzR is the same as that of type 1 rhodopsins, including XeR, and that the H+ is transported from the N-terminal side to the C-terminal side. The formation of an M intermediate, representing the deprotonation of RSB, and the concomitant H+ release and uptake suggest that the H+ of RSB is directly involved in the transport (Fig. 4). In general, the counterion on the extracellular side of RSB works as an H+ acceptor in outward H+ pumps, such as BR, PR, and so on (1). In contrast, cytoplasmic H+-accepting acidic amino acid residues play a critical role in artificial and natural inward H+ pumps (9, 10, 32). Fifty-eight of 63 SzRs shown in fig. S1 have a highly conserved glutamate or aspartate at the position of BR Asp96. The loss of function for the mutation of this residue to untitratable residue (SzR1 E81Q, Fig. 6A) indicates that it either works as a cytoplasmic H+ acceptor or forms part of the H+ release pathway. FTIR spectroscopy revealed the transient protonation of a cytoplasmic H+ acceptor (Asp216) in PoXeR (9). In contrast, no C=O stretching vibration of protonated carboxylic acid appeared in the FTIR spectra for any of the four intermediates in the SzR1 photocycle (Fig. 5A). This suggests that Glu81 does not accept an H+ from the RSB; however, on the basis of the complete loss of function of the E81Q mutant, this discrepancy is kinetically derived, that is, the rate of H+ release from Glu81 to the cytoplasmic milieu is faster than the H+ transfer from the RSB, such that the H+ is not metastably trapped by Glu81. The role of internal water molecule in inward H+ pump function is unclear, and it would be revealed by further spectroscopic and structural studies in the future. In particular, time-resolved vibrational spectroscopy and structural analysis will provide new structural insights, such as different retinal configuration, protonated water cluster, and so on, which can only be observed through more sophisticated room temperature measurements (33–37).
Two inward H+ transport models of SzR (SzR1) and XeR (PoXeR) were compared (Fig. 6D). The most prominent difference between these models is the kinetically untrapped and trapped H+ release in the cytoplasmic side, as discussed above. While H+ uptake from the extracellular side of SzR is not revealed in detail in this study, there is no specific extracellular H+ donor in PoXeR, and an H+ is directly taken up from the extracellular milieu simultaneously with a thermal isomerization of the retinal chromophore (24). On the extracellular side of the putative transmembrane region of SzR, only Glu2 and Glu3 are conserved, such that they are possible H+ donors to the RSB. The glutamine mutations, however, did not significantly impair H+ transport activity (Fig. 6A) and suggest that SzR also directly takes H+ from the extracellular side without any specific H+ donor.
We would like to mention another similarity between SzR and XeR. In XeR, the secondary counter ion in many microbial rhodopsins, that is, BR Asp212, is replaced with a proline, and a new negative charge was placed on the cytoplasmic side of the same helix (Fig. 6D, right). In SzR, a similar rearrangement of the negative charge took place in the helix C. The negative charge at the BR Asp85 position was lost, and a new negative charge was formed at BR Asp96 (Fig. 6D, left). These findings indicate that the one-extracellular-and-one-cytoplasmic negative charge configuration is a critical element of inward H+ pump of rhodopsins, which evolved convergently in both SzR and XeR (Fig. 6C).
SzRs contain a conserved cysteine in helix C, which is homologous to that in ChR, Rh-GC, and Rh-PDE. While this cysteine is critical for inward H+ transport in SzR, no alteration of hydrogen bonding strength of its S-H group was observed during the photocycle (Fig. 5B). This is different from ChR and Rh-PDE, in which we observed a weakening of the hydrogen bonding in the S-H group upon photoexcitation. This difference in hydrogen bonding suggests that the hydrogen bonding network and/or helical movement during the photocycle in SzR is different from those in ChR and Rh-PDE.
The arginine at the BR Arg82 position is one of most highly conserved residues not only in typical microbial rhodopsins but also in HeRs (1, 5). The mutation of BR Arg82 to alanine results in a rate of H+ release that is at least 20-fold slower than that in BR wild type (WT) (38). Homologous arginine in other rhodopsins also plays a critical role in their function (9, 39, 40). In the photocycle of BR, the orientation and hydrogen bonding of Arg82 are altered (41). This arginine is also important for the inward H+-pump function of SzR (Fig. 6A), and its structure in both dark and light conditions will need to be investigated in future studies to understand its mechanism.
Inward H+-pumping rhodopsins have been previously used as a new optogenetic tool to induce neural action potential in reaction to light (10). The time constant of the shutoff of SzR3 (3 to 10 ms) is shorter than the photocycle of NsXeR (27 ms) (10), which can induce neural firing at 40 Hz. This suggests that SzR3 also has a potential to be applied in optogenetics to investigate fast-firing neural circuits; however, this will need to be investigated in the future.
MATERIALS AND METHODS
Mutagenesis and protein expression
The synthesized genes of SzRs from Asgardarchaeota [GenBank accession numbers: TFG21677 (SzR TE_5_00977), TFG18381 (SzR1), TFG03937 (SzR TE_5S_00009), MK463862 (SzR TE_8S_00242), TFF95899 (SzR3), and MK463863 (SzR un_Tekir_02407)], SzR2, and Heimdallarchaeia rhodopsin [GenBank accession number: GCA_002728275.1 (Heimdallarchaeia rhodopsin RS678_01120)], codon-optimized for E. coli (Genscript, NJ), were incorporated into a pET21a(+) vector (Novagen, Merck KGaA, Germany). Site-directed mutagenesis was conducted using a QuikChange site-directed mutagenesis kit (Agilent, CA, USA). The sequences of the primers used are listed in table S4. The plasmids carrying the genes of SzRs and their mutants were transformed into the E. coli C43(DE3) strain (Lucigen, WI, USA). The protein expression was induced using 1 mM isopropyl β-d-1-thiogalactopyranoside in the presence of 10 μM all-trans retinal for 4 hours.
Ion transport assay of SzRs in E. coli cells
To assay the ion transport activity in E. coli cells, the cells carrying the expressed rhodopsin were washed three times and resuspended in unbuffered 100 mM NaCl. A cell suspension of 7.5 ml at OD660 (optical density at 660 nm) = 2 was placed in the dark in a glass cell at 20°C and illuminated at λ > 500 nm from the output of a 1-kW tungsten-halogen projector lamp (Master HILUX-HR, Rikagaku, Japan) through a long pass filter (Y-52; AGC Techno Glass, Japan) and a heat-absorbing filter (HAF-50S-50H; SIGMAKOKI, Japan). The light-induced changes in pH were measured using a pH electrode (9618S-10D; HORIBA, Japan). The measurements were repeated under the same conditions after the addition of 10 μM CCCP (carbonyl cyanide m-chlorophenylhydrazone). To qualitatively compare the ion transport activity of SzR1 and SzR2 and the mutants, the amount of the protein was determined by measuring the near-UV absorption of retinal oxime generated by the hydrolysis reaction between the RSB in the proteins and hydroxylamine. Briefly, the E. coli cells expressing the rhodopsins were washed with a solution containing 100 mM NaCl and 50 mM Na2HPO4 (pH 7) three times. The washed cells were treated with 1 mM lysozyme for 1 hour and then disrupted by sonication. To solubilize the rhodopsins, 3% DDM was added and the samples were stirred overnight at 4°C. The rhodopsins were bleached with 500 mM hydroxylamine and subjected to illumination of yellow light (λ > 500 nm) from the output of a 1-kW tungsten-halogen projector lamp (Master HILUX-HR, Rikagaku, Japan) through a glass filter (Y-52, AGC Techno Glass, Japan) and a heat-absorbing water bottle. The absorption change upon the bleaching of rhodopsin by hydroxylamine hydrolysis reaction and the formation of retinal oxime was measured using a UV-vis spectrometer (V-730, JASCO, Japan). The relative ion transport activities were determined by normalizing the initial slope of the light-induced pH change with the relative amount of expressed proteins.
Expression plasmids for mammalian cells
Synthesized genes encoding SzRs were subcloned into a phKR2-3.0-eYFP plasmid [gifted by H. Yawo (University of Tokyo, Japan)] using an In-Fusion HD cloning kit (Takara Bio, Japan). For cytochemistry, the plasmid containing the SzR3 gene was inserted N-QKLISEEDL-C (10 amino acids, c-Myc epitope tag) in the C terminus of eYFP using inverse polymerase chain reaction. The oligonucleotides used for the reaction are listed in table S5. All the constructs were verified by DNA sequencing.
Cell culture
The electrophysiological assays of SzRs were performed using ND7/23 cells, that is, hybrid cell lines derived from neonatal rat dorsal root ganglia neurons fused with mouse neuroblastoma (42). ND7/23 cells were purchased from DS Pharma Biomedical Co. Ltd. (Osaka, Japan) and cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) (Wako, Osaka, Japan) supplemented with 5% fetal bovine serum at 37°C and incubated in 5% CO2. For the electrophysiological recordings, the expression plasmids were transiently transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. For cytochemistry, the expression plasmids were transiently transfected using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Cells were supplemented with 1 μM all-trans retinal (Sigma-Aldrich, St. Louis, MO, USA) for the electrophysiological recordings or 2.5 μM all-trans retinal for the cytochemical assay after transfection. Electrophysiological recordings and cytochemical assay were then conducted 24 to 36 hours after the transfection. Successfully transfected cells were identified by eYFP fluorescence under a microscope.
Electrophysiological recordings
Whole-cell patch clamp recordings on ND7/23 cells were performed using an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA, USA). Continuous light was illuminated using collimated light-emitting diodes (LEDs) (part no. LCS-0480-03-22, Mightex, CA, USA) connected to an IX-70 inverted microscope (Olympus, Tokyo, Japan). For an action spectrum measurement, continuous light by opto-spectrum generator L12194-00-39070 (Hamamatsu Photonics, Shizuoka, Japan) was introduced into an IX-70 microscope via a light guide. Illumination was controlled by a mechanical shutter LS6S (Vincent Associates, Rochester, NY, USA). The light power was directly measured using objective lens microscopy (LP1, Sanwa Electric Instruments Co. Ltd., Tokyo, Japan). Glass pipettes were fabricated using a micropipette puller, PC-100 (Narishige, Tokyo, Japan), and fire polished using a micro forge, MF-830 (Narishige, Tokyo, Japan). The resistance of the pipettes was between 2.0 and 3.0 megaohms, and the pipette electrode was controlled by a micromanipulator, uMp-3 (Sensapex, Oulu, Finland). Traces were recorded at 10 kHz and filtered to 2 kHz using the internal circuit of the amplifier. Data acquisition and shutter triggering were performed and filtered using pClamp 9 software via a Digidata 1320 (Molecular Devices, Sunnyvale, CA, USA). The data were analyzed using Clampfit 10 (Molecular Devices, Sunnyvale, CA, USA) and Origin 7 (Origin Lab Corporation, Northampton, MA, USA).
The standard external solution contained 140 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 2 mM KCl, and 10 mM Hepes-NaOH (pH 7.2). The high-pH solution contained 140 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 2 mM KCl, and 10 mM glycine (pH 9.0). The standard internal solution contained 110 mM NaCl, 2 mM MgCl2, 1 mM CaCl2, 5 mM KCl, 10 mM EGTA, and 10 mM Hepes-NaOH (pH 7.2). The osmolality of the solutions was adjusted to 300 mOsm by adding the appropriate amount of sucrose.
Cytochemistry
Live cultured ND7/23 cells on glass coverslips were washed with phosphate-buffered saline (PBS). Two cell samples were changed from a cultured medium to nonserum DMEM and reacted with rabbit anti–c-Myc primary antibody (C3956; Sigma-Aldrich, St. Louis, MO, USA) at 1:500 dilution for 60 min under a 5% CO2 atmosphere at 37°C. The cells were washed with PBS twice before fixing with 4% paraformaldehyde phosphate buffer solution (Nacalai, Kyoto, Japan) for 15 min at room temperature. The cells were washed with PBS three times before labeling with goat anti-rabbit IgG secondary antibody Alexa Fluor 594 (A-11037; Thermo Fisher Scientific, Waltham, MA, USA) at 1:200 dilution for 2 hours at room temperature. One sample was fixed in 4% paraformaldehyde phosphate buffer solution for 15 min at room temperature. The cells were washed with PBS three times and then permeabilized with 0.5% Triton X-100 for 15 min at room temperature. The cells were treated with blocking buffer consisting of 3% goat serum for 60 min at room temperature. Then, the cells were washed with PBS three times and labeled with goat anti-rabbit IgG secondary antibody Alexa Fluor 594. After a final wash with PBS, the coverslips were mounted on glass slides with ProLong Diamond Antifade Mountant (Thermo Fisher Scientific, MA, USA). Localization was assessed using a IX83/FV3000 confocal laser scanning microscope (Olympus, Tokyo, Japan) equipped with a PlanApo N 60×/1.42 numerical aperture (NA) oil objective (Olympus, Tokyo, Japan), and a Z-drift compensator system (IX3-ZDC2, Olympus, Tokyo, Japan). The captured images were analyzed with Fiji software (43).
Protein purification
The expressed SzR1 and SzR2 were purified from E. coli cells. The cells were disrupted using a French Press (Ohtake, Japan), and the membrane fraction was collected by ultracentrifugation (125,000g, 1 hour). The protein was solubilized with 2% DDM (D310, Anatrace, OH) in the presence of 300 mM NaCl, 5 mM imidazole, and 50 mM MES (pH 6.5). After Co2+–NTA (nitrilotriacetic acid) affinity chromatography, the collected fractions were dialyzed using a solution containing 0.1% DDM, 100 mM NaCl, and 50 mM tris-HCl (pH 8.0) to remove the imidazole used for the column elution.
CD spectroscopy measurement
For the CD spectroscopy, the rhodopsins were solubilized in 0.1% DDM, 100 mM NaCl, and 50 mM tris-HCl (pH 8.0). The concentration of rhodopsin was adjusted to an absorption of 0.8 to 0.9 for the visible absorption peak of retinal. CD spectra were obtained using a CD spectrometer (J-1500, JASCO, Tokyo, Japan). The spectra were scanned 24 times at 300 to 750 nm to obtain higher signal-to-noise (S/N) ratios at a scan rate of 200 nm/min at 25°C.
High-speed AFM imaging
For the high-speed AFM imaging, SzR1 was reconstituted in a lipid bilayer according to the nanodisc protocol (44). Briefly, SzR1 (~5 nM) solubilized in 0.1% DDM, membrane scaffold proteins (MSP1E3D1) (M7074; Sigma-Aldrich, MO, USA) (~30 μM), and asolectin (11145; Sigma-Aldrich, MO) (~120 μg) were mixed in a solution containing 20 mM Hepes (pH 7.4) and 100 mM NaCl. The mixture was incubated for 1 hour at 4°C and then Bio-Beads (SM-2, Bio-Rad, CA, USA) (60 mg) were added to remove the detergent. After mixing overnight at 4°C, the Bio-Beads were removed by centrifugation at 14,000 rpm and 4°C for 1 min. The nanodisc products were purified by a 0.45-μm syringe filter (Merck Millipore, MA, USA).
Laboratory-built high-speed AFM operated in tapping mode was used (45) to image SzR1 in the nanodisc. Cantilever deflection was detected using an optical beam deflection method with an infrared laser (780 nm), which was focused onto the back side of a miniature cantilever (BL-AC7DS-KU4, Olympus, Japan) through a 20× objective lens (CFI S Plan Fluor ELWD 20×, Nikon, Japan). Its spring constant, resonant frequency, and quality factor in an aqueous solution were ~0.2 N/m, ~1 MHz, and ~2, respectively. An amorphous carbon tip was grown on the original bird-beak–like tip by electron beam deposition and then sharpened to a ~4-nm radius by plasma etching under argon gas. In the tapping-mode imaging, the free oscillation amplitude was ~1 nm, and the set-point amplitude was set to be 90% of the free amplitude to obtain a tapping force less than 30 pN. All AFM observations were performed in a solution containing 20 mM tris-HCl (pH 8.0) and 100 mM NaCl at room temperature.
HPLC analysis of retinal configuration
The high-performance liquid chromatograph was equipped with a silica column (6.0 × 150 mm; YMC-Pack SIL, YMC, Japan), a pump (PU-2080, JASCO, Japan), and a UV-vis detector (UV-2070, JASCO, Japan). The solvent was composed of 12% (v/v) ethyl acetate and 0.12% (v/v) ethanol in hexane with a flow rate of 1.0 ml min−1. Retinal oxime was formed by a hydrolysis reaction with the sample in 100-μl solution at 0.1 mg ml−1 protein concentration and 50-μl hydroxylamine solution at 1 M at 0°C. To ensure all the protein molecules reacted completely, 300 μl of methanol was added to denature the proteins. For light-adapted SzR1, the sample solution was illuminated with λ > 500 nm light (Y-52, AGC Techno Glass, Japan) for 1 min before denaturation and extraction. Then, the retinal oxime was extracted using hexane, and 300 μl of solution was injected into the HPLC system. The molar composition of the retinal isomers was calculated from the areas of the corresponding peaks in the HPLC patterns. The assignment of the peaks was performed by comparing them with the HPLC pattern from retinal oximes of authentic all-trans, 13-cis, and 11-cis retinals. To estimate the experimental error, three independent measurements were carried out.
Laser flash photolysis
For the laser flash photolysis measurement, SzR1 and SzR2 were purified and reconstituted into a mixture of POPE (Avanti Polar Lipids, AL, USA) and POPG (sodium salt) (Avanti Polar Lipids, AL, USA) (molar ratio, 3:1) with a protein-to-lipid molar ratio of 1:50 in 100 mM NaCl and 20 mM tris-HCl (pH 8.0). The absorption of the protein solution was adjusted to 0.8 to 0.9 (total protein concentration, ~0.15 mg ml−1) at an excitation wavelength of 532 nm. The sample was illuminated with a beam of second harmonics of a nanosecond-pulsed Nd3+-YAG laser (λ = 532 nm, 3 mJ pulse−1, 2 Hz) (INDI40, Spectra-Physics Inc., CA, USA). The time evolution of transient absorption change was obtained by observing the intensity change of an output of an Xe arc lamp (L9289-01, Hamamatsu Photonics, Japan), monochromated by a monochrometer (S-10, Soma Optics Ltd., Japan), and passed through the sample, after photoexcitation by a photomultiplier tube (R10699, Hamamatsu Photonics, Japan) equipped with a notch filter (532 nm; bandwidth, 17 nm; Semrock, NY, USA) to remove the scattered pump pulse. Transient absorption spectra were reconstructed from the time evolution of the change in transient absorption at various wavelengths from 360 to 690 nm at 10-nm intervals. To increase the S/N ratio, 100 spectra were averaged. The signals were global fitted with a multiexponential function to obtain the lifetimes of each photointermediate.
The assignment of H+ release and uptake by rhodopsin was carried out by observing the transient absorption change of the pH-indicating dye, cresol red (5). To minimize the buffering effect by the solvent, any buffer was removed from the solvent, and the protein was solubilized in 0.1% DDM and 100 mM NaCl. The pH was adjusted to about 8.1, close to the pKa of cresol red, by adding NaOH. The transient absorption change of cresol red was obtained by subtracting the transient absorption signals at 429 and 573 nm (representing the protonated and deprotonated form of cresol red, respectively) obtained without cresol red from those obtained with 50 μM cresol red.
Light-induced low-temperature FTIR spectroscopy
The purified proteins of SzR1 were reconstituted into a mixture of POPE and POPG membranes (molar ratio, 3:1) with a protein-to-lipid molar ratio of 1:20 by removing DDM using Bio-Beads (SM-2; Bio-Rad, CA, USA). The reconstituted samples were washed three times with 1 mM NaCl and 2 mM tris-HCl (pH 8.0). The pellet was resuspended in the same buffer, where the concentration was adjusted to make the intensity of amide I ~0.7. A 60-μl aliquot was placed onto a BaF2 window and dried gently at 4°C. The films were then rehydrated with 2 μl of H2O, or D2O, and allowed to stand at room temperature for 15 min to complete the hydration. The sample was then placed and cooled in an Oxford Optistat DN2 cryostat mounted in a Cary670 spectrometer (Agilent Technologies, Japan). For the measurement of the high frequency of the K and M intermediates, sample hydration was controlled using 30% (v/v) d-glycerol D2O, or D218O, at an O-D band intensity of around 1.
For the formation of the K intermediate, samples of SzR1 were illuminated with 520-nm light (interference filter) from a 1-kW tungsten-halogen projector lamp (Rikagaku, Japan) for 2 min at 110 K. The K intermediate was photo reversed with λ > 590 nm light (R-61 cutoff filter, Toshiba, Japan) for 1 min, followed by illumination with 540-nm light. For the formation of the L intermediate, samples of SzR1 were illuminated with λ > 520 nm light (O-54 cutoff filter, Toshiba, Japan) from a 1-kW halogen-tungsten lamp for 1 min at 190 K. The L intermediate was photo reversed with 460-nm light (interference filter) for 2 min, followed by illumination with λ > 520 nm light. For the formation of the L/M intermediate, samples of SzR1 were illuminated with λ > 520 nm light (O-54 cutoff filter, Toshiba, Japan) from a 1-kW halogen-tungsten lamp for 1 min at 210 K. The L/M intermediate was photo reversed with the 460-nm light (interference filter) for 2 min and 400-nm light (interference filter) for 2 min, followed by illumination with λ > 520 nm light. For the formation of the M intermediate, samples of SzR1 were illuminated with λ > 500 nm light (Y-52 cutoff filter, Toshiba, Japan) from a 1-kW halogen-tungsten lamp for 1 min at 230 K. The M intermediate was photo reversed with the 400-nm light (interference filter) for 2 min, followed by illumination with λ > 500 nm light. For each measurement, 128 interferograms were accumulated with 2-cm−1 spectral resolution, and 10 to 95 identical recordings were averaged for each measurement.
Light-induced low-temperature UV-vis spectroscopy
The sample film preparation, film hydration with H2O, and illumination method to measure K intermediate were the same as FTIR spectroscopy. The sample was placed and cooled in an Optistat DN cryostat (Oxford Instruments, Abingdon, UK) mounted in a UV-vis spectrometer (V-550, JASCO, Japan).
Supplementary Material
Acknowledgments
We thank M. Iwatani for technical support and S. Tsukiji and T. Yoshii for help with confocal microscopy. Funding: This work was supported by the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (KAKENHI), Japan (grant numbers 17H03007 to K.I., 19H05389, 18H04512, and 18H01837 to T.U., 18K06109 to S.P.T., and 15H02391 and 19H04959 to H.K.) and by the Japan Science and Technology Agency (JST), PRESTO, Japan (grant numbers JPMJPR15P2 to K.I. and JPMJPR1688 to S.P.T.). O.B. received support from the Louis and Lyra Richmond Memorial Chair in Life Sciences. R.G. was supported by the research grant 17-04828S (Grant Agency of the Czech Republic). P.-A.B. was supported by the research grant PN-III-P4-ID-PCE-2016-0303 (Romanian National Authority for Scientific Research). H.L.B. was supported by the research grants PN-III-P4-ID-PCE-2016-0303 (Romanian National Authority for Scientific Research) and STAR-UBB Advanced Fellowship-Intern (Babeș-Bolyai University, Cluj-Napoca, Romania). A.-S.A. was supported by the research grants 17-04828S (Grant Agency of the Czech Republic) and MSM200961801 (Academy of Sciences of the Czech Republic). Author contributions: K.I., R.G., O.B., and H.K. contributed to the study design. R.N. constructed the DNA plasmids of mutant proteins and introduced them into E. coli cells. M.K. and R.N. conducted the pump activity assay of SzR WTs and mutants in E. coli cells. S.P.T. performed the electrophysiological measurement of inward H+ transport of SzR in mammalian cells. S.H. and S.P.T. performed fluorescence immunostaining microscopy to determine the molecular orientation of SzR in mammalian cells. M.S. purified the SzR proteins from E. coli cells and performed CD spectroscopy, HPLC analysis, and the pH titration experiment. T.U. and H.W. performed HS-AFM observation of SzR in lipid membrane. S.T. measured the FTIR spectra. K.I. and M.S. conducted the transient absorption measurement and analysis of the kinetic data. K.I. conducted the phylogenetic analysis of rhodopsins. R.G., A.-S.A., P.-A.B., and H.L.B. identified SzR sequences and performed initial sequence analyses. K.I., O.B., and H.K. wrote the paper. All authors discussed and commented on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/15/eaaz2441/DC1
REFERENCES AND NOTES
- 1.Ernst O. P., Lodowski D. T., Elstner M., Hegemann P., Brown L. S., Kandori H., Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem. Rev. 114, 126–163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Philosof A., Béjà O., Bacterial, archaeal and viral-like rhodopsins from the Red Sea. Environ. Microbiol. Rep. 5, 475–482 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Mukherjee S., Hegemann P., Broser M., Enzymerhodopsins: Novel photoregulated catalysts for optogenetics. Curr. Opin. Struct. Biol. 57, 118–126 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Govorunova E. G., Sineshchekov O. A., Hemmati R., Janz R., Morelle O., Melkonian M., Wong G. K.-S., Spudich J. L., Extending the time domain of neuronal silencing with cryptophyte anion channelrhodopsins. eNeuro 5, ENEURO.0174-18.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pushkarev A., Inoue K., Larom S., Flores-Uribe J., Singh M., Konno M., Tomida S., Ito S., Nakamura R., Tsunoda S. P., Philosof A., Sharon I., Yutin N., Koonin E. V., Kandori H., Béjà O., A distinct abundant group of microbial rhodopsins discovered using functional metagenomics. Nature 558, 595–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spang A., Saw J. H., Jørgensen S. L., Zaremba-Niedzwiedzka K., Martijn J., Lind A. E., van Eijk R., Schleper C., Guy L., Ettema T. J. G., Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spang A., Eme L., Saw J. H., Caceres E. F., Zaremba-Niedzwiedzka K., Lombard J., Guy L., Ettema T. J. G., Asgard archaea are the closest prokaryotic relatives of eukaryotes. PLoS Genet. 14, e1007080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulzu P.-A., Andrei A.-Ş., Salcher M. M., Mehrshad M., Inoue K., Kandori H., Béjà O., Ghai R., Banciu H. L., Casting light on Asgardarchaeota metabolism in a sunlit microoxic niche. Nat. Microbiol. 4, 1129–1137 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Inoue K., Ito S., Kato Y., Nomura Y., Shibata M., Uchihashi T., Tsunoda S. P., Kandori H., A natural light-driven inward proton pump. Nat. Commun. 7, 13415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shevchenko V., Mager T., Kovalev K., Polovinkin V., Alekseev A., Juettner J., Chizhov I., Bamann C., Vavourakis C., Ghai R., Gushchin I., Borshchevskiy V., Rogachev A., Melnikov I., Popov A., Balandin T., Rodriguez-Valera F., Manstein D. J., Bueldt G., Bamberg E., Gordeliy V., Inward H+ pump xenorhodopsin: Mechanism and alternative optogenetic approach. Sci. Adv. 3, e1603187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue S., Yoshizawa S., Nakajima Y., Kojima K., Tsukamoto T., Kikukawa T., Sudo Y., Spectroscopic characteristics of Rubricoccus marinus xenorhodopsin (RmXeR) and a putative model for its inward H+ transport mechanism. Phys. Chem. Chem. Phys. 20, 3172–3183 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Sunagawa S., Coelho L. P., Chaffron S., Kultima J. R., Labadie K., Salazar G., Djahanschiri B., Zeller G., Mende D. R., Alberti A., Cornejo-Castillo F. M., Costea P. I., Cruaud C., d'Ovidio F., Engelen S., Ferrera I., Gasol J. M., Guidi L., Hildebrand F., Kokoszka F., Lepoivre C., Lima-Mendez G., Poulain J., Poulos B. T., Royo-Llonch M., Sarmento H., Vieira-Silva S., Dimier C., Picheral M., Searson S., Kandels-Lewis S.; Tara Oceans coordinators, Bowler C., de Vargas C., Gorsky G., Grimsley N., Hingamp P., Iudicone D., Jaillon O., Not F., Ogata H., Pesant S., Speich S., Stemmann L., Sullivan M. B., Weissenbach J., Wincker P., Karsenti E., Raes J., Acinas S. G., Bork P., Structure and function of the global ocean microbiome. Science 348, 1261359 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Brum J. R., Ignacio-Espinoza J. C., Roux S., Doulcier G., Acinas S. G., Alberti A., Chaffron S., Cruaud C., de Vargas C., Gasol J. M., Gorsky G., Gregory A. C., Guidi L., Hingamp P., Iudicone D., Not F., Ogata H., Pesant S., Poulos B. T., Schwenck S. M., Speich S., Dimier C., Kandels-Lewis S., Picheral M., Searson S.; Tara Oceans Coordinators, Bork P., Bowler C., Sunagawa S., Wincker P., Karsenti E., Sullivan M. B., Patterns and ecological drivers of ocean viral communities. Science 348, 1261498 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Hingamp P., Grimsley N., Acinas S. G., Clerissi C., Subirana L., Poulain J., Ferrera I., Sarmento H., Villar E., Lima-Mendez G., Faust K., Sunagawa S., Claverie J.-M., Moreau H., Desdevises Y., Bork P., Raes J., de Vargas C., Karsenti E., Kandels-Lewis S., Jaillon O., Not F., Pesant S., Wincker P., Ogata H., Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 7, 1678–1695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philosof A., Yutin N., Flores-Uribe J., Sharon I., Koonin E. V., Béjà O., Novel abundant oceanic viruses of uncultured marine group II euryarchaeota. Curr. Biol. 27, 1362–1368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nack M., Radu I., Gossing M., Bamann C., Bamberg E., von Mollard G. F., Heberle J., The DC gate in Channelrhodopsin-2: Crucial hydrogen bonding interaction between C128 and D156. Photochem. Photobiol. Sci. 9, 194–198 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Scheib U., Stehfest K., Gee C. E., Körschen H. G., Fudim R., Oertner T. G., Hegemann P., The rhodopsin–guanylyl cyclase of the aquatic fungus Blastocladiella emersonii enables fast optical control of cGMP signaling. Sci. Signal. 8, rs8 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Watari M., Ikuta T., Yamada D., Shihoya W., Yoshida K., Tsunoda S. P., Nureki O., Kandori H., Spectroscopic study of the transmembrane domain of a rhodopsin–phosphodiesterase fusion protein from a unicellular eukaryote. J. Biol. Chem. 294, 3432–3443 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue K., Kato Y., Kandori H., Light-driven ion-translocating rhodopsins in marine bacteria. Trends Microbiol. 23, 91–98 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Inoue K., Tsukamoto T., Shimono K., Suzuki Y., Miyauchi S., Hayashi S., Kandori H., Sudo Y., Converting a light-driven proton pump into a light-gated proton channel. J. Am. Chem. Soc. 137, 3291–3299 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Fudim R., Szczepek M., Vierock J., Vogt A., Schmidt A., Kleinau G., Fischer P., Bartl F., Scheerer P., Hegemann P., Design of a light-gated proton channel based on the crystal structure of Coccomyxa rhodopsin. Sci. Signal. 12, eaav4203 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Shibata M., Inoue K., Ikeda K., Konno M., Singh M., Kataoka C., Abe-Yoshizumi R., Kandori H., Uchihashi T., Oligomeric states of microbial rhodopsins determined by high-speed atomic force microscopy and circular dichroic spectroscopy. Sci. Rep. 8, 8262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Druckmann S., Ottolenghi M., Pande A., Pande J., Callender R. H., Acid-base equilibrium of the Schiff base in bacteriorhodopsin. Biochemistry 21, 4953–4959 (1982). [DOI] [PubMed] [Google Scholar]
- 24.Inoue K., Tahara S., Kato Y., Takeuchi S., Tahara T., Kandori H., Spectroscopic study of proton-transfer mechanism of inward proton-pump rhodopsin, Parvularcula oceani xenorhodopsin. J. Phys. Chem. B 122, 6453–6461 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Aton B., Doukas A. G., Callender R. H., Becher B., Ebrey T. G., Resonance Raman studies of the purple membrane. Biochemistry 16, 2995–2999 (1977). [DOI] [PubMed] [Google Scholar]
- 26.Rothschild K. J., Marrero H., Braiman M., Mathies R., Primary photochemistry of bacteriorhodopsin: Comparison of Fourier transform infrared difference spectra with resonance Raman spectra. Photochem. Photobiol. 40, 675–679 (1984). [DOI] [PubMed] [Google Scholar]
- 27.Lórenz-Fonfría V. A., Muders V., Schlesinger R., Heberle J., Changes in the hydrogen-bonding strength of internal water molecules and cysteine residues in the conductive state of channelrhodopsin-1. J. Chem. Phys. 141, 22D507 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Ito S., Kato H. E., Taniguchi R., Iwata T., Nureki O., Kandori H., Water-containing hydrogen-bonding network in the active center of channelrhodopsin. J. Am. Chem. Soc. 136, 3475–3482 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Muroda K., Nakashima K., Shibata M., Demura M., Kandori H., Protein-bound water as the determinant of asymmetric functional conversion between light-driven proton and chloride pumps. Biochemistry 51, 4677–4684 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Ito S., Sugita S., Inoue K., Kandori H., FTIR analysis of a light-driven inward proton-pumping rhodopsin at 77 K. Photochem. Photobiol. 93, 1381–1387 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Petrovskaya L. E., Lukashev E. P., Chupin V. V., Sychev S. V., Lyukmanova E. N., Kryukova E. A., Ziganshin R. H., Spirina E. V., Rivkina E. M., Khatypov R. A., Erokhina L. G., Gilichinsky D. A., Shuvalov V. A., Kirpichnikov M. P., Predicted bacteriorhodopsin from Exiguobacterium sibiricum is a functional proton pump. FEBS Lett. 584, 4193–4196 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Kawanabe A., Furutani Y., Jung K.-H., Kandori H., Engineering an inward proton transport from a bacterial sensor rhodopsin. J. Am. Chem. Soc. 131, 16439–16444 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Doig S. J., Reid P. J., Mathies R. A., Picosecond time-resolved resonance Raman spectroscopy of bacteriorhodopdn's J, K, and KL intermediates. J. Phys. Chem. 95, 6372–6379 (1991). [Google Scholar]
- 34.Garczarek F., Gerwert K., Functional waters in intraprotein proton transfer monitored by FTIR difference spectroscopy. Nature 439, 109–112 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Gerwert K., Freier E., Wolf S., The role of protein-bound water molecules in microbial rhodopsins. Biochim. Biophys. Acta 1837, 606–613 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Lórenz-Fonfría V. A., Kandori H., Spectroscopic and kinetic evidence on how bacteriorhodopsin accomplishes vectorial proton transport under functional conditions. J. Am. Chem. Soc. 131, 5891–5901 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Nango E., Royant A., Kubo M., Nakane T., Wickstrand C., Kimura T., Tanaka T., Tono K., Song C., Tanaka R., Arima T., Yamashita A., Kobayashi J., Hosaka T., Mizohata E., Nogly P., Sugahara M., Nam D., Nomura T., Shimamura T., Im D., Fujiwara T., Yamanaka Y., Jeon B., Nishizawa T., Oda K., Fukuda M., Andersson R., Båth P., Dods R., Davidsson J., Matsuoka S., Kawatake S., Murata M., Nureki O., Owada S., Kameshima T., Hatsui T., Joti Y., Schertler G., Yabashi M., Bondar A.-N., Standfuss J., Neutze R., Iwata S., A three-dimensional movie of structural changes in bacteriorhodopsin. Science 354, 1552–1557 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Balashov S. P., Govindjee R., Kono M., Imasheva E., Lukashev E., Ebrey T. G., Crouch R. K., Menick D. R., Feng Y., Effect of the arginine-82 to alanine mutation in bacteriorhodopsin on dark adaptation, proton release, and the photochemical cycle. Biochemistry 32, 10331–10343 (1993). [DOI] [PubMed] [Google Scholar]
- 39.Inoue K., Ono H., Abe-Yoshizumi R., Yoshizawa S., Ito H., Kogure K., Kandori H., A light-driven sodium ion pump in marine bacteria. Nat. Commun. 4, 1678 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Ikeura Y., Shimono K., Iwamoto M., Sudo Y., Kamo N., Arg-72 of pharaonis phoborhodopsin (sensory rhodopsin II) is important for the maintenance of the protein structure in the solubilized states. Photochem. Photobiol. 77, 96–100 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Tanimoto T., Shibata M., Belenky M., Herzfeld J., Kandori H., Altered hydrogen bonding of Arg82 during the proton pump cycle of bacteriorhodopsin: A low-temperature polarized FTIR spectroscopic study. Biochemistry 43, 9439–9447 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Wood J. N., Bevan S. J., Coote P. R., Dunn P. M., Harmar A., Hogan P., Latchman D. S., Morrison C., Rougon G., Theveniau M., Wheatley S., Novel cell lines display properties of nociceptive sensory neurons. Proc. Biol. Sci. 241, 187–194 (1990). [DOI] [PubMed] [Google Scholar]
- 43.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bayburt T. H., Grinkova Y. V., Sligar S. G., Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett. 2, 853–856 (2002). [Google Scholar]
- 45.Uchihashi T., Kodera N., Ando T., Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat. Protoc. 7, 1193–1206 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/15/eaaz2441/DC1


