Abstract
The majority of the human genome does not encode proteins. Many of these noncoding regions contain important regulatory sequences that control gene expression. To date, most studies have focused on activators such as enhancers, but regions that repress gene expression—silencers—have not been systematically studied. We have developed a system that identifies silencer regions in a genome-wide fashion on the basis of silencer-mediated transcriptional repression of caspase 9. We found that silencers are widely distributed and may function in a tissue-specific fashion. These silencers harbor unique epigenetic signatures and are associated with specific transcription factors. Silencers also act at multiple genes, and at the level of chromosomal domains and long-range interactions. Deletion of silencer regions linked to the drug transporter genes ABCC2 and ABCG2 caused chemo-resistance. Overall, our study demonstrates that tissue-specific silencing is widespread throughout the human genome and likely contributes substantially to the regulation of gene expression and human biology.
Less than 2% of the 3 billion base pairs in the human genome codes for proteins1,2. The majority is non-protein-coding, and includes repeat regions, noncoding RNAs, gene introns and other intergenic regions1. Individual laboratories as well as large consortia such as ENCODE (Encyclopedia of DNA Elements) and Roadmap Epigenomics have made enormous contributions to annotating the noncoding genome with epigenetic modifications and transcription factor binding sites3–8. Based on the profiling of epigenetic modifications, the human genome can be categorized into distinct functional units such as enhancers, insulators and promoters9,10. Disruption of noncoding regulatory regions has been shown to have deleterious effects and cause cancer11–14. However, most research has focused on defining enhancers, insulators and promoters15–19. Although silencers are an important class of regulatory elements20, thus far, most studies have been performed on identifying and characterizing individual silencer regions (e.g. those that regulate CD4 gene transcription during T cell development21–25), and high-throughput methods have not been described to systematically identify genomic silencers. As such, we do not know how many silencers exist, how they operate at the level of chromosomal domains or whether they exhibit ubiquitous or cell-type-specific activity.
Here we present a method to identify and define the function of silencer regions in a systematic and high-throughput fashion. This method measures the repressive ability of silencer elements (ReSE) by screening for genomic fragments that repress the transcription of an inducible cell death protein. Using this method, we identified more than 5,000 tissue-specific candidate silencer elements in the human genome. These silencers are preferentially located in repressed and/or weakly transcribed regions and share unique epigenetic marks. They operate in topologically associating domains (TADs) and participate in long-range interactions. Deletion of silencers linked to drug transporter genes led to transcriptional up-regulation of these genes and promoted chemotherapy resistance, suggesting that genetic variation in silencer regions may impact both biology and personalized medicine.
Results
Identification of silencers using ReSE
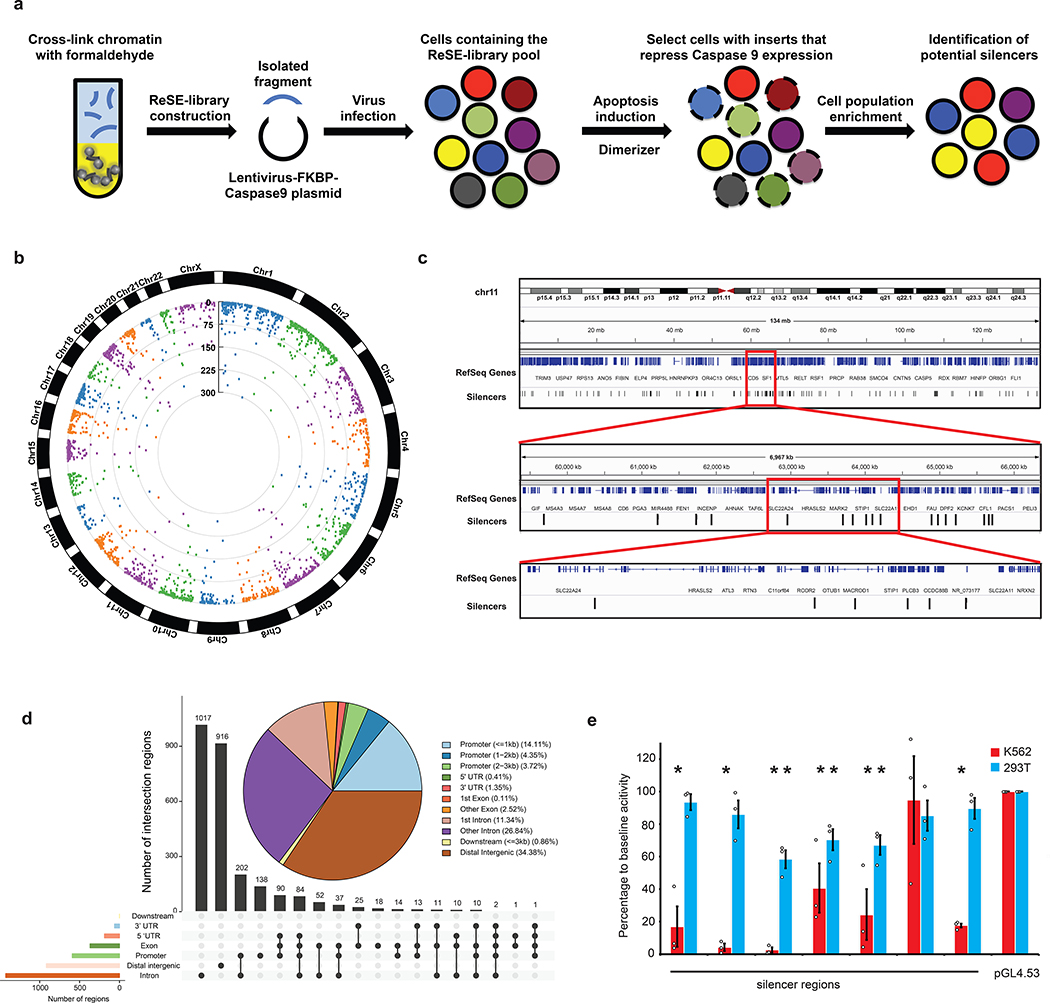
To systematically discover silencer regions in the human genome, a high-throughput ReSE lentiviral screen system was developed (Fig. 1a). In this system, genomic regions are cloned upstream of the EF-1α promoter that drives the expression of a modified caspase 9 fused to an FK506 binding protein (FKBP-Casp9)26,27. Upon the addition of a dimerizer molecule AP20187, the expressed caspase 9 is activated to induce apoptosis26,27. The system was designed such that if silencers are inserted, they will repress the transcription of the FKBP-Casp9 gene in the cells, and these cells will not undergo apoptosis. Surviving cells are then expanded and candidate inserts sequenced and mapped to the genome. This method allows for the systematic identification of silencer regions.
Figure 1. Identification of silencers using the ReSE screen.
(a) Outline of the screen design. Candidate sequences are cloned in front of the EF-1α promoter that drives the expression of a modified caspase 9 fused to an FK506 binding protein (FKBP-Casp9). Upon addition of a dimerizer molecule, the expressed caspase 9 is activated to induce apoptosis, unless a potential silencer fragment is inserted. Enriched fragments representing potential silencers are identified when compared to sequences from the cells not treated with the dimerizer.
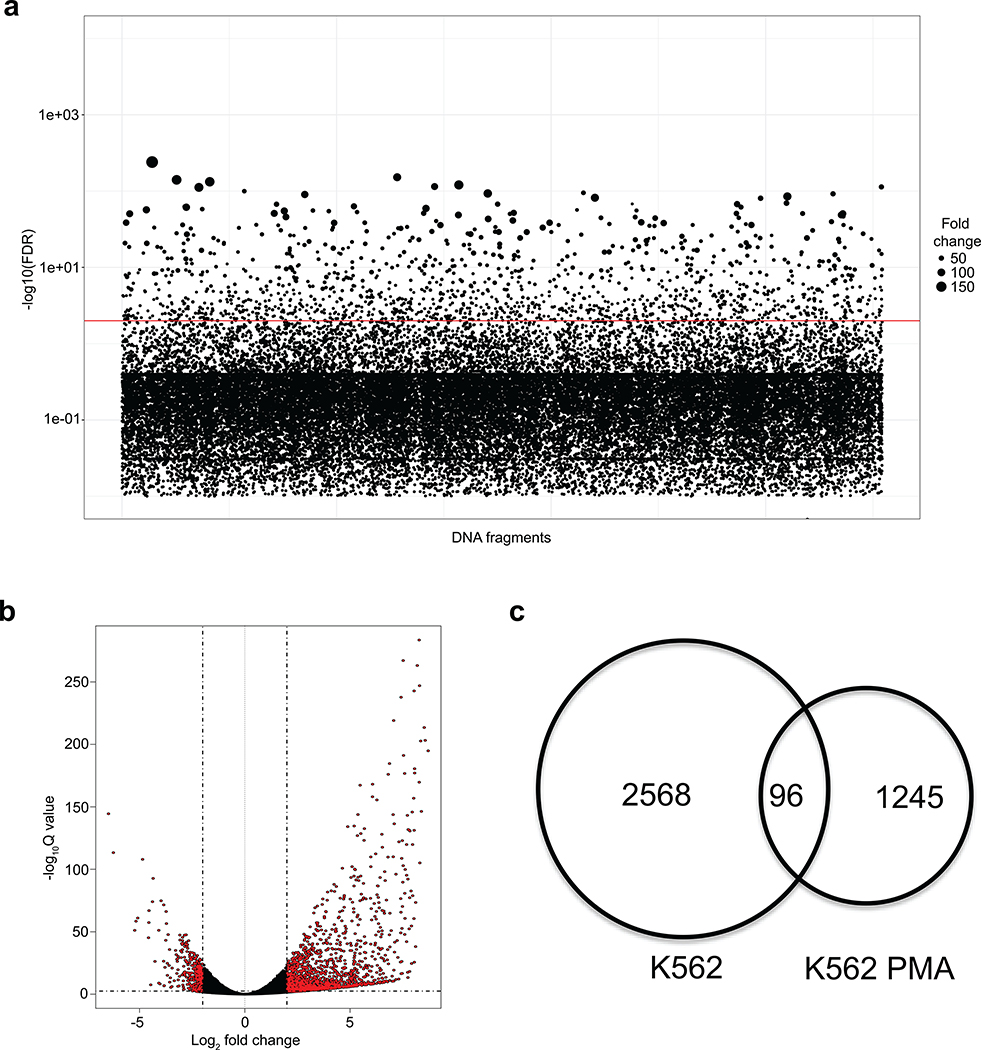
(b) ReSE screen results from two biological replicate experiments in K562 cells. Each dot represents one tested fragment. The position of the dot indicates the location of each fragment within the respective chromosome. The height of the dot indicates the –log10(FDR) of enrichment of the ReSE fragments after the induction of apoptosis compared with the cells not treated with dimerizer. The cutoff value of FDR is 0.01. See Methods for detailed information.
(c) Snapshot of distribution of silencers in K562 cells. Chromosome 11 is used as an example.
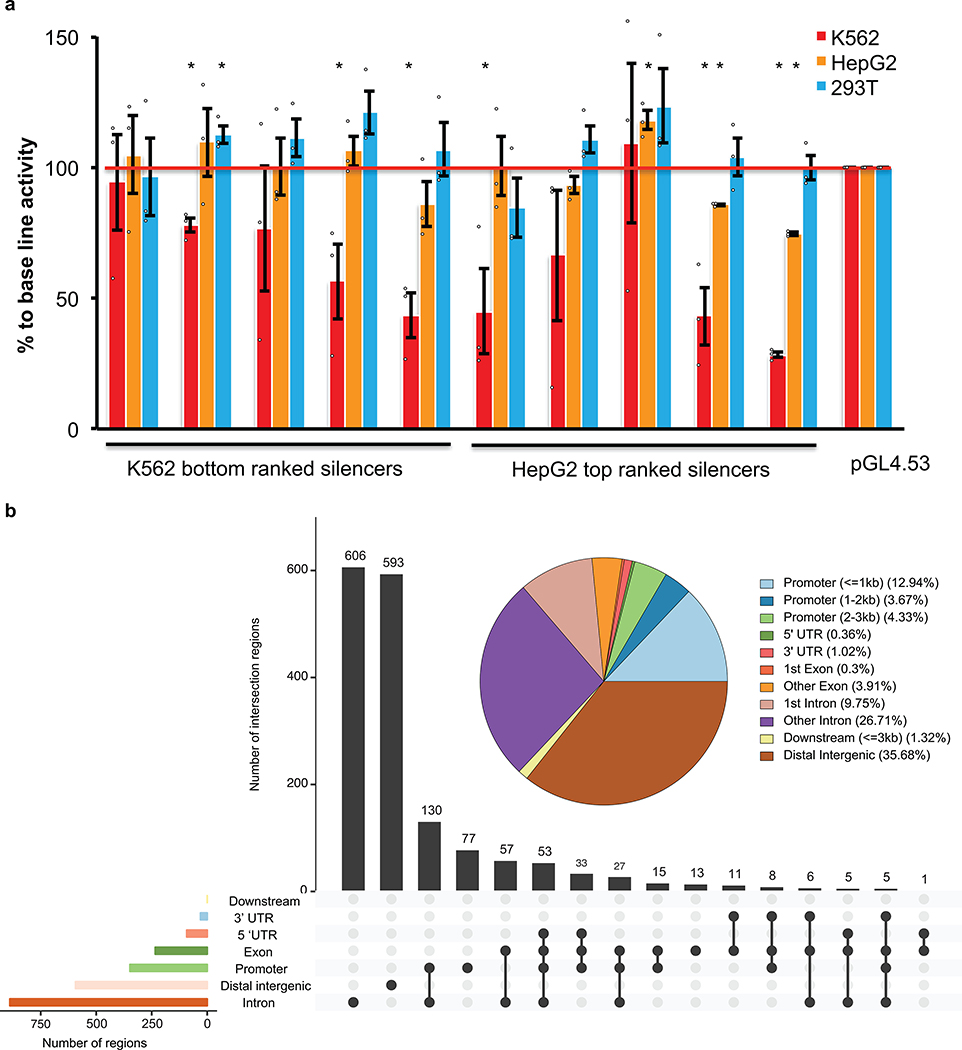
(d) Distribution of significantly enriched silencer regions from K562 cells in the genome. The pie chart indicates the distribution of silencers in genomic features. The bar plot indicates the distribution of silencers in overlapped annotation features in the genome.
(e) Luciferase assays to determine the repressive activity of identified silencers from K562 cells. Silencer regions were cloned by PCR from the genomic DNA of K562 cells, and then inserted upstream of the promoter of a luciferase reporter plasmid pGL4.53. 293T cells were used as the control cell line to control for cell-type-dependent repressive activity, and empty pGL4.53 plasmid was used as the control for baseline luciferase activity. The y-axis represents the percentage of luciferase activity compared to pGL4.53 empty plasmids in the respective cells. (n = 3 biological independent samples; the bars show the mean value ± S.E.M; * P value < 0.05, calculated using two-sided Student’s t test).
All exact P values are provided in the Source Data.
Presently, it is difficult to screen the entire human genome with small genomic fragments in a lentiviral assay. Therefore, an enrichment strategy was used. It has been shown that 94.4% of the combined transcription factor ChIP-seq peaks from the ENCODE project fall within accessible regions28. Many of these transcription factors are associated with transcriptionally repressive activities. Therefore, we expected that at least some silencers might lie in accessible chromatin regions, as shown for the regulation of the CD4 gene21–25. These silencers would likely harbor regulatory proteins rather than simply be regions that are globally repressed through general heterochromatin mechanisms.
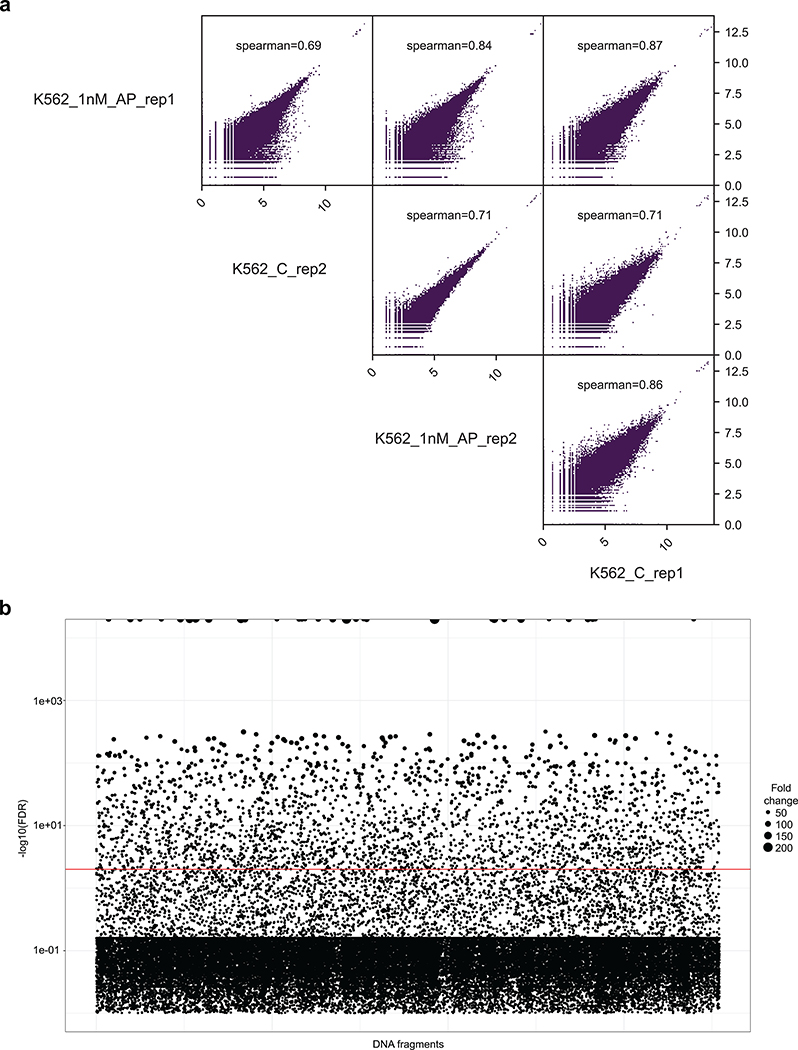
Accessible chromatin regions enriched by FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements)29 from chronic myeloid leukemia K562 cells were isolated to construct the ReSE screen library (Supplementary Fig. 1). Briefly, accessible chromatin regions of ~200 base pairs (bp) prepared from K562 cells were cloned into the ReSE lentiviral plasmids, and a library of more than 177,000 independent regions (covering 1% of the human genome; distribution in Supplementary Fig. 2) was constructed and used as the screening library. The library was transduced into K562 cells in two independent replicate experiments, and AP20187 added to induce apoptosis (Supplementary Fig. 3, more details in Method). The surviving cells were grown and the inserts were sequenced before and after selection. The screen has considerable background cell survival during the initial puromycin selection and the subsequent apoptosis induction. Therefore the fold enrichments are variable due to this and the low read counts (see Methods). Nonetheless, the results from the replicate experiments correlated (Extended Data Fig. 1a), although only a small percentage of the potential fragments were consistently enriched between replicates when fold-change was considered (Supplementary Fig. 4).
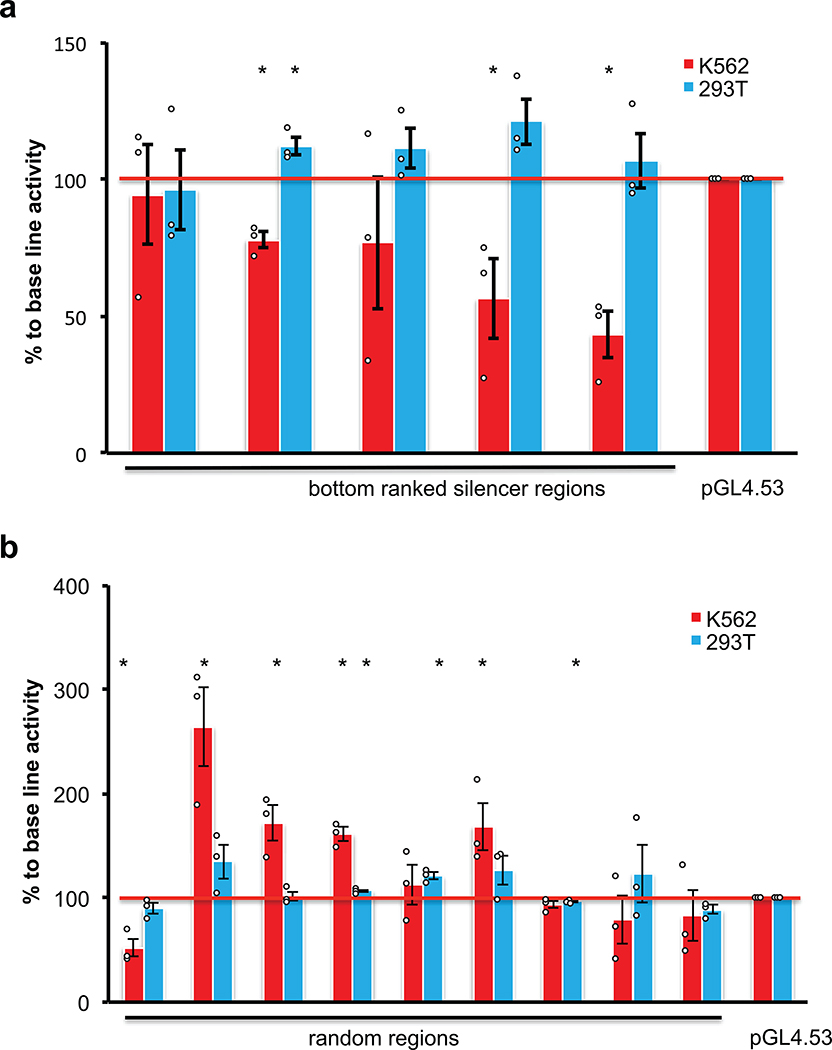
To reliably identify significantly enriched silencers, an algorithm based on a negative binomial (NB) model was adapted30, as used previously in CRISPR screen and other RNA-seq differential analyses30–32. This led to the identification of 2,664 potential silencer regions with an FDR cutoff of 0.01 in K562 cells (Fig. 1b and c, Extended Data Fig. 1b, Supplementary Fig. 5 and Supplementary Table 1). The majority of these potential silencer regions were in promoter regions, introns and intergenic regions (Fig. 1d). To validate the transcriptional repressive activity of the identified silencer regions, seven regions identified from the screen with the lowest FDR were cloned upstream of a luciferase plasmid with a phosphoglycerate kinase (PGK) promoter. Very strong repressive activity was observed from the silencer fragments in K562 cells in 6 of 7 cases (Fig. 1e). In addition, testing of five silencers from the bottom of our silencer list revealed that three showed significant repressive activity (Extended Data Fig. 2a), suggesting that the high threshold used for calling positives (FDR 0.01) was adequate. The majority of randomly selected regions (eight of nine) from the library did not show repressive ability (Extended Data Fig. 2b). These data indicate that most fragments identified in our screen are silencers, and in addition suggest that the activity of most silencers was not limited to the specific promoter used in the initial screen (i.e. the EF-1α promoter in Fig. 1a).
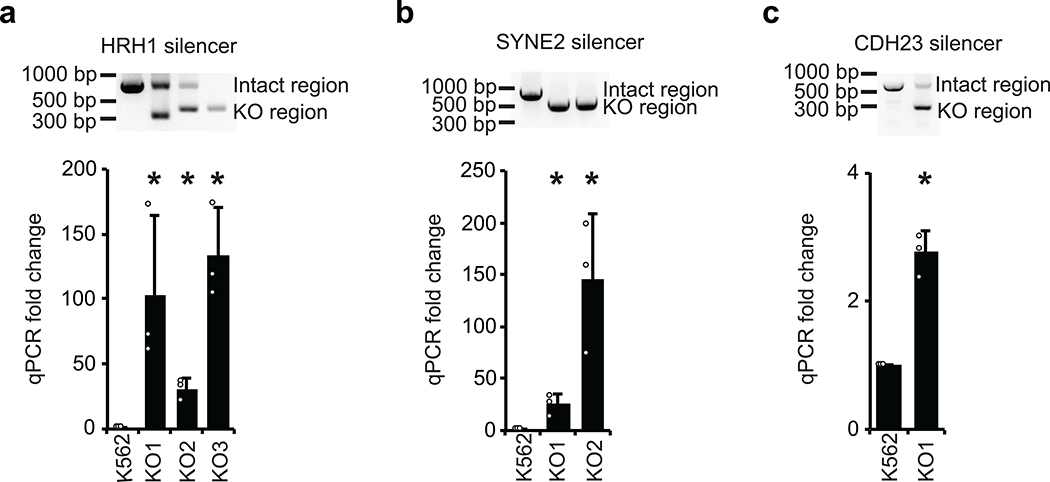
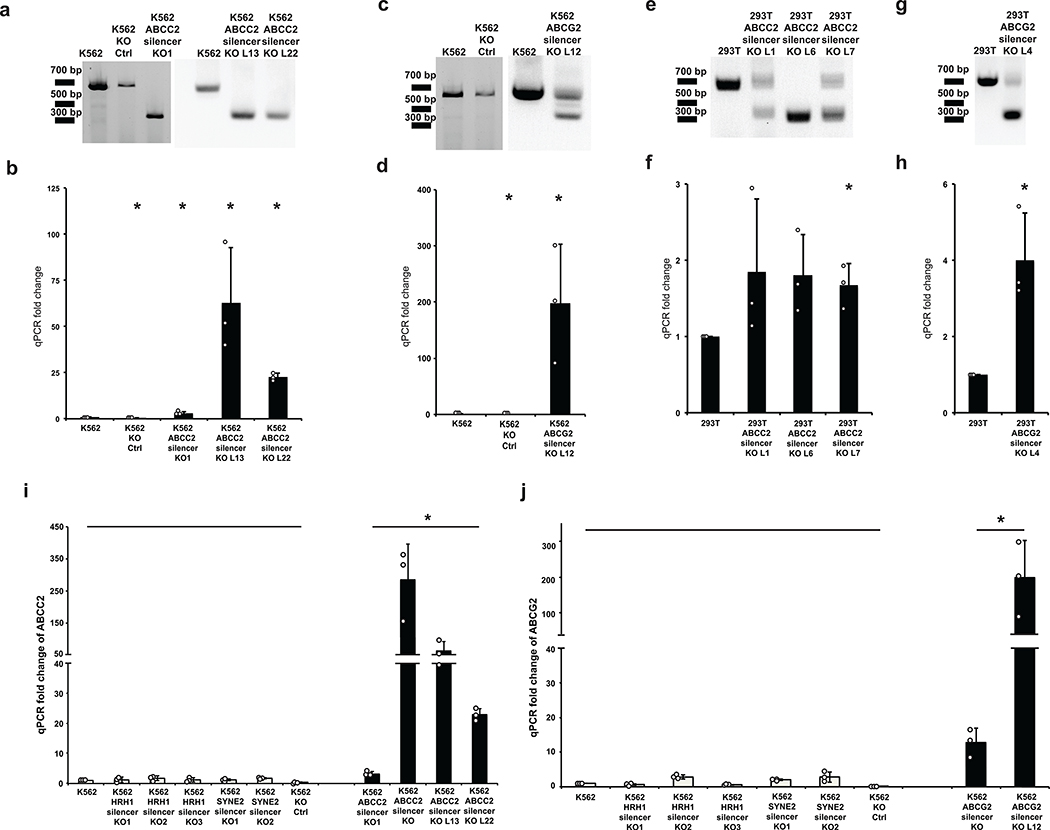
To test if the silencers can repress their native endogenous genes, three silencer regions from Figure 1e were deleted from their native loci. Silencer regions located in the intron regions of genes HRH1, SYNE2 and CDH23 were deleted using paired CRISPR guide RNAs that targeted both sides of the individual silencer region. K562 silencer-knockout clones were isolated, and significant up-regulation of the respective genes was observed in each case (Extended Data Fig. 3). These data indicate that silencers identified in the ReSE screen repress the transcription of their nearby endogenous genes.
ReSE identifies tissue-specific and conserved silencers
Since large-scale analysis of silencers has not been performed previously, we wanted to determine if they were common across cell types or whether they function in a tissue-specific manner, similar to enhancers33,34. We used the same screening library to test if a different pool of silencers was enriched during differentiation of K562 cells; the rationale was that if most silencers are common across cell types we would isolate many of the same DNA sequences found in the K562 screen. If the silencers are cell-type-specific, the overlap should be modest. K562 cells were treated with PMA to induce megakaryocytic differentiation. Repeating the ReSE screen in these PMA-treated cells identified a different set of 1,245 silencers compared to those identified in the original K562 cells (Extended Data Fig. 4 and Supplementary Table 2). This result suggests that silencers may function in a tissue-specific manner.
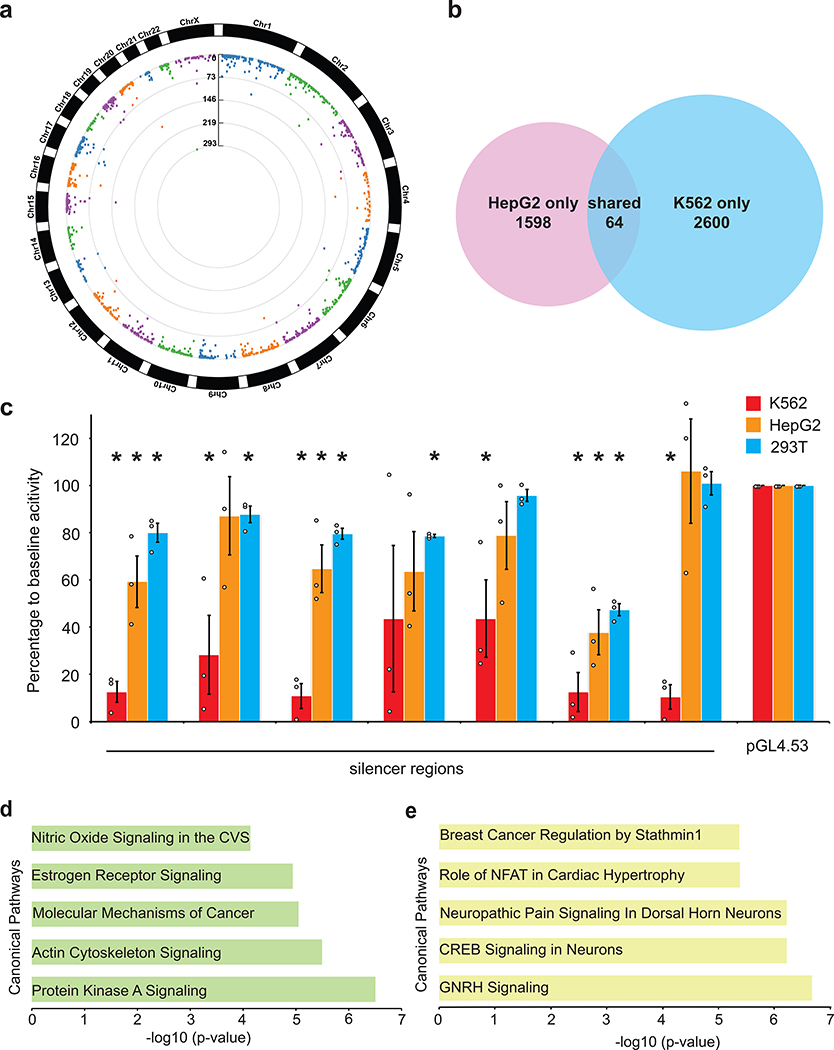
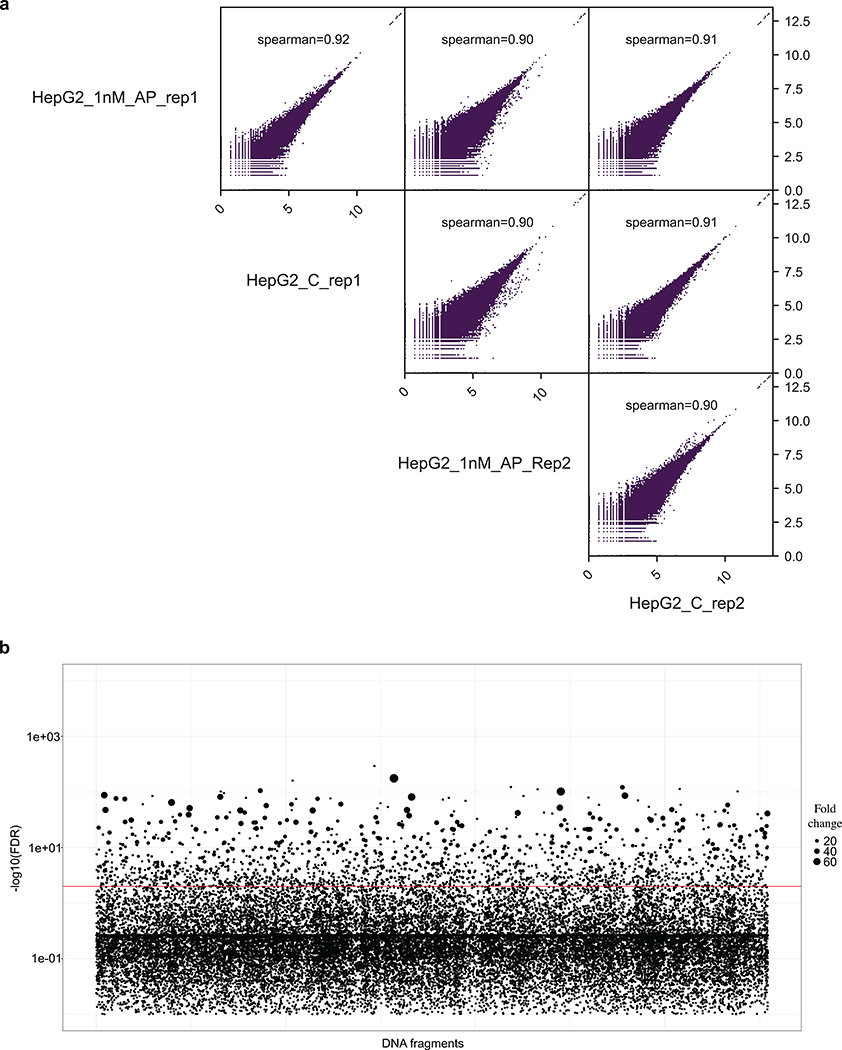
To further test this observation, we repeated the ReSE screen on HepG2 cells that are of hepatocyte origin using the same ReSE library made from the K562 FAIRE enrichment. Again the rationale was that if the two cell types shared common silencers there would be substantial overlap in the silencers between both cell types. Two independent biological ReSE screens of HepG2 cells led to the identification of 1,662 potential silencer regions with FDR of 0.01 (Fig. 2a, Extended Data Fig. 5 and 6, Supplementary Fig. 6, and Supplementary Table 3). Although these silencer regions shared a similar overall genomic distribution with K562 cells (Extended Data Fig. 6b), only a small fraction (less than 2% of the total) of the silencer regions was shared between these two cell lines, indicating that the majority of the silencers that we identified, similar to enhancers, may exert their function in a tissue-specific fashion (Fig. 2b and Supplementary Fig. 6b). However, as we applied a very stringent FDR cutoff to identify silencers in the respective cell lines, we may underestimate the percentage of conserved silencers among different tissues. Nonetheless, the data suggest that many silencers may be tissue-specific.
Figure 2. Conserved and tissue-specific distribution of silencers.
(a) ReSE screen results from two biological replicate experiments in HepG2 cells. Each dot represents one tested fragment. The position of the dot indicates the location of each fragment within the respective chromosome. The height of the dot indicates the –log10(FDR) of enrichment of the ReSE fragments after induction of apoptosis compared with the cells not treated with dimerizer. The cutoff value of FDR is 0.01. See Methods for detailed information.
(b) Comparison of silencer regions identified from K562 and HepG2 cells. Overlapping was not random as determined by permutation tests (n of testing = 20,000, adjusted P value = 0.00005).
(c) Luciferase assays to determine the repressive activity of shared silencers from K562 and HepG2 cells. Silencer regions were cloned by PCR from the genomic DNA of K562 cells into a luciferase reporter plasmid pGL4.53. 293T cells were used as the control cell line and empty pGL4.53 plasmid was used as the control for baseline luciferase activity. The y-axis represents the percentage of luciferase activity compared to pGL4.53 empty plasmids in the respective cells. (n = 3 biological independent samples; the bars show the mean value ± S.E.M; * P value < 0.05, calculated using two-sided Student’s t test).
(d) Top canonical pathways enriched in gene sets containing silencers in K562 cells (CVS, cardiovascular system).
(e) Top canonical pathways enriched in gene sets containing silencers in HepG2 cells.
All exact P values are provided in the Source Data.
We next directly investigated whether the small percentage of the shared silencers found in K562 and HepG2 cells might be ubiquitous silencers and act in different cell types. To examine this possibility, 7 of the silencer regions shared by both K562 and HepG2 screens were tested using the luciferase assay and 3 were found to be repressive in both cell types (Fig. 2c). In addition, repression activity was also observed for 5 out of the 7 common regions tested in 293T cells, suggesting these regions may be cell-type-independent silencers. It should be noted that silencers were called using a stringent algorithm within individual cell lines and the false negative rate may be higher when comparing silencers among cell lines; some top ranked silencers from HepG2 also showed some repressive activity in K562 cells (Extended Data Fig. 6a). We also noted that the false discovery rate is higher in HepG2 cells, presumably because the screen library was made from FAIRE regions of K562 cells and we may not have identified the strongest silencers in HepG2 cells. Overall, our current data suggest that the majority of silencers may be cell-type-specific with a small number that operates in two or more cell lines (Fig. 1e, Extended Data Fig. 2a and Extended Data Fig. 6a).
Since the majority of silencers identified by ReSE screen may function in a tissue-specific manner, we further tested if silencers associate with genes in unique pathways. Pathway analyses using Ingenuity Pathway Analysis (IPA) revealed unique pathways with strong confidence (i.e. lower P value) for the proximal genes associated with silencers identified from the different cell types. We employed two different methods to identify proximal genes that may be regulated by silencers in K562 and HepG2 cells: 1) the presence of silencers only in the promoter regions (10 kb surrounding transcription starting sites [TSS], Supplementary Tables 4 and 5); 2) the presence of silencers in both promoter regions (1 kb surrounding TSS that is more stringent definition)35,36 and gene bodies, since many silencers were enriched in the intron regions (Fig. 2d and e, Supplementary Tables 6 and 7). When the presence of silencers in both promoters and gene bodies are considered, in K562 cells, protein kinase A signaling and actin cytoskeleton signaling pathways were among the top pathways enriched for silencer associated genes (Fig. 2d and Supplementary Table 6). Since K562 cells are a myeloid cell line growing in suspension culture, it is likely that the actin cytoskeleton signaling pathway is repressed or poised for activation as compared to adherent cells37. In contrast, neuronal pathways and cardiac pathways were the top genes that lay near silencers isolated from HepG2 cells (Fig. 2e and Supplementary Table 7). Such pathways are also expected to be repressed, since HepG2 cells were derived from a hepatic cell lineage.
Silencers consist of unique genetic and epigenetic signatures
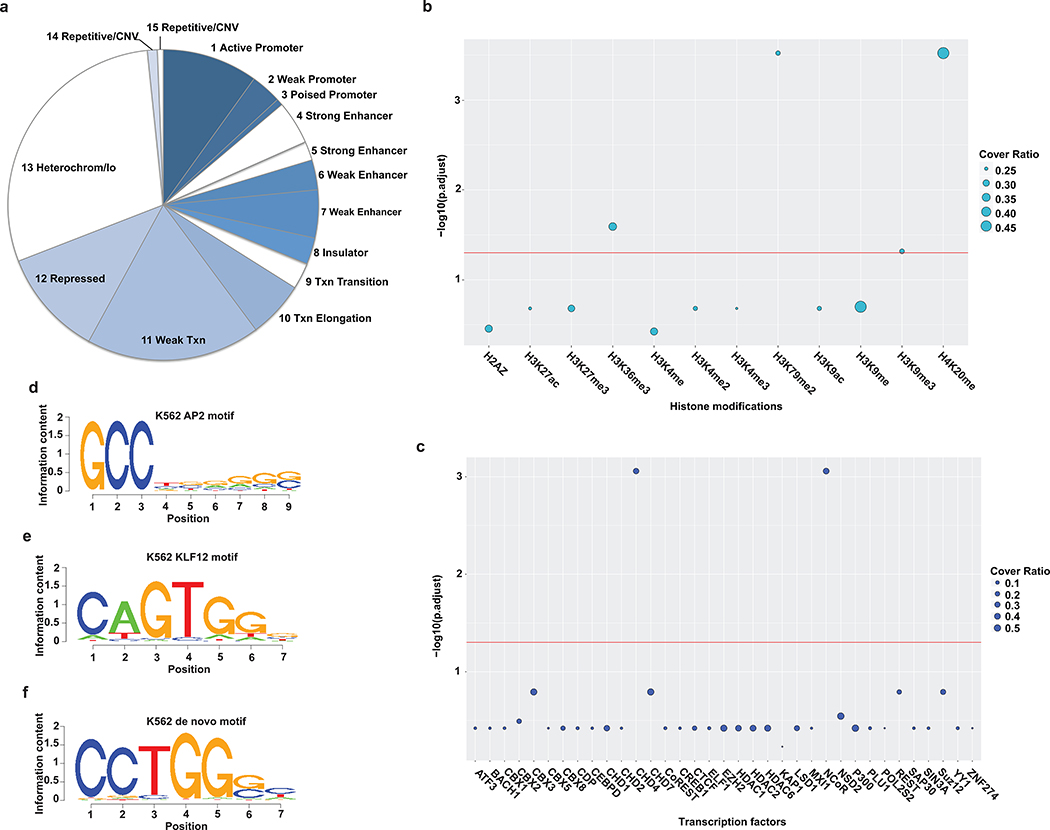
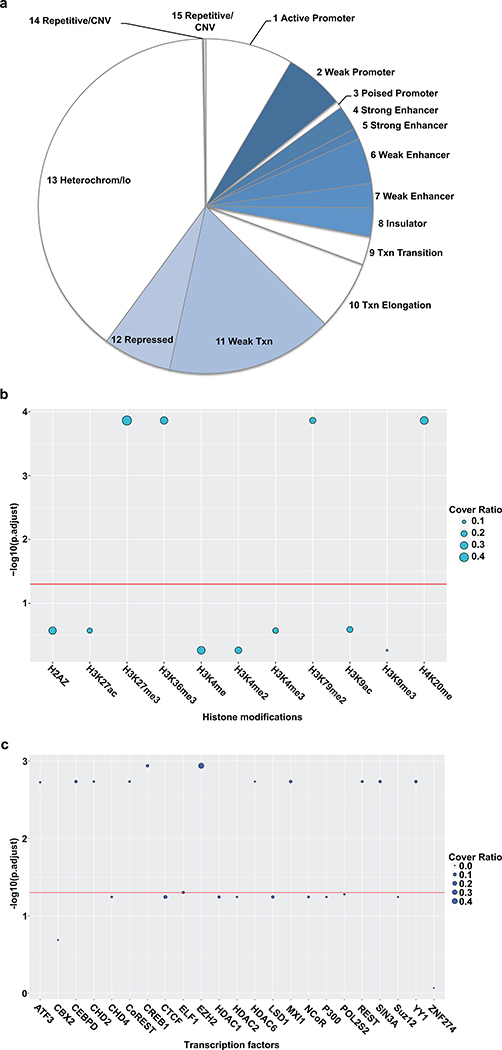
Functional regulatory elements are usually present in defined chromatin states10,38. For instance, enhancer regions often are marked by modifications such as H3K4me1 and H3K27ac10. To determine the chromatin states of silencer regions ReSE identified, the recovered regions from K562 and HepG2 were first classified based on the ENCODE chromatin definitions10,39. More than one quarter of the silencers were enriched in the weak transcription chromatin state (P value < 2.2 × 10−16, one-sided binomial test using the screen library as the background) or repressed state (P value = 3.325 × 10−5, one-sided binomial test) (Fig. 3a and Extended Data Fig. 7a). This indicates that silencers may be associated with specific chromatin modifications that might be necessary to exert their silencing function.
Figure 3. Unique signatures in silencer regions.
(a) Distribution of silencer regions in respective chromatin states in K562 cells. (Txn, transcription; Repressed, Polycomb repressed; lo, low signal; CNV, copy number variation). Colored parts indicate chromatin states that are significantly enriched compared to the library background distribution (P value < 0.001, one-sided binomial test).
(b) Association of histone modifications with silencers in K562 cells. The y-axis indicates the significance of the enrichment of histone modifications with silencer fragments. The size of the circle indicates the ratio of silencers covered by the respective histone modification (scale is 0 to 1). The enrichment analysis is based on permutation tests using 20,000 random permutations. The P value was calculated and multiple comparison corrections were computed using the Benjamini-Hochberg procedure. The red line shows the cutoff of the adjusted P value < 0.05.
(c) Association of transcription factors with silencers in K562 cells. The y-axis indicates the significance of the enrichment of transcription factors with silencer fragments. The size of the circle indicates the ratio of silencers covered by the respective transcription factor (scale is 0 to 1). The enrichment analysis is based on permutation tests using 20,000 random permutations. The P value was calculated and multiple comparison corrections were computed using the Benjamini-Hochberg procedure. The red line shows the cutoff of the adjusted P value < 0.05.
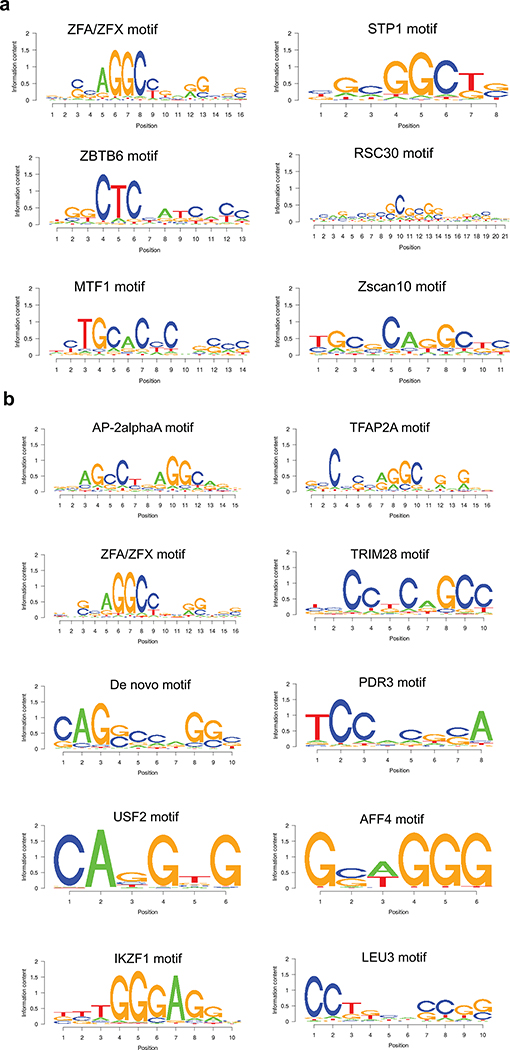
(d,e) Example of top known motifs present in the silencer regions in K562 cells.
(f) Example of top de novo motif present in the silencer regions identified from K562 cells.
All exact P values are provided in the Source Data.
To further examine the epigenetic marks and transcription factors that may be enriched in the silencer regions, a permutation-based test was performed to associate silencer regions with available datasets from the ENCODE project3,40. When histone marks were analyzed, H4K20me modified chromatin was significantly co-associated with silencers from both K562 and HepG2 cells (Fig. 3b and Extended Data Fig. 7b). The role of H4K20me modification is still complex as both transcriptional repression and activation have been associated with this mark41. Interestingly, different heterochromatin histone marks, each associated with gene inactivation, were found to associate with silencers isolated from K562 (H3K9me3; Fig. 3b) and HepG2 cells (H3K27me3; Extended Data Fig. 7b). This difference may be due to the fact that the initial silencer screen library was constructed from K562 cells, and these silencers may represent a different pool in HepG2 cells. In addition to histone marks related to silencing, H3K36me3 and H3K79me2, both active histone marks, were also significantly associated with silencers. It has been suggested that when combined with other histone marks, active marks like H3K36me3 can be found in heterochromatin regions42. This finding is also consistent with the observation that the identified silencers are enriched in weak transcription chromatin states (Fig. 3a and Extended Data Fig. 7a). We note that the selected silencer regions that were targeted for knockout in K562 cells (Extended Data Fig. 3) all showed the presence of H4K20me modification (Supplementary Fig. 7), further indicating that in these representative silencer regions, this mark may be associated with gene silencing (in Fig. 3b). In addition, these silencers also showed overlap with H3K27me3 or H3K9me3 (Supplementary Fig. 7), indicating either a weak transcription chromatin state or a repressed state (Fig. 3a).
Regulatory proteins that participate in repression might be enriched in silencers. Therefore, a permutation-based co-association test was performed between silencers and regions bound by transcription factors in both K562 and HepG2 cells3,40. In K562 cells, CHD4 and NCoR were significantly enriched in silencer regions (adjusted P value = 0.0008; Fig. 3c). A different set of transcription factors, e.g. EZH2 and REST, were enriched in HepG2 cells (Extended Data Fig. 7c). This likely reflects the cell-type specificity of silencers, as different cell types presumably use different proteins to regulate silencers. While it is likely that different cell types may employ different proteins for silencing, it is also possible that other unidentified silencer proteins are common to different cell types. It is worth noting that among the transcription factors tested, KAP1 and ZNF274 were not associated with silencers identified using our open chromatin screen. This is likely because they are primarily associated with inaccessible chromatin43, which is less likely enriched in the screen library. As controls, p300, which is usually associated with transcription activation44 or Pol2S2 (phosphorylation of Serine 2 on CTD of RNA polymerase II), which is usually associated with active transcription45, were also not enriched with silencers.
To identify other potential novel factors that may recognize the silencer regions identified by our ReSE screen, motif analyses were performed using SeqPos46. The top known motif identified in both K562 and HepG2 cells was the AP2 binding domain (Fig. 3d and Extended Data Fig. 8; with P values = 10−358 and 10−225 respectively). The role of AP2 family members in transcription repression has been shown previously47,48. However, a general repressive activity of AP2 in different tissues has not been established. Furthermore, the motif of another known transcription repressor KLF12 was also enriched in K562 cells (Fig. 3e)49. In addition to motifs with known binding factors, many de novo motifs were also identified in both K562 (Fig. 3f) and HepG2 cells (Extended Data Fig. 8b). These motifs are GC-rich, similar to AP2 motifs. These data indicate that there may be unique factors that function in silencer regions that are yet to be identified.
Silencers regulate proximal endogenous genes to promote chemoresistance
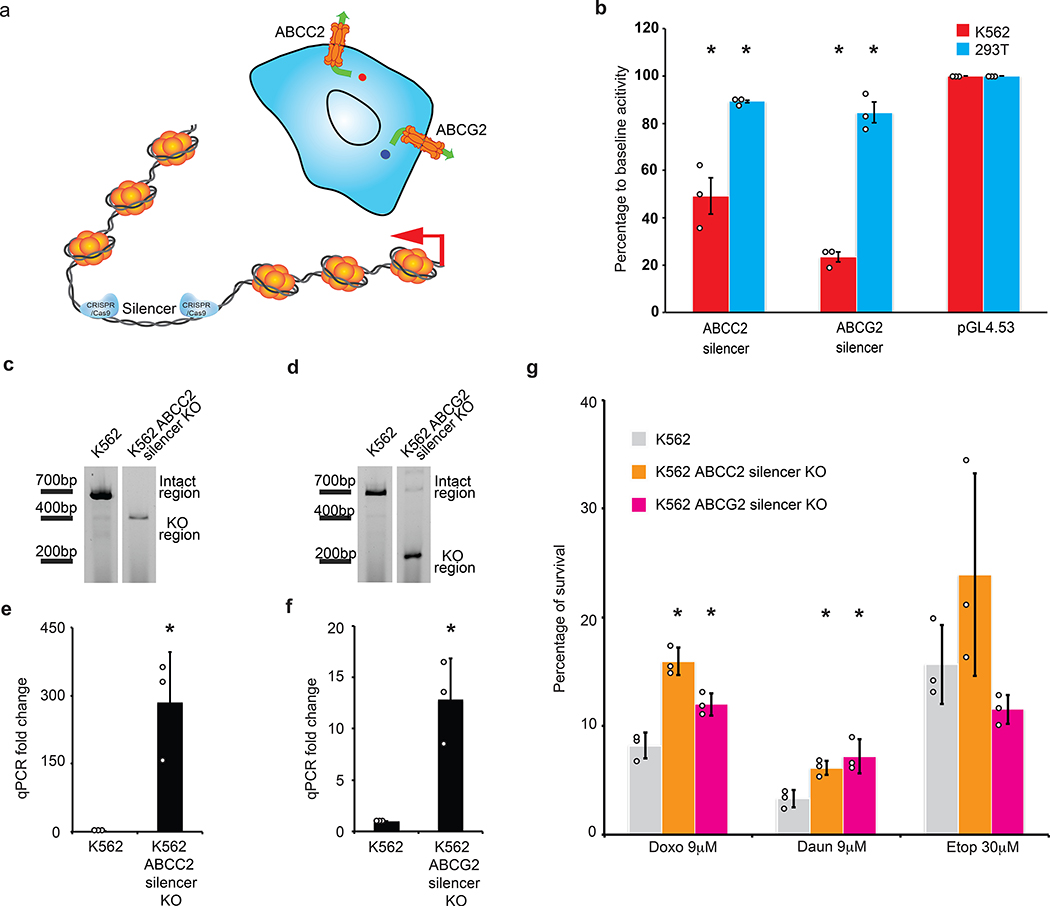
We next examined if the silencer regions identified by the ReSE screen have direct biological effects. Pathway analyses based on genes that harbor silencers both in the promoter and gene body regions (Fig. 2d and Supplementary Table 6) revealed that silencers were enriched in genes for an ABC drug transporter pathway in K562 cells (Supplementary Fig. 8, -log(P value) = 2.671). Within the intron regions of two drug transporter genes ABCC2 (on chromosome 10) and ABCG2 (on chromosome 4) there exist two potential silencers (Supplementary Fig. 9). It is possible that these silencers affect the transcription of these genes and thus participate in response to drug treatment (Fig. 4a). Luciferase assays showed that these silencer regions have repressive activity and repressed the transcription of the pGL4.53 reporter by 50% (ABCC2 locus silencer) and 80% (ABCG2 locus silencer) (Fig. 4b).
Figure 4. Drug resistance regulated by silencer regions.
(a) The ABCC2 and ABCG2 genes contain silencers. The upper panel shows the primary known function of ABCC2 and ABCG2 in cells, which is to excrete drugs from cells. The lower panel shows the main strategy to use CRISPR/Cas9 to remove silencers from the genome and test their function endogenously.
(b) Luciferase assays to determine the repressive activity of silencers from within ABCC2 and ABCG2 genes in K562 cells. 293T cells were used as the control cell line and empty pGL4.53 plasmid was used as the control for baseline luciferase activity. The y-axis represents the percentage of luciferase activity compared to pGL4.53 empty plasmids in the respective cells. (n = 3 biological independent samples; the bars show the mean value ± S.E.M.; P value < 0.05, calculated using two-sided Student’s t test).
(c) Knocking out silencer from ABCC2 gene using CRISPR/Cas9. PCR result showing the removal of the silencer from the ABCC2 gene of two K562 cell clones, which is the representative result of 2 experiments. The blots were cropped. Full scans of the blots are shown in Source Data Full Gel Figure 4.
(d) Knocking out silencer from ABCG2 gene using CRISPR/Cas9. PCR results showing the removal of the silencer from the ABCG2 gene of one K562 cell clone, which is the representative result of 2 experiments. The blots were cropped. Full scans of the blots are shown in Source Data Full Gel Figure 4.
(e) ABCC2 gene expression quantified by qPCR. (n = 3 biological replicates of the same clones; the bars show the mean value ± S.D.; * P value < 0.05, calculated using two-sided Student’s t test).
(f) ABCG2 gene expression quantified by qPCR. (n = 3 biological replicates of the same clones; the bars show the mean value ± S.D.; * P value < 0.05, calculated using two-sided Student’s t test).
(g) Drug resistance promoted by the up-regulation of ABCC2 and ABCG2 genes after silencer removal. (n = 3 biological replicates of the same clones; the bars show the mean value ± S.D.; * P value < 0.05, calculated using two-sided Student’s t test; Doxo: doxorubicin; Daun: daunorubicin and Etop: etoposide).
All exact P values are provided in the Source Data.
The silencers in the ABCC2 and ABCG2 loci were targeted with flanking CRISPR guide RNAs to delete the regions from the genome (Fig. 4a), and K562 cell clones with the complete knockout of the silencer within ABCC2 (Fig. 4c) and ABCG2 (Fig. 4d) were derived. Transcription of the ABCC2 and ABCG2 genes was significantly up-regulated in these clones, and such up-regulation was unrelated to puromycin selection since control knockout clones did not show this strong increase (Fig. 4e and f; additional clones and comparison between all individual clones with control knockout clones in Extended Data Fig. 9). In contrast, knocking out the same silencer regions did not induce significant up-regulation of ABCC2 gene in 293T cells or only modestly up-regulated the ABCG2 gene in 293T cells (Extended Data Fig. 9), in accordance with the luciferase assay in Figure 4b, further indicating that silencers function in a tissue-specific manner. As a result of the transcriptional up-regulation of drug transporters, these knockout clones are both more resistant to chemotherapeutic drug treatments compared to the parental cell line (Fig. 4g), suggesting that silencer regions may affect drug sensitivity.
When the local epigenetic modifications were examined, the silencer region in the ABCC2 gene was marked with H2A.Z and H3K27ac (Supplementary Fig. 9), whereas the silencer region in ABCG2 overlapped with H3K27me3 and the H4K20me modification resided nearby (Supplementary Fig. 9). The latter mark is consistent with the results presented in Figure 3b and Supplementary Figure 7. In addition, the gene bodies of both ABCC2 and ABCG2 were largely covered by H3K27me3 modification, indicating a repressed state of these genes. These two silencer regions were not covered by the H4K20me mark (although the silencer in the ABCG2 locus showed H4K20me in close proximity).
Silencers regulate transcription in the 3D genome
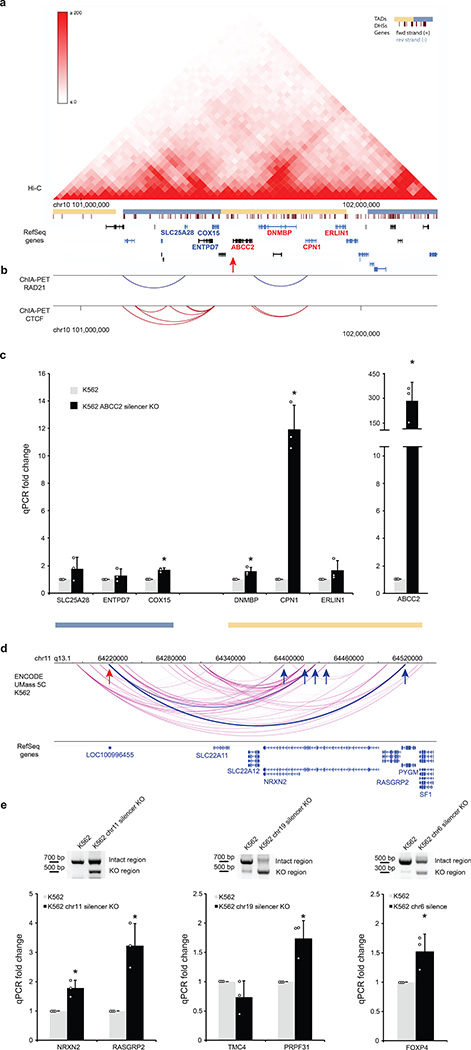
Cis-regulatory elements often regulate not only a single gene, but a group of genes within a topologically associating domain (TAD)50,51. To test if this occurs for the ReSE-identified silencers, Hi-C data from K562 cells were integrated to define the different domains surrounding the silencer within the ABCC2 gene52,53 (Fig. 5a). Hi-C data indicate that one TAD contains the silencer within the ABCC2 gene in K562 cells (Fig. 5a). Furthermore, in this TAD a chromatin loop connects both the ABCC2 and CPN1 genes (Fig. 5b), as defined by ChIA-PET assays targeting RAD21 and CTCF54. This result raises the possibility that the silencer within the ABCC2 gene may also regulate the CPN1 gene.
Figure 5. Regulation of distal genes by silencers.
(a) TADs identified by Hi-C data. Arrow indicates the relative location of the silencer. Yellow and blue bars indicate distinct TADs. DHS are DNase I hypersensitive sites.
(b) ChIA-PET interactions identified using RAD21 (blue, component of cohesin complex) and CTCF (red) surrounding the ABCC2 gene.
(c) Transcription of genes within different TADs quantified by qPCR. The blue bar indicates genes that do not reside in the same TAD with the ABCC2 gene. The yellow bar indicates genes that reside in the same TAD as the ABCC2 gene. (n = 3 biological replicates of the same clones; the bars show the mean value ± S.D.; * P value < 0.001, calculated using two-sided Student’s t test)
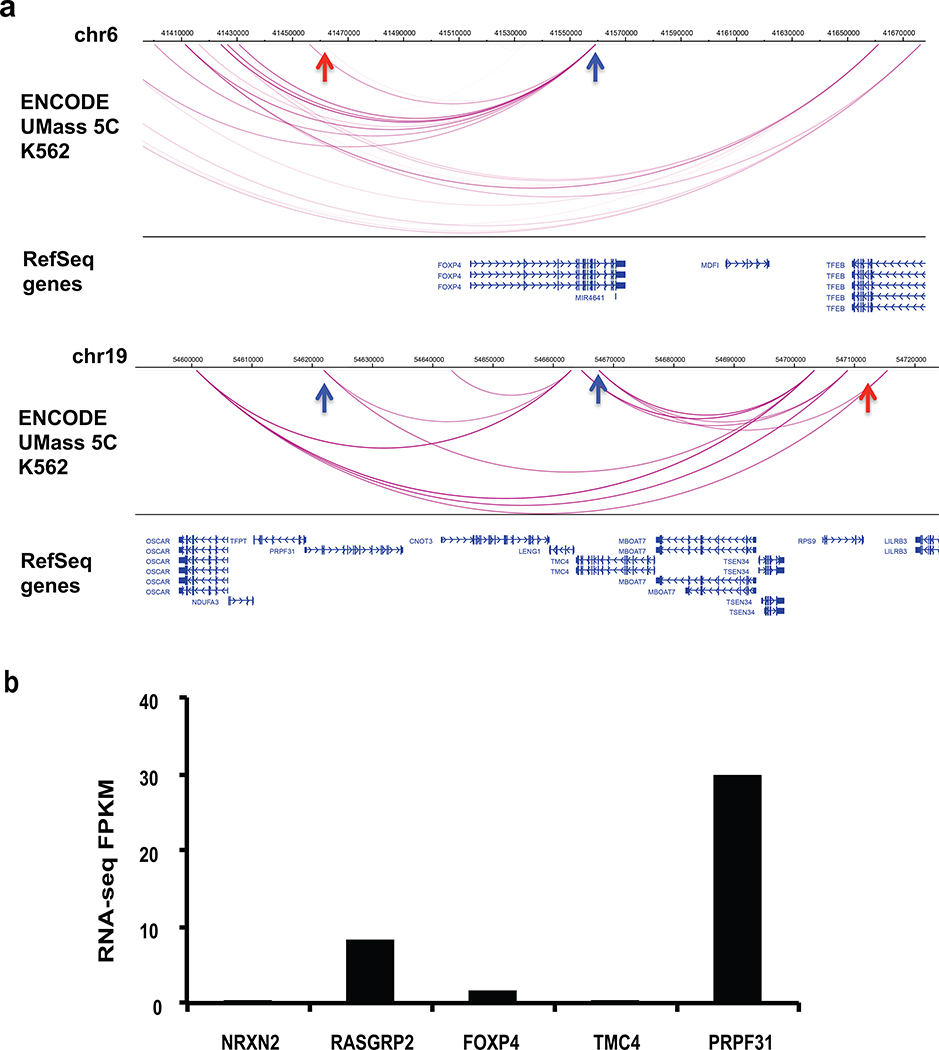
(d) Direct interaction of silencers with promoters of distal genes from 5C analysis. Red arrow indicates the silencer region, and blue arrow shows the significant interaction region with genes. Significant interaction arcs are highlighted in blue.
(e) Gene up-regulation by removing promoter-interacting silencers from the endogenous regions. Paired CRISPR guide RNAs were designed to target both sides of the selected silencers identified in (d) and Extended Data Figure 10a. Single clones were selected and verified by PCR (upper panel), which is the representative result of 2 experiments. The blots were cropped. Full scans of the blots are shown in Source Data Full Gel Figure 5. The expression of respective genes with promoter-silencer interactions was quantified by qPCR (lower panel). (n = 3 biological replicates of the same clones; the bars show the mean value ± S.D.; * P value < 0.05, calculated using two-sided Student’s t test).
All exact P values are provided in the Source Data.
Therefore transcriptional changes of genes from two topologically distinct domains were tested between control and knockout cell lines using qPCR assays. We found that similar to enhancers51, silencers also acted on genes within the same chromatin-loop domain, as the CPN1 gene was significantly up-regulated in the K562 ABCC2 silencer KO cell line (12-fold, Fig. 5c, right with yellow bar), in which the ABCC2 gene was also strongly up-regulated (300-fold; Fig. 4d and 5c). Strong gene up-regulation was confined to only these two genes within the TAD (Fig. 5a, yellow domain containing ABCC2 gene), in contrast to the genes located in a nearby distinct TAD (Fig. 5c, left with blue bar).
In order to globally identify distal genes that may be regulated by the silencers, capture Hi-C data that profiled interactions with 31,253 promoter regions from human primary blood cells55 were integrated with silencers identified from K562 cells. Since K562 cells resemble a common myeloid progenitor origin that is similar to many of the cell types in blood and can be differentiated into many of these cell types, the whole-blood data should contain a lot of these regulatory regions. Silencer regions identified by ReSE in K562 cells interacted with approximately 4,000 promoter regions (Supplementary Table 8, permutation test adjusted P value = 4.99 × 10−5, n = 20,000), suggesting that ReSE silencers can directly interact with many promoter regions.
To directly test the effect of promoter-silencer interactions in K562 cells, chromosome conformation capture carbon copy (5C) data from ENCODE that reported long-range interactions between promoter regions and distal elements was examined56. However, these 5C experiments only targeted 1% of the genome56. We intersected 5C data with the silencer regions identified in K562 cells and found five genes that directly interact with silencer regions (Fig. 5d and Extended Data Fig. 10a). Among these interactions, three genes NRXN2, TMC4 and FOXP4 showed extremely low expression (FPKM < 2) based on RNA-seq data in K562 cells57 (Fig. 5d and Extended Data Fig. 10b) and an additional gene RASGRP2 gene showed low expression (FPKM < 10) (Fig. 5d and Extended Data Fig. 10b). When these silencers were removed individually with flanking CRISPR guide RNAs in K562 cells, 4 out of 5 genes were up-regulated significantly in the respective clones (Fig. 5e). These data further support the suggestion that silencers may also interact with and regulate distal genes.
Discussion
Although only approximately 2% of the human genome contains coding sequences that can be translated into proteins, many noncoding regions contain unique sequences that can be recognized by chromatin modifiers and transcription factors, as candidate regulatory elements. Systematic analysis of promoters and enhancers has been performed previously15–19, however a global analysis of silencers has not been described. In our study, a robust screening system, ReSE, was developed to systematically identify silencer elements in the human genome. ReSE utilizes a lentiviral system to test the ability of candidate genomic fragments to repress the caspase-based “kill switch” for the enrichment of potential silencers. In principle, other plasmid-based reporter assays normally used for assessing enhancers and promoters could also be used to evaluate silencer activity. However, these systems rely on RNA-seq and therefore may be better suited to evaluate activation rather than repression. Despite the fact that fewer genomic regions are individually assayed in the lentiviral system than in other plasmid-based reporter assays, the ReSE lentiviral method can be used to directly select for regions of interest, and in addition it can overcome the plasmid-transfection-related systematic errors that have been realized recently58. Nonetheless, akin to other genome-wide screen systems that are intrinsically noisy, ReSE is also limited by false positive and negative discovery. Therefore multiple biological replicates are recommended for the screen to increase the statistical power. Although silencer regions may exist in different genomic regions with distinct chromatin structures, to prioritize the testing regions in the human genome, we analyzed open chromatin regions, as these regions are accessible for regulatory factors and might directly exert repressive functions rather than passively be silenced through repressive heterochromatin. The ReSE screen reliably identified silencer fragments in different cell lineages. Our results suggest that many of the silencers that we identified may function in a tissue-specific manner. Nonetheless, it is possible that a large and common pool of shared silencers exists in different tissues, as the current ReSE screen library was derived only from FAIRE regions of K562 cells and silencers were identified using a stringent cutoff. These data indicate that silencers may play an important role in development and regulate tissue differentiation. Unique motifs are present in the silencers that could potentially be recognized by specific factors to exert repressive functions.
Although the majority of the silencers may be present in the transcriptionally inactive states, presumably some accessible DNA still exists in these chromatin regions to be isolated using FAIRE59,60. In agreement with this interpretation, silencers were found in the vicinity of genes that are poised for responses or repressed during differentiation. Silencers may possess a unique combination of histone signatures (Fig. 3b, Extended Data Fig. 7b, and Supplementary Fig. 7 and 9). Such chromatin states may be the result of the recognition of silencers by repressive transcription factors. Since the current ReSE screen was focused on accessible chromatin regions with a fixed testing size (around 200 bp), the resulting signatures, such as histone modifications, transcription factor associations and sequence motifs, may also be biased towards a particular class of silencers using this library. It is possible that there are many different transcription factors recognizing their respective sub-clusters of silencers within distinct cell types (Fig. 3c and Extended Data Fig. 7c). Future genome-wide analysis of silencers is required to provide a clearer picture of all possible silencing signatures. Systematic identification of silencers in this way helps to further our understanding of the relationship between repressed chromatin states and gene silencing.
Thus far, investigations of drug responses or disease progression often focus on the coding regions of genes, or known regulatory regions such as promoters and enhancers36,61–64. For instance, a recent CRISPR activation screen targeting promoter regions led to the identification of ABCG2 as an important drug-resistant player in K562 cells65. We found that deleting silencers within drug transporter genes ABCC2 and ABCG2 also led to the up-regulation of these genes and chemotherapeutic drug resistance, suggesting silencer-mediated transcription repression may be another layer of regulation contributing to important medical phenotypes. Thus, it is expected that phenotype-associated genetic variants in silencers may affect drug responses (Supplementary Fig. 10), disease initiation and progression, and be considered as candidates for precision medicine. Furthermore, many diseases are caused by haploinsufficiency or insufficient gene expression. This can be effectively rescued by the newly developed CRISPR/dCas9-mediated activation technology, either by targeting the promoter or enhancer regions of the relevant genes66–68. However, unlike the CRISPR/Cas9-mediated genomic editing/correction that requires only transient expression of the CRISPR/Cas936,69, the activation system requires constant expression of the CRISPR/dCas9. Therefore such regulation is often reversible68,70, which may not be ideal for future applications in human diseases. As shown in our data, genomic editing of the silencer regions can lead to gene up-regulation. Therefore, inactivating silencer regions could be complementary to CRISPR/dCas9-mediated activation system to treat many diseases. As such, systematic identification of silencers in the genome using the ReSE screen may not only provide insights into the biology of the genome, but also assist in personalized medicine.
Methods
Cell culture
K562 cells were cultured in RPMI 1640 with L-glutamine, 10% FBS and Pen-Strep. HepG2 cells were cultured in DMEM, 10% FBS and Antibiotic-Antimycotic. Cell density and culture conditions were maintained according to the ENCODE Cell Culture Guidelines.
Library construction
FAIRE was performed using K562 cells as previously described29,71,72. Briefly, 5 × 107 K562 cells were fixed with a final concentration of 1% formaldehyde for 5 minutes. 2.5 M glycine was added to a final concentration of 125 mM, and cells were incubated for 5 minutes at room temperature while shaking. Cells were then lysed in 5 ml of Lysis Buffer 1 (50 mM HEPES-KOH, pH 7.5,140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) and rocked at 4°C for 10 minutes. The tubes were subsequently centrifuged at 1,300 g for 5 minutes at 4°C and the supernatant was removed. The pellet was suspended in 5 ml of Lysis Buffer 2 (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA) and rocked at room temperature for 10 minutes, centrifuged at 1,300 g for 5 minutes at 4°C and the supernatant was removed. The pellet was then suspended in 2 ml of Lysis Buffer 3 (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-Deoxycholate, 0.5% N-lauroylsarcosine). Cells were then sonicated in bioruptor tubes with sonication beads for 16 cycles for 30 seconds each followed by 30 seconds incubation periods at 4°C. The tubes were centrifuged at 3,000 rpm for 5 minutes at 4°C and an equal volume of phenol/chloroform (phenol, chloroform, and isoamyl alcohol 25:24:1 saturated with 10 mM Tris, pH 8.0, 1 mM EDTA) was added to the lysate and the aqueous phase was separated with phase lock gel. DNA from aqueous phase was then precipitated with ethanol at −80°C. The pelleted DNA was reverse cross-linked and processed according to the Illumina sequencing library preparation protocol. After Illumina adapters were ligated, fragments were size-selected for 200 bp. PCR procedures using Phusion High-Fidelity PCR 2× Master Mix and PCR primer 1.0 and 2.0 for Illumina TruSeq adapters were: 98°C for 30 s; 12 cycles of (98°C for 10 s; 65°C for 30 s; 72°C for 30 s); and 72°C for 5 min. Half of the fragments were further processed for next-generation sequencing using Illumina MiSeq platform to confirm the FAIRE process enriched the proper open chromatin regions. The other half was amplified using primers containing additional sequences by PCR for downstream Gibson assembly. Primer sequences for using Phusion High-Fidelity PCR 2× Master were: ACTCCTTTCAAGACCTAGCTCAAGCAGAAGACGGCATACGAGAT; ATCCAGTTTGGTTAATTAAGAATGATACGGCGACCACCGAGATCTACACTCTTTCCCTAC. PCR procedures were: 98°C for 30 s; 8 cycles of (98°C for 10 s; 65°C for 30 s; 72°C for 30 s); and 72°C for 5 min. These fragments were then gel-purified.
The ReSE screen lentivirus vector pLenti-FKBP-delCasp9-Puro was designed based on plasmids from Addgene (Plasmid #15567 and #52961)27,73. EF-1α, a human constitutive promoter was used to drive the expression of FKBP-Casp9, and the UbC promoter was used to drive the expression of a puromycin-resistance gene. We reasoned that strong silencer activity would have limited effects on the virus packaging and subsequent puromycin selection, as shown in a retrospective experiment that virus titer and the subsequent puromycin selection were not affected by silencer insertions in the screen plasmid (Supplementary Fig. 11). However, we cannot rule out that there might exist “super-silencer” fragments that would affect the virus production or puromycin expression. pLenti-FKBP-delCasp9-Puro plasmids were digested with BsmBI enzyme and gel-purified. The FAIRE fragments were then inserted into the digested plasmids, 15 bp upstream of the EF-1α using Gibson Assembly. The rationale was to identify a class of strong and more general silencers that are able to repress transcription upstream of the constitutive promoter EF-1α. The assembly mix was made using 50 ng of insert DNA, 50 ng of digested plasmids, and 10 μl of 2× Gibson Assembly Master Mix to produce a final volume of 20 μl. The assembly mix was incubated at 50°C for 60 min. Then 2 μl of the mix was electroporated into 25 μl of Endura electrocompetent cells to test the transformation efficiency. The electroporation was scaled to reach approximately 160,000 colonies which were plated on 4 245-mm Petri dishes with 100 μg/ml carbenicillin. Colonies were then scraped and plasmid DNA extracted using Qiagen Maxiprep Kit.
Lentivirus production and infection
293T cells were grown in 5 T175 flasks at 50% confluency before transfection. For each flask of 293T cells grown in 25 ml of fresh medium. 15 μg of library plasmids, 10 μg of psPAX2, 5 μg of pCMV-VSV-G and 90 μl of X-tremeGENE 9 DNA Transfection Reagent were mixed in 1 ml serum-free medium and used for transfection. Fresh medium was added the next day after transfection. Media supernatant containing virus particles was collected from the 2nd and 3rd day after transfection, pooled and further concentrated using Lenti-X according to the manufacturer’s protocol. Virus titer was then determined by making serial (10−3 to 10−10) dilutions of 4 μl of frozen virus supernatant in media containing 8 μg/ml of polybrene to infect 293T cells. Two days after infection, cells were selected with 2 μg/ml puromycin for an additional 7 days. The virus titer was then calculated based on the survival colonies and the related dilution. K562 cells and HepG2 cells were then infected with the same virus library at MOI 0.5 by spin infection. For spin infection, 3 × 106 cells in each well of a 12-well plate were infected in 1 ml medium containing 8 μg/ml of polybrene. In total, 4 plates were used for each infection to analyze a total of 1.5 × 108 cells. Two days after infection, cells were selected by 2 mg/ml puromycin for another 5 days. For each biological replicate experiment of both K562 and HepG2 cells, the infection was repeated and cells were infected with lentivirus from the same pool of virus containing the same library content.
Silencer screen
After puromycin selection, 3.5 × 108 K562 cells were frozen as non-treated control. A separate aliquot of 3.5 × 108 cells were treated with 1 nM of AP20187 for 18 h to induce apoptosis. Then dead cells were removed with Dead Cell Removal Kit from Miltenyi Biotec. In the screen of K562 cells, we retrieved 45.6% of live cells compared to the original input cell number, after 18 h of AP20187 treatment. If cell growth of the live cells during this 18-h period is also considered, the real survival rate should be around 30.4% (considering the normal doubling time of live K562 cells is 24 hours). In addition, there are also some other scenarios, for example, although cells with virus infection survived puromycin selection (as the expression of puromycin-resistance gene is under another independent promoter UbC promoter), the expression of FKBP-Casp9 was silenced by other machinery within the cells; Or cells were still in the early stage of apoptosis, and were not removed by the live cell isolation method. These could be the reason of higher survival rates. Many such false positive regions were removed during biological repeat experiments. Live cells were further grown for another 5 days. Genomic DNA from 3.5 × 108 cells of non-treated control cells or post apoptosis-induction cells was isolated using QIAamp DNA Blood Maxi Kit, with 2 columns per treatment. For the K562 cell differentiation test, the same batches of cells that were analyzed as biological replicates were recovered from cells frozen in 10% DMSO. Cells were then differentiated with 10 nM PMA (phorbol 12-myristate 13-acetate) for 2 days. Cells were divided into differentiated non-treated control cells and the other half of cells were treated with 1nM AP20187 for 18 h to induce apoptosis. Dead cells were cleared as described previously. For HepG2 cells, the experiment procedures were similar except that dead cells were removed by removing the media, since HepG2 cells are adherent cells and live cells remained attached to the tissue culture flasks.
Library sequencing and analyses
Genomic DNA containing the ReSE lentivirus inserts was amplified by PCR using Illumina PCR primer 1.0 and 2.0. For each 100 μl PCR reaction, 10 μg of genomic DNA, 20 μl of 5× Phusion HF Buffer, 2 μl of 10 mM dNTP, 2.5 μl of Phusion polymerase, and 5 μl of 25 μM 1.0 primer and 5 μl of 25 μM 2.0 primer were used. For each treatment sample, 16 reactions were prepared and pooled. PCR procedures were: 98°C for 30 s; 20 cycles of (98°C for 10 s; 65°C for 30 s; 72°C for 30 s); and 72°C for 5 min. PCR products were then size-selected and purified. Final products were sequenced by Illumina MiSeq or Hiseq4000 platform. Sequence reads were aligned using Bowtie to hg19. Approximately 100,000 regions of the 177,000 regions within the library were estimated to be well covered in the screen. A GFF file was made from aligned reads pooled from all experiments. Then read counts (quality ≥ 30) were calculated using HTSeq74. Final enrichment was calculated by MAGeCK30, with two biological replicates for each condition. Briefly, read counts derived from HTSeq of different samples were first median-normalized to adjust for the effect of library sizes and read count distributions. Then the variance of read counts was estimated by sharing information across features, and a negative binomial (NB) model was used to test whether fragment abundance differs significantly between post apoptosis-induction replicates and control replicates. P values were calculated from the NB model using a modified robust ranking aggregation algorithm30. FDR was then computed from the empirical permutation P values using the Benjamini-Hochberg procedure30. As fold enrichments are only semi-quantitative, fragments with an FDR lower than 0.01 were considered as significant hits for downstream analyses, and the list of silencers was sorted based on FDR value from low to high.
Luciferase assay
Candidate silencer sequences were amplified with primers containing a homologous arm using PCR from the genomic DNA of K562 cells. These fragments were then inserted in front of the PGK promoter of the luciferase plasmid pGL4.53 (Promega) using Gibson assembly. Cells were then co-transfected with the pRL-CMV Renilla reporter vector and the pGL4.53 vector with the silencer sequence inserted. The luciferase assay was performed using the Dual-Glo Luciferase Assay Kit from Promega according to the manufacturer’s protocol. Original luciferase plasmid without any insertion was used as the control. All luciferase assays were from 3 independent transfections done on different days. All tested regions and associated primers are listed in Supplementary Table 9.
Pathway analyses
Proximal genes around silencers were defined as 1) the presence of silencers only in the promoter regions (10 kb surrounding transcription starting sites [TSS]); or 2) the presence of silencers in both promoter regions (1 kb surrounding TSS) and gene bodies. Pathway analyses were performed using proximal genes with Ingenuity Pathway Analysis (IPA).
CRISPR/Cas9-guided silencer knock-out
Guide RNAs targeting the 5-prime and 3-prime ends of the silencer were designed using http://crispr.mit.edu/. The guide RNA sequence was cloned into the PX459V2 plasmid containing the guide RNA scaffold and Cas9 sequence. Two CRISPR/Cas9 plasmids targeting both the 5-prime and 3-prime ends of the silencer were co-transfected into the cells. Cells were then selected for successful transfection using puromycin. Single clones of cells were picked and verified using PCR and Sanger sequencing. Gene expression of target genes was quantified using qPCR, and normalized to the expression of the housekeeping gene GAPDH. All tested regions and associated primers were listed in Supplementary Table 9.
Downstream informatic analyses
Genomic annotation of silencer regions was analyzed using the R packages ChIPseeker and CEAS40,75. Motif analyses were performed using Cistrome46. For Motif analyses, only the silencers outside the promoter regions were used for the analyses to reduce bias from motif-rich promoter regions, though we did not observe major differences using all silencers or only the silencers outside of the promoter regions for motif analyses. Region intersections, comparisons, binomial test and other downstream analyses were calculated using R, ChIPpeakAnno, Galaxy or Cistrome46,76,77. Enrichment of chromatin states was calculated using a one-sided binomial test against the whole ReSE library as the background. ChIP-seq data from ENCODE and dbSNP147 data were downloaded from the UCSC Genome Browser Database with the hg19 genome assembly78. Association of histone modifications and transcription factor binding regions with silencers was calculated using the R package ChIPseeker40. The enrichment analysis is based on permutation tests using 20,000 random permutations. The P value was then calculated and multiple comparison corrections were computed using the Benjamini-Hochberg procedure for the adjusted P value40. For comparing silencers with capture Hi-C data from human primary blood cells55 or 5C data from K562 cells56, silencer regions identified from the ReSE screen in K562 cells were intersected with the distal regions that interacted with the promoter regions from the respective studies. Hi-C, ChIA-PET and 5C data of K562 cells were used and visualized using genome browsers52–54,56,79.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The sequencing data are available at GEO under the accession number: GSE108536.
Extended Data
Extended Data Fig. 1: ReSE screen results from K562 cells.
(a) The genome is split into bins of 1000 bp. For each bin, the number of reads found in each sample is counted. All regions were included. Spearman correlation was used to calculate the correlation efficiency among all 4 samples.
(b) ReSE screen results from two biological replicate experiments in K562 cells. Each circle represents one tested fragment. The y-axis indicates the FDR of enrichment of the ReSE fragments after induction of apoptosis compared with untreated control cells. The size of the circle indicates the fold enrichment of ReSE fragment counts of apoptosis-induction samples over untreated control after normalization. The red line indicates the cutoff value of FDR at 0.01.
Extended Data Fig. 2: Luciferase assays to determine the repressive activity of regions from K562 cells.
Silencer regions ranked on the bottom of the list based on the FDR value (a) or random regions from the screen (b) were cloned by PCR from the genomic DNA of K562 cells, and then inserted upstream of the promoter of a luciferase reporter plasmid pGL4.53. The 293T cells were used as the control cell line to control for cell-type dependent repressive activity, and the empty pGL4.53 plasmid was used as the control for the baseline luciferase activity. The Y-axis represents the percentage of luciferase activity compared to the pGL4.53 empty plasmids in the respective cells (n = 3 biological independent samples; the bars show the mean value ± S.E.M; *P value < 0.05, calculated using two-sided Student’s t test).
All exact P values are provided in the Source Data.
Extended Data Fig. 3: Gene up-regulation by removing silencers from endogenous regions in K562 cells.
Three silencers from (Figure 1e) are in the intron regions of genes HRH1 (a), SYNE2 (b) and CDH23 (c). Paired CRISPR guide RNAs were designed to target both sides of the selected silencer in order to remove it from the endogenous locus. Single clones were selected and verified by PCR (upper panel), which is the representative result of 2 experiments. The expression of the respective genes was quantified by qPCR (lower panel). (n = 3 biological replicates of the same clones; the bars show the mean value ± S.D.; *P value < 0.05, calculated using two-sided Student’s t test).
All exact P values are provided in the Source Data. The blots were cropped. Full scans of the blots are shown in Source Data Full Gel Extended Data Figure 3.
Extended Data Fig. 4: ReSE screen results from K562 cells differentiated with PMA.
(a) ReSE screen results from two biological replicate experiments in K562 cells treated with PMA. Each circle represents one tested fragment. The y-axis indicates the FDR of the enrichment of the ReSE fragments after induction of apoptosis compared with untreated control cells. The size of the circle indicates the fold enrichment of the ReSE fragment counts of the apoptosis-induction samples over the untreated control after normalization. The red line indicates the cutoff value of FDR at 0.01.
(b) RNA-seq experiments were performed on the cell subsamples from (a). DEseq2 was used to calculate the genes deregulated in the cells with PMA treatment compared to the non-treated K562 cells. Around 4666 genes were changed at least 2 fold with the adjusted P value less than 0.01 during the PMA treatment (red dots). Two biological replicates were included for each condition. (c) Comparison of silencer regions identified from K562 cells and K562 cells differentiated by PMA. The overlapping was not random as determined by permutation tests (n = 20,000, adjust P value = 0.00005).
Extended Data Fig. 5: ReSE screen results from HepG2 cells.
(a) The genome is split into bins of 1000 bp. For each bin, the number of reads found in each sample is counted. All regions were included. Spearman correlation was used to calculate the correlation efficiency among all 4 samples. (b) ReSE screen results from two biological replicate experiments in HepG2 cells. Each circle represents one tested fragment. The y-axis indicates the FDR of the enrichment of the ReSE fragments after induction of apoptosis compared with the untreated control cells. The size of the circle indicates the fold enrichment of ReSE fragment counts of the apoptosis-induction samples over the untreated control after normalization. The red line indicates the cutoff value of FDR at 0.01.
Extended Data Fig. 6: Silencer regions identified from HepG2 cells.
(a) Luciferase assays to determine the repressive activity of silencers from K562 and HepG2 cells. Silencer regions were cloned by PCR from the genomic DNA of K562 cells into a luciferase reporter plasmid pGL4.53. The 293T cells were used as the control cell line and the empty pGL4.53 plasmid was used as the control for the baseline luciferase activity. The Y-axis represents the percentage of luciferase activity compared to the pGL4.53 empty plasmids in the respective cells. Part of the data from K562 bottom ranked silencers were also used in Extended Data Figure 2 (n = 3 biological independent samples; the bars show the mean value ± S.E.M; *P value < 0.05, calculated using two-sided Student’s t test). (b) Distribution of the significantly enriched silencer regions from HepG2 cells in the genome. The pie chart indicates the distribution of silencers in genomic features. The bar plot indicates the distribution of silencers in the overlapped annotation features in the genome.
All exact P values are provided in the Source Data.
Extended Data Fig. 7: Unique signatures in silencer regions from HepG2 cells.
(a) Distribution of silencer regions in the respective chromatin states in HepG2 cells. (Txn, transcription; Repressed, Polycomb repressed; lo, low signal; CNV, copy number variation). Colored parts indicate the chromatin states that are significantly enriched compared to the library background distribution (P value < 0.001, one-sided binomial test). (b) Association of histone modifications with silencers in HepG2 cells. The y-axis indicates the significance of the enrichment of histone modifications with silencer fragments. The size of the circle indicates the ratio of silencers covered by the respective histone modification (scale is 0 to 1). The enrichment analysis is based on permutation tests using 20,000 random permutations. The P value was calculated and multiple comparison corrections were computed using the Benjamini-Hochberg procedure. The red line shows the cutoff of the adjusted P value < 0.05. (c) Association of transcription factors with silencers in HepG2 cells. The y-axis indicates the significance of the enrichment of transcription factors with silencer fragments. The size of the circle indicates the ratio of silencers covered by the respective transcription factor (scale is 0 to 1). The enrichment analysis is based on permutation tests using 20,000 random permutations. The P value was calculated and multiple comparison corrections were computed using the Benjamini-Hochberg procedure. The red line shows the cutoff of the adjusted P value < 0.05.
All exact P values are provided in the Source Data.
Extended Data Fig. 8: Additional top motifs present in the silencer regions from K562 or HepG2 cells.
Related to Figure 3d, e and f. Top motifs identified in the silencers regions from K562 (a) or HepG2 (b) cells are shown.
Extended Data Fig. 9: Additional silencer knockout clones from K562 and 293T cells.
Related to Figure 4c, d, e and f. Additional clones where silencer was knocked out from the ABCC2 gene (a) or ABCG2 gene (c) in K562 cells were generated, and confirmed by PCR. The expressions of ABCC2 gene (b) and ABCG2 gene (d) of the respective cells were quantified by qPCR. Clones where silencer was knocked out from the ABCC2 gene (e) or ABCG2 gene (g) in 293T cells were also generated, and confirmed by PCR. The expressions of ABCC2 gene (f) and ABCG2 gene (h) of the respective cells were quantified by qPCR (For all qPCR experiments, n = 3 biological replicates of the same clones; the bars show the mean value ± S.D.; *P value < 0.05, calculated using two-sided Student’s t test. All PCR experiments are the representative result of 2 experiments). (i and j) Silencer knockout clones (from Extended Data Figure 3) were used as additional control knockout clones, since the targeted deletion regions were completely different from the ABCC2 or ABCG2 silencer regions. The expression of ABCC2 (i) and ABCG2 (j) genes of these clones was quantified by qPCR. The expression data of all individual clones were grouped as the control set, and compared to that of all the ABCC2 or ABCG2 silencer knockout clones (for each individual clone, n = 3 biological replicates of the same clones; the bars show the mean value ± S.D.). The ABCC2 and ABCG2 genes were significantly up-regulated in ABCC2 silencer or ABCG2 silencer knockout clones respectively (*P value < 0.05, calculated using one-sided Student’s t test).
All exact P values are provided in the Source Data. The blots were cropped. Full scans of the blots are shown in Source Data Full Gel Extended Data Figure 9.
Extended Data Fig. 10: Long-range actions of silencers with distal genes.
(a) Related to Figure 5d. Direct interactions of silencers with the promoters of distal genes as identified by the 5C data from K562 cells. The red arrow indicates the silencer region, and the blue arrow shows the interaction region with the gene. Interaction arcs are highlighted in blue.
(b) Related to Figure 5e. K562 RNA-seq data is shown for genes that interact with silencers as identified by the 5C data from K562 cells. Source Data.
Supplementary Material
Acknowledgements
We thank Joshua Gruber, Xiao Li, Emma Monte, Kevin Van Bortle, Chao Jiang and Minyi Shi at Stanford University; Lennert Janssen, Yufeng Li, Leonie Hamoen and Almira Henic at Leiden University Medical Center for many helpful discussions and inputs regarding the manuscript. This work was supported by the grant from the National Institutes of Health (NIH) 3U54HG00699604S1 to M.P.S. This work used the Genome Sequencing Service Center of Stanford Center for Genomics and Personalized Medicine Sequencing Center, supported by the grant award NIH S10OD020141. B.P. was partially supported by the LUMC Gisela Thier Fellowship and KWF/Alpe Young Investigator Grant 11707.
Footnotes
Competing interests
M.P.S. is a founder and member of the science advisory board of Personalis, SensOmics, Qbio, January, Akna and Filtricine, and a science advisory board member of Genapsys and Epinomics.
References
- 1.Venter JC et al. The Sequence of the Human Genome. Science 291, 1304–1351, (2001). [DOI] [PubMed] [Google Scholar]
- 2.Lander ES et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921, (2001). [DOI] [PubMed] [Google Scholar]
- 3.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerstein MB et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neph S et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489, 83–90, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djebali S et al. Landscape of transcription in human cells. Nature 489, 101–108, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thurman RE et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roadmap Epigenomics C et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filion GJ et al. Systematic Protein Location Mapping Reveals Five Principal Chromatin Types in Drosophila Cells. Cell 143, 212–224, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst J et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katainen R et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet 47, 818–821, (2015). [DOI] [PubMed] [Google Scholar]
- 12.Lovén J et al. Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell 153, 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khurana E et al. Role of non-coding sequence variants in cancer. Nat Rev Genet 17, 93–108, (2016). [DOI] [PubMed] [Google Scholar]
- 14.Flavahan WA et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melnikov A et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotech 30, 271–277, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwasnieski JC, Fiore C, Chaudhari HG & Cohen BA High-throughput functional testing of ENCODE segmentation predictions. Genome Research, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnold CD et al. Genome-Wide Quantitative Enhancer Activity Maps Identified by STARR-seq. Science 339, 1074–1077, (2013). [DOI] [PubMed] [Google Scholar]
- 18.Zabidi MA et al. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518, 556–559, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Arensbergen J et al. Genome-wide mapping of autonomous promoter activity in human cells. Nat Biotech 35, 145–153, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maston GA, Evans SK & Green MR Transcriptional regulatory elements in the human genome. Annual review of genomics and human genetics 7, 29–59, (2006). [DOI] [PubMed] [Google Scholar]
- 21.Siu G, Wurster AL, Duncan DD, Soliman TM & Hedrick SM A transcriptional silencer controls the developmental expression of the CD4 gene. Embo j 13, 3570–3579, (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawada S, Scarborough JD, Killeen N & Littman DR A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 77, 917–929, (1994). [DOI] [PubMed] [Google Scholar]
- 23.Zou Y-R et al. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nature Genetics 29, 332, (2001). [DOI] [PubMed] [Google Scholar]
- 24.Taniuchi I et al. Differential Requirements for Runx Proteins in CD4 Repression and Epigenetic Silencing during T Lymphocyte Development. Cell 111, 621–633, (2002). [DOI] [PubMed] [Google Scholar]
- 25.Taniuchi I, Sunshine MJ, Festenstein R & Littman DR Evidence for Distinct CD4 Silencer Functions at Different Stages of Thymocyte Differentiation. Molecular Cell 10, 1083–1096, (2002). [DOI] [PubMed] [Google Scholar]
- 26.Pang B et al. Direct Antigen Presentation and Gap Junction Mediated Cross-Presentation during Apoptosis. The Journal of Immunology 183, 1083–1090, (2009). [DOI] [PubMed] [Google Scholar]
- 27.Straathof KC et al. An inducible caspase 9 safety switch for T-cell therapy. Blood 105, 4247–4254, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thurman RE et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giresi PG, Kim J, McDaniell RM, Iyer VR & Lieb JD FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Research 17, 877–885, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biology 15, 554, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Y, Schafer DW, Cumbie JS & Chang JH The NBP negative binomial model for assessing sifferential gene expression from RNA-seq. Stat. Appl. Genet. Mol. Biol 10.2202/1544-6115.1637 (2011). [DOI] [Google Scholar]
- 33.Visel A et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonn S et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44, 148–156, (2012). [DOI] [PubMed] [Google Scholar]
- 35.Dahlman JE et al. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nature biotechnology 33, 1159–1161, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konermann S et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calderwood DA, Shattil SJ & Ginsberg MH Integrins and Actin Filaments: Reciprocal Regulation of Cell Adhesion and Signaling. Journal of Biological Chemistry 275, 22607–22610, (2000). [DOI] [PubMed] [Google Scholar]
- 38.Shlyueva D, Stampfel G & Stark A Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15, 272–286, (2014). [DOI] [PubMed] [Google Scholar]
- 39.Ernst J & Kellis M ChromHMM: automating chromatin state discovery and characterization. Nature methods 9, 215–216, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu G, Wang LG & He QY ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383, (2015). [DOI] [PubMed] [Google Scholar]
- 41.Beck DB, Oda H, Shen SS & Reinberg D PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes & Development 26, 325–337, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chantalat S et al. Histone H3 trimethylation at lysine 36 is associated with constitutive and facultative heterochromatin. Genome Research 21, 1426–1437, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thurman RE et al. The accessible chromatin landscape of the human genome. Nature 489, 75, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visel A et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn SH, Kim M & Buratowski S Phosphorylation of Serine 2 within the RNA Polymerase II C-Terminal Domain Couples Transcription and 3′ End Processing. Molecular Cell 13, 67–76, (2004). [DOI] [PubMed] [Google Scholar]
- 46.Liu T et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biology 12, R83–R83, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang JG, DeFrances MC, Machen J, Johnson C & Zarnegar R The repressive function of AP2 transcription factor on the hepatocyte growth factor gene promoter. Biochemical and biophysical research communications 272, 882–886, (2000). [DOI] [PubMed] [Google Scholar]
- 48.Eckert D, Buhl S, Weber S, Jäger R & Schorle H The AP-2 family of transcription factors. Genome Biology 6, 246, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roth C, Schuierer M, Günther K & Buettner R Genomic Structure and DNA Binding Properties of the Human Zinc Finger Transcriptional Repressor AP-2rep (KLF12). Genomics 63, 384–390, (2000). [DOI] [PubMed] [Google Scholar]
- 50.Bonev B & Cavalli G Organization and function of the 3D genome. Nat Rev Genet 17, 661–678, (2016). [DOI] [PubMed] [Google Scholar]
- 51.Krijger PHL & de Laat W Regulation of disease-associated gene expression in the 3D genome. Nat Rev Mol Cell Biol 17, 771–782, (2016). [DOI] [PubMed] [Google Scholar]
- 52.Rao SS et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y et al. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biology 19, 151, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heidari N et al. Genome-wide map of regulatory interactions in the human genome. Genome Res 24, 1905–1917, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Javierre BM et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 167, 1369–1384.e1319, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanyal A, Lajoie BR, Jain G & Dekker J The long-range interaction landscape of gene promoters. Nature 489, 109, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hart T, Komori HK, LaMere S, Podshivalova K & Salomon DR Finding the active genes in deep RNA-seq gene expression studies. BMC Genomics 14, 778, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muerdter F et al. Resolving systematic errors in widely used enhancer activity assays in human cells. Nature Methods 15, 141, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang YM et al. Correlation between DNase I hypersensitive site distribution and gene expression in HeLa S3 cells. PloS one 7, e42414, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bell O, Tiwari VK, Thomä NH & Schübeler D Determinants and dynamics of genome accessibility. Nature Reviews Genetics 12, 554, (2011). [DOI] [PubMed] [Google Scholar]
- 61.Evers B et al. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nature biotechnology 34, 631, (2016). [DOI] [PubMed] [Google Scholar]
- 62.Morgens DW, Deans RM, Li A & Bassik MC Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nature biotechnology 34, 634, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wijdeven RH et al. Genome-Wide Identification and Characterization of Novel Factors Conferring Resistance to Topoisomerase II Poisons in Cancer. Cancer Research 75, 4176–4187, (2015). [DOI] [PubMed] [Google Scholar]
- 64.Korkmaz G et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nature biotechnology 34, 192, (2016). [DOI] [PubMed] [Google Scholar]
- 65.Boettcher M et al. Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nature biotechnology 36, 170, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao H-K et al. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 171, 1495–1507.e1415, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matharu N et al. CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science 363, eaau0629, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Komor AC, Badran AH & Liu DR CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 168, 20–36, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ma H et al. Correction of a pathogenic gene mutation in human embryos. Nature 548, 413, (2017). [DOI] [PubMed] [Google Scholar]
- 70.Gilbert Luke A. et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 71.Pang B et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nature Communications 4, 1908, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giresi PG & Lieb JD Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements). Methods (San Diego, Calif.) 48, 233–239, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anders S, Pyl PT & Huber W HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ji X, Li W, Song J, Wei L & Liu XS CEAS: cis-regulatory element annotation system. Nucleic Acids Research 34, W551–W554, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu LJ et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Giardine B et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Research 15, 1451–1455, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karolchik D, Hinrichs AS & Kent WJ The UCSC genome browser. Curr. Protoc. Bioinformatics 28, 1.4 (2009). [DOI] [PubMed] [Google Scholar]
- 79.Zhou X & Wang T Using the Wash U epigenome browser to examine genome-wide sequencing data. Curr. Protoc. Bioinformatics 40, 10.10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data are available at GEO under the accession number: GSE108536.