Bacterial MCPs have a number of potential biotechnology applications and have been linked to bacterial pathogenesis, cancer, and heart disease. Glycyl radical MCPs are a large but understudied class of bacterial MCPs. Here, we show that uropathogenic E. coli CFT073 uses a glycyl radical MCP for 1,2-PD fermentation, and we conduct a comprehensive genetic analysis of the genes involved. Studies suggest significant functional differences between the glycyl radical MCP of E. coli CFT073 and better-studied MCPs. They also provide a foundation for building a deeper general understanding of glycyl radical MCPs in an organism where sophisticated genetic methods are available.
KEYWORDS: microcompartment; carboxysome; 1,2-propanediol; Escherichia coli CFT073; glycyl radical diol dehydratase; E. coli CFT073; diol dehydratase; glycyl radical
ABSTRACT
Bacterial microcompartments (MCPs) are widespread protein-based organelles composed of metabolic enzymes encapsulated within a protein shell. The function of MCPs is to optimize metabolic pathways by confining toxic and/or volatile pathway intermediates. A major class of MCPs known as glycyl radical MCPs has only been partially characterized. Here, we show that uropathogenic Escherichia coli CFT073 uses a glycyl radical MCP for 1,2-propanediol (1,2-PD) fermentation. Bioinformatic analyses identified a large gene cluster (named grp for glycyl radical propanediol) that encodes homologs of a glycyl radical diol dehydratase, other 1,2-PD catabolic enzymes, and MCP shell proteins. Growth studies showed that E. coli CFT073 grows on 1,2-PD under anaerobic conditions but not under aerobic conditions. All 19 grp genes were individually deleted, and 8/19 were required for 1,2-PD fermentation. Electron microscopy and genetic studies showed that a bacterial MCP is involved. Bioinformatics combined with genetic analyses support a proposed pathway of 1,2-PD degradation and suggest that enzymatic cofactors are recycled internally within the Grp MCP. A two-component system (grpP and grpQ) is shown to mediate induction of the grp locus by 1,2-PD. Tests of the E. coli Reference (ECOR) collection indicate that >10% of E. coli strains ferment 1,2-PD using a glycyl radical MCP. In contrast to other MCP systems, individual deletions of MCP shell genes (grpE, grpH, and grpI) eliminated 1,2-PD catabolism, suggesting significant functional differences with known MCPs. Overall, the studies presented here are the first comprehensive genetic analysis of a Grp-type MCP.
IMPORTANCE Bacterial MCPs have a number of potential biotechnology applications and have been linked to bacterial pathogenesis, cancer, and heart disease. Glycyl radical MCPs are a large but understudied class of bacterial MCPs. Here, we show that uropathogenic E. coli CFT073 uses a glycyl radical MCP for 1,2-PD fermentation, and we conduct a comprehensive genetic analysis of the genes involved. Studies suggest significant functional differences between the glycyl radical MCP of E. coli CFT073 and better-studied MCPs. They also provide a foundation for building a deeper general understanding of glycyl radical MCPs in an organism where sophisticated genetic methods are available.
INTRODUCTION
Bacterial microcompartments (MCPs) are proteinaceous organelles produced by hundreds of species of bacteria (1–6). They are polyhedral in shape, about 100 to 150 nm in diameter, and are built from tens of thousands of protein subunits of 10 to 20 different types. In the cases that have been best studied, MCPs consist of a selectively permeable protein shell that encapsulates sequentially acting enzymes together with their substrates. The proposed function of MCPs is to optimize metabolic pathways that have toxic or volatile intermediates. Such intermediates are produced within the MCP and then further metabolized into compounds that can safely exit into the cytoplasm of the cell. In the cytoplasm, MCP products are metabolized to provide the cell with energy, carbon, or carbon and energy for growth. Overall, compartmentalization of specific pathways within MCPs mitigates damage to DNA and other cytoplasmic components by toxic metabolic intermediates (7–11). It also minimizes the loss of small nonpolar intermediates (such as short-chain aldehydes) that readily diffuse across the cell envelope but are retained by the protein shells of MCPs (6, 8, 12). Moreover, MCPs concentrate enzymes together with their substrates, increasing reaction rates, as exemplified by the carboxysome MCP, which enhances CO2 fixation (6).
Bacterial MCPs are functionally diverse and have potential importance in human health and biotechnology. Studies have indicated that MCPs are involved in ten or more metabolic processes, which include carbon dioxide fixation as well as the catabolism of 1,2-propanediol, ethanolamine, choline, glycerol, rhamnose, fucose, fucoidan, and aminoacetone (12–21). MCPs are associated with pathogenesis in Salmonella and Listeria (22–25) and might be linked to heart disease and cancer in humans due to their metabolic roles in the gut environment (26–29). In addition, MCP components are being developed as nanomaterials for the production of protein-based containers for use in renewable chemicals production, drug delivery, and the expression of toxic proteins (30–40).
In most cases, the genes for MCPs are found in large operons/gene clusters that are sufficient for MCP formation (1–6, 41, 42). The enzymes encoded by these clusters vary according to the MCP substrate metabolized, whereas the proteins used to build the shells are generally conserved. The vertices of MCP shells are formed by bacterial microcompartment vertex (BMV) proteins (43). The facets of the shells are assembled from a family of conserved proteins known as bacterial microcompartment (BMC) domain proteins (44). MCP shell facets are usually composed of 3 to 8 different types of BMC domain proteins (5, 41). Diverse BMC domain proteins share a conserved flat hexagonal shape and tessellate into the mixed sheets that form the facets of the MCP. Functional diversification among BMC proteins is thought to determine the selective permeability of the shell and optimize MCP function in diverse host contexts.
Despite their widespread occurrence and diversity, only three types of MCPs have been studied in any detail. These include the 1,2-propanediol utilization (Pdu) MCP, the ethanolamine utilization (Eut) MCP, and the carboxysome. The Pdu and Eut MCPs mediate catabolism of 1,2-propanediol (1,2-PD) and ethanolamine by pathways that require coenzyme B12 (4, 5, 41). The carboxysome enhances autotrophic CO2 fixation by optimizing ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) activity (6). A widespread but understudied group of MCPs (associated with glycyl radical enzymes) is termed glycyl radical MCPs (3, 41). Glycyl radical MCPs associated with choline trimethylamine (TMA) lyase mediate anaerobic choline degradation in varied organisms (45–47). Glycyl radical MCPs associated with glycyl radical diol dehydratases (GR-DDH) mediate the degradation of 1,2-PD, fucose, rhamnose, and possibly glycerol by pathways that are independent of coenzyme B12 (19, 48–50). Recent studies partially characterized the type 3 glycyl radical MCP loci of Rhodopseudomonas palustris and Rhodobacter capsulatus (48, 49). Results indicated that these loci mediate 1,2-PD degradation (48, 49); however, relatively limited genetic studies were performed, and MCP formation was not demonstrated (48, 49). Here, we establish that Escherichia coli CFT073 uses an MCP for fermentation of 1,2-PD, and we perform a comprehensive genetic analysis to gain insights into its functions and mechanisms. Electron microscopy, genetic, and physiological studies establish that an MCP is used for 1,2-PD fermentation. Genetic and bioinformatic studies support a proposed pathway for 1,2-PD degradation, define the regulation of the system, address the distribution of glycyl radical MCPs among E. coli strains, and suggest significant functional differences between the glycyl radical MCP of E. coli CFT073 and better-studied MCPs. In accordance with prior work, we refer to the genes used for formation of this MCP as grp (glycyl radical propanediol) and the MCP as the Grp MCP (3).
RESULTS
The E. coli CFT073 grp gene cluster.
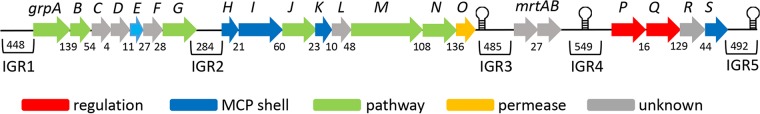
Bioinformatic studies conducted here and previously (41) identified a large gene cluster in E. coli CFT073 that appears to encode a type 3 glycyl radical MCP used for the degradation of 1,2-PD or a related substrate(s) (Fig. 1). This gene cluster (the grp cluster) contains 21 genes transcribed in the same direction. Sequence analyses indicate that grp genes encode enzymes for the metabolism of 1,2-PD to propionate and 1-propanol (including GR-DDH), five MCP shell proteins, and a number of proteins of unknown function. These analyses suggest that E. coli CFT073 uses an MCP to metabolize 1,2-PD as shown in Fig. 2.
FIG 1.
The grp gene cluster of E. coli CFT073. The grp gene cluster of E. coli CFT073 includes 21 genes transcribed in the same direction. General gene functions are indicated by color. The green-colored genes encode all the enzymes needed for the conversion of 1,2-PD to 1-propanol and propionate (as shown in Fig. 2), including a glycyl radical diol dehydratase (grpM). The numbers beneath genes indicate the size of intergenic regions (IGRs) in base pairs. The largest internal IGRs are numbered 2 through 4. The graphical stem-loop structures indicate the locations of possible intrinsic terminators predicted by RegRNA 2.0 software.
FIG 2.
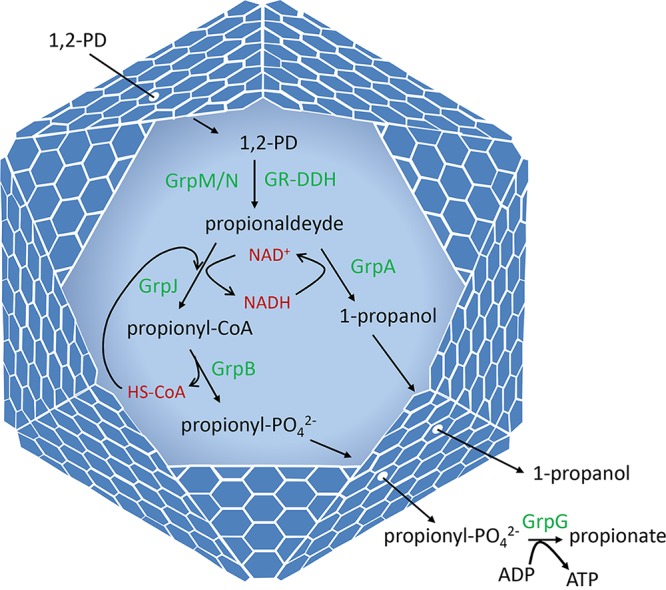
Model for 1,2-PD degradation by the Grp MCP of E. coli CFT073. The Grp MCP allows E. coli CFT073 to grow by 1,2-PD fermentation. 1,2-PD diffuses through the protein shell of the MCP and enters the lumen, where it is converted to propionaldehyde by a glycyl radical diol dehydratase (GrpM) supported by its activating enzyme (GrpN). Propionaldehyde is further metabolized to 1-propanol and propionate by the GrpJ aldehyde dehydrogenase, the GrpA alcohol dehydrogenase, the GrpB phosphotransacylase, and the GrpG propionate kinase. This pathway generates ATP, which is used to support growth. Based on analogy with other MCPs, its likely function is to sequester propionaldehyde to prevent toxicity and/or diffusive loss through the cell envelope. Also based on analogy, the GrpJ aldehyde dehydrogenase and the grpA 1-propanol dehydrogenase are likely required for recycling coenzyme A (HSCoA) and NAD+, respectively, internally within the MCP.
Further sequence analyses showed that the grp cluster contains three relatively large internal intergenic regions (IGR2, IGR3, and IGR4) that are ≥284 bp in length (Fig. 1). Since large IGRs are uncommon in bacterial operons/gene clusters, we examined these IGRs for open reading frames (ORFs) that might have been overlooked by prior annotation or were perhaps missed due to sequencing errors. IGR2, IGR3, and IGR4 were amplified by PCR and resequenced. The sequences of all three IGRs matched the genome sequence of E. coli CFT073 available through the NCBI database (GenBank accession number NC_004431). To search for ORFs that may have been missed, the internal IGRs and their flanking sequences were analyzed using BLASTX (51), PSI-BLAST (52), GeneMark.hmm (53), and the ExPASy translate tool. IGR2 contains a small ORF (ORFA) of 171 bp (56 amino acids), but this ORF was not identified as a gene by GeneMark.hmm trained with the E. coli CFT073 sequences, and PSI-BLAST found no homologs of this ORF in the NCBI nonredundant database. Thus, bioinformatic analyses suggest that the small ORF located between the grpG and grpH genes is not translated. IGR3 also contains a small ORF (ORFB; 201 bp, 67 amino acids) transcribed in the direction opposite to the grp genes. This ORF is not found by GeneMark.hmm, but it is conserved in many strains of E. coli, so translation of this ORF is an open question. Lastly, IGR4 contains three small ORFs. Two (ORFC and ORFD) are unlikely to be translated based on the criteria described above. The third (ORFE) is adjacent to mrtB (hypothetical maturase/reverse transcriptase gene) and encodes a protein with homology to MrtB. However, ORFE has no start codon, and resequencing verified the “UAG” stop codon of mrtB. This raises the possibility that the UAG stop codon of mrtB is translated at some frequency or that the mrtB gene is nonfunctional and degenerating. These ORF searches are summarized in Table S1 in the supplemental material.
As mentioned above, the grp gene cluster includes three IGRs of ≥284 bp (Fig. 1). In addition, there are IGRs of 108, 136, and 129 bp just upstream of the grpN, grpO, and grpR genes. These intergenic sequences are long enough to include elements that influence gene expression, raising the possibility that the grp cluster includes more than one operon and/or is regulated in a complex manner. We also analyzed the grp gene cluster with RegRNA 2.0 (54) and found three predicted intrinsic terminators; two flank the mrtA-mrtB genes (GCCAGCAGTGTCTGCCTGCTGGC and CGCCAGTAACTGGC) and one is located just past the end of the grp locus (TGCCGCGGTAACAATTTCTTTATCGCGGCA) (Fig. 1).
We note that bioinformatic analyses showed that the grp locus contains two genes with homology to reverse transcriptase maturase genes (mrtA and mrtB). These genes have no known role in 1,2-PD metabolism and may be remnants of horizontal gene transfer, as further considered in the Discussion. Overall, the bioinformatic analyses described above suggest that the grp gene cluster of E. coli CFT073 encodes a glycyl radical MCP used for 1,2-PD degradation by the pathway shown in Fig. 2. These analyses also suggest that the grp locus might have complex regulation and that it might contain genes associated with horizontal gene transfer (mrtAB).
E. coli CFT073 degrades 1,2-PD by fermentation.
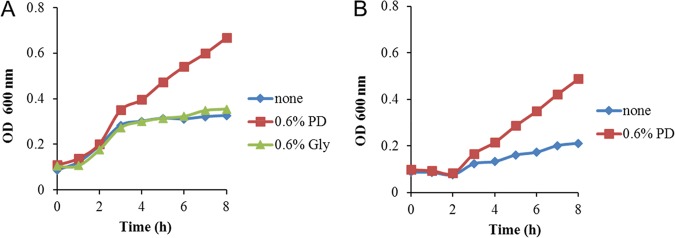
The bioinformatic studies described above suggested that the grp cluster of E. coli CFT073 encodes a glycyl radical MCP used for the degradation of 1,2-PD. To test this, growth studies were conducted. E. coli CFT073 was unable to use 1,2-PD as a sole carbon and energy source under aerobic or anaerobic conditions, with or without vitamin B12 supplementation (GR-DDH enzymes do not require coenzyme B12, but other pathways of 1,2-PD degradation are B12 dependent). In contrast, under anaerobic conditions, 1,2-PD stimulated growth of E. coli CFT073 in medium supplemented with fumarate or small amounts of yeast extract (0.2%) (Fig. 3). This indicated that E. coli CFT073 fermented 1,2-PD, producing ATP that stimulated growth. It is likely that the yeast extract provided biosynthetic precursors, since there are no known pathways in E. coli that convert intermediates of 1,2-PD degradation to biosynthetic building blocks. The fumarate used in some experiments may have provided biosynthetic precursors or served as a terminal electron acceptor or both. The studies done here do not distinguish among these possibilities.
FIG 3.
Growth response of E. coli CFT073 to 1,2-propanediol. 1,2-Propanediol stimulated growth of E. coli CFT073 under anaerobic conditions on minimal medium containing 0.2% yeast extract (A) or 20 mM fumarate (B). Cells were grown anaerobically in sealed serum tubes. Cell growth was measure by monitoring the optical density at 600 nm (OD600) over time.
Further growth studies showed that E. coli CFT073 was unable to use 1,2-PD for anaerobic respiration with dimethyl sulfoxide (DMSO), trimethylamine N-oxide (TMAO), or nitrate. Glycerol was used as a positive control, and E. coli CFT073 grew well by anaerobic respiration of glycerol with nitrate, TMAO, or DMSO as terminal electron acceptors. We also found that glycerol did not stimulate growth of E. coli CFT073 on medium supplemented with 0.2% yeast extract. Thus, overall, growth studies indicated that E. coli CFT073 degrades 1,2-PD by fermentation but does not ferment glycerol (a possible substrate for GR-DDH).
1,2-PD metabolism in the ECOR collection.
The E. coli Reference (ECOR) collection is a set of 72 natural isolates of E. coli intended to represent the genetic diversity of the species (55). This collection was tested for strains capable of 1,2-PD degradation using MacConkey–1,2-PD indicator medium. On this medium, positive strains are red due to acid production from 1,2-PD. Results indicated that nine ECOR strains (12.5%) were able to degrade 1,2-PD. ECOR strains 24, 49, 50, 59, 61, and 62 gave clearly positive results, while ECOR strains 35, 36, and 55 gave weak positive results. These ECOR strains did not require added vitamin B12 for 1,2-PD degradation, indicating that 1,2-PD degradation was B12 independent (56). Prior studies found that about 20% of strains in the ECOR collection could degrade 1,2-PD in a B12-independent manner (including all of the strains reported as 1,2-PD positive here) (56). Despite numerous retests on MacConkey–1,2-PD plates, we were unable to replicate those studies. The reason why prior tests found more 1,2-PD positive strains will require further study.
The nine 1,2-PD-positive ECOR strains identified here were tested for the presence of the grp gene cluster by PCR. This was done by using a PCR primer pair that amplified across the grpKLM genes, which encode three proteins characteristic of the grp gene cluster (an MCP shell protein, a protease-like protein, and GR-DDH). For 8/9 1,2-PD-positive ECOR strains, an amplicon was obtained that had the expected size and that was >98% identical in DNA sequence to the corresponding region from E. coli CFT073. The exception was ECOR 55, for which no PCR product was obtained. Thus, it is likely that ECOR strains 24, 49, 50, 59, 61 62, 35, and 36 (and possibly 55) have a grp gene cluster that allows the B12-independent degradation of 1,2-PD.
We also note that MacConkey–1,2-PD agar provides a simple test for bacterial 1,2-PD degradation that should prove helpful to varied studies of 1,2-PD metabolism. Even though 1,2-PD metabolism occurs only in the absence of oxygen, the MacConkey–1,2-PD plates were incubated under aerobic conditions because as colonies expand their centers become anaerobic.
E. coli CFT073 produces bacterial microcompartments in the presence of 1,2-PD.
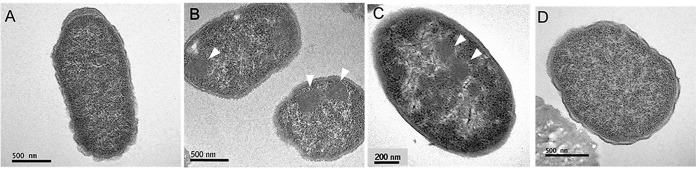
The presence of five MCP shell genes in the grp gene cluster of E. coli CFT073 suggested that a bacterial MCP might be involved in 1,2-PD degradation in this organism. To test this, E. coli CFT073 was grown anaerobically on lysogeny broth (LB) with and without 1,2-PD, and thin sections were prepared and viewed by transmission electron microscopy. Large protein complexes similar in size and appearance to bacterial MCPs were observed in cells grown in the presence of 1,2-PD, but similar complexes were not seen in cells grown without 1,2-PD supplementation (Fig. 4). We also found that a grpP deletion did not form MCPs under conditions where MCPs were seen in wild-type E. coli CFT073. As described below, grpP is a histidine kinase required for induction of the grp operon. This indicated that a grp gene was required for MCP formation.
FIG 4.
Electron microscopy of E. coli CFT073 grown in the presence and absence of 1,2-PD. (A to C) E. coli CFT073 grown in the absence of 1,2-PD (A), the presence of 1,2-PD (B), and the presence of 1,2-PD (higher magnification (C)). (D) A grpP mutant (unable to express grp genes) grown in the presence of 1,2-PD. Arrowheads point to bacterial MCPs.
Genes in the grp cluster are required for 1,2-PD degradation by E. coli CFT073.
Each gene of the grp cluster was individually deleted by linear recombination of PCR products (57). Deletions were designed to leave the ribosome binding region (∼20 bp) of the downstream gene intact (57). The target gene was initially replaced with a chloramphenicol or kanamycin resistance marker that was removed with the Flp recombinase, leaving behind an frt site, as described previously (57). This method is designed to produce nonpolar deletions (57).
For each mutant, minimal medium and MacConkey–1,2-PD plates were used to test for 1,2-PD degradation. Growth curves on minimal medium are shown in Fig. S1. Results are summarized in Table 1. Deletions of, grpE, grpH, grpI, grpJ, grpM, grpN, grpP, and grpQ genes eliminated 1,2-PD utilization in anaerobic liquid cultures and on MacConkey–1,2-PD indicator plates. Individual deletions of seven additional grp genes (grpA grpB, grpC, grpG, grpK, grpL, and grpS) partially impaired 1,2-PD utilization in anaerobic liquid cultures and on MacConkey–1,2-PD indicator plates. In contrast, individual deletions of four grp genes (grpD, grpF, grpO, and grpR) had no detectable effect on 1,2-PD degradation in liquid cultures or on MacConkey plates. Similarly, a double deletion of both the mrtA are mrtB genes (which are flanked by grp genes) (Fig. 1) had no discernible effects on 1,2-PD degradation by E. coli CFT073. These studies demonstrated that grp genes were required for 1,2-fermenation by E. coli CFT073, supported the proposed pathway shown in Fig. 2, and suggested functional differences between the Grp MCP and other better-studied MCPs, as further addressed in the Discussion.
TABLE 1.
Growth of grp deletion mutants on 1,2-PD
| Strain or deletion mutant description | Closest homolog of deleted gene product | 1,2-PD utilizationb | Protein product of deleted gene (GenPept accession no.) |
|---|---|---|---|
| WTa | +++ | ||
| ΔgrpA | Alcohol dehydrogenase | +−− | WP_000135343.1 |
| ΔgrpB | Phosphotransacylase | +−− | WP_001109982.1 |
| ΔgrpC | EutJ-like chaperone of unknown function | ++− | WP_001332070.1 |
| ΔgrpD | Hypothetical flavoprotein | +++ | WP_001329683.1 |
| ΔgrpE | EutN MCP vertex protein | − | WP_000280292.1 |
| ΔgrpF | Heme-binding protein of unknown function | +++ | WP_001301167.1 |
| ΔgrpG | Propionate kinase | ++− | WP_001362987.1 |
| ΔgrpH | Shell hexamer | − | WP_001301171.1 |
| ΔgrpI | Shell trimer (stacked/gated) | − | WP_001205499.1 |
| ΔgrpJ | Aldehyde dehydrogenase | − | WP_000997839.1 |
| ΔgrpK | Shell hexamer with FeS cluster | +/− | WP_000746009.1 |
| ΔgrpL | Putative protease of unknown function | +/− | WP_000722279.1 |
| ΔgrpM | Glycyl radical diol dehydratase (GR-DDH) | − | WP_000890295.1 |
| ΔgrpN | grpM activating enzyme | − | WP_001044978.1 |
| ΔgrpO | Permease | +++ | WP_001086421.1 |
| ΔmtrA | Maturase-reverse transcriptase | +++ | AAN82974.1 |
| ΔmtrB | Maturase-reverse transcriptase | +++ | WP_000237741.1 |
| ΔgrpP | Histidine kinase | − | AAN82979.1 |
| ΔgrpQ | DNA-binding response regulator | − | WP_000494233.1 |
| ΔgrpR | Methionine adenosyltransferase | +++ | WP_096489982.1 |
| ΔgrpS | Shell hexamer with C-terminal extension | ++− | WP_000015571.1 |
| ΔmrtAB | Maturase-reverse transcriptase | +++ |
WT, wild-type E. coli CFT073.
Qualitative estimate of growth based on the growth curves shown in Fig. S1. A minus sign indicates no growth. One, two, or three plus signs indicate slow, medium, or normal growth, respectively.
Complementation studies.
Complementation studies were performed on the 13 grp mutants that had clear phenotypes in the growth tests (ΔgrpA, ΔgrpB, ΔgrpE, ΔgrpG, ΔgrpH, ΔgrpI, ΔgrpJ, ΔgrpK, ΔgrpL, ΔgrpM, ΔgrpN, ΔgrpP, and ΔgrpQ) (Fig. S2). For these studies, we tested whether expression of the corresponding gene from plasmid pLac22 restored growth stimulation by 1,2-PD in liquid cultures. For 10/13 mutants (ΔgrpB, ΔgrpE, ΔgrpJ, ΔgrpK, ΔgrpG, ΔgrpL, ΔgrpM, ΔgrpN, ΔgrpP, and ΔgrpQ), full complementation was observed. This indicated that the ΔgrpB, ΔgrpE, ΔgrpG, ΔgrpJ, ΔgrpK, ΔgrpL, ΔgrpM, ΔgrpN, ΔgrpP, and ΔgrpQ mutants are nonpolar and that the impaired growth of each mutant on 1,2-PD was a consequence of grp mutation being tested. On the other hand, complementation of grpA (which encodes a putative alcohol dehydrogenase) was partial (Fig. S1). Two independent mutants were tested, with similar results. The DNA sequences of both mutants and of the grpA gene cloned into pLac22 (the complementation vector) were confirmed. Hence, the reason for partial complementation is uncertain, but it could be due to poor production of the GrpA protein. Somewhat surprisingly, the grpH and grpI deletions were not complemented by the corresponding genes expressed from pLac22. Two independent alleles of each deletion were tested. DNA sequences of clones and mutants were verified. Prior studies showed that complementation of MCP shell gene mutations can be difficult due to the effects of gene dosage or chromosomal position (58). The vector used for these studies (pLac22) allows regulated gene expression by isopropyl-β-d-1-thiogalactopyranoside (IPTG), and five different IPTG levels were tested. Thus, we tentatively suggest that the functions of grpH and grpI genes are influenced by genomic position, as was shown for the pduJ MCP shell gene (59).
The grp gene cluster of E. coli CFT073 is induced by 1,2-propanediol.
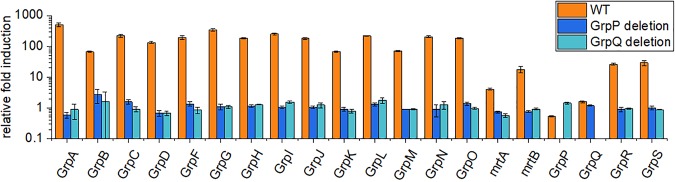
Real-time quantitative PCR (RT-qPCR) was used to measure transcription of the grp operon in wild-type E. coli CFT073. Results showed that the grp operon was induced by 1,2-PD under anaerobic conditions (Fig. 5). The various grp genes were induced by 1,2-PD from 20- to 500-fold, with the exceptions of grpP and grpQ, which were not induced (Fig. 5). This suggests that a 1,2-PD responsive promoter is located upstream of grpA in IGR1. The finding that grpP and grpQ were not induced by 1,2-PD suggests that a transcriptional terminator is present in the intergenic region upstream of grpP. An intrinsic terminator was detected bioinformatically (by RegRNA 2.0) 170 bp upstream of grpP (CGCCAGTAACTGGCG). Presumably, this terminator accounts for why 1,2-PD induced the upstream genes of the grp locus but not the grpP and grpQ genes. These results also suggest that a regulated promoter is located upstream of grpR and grpS, since these two genes are both induced >20-fold by 1,2-PD supplementation despite the transcriptional terminator located upstream of grpP (Fig. 1). This promoter could possibly be located in the 128 bp IGR just upstream of the grpR gene.
FIG 5.
Regulation of the grp genes of E. coli CFT073. RT-qPCR was used to measure the relative fold induction of each grp gene by 1,2-PD in wild-type CFT073 E. coli and in mutants where grpP or grpQ was deleted. Results shown are the average from three replicates, with induction calculated using the threshold cycle (ΔΔCT) method with the gapA gene as the standard. The error bars represent one standard deviation. The growth conditions and RT-qPCR methods are described in Materials and Methods.
grpP and grpQ genes are required for induction for the grp gene cluster.
GrpP and GrpQ have sequence similarity to histidine kinases and response regulators, respectively, suggesting a possible role in regulating transcription of grp genes. RT-qPCR showed that deletion of either grpP or grpQ prevented the induction of grp genes by 1,2-PD (Fig. 5), suggesting that they are a two-component regulatory system that mediates induction of the grp genes in response to 1,2-PD. This idea is supported by growth and complementation studies that showed that grpP and grpQ were required for 1,2-PD degradation (Table 1; see also Fig. S1 and S2 in the supplemental material), as well as by electron microscopy, which showed grpP mutants were unable to form MCPs when growing on 1,2-PD (Fig. 4).
DISCUSSION
Prior bioinformatic and structural analyses placed glycyl radical MCPs into five classes (GRM1 to GRM5) (3, 41). The Grp MCP of E. coli CFT073 was classified as a GRM3. Previous in vitro studies of three key enzymes encoded by a GRM3 locus in R. palustris indicated that it was used for 1,2-PD metabolism (49), which is consistent with the studies presented here that showed that the grp locus of E. coli CFT073 is used for 1,2-PD fermentation. Prior studies of a GRM3 locus of Rhodobacter capsulatus showed that it mediated the metabolism of 1,2-PD during anaerobic respiration on glucose-pyruvate with DMSO (48). High-performance liquid chromatography (HPLC) identified propionate, 1-propanol, and propionaldehyde as metabolic products and was used to show that deletion of GR-DDH eliminated 1,2-PD metabolism (48). These results are consistent with the pathway proposed in Fig. 2. However, in R. capsulatus 1,2-PD metabolism had the unexpected effect of inhibiting cell growth and it was suggested that wild-type R. capsulatus might not assemble an intact MCP shell, resulting in propionaldehyde toxicity (48). Thus, the function of the GRM3 in R. capsulatus is currently enigmatic. In contrast, here we showed that a GRM3 of E. coli CFT073 is used for the fermentation of 1,2-PD. We also expanded on prior studies in R. palustris and R. capsulatus by performing a comprehensive genetic analysis of the genes involved in 1,2-PD fermentation by E. coli CFT073 and by establishing experimentally that an MCP was involved. These results are further discussed below.
Genetic and bioinformatic analysis indicated that E. coli CFT073 ferments 1,2-PD by the pathway shown in Fig. 2. Bioinformatic analyses performed here and previously (41) showed the grp locus encodes homologs of all the pathway enzymes shown in Fig. 2, including GR-DDH (GrpM) and its activating enzyme (GrpN), as well as propionaldehyde dehydrogenase (GrpJ), phosphotransacylase (GrpB), 1-propanol dehydrogenase (GrpA), and propionate kinase (GrpG). This pathway is very similar to the 1,2-PD degradative pathway of Salmonella, except that a GR-DDH (coenzyme B12 independent) replaces the coenzyme B12-dependent DDH found in Salmonella. The pathway proposed in Fig. 2 is further supported by analysis of deletion mutants that removed each pathway enzyme individually. Deletions of the GrpM GR-DDH, the GrpN GR-DDH activating enzyme, and GrpJ aldehyde dehydrogenase eliminated growth stimulation by 1,2-PD, consistent with their roles as pathway enzymes. Deletion of the GrpA Adh and the GrpB phosphotransacylase substantially impaired 1,2-PD catabolism but did not eliminate it. Similar partial phenotypes were seen when genes encoding the analogous enzymes of the Salmonella B12-dependent pathway of 1,2-PD degradation (PduQ and PduL) were deleted (60, 61). In this case, the reason for partial phenotypes was that although the Salmonella Pdu enzymes are redundant with housekeeping enzymes (and should have little to no phenotype) they are still needed to recycle cofactors internally within the Pdu MCP (60, 61). Hence, we propose that GrpA and GrpB are pathway enzymes that play an important role recycling of cofactors internally with the Grp MCP. Results also showed that the grpG Prk was mildly impaired for 1,2-PD degradation. For the Pdu pathway of Salmonella, deletion of the gene that encodes Prk (pduW) has no effect on 1,2-PD degradation (62). The Salmonella Prk is thought to be redundant with the housekeeping acetate kinase (which uses both acetate and propionate as substrates). It seems likely to us that GrpG is a pathway enzyme that is partially redundant with the housekeeping acetate kinase. Thus, overall, the bioinformatic and genetic analyses of grp genes described above support the 1,2-PD degradative pathway shown in Fig. 2 and suggest internal cofactor recycling.
Several studies indicated that a bacterial MCP is used for 1,2-PD degradation by E. coli CFT073. Electron microscopy showed that large complexes similar in size and shape to bacterial MCPs were produced during growth of E. coli CFT073 on 1,2-PD but not during growth on other substrates (Fig. 4). In addition, MCPs were absent from cells where induction of the grp operon was prevented by deletion of the GrpP histidine kinase. Bioinformatic analyses performed here and previously (3, 41) identified five grp genes that encoded homologs of microcompartment shell proteins (grpE, grpH, grpI, grpK, and grpS). Hence, several independent lines of evidence indicate that the grp locus of E. coli CFT073 encodes a glycyl radical MCP used for 1,2-PD fermentation. Based on analogy with the 1,2-PD, ethanolamine, and carboxysome MCPs, possible functions of the Grp MCP are to increase pathway flux and/or to sequester propionaldehyde to prevent toxicity and carbon loss (7–11); however, alternative/additional functions are suggested by studies reported here.
The genetic analyses reported here suggested significant functional differences between the Grp MCP and other MCPs that have been studied. The grp locus of E. coli CFT073 includes five genes that encode homologs of proteins that make up the shells of MCPs (grpE, grpH, grpI, grpK, and grpS). Individual deletions of the grpH, grpI, or grpE genes prevented growth stimulation by 1,2-PD. This result was unexpected. For the Eut and Pdu MCPs, shell gene deletions lead to temporary growth arrest (due to aldehyde toxicity) or have no effect on growth, depending on culture conditions (7–9). In the case of the choline utilization (Cut) MCP, disruption of the MCP shell does not impair growth on choline under standard anaerobic growth conditions (47). The Cut MCP is proposed to protect cells from DNA damage by acetaldehyde and to minimize the loss of acetaldehyde, and neither condition is expected to impair growth under standard anaerobic culture conditions (47). Hence, the finding that the shell of the Grp MCP is required for fermentation of 1,2-PD contrasts with those of studies done on other MCPs and suggests that the Grp MCP has a substantially different function than those of previously studied MCPs. This is of particular interest since the function of a Grp-MCP has not been determined experimentally in any organism. One interesting possibility is that the shell of the Grp MCP is required for the activity or stability of its associated 1,2-PD degradative enzymes, although more work will be needed to sort this out.
We note that individual deletions of two shell genes, grpS and grpK, only partially impaired growth of E. coli CFT073 on 1,2-PD. Presumably, some shell function remains when the GrpS and GrpK shell proteins are eliminated. A similar situation is seen for the Pdu and Eut MCPs, where deletion of minor abundance shell proteins has little effect on MCP function (8, 58). In cases where shell gene deletions lack significant phenotypes, their encoded proteins are thought to have specialized roles that are required under specific environmental conditions (58).
The grp operon encodes a number of proteins of unknown/uncertain function, including GrpC (putative eutJ-like chaperone), GrpD (possible flavoprotein), GrpF (heme-binding protein), GrpL (protease), GrpO (permease), MrtA and MrtB (maturase-reverse transcriptase), and GrpR (methionine adenosyltransferase). Deletion of grpC, grpD, grpF, grpO, grpR, or mrtAB individually had little effect on growth of E. coli CFT073 on 1,2-PD. However, in conjunction with prior work, we can make inferences about their possible functions and suggest reasons why deletion mutants did not have apparent phenotypes. GrpR, a putative methionine adenosyltransferase, might be used to augment the cellular supply of S-adenosylmethioine, which is needed for activation/reactivation of GR-DDH. However, GrpR might only be needed when GR-DDH inactivation rates are high, such as those under microaerobic conditions. Similarly, the GrpD flavoprotein could be an electron donor for the GrpN GR-DDH activase/reactivase and perhaps only needed when the rate of inactivation of GR-DDH is high. Presumably, the GrpO permease is used to facilitate diffusion of 1,2-PD across the cytoplasmic membrane. However, given the structural similarities between 1,2-PD and glycerol, this protein might be redundant with the GlpF glycerol facilitator encoded outside the grp locus. In the case of the GrpC EutJ-like chaperone and the GrpF heme-binding proteins, homologs are widely found in MCP gene clusters, suggesting a role in MCP assembly or function. However, deletion of the grpC and grpF genes did not result in substantial phenotypes, suggesting that their functions may depend on environmental conditions. Deletion of the mrtA and mrtB genes, which encode a maturase-reverse transcriptase homolog (MrtAB), also had no effect on the growth of E. coli CFT073 on 1,2-PD. Since genes related to mrtAB mediate the movement of mobile genetic elements (63), the mrtAB genes may have had a role in horizontal gene transfer into E. coli CFT073 rather than a direct role in 1,2-PD metabolism. Deletion of one gene of unknown function (grpL) significantly impaired growth of E. coli CFT073 on 1,2-PD but did not eliminate growth. The GrpL protein has homology to proteases. By analogy with viral capsid assembly (64), the GrpL protease might be needed for maturation of the MCP shell. This would be unprecedented among bacterial MCPs.
Studies also showed that the grpP and grpQ genes are essential for growth of E. coli CFT073 on 1,2-PD, and are required for induction of the grp locus by 1,2-PD. GrpP and GrpQ have homology to histidine kinases and response regulators, respectively. The N-terminal region of the GrpP histidine kinase is related in sequence to the PocR protein of Salmonella, which induces the Pdu MCP in response to 1,2-PD (65, 66). In addition, recent studies showed that a homologous two-component system regulates the GRM3 locus of R. capsulatus (48). Thus, it is likely that GrpPQ is a two-component system that induces grp genes in response to 1,2-PD.
Finally, to better evaluate the distribution of Grp MCPs among E. coli strains, we screened the ECOR collection using MacConkey agar supplemented with 1,2-PD. The ECOR collection consists of 72 E. coli strains that are representative of the genetic variation of the species as a whole (55). Nine of 72 ECOR strains were found to degrade 1,2-PD without added vitamin B12. PCR followed by DNA sequencing showed that eight of nine 1,2-PD-positive ECOR strains carried genes characteristic of the grp locus, which include grpM (encodes GR-DDH), grpK (encodes a hexameric shell protein), and grpL (encodes a putative protease). Two of the 1,2-PD-positive ECOR strains were isolated from patients with urinary tract infections (UTI), and seven were from feces of healthy individuals. E. coli CFT073, which was studied here, is a uropathogenic strain. Although 1,2-PD degradation by the vitamin B12-dependent pathway has been linked to pathogenesis in Salmonella and Listeria (22–25), to our knowledge, a link between 1,2-PD metabolism via the Grp MCP and pathogenesis has not yet been found.
MATERIALS AND METHODS
Chemicals and reagents.
Antibiotics were from Sigma Chemical Company (St. Louis, MO). Choice Taq Blue master mix was from Denville Scientific (Holliston, MA). KOD Hot Start master mix was from EMD Millipore (Billerica, MA). Isopropyl-β-d-1-thiogalactopyranoside (IPTG) was from Diagnostic Chemicals Limited (Charlotteville, PEI, Canada). The restriction enzymes, HiFi DNA assembly master mix, and T4 DNA ligase were from New England Biolabs (Beverly, MA).
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table S2 in the supplemental material. Biosafety level 2 (BSL2) precautions were used for handling all strains. The rich media used were lysogeny broth (LB) medium, also known as Luria-Bertani medium (Becton, Dickinson and Company, Franklin Lakes, NJ) (67) and Terrific Broth (MP Biomedicals, Solon, OH). MacConkey–1,2-PD indicator plates were prepared using Difco MacConkey agar base supplemented with 1% 1,2-PD. The liquid medium used for growth curves and complementation studies was no-carbon-E (NCE) medium (68) supplemented with 1 mM MgSO4, 50 μM ferric citrate, 125 mM NaCl, 1× BME vitamins (catalog no. B6891; Sigma), and one or more of the following at the concentrations indicated in the text: yeast extract, 1,2-PD, disodium fumarate (pH 7.0), and glycerol. The growth studies shown in Fig. 2 were performed using 18- by 150-mm serum tubes sealed with butyl rubber stoppers as described previously (47). All other growth curves were determined using a Synergy HT microplate reader (BioTek, Winooski, VT) as described previously (69), with the modification that the microplate reader was placed inside an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) having an atmosphere of 90% N2, 5% CO2, and 5% H2. For growth and complementation studies, microplate cultures were inoculated to 2% with an aerobic LB overnight culture supplemented with 50 μg/ml ampicillin as appropriate.
Construction of chromosomal mutations.
Chromosomal deletions were made by linear recombination of PCR products as described previously (57, 69). Kanamycin resistance was selected, and the resistance markers were removed using the Flp recombinase (57). This leaves behind an 82-nucleotide scar (an frt site). All mutations were verified by PCR amplification across the scar followed by DNA sequencing.
Electron microscopy.
For each strain, an aerobic overnight culture was prepared on LB medium supplemented with 10 mM MgSO4. The overnight culture was placed in the anaerobic chamber for ∼1 h, and then 30 μl was used to inoculate 3 ml of double-strength LB supplemented with 10 mM MgSO4, 50 μM Fe-citrate, and 1% 1,2-PD as appropriate. The resulting culture was incubated for 8 h at 37°C, statically, in the anaerobic chamber. A 200-μl aliquot was used to inoculate 20 ml of double-strength LB supplemented with 10 mM MgSO4, 50 μM Fe-citrate, and 1% 1,2-PD as needed. These cultures were incubated under anaerobic conditions, statically, at 37°C to an optical density at 600 nm (OD600) of 1.2 to 1.5 (∼16 h). Cultures were pipetted into 50-ml polycarbonate centrifuge tubes and tightly sealed with screw caps having rubber O rings. Cells were centrifuged at 5,000 × g for 5 min, then returned to the anaerobic chamber. The supernatant was decanted to a waste container. Two percent glutaraldehyde (1 ml) was added, and the suspension was mixed by pipetting up and down. Cells were transferred to a 1.5-ml microcentrifuge tube, removed from the anaerobic chamber, and incubated at room temperature for 1 h. Cells were pelleted by centrifugation for 5 min at 5,000 × g. The supernatant was decanted. Two percent glutaraldehyde (1 ml) was added, followed by pipetting up and down to mix. The suspension was incubated at room temperature overnight, then washed by pelleting three times and resuspending in 0.1 M cacodylate buffer (pH 7.2). From this point, imbedding, sectioning, and electron microscopy were carried out as described previously (69).
Molecular biology methods.
Agarose gel electrophoresis, plasmid purification, PCR, restriction digests, ligation reactions, and electroporation were carried out using standard protocols as described previously (13, 70). Taq or Phusion DNA polymerase (New England Biolabs) were used for amplification of chromosomal DNA and for colony PCR. KOD DNA polymerase was used for amplification of plasmid templates. Plasmid DNA was purified using Qiagen products (Qiagen, Chatsworth, CA) according to the manufacturer’s instructions. Following restriction digestion or PCR amplification, DNA was purified using Wizard PCR Preps (Promega, Madison, WI) or Qiagen gel extraction kits. For ligation of DNA fragments, T4 DNA ligase or NEBuilder HiFi DNA assembly master mix was used according to the manufacturer’s instructions (New England Biolabs).
Complementation studies.
Genes used for complementation were cloned into plasmid pLac22, which allows for tight regulation of cloned genes by IPTG (71). Chromosomal genes were amplified using Phusion or Taq DNA polymerase and cloned between the BglII and HindIII sites of pLac22 by Gibson assembly. PCR primers with 20 bp of homology to the vector were used to make inserts for Gibson assembly, which was done with retention of the BglII site to provide correct placement of the ribosome binding site. Primers for Gibson assembly were designed using SnapGene software (GSL Biotech). Positive clones were identified by colony PCR, and the DNA sequences of all clones were verified. Growth curves for testing complementation were done in 48-well microplates as described above using IPTG at 0, 10, 20, 50, 100, and 200 μM.
Amplification and sequencing of the grpKLM genes of ECOR strains.
The grpKLM genes of selected ECOR strains were amplified by colony PCR using Choice Taq Blue master mix and primers TGCCGGTTCCGGATCTCTAT and TAATCGCGGGCAGTTGTTCT which were also used for sequencing of the PCR products.
RT-qPCR for measuring grp expression.
Cells from E. coli strain CFT073 were grown overnight anaerobically in LB medium in the presence or absence of 1,2-PD. A fresh 20-ml flask of LB was inoculated at 1% and grown anaerobically for 3.5 h with 1,2-PD at 37°C and then harvested. Total RNA was extracted using the RNeasy Protect bacteria minikit (Qiagen) and converted to cDNA with EcoDry Premix (TaKaRa) following the manufacturer’s instructions. Genomic DNA was cleaned up using SYBR green PCR master mix (Applied Biosystems). Samples (50-μl volume) were prepared in MicroAmp optical strips (Applied Biosystems) with three replicates per gene of interest. Three replicates of housekeeping gene gapA were collected in parallel as an internal standard. Samples were analyzed using a StepOnePlus RT-PCR instrument (Thermo Fisher) using the comparative threshold cycle (ΔΔCT) experimental protocol, which was comprised of 40 cycles of 15 s at 95°C followed by 60 s at 60°C, with a final melt curve step performed under the same conditions. An identical procedure was utilized for ΔgrpP and ΔgrpQ strains. Data were analyzed using the ΔΔCT method, and the standard error was reported for each gene of interest.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant AI081146 from the National Institutes of Health to T.A.B.
We thank Harry L. T. Mobley and Stephanie Himpsl for kindly providing E. coli CFT073. We also thank the ISU DNA Sequencing and Synthesis Facility for assistance with DNA analyses and the ISU Microscopy and Nanoimaging facility for help with electron microscopy.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Abdul-Rahman F, Petit E, Blanchard JL. 2013. The distribution of polyhedral bacterial microcompartments suggests frequent horizontal transfer and operon reassembly. J Phylogen Evolution Biol 1:4. [Google Scholar]
- 2.Axen SD, Erbilgin O, Kerfeld CA. 2014. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput Biol 10:e1003898. doi: 10.1371/journal.pcbi.1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorda J, Lopez D, Wheatley NM, Yeates TO. 2013. Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Sci 22:179–195. doi: 10.1002/pro.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobik TA, Lehman BP, Yeates TO. 2015. Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol Microbiol 98:193–207. doi: 10.1111/mmi.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowdhury C, Sinha S, Chun S, Yeates TO, Bobik TA. 2014. Diverse bacterial microcompartment organelles. Microbiol Mol Biol Rev 78:438–468. doi: 10.1128/MMBR.00009-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rae BD, Long BM, Badger MR, Price GD. 2013. Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol Mol Biol Rev 77:357–379. doi: 10.1128/MMBR.00061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havemann GD, Sampson EM, Bobik TA. 2002. PduA is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J Bacteriol 184:1253–1261. doi: 10.1128/jb.184.5.1253-1261.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penrod JT, Roth JR. 2006. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol 188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sampson EM, Bobik TA. 2008. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol 190:2966–2971. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinsmade SR, Paldon T, Escalante-Semerena JC. 2005. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J Bacteriol 187:8039–8046. doi: 10.1128/JB.187.23.8039-8046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rondon RR, Horswill AR, Escalante-Semerena JC. 1995. DNA polymerase I function is required for the utilization of ethanolamine, 1,2-propanediol, and propionate by Salmonella typhimurium LT2. J Bacteriol 177:7119–7124. doi: 10.1128/jb.177.24.7119-7124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price GD, Badger MR. 1989. Isolation and characterization of high CO2-requiring-mutants of the cyanobacterium Synechococcus PCC7942: two phenotypes that accumulate inorganic carbon but are apparently unable to generate CO2 within the carboxysome. Plant Physiol 91:514–525. doi: 10.1104/pp.91.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J Bacteriol 181:5967–5975. doi: 10.1128/JB.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol 181:5317–5329. doi: 10.1128/JB.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erbilgin O, McDonald KL, Kerfeld CA. 2014. Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl Environ Microbiol 80:2193–2205. doi: 10.1128/AEM.03887-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sriramulu DD, Liang M, Hernandez-Romero D, Raux-Deery E, Lünsdorf H, Parsons JB, Warren MJ, Prentice MB. 2008. Lactobacillus reuteri DSM 20016 produces cobalamin-dependent diol dehydratase in metabolosomes and metabolizes 1,2-propanediol by disproportionation. J Bacteriol 190:4559–4567. doi: 10.1128/JB.01535-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shively JM, Ball F, Brown DH, Saunders RE. 1973. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- 18.Talarico TL, Casas IA, Chung TC, Dobrogosz WJ. 1988. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob Agents Chemother 32:1854–1858. doi: 10.1128/aac.32.12.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petit E, LaTouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, Deshpande S, Haas K, Alvelo-Maurosa JG, Wardman C, Schnell DJ, Leschine SB, Blanchard JL. 2013. Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One 8:e54337. doi: 10.1371/journal.pone.0054337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallette E, Kimber MS. 2018. Structural and kinetic characterization of (S)-1-amino-2-propanol kinase from the aminoacetone utilization microcompartment of Mycobacterium smegmatis. J Biol Chem 293:19909–19918. doi: 10.1074/jbc.RA118.005485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallette E, Kimber MS. 2018. Structure and kinetics of the S-(+)-1-amino-2-propanol dehydrogenase from the RMM microcompartment of Mycobacterium smegmatis. Biochemistry 57:3780–3789. doi: 10.1021/acs.biochem.8b00464. [DOI] [PubMed] [Google Scholar]
- 22.Buchrieser C, Listeria Consortium, Rusniok C, Kunst F, Cossart P, Glaser P. 2003. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol Med Microbiol 35:207–213. doi: 10.1016/S0928-8244(02)00448-0. [DOI] [PubMed] [Google Scholar]
- 23.Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol 188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faber F, Thiennimitr P, Spiga L, Byndloss MX, Litvak Y, Lawhon S, Andrews-Polymenis HL, Winter SE, Bäumler AJ. 2017. Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLoS Pathog 13:e1006129. doi: 10.1371/journal.ppat.1006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. 2013. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlsson FH, Fak F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Backhed F, Nielsen J. 2012. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun 3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bae S, Ulrich CM, Neuhouser ML, Malysheva O, Bailey LB, Xiao L, Brown EC, Cushing-Haugen KL, Zheng Y, Cheng TY, Miller JW, Green R, Lane DS, Beresford SA, Caudill MA. 2014. Plasma choline metabolites and colorectal cancer risk in the Women’s Health Initiative Observational Study. Cancer Res 74:7442–7452. doi: 10.1158/0008-5472.CAN-14-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim EY, Tullman-Ercek D. 2013. Engineering nanoscale protein compartments for synthetic organelles. Curr Opin Biotechnol 24:627–632. doi: 10.1016/j.copbio.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Tsai SJ, Yeates TO. 2011. Bacterial microcompartments insights into the structure, mechanism, and engineering applications. Prog Mol Biol Transl Sci 103:1–20. doi: 10.1016/B978-0-12-415906-8.00008-X. [DOI] [PubMed] [Google Scholar]
- 32.Held M, Kolb A, Perdue S, Hsu SY, Bloch SE, Quin MB, Schmidt-Dannert C. 2016. Engineering formation of multiple recombinant Eut protein nanocompartments in E. coli. Sci Rep 6:24359. doi: 10.1038/srep24359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quin MB, Perdue SA, Hsu SY, Schmidt-Dannert C. 2016. Encapsulation of multiple cargo proteins within recombinant Eut nanocompartments. Appl Microbiol Biotechnol 100:9187–9200. doi: 10.1007/s00253-016-7737-8. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez-Esquer CR, Newnham SE, Kerfeld CA. 2016. Bacterial microcompartments as metabolic modules for plant synthetic biology. Plant J 87:66–75. doi: 10.1111/tpj.13166. [DOI] [PubMed] [Google Scholar]
- 35.Yung MC, Bourguet FA, Carpenter TS, Coleman MA. 2017. Re-directing bacterial microcompartment systems to enhance recombinant expression of lysis protein E from bacteriophage ϕX174 in Escherichia coli. Microb Cell Fact 16:71. doi: 10.1186/s12934-017-0685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferlez B, Sutter M, Kerfeld CA. 2019. A designed bacterial microcompartment shell with tunable composition and precision cargo loading. Metab Eng 54:286–291. doi: 10.1016/j.ymben.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee MJ, Palmer DJ, Warren MJ. 2019. Biotechnological advances in bacterial microcompartment technology. Trends Biotechnol 37:325–336. doi: 10.1016/j.tibtech.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Nichols TM, Kennedy NW, Tullman-Ercek D. 2019. Cargo encapsulation in bacterial microcompartments: methods and analysis. Methods Enzymol 617:155–186. doi: 10.1016/bs.mie.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang G, Johnston T, Quin MB, Schmidt-Dannert C. 2019. Developing a protein scaffolding system for rapid enzyme immobilization and optimization of enzyme functions for biocatalysis. ACS Synth Biol 8:1867–1876. doi: 10.1021/acssynbio.9b00187. [DOI] [PubMed] [Google Scholar]
- 40.Uddin I, Frank S, Warren MJ, Pickersgill RW. 2018. A generic self-assembly process in microcompartments and synthetic protein nanotubes. Small 14:e1704020. doi: 10.1002/smll.201704020. [DOI] [PubMed] [Google Scholar]
- 41.Zarzycki J, Erbilgin O, Kerfeld CA. 2015. Bioinformatic characterization of glycyl radical enzyme-associated bacterial microcompartments. Appl Environ Microbiol 81:8315–8329. doi: 10.1128/AEM.02587-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bobik TA. 2006. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol 70:517–525. doi: 10.1007/s00253-005-0295-0. [DOI] [PubMed] [Google Scholar]
- 43.Wheatley NM, Gidaniyan SD, Liu Y, Cascio D, Yeates TO. 2013. Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein Sci 22:660–665. doi: 10.1002/pro.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeates TO, Jorda J, Bobik TA. 2013. The shells of BMC-type microcompartment organelles in bacteria. J Mol Microbiol Biotechnol 23:290–299. doi: 10.1159/000351347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Craciun S, Balskus EP. 2012. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci U S A 109:21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jameson E, Fu T, Brown IR, Paszkiewicz K, Purdy KJ, Frank S, Chen Y. 2016. Anaerobic choline metabolism in microcompartments promotes growth and swarming of Proteus mirabilis. Environ Microbiol 18:2886–2898. doi: 10.1111/1462-2920.13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herring TI, Harris TN, Chowdhury C, Mohanty SK, Bobik TA. 2018. A bacterial microcompartment is used for choline fermentation by Escherichia coli 536. J Bacteriol 200. doi: 10.1128/JB.00764-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schindel HS, Karty JA, McKinlay JB, Bauer CE. 2019. Characterization of a glycyl radical enzyme bacterial microcompartment pathway in Rhodobacter capsulatus. J Bacteriol 201. doi: 10.1128/JB.00343-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zarzycki J, Sutter M, Cortina NS, Erb TJ, Kerfeld CA. 2017. In vitro characterization and concerted function of three core enzymes of a glycyl radical enzyme - associated bacterial microcompartment. Sci Rep 7:42757. doi: 10.1038/srep42757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott KP, Martin JC, Campbell G, Mayer CD, Flint HJ. 2006. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium Roseburia inulinivorans. J Bacteriol 188:4340–4349. doi: 10.1128/JB.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. 2013. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics 14(Suppl 2):S4. doi: 10.1186/1471-2105-14-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ochman H, Selander RK. 1984. Standard reference strains of Escherichia coli from natural populations. J Bacteriol 157:690–693. doi: 10.1128/JB.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawrence JG, Roth JR. 1996. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics 142:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng S, Sinha S, Fan C, Liu Y, Bobik TA. 2011. Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella. J Bacteriol 193:1385–1392. doi: 10.1128/JB.01473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chowdhury C, Chun S, Sawaya MR, Yeates TO, Bobik TA. 2016. The function of the PduJ microcompartment shell protein is determined by the genomic position of its encoding gene. Mol Microbiol 101:770–783. doi: 10.1111/mmi.13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng S, Fan C, Sinha S, Bobik TA. 2012. The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLoS One 7:e47144. doi: 10.1371/journal.pone.0047144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y, Jorda J, Yeates TO, Bobik TA. 2015. The PduL phosphotransacylase is used to recycle coenzyme A within the Pdu microcompartment. J Bacteriol 197:2392–2399. doi: 10.1128/JB.00056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palacios S, Starai VJ, Escalante-Semerena JC. 2003. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J Bacteriol 185:2802–2810. doi: 10.1128/jb.185.9.2802-2810.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martínez-Rodríguez L, García-Rodríguez FM, Molina-Sánchez MD, Toro N, Martínez-Abarca F. 2014. Insights into the strategies used by related group II introns to adapt successfully for the colonisation of a bacterial genome. RNA Biol 11:1061–1071. doi: 10.4161/rna.32092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson JE. 2010. Virus particle maturation: insights into elegantly programmed nanomachines. Curr Opin Struct Biol 20:210–216. doi: 10.1016/j.sbi.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bobik TA, Ailion M, Roth JR. 1992. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol 174:2253–2266. doi: 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rondon MR, Escalante-Semerena JC. 1996. In vitro analysis of the interactions between the PocR regulatory protein and the promoter region of the cobalamin biosynthetic (cob) operon of Salmonella typhimurium LT2. J Bacteriol 178:2196–2203. doi: 10.1128/jb.178.8.2196-2203.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. doi: 10.1128/JB.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berkowitz D, Hushon JM, Whitfield HJ Jr, Roth J, Ames BN. 1968. Procedure for identifying nonsense mutations. J Bacteriol 96:215–220. doi: 10.1128/JB.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sinha S, Cheng S, Sung YW, McNamara DE, Sawaya MR, Yeates TO, Bobik TA. 2014. Alanine scanning mutagenesis identifies an asparagine-arginine-lysine triad essential to assembly of the shell of the Pdu microcompartment. J Mol Biol 426:2328–2345. doi: 10.1016/j.jmb.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 71.Warren JW, Walker JR, Roth JR, Altman E. 2000. Construction and characterization of a highly regulable expression vector, pLAC11, and its multipurpose derivatives, pLAC22 and pLAC33. Plasmid 44:138–151. doi: 10.1006/plas.2000.1477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.