Chlamydia trachomatis is an obligate intracellular pathogen, causing sexually transmitted diseases and trachoma. The study of chlamydial physiology is important for developing novel therapeutic strategies for these diseases. Chlamydiae divide by a unique MreB-dependent polarized cell division process. In this study, we investigated unique properties of chlamydial MreB and observed that chlamydial species harbor an extended N-terminal region possessing amphipathicity. MreB formed a ring at the septum, like FtsZ in Escherichia coli, and its localization was dependent upon the amphipathic nature of its extended N terminus. Furthermore, this region is crucial for the interaction of MreB with cell division proteins. Given these results, chlamydial MreB likely functions at the septum as a scaffold for divisome proteins to regulate cell division in this organism.
KEYWORDS: Chlamydia, cell division, MreB, FtsZ, amphipathic helix, polarized division
ABSTRACT
Chlamydiae lack the conserved central coordinator protein of cell division FtsZ, a tubulin-like homolog. Current evidence indicates that Chlamydia uses the actin-like homolog, MreB, to substitute for the role of FtsZ in a polarized division mechanism. Interestingly, we observed MreB as a ring at the septum in dividing cells of Chlamydia. We hypothesize that MreB, to substitute for FtsZ in Chlamydia, must possess unique properties compared to canonical MreB orthologs. Sequence differences between chlamydial MreB and orthologs in other bacteria revealed that chlamydial MreB possesses an extended N-terminal region, harboring predicted amphipathicity, as well as the conserved amphipathic helix found in other bacterial MreBs. The conserved amphipathic helix-directed green fluorescent protein (GFP) to label the membrane uniformly in Escherichia coli but the extended N-terminal region did not. However, the extended N-terminal region together with the conserved amphipathic region directed GFP to restrict the membrane label to the cell poles. In Chlamydia, the extended N-terminal region was sufficient to direct GFP to the membrane, and this localization was independent of an association with endogenous MreB. Importantly, mutating the extended N-terminal region to reduce its amphipathicity resulted in the accumulation of GFP in the cytosol of the chlamydiae and not in the membrane. The N-terminal domain of MreB was not required for homotypic interactions but was necessary for interactions with cell division components RodZ and FtsK. Our data provide mechanistic support for chlamydial MreB to serve as a substitute for FtsZ by forming a ringlike structure at the site of polarized division.
IMPORTANCE Chlamydia trachomatis is an obligate intracellular pathogen, causing sexually transmitted diseases and trachoma. The study of chlamydial physiology is important for developing novel therapeutic strategies for these diseases. Chlamydiae divide by a unique MreB-dependent polarized cell division process. In this study, we investigated unique properties of chlamydial MreB and observed that chlamydial species harbor an extended N-terminal region possessing amphipathicity. MreB formed a ring at the septum, like FtsZ in Escherichia coli, and its localization was dependent upon the amphipathic nature of its extended N terminus. Furthermore, this region is crucial for the interaction of MreB with cell division proteins. Given these results, chlamydial MreB likely functions at the septum as a scaffold for divisome proteins to regulate cell division in this organism.
INTRODUCTION
Bacteria within the genus Chlamydia are obligate intracellular pathogens that cause diverse diseases in humans and animals. These Gram-negative cocci differentiate between two morphologically and functionally distinct cellular forms during their developmental cycle: the elementary body (EB) and the reticulate body (RB) (1). The EB is the infectious but nondividing form, whereas the RB is the dividing but noninfectious form. After entering the host cell, Chlamydia remains within a membrane-bound parasitic organelle, termed an inclusion (2). RBs undergo multiple rounds of cell division until they engage a secondary differentiation program and convert to EBs, which are subsequently released from the host cell to propagate the infection.
In evolving to obligate intracellular dependence, Chlamydia has significantly reduced its genome, eliminating ftsZ (3), the conserved tubulin-like cell division coordinator of binary fission (4). However, Chlamydia harbors rod-shape-determining proteins associated with peptidoglycan synthesis in the lateral cell wall of bacilli. That Chlamydia has retained these genes is unusual, since these are coccoid bacteria and synthesize peptidoglycan only during division (5). The absence of FtsZ in chlamydiae suggests that they may not divide by binary fission. Recent work from our labs supports this. We employed both live- and fixed-cell imaging to localize various markers, including a division protein (FtsQ) (6), during EB-to-RB differentiation and the first division of the RB. Interestingly, we observed that chlamydiae are highly polarized throughout this time and that division itself occurs via a polarized process (7). This polarized budding process remains the primary mode of division even at later times during the developmental cycle (J. V. Cox, Y. M. AbdelRahman, and S. P. Ouellette, unpublished data).
In 2012, we hypothesized and presented evidence that Chlamydia coopted the rod-shape-determining protein MreB to function as the central coordinator of division (8). To perform this function, we hypothesize that chlamydial MreB must possess unique properties. Many MreB homologs harbor a conserved N-terminal amphipathic helix that facilitates MreB association with the inner membrane (9). At the membrane, it engages peptidoglycan machinery while forming short moving filaments (10–12). These filaments move around the rod-shaped bacterium and are excluded (or move randomly) in areas of high membrane curvature (13), as occurs at the poles of the bacterium. As chlamydiae are coccoid, their membranes display uniform membrane curvature; thus, how MreB can be spatially localized to form filaments to direct cell division is an interesting question.
Here, we investigated the unique structural features of chlamydial MreB that allow it to substitute for FtsZ. Chlamydial MreB forms rings at the polarized division site, similar to FtsZ rings in binary fission, and bioinformatics analyses indicate that chlamydial MreB orthologs harbor an extended N-terminal region (amino acids [aa] 1 to 23) in addition to the conserved amphipathic helix. The aa 1 to 23 residues also encode predicted amphipathicity. We confirmed that the conserved amphipathic helix, but not aa 1 to 23, is important for directing membrane localization of green fluorescent protein (GFP) in Escherichia coli. Nevertheless, aa 1 to 23 were sufficient to direct GFP to the membrane in Chlamydia, and this localization was abolished when mutations were introduced in aa 1 to 23 to reduce its amphipathicity. Conversely, inhibiting endogenous MreB activity by treatment with the inhibitor A22 did not affect the membrane localization of the GFP fusions, indicating that its localization was not governed by interactions with endogenous MreB. Finally, we determined that the extended N-terminal region of chlamydial MreB is crucial for its interactions with cell division proteins but not its ability to form homo-oligomers. Our data indicate the N-terminal region of chlamydial MreB is important for its membrane localization and interaction with elements of the divisome. These properties likely allow it to function in a manner distinct from that of other MreB orthologs and to regulate the cell division process in Chlamydia.
RESULTS
Chlamydial MreB_6×H localizes as a ring at the division site.
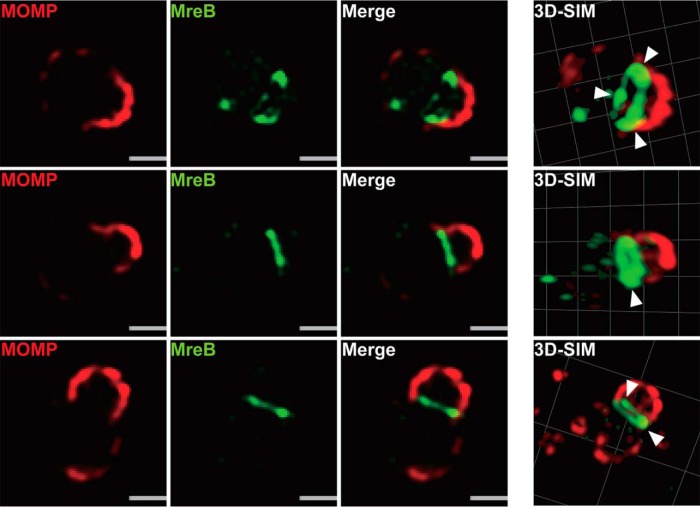
Given our hypothesis for the role of chlamydial MreB in directing polarized division, we examined its localization during the first division of an RB. We were unable to detect endogenous MreB with the antibody previously used to image the distribution of MreB in Chlamydia (14). We then attempted to image wild-type and various truncation mutants of MreB using a GFP sandwich (SW) fusion. In E. coli, an MreB_mCherrySW was reported to be functional (15) (N- or C-terminal fluorescent protein fusions with MreB display artifactual localization [16]). However, when we inducibly expressed MreB_GFPSW in Chlamydia trachomatis L2, bacterial growth was significantly inhibited even with low levels of expression, as observed by the presence of a small number of enlarged bacteria (see Fig. S1 in the supplemental material). More importantly, the fusion protein did not localize to the membrane as reported for the mCherry sandwich fusion (Fig. S1) (14). The reasons for this are not clear but could possibly be due to the ability of GFP to dimerize, even as a fusion. We therefore opted to construct an MreB fusion with a small C-terminal hexahistidine tag (MreB_6×H). The human epithelial cell line, HeLa, was infected with a transformant of C. trachomatis L2 carrying a plasmid to inducibly express MreB_6×H. At 6 h postinfection (hpi), expression of MreB_6×H was induced with anhydrotetracycline (aTc), and cells were fixed at 10.5 hpi, a time when the nascent RB has begun its first division (7). As seen in Fig. 1, we observed using superresolution structured illumination microscopy (SIM) the polar localization of the major outer membrane protein (MOMP) in the budding daughter cell, as previously reported (7). Consistent with its proposed role in division (8), MreB_6×H was localized in a band-like structure across the septum and not in puncta as previously reported (14, 17). Interestingly, when three-dimensional (3D) reconstructions were assembled from the SIM images, we observed that MreB_6×H formed ringlike structures at the septum with areas of more intense signal (Fig. 1, arrowheads). These areas of more intense signal may correspond to the puncta previously observed (14, 17).
FIG 1.
Localization of chlamydial MreB_6×H in C. trachomatis using structured illumination microscopy (SIM). C. trachomatis without plasmid (−pL2) was transformed with an anhydrotetracycline (aTc)-inducible vector encoding chlamydial MreB with a six-histidine (6×H) tag at the C terminus. HeLa cells were infected with this strain and chlamydial MreB_6×H expression was induced with 10 nM aTc at 6 hpi. At 10.5 hpi, the infected cells were fixed (3.2% formaldehyde, 0.022% glutaraldehyde in PBS) for 2 min and permeabilized with 90% methanol (MeOH) for 1 min. The sample was stained for major outer membrane protein (MOMP; red) and chlamydial MreB (green). Three representative images are displayed. The arrowheads indicate regions of more intense fluorescence. SIM images were acquired on a Zeiss ELYRA PS.1 superresolution microscope. Bars, 0.5 μm.
Chlamydial MreB harbors an amphipathic helix and an extended N-terminal region conserved in Chlamydia.
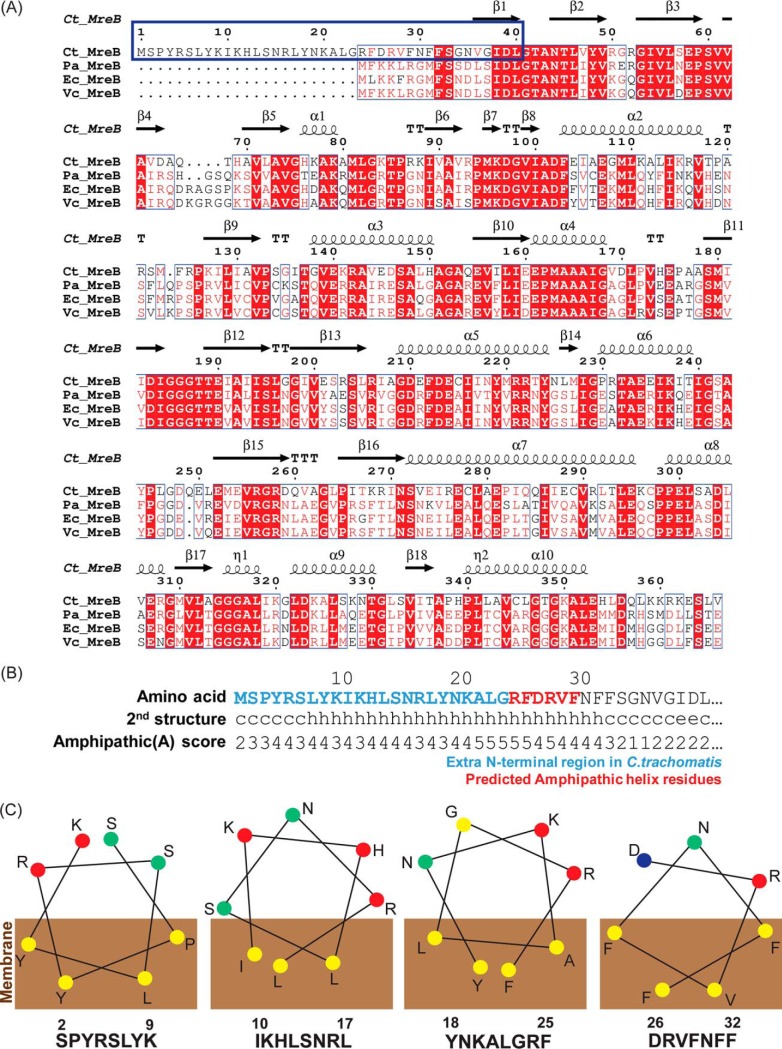
In many Gram-negative bacteria, there is an amphipathic helix at the N terminus of MreB, and the helix is important for the membrane localization of MreB (9). This localization is crucial for the role of MreB in directing peptidoglycan (PG) synthesis during lateral cell wall growth (9, 18). To confirm whether chlamydial MreB also has these features, we predicted its secondary structure and amphipathic regions using bioinformatics tools (Fig. 2). The alignment data show that chlamydial MreB harbors an extended N-terminal region (amino acids [aa] 1 to 23), which other Gram-negative bacteria lack (Fig. 2A). Importantly, this extended N-terminal region is conserved among Chlamydia spp. (see Fig. S2). We attempted to determine if extended N termini were present in other bacterial species by BLAST homology searches excluding chlamydiae. Only five examples of uncharacterized “Candidatus” species (Microgemates, Woesebacteria, Aerophobetes, Omnitrophica, and WWE3) containing an extended N terminus were found, with two of these possessing additional methionine residues that could potentially serve as the true translational start site. Given the possibility of misannotation and the general lack of information about these bacteria, it is likely that the extended N-terminal region may be unique to Chlamydia and closely related bacteria.
FIG 2.
Bioinformatics analysis of chlamydial MreB. Chlamydial MreB has unique structural features based on bioinformatics analyses. (A) Protein sequence alignment of chlamydial and other bacterial MreBs. The blue box represents the extended N terminus and predicted amphipathic helix of chlamydial MreB. The alignment was performed with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and represented with ESPript 3.0 (http://espript.ibcp.fr). (B) The amphipathic score and the predicted secondary structure of the N terminus of chlamydial MreB. The prediction of amphipathicity was performed by using AMPHIPASEEK. The blue and red residues represent the extended N terminus and predicted amphipathic helix, respectively. (C) The amphipathicity of the fragments of the N-terminal region of chlamydial MreB. These amphipathic structures were predicted by an online helical wheel program, named Helixator (http://www.tcdb.org/progs/helical_wheel.php). The circles represent the indicated amino acid residues: yellow, nonpolar residue; green, uncharged polar residue; red, basic polar residue; blue, acidic polar residue.
Following the extended N-terminal region, there is a conserved amphipathic helix, from R24 to F29 (Fig. 2B), that aligns with the amphipathic helices of the other bacteria (Fig. 2A). A high amphipathic score (A-score) extends to residue F32 in chlamydial MreB. Interestingly, aa 1 to 23 also have a high A-score, though the region was not predicted to form an amphipathic helix (Fig. 2B). Similar results were seen with the more divergent Waddlia MreB homolog (Fig. S2B). Nevertheless, performing a helical wheel analysis with 8 amino acid windows revealed that each window shows amphipathicity (Fig. 2C). Based on these observations, we conclude that the extended N-terminal region of chlamydial MreB may possess membrane-associating properties.
Chlamydial MreB is unable to complement an mreB-deficient mutant of E. coli.
The bioinformatics data suggested that chlamydial MreB harbors unique structural features absent in other bacterial MreBs (Fig. 2A). To assess the activities associated with these unique structural elements, we introduced chlamydial MreB into an mreB-deficient mutant of E. coli. As mreB is an essential gene in E. coli, Bendezu and de Boer made the strain P2733, which has a deletion in the mreB gene and is conditionally viable by the overexpression of the ftsQAZ operon (19). This strain has been used for complementation assays and grows as coccoid bacteria, since MreB is necessary for the maintenance of rod shape (19). To test whether chlamydial MreB (CtrMreB) is capable of complementing the mreB-deficient mutant, P2733 was transformed with an arabinose-inducible vector harboring chlamydial mreB. Cell shape was compared after inducing MreB expression in comparison to that from an empty vector negative control and an E. coli MreB (EcMreB) positive control (see Fig. S3A and B). When EcMreB was expressed from an arabinose-inducible promoter in the P2733 strain, the E. coli cells began to adopt a rod shape, consistent with other reports (15, 19, 20). When chlamydial MreB was induced, the morphology of P2733 was unchanged compared to that of uninduced or empty vector controls, indicating that chlamydial MreB does not complement the mreB-deficient mutant of E. coli (Fig. S3C). To determine whether the inability of chlamydial MreB to complement was due to its extended N-terminal region, we performed the same experiment with ΔN22 chlamydial MreB, which more closely resembles E. coli MreB in size and characteristics at the N terminus. We observed that ΔN22 chlamydial MreB also does not complement the mreB-deficient E. coli, suggesting that the extended N-terminal region is not the only reason for the failure to complement (Fig. S3D; see also Discussion). Chlamydial MreB expression was confirmed in these experiments by Western blotting (Fig. S3E).
The amphipathic helix, but not the extended N-terminal region, of chlamydial MreB is sufficient to direct GFP to the membrane in E. coli.
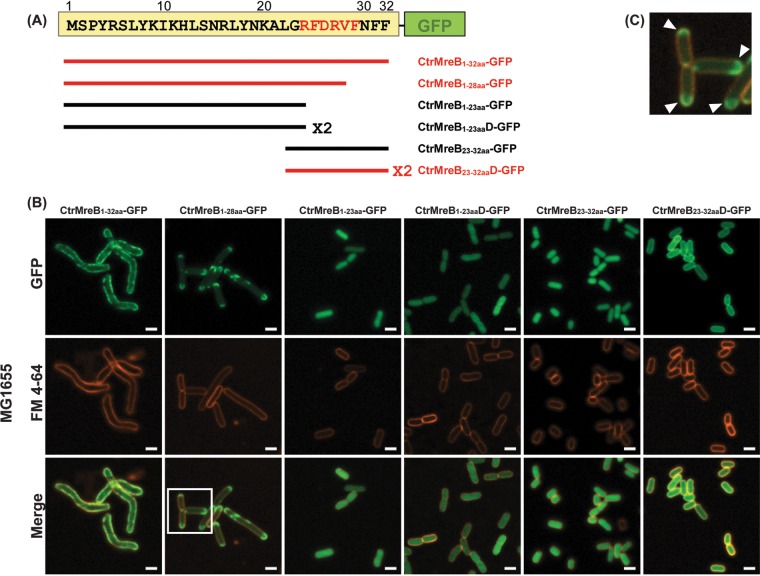
We hypothesized that, because of their amphipathicity, the unique sequence elements at the N terminus of chlamydial MreB could direct the membrane localization of MreB (Fig. 1). To test this, we performed a series of localization studies in E. coli using the N-terminal regions of chlamydial MreB (CtrMreB) fused to GFP. A similar strategy was used to demonstrate that two copies of the amphipathic helix of E. coli MreB (EcMreB) were sufficient to direct GFP to the membrane (9). We made various fusions in which either one or two copies of portions of the N-terminal region of chlamydial MreB were fused to GFP (illustrated in Fig. 3A). We then observed the localization of these proteins in both wild-type (MG1655) and MreB-deficient (P2733) E. coli (Fig. 3B and S4). We observed that a single copy of aa 1 to 32 (MreB1-32aa), comprising all high-A-score residues, was sufficient to direct GFP to the membrane. Interestingly, aa 1 to 28 (MreB1-28aa), comprising both the predicted amphipathic helix and the extended N terminus, were also sufficient to direct GFP to the membrane, but in wild-type cells, its localization was primarily restricted to the poles where the overall membrane curvature is similar to that of a coccoid bacterium (Fig. 3C). Consistent with what has been observed in E. coli, two copies of aa 23 to 32 (MreB23-32aa), comprising the predicted amphipathic helix, directed GFP to the membrane. Contrary to our prediction, one or two copies of aa 1 to 23 (MreB1-23aa) were not able to direct GFP to the membrane in E. coli even though the region has a high A-score (Fig. 2B).
FIG 3.
Localization in E. coli of various N-terminal regions of chlamydial MreB fused to GFP. Wild-type MG1655 E. coli was transformed with the arabinose-inducible vectors encoding GFP fusions with various N-terminal regions of chlamydial MreB. Stationary-phase cultures strains were diluted 1:100 in LB medium containing 34 μg/ml chloramphenicol and cultured for 2 h. The cells were then induced or not with 0.05% (wt/vol) arabinose and cultured for 2 h more. To stain the membrane, SynaptoRed-C2 (FM4-64) was added to the samples at a final concentration of 1.5 μM. After 15 min, 4 μl of each culture was spotted on a 1% LB agar pad and covered with a coverslip. The images were acquired on a Zeiss Imager.Z2 equipped with an Apotome2 using a 100× lens objective. (A) The structure of the various N-terminal MreB-GFP fusion peptides. In the boxed region containing the N terminus of chlamydial MreB, red residues represent the predicted amphipathic helix residues. Red constructs show the membrane localization and black constructs show cytosolic localization. (B) The localization of various N-terminal regions of chlamydial MreB fused to GFP in MG1655. Note the polar membrane localization of the MreB1-28aa-GFP. Bars, 2 μm. (C) A zoomed image of MreB1-28aa-GFP localization represented by the box in panel B. The arrowheads represent the polar membrane localization of the MreB1-28aa-GFP.
The extended N-terminal region of chlamydial MreB is sufficient to direct GFP to the membrane in C. trachomatis L2.
We next asked the question whether aa 1 to 23, alone or in combination with the predicted amphipathic region (aa 24 to 28), could direct GFP to the membrane in Chlamydia. The MreB1-23aa-GFP or MreB1-28aa-GFP fusions were moved to an inducible chlamydial expression plasmid and transformed into C. trachomatis L2. These transformants were then used to infect HeLa cells, and construct expression was induced at either 6 or 16 hpi. Infected cells were fixed and imaged at 10.5 or 20 hpi (Fig. 4). GFP alone is a cytosolic protein when expressed in chlamydiae (Fig. 4A). For both fusion proteins, GFP fluorescence was observed at membrane sites (Fig. 4B and C, arrowheads in panel B indicate individual bacteria with membrane localization of GFP) at both time points examined.
FIG 4.
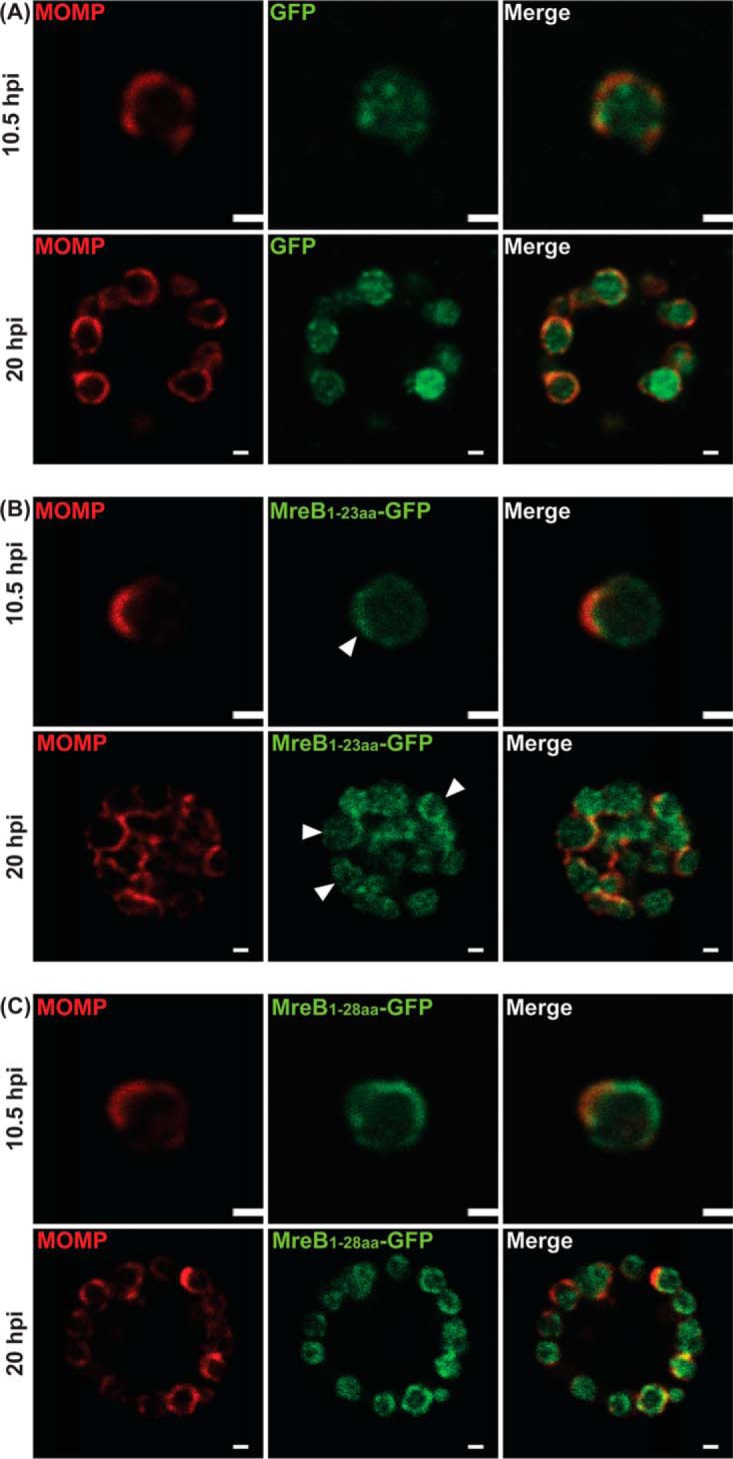
Localization in C. trachomatis of various N-terminal regions of chlamydial MreB fused to GFP. C. trachomatis serovar L2 without plasmid (−pL2) was transformed with anhydrotetracycline (aTc)-inducible vectors encoding GFP (A), chlamydial MreB1-23aa-GFP fusion peptide (B), or chlamydial MreB1-28aa-GFP fusion peptide (C). HeLa cells were infected with the indicated strains, and expression of the GFP fusions was induced at 6 hpi or 16 hpi with 10 nM aTc. At 10.5 hpi or 20 hpi, the samples were fixed (3.2% formaldehyde, 0.022% glutaraldehyde in 1× PBS) for 2 min and permeabilized with 90% methanol (MeOH) for 1 min. These samples were stained for major outer membrane protein (MOMP; red) with GFP imaged in green. The arrowheads represent the MreB1-23aa-GFP localized at the membrane. Images were acquired on a Zeiss LSM 800 confocal microscope with 63× lens objective. Bars, 0.5 μm (10.5 hpi) or 1 μm (20 hpi).
The N-terminal region of MreB from C. trachomatis has two leucine residues (L13 and L22) encoded by TTG. As bacteria can use UUG and GUG as alternative start codons (21), we performed a similar series of experiments using the N-terminal region of MreB from Chlamydia suis, which does not use UUG codons for its homologous leucine residues. In all cases tested, the N-terminal residues of C. suis MreB (fused to GFP) exhibited the same localization patterns as those observed for the C. trachomatis N-terminal MreB-GFP fusions (see Fig. S5). Based on these data, we conclude that it is unlikely the MreB of C. trachomatis bypasses the predicted AUG start codon in favor of downstream alternative start codons.
The localization of MreB1-23aa-GFP or MreB1-28aa-GFP in C. trachomatis L2 is independent of endogenous MreB.
Importantly, the MreB1-23aa-GFP and MreB1-28aa-GFP localization profiles were distinct from that of full-length MreB (Fig. 1 and 5), suggesting their localization profiles are not dependent on interactions with endogenous (i.e., chromosomally encoded) MreB. To test this, we used the bacterial adenylate cyclase-based two hybrid (BACTH) assay, which is based on the reconstitution of enzyme activity by two interacting proteins that bring catalytic adenylate cyclase fragments (T25 and T18) into close proximity. We performed BACTH assays with full-length MreB and aa 1 to 32 of MreB and observed no interaction (data not shown). To further test a role for interactions with endogenous MreB, we expressed the GFP fusion proteins or MreB_6×H and then treated the cultures with A22, an MreB-specific antibiotic, to depolymerize MreB. Under these conditions, we observed the localization of the GFP fusion proteins at the membrane, whereas the membrane-associated MreB_6×H was significantly reduced (Fig. 5). Based on these data, we conclude that the membrane localization of the GFP fusion proteins is caused by the amphipathic nature of the N terminus and not its interaction with endogenous MreB.
FIG 5.
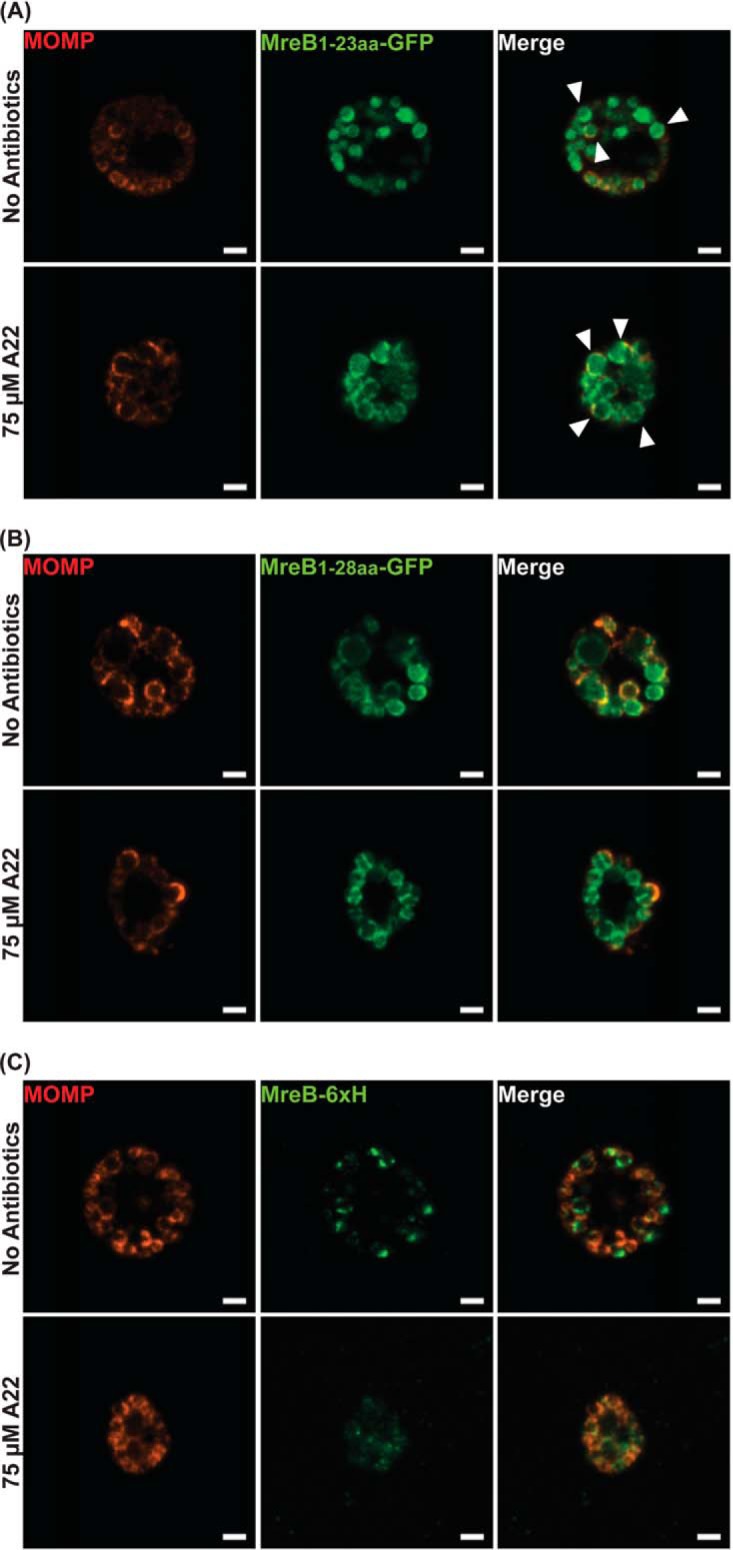
Localization of various N-terminal regions of chlamydial MreB fused to GFP in A22-treated C. trachomatis. C. trachomatis without plasmid (−pL2) was transformed with anhydrotetracycline (aTc)-inducible vectors encoding chlamydial MreB1-23aa-GFP fusion peptide (A), chlamydial MreB1-28aa-GFP fusion peptide (B), or chlamydial MreB_6×H (C). At 16 hpi, expression of the constructs was induced with 10 nM aTc, and at 18 hpi, 75 μM A22 was added to disrupt MreB localization. The samples were fixed at 20 hpi with 1× DPBS containing 3.2% formaldehyde and 0.022% glutaraldehyde for 2 min. Afterwards, the samples were permeabilized with 90% methanol for 1 min. These samples were stained for major outer membrane protein (MOMP; red). The arrowheads show the membrane localization of the MreB1-23aa-GFP peptide. Images were acquired on a Zeiss Imager.Z2 equipped with an Apotome2 using a 100× lens objective. Bars, 2 μm.
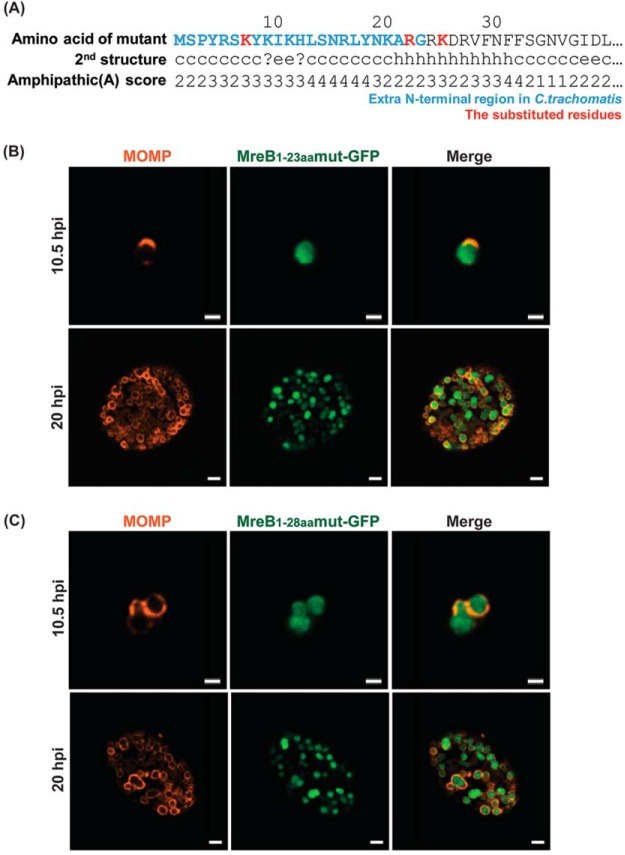
Disrupting the amphipathicity of the extended N-terminal region of MreB prevents its ability to direct GFP to the membrane in C. trachomatis L2.
To further examine whether the membrane localization of MreB1-23aa-GFP and MreB1-28aa-GFP was dependent on their amphipathicity, we created the mutations L7K, L22R, and F25K. These mutations were predicted to diminish the amphipathicity of this region (Fig. 6A). The mutations within the N-terminal region were incorporated into the MreB1-23aa-GFP and MreB1-28aa-GFP fusion constructs, transformed into C. trachomatis L2, and inducibly expressed. As demonstrated in Fig. 6B and C, we observed the cytosolic localization of the fusion peptides. Therefore, from the combination of these data (Fig. 4 and 6), we conclude that the amphipathic properties of the extended N-terminal region of chlamydial MreB are sufficient to direct GFP to the membrane in Chlamydia.
FIG 6.
Localization in C. trachomatis of various mutated N-terminal regions of chlamydial MreB fused to GFP. (A) Mutations were introduced into the N-terminal region of chlamydial MreB and modeled for their effect on amphipathicity. L7K, L22R, and F25K (red residues) mutations disrupted the predicted amphipathicity (see Fig. 2 for comparison). (B) C. trachomatis serovar L2 without plasmid (−pL2) was transformed with anhydrotetracycline (aTc)-inducible vectors encoding the mutated chlamydial MreB1-23aa-GFP or MreB1-28aa-GFP fusion peptide. HeLa cells were infected with the indicated strains, and expression of the GFP fusions was induced at 6 hpi or 16 hpi with 10 nM aTc. At 10.5 hpi or 20 hpi, the samples were fixed (3.2% formaldehyde, 0.022% glutaraldehyde in 1× PBS) for 2 min and permeabilized with 90% methanol (MeOH) for 1 min. These samples were stained for major outer membrane protein (MOMP; red) with GFP imaged in green. Images were acquired on a Zeiss Imager.Z2 equipped with an Apotome2 using a 100× lens objective. Bars, 1 μm (10.5 hpi) or 2 μm (20 hpi).
The extended N-terminal region is dispensable for MreB homo-oligomerization but essential for interactions with other division proteins.
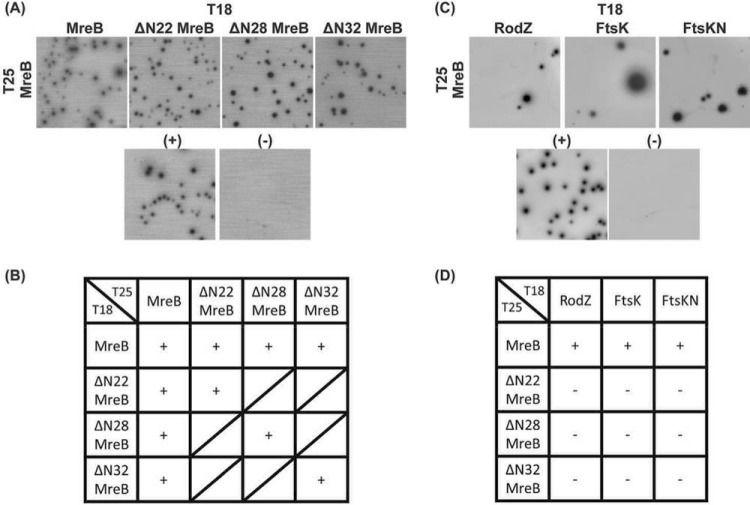
We next asked whether the N-terminal region is necessary for MreB homo-oligomerization. In E. coli, MreB interacts with itself to form short filaments (10). In addition, since MreB participates in both cell shape determination and cell division, it interacts with diverse proteins related to cell division and peptidoglycan synthesis (15, 20, 22). These features are shared with chlamydial MreB, which interacts with itself and with several proteins such as the cytoskeletal protein RodZ and the cell division components FtsK and FtsQ (6, 8, 22). Based on these previous reports, we hypothesized that the extended N-terminal region is important for these interactions. To test this, we used the bacterial adenylate cyclase-based two hybrid (BACTH) assay. We designed BACTH constructs encoding full-length or truncated chlamydial MreB mutants and then performed the interaction assays on minimal medium supplemented with maltose (Fig. 7). Importantly, when the BACTH is performed on minimal medium, growth occurs only when two proteins interact, as this allows for the regeneration of cAMP that is necessary to support growth on maltose. The N-terminal truncations of MreB interacted with full-length MreB (Fig. 7A; summarized in Fig. 7B), providing a potential explanation for the localization data of MreB_6×H truncations in Chlamydia (see Fig. S6). However, these truncated MreBs did not interact with chlamydial RodZ, full-length FtsK, or the N-terminal region of FtsK lacking its ATPase domain, whereas full-length MreB did interact with these constructs (Fig. 7C; summarized in Fig. 7D) (8, 23). From these data, we conclude that the extended N-terminal region is dispensable for MreB homo-oligomerization but is critical for interacting with other proteins involved in the cell division process. These data indicate the importance of localizing MreB to the membrane for interactions with the membrane-associated divisome components.
FIG 7.
BACTH assay to test chlamydial protein-protein interactions of full-length and truncated MreB with divisome components. E. coli DHT1 (Δcya) was cotransformed with vectors encoding the indicated genes fused to the T25 and T18 catalytic domains of the Bordetella pertussis adenylate cyclase. Transformants were plated on M63 minimal medium plates containing 50 μg/ml ampicillin, 25 μg/ml kanamycin, 0.5 mM IPTG, 40 μg/ml X-Gal, 0.04% Casamino Acids, and 0.2% maltose. The plates were incubated at 30°C for 5 to 7 days. (A and B) BACTH assay of the interaction between full-length and truncated chlamydial MreBs. (C and D) BACTH assay of the interaction between full-length or truncated chlamydial MreBs and previously described chlamydial cell division components FtsK, the N-terminal domain of FtsK (FtsKN), and RodZ. A positive control [(+)] is the interaction between T25-Zip and T18-Zip, the GCN4 leucine zipper motif, and a negative control [(−)] is the lack of interaction between T25 and the mixture of T18-MreB and T18-ΔN22 MreB. These tests were performed a minimum of two times. +, interaction; −, no interaction; ∕, not tested.
DISCUSSION
MreB is a well-characterized rod-shape-determining protein, which is conserved in most bacilli (19). When the mreB gene is deleted, the cell shape is changed from bacillus to coccoid in E. coli (19). MreB participates in organizing peptidoglycan (PG) synthesis by interacting with other proteins, such as MreC and RodZ, to direct Pbp2 and RodA activity at the membrane (20, 24). Recently, MreB has been observed as a dynamic cytoskeletal protein forming short filaments that rotate around the membrane perpendicularly to the longitudinal axis of the cell (11). This motion is driven by PG synthesis and is critical for cell elongation and maintenance of rod shape (25). Given these characteristics and properties of MreB, it is surprising that Chlamydia, a Gram-negative coccoid bacterium that lacks PG in its cell wall, harbors MreB. To date, we have been unable to complement any structural components of the E. coli divisome (e.g., FtsQ [6], Mre system [MreB {this study}], or RodZ [23]) using chlamydial orthologs, which indicates the importance of recapitulating all necessary interactions for efficient complementation. In contrast, cytosolic components of the PG synthesis pathway possessing enzymatic activity have been shown to complement E. coli conditional mutants (26–28). These data are consistent with the notion that the PG synthesis machinery is conserved in Chlamydia but that the mechanisms for spatially regulating its synthesis are not conserved. These observations further suggest that the maintenance of cell morphology may not be the primary function of chlamydial MreB (5, 29). Rather, given the function of chlamydial PG strictly in cell division, chlamydial MreB may primarily function in this process.
Since Chlamydia lacks the conserved cell division-organizing protein FtsZ, we have hypothesized and presented evidence that MreB is a functional substitute for FtsZ (6–8, 23). However, this raises the intriguing question of how MreB, which is highly conserved between Chlamydia and phylogenetically unrelated bacteria, can serve as the divisome organizer. Interestingly, when MreB is reintroduced into MreB-depleted cells in Bacillus subtilis, a budding shape is observed that is enriched in MreB (13). This eventually leads to recruitment of cell wall machinery and formation of a rod shape. However, in Chlamydia, which also undergoes a budding-like polarized division, this process is spatially restricted; thus, what prevents Chlamydia from continuing to synthesize PG to produce a rod shape? One hypothesis is that chlamydial MreB exhibits a restricted distribution that is at least in part dependent upon its unique structural features, and we have shown here that the unique N-terminal region of chlamydial MreB can facilitate additional membrane association of this cytoskeletal protein.
When initiating this study, our goal was to capture MreB localization at high resolution and at early stages of division, since it becomes more difficult to resolve individual organisms within the inclusion as RBs multiply. However, we were unable to detect endogenous MreB using the previously reported antibody (14). We therefore took advantage of genetic tools that have been recently developed for Chlamydia to transform this bacterium with a plasmid construct encoding an inducible MreB_6×H. Using this strategy, we detected MreB_6×H localized at the division septum during the polarized division of the first RB. More interestingly, MreB formed a ring at the septum in the polarized dividing cell that resembled the FtsZ ring at the septum of E. coli dividing by binary fission (Fig. 1). The septal MreB ring is very similar to the PG ring previously observed in Chlamydia (5). We speculate that the MreB puncta previously observed may be due to the low affinity of the MreB antibody, as we were also unable to detect MreB at later stages during infection using this reagent. However, we also observed areas along the MreB ring of more intense staining (Fig. 1, arrowhead). These regions may be the puncta reported previously and may represent areas of active PG synthesis. Alternatively, the puncta previously described may represent an early stage of division before ring formation. However, to date, we have been unable to image both PG and MreB_6×H, perhaps due to the aldehyde fixation methods required to preserve the budding morphology of chlamydiae. Given the effects of inhibiting MreB activity on chlamydial division and PG synthesis (7, 8, 17), these data further suggest that chlamydial MreB likely has a scaffolding function for cell divisome proteins to direct PG synthesis in a manner similar to the function of FtsZ in E. coli (30).
Many Gram-negative MreB homologs possess an amphipathic helix at their N terminus that allows MreB to associate with the inner membrane (9). Chlamydia species have conserved this feature. The most striking difference between the chlamydial species and other “canonical” MreB homologs is the presence of an extended N terminus of 23 amino acids (aa 1 to 23). Amphipathic prediction algorithms suggested this region possesses amphipathicity. Interestingly, canonical MreB filaments are excluded from areas of high membrane curvature in bacilli that are typically enriched in anionic phospholipids such as cardiolipin (i.e., the cell poles [31]), but Chlamydia is coccoid, with its membrane displaying curvature similar to the poles of bacilli. Therefore, we hypothesized that the additional residues at the N terminus of chlamydial MreB may allow it to associate more efficiently with membranes that exhibit curvature. In addition, these residues may be critical for establishing polarity in this organism, as we previously observed that inhibiting MreB activity with A22 or MP265 not only blocks cell division but also depolarizes the RB (7, 8). Indeed, we observed here that A22 treatment dissociates MreB_6×H from the membrane (Fig. 5).
The amphipathic helix of canonical Gram-negative MreB homologs is critical for the association of MreB with membranes. This was demonstrated by two means: expressing two copies of the helix in tandem fused to the N terminus of GFP and deleting the helix from MreB and assessing its localization (9). In the former, GFP was directed to the inner membrane (9). In the latter, MreB no longer localized as membrane patches but rather as cytosolic aggregates in E. coli (9). Using the former approach, we observed that two copies, but not one copy, of the chlamydial amphipathic helix were sufficient to direct MreB to the membrane, consistent with what has been observed in E. coli (Fig. 3B). However, the additional N-terminal region of chlamydial MreB (aa 1 to 23) together with a single copy of the conserved amphipathic region (aa 24 to 28) directed GFP to the membrane (Fig. 3B). Intriguingly, the membrane localization was restricted to the cell polar region but did not appear to be an inclusion body, since in an mreB-deficient mutant E. coli, which has a spherical morphology, the peptide fluorescence was more uniformly distributed in the membrane (see Fig. S4 in the supplemental material). Whether this region recognizes specific lipid moieties that directs its polar localization is under investigation.
We conclude that the additional N-terminal region of chlamydial MreB harbors a membrane-targeting function. Indeed, this was supported by (i) the ability of this region to direct GFP to the membrane when expressed in Chlamydia and (ii) the loss of GFP membrane localization when the amphipathicity of the N-terminal region was disrupted by mutagenesis. We excluded any possible effects of alternative start codons encoded by leucine residues by replicating our results using the N terminus of C. suis, which does not harbor any UUG codons for leucine in this region (Fig. S5). We propose that chlamydial MreB, with both its extended N terminus and amphipathic helix, may have enhanced affinity for membranes and is critical for it to form ring structures associated with regions of high membrane curvature.
As Chlamydia is an obligate intracellular bacterium, it is time-consuming and difficult to genetically modify its genes in a targeted manner, and there is a high failure rate in doing so. Ideally, we would create a conditional knockout such that we could express truncated mutants of chlamydial MreB in the absence of the chromosomal full-length copy. We did attempt to localize truncation mutants of MreB in Chlamydia, but only the mutant lacking all predicted amphipathicity showed reduced membrane localization (Fig. S6). Not surprisingly, we detected interactions between the MreB truncations and the full-length MreB; thus, one interpretation is that the truncations, when expressed in Chlamydia, interacted with full-length MreB (Fig. 7), which is itself at the membrane. Furthermore, the N-terminal region is important for interactions with other proteins related to cell division and PG synthesis. When the region was deleted, chlamydial MreB no longer interacted with RodZ and FtsK by bacterial two-hybrid analysis (Fig. 7). One possible explanation of these data is that the truncated MreB isoforms do not associate with the membrane efficiently enough to interact with the membrane proteins RodZ and FtsK. Nevertheless, these data suggest the extended N-terminal region may function as a scaffolding platform for MreB to interact with other proteins related to cell division and may be crucial for the cell divisome formation at the septum, like FtsZ.
Outstanding questions remain: how is polarity established in Chlamydia? Is it MreB dependent or are other factors involved? Is MreB actively excluded from other parts of the membrane by an unknown system similar to the function of the MinCDE system in restricting FtsZ to division septa during binary fission? In this study, we performed the first systematic investigation of chlamydial MreB and its function as a cell division coordinator. Our data suggest that chlamydial MreB has multiple membrane-associating domains that may promote its assembly at the site of polarized division. This in turn may allow it to functionally substitute for the role of FtsZ in organizing the division septum.
MATERIALS AND METHODS
Organisms and cell culture.
McCoy (kind gift of Harlan Caldwell) and HeLa (ATCC, Manassas, VA) cells were cultured at 37°C with 5% CO2 in Dulbecco’s modified eagle medium (DMEM; Invitrogen, Waltham, MA) containing 10% fetal bovine serum (FBS; HyClone, Logan, UT) and 10 μg/ml gentamicin (Gibco, Waltham, MA). Chlamydia trachomatis serovar L2 (strain 434/Bu) lacking the endogenous plasmid (−pL2) was infected and propagated in McCoy cells for use in transformations. HeLa cells were infected with chlamydial transformants in DMEM containing 10% FBS, 10 μg/ml gentamicin, 1 U/ml penicillin G, and 1 μg/ml cycloheximide. All cell cultures and chlamydial stocks were routinely tested for Mycoplasma contamination using the Mycoplasma PCR detection kit (Sigma, St. Louis, MO). For E. coli, wild-type MG1655 and the mreB-deficient strain P2733 (kind gift of Piet de Boer) were cultured at 37°C with 225 rpm shaking in lysogeny broth (LB) medium containing no or several antibiotics as indicated: 50 μg/ml spectinomycin, 34 μg/ml chloramphenicol, or 25 μg/ml tetracycline. All chemicals and antibiotics were obtained from Sigma unless otherwise noted.
Cloning.
The plasmids and primers used in this study are described in Table S1 in the supplemental material. The wild-type and truncated Chlamydia trachomatis mreB genes were amplified by PCR with Phusion DNA polymerase (NEB, Ipswich, MA) using 100 ng C. trachomatis L2 genomic DNA as a template. Some gene segments were directly synthesized as a gBlock fragment (Integrated DNA Technologies, Coralville, IA). The PCR products were purified using a PCR purification kit (Qiagen, Hilden, Germany). The HiFi Assembly reaction master mix (NEB) was used according to the manufacturer’s instructions in conjunction with plasmids pASK-GFP-mKate2-L2 (kind gift of P. Scott Hefty) cut with FastDigest AgeI and EagI (Thermo Fisher, Waltham, MA), pBOMB4-Tet (kind gift of Ted Hackstadt) cut with EagI and KpnI, pKT25 or pUT18C cut with BamHI and EcoRI, or pBAD33 cut with XbaI and SalI depending on the construct being prepared. All plasmids were also dephosphorylated with FastAP (Thermo Fisher). The products of the HiFi reaction were transformed into either NEB-10beta (for chlamydial transformation plasmids) or DH5α (Iq) competent cells (NEB), plated on appropriate antibiotics, and plasmids were subsequently isolated from individual colonies grown overnight in LB by using a miniprep kit (Qiagen). For chlamydial transformation, the constructs were transformed into dam− dcm− competent cells (NEB) and purified as demethylated constructs.
Bioinformatics analysis.
Sequences for Chlamydia trachomatis serovar L2/434, Pseudomonas aeruginosa (PAO1), and Vibrio cholerae (O395) were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/) and, for E. coli MG1655, from Ecocyc database (https://ecocyc.org/) (32). Searches for other extended N termini in MreB orthologs was performed by BLASTing the 150 N-terminal amino acids of C. trachomatis serovar L2/434 MreB against all nonredundant sequences in the database while excluding “Chlamydiae (taxid: 204428).” Please note there is an error in chlamydial protein BLAST searches due to a contaminant chlamydial sequence in the Bacteroides fragilis S6L5 strain (i.e., any chlamydial sequence generates a hit in this strain). Protein sequence alignment was performed using Clustal Omega website (https://www.ebi.ac.uk/Tools/msa/clustalo/) (33) and the ESPript3 program (http://espript.ibcp.fr) (34). Helical wheels were made by using Helixator (http://www.tcdb.org/progs/helical_wheel.php) (35). Amphipathic helixes were predicted by using Amphipaseek program (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_amphipaseek.html) (36).
Transformation of Chlamydia trachomatis.
McCoy cells were plated in a six-well plate the day before beginning the transformation procedure. Chlamydia trachomatis serovar L2 without plasmid (−pL2) (kind gift of Ian Clarke) was incubated with 2 μg demethylated plasmid in Tris-CaCl2 buffer (10 mM Tris-Cl, 50 mM CaCl2) for 30 min at room temperature. During this step, the McCoy cells were washed with 2 ml Hanks’ balanced salt solution (HBSS) medium containing Ca2+ and Mg2+ (Gibco). After that, McCoy cells were infected with the transformants in 2 ml HBSS per well. The plate was centrifuged at 400 × g for 15 min at room temperature and incubated at 37°C for 15 min. The inoculum was aspirated, and DMEM containing 10% FBS and 10 μg/ml gentamicin was added per well. At 8 h postinfection (hpi), the medium was changed to medium containing 1 μg/ml cycloheximide and 1 or 2 U/ml penicillin G, and the plate was incubated at 37°C until 48 hpi. At 48 hpi, the transformants were harvested and used to infect a new McCoy cell monolayer. These harvest and infection steps were repeated every 48 hpi until mature inclusions were observed.
Complementation assay.
The arabinose-inducible pBAD33 vectors encoding nothing, E. coli MreB, chlamydial MreB, or ΔN22 chlamydial MreB were transformed into mreB mutant strains (P2733; kind gift of Piet de Boer). Chemically competent cells of each strain were prepared using standard techniques with CaCl2. The strains were cultured overnight, diluted 1/50 in LB medium containing chloramphenicol (34 μg/ml), tetracycline (25 μg/ml), or spectinomycin (50 μg/ml) and cultured for 2 h when 0.01% arabinose or 0.4% glucose was added as inducer or control, respectively. After 2 h and 6 h of induction, 4 μl of the cells was mounted on a 1% LB agar pad and covered with a coverslip. The morphologies of the strains were observed by a Zeiss Imager.Z2 equipped with an Apotome2 using a 100× lens objective.
The localization of chlamydial MreB fusion proteins in E. coli.
The E. coli MG1655 wild-type and ΔmreB mutant (P2733) transformed with the arabinose-inducible pBAD33G vectors encoding GFP with various MreB N-terminal peptides were cultured at 37°C with 255-rpm shaking overnight. Overnight cultures were diluted 1:100 (MG1655) or 1:50 (P2733) in LB medium containing appropriate antibiotics and cultured at 37°C with 255-rpm shaking. After 2 h of shaking, the cells were induced, or not, with 0.05% (wt/vol) arabinose and cultured for another 2 h. Before mounting the samples, SynaptoRed-C2 (FM4-64; Cayman Chemical, Ann Arbor, MI) was added in the samples to a final concentration 1.5 μM to stain the membrane. After 15 min, 4 μl of each culture was placed on a 1% LB agar pad and covered with a coverslip. The samples were observed with a Zeiss Imager.Z2 equipped with an Apotome2 using a 100× lens objective.
Indirect immunofluorescence microscopy.
HeLa cells were seeded in 24-well plates on coverslips at a density of 105 cells per well the day before infection. Chlamydial strains expressing wild-type or truncated MreBs with a six-histidine tag at the C terminus were used to infect HeLa cells in DMEM containing penicillin G and cycloheximide. At 6 hpi, 10 nM anhydrotetracycline (aTc) was added. At 10.5 hpi, the coverslips of infected cells were washed with Dulbecco’s phosphate-buffered saline (DPBS) and fixed with fixing solution (3.2% formaldehyde and 0.022% glutaraldehyde in DPBS) for 2 min. The samples were then washed three times with DPBS and permeabilized with ice-cold 90% methanol for 1 min. Afterwards, the fixed cells were labeled with primary antibodies, including goat anti-major outer membrane protein (anti-MOMP; Meridian, Memphis, TN), rabbit anti-MreBCT antibody (custom antipeptide antibody directed against the C terminus of C. trachomatis serovar L2 MreB; Thermo Fisher), rabbit anti-Hsp60 (kind gift of Rick Morrison), and mouse and rabbit anti-six histidine tags (GenScript, Nanjing, China, and Abcam, Cambridge, UK, respectively). Secondary antibodies, donkey anti-goat antibody (594), donkey anti-rabbit antibody (647), and donkey anti-mouse antibody (488), were used to visualize the primary antibodies. The secondary antibodies were obtained from Invitrogen. Coverslips were observed by using either a Zeiss LSM 800 confocal microscope or a superresolution SIM scope (Zeiss ELYRA PS.1).
BACTH assay.
Competent DHT1 E. coli, an adenylate cyclase-deficient strain, was cotransformed with pKT25 and pUT18C vectors carrying the genes of interest or empty vectors and spread on M63 minimal medium plates containing 50 μg/ml ampicillin, 25 μg/ml kanamycin, 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), 40 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), 0.04% Casamino Acids, and 0.2% maltose. The plates were incubated at 30°C for 5 to 7 days. Blue colonies indicated positive interactions, whereas no growth or small white colonies indicated no interactions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lisa Rucks for critical review of the manuscript. We also thank the following individuals for providing reagents used in this study: Harlan Caldwell (National Institutes of Health), Ian Clarke (University of Southampton), Piet de Boer (Case Western University), Ted Hackstadt (Rocky Mountain Labs/NIH), P. Scott Hefty (University of Kansas), and Rick Morrison (University of Arkansas for Medical Sciences).
This work was supported by a grant from the National Institute for General Medical Science (R35GM124798-01) within the National Institutes of Health to S.P.O. The University of Nebraska Medical Center Advanced Microscopy Core Facility receives partial support from the National Institute for General Medical Science INBRE-P20 GM103427 and COBRE-P30 GM106397 grants, as well as support from the National Cancer Institute (NCI) for The Fred & Pamela Buffett Cancer Center support grant P30 CA036727 and the Nebraska Research Initiative.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Abdelrahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol Rev 29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Moore ER, Ouellette SP. 2014. Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins. Front Cell Infect Microbiol 4:157. doi: 10.3389/fcimb.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 4.Bi EF, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 5.Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, Maurelli AT. 2014. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506:507–510. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ouellette SP, Rueden KJ, AbdelRahman YM, Cox JV, Belland RJ. 2015. Identification and partial characterization of potential FtsL and FtsQ homologs of Chlamydia. Front Microbiol 6:1264. doi: 10.3389/fmicb.2015.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdelrahman Y, Ouellette SP, Belland RJ, Cox JV. 2016. Polarized cell division of Chlamydia trachomatis. PLoS Pathog 12:e1005822. doi: 10.1371/journal.ppat.1005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouellette SP, Karimova G, Subtil A, Ladant D. 2012. Chlamydia co-opts the rod shape-determining proteins MreB and Pbp2 for cell division. Mol Microbiol 85:164–178. doi: 10.1111/j.1365-2958.2012.08100.x. [DOI] [PubMed] [Google Scholar]
- 9.Salje J, van den Ent F, de Boer P, Löwe J. 2011. Direct membrane binding by bacterial actin MreB. Mol Cell 43:478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srinivasan R, Mishra M, Murata-Hori M, Balasubramanian MK. 2007. Filament formation of the Escherichia coli actin-related protein, MreB, in fission yeast. Curr Biol 17:266–272. doi: 10.1016/j.cub.2006.11.069. [DOI] [PubMed] [Google Scholar]
- 11.van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z. 2011. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci U S A 108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strahl H, Burmann F, Hamoen LW. 2014. The actin homologue MreB organizes the bacterial cell membrane. Nat Commun 5:3442. doi: 10.1038/ncomms4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussain S, Wivagg CN, Szwedziak P, Wong F, Schaefer K, Izoré T, Renner LD, Holmes MJ, Sun Y, Bisson-Filho AW, Walker S, Amir A, Löwe J, Garner EC. 2018. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. Elife 7:e32471. doi: 10.7554/eLife.32471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kemege KE, Hickey JM, Barta ML, Wickstrum J, Balwalli N, Lovell S, Battaile KP, Hefty PS. 2015. Chlamydia trachomatis protein CT009 is a structural and functional homolog to the key morphogenesis component RodZ and interacts with division septal plane localized MreB. Mol Microbiol 95:365–382. doi: 10.1111/mmi.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swulius MT, Jensen GJ. 2012. The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-terminal yellow fluorescent protein tag. J Bacteriol 194:6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liechti G, Kuru E, Packiam M, Hsu YP, Tekkam S, Hall E, Rittichier JT, VanNieuwenhze M, Brun YV, Maurelli AT. 2016. Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog 12:e1005590. doi: 10.1371/journal.ppat.1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Arellano-Santoyo H, Combs PA, Shaevitz JW. 2010. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. Proc Natl Acad Sci U S A 107:9182–9185. doi: 10.1073/pnas.0911517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bendezu FO, de Boer PA. 2008. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol 190:1792–1811. doi: 10.1128/JB.01322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenton AK, Gerdes K. 2013. Direct interaction of FtsZ and MreB is required for septum synthesis and cell division in Escherichia coli. EMBO J 32:1953–1965. doi: 10.1038/emboj.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy JE, Brimacombe R. 1994. Prokaryotic translation: the interactive pathway leading to initiation. Trends Genet 10:402–407. doi: 10.1016/0168-9525(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 22.Kruse T, Bork-Jensen J, Gerdes K. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol 55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- 23.Ouellette SP, Rueden KJ, Gauliard E, Persons L, de Boer PA, Ladant D. 2014. Analysis of MreB interactors in Chlamydia reveals a RodZ homolog but fails to detect an interaction with MraY. Front Microbiol 5:279. doi: 10.3389/fmicb.2014.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Ent F, Johnson CM, Persons L, de Boer P, Löwe J. 2010. Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J 29:1081–1090. doi: 10.1038/emboj.2010.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgenstein RM, Bratton BP, Nguyen JP, Ouzounov N, Shaevitz JW, Gitai Z. 2015. RodZ links MreB to cell wall synthesis to mediate MreB rotation and robust morphogenesis. Proc Natl Acad Sci U S A 112:12510–12515. doi: 10.1073/pnas.1509610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hesse L, Bostock J, Dementin S, Blanot D, Mengin-Lecreulx D, Chopra I. 2003. Functional and biochemical analysis of Chlamydia trachomatis MurC, an enzyme displaying UDP-N-acetylmuramate:amino acid ligase activity. J Bacteriol 185:6507–6512. doi: 10.1128/jb.185.22.6507-6512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy AJ, Maurelli AT. 2005. Characterization of Chlamydia MurC-Ddl, a fusion protein exhibiting d-alanyl-d-alanine ligase activity involved in peptidoglycan synthesis and d-cycloserine sensitivity. Mol Microbiol 57:41–52. doi: 10.1111/j.1365-2958.2005.04661.x. [DOI] [PubMed] [Google Scholar]
- 28.Patin D, Bostock J, Blanot D, Mengin-Lecreulx D, Chopra I. 2009. Functional and biochemical analysis of the Chlamydia trachomatis ligase MurE. J Bacteriol 191:7430–7435. doi: 10.1128/JB.01029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox A, Rogers JC, Gilbart J, Morgan S, Davis CH, Knight S, Wyrick PB. 1990. Muramic acid is not detectable in Chlamydia psittaci or Chlamydia trachomatis by gas chromatography-mass spectrometry. Infect Immun 58:835–837. doi: 10.1128/IAI.58.3.835-837.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hale CA, de Boer PA. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 31.Kawazura T, Matsumoto K, Kojima K, Kato F, Kanai T, Niki H, Shiomi D. 2017. Exclusion of assembled MreB by anionic phospholipids at cell poles confers cell polarity for bidirectional growth. Mol Microbiol 104:472–486. doi: 10.1111/mmi.13639. [DOI] [PubMed] [Google Scholar]
- 32.Keseler IM, Mackie A, Santos-Zavaleta A, Billington R, Bonavides-Martínez C, Caspi R, Fulcher C, Gama-Castro S, Kothari A, Krummenacker M, Latendresse M, Muñiz-Rascado L, Ong Q, Paley S, Peralta-Gil M, Subhraveti P, Velázquez-Ramírez DA, Weaver D, Collado-Vides J, Paulsen I, Karp PD. 2017. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res 45:D543–D550. doi: 10.1093/nar/gkw1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saier MH Jr, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. 2016. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res 44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sapay N, Guermeur Y, Deleage G. 2006. Prediction of amphipathic in-plane membrane anchors in monotopic proteins using a SVM classifier. BMC Bioinformatics 7:255. doi: 10.1186/1471-2105-7-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.