Abstract
Objective:
Our objective was to investigate the mechanisms that govern natural killer (NK) cell responses to HIV, with a focus on specific receptor-ligand interactions involved in HIV recognition by NK cells.
Design and Methods:
We first performed a mass cytometry-based screen of NK cell receptor expression patterns in healthy controls and HIV+ individuals. We then focused mechanistic studies on the expression and function of T cell immunoreceptor with Ig and ITIM domains (TIGIT).
Results:
The mass cytometry screen revealed that TIGIT is upregulated on NK cells of untreated HIV+ women, but not in antiretroviral-treated women. TIGIT is an inhibitory receptor that is thought to mark exhausted NK cells; however, blocking TIGIT did not improve anti-HIV NK cell responses. In fact, the TIGIT ligands CD112 and CD155 were not upregulated on CD4+ T cells in vitro or in vivo, providing an explanation for the lack of benefit from TIGIT blockade. TIGIT expression marked a unique subset of NK cells that express significantly higher levels of NK cell activating receptors (DNAM-1, NTB-A, 2B4, CD2) and exhibit a mature/adaptive phenotype (CD57hi, NKG2Chi, LILRB1hi, FcRγlo, Syklo). Furthermore, TIGIT+ NK cells had increased responses to mock-infected and HIV-infected autologous CD4+ T cells, and to PMA/ionomycin, cytokine stimulation and the K562 cancer cell line.
Conclusions:
TIGIT expression is increased on NK cells from untreated HIV+ individuals. Although TIGIT does not participate directly to the response to HIV-infected cells, it marks a population of mature/adaptive NK cells with increased functional responses.
Keywords: TIGIT, NK cells, HIV, adaptive, mature
INTRODUCTION
Natural killer (NK) cells are among the first responders to viral infections and can swiftly recognize and kill virus-infected cells [1]. These responses are traditionally thought to be non-specific, as NK cell function is mediated by the expression of germ-line encoded receptors rather than antigen-specific receptors [2]. However, recent evidence has revealed that NK cells are capable of antigen-specific adaptive responses against viruses, such as cytomegalovirus (CMV), Epstein-Barr Virus, Varicella Zoster Virus, and influenza virus [1,3–9].
HIV infection profoundly alters the NK cell compartment, with expansion of a CD56−CD16+ subpopulation [10,11], downregulation of several activating NK cell receptors, including FcRγ (FcεRIγ), NKp30, and NKp46 [12–14], and impairment of NK cell function [15–18]. NK cells also play a critical role in the immune response against HIV [19,20]. NK cells may protect from HIV acquisition, as increased NK cell activity is associated with lower risk of acquiring HIV in highly exposed individuals [21–23]. Consistent with this, genotypes of specific NK cell receptors and human leukocyte antigens (HLA) are enriched in exposed seronegative individuals [21,24–26]. The expression of specific NK cell receptor/HLA class I ligand pairs (KIR3DL1/KIR3DS1 with HLABw4-80I) is also associated with slower disease progression [27,28] and NK cells from long term non-progressors show increased function compared to HIV+ typical progressors [12,29]. Altogether, these data raise the possibility that specific NK receptor-ligand interactions may contribute to HIV control and could be used as targets to improve HIV-specific NK cell function.
T cell immunoreceptor with Ig and ITIM domains (TIGIT) is an inhibitory receptor expressed on T cells and NK cells [30,31]. The TIGIT inhibitory signal is mediated by ligation with its high affinity ligand, the poliovirus receptor (CD155 or PVR), and its low affinity ligand CD112 (Nectin-2 or PVRL2) [30,32,33]. TIGIT has been associated with CD4+ T cell, CD8+ T cell and NK cell exhaustion both in the setting of chronic viral infections and malignancy [31,34–40]. Blockade of the TIGIT/CD155/CD112 pathway to improve the function of T cells and NK cells against solid cancers is currently under investigation [41,42].
In HIV-1 infection, TIGIT marks exhausted T cells, correlates with disease progression and is decreased on CD4+ T cells from elite controllers [43,44]. Early initiation of antiretroviral treatment (ART) in HIV-infected individuals does not return TIGIT/CD155 to normal levels on CD8+ T cells [37]. Additionally, co-expression of TIGIT with immune checkpoint inhibitor PD-1 marks CD4+ T cells harboring latent virus [45–47]. These data suggest that the TIGIT/CD155/CD112 pathway in T cells could contribute to HIV pathogenesis.
In NK cells, less is known regarding the role of TIGIT during HIV infection. A recent report showed that TIGIT is increased on NK cells from HIV-1 infected patients and that TIGIT blockade improves NK cell responses to cytokines ex-vivo [48], consistent with the idea that TIGIT may represent a marker of NK cell exhaustion. Here, we used blood samples from Beninese women to study the effect of HIV infection on the NK cell compartment, with a particular focus on TIGIT expression and function.
METHODS:
Study subjects and sample processing
Cryopreserved peripheral blood mononuclear cells (PBMCs) were obtained from a study of commercial sex workers in Cotonou, Benin. HIV-1-infected women were enrolled from a sex-worker clinic and HIV-1-uninfected women were enrolled from a general health clinic, as described previously [49,50]. PBMCs were obtained from twenty untreated HIV-infected women, twenty HIV-infected women receiving ART and ten healthy women (Table S1). Written informed consent was obtained from all subjects. The study was approved by the Comité National Provisoire d’Éthique de la Recherche en Santé in Cotonou and the Centre Hospitalier de l’Université de Montréal (CHUM) Research Ethics Committees.
For in vitro studies using healthy donors, leukoreduction system chambers were purchased from the Stanford Blood Bank. PBMCs were purified using Ficoll density gradient centrifugation and cryopreserved in fetal bovine serum (FBS) with 10% DMSO.
Cell isolation
For profiling PBMCs from Beninese women, 1x106 PBMCs were stained for mass cytometry with the ligand antibody panel (Table S2). NK cells were purified from PBMCs by magnetic-activated isolation via negative selection (Miltenyi) and stained with the NK cell antibody panel (Table S3, Figure S2a).
For the in vitro co-cultures, cryopreserved PBMCs from healthy donors were used to purify CD4+ T cells and NK cells by magnetic-activated isolation via negative selection (Miltenyi).
Antibody conjugation, mass cytometry staining and data acquisition
Antibodies were conjugated using MaxPar® ×8 labeling kits (Fluidigm). To ensure antibody stability over time, antibody panels were lyophilized into single-use pellets prior to use (Biolyph). Cells were stained for mass cytometry as described previously [51,52] and acquired on a Helios mass cytometer (Fluidigm).
In vitro HIV infection
Given that the majority of HIV strains circulating in Benin are clade A or G, the clade A early HIV-1 strain Q23-FL was chosen for in vitro infection. Replication competent Q23-FL was made by transfection and titrated as described [51]. CD4+ T cells were activated for 2 days with plate-bound anti-CD3 (clone OKT3, eBioscience, 10 μg/ml), soluble anti-CD28/CD49d (clones L293/L25, BD Biosciences, 1 μg/ml each) and phytohemagglutinin (eBioscience, 2.5 μg/ml). CD4+ T cells were infected overnight with Q23-FL via ViroMag R/L magnetofection (Oz Biosciences) at a multiplicity of infection of 20. HIV infection was as measured by intracellular p24.
TIGIT blockade assay
Cells were cultured in RPMI media containing 10% FBS (Thermo Fisher Scientific) and 1% penicillin/streptomycin/amphotericin (Thermo Fisher Scientific) (RP10). NK cells were incubated in 24-well plates at a concentration of 2x106 cells/ml in for 3 days at 37°C with 5% CO2, with addition of rhIL-2 (R&D Systems, 300 IU/ml). After 3 days, NK cells were washed with fresh media and plated in 96-well plates (80,000-100,000 cells/well). Blocking mIgG1κ anti-hTIGIT antibody (eBioscience, clone MBSA43, 10 μg/ml) or a mIgG1κ isotype control (eBioscience, clone P3.6.2.8.1, 10 μg/ml) were added. Cells were incubated at 4°C for 20 minutes, washed with PBS + 2% FBS and resuspended in media.
CD112 and CD155 mRNA expression levels
RNA from resting, mock-infected or HIV-infected CD4+ T cells was isolated using the RNeasy Mini Kit with QIAshredder columns (Qiagen). cDNA was produced using the SuperScript III First Strand Synthesis Kit (Thermo Fisher Scientific). Quantitative PCR (qPCR) was achieved with Taqman Universal Master Mix (Applied Biosystems) with the following Taqman Assays, using FAM-MGB probes: Nectin-2/CD112 (Thermo Fisher Scientific), PVR/CD155 (Thermo Fisher Scientific), and Human GAPDH (Applied Biosystems). qPCR was run on the Applied Biosystems Step One Plus Real Time PCR System. Transcription levels were normalized to GAPDH. mRNA fold change expression was calculated using the double delta Ct analysis method with resting CD4+ T cells as the control group [53].
NK cell:CD4+ T cell co-cultures, cytokine stimulation and determination of NK cell function
All cells were cultured in RP10. NK cells (80,000-100,000/well) were incubated with HIV-infected or mock-infected autologous CD4+ T cells (320,000/well at a 1:4 effector:target ratio), 0.4 μl/200μl of cell stimulation cocktail (phorbol 12-myristate 13-acetate (PMA) and ionomycin, eBioscience), a cocktail of IL-12, IL-15 and IL-18 (rhIL-12, R&D Systems, 5ng/ml; rhIL-15, Pepro-Tech, 20ng/ml; rhIL-18, R&D Systems, 0.5 μg/ml), or with the cancer cell line K562 (ATCC, used at passage 2-10, 300,000 cells/well at a 1:3 effector:target ratio). Cells were incubated for 4 hours at 37°C with 5% CO2, in the presence of brefeldin, monensin (eBioscience) and anti-CD107a-APC-H7 antibody (BD Biosciences, Clone H4A3). After incubation, cells were stained with LIVE/DEAD™ Fixable Yellow Staining Kit (Thermo Fisher Scientific) for 20 minutes at room temperature, then fixed (BD FACS Lyse), permeabilized (BD FACS Perm II) and stained with anti-IFN-γ-V450 (BD Biosciences, Clone B27), anti-TIGIT-APC (eBioscience, Clone MBSA43), anti-CD3-PerCP-Cy5.5 (Biolegend, Clone UCHT1), anti-HIVp24-FITC (Beckman Coulter, Clone KC57) for 30 minutes at 4°C. Data was acquired on a MACSQuant® Analyzer Flow Cytometer (Miltenyi).
Data analysis
Bead normalization was performed before downstream analyses (https://github.com/ParkerICI/premessa) [54]. Data were visualized using FlowJo v10.5.3 and FAS-L, Ki-67 and CXCR6 were excluded from subsequent analyses due to poor staining. The original data is available at ImmPort (https://www.immport.org) under study accession SDY1535. Serial negative gating was performed to identify NK cells from any contaminating cells remaining after purification (Figure S1a). Samples with cell number <1000 were excluded from analyses; The open source statistical package R (https://www.r-project.org/) was used for all statistical analyses [55]. Normalized signal intensities were transformed using the asinh function with a cofactor of 5 prior to generalized linear model (GLM) analysis, multidimensional scaling (MDS) and Uniform Manifold Approximation and Projection (UMAP) visualization. For the GLM analysis and MDS projection, we used the custom-made package “CytoGLMM” [56,57]. For comparison between groups, we used the Wilcoxon rank-sum test. When appropriate, p-values were adjusted for multiple comparisons using the Benjamini-Hochberg method. UMAP embeddings were calculated using the R package “uwot”, with n_neighbor = 5 and min_dist = 0.2 [58]. Embeddings were visualized in Cytobank (www.cytobank.org).
RESULTS
TIGIT is upregulated on NK cells from untreated HIV-infected women
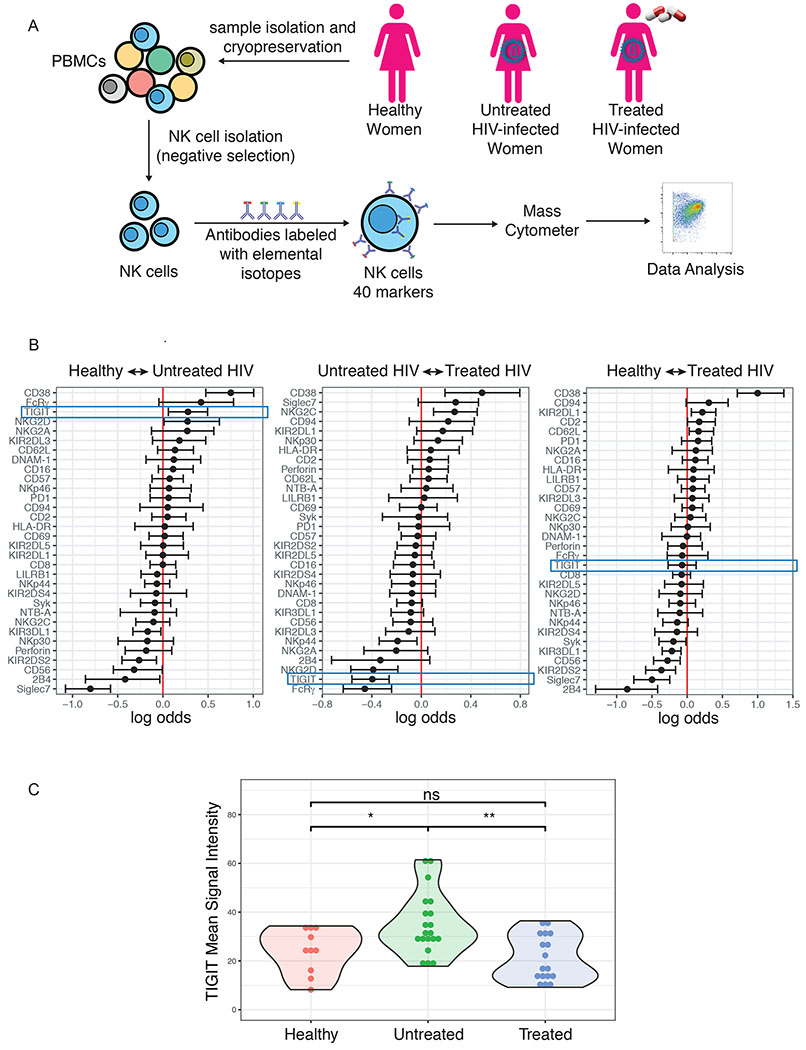
To investigate the effect of HIV-1 infection on NK cells, we used CyTOF to profile the NK cells of twenty untreated HIV-1-infected women, twenty HIV-1 infected women on ART and ten healthy women (Figure 1a). A generalized linear model with bootstrap resampling revealed that HIV-1 infection is associated with a profound alteration of the NK cell phenotype, with higher levels of CD38 and lower levels of Siglec-7 predicting chronic HIV-1 infection (Figure 1b), as previously described [59,60]. This analysis also confirmed that TIGIT is upregulated in untreated HIV-1-infected individuals [48]. Additionally, our data revealed that TIGIT is not elevated on NK cells from antiretroviral-treated HIV-1-infected women (Figure 1b and 1c). We performed correlations with CD4+ T cell counts and found that TIGIT levels did not correlate with CD4+ T cell counts (Figure S3).
Figure 1: TIGIT is upregulated on NK cells from untreated HIV-infected women.
(A) Schematic of study design and experiment. (B) A generalized linear model with bootstrap resampling was used to identify markers predictive of the study groups. Log-odds are logarithm of ratios of the probability that a cell belongs to each study subject. An increase in the parameter coefficient corresponds to the strength of the classification power, with the 95% confidence interval represented by line surrounding the point estimate. (C) Mean Signal Intensity of TIGIT on NK cells from healthy women (n=10), untreated HIV+ women (n=20), treated HIV+ women (n=17). * = unadjusted p-value≤0.05, ** = unadjusted p-value≤0.01, ns = non-significant.
TIGIT expression is increased in CD56dim and CD56− NK cells from untreated HIV-1+ women
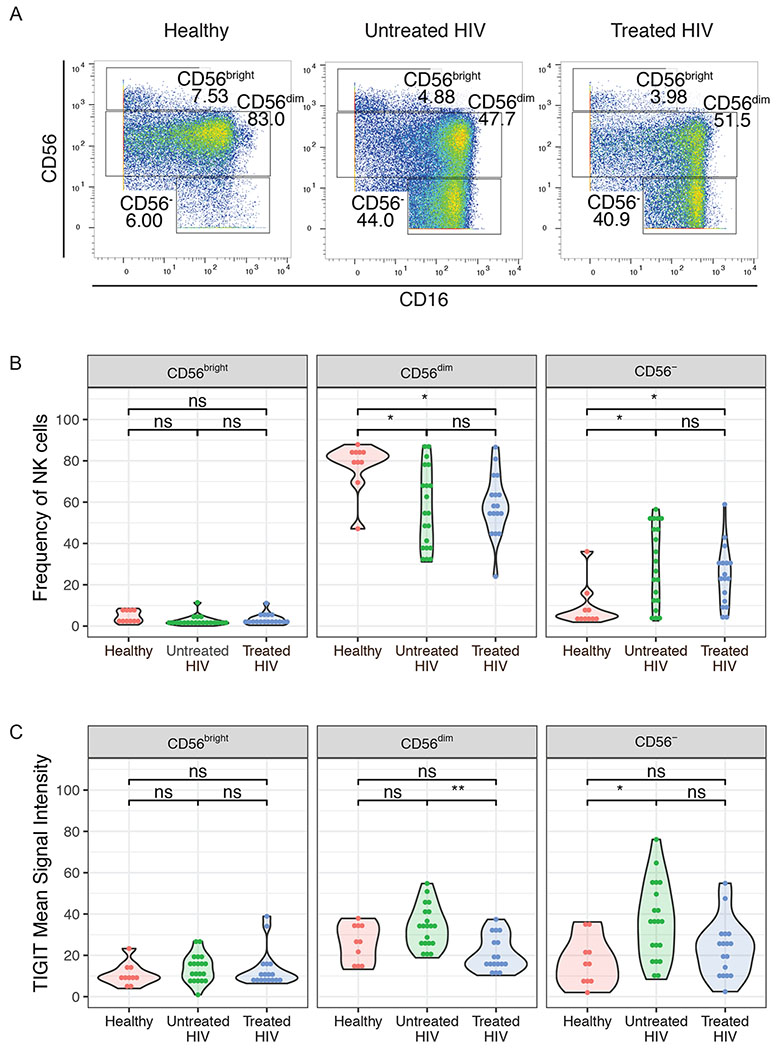
To explore the nature of this increased TIGIT expression, we evaluated TIGIT expression within different NK cell subpopulations. NK cells are classically divided into three distinct subpopulations: CD56bright, which secrete cytokines at high levels, CD56dim, a more cytotoxic subpopulation, and CD56−, a subpopulation of NK cells that are thought to be functionally impaired and are expanded during chronic viral infections, including HIV [10,61–63]. Here we confirmed that CD56− NK cells are increased in HIV-infected individuals, regardless of treatment status (Figure 2a and 2b). Additionally, TIGIT expression was higher on CD56− NK cells from untreated HIV-1 infected women compared to healthy controls, and on the CD56dim subpopulation in untreated HIV-1+ women compared to treated women (Figure 2c).
Figure 2. TIGIT expression is increased in CD56dim and CD56− NK cells of untreated HIV-1+ women.
(A) Representative CyTOF plots of gating of NK cells based on CD56 expression. (B) Frequency of CD56bright, CD56dim and CD56− NK cells in healthy women (n=10), untreated HIV+ women (n=20), treated HIV+ women n=17). (C) Mean Signal Intensity of TIGIT on NK cell subsets from healthy women (n=10), untreated HIV+ women (n=20), treated HIV+ women (n=17). * = adjusted p-value≤0.05, ** = adjusted p-value≤0.01, ns = non-significant.
TIGIT blockade does not alter NK cell response to HIV-infected cells
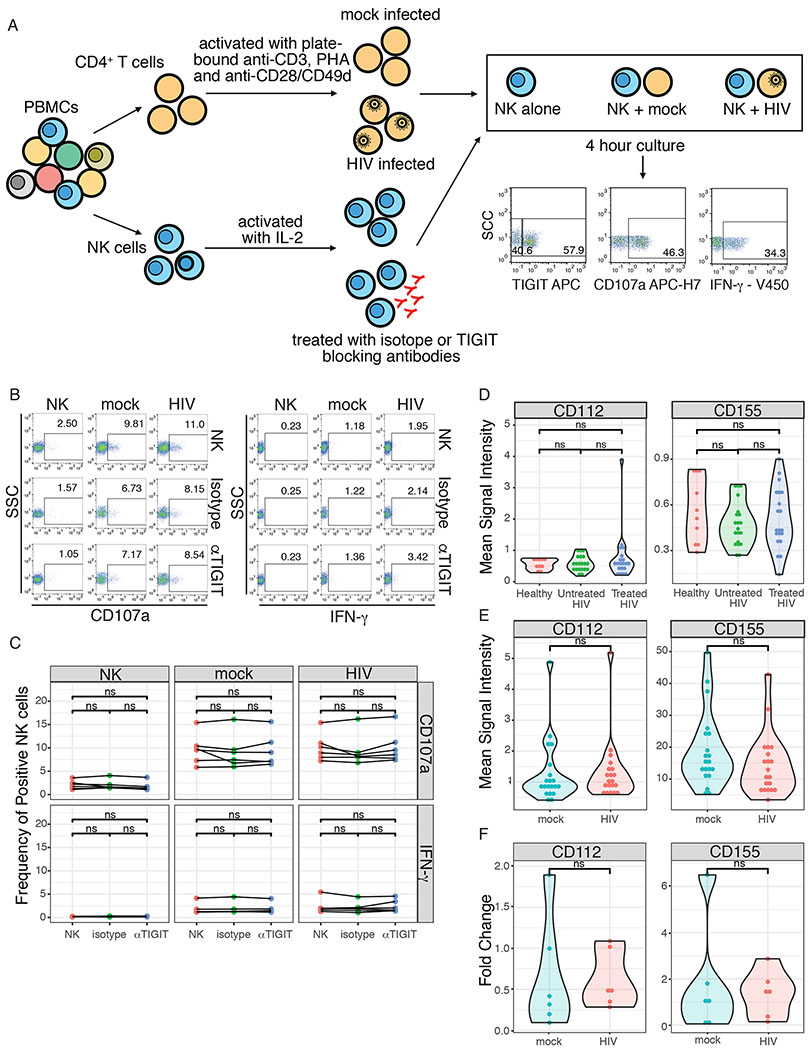
TIGIT blockade increases ex vivo NK cell responses to cytokines in HIV-1 infected men [48] and elicits improved anti-tumor responses in mouse models [35]. To explore the effect of TIGIT blockade on NK cell responses against HIV in vitro, we treated NK cells from healthy donors with an anti-TIGIT monoclonal antibody and measured NK cell responses (CD107a and IFN-γ) by flow cytometry after incubation with mock-infected or HIV-infected autologous CD4+ T cells (Figure 3a). Blockade of TIGIT did not improve NK cell responses, when compared to untreated NK cells or cells treated with an isotype control (Figures 3b and 3c). This lack of effect was not due to inadequate blockade, as <10% of NK cells could bind the anti-TIGIT antibody at the end of co-culture, suggesting continued binding of the blocking antibody (Figure S4).
Figure 3. TIGIT blockade does not alter HIV-specific NK cell responses to HIV and TIGIT ligands are not upregulated by during HIV infection.
(A) Schematic of TIGIT blockade assay and NK cell:CD4+ T cell co-cultures. (B) Representative flow cytometry plots of CD107a and IFN-γ production, expressed as frequency of positive cells, by NK cells alone (NK) or after co-culture with mock-infected (mock) or HIV-infected (HIV) autologous CD4+ T cells. Cells were incubated alone (NK), pre-treated with isotype control (Isotype) or a blocking anti-TIGIT antibody (αTIGIT). (C) Summary plot of frequency of CD107a+ and IFN-γ+ NK cells after 4 hour co-culture with mock-infected or HIV-infected autologous CD4+ T cells. (D) Mean Signal Intensity of CD112 and CD155 expression measured by mass cytometry in healthy women (n=9), untreated HIV+ women (n=20), treated HIV+ women (n=17). (E) Mean Signal Intensity of CD112 and CD155 expression measured by mass cytometry in mock-infected and HIV-infected CD4+ T cells. (F) Fold change in the NECTIN2 (CD112) and PVR (CD155) transcript levels, normalized to GAPDH, in mock-infected and HIV-infected CD4+ T cells, relative to resting CD4+ T cells. ns = non-sigificant.
TIGIT ligands are not substantially upregulated by HIV infection
The TIGIT inhibitory signal is mediated by ligation with CD155 (high affinity) or CD112 (low affinity) [30,32]. We therefore measured the expression of CD155 and CD112 on HIV-infected CD4+ T cells ex vivo and in vitro. CD155 and CD112 were both expressed at low levels on CD4+ T cells from HIV+ women and there was no significant difference in expression levels between healthy women, untreated HIV+ women and treated HIV+ women (Figures 3d and S2b). CD155 was not significantly induced in CD4+ T cells infected with HIV in vitro (Figures 3e and S2c). A median of 36% of cells expressed p24 in vitro and CD155 was not differentially expressed between mock-infected and p24+CD4+ T cells. While CD112 expression was significantly higher in p24+CD4+ T cells, its overall expression level was extremely low with a median of only 1.6% of cells expressing it (Figure S5). Finally, CD112 and CD115 transcription levels did not differ between mock-infected and HIV-infected CD4+ T cells (Figure 3f) with a median fold change of HIV relative to mock-infected CD4+ T cells 1.15 (interquartile range = 0.94-2.4) for CD155 and 1.07 (interquartile range = 0.93-3.13) for CD112.
TIGIT expression is increased in adaptive/mature NK cells
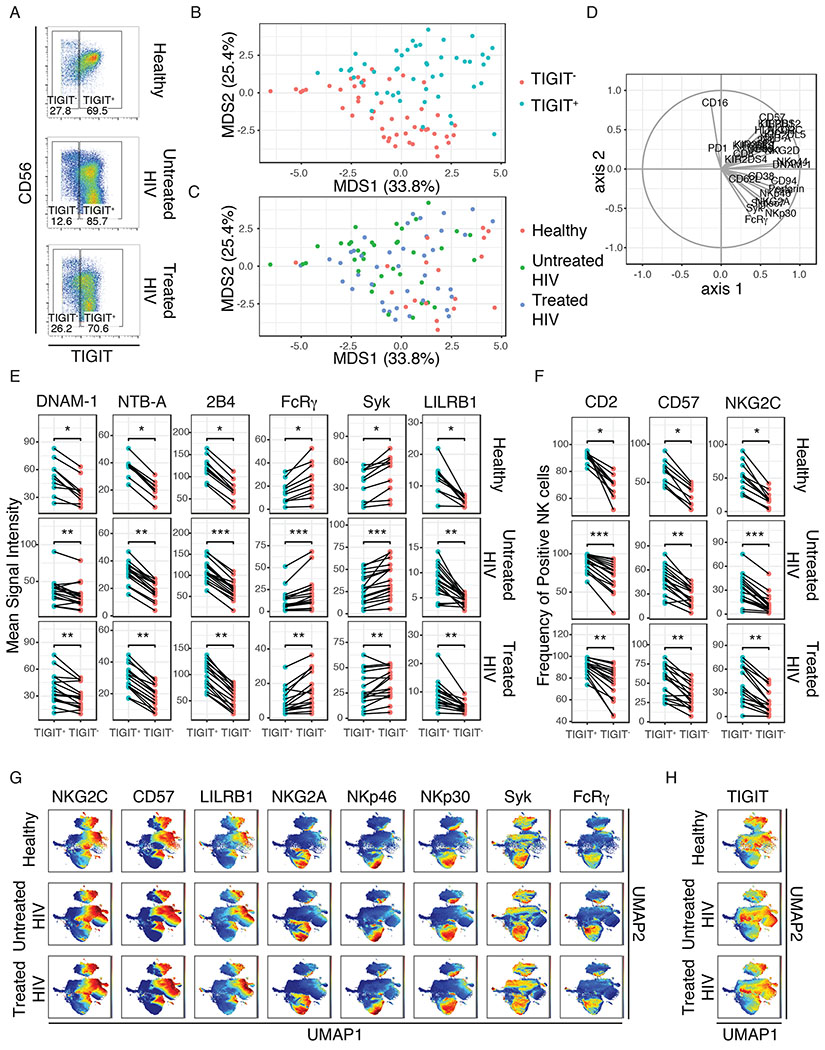
To characterize NK cells with increased TIGIT expression, we gated on TIGIT and compared TIGIT+ and TIGIT− NK cells from our Beninese women (Figure 4a). Using MDS, we found that TIGIT expression marks a distinct population of NK cells that was not explained by HIV infection or treatment status (Figure 4b–d). We then focused our analysis on features distinguishing TIGIT+ and TIGIT− NK cells. We found that TIGIT+ NK cells express higher levels of activating markers (DNAM-1, NTB-A, 2B4, CD2) and exhibit an adaptive/mature phenotype (CD57hi, NKG2Chi, LILRB1 (ILT2, CD85j)hi, FcRγlo, Syklo) (Figures 4e and 4f). To follow-up on this finding, we used UMAP to visualize NK cell subsets and found that adaptive/mature NK cells (CD57hi, NKG2Chi, LILRB1hi, NKG2Alo, NKp46lo, NKp30lo, FcRγlo, Syklo) were separated on the right side of the UMAP1 axis (Figure 4g). Additionally, when we colored the UMAP plot by TIGIT expression, we found that TIGIT co-localized with markers of adaptive/mature NK cells (Figure 4h).
Figure 4. TIGIT expression is increased in adaptive/mature NK cells.
(A) Representative CyTOF plots of gating for TIGIT+ and TIGIT− NK cells from healthy women (n=9), untreated HIV+ women (n=17), treated HIV+ women (n=16). (B) Multidimensional Scaling (MDS) of TIGIT+ and TIGIT− NK cells colored by TIGIT expression. (C) MDS of TIGIT+ and TIGIT− NK cells colored by group (Healthy, Untreated HIV and Treated HIV). (D) Vectors driving the variance of the MDS. (E) Mean Signal Intensity of DNAM-1, NTB-A, 2B4, FcRγ, Syk and LILRB1 expression in TIGIT+ and TIGIT− NK cells from healthy women, untreated HIV+ women and treated HIV+ women. (F) Frequency of CD2+, CD57+ and NKG2C+ NK cells from TIGIT+ and TIGIT− NK cells from healthy women, untreated HIV+ women and treated HIV+ women. (G) UMAP plot of pooled NK cells from healthy women, untreated HIV+ women and treated HIV+ women colored by markers of adaptive/mature NK cells (NKG2C, CD57, LILRB1, NKG2A, NKp46, NKp30, Syk, FcRγ) . (H) UMAP plot of pooled NK cells from healthy women, untreated HIV+ women and treated HIV+ women, colored by TIGIT expression. * = adjusted p-value≤0.05, ** = adjusted p-value≤0.01, *** = adjusted p-value≤0.001.
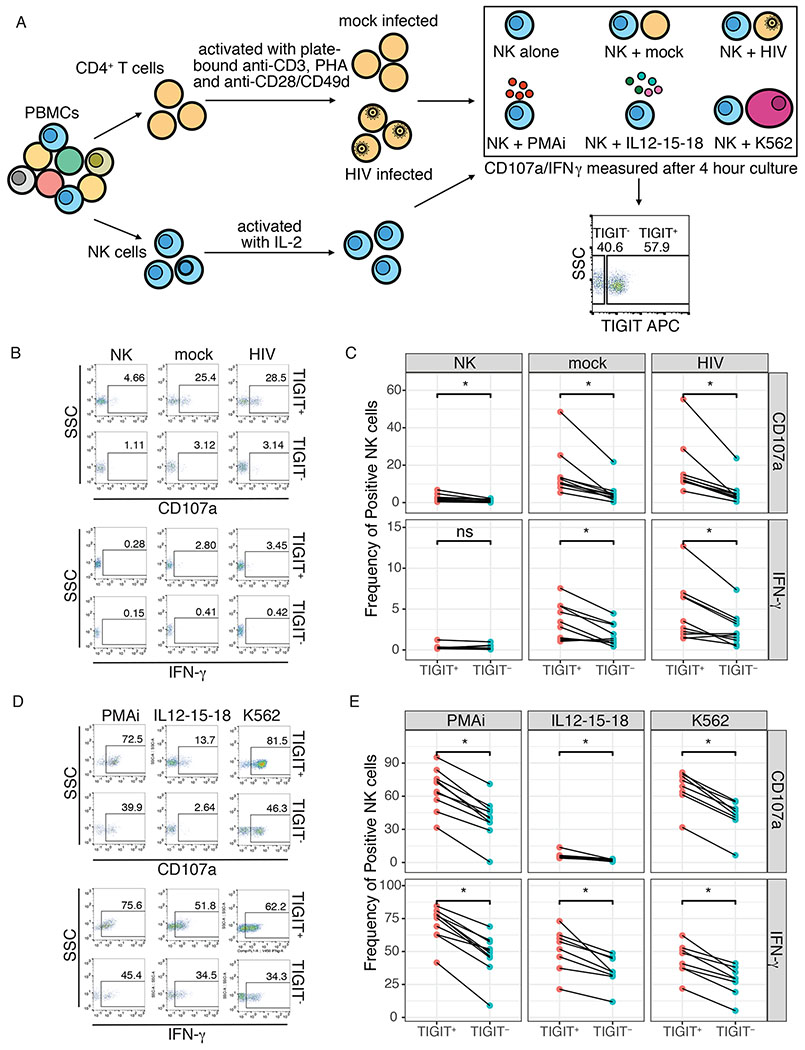
TIGIT expression is associated with increased NK cell responses
As mature NK cells are typically associated with higher functional activity, we sought to better characterize the function of TIGIT+ and TIGIT− NK cells. We isolated NK cells from healthy donors and cultured them with mock-infected or HIV infected autologous CD4+ T cells (Figure 5a). We then measured the functional responses (CD107a and IFN-γ) of TIGIT+ and TIGIT− NK cells by flow cytometry (Figures 5b–e). We found that TIGIT + NK cells had increased responses to mock-infected and HIV-infected autologous CD4+ T cells (Figures 5b and 5c). To better define whether this difference was specific to HIV, we measured the responses to PMA/ionomycin, a cocktail of IL-12, IL-15 and IL-18, and the leukemia cell line K562 (Figures 5a, 5d and 5e). TIGIT+ NK cells also exhibited increased responses to PMA/ionomycin, cytokines and tumor cells (Figures 5d and 5e).
Figure 5. TIGIT expression is associated with increased NK cell responses.
(A) Schematic of CD4+ T cell:NK cell co-cultures or stimulation and representative flow cytometry plot of gating of TIGIT+ and TIGIT− NK cells. (B) Representative flow cytometry plots of CD107a and IFN-γ production, expressed as frequency of positive cells, by TIGIT+ and TIGIT− NK cells after 4 hour co-culture with mock-infected (mock) or HIV-infected (HIV) autologous CD4+ T cells. (C) Summary plot of the frequency of TIGIT-gated CD107a+ and IFN-γ+ NK cells after 4 hour co-culture with mock-infected or HIV-infected autologous CD4+ T cells. (D) Representative flow cytometry plots of CD107a and IFN-γ production by TIGIT+ and TIGIT− NK cells after 4 hour stimulation with PMA/ionomycin (PMA/i), a cocktail of IL12, IL-15 and IL-18 (IL12-15-18) or the leukemia cell line K562 (K562). (E) Summary plot of the frequency of TIGIT-gated CD107a+ and IFN-γ+ NK cells after 4 hour stimulation with PMA/ionomycin (PMA/i), a cocktail of IL12, IL-15 and IL-18 (IL12-15-18) or the leukemia cell line K562 (K562). * = adjusted p-value≤0.05, ns = non-sigificant.
DISCUSSION
NK cells undergo significant changes during chronic HIV infection [16,64], with expansion of a hypofunctional CD56−CD16+ NK cell subpopulation [10,11], downregulation of several activating NK cell receptors [12–14], and adaptive reconfiguration [65]. Some of these changes are not observed after treatment or in the setting of natural HIV control [66], indicating that diminished NK cell function could contribute to disease pathogenesis. To better understand the role of different NK cell receptors in the pathogenesis of HIV, we profiled the NK cells from infected and uninfected HIV+ women. A particularly striking finding was that TIGIT is upregulated in untreated HIV infection, but not in the setting of ART. TIGIT is an inhibitory receptor that has been associated with hypofunctional/exhausted NK cells in cancer [31,35]. TIGIT is also associated with exhausted CD8+ T cells in hematologic malignancy and in HIV infection [36,39,43], making it an interesting potential target for immunomodulation in the setting of HIV infection. TIGIT upregulation on NK cells from HIV-infected men has been recently described [48]. Here, we demonstrate that TIGIT is upregulated in HIV-infected women and that this upregulation does not occur in women receiving ART. Further, TIGIT upregulation is more prominent on CD56−/CD16+ NK cells, a subpopulation thought to be functionally exhausted [10,11].
Several studies have explored the use of TIGIT blockade to rescue T and NK cell function. Blocking TIGIT is associated with improved anti-tumoral response in tumor-bearing mice and in patients with colorectal cancer [35], and with improved NK cell responses to cytokine stimulation in HIV+ patients [48]. However, we found that TIGIT blockade does not improve NK cell responses to HIV-infected CD4+ T cells in healthy donors. The lack of a benefit from TIGIT blockade may be explained by the fact that TIGIT is unlikely to be engaged by its ligands during NK cell recognition of HIV-infected cells. We found that CD155 is not upregulated on CD4+ T cells actively infected in vitro, consistent with previous reports [67–69]. In our study, CD155 is also not upregulated on CD4+ T cells from HIV-infected women. This differs from the findings of Yin et al. [48], who reported elevated CD155 expression on CD4+ T cells from HIV-infected men, though with extremely low expression levels. Notably, Yin et al. did not observe improvement in NK cell responses by blocking CD155, suggesting that its engagement was not contributing to NK cell suppression. While we showed that CD112 is upregulated on CD4+ T cells infected in vitro, only a median 1.6% of cells express this receptor and upregulation is not observed in vivo, calling into question the biological significance of this modest upregulation. Together, the data suggest that the TIGIT/CD155/CD112 pathway may not be directly involved in the regulation of NK cell responses to HIV-infected cells.
While TIGIT itself may not play a key role in immune response to HIV, we noted that TIGIT marks a distinct NK subpopulation, co-expressing several activation markers (DNAM-1, NTB-A, 2B4, CD2). Co-expression of DNAM-1 is of particular interest because it shares the ligands CD112 and CD155 with TIGIT [70] and is associated with NK cell memory to murine CMV [71]. TIGIT outcompetes DNAM-1 for binding to CD155 [72]. However, this interaction is unlikely to be relevant in the context of HIV infection, as we show that HIV does not upregulate CD155 in CD4+ T cells. In our cohort, TIGIT+ NK cells also exhibit a more adaptive/mature phenotype (CD57hi, NKG2Chi, LILRB1hi, FcRγlo, Syklo) and adaptive NK cells demonstrated higher TIGIT expression in unsupervised clustering via UMAP. This is consistent with the notion that NK cells undergo adaptive reconfiguration during HIV infection [65]. Notably, Sarhan et al. demonstrated that adaptive NK cells from healthy donors express TIGIT at lower levels [73]. Our contrasting finding could reflect unique differentiation pathways driven by HIV infection or a differential downregulation of TIGIT in response to cytokine stimulation, given that Sarhan et al. evaluated cytokine-treated NK cells rather than PBMCs directly ex vivo. Importantly, NKG2C+CD57+ adaptive NK cells are typically expanded in response to human CMV infection, making it difficult to distinguish CMV-related from HIV-related NK phenotypic changes. Unfortunately, CMV status is not available from our study, but the CMV seroprevalence in Benin is extremely high [74]. Thus, although we cannot exclude an effect of CMV in our cohort, we can reasonably assume that most of our women are CMV+ and that TIGIT upregulation in untreated HIV+ women is thus due to the direct effect of HIV infection rather than CMV, which both HIV-infected and uninfected women are likely to have.
Adaptive NK cells show increased responses to FcγRIIIa triggering [75–77], are expanded in HIV-infected patients and robustly respond to HIV peptides [14]. Here we demonstrate that TIGIT+ NK cells have higher functional activity against mock-infected and HIV-infected CD4+ T cells, compared to TIGIT− NK cells. Further, higher functional responses also occur in the presence of non-HIV stimuli. This increased activity could be explained by the co-expression of activating receptors, which may overwhelm the inhibitory signal provided by TIGIT, or by changes in the activation threshold during adaptive reconfiguration. Alternatively, a recent report showed that TIGIT could contribute to NK cell licensing [78], suggesting that the presence of TIGIT may be required during NK cell maturation to ensure the development of optimal effector function.
Some limitations should be noted in our study. First, the limited sample availability only allowed us to profile NK cells from a small sample size of female subjects. To overcome this, functional experiments were done on both female and male subjects. Additionally, we did not have sufficient samples from viremic HIV+ patients, thus functional experiments were carried on healthy, CMV-negative, HIV-negative subjects. Healthy individuals have lower TIGIT levels, potentially hindering the effect of TIGIT blockade. However, NK cells were treated with IL-2, which increases TIGIT expression (Figure S6), thus providing sufficient substrate for the blocking assays.
In summary, our work demonstrates that TIGIT is overexpressed on NK cells of untreated HIV+ women, but fully corrected by ART. We find that TIGIT does not directly participate in the NK cell response to HIV-infected cells, but rather marks a mature NK cell subpopulation with adaptive features that is enhanced in functional responses to virus-infected cells, tumors, and cytokine stimulation. These results need to be confirmed in independent cohorts that evaluate both the direct functional contributions of TIGIT and the role of ART in regulating TIGIT expression.
Supplementary Material
Figure S1. Gating Schemes. (A) Gating scheme used for negative gating of NK cells by CyTOF. NK cells were already purified with magnetic bead isolation prior to staining. Negative gating was performed to ensure further NK cell purity for downstream analyses. (B) Gating scheme used for analysis of CD4+ T cells by CyTOF. (C) Gating scheme used for analysis of NK cells and CD4+ T cells by flow cytometry.
Figure S2. Gating strategy. (A) Representative CyTOF plots of the expression of NK cell markers from one of the treated HIV+ individuals. (B) Representative CyTOF plot of CD4+ T cell expression of CD112 and CD155 from one healthy woman, one untreated HIV+ woman and one treated HIV+ woman. (C) Representative CyTOF plots of CD112 and CD155 expression on mock-infected or HIV-infected CD4+ T cells. (D) Representative flow cytometry plot of p24 expression in mock-infected and HIV-infected CD4+ T cells. The median infection level for the blocking assay was 31.4% (range: 14.5-55.5%). The median infection levels for the CyTOF experiments was 35.9% (range: 16.5-74.7%). The median infection level for the TIGIT+ vs TIGIT− comparison was 38.0% (range: 14.6%-70.5%)..
Figure S3. TIGIT expression level does not correlate with CD4+ T cell counts. (A) Correlation between TIGIT Mean Signal Intensity and CD4+ T cell count in all HIV+ women. (B) Correlation between TIGIT Mean Signal Intensity and CD4+ T cell count in untreated HIV+ women. (C) Correlation between TIGIT Mean Signal Intensity and CD4+ T cell count in treated HIV+ women. All correlations were calculated using Spearman’s Correlation.
Figure S4. TIGIT blockade is effective in preventing further staining of TIGIT on NK cells. (A) Representative flow plots of TIGIT expression without antibody treatment (control), after treatment with mIgG1k isotype control (isotype), or after treatment with anti-TIGIT blocking antibody (TIGIT block). (B) Summary data of TIGIT expression by flow cytometry without antibody treatment (control), after treatment with mIgG1k isotype control (isotype), or after treatment with anti-TIGIT blocking antibody (TIGIT block). * = p-value<0.05, ns = non-significant.
Figure S5. TIGIT ligands expression in HIV-infected CD4+ T cells. Summary data of mean signal intensity of CD112 and CD155 on mock-infected CD4+ T cells (mock) and HIV-infected p24+CD4+ T cells (HIV_p24+). ** = p-value<0.01, ns = non-significant.
Figure S6. IL-2 upregulates TIGIT expression on purified NK cells. Summary data of frequency of TIGIT+ on resting NK cells and IL-2 stimulated NK cells from healthy donors. * = p-value<0.05.
ACKNOWLEDGEMENTS
We are grateful to the Beninese study participants. We are indebted to N. Geraldo, A. Gabin, C. Assogba and C. Agossa-Gbenafa for their clinical expertise, to M. Massinga-Loembe, G. Ahotin, L.Djossou, and E. Goma for their technical assistance and to G. Batona and other field workers who helped with recruitment of commercial sex workers.
Disclosures/Grant Support: This work was supported by: NIH Ruth L. Kirschstein Institutional National Research Service Award T32AI007502 and TL1TR001084 (EV), NIH/NIAID K08 K08AI138640 (EV), ITI/Bill & Melinda Gates Foundation Pilot Grant (CAB), NIH/NIAID DP2 AI1219301 (CAB), NIH/NIDA Avant Garde Award for HIV Research DP1 DA045089 (CAB), Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases (CAB), Grant # PJT-148529 from the Canadian Institutes of Health Research and by the Réseau SIDA from the Fonds de Recherche du Québec en Santé (MR). CAB is an investigator of the Chan Zuckerberg Biohub. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
REFERENCES:
- 1.Lam VC, Lanier LL. NK cells in host responses to viral infections. Current Opinion in Immunology. 2017; 44:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanier LL, Phillips JH, Hackett J Jr, Tutt M, Kumar V. Natural killer cells: definition of a cell type rather than a function. J Immunol 1986; 137:2735–2739. [PubMed] [Google Scholar]
- 3.Paust S, Gill HS, Wang B-Z, Flynn MP, Moseman EA, Senman B, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol 2010; 11:1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paust S, Blish CA, Reeves RK. Redefining Memory: Building the Case for Adaptive NK Cells. J Virol 2017; 91. doi: 10.1128/JVI.00169-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paust S, von Andrian UH. Natural killer cell memory. Nat Immunol 2011; 12:500–508. [DOI] [PubMed] [Google Scholar]
- 6.Cerwenka A, Lanier LL. Natural killer cell memory in infection, inflammation and cancer. Nat Rev Immunol 2016; 16:112–123. [DOI] [PubMed] [Google Scholar]
- 7.O’Sullivan TE, Sun JC, Lanier LL. Natural Killer Cell Memory. Immunity. 2015; 43:634–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikzad R, Angelo LS, Aviles-Padilla K, Le DT, Singh VK, Bimler L, et al. Human natural killer cells mediate adaptive immunity to viral antigens. Sci Immunol 2019; 4. doi: 10.1126/sciimmunol.aat8116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaulieu AM. Memory responses by natural killer cells. Journal of Leukocyte Biology. 2018; 104:1087–1096. [DOI] [PubMed] [Google Scholar]
- 10.Hu P-F, Hultin LE, Hultin P, Hausner MA, Hirji K, Jewett A, et al. Natural Killer Cell Immunodeficiency in HIV Disease is Manifest by Profoundly Decreased Numbers of CD16 CD56 Cells and Expansion of a Population of CD16dim CD56- Cells with Low Lytic Activity. Journal of Acquired Immune Deficiency Syndromes & Human Retrovirology. 1995; 10:331–340. [PubMed] [Google Scholar]
- 11.Mavilio D, Lombardo G, Benjamin J, Kim D, Follman D, Marcenaro E, et al. Characterization of CD56-/CD16+ natural killer (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci U S A 2005; 102:2886–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vieillard V, Fausther-Bovendo H, Samri A, Debré P, French Asymptomatiques à Long Terme (ALT) ANRS-CO15 Study Group. Specific phenotypic and functional features of natural killer cells from HIV-infected long-term nonprogressors and HIV controllers. J Acquir Immune Defic Syndr 2010; 53:564–573. [DOI] [PubMed] [Google Scholar]
- 13.Mavilio D, Benjamin J, Daucher M, Lombardo G, Kottilil S, Planta MA, et al. Natural killer cells in HIV-1 infection: Dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proceedings of the National Academy of Sciences. 2003; 100:15011–15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J, Amran FS, Kramski M, Angelovich TA, Elliott J, Hearps AC, et al. An NK Cell Population Lacking FcRγ Is Expanded in Chronically Infected HIV Patients. J Immunol 2015; 194:4688–4697. [DOI] [PubMed] [Google Scholar]
- 15.Björkström NK, Ljunggren H-G, Sandberg JK. CD56 negative NK cells: origin, function, and role in chronic viral disease. Trends Immunol 2010; 31:401–406. [DOI] [PubMed] [Google Scholar]
- 16.Scott-Algara D, Paul P. NK cells and HIV infection: lessons from other viruses. Curr Mol Med 2002; 2:757–768. [DOI] [PubMed] [Google Scholar]
- 17.Ahmad A, Morisset R, Thomas R, Menezes J. Evidence for a defect of antibody-dependent cellular cytotoxic (ADCC) effector function and anti-HIV gp120/41-specific ADCC-mediating antibody titres in HIV-infected individuals. J Acquir Immune Defic Syndr 1994; 7:428–437. [PubMed] [Google Scholar]
- 18.Fehniger TA, Herbein G, Yu H, Para MI, Bernstein ZP, O’Brien WA, et al. Natural killer cells from HIV-1+ patients produce CC chemokines and inhibit HIV-1 infection. The Journal of Immunology 1998; 161:6433–6438. [PubMed] [Google Scholar]
- 19.Borrow P, Bhardwaj N. Innate immune responses in primary HIV-1 infection. Curr Opin HIV AIDS 2008; 3:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scully E, Alter G. NK Cells in HIV Disease. Curr HIV/AIDS Rep 2016; 13:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott-Algara D, Truong LX, Versmisse P, David A, Luong TT, Nguyen NV, et al. Cutting edge: increased NK cell activity in HIV-1-exposed but uninfected Vietnamese intravascular drug users. J Immunol 2003; 171:5663–5667. [DOI] [PubMed] [Google Scholar]
- 22.Montoya CJ, Velilla PA, Chougnet C, Landay AL, Rugeles MT. Increased IFN-γ production by NK and CD3+/CD56+ cells in sexually HIV-1-exposed but uninfected individuals. Clin Immunol 2006; 120:138–146. [DOI] [PubMed] [Google Scholar]
- 23.Ravet S, Scott-Algara D, Bonnet E, Tran HK, Tran T, Nguyen N, et al. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood 2007; 109:4296–4305. [DOI] [PubMed] [Google Scholar]
- 24.Jennes W, Verheyden S, Mertens JW, Camara M, Seydi M, Dieye TN, et al. Inhibitory KIR/HLA incompatibility between sexual partners confers protection against HIV-1 transmission. Blood 2013; 121:1157–1164. [DOI] [PubMed] [Google Scholar]
- 25.Jennes W, Verheyden S, Demanet C, Adjé-Touré CA, Vuylsteke B, Nkengasong JN, et al. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J Immunol 2006; 177:6588–6592. [DOI] [PubMed] [Google Scholar]
- 26.Merino A, Malhotra R, Morton M, Mulenga J, Allen S, Hunter E, et al. Impact of a functional KIR2DS4 allele on heterosexual HIV-1 transmission among discordant Zambian couples. J Infect Dis 2011; 203:487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Y, Chen O, Cui C, Zhao B, Han X, Zhang Z, et al. KIR3DS1/L1 and HLA-Bw4-80I are associated with HIV disease progression among HIV typical progressors and long-term nonprogressors. BMC Infect Dis 2013; 13:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 2007; 39:733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Connor GM, Holmes A, Mulcahy F, Gardiner CM. Natural Killer cells from long-term non-progressor HIV patients are characterized by altered phenotype and function. Clin Immunol 2007; 124:277–283. [DOI] [PubMed] [Google Scholar]
- 30.Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol 2009; 10:48–57. [DOI] [PubMed] [Google Scholar]
- 31.Stanietsky N, Simic H, Arapovic J, Toporik A, Levy O, Novik A, et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci U S A 2009; 106:17858–17863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boles KS, Vermi W, Facchetti F, Fuchs A, Wilson TJ, Diacovo TG, et al. A novel molecular interaction for the adhesion of follicular CD4 T cells to follicular DC. Eur J Immunol 2009; 39:695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Xia P, Du Y, Liu S, Huang G, Chen J, et al. T-cell Immunoglobulin and ITIM Domain (TIGIT) Receptor/Poliovirus Receptor (PVR) Ligand Engagement Suppresses Interferon-γ Production of Natural Killer Cells via β-Arrestin 2-mediated Negative Signaling. Journal of Biological Chemistry. 2014; 289:17647–17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doherty M Faculty of 1000 evaluation for TIGIT Marks Exhausted T Cells, Correlates with Disease Progression, and Serves as a Target for Immune Restoration in HIV and SIV Infection. F1000 - Post-publication peer review of the biomedical literature. 2016. doi: 10.3410/f.726060985.793518814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q, Bi J, Zheng X, Chen Y, Wang H, Wu W, et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat Immunol 2018; 19:723–732. [DOI] [PubMed] [Google Scholar]
- 36.Josefsson SE, Beiske K, Blaker YN, Førsund MS, Holte H, Østenstad B, et al. TIGIT and PD-1 Mark Intratumoral T Cells with Reduced Effector Function in B-cell Non-Hodgkin Lymphoma. Cancer Immunol Res 2019; 7:355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tauriainen J, Scharf L, Frederiksen J, Naji A, Ljunggren H-G, Sönnerborg A, et al. Perturbed CD8+ T cell TIGIT/CD226/PVR axis despite early initiation of antiretroviral treatment in HIV infected individuals. Sci Rep 2017; 7:40354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong Y, Zhu L, Schell TD, Zhang J, Claxton DF, Ehmann WC, et al. T-Cell Immunoglobulin and ITIM Domain (TIGIT) Associates with CD8+ T-Cell Exhaustion and Poor Clinical Outcome in AML Patients. Clin Cancer Res 2016; 22:3057–3066. [DOI] [PubMed] [Google Scholar]
- 39.Johnston RJ, Yu X, Grogan JL. The checkpoint inhibitor TIGIT limits antitumor and antiviral CD8+ T cell responses. Oncoimmunology 2015; 4:e1036214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F, Hou H, Wu S, Tang Q, Liu W, Huang M, et al. TIGIT expression levels on human NK cells correlate with functional heterogeneity among healthy individuals. Eur J Immunol 2015; 45:2886–2897. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez-Correa B, Lopez-Sejas N, Duran E, Labella F, Alonso C, Solana R, et al. Modulation of NK cells with checkpoint inhibitors in the context of cancer immunotherapy. Cancer Immunol Immunother 2019; 68:861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu F, Sunderland A, Zhou Y, Schulick RD, Edil BH, Zhu Y. Blockade of CD112R and TIGIT signaling sensitizes human natural killer cell functions. Cancer Immunol Immunother 2017; 66:1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chew GM, Fujita T, Webb GM, Burwitz BJ, Wu HL, Reed JS, et al. TIGIT Marks Exhausted T Cells, Correlates with Disease Progression, and Serves as a Target for Immune Restoration in HIV and SIV Infection. PLoS Pathog 2016; 12:e1005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noyan K, Nguyen S, Betts MR, Sönnerborg A, Buggert M. Human Immunodeficiency Virus Type-1 Elite Controllers Maintain Low Co-Expression of Inhibitory Receptors on CD4+ T Cells. Front Immunol 2018; 9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pardons M, Baxter AE, Massanella M, Pagliuzza A, Fromentin R, Dufour C, et al. Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PLoS Pathog 2019; 15:e1007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Llewellyn GN, Seclén E, Wietgrefe S, Liu S, Chateau M, Pei H, et al. Humanized Mouse Model of HIV-1 Latency with Enrichment of Latent Virus in PD-1 and TIGIT CD4 T Cells. Journal of Virology. 2019; 93. doi: 10.1128/jvi.02086-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fromentin R, Bakeman W, Lawani MB, Khoury G, Hartogensis W, DaFonseca S, et al. CD4+ T Cells Expressing PD-1, TIGIT and LAG-3 Contribute to HIV Persistence during ART. PLoS Pathog 2016; 12:e1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yin X, Liu T, Wang Z, Ma M, Lei J, Zhang Z, et al. Expression of the Inhibitory Receptor TIGIT Is Up-Regulated Specifically on NK Cells With CD226 Activating Receptor From HIV-Infected Individuals. Frontiers in Immunology. 2018; 9. doi: 10.3389/fimmu.2018.02341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sabourin-Poirier C, Fourcade L, Chagnon-Choquet J, Labbé A-C, Alary M, Guédou F, et al. Blood B Lymphocyte Stimulator (BLyS)/BAFF levels may reflect natural immunity to HIV in highly exposed uninfected Beninese Commercial Sex Workers. Sci Rep 2016; 6:32318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thibodeau V, Lajoie J, Labbé A-C, Zannou MD, Fowke KR, Alary M, et al. High level of soluble HLA-G in the female genital tract of Beninese commercial sex workers is associated with HIV-1 infection. PLoS One 2011; 6:e25185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauss-Albee DM, Fukuyama J, Liang EC, Yao Y, Jarrell JA, Drake AL, et al. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci Transl Med 2015; 7:297ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vendrame E, Fukuyama J, Strauss-Albee DM, Holmes S, Blish CA. Mass cytometry analytical approaches reveal cytokine-induced changes in natural killer cells. Cytometry B Clin Cytom 2017; 92:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 54.Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, et al. Normalization of mass cytometry data with bead standards. Cytometry A 2013; 83:483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: 2012 URL http://wwwR-projectorg 2018. [Google Scholar]
- 56.Seiler C, Kronstad LM, Simpson LJ, Le Gars M, Vendrame E, Blish CA, et al. Uncertainty Quantification in Multivariate Mixed Models for Mass Cytometry Data. arXiv [stat.AP]. 2019http://arxiv.org/abs/1903.07976 [Google Scholar]
- 57.Kronstad LM, Seiler C, Vergara R, Holmes SP, Blish CA. Differential Induction of IFN-α and Modulation of CD112 and CD54 Expression Govern the Magnitude of NK Cell IFN-γ Response to Influenza A Viruses. J Immunol 2018; 201:2117–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv [stat.ML]. 2018http://arxiv.org/abs/1802.03426 [Google Scholar]
- 59.Lichtfuss GF, Cheng W-J, Farsakoglu Y, Paukovics G, Rajasuriar R, Velayudham P, et al. Virologically suppressed HIV patients show activation of NK cells and persistent innate immune activation. J Immunol 2012; 189:1491–1499. [DOI] [PubMed] [Google Scholar]
- 60.Brunetta E, Fogli M, Varchetta S, Bozzo L, Hudspeth KL, Marcenaro E, et al. The decreased expression of Siglec-7 represents an early marker of dysfunctional natural killer-cell subsets associated with high levels of HIV-1 viremia. Blood 2009; 114:3822–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagler A, Lanier LL, Cwirla S, Phillips JH. Comparative studies of human FcRIII-positive and negative natural killer cells. J Immunol 1989; 143:3183–3191. [PubMed] [Google Scholar]
- 62.Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, et al. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood 2001; 97:3146–3151. [DOI] [PubMed] [Google Scholar]
- 63.Gonzalez VD, Falconer K, Björkström NK, Blom KG, Weiland O, Ljunggren H-G, et al. Expansion of Functionally Skewed CD56-Negative NK Cells in Chronic Hepatitis C Virus Infection: Correlation with Outcome of Pegylated IFN-α and Ribavirin Treatment. The Journal of Immunology 2009; 183:6612–6618. [DOI] [PubMed] [Google Scholar]
- 64.Mikulak J, Oriolo F, Zaghi E, Di Vito C, Mavilio D. Natural killer cells in HIV-1 infection and therapy. AIDS 2017; 31:2317–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peppa D, Pedroza-Pacheco I, Pellegrino P, Williams I, Maini MK, Borrow P. Adaptive Reconfiguration of Natural Killer Cells in HIV-1 Infection. Front Immunol 2018; 9:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alter G, Teigen N, Davis BT, Addo MM, Suscovich TJ, Waring MT, et al. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood 2005; 106:3366–3369. [DOI] [PubMed] [Google Scholar]
- 67.Davis ZB, Sowrirajan B, Cogswell A, Ward JP, Planelles V, Barker E. CD155 on HIV-Infected Cells Is Not Modulated by HIV-1 Vpu and Nef but Synergizes with NKG2D Ligands to Trigger NK Cell Lysis of Autologous Primary HIV-Infected Cells. AIDS Research and Human Retroviruses. 2017; 33:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matusali G, Potestà M, Santoni A, Cerboni C, Doria M. The human immunodeficiency virus type 1 Nef and Vpu proteins downregulate the natural killer cell-activating ligand PVR. J Virol 2012; 86:4496–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tremblay-McLean A, Bruneau J, Lebouché B, Lisovsky I, Song R, Bernard NF. Expression Profiles of Ligands for Activating Natural Killer Cell Receptors on HIV Infected and Uninfected CD4+ T Cells. Viruses 2017; 9:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tahara-Hanaoka S Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112). International Immunology. 2004; 16:533–538. [DOI] [PubMed] [Google Scholar]
- 71.Nabekura T, Kanaya M, Shibuya A, Fu G, Gascoigne NRJ, Lanier LL. Costimulatory molecule DNAM-1 is essential for optimal differentiation of memory natural killer cells during mouse cytomegalovirus infection. Immunity 2014; 40:225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu X, Harden K. C Gonzalez L, Francesco M, Chiang E, Irving B, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol 2009; 10:48–57. [DOI] [PubMed] [Google Scholar]
- 73.Sarhan D, Cichocki F, Zhang B, Yingst A, Spellman SR, Cooley S, et al. Adaptive NK Cells with Low TIGIT Expression Are Inherently Resistant to Myeloid-Derived Suppressor Cells. Cancer Research. 2016; 76:5696–5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Paschale M, Ceriani C, Cerulli T, Cagnin D, Cavallari S, Cianflone A, et al. Antenatal screening for Toxoplasma gondii, Cytomegalovirus, rubella and Treponema pallidum infections in northern Benin. Trop Med Int Health 2014; 19:743–746. [DOI] [PubMed] [Google Scholar]
- 75.Rölle A, Brodin P. Immune Adaptation to Environmental Influence: The Case of NK Cells and HCMV. Trends Immunol 2016; 37:233–243. [DOI] [PubMed] [Google Scholar]
- 76.Foley B, Cooley S, Verneris MR, Curtsinger J, Luo X, Waller EK, et al. Human Cytomegalovirus (CMV)-Induced Memory-like NKG2C+ NK Cells Are Transplantable and Expand In Vivo in Response to Recipient CMV Antigen. The Journal of Immunology 2012; 189:5082–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wagner JA, Fehniger TA. Human Adaptive Natural Killer Cells: Beyond NKG2C. Trends Immunol. 2016; 37:351–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He Y, Peng H, Sun R, Wei H, Ljunggren H-G, Yokoyama WM, et al. Contribution of inhibitory receptor TIGIT to NK cell education. J Autoimmun 2017; 81:1–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Gating Schemes. (A) Gating scheme used for negative gating of NK cells by CyTOF. NK cells were already purified with magnetic bead isolation prior to staining. Negative gating was performed to ensure further NK cell purity for downstream analyses. (B) Gating scheme used for analysis of CD4+ T cells by CyTOF. (C) Gating scheme used for analysis of NK cells and CD4+ T cells by flow cytometry.
Figure S2. Gating strategy. (A) Representative CyTOF plots of the expression of NK cell markers from one of the treated HIV+ individuals. (B) Representative CyTOF plot of CD4+ T cell expression of CD112 and CD155 from one healthy woman, one untreated HIV+ woman and one treated HIV+ woman. (C) Representative CyTOF plots of CD112 and CD155 expression on mock-infected or HIV-infected CD4+ T cells. (D) Representative flow cytometry plot of p24 expression in mock-infected and HIV-infected CD4+ T cells. The median infection level for the blocking assay was 31.4% (range: 14.5-55.5%). The median infection levels for the CyTOF experiments was 35.9% (range: 16.5-74.7%). The median infection level for the TIGIT+ vs TIGIT− comparison was 38.0% (range: 14.6%-70.5%)..
Figure S3. TIGIT expression level does not correlate with CD4+ T cell counts. (A) Correlation between TIGIT Mean Signal Intensity and CD4+ T cell count in all HIV+ women. (B) Correlation between TIGIT Mean Signal Intensity and CD4+ T cell count in untreated HIV+ women. (C) Correlation between TIGIT Mean Signal Intensity and CD4+ T cell count in treated HIV+ women. All correlations were calculated using Spearman’s Correlation.
Figure S4. TIGIT blockade is effective in preventing further staining of TIGIT on NK cells. (A) Representative flow plots of TIGIT expression without antibody treatment (control), after treatment with mIgG1k isotype control (isotype), or after treatment with anti-TIGIT blocking antibody (TIGIT block). (B) Summary data of TIGIT expression by flow cytometry without antibody treatment (control), after treatment with mIgG1k isotype control (isotype), or after treatment with anti-TIGIT blocking antibody (TIGIT block). * = p-value<0.05, ns = non-significant.
Figure S5. TIGIT ligands expression in HIV-infected CD4+ T cells. Summary data of mean signal intensity of CD112 and CD155 on mock-infected CD4+ T cells (mock) and HIV-infected p24+CD4+ T cells (HIV_p24+). ** = p-value<0.01, ns = non-significant.
Figure S6. IL-2 upregulates TIGIT expression on purified NK cells. Summary data of frequency of TIGIT+ on resting NK cells and IL-2 stimulated NK cells from healthy donors. * = p-value<0.05.