Abstract
It is important to evaluate the clinical importance of both CD8 T cells and CD4 T cells expression simultaneously because they have crucial networks in tumour targeting immune responses. In 97 RCC patients, RNA sequencing and gene set enrichment analysis of both CD8 and CD4 T cells based on the expression levels of PD-1 and TIM-3 implied that the populations of PD-1+TIM-3+ CD8 T cells and PD-1lowTIM-3 + CD4 T cells were characterized as exhausted CD8 T cells and regulatory CD4 T cells, respectively. These populations of CD4 and CD8 T cells were significantly upregulated in the patients with RCC of higher WHO/ISUP grade (grades 3, 4) (P < 0.001). Moreover, the cytokine productivities of each population in both CD4 and CD8 T cells of the higher-grade patients were significantly lower than those of the lower-grade patients (P < 0.05). Multivariate analysis showed the prognosis of patients with metastatic RCC of higher WHO/ISUP grade treated by nivolumab to be significantly worse than that of patients with lower grade (P = 0.026). This study showed that tumour grade significantly correlated with dysfunction of both CD4+ and CD8+ TILs and the efficacy of nivolumab treatment.
Subject terms: Cancer microenvironment, Renal cell carcinoma
Introduction
Recently, cancer immunotherapy including monotherapy1 or combination therapy with anti-PD-1 antibody2 showed better therapeutic effect than molecular-targeted therapies for metastatic renal cell carcinoma (RCC) patients. However, the therapeutic effect of anti-PD-1 antibody monotherapy against RCC is limited, and both the establishment of biomarkers to predict efficacy and the development of new therapeutic target factors are necessary using tissue infiltrating lymphocytes (TILs).
Recently, subpopulations of CD4 and CD8 T cells stratified by PD-1 expression in the tumour microenvironment were reported to be important prognostic factors in cancer immunotherapy3–5. The transcriptomic data of TILs were published based not only on a single cell6,7 but also on the intensity of PD-1 expression3, and high expression of PD-1 was identified as a predictive marker in non-small cell lung cancer patients. Analysis of TILs according to the expression pattern of multiple immunocheckpoint (IC) molecules revealed the presence of terminally differentiated and stem like CD8 T cells in RCC patients8. However, few studies have simultaneously evaluated the functions of both CD8 T cells and CD4 T cells, which have crucial networks in establishing immune status in tumour microenvironments9 and tumour targeting immune responses10–12.
In this study, we performed RNA sequencing of both CD8 T cells and CD4 T cells based on the expression patterns of PD-1 and TIM-3 and aimed to clarify those characteristics and the differences stratified by tumour grade in RCC patients. In addition, we also evaluated the relationship between tumour grade and the efficacy of nivolumab monotherapy.
Results
Patient characteristics
Clinical characteristics of the 97 patients are listed in Supplementary Table 1. The histology of 86 of the patients (88.7%) was clear cell RCC. Twenty-nine patients (29.9%) had a higher World Health Organisation (WHO)/International Society of Urological Pathology (ISUP) grade of RCC (Grade 3 or 4).
Characteristics of the 20 patients treated with anti PD-1 antibody monotherapy are shown in Supplementary Table 2. Eleven patients (55.0%) had a higher WHO/ISUP grade, and seven patients (35.0%) were in the poor Memorial Sloan-Kettering Cancer Center (MSKCC) risk group. Most of the patients (70.0%) had received one or two prior therapies before nivolumab treatment, and none had been treated previously with IC inhibitors.
Difference in expression patterns of PD-1 and TIM-3 by sample types among PBMCs, NILs, and TILs
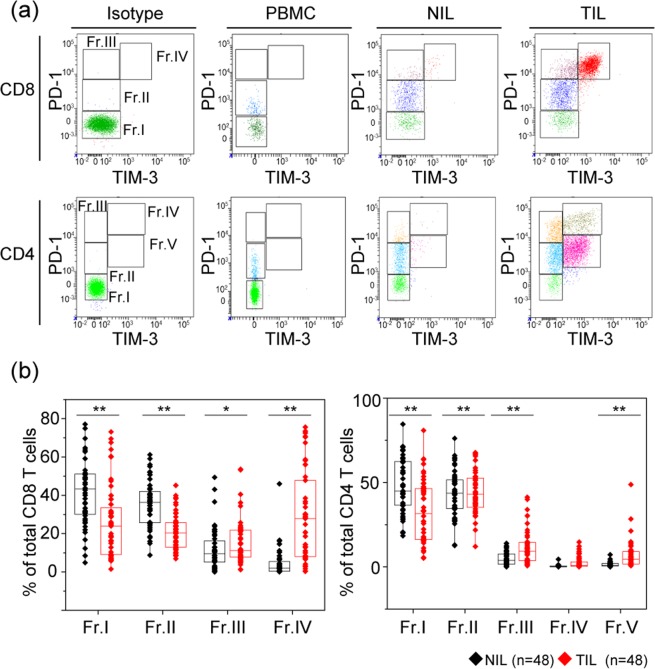
The representative expression patterns of PD-1 and TIM-3 on both CD4 T cells and CD8 T cells of peripheral blood mononuclear cells (PBMCs), adjacent normal tissue-infiltrating lymphocytes (NILs), and TILs are shown in Fig. 1a. In CD8 T cells, the intensity of PD-1 expression was highest in TILs, followed by NILs and PBMCs, and the expression of TIM-3 was found only with high expression of PD-1. Unlike other carcinomas, there are few cells with high TIM-3 and low PD-1 expression in RCC. In CD4 T cells, the intensity of PD-1 expression was highest in TILs, followed by NILs and PBMCs, and the populations with high TIM-3 expression were classified into two groups based on the intensity of PD-1 expression.
Figure 1.
Co-expression pattern of PD-1 and TIM-3 on both CD8 and CD4 T cells of peripheral blood mononuclear cells (PBMC), adjacent normal tissue-infiltrating lymphocytes (NILs), and tumour tissue-infiltrating lymphocytes (TILs) in 48 patients with renal cell carcinoma. (a) Gating strategy to identify subsets of CD8 and CD4 T cells within PBMC, NILs, and TILs. CD8 T cells were classified into four groups (Fr. I: PD-1-TIM-3−, Fr. II: PD-1lowTIM-3−, Fr. III: PD-1+TIM-3−, Fr. IV: PD-1+TIM-3+), and CD4 T cells were classified into 5 groups (Fr. I: PD-1-TIM-3−, Fr. II: PD-1lowTIM-3−, Fr. III: PD-1+TIM-3−, Fr. IV: PD-1+TIM-3+, Fr. V: PD-1lowTIM-3+). (b) Expression ratios of each fraction on both CD4 and CD8 T cells were compared among NILs and TILs simultaneously extracted from the same patients. Comparison of two sample types was performed by Mann-Whitney U test. The central tendency of the box plot shows the median of each group, and the upper and lower ranges of the box plot show the 25th and 75th percentiles of each data set, respectively. **P < 0.01, *P < 0.05.
From the above results, CD8 T cells were fractionated into four groups (Fr. I: PD-1-TIM-3−, Fr. II: PD-1lowTIM-3−, Fr. III: PD-1highTIM-3−, and Fr. IV: PD-1highTIM-3+), and CD4 T cells were fractionated into 5 groups (Fr. I: PD-1-TIM-3−, Fr. II: PD-1lowTIM-3−, Fr. III: PD-1highTIM-3−, Fr. IV: PD-1highTIM-3+, and Fr. V: PD-1lowTIM-3+).
In 48 patients whose TILs and NILs were sampled simultaneously, the frequencies of NILs and TILs were significantly different in all fractions of CD8 T cells and all fractions but Fr. IV of CD4 T cells (Fig. 1b). Therefore, these 8 fractions showing a significant difference between NILs and TILs were examined in the subsequent analysis. In some patients, CD8+ CD103 + T cells, defined as tissue-resident memory T (Trem) cells13, were expressed in NILs and TILs (Supplementary Fig. 1).
Significant difference in characteristics within CD8 TILs based on expression patterns of PD-1 and TIM-3
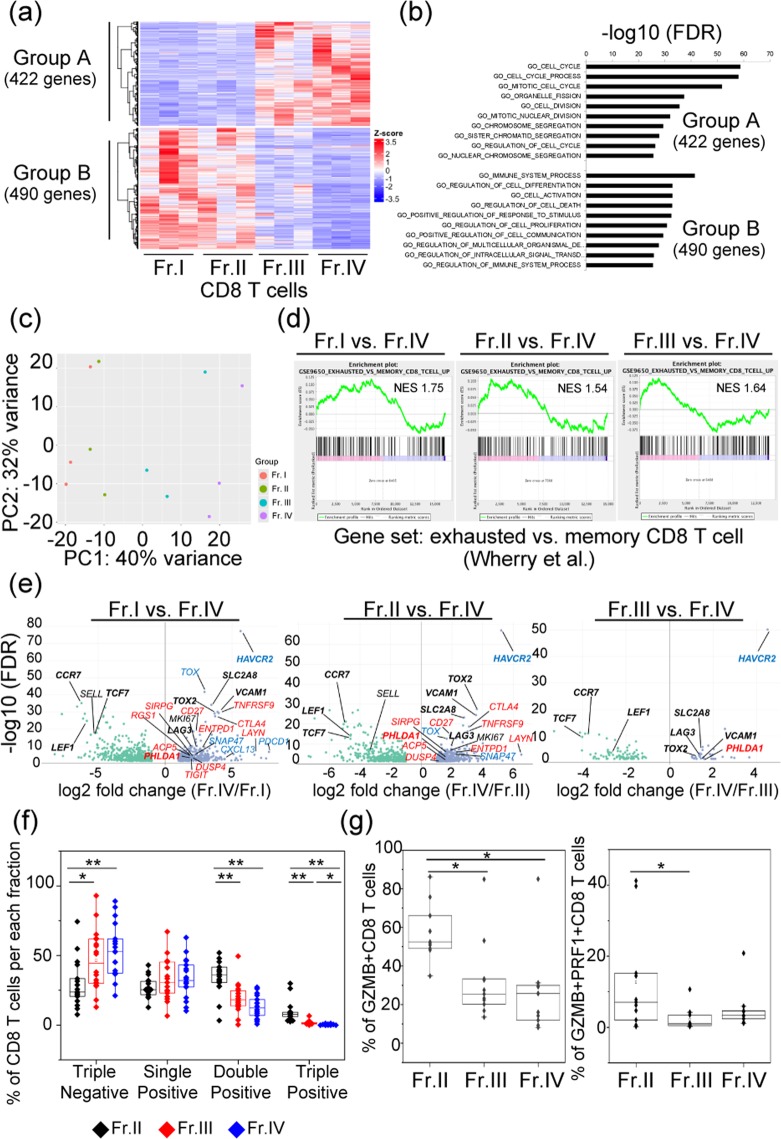
Transcriptomic analysis of each fraction performed by RNA sequencing using 12 fractions and hierarchical clustering analysis using 912 genes mainly revealed two clusters composed of two groups (group A: Fr. I/Fr. II and group B: Fr. III/Fr. IV) (Fig. 2a). Pathway enrichment analysis showed that 422 genes of group A were involved in cell cycle regulation and proliferation and were already reported to correlate with high expression of PD-1 in TILs in lung cancer3, and 490 genes of group B were involved in immune system processes (Fig. 2b). Principal component analysis (PCA) of these genes revealed that all samples of Fr. I and Fr. II were located nearby each other, and all samples of Fr. III and Fr. IV were located closer to, rather than farther from, each other (Fig. 2c). Gene set enrichment analysis (GSEA) of published data sets from CD8 T cell exhaustion during chronic viral infection14 was performed for each fraction, and the exhaustion-related genes were significantly enriched in Fr. IV compared with those in the other fractions (Fig. 2d).
Figure 2.
Gene expression profile and functional analysis of CD8 tumour tissue-infiltrating lymphocytes (TILs) based on the expression patterns of PD-1 and TIM-3. (a) Clustering analysis for genes distinguishing four subsets of CD8 TILs extracted from three patients with renal cell carcinoma. In total, 912 genes were significantly upregulated or downregulated (log2 fold change > [1], false discovery rate [FDR] < 0.05) as indicated by at least one group comparison. (b) Biological processes (Gene Ontology [GO] terms) enriched in group A and group B. (c) Principal component (PC) analysis of four subsets of CD8 T cells based on the expression patterns of PD-1 and TIM-3. (d) Gene set enrichment analysis (GSEA) of published data sets of biological processes (GO terms) enriched in group A and group B14. Statistical significance was determined by permutation testing with normalized enrichment score (NES). (e) Volcano plot of up- or downregulated genes between Fr. I vs. Fr. IV, Fr. II vs. Fr. IV, and Fr. III vs. Fr. IV. Genes specific only to exhausted T cells (blue) and to both exhausted CD8 T cells and CD4 regulatory T cells (Tregs) (red)7 are shown, and bold characters indicate the genes commonly having significant differences between these three combinations. (f) Comparison of multiple cytokine productivity (IFNγ, TNFα, and IL-2) of CD8 TILs from 18 patients in Fr. II, Fr. III, and Fr. IV among three sample types. (g) Cytotoxic functionality producing GZMB and both GZMB and PRF1 were compared within three fractions by Kruskal-Wallis analysis, and then a comparison of each sample type was performed by Bonferroni-corrected Mann-Whitney U test. The central tendency of the box plot indicates the median of each group, and the upper and lower ranges of the box plot show the 25th and 75th percentiles of each data set, respectively. **P < 0.01, *P < 0.05.
Compared with Fr. I and Fr. II, suppressive immune-related genes such as LAG3, CTLA4, and HAVCR2 were significantly upregulated in Fr. IV (Fig. 2e). Especially, among 21 genes specific to exhausted T cells identified by single-cell RNA sequencing7, 16 genes (76.2%) were upregulated in Fr. IV compared with 13 genes (61.9%) upregulated in Fr. I and Fr. II. Also 9 of 12 genes reported to be commonly elevated in both exhausted CD8 T cells and CD4 regulatory T cells (Tregs)7 were upregulated in Fr. IV compared to both Fr. I and Fr. II. Only expression levels of HAVCR2 and PHLDA1 among the above 21 genes were upregulated when comparing Fr. III and Fr. IV. Fourteen genes including LAG3 and TOX2 were consistently upregulated in Fr. IV compared to Frs. I, II, and III. Moreover, genes such as TCF7, CCR7, and LEF1 specific to naïve CD8 T cells identified by single-cell RNA sequencing7 were consistently downregulated in Fr. IV compared to Frs. I, II, and III (Fig. 2e).
TCR clonal analysis showed that diversities of T-cell receptor (TCR) clonotypes of both α chain (TCRA) and β chain (TCRB) were the lowest in Fr. IV among the 4 populations although the difference was not significant (Supplementary Fig. 2a,b). Clonotypes with specific proportions tended to be higher in Fr. IV among 3 patients (Supplementary Fig. 2c), and some clonotypes of TCRA and TCRB were shared among the 4 fractions within each individual (Supplementary Fig. 2d). There were no differences in the proportion of clonotypes according to the expression of CD103 as a feature of Trem cells (Supplementary Figs. 1 and 2c).
The expression ratio of CD8 T cells producing all three cytokines (IL-2, TNFα, and IFNγ) in Fr. IV was significantly lower than that of Fr. II and Fr. III (Fig. 2f). In addition, the expression ratio of CD8 T cells producing either of two kinds of cytokines was significantly higher in Fr. II compared to Fr. III and Fr. IV, and the expression ratio of CD8 T cells producing no cytokines was higher in Fr. III and Fr. IV compared to Fr. II.
In terms of cytotoxic functionality, the expression ratio of CD8 T cells producing GZMB was significantly higher in Fr. II compared to Fr. III and Fr. IV (P = 0.018), and that of CD8 T cells producing both GZMB and PRF1 was higher in Fr. II compared to Fr. III and Fr. IV in 11 patients (P = 0.013) (Fig. 2g).
The above results showed that depending on the expression of PD-1 and TIM-3, the lineage of CD8 T cells correlated significantly with the dysfunction of these cells. Especially, CD8+ PD-1highTIM-3+ was the most functionally impaired population within the four groups.
Significant difference in characteristics within CD4 TILs based on expression patterns of PD-1 and TIM-3
As with the CD8 T cells, transcriptomic analysis of each fraction was performed by RNA sequencing using 12 fractions, except for three fractions of Fr. IV, and hierarchical clustering analysis using 1446 genes mainly revealed two clusters that were classified into two groups (group A: Fr. I/Fr. II/Fr. III and group B: Fr. V) (Fig. 3a). Pathway enrichment analysis revealed 714 genes of group A that were involved in immune responses and were already reported to correlate with CD4 Tregs15,16 (Fig. 3b). PCA of these genes revealed that three samples of Fr. V were located apart from the other 9 samples of Fr. I/Fr. II and Fr. III (Fig. 3c). GSEA of published gene data sets specific to CD4 Tregs17 was performed among each fraction, and the Treg-specific genes were significantly enriched in Fr. V compared to Frs. I, II, and III (Fig. 3d).
Figure 3.
Gene expression profile and functional analysis of CD4 tumour tissue-infiltrating lymphocytes (TILs) based on the expression patterns of PD-1 and TIM-3. (a) Clustering analysis for genes distinguishing four subsets except for Fr. IV within five fractions of CD4 TILs extracted from three patients with renal cell carcinoma. In total, 1446 genes were significantly upregulated or downregulated (log2 fold change > [1], false discovery rate [FDR] < 0.05) as indicated by at least one group comparison. (b) Biological processes (Gene Ontology [GO] terms) enriched in group A and group B. (c) Principal component (PC) analysis for four subsets of CD4 T cells based on the expression patterns of PD-1 and TIM-3. (d) Gene set enrichment analysis (GSEA) of published data sets of biological processes (GO terms) enriched in group A and group B17. Statistical significance was determined by permutation testing with normalized enrichment score (NES). (e) Volcano plot of up- or downregulated genes between Fr. I vs. Fr. IV, Fr. II vs. Fr. IV, and Fr. III vs. Fr. IV. Genes specific only to CD4 regulatory T cells (Tregs) (blue) and to both exhausted CD8 T cells and Tregs (red)7 are shown, and bold characters indicate the genes commonly having significant differences between these three combinations. (f) Gene expression of FOXP3 among each fraction. (g) Comparison of the expressions of CD4 + CD25high T cells, previously defined as CD4 Tregs, is stratified by each fraction. (h) Comparison of multiple cytokine productivity (IFNγ, TNFα, and IL-2) of CD4 TILs from 18 patients in Fr. II, Fr. III, and Fr. V. among three sample types was performed by Kruskal-Wallis analysis, and then a comparison of each sample type was performed by Bonferroni-corrected Mann-Whitney U test. The central tendency of the box plot indicates the median of each group, and the upper and lower ranges of the box plot show the 25th and 75th percentiles of each data set, respectively. **P < 0.01.
Compared with that in Frs. I, II, and III, FOXP3 was significantly upregulated in Fr. V along with 13 of 20 genes characterized as CD4 Treg-specific genes7 (Fig. 3e,f). Also 8 of 12 genes reported to be commonly elevated in both exhausted CD8 T cells and CD4 Tregs7 were upregulated in Fr. V compared that in to Frs. I, II, and III. Moreover, genes such as TCF7 and CCR7, which were enriched in naïve CD4 T cells identified by single-cell RNA sequencing7, were consistently downregulated in Fr. V compared to that in Frs. I, II, and III.
These results indicated that CD4 Tregs were enriched in the Fr. V population. Actually, the population of CD4 + CD25high T cells in Fr. V, which were reported to be the major population of CD4 Tregs in RCC18, was significantly higher compared to those in Fr. I/Fr. II and Fr. III (Fig. 3g).
TCR clonal analysis showed that diversities of TCR clonotypes of both TCRA and TCRB were the lowest in Fr. III and Fr. V among the 4 populations although the difference was not significant (Supplementary Fig. 3a,b). Clonotypes with specific proportions tended to be higher in Fr. III of 2 patients (Supplementary Fig. 3c), and none of the clonotypes of TCRA were shared among all 4 fractions although some clonotypes of TCRA and TCRB were shared among Fr. I, Fr. II, and Fr. III within each individual (Supplementary Fig. 3d).
The expression ratio of CD4 T cells producing all three cytokines (IL-2, TNFα, and IFNγ) in Fr. V was significantly lower than that of Fr. II and Fr. III (Fig. 3h). In addition, the expression ratio of CD4 T cells producing either of two kinds of cytokines was significantly higher in Fr. II and Fr. III compared to Fr. V, and the expression ratio of CD4 T cells producing no cytokines was higher in Fr. V compared to Fr. II and Fr. III (Fig. 3h).
The above results showed that one specific population of CD4 T cells (CD4 + PD-1lowTIM-3+) correlated significantly with dysfunction of CD4 T cells that was similar to that in CD4 Tregs.
Significant correlation of tumour grade with dysfunctional populations in both CD4 and CD8 T cells
Previously, we reported that immunological classification based on IC molecules was significantly correlated with tumour grade in RCC patients18. Stratified by tumour grade, the expression ratio of Fr. IV in CD8 T cells and Fr. V in CD4 T cells, which were both defined as dysfunctional populations in each T cell, were significantly upregulated in the patients with higher tumour grade (WHO/ISUP grades 3 and 4) (n = 29) rather than lower tumour grade (WHO/ISUP grades 1 and 2) (n = 68) (Fr. IV in CD8 T cells: P = 0.030, Fr. V in CD4 T cells: P = 0.029) (Fig. 4a,b).
Figure 4.

The relationship between total dysfunction of CD8 and CD4 T cells and tumour grade. (a) Comparison of the expression ratio of each fraction of CD8 T cells stratified by tumour grade. (b) Comparison of the expression ratio of each fraction of CD4 T cells stratified by tumour grade. For this panel, the comparison was performed by Mann-Whitney U test. The central tendency of the box plot shows the median of each group, and the upper and lower ranges of the box plot show the 25th and 75th percentiles of each data set, respectively. *P < 0.05. (c) The ratio of the patients stratified by expression pattern of CD4 T cells and CD8 T cells with dysfunctional populations according to tumour grade. Dysfunctional populations were divided into two groups based on the median expression level (Fr. IV of CD8 T cells: 20.3%, Fr. V of CD4 T cells: 2.79%). (d) The ratio of the patients with high expression ratio of both CD4 T cells and CD8 T cells with dysfunctional populations according to tumour grade. ***P < 0.001. (e) Comparison of multiple cytokine productivity (IFNγ, TNFα, and IL-2) of CD8 TILs from 18 patients stratified by tumour grade within Fr. II, Fr. III, and Fr. V. (f) Comparison of multiple cytokine productivity (IFNγ, TNFα, and IL-2) of CD4 TILs from 18 patients stratified by tumour grade within Fr. II, Fr. III, and Fr. IV. **P < 0.01, *P < 0.05. (g) Cytotoxic functionality producing GZMB and both GZMB and PRF1 in Fr. II of CD8 T cells is compared according to tumour grade. Comparison of two sample types was performed by Mann-Whitney U test. The central tendency of the box plot indicates the median of each group, and the upper and lower ranges of the box plot show the 25th and 75th percentiles of each data set, respectively. *P < 0.05.
When these dysfunctional populations were divided into two groups based on the median expression level (Fr. IV of CD8 T cells: 20.3%, Fr. V of CD4 T cells: 2.79%), 20% (1/5) of the patients with WHO/ISUP grade 1 and 23.8% (15/63) with WHO/ISUP grade 2 had higher expressions of both populations. In contrast, 45% (9/20) of the patients with WHO/ISUP grade 3 and 77.7% (7/9) with WHO/ISUP grade 4 had higher expressions of both populations (Fig. 4c). Overall, the ratio of patients with higher dysfunctional populations of both CD4 T and CD8 T cells was significantly higher in those with higher (16/29, 55.2%) rather than lower tumour grade (16/58, 23.5%) (P = 0.004) (Fig. 4d). In multivariate analysis along with other clinical items including tumour stage, only tumour grade significantly correlated with higher dysfunctional populations of both CD4 T and CD8 T cells (odds ratio: 3.48, 95% confidence interval [CI]: 1.39–8.73, P = 0.008) (Table 1).
Table 1.
Univariate and multivariate analysis of logistic regression of clinical characteristics correlated with high expression of both CD8 and CD4 dysfunctional populations.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | |
| Age (<66 vs. ≥66 years) | 0.39 | 0.08–1.88 | 0.239 | |||
| Gender (Male vs. Female) | 1.72 | 0.68–4.34 | 0.252 | |||
| WHO/ISUP grade (1, 2 vs. 3, 4) | 3.48 | 1.39–8.73 | 0.008 | 3.48 | 1.39–8.73 | 0.008 |
| Clinical T stage (I, II vs. III, IV) | 1.66 | 0.69–4.03 | 0.261 | |||
| Coagulative necrosis (absence vs. presence) | 1.62 | 0.63–4.72 | 0.319 | |||
| Histological type (clear cell vs. non clear cell) | 0.50 | 0.10–2.51 | 0.399 | |||
| Lymphovascular invasion (No vs. Yes) | 2.37 | 0.97–5.82 | 0.059 | |||
| NLR (<3.45 vs. ≥3.45) | 3.17 | 1.31–7.68 | 0.011 | 0.106 | ||
| PLR (<1.61 vs. ≥1.61) | 3.33 | 1.35–8.21 | 0.009 | 0.121 | ||
| CRP (<1 mg/dL vs. ≥1 mg/dL) | 2.29 | 0.82–6.40 | 0.114 | |||
Abbreviations: OR = odds ratio, CI = confidence interval, NLR = neutrophil to lymphocyte ratio, PLR = platelet to lymphocyte ratio, CRP = C reactive protein.
Cytokine productivities of both CD4 and CD8 T cells decreased significantly in the patients with higher versus lower tumour grade
Interestingly, we noticed heterogeneity in the results of the ability to produce cytokines even within the same fraction (Figs. 2f and 3h). The results of the multi-functionality assay stratified by tumour grade showed that in CD8 T cells, the abilities to produce a single cytokine in Fr. II and Fr. III, and two kinds of cytokines in Fr. III, were significantly higher in the patients with lower (n = 11) rather than higher tumour grade (n = 7) (P < 0.05) (Fig. 4e). In contrast, there was no significant difference in cytokine productivity depending on tumour grade in Fr. IV of CD8 T cells. However, in the CD4 T cells, the abilities to produce two kinds of cytokines in Frs. II, III, and V and three kinds of cytokines in Frs. III and V were significantly greater in the patients with lower rather than higher tumour grade (P < 0.05) (Fig. 4f).
Within 12 sets of TILs from the patients with higher dysfunctional populations of both CD4 T and CD8 T cells, cytokine productivities of only CD4 T cells (two kinds of cytokines in Fr. II [P = 0.030] and three kinds of cytokines in Fr. III [P = 0.030] and Fr. V [P = 0.048]) were significantly greater in the patients with lower rather than higher tumour grade (Supplementary Fig. 4).
With regard to cytotoxic functionality, there was no significant difference in the expression ratio of Fr. II of CD8 T cells producing granzyme B (GZMB) depending on the tumour grade. However, the expression ratio of Fr. II of CD8 T cells producing both GZMB and perforin (PRF1) was greater in the patients with lower rather than higher tumour grade (P = 0.017) (Fig. 4g). Within 9 sets of TILs from the patients with higher dysfunctional populations of both CD4 T and CD8 T cells, there was no significant difference of cytotoxic functionality between lower and higher tumour grade.
These results suggested that tumor grade correlated not only with the induction of dysfunctional populations in CD8 T cells and CD4 Tregs but also with the decline of cytokine productivities of both CD8 and CD4 TILs.
Significant correlation of tumour grade with prognosis of RCC patients treated with nivolumab monotherapy
Among the 97 patients, 10 were treated with nivolumab monotherapy as second-line or higher treatments (Supplementary Fig. 5a). Five patients (50%) had higher expression of both Fr. IV of CD8 T cells and Fr. V of CD4 T cells, and 6 patients had higher tumour grade. Tumour grade could have stratified the prognosis of cancer-specific survival (CSS) better than the expression of Fr. IV of CD8 T cells and Fr. V of CD4 T cells (Supplementary Fig. 5b).
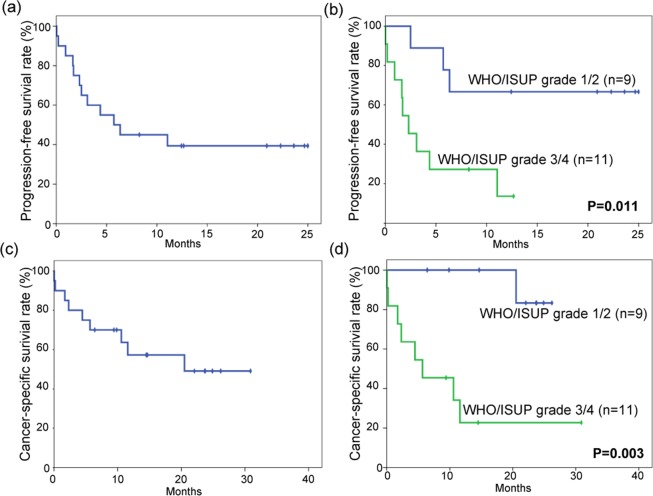
Finally, 20 RCC patients treated with nivolumab monotherapy as second-line or higher treatments were evaluated for therapeutic effect stratified by tumour grade (Supplementary Table 2). The median progression-free survival (PFS) of all patients was 6.35 months (Fig. 5a). In the subgroup analyses, PFS of the patients with lower-grade tumours (median PFS: not reached, n = 9) was better than that of patients with higher-grade tumours (median PFS: 2.3 months, n = 11) (P = 0.011) (Fig. 5b).
Figure 5.
Probability estimates of progression-free survival time of patients with metastatic renal cell carcinoma treated with nivolumab (a) (n = 20) and stratification by tumour grade (b). Also shown are the probability estimates of cancer-specific survival time of the same patients (c) (n = 20) and stratification by tumour grade (d). Statistical analysis was performed by log-rank test.
The median CSS of all patients was 20.5 months (Fig. 5c). Subgroup analyses showed CSS of the patients with lower-grade tumours (median CSS: not reached, n = 9) to be better than that of patients with higher-grade tumours (n = 11) (median CSS: 5.66 months, P = 0.003) (Fig. 5d).
Multivariate Cox regression analysis identified higher tumour grade (hazard ratio [HR]: 4.63, 95% CI: 1.10–19.45, P = 0.036) to be an independent predictor of inferior PFS time along with MSKCC risk classification (P = 0.002) (Table 2). Similarly, higher tumour grade was also identified as an independent predictor of CSS time (HR: 22.49, 95% CI: 1.44–350.42, P = 0.026) along with MSKCC risk classification (P = 0.006) (Table 3).
Table 2.
Univariate and multivariate analysis of prognostic factors for progression-free survival of metastatic RCC patients treated by nivolumab monotherapy.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age (<66 vs. ≥66 years) | 1.08 | 0.35–3.35 | 0.901 | |||
| Gender (Male vs. Female) | 2.18 | 0.64–7.50 | 0.216 | |||
| WHO/ISUP grade (1, 2 vs. 3, 4) | 4.80 | 1.27–18.10 | 0.020 | 4.63 | 1.10–19.45 | 0.036 |
| Coagulative necrosis (absence vs. presence) | 1.39 | 0.44–4.42 | 0.576 | |||
|
MSKCC risk classification (Good/Intermediate vs. Poor) |
8.83 | 2.47–31.60 | 0.001 | 9.05 | 2.22–36.86 | 0.002 |
| Number of prior therapies (1 vs. ≥2) | 0.66 | 0.21–2.08 | 0.479 | |||
| NLR (<3.45 vs. ≥3.45) | 2.98 | 0.89–10.03 | 0.078 | |||
| PLR (<1.61 vs. ≥1.61) | 2.27 | 0.68–7.59 | 0.185 | |||
| CRP (<1 mg/dL vs. ≥1 mg/dL) | 2.69 | 0.80–9.00 | 0.109 | |||
Abbreviations: RCC = renal cell carcinoma, HR = hazard ratio, CI = confidence interval, MSKCC = Memorial Sloan Kettering Cancer Center, NLR = neutrophil to lymphocyte ratio, PLR = platelet to lymphocyte ratio, CRP = C reactive protein.
Table 3.
Univariate and multivariate analysis of prognostic factors for cancer-specific survival of metastatic RCC patients treated by nivolumab monotherapy.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Age (<66 vs. ≥66 years) | 1.33 | 0.36–4.99 | 0.670 | |||
| Gender (Male vs. Female) | 1.98 | 0.49–8.07 | 0.341 | |||
| WHO/ISUP grade (1, 2 vs. 3, 4) | 13.9 | 1.64–117.4 | 0.016 | 22.49 | 1.44–350.4 | 0.026 |
| Coagulative necrosis (absence vs. presence) | 1.39 | 0.37–5.27 | 0.630 | |||
|
MSKCC risk classification (Good/Intermediate vs. Poor) |
14.8 | 2.97–73.66 | 0.001 | 21.14 | 2.41–185.2 | 0.006 |
| Number of prior therapies (1 vs. ≥2) | 0.52 | 0.14–1.98 | 0.338 | |||
| NLR (<3.45 vs. ≥3.45) | 3.05 | 0.75–12.36 | 0.119 | |||
| PLR (<1.61 vs. ≥1.61) | 1.88 | 0.47–7.52 | 0.374 | |||
| CRP (<1 mg/dL vs. ≥1 mg/dL) | 2.79 | 0.68–11.41 | 0.153 | |||
Abbreviations: RCC = renal cell carcinoma, HR = hazard ratio, CI = confidence interval, MSKCC = Memorial Sloan Kettering Cancer Center, NLR = neutrophil to lymphocyte ratio, PLR = platelet to lymphocyte ratio, CRP = C reactive protein.
Discussion
The evolution of cancer immunotherapy using anti-PD-1 antibody has been remarkable for various cancer types including RCC1,2. Several combination therapies targeting PD-12,19,20 showed better therapeutic effect compared to conventional molecular-targeted therapies, and the first combination was approved as first-line therapy for patients with metastatic or advanced RCC21. However, no predictive or prognostic biomarkers including PD-L1 expression were established in these randomized phase III studies in RCC1,2, unlike the other cancers22–24. One reason might be that compared with other cancer types25, the number of TILs was originally a poor prognostic factor in RCC, and PD-L1 expression was a significant prognostic factor in RCC patients26. Therefore, other types of markers focusing on the functions of TILs should be identified in RCC.
This is the first study, to our knowledge, to show transcriptome data using RNA-Seq based on TIM-3 expression in RCC patients with high PD-1 expression although there was a recently published study on transcriptome data based on the intensity of PD-1 in non-small lung cancer patients3. In CD8 T cells, the population of CD8+ PD-1highTIM-3+ had the lowest function among the four groups analysed, and this population was significantly upregulated in the patients with higher rather than lower tumour grade as previously reported9,27. In the present study, there was only slight functional difference between the presence and absence of TIM-3 expression on CD8 T cells highly expressing PD-1. However, both TOX and TOX2, which were recently reported to impose CD8+ T cell exhaustion5, were significantly upregulated in CD8+ PD-1highTim-3+ compared to the other populations. So, upregulation of CD8+ PD-1highTim-3+ in the tumour microenvironment could potentially become an important predictive factor in cancer immunotherapy. In CD4 T cells, the population of CD4 + PD-1lowTIM-3+ had the lowest function among the four groups analysed. In previously published data on the immune landscape in RCC, there was one distinct population of CD4 T cells that showed mid-level expression of PD-1/TIM-3 and high-level expression of FOXP3. This population was defined as Tregs and was upregulated at an advanced stage compared to an early stage, which agreed with our data9. Recently, PD-1 + CD4 Tregs in the tumour microenvironment were suggested to induce hyper-progressive disease after anti PD-1 monotherapy4. So, upregulation of CD4 + PD-1lowTIM-3+ in the tumour microenvironment also could potentially become an important predictive factor along with CD8+ PD-1highTIM-3+ cells.
Interestingly, the present study showed the presence of functional heterogeneity even in the same phenotypic populations of both CD4 T cells and CD8 T cells, and it was dependent on tumour grade. Especially, the functional differences between the lower- and higher-grade tumours were remarkable in the CD8+ PD-1highTIM-3− cells and CD4 + PD-1highTIM-3− cells. In CD8+ T cells, CD8+ PD-1+TIM-3− cells were reported to contain stem-like CD8 T cells with higher proliferative potential. This study supposed that stem-like CD8 T cells might be more abundant in the lower-grade tumours though we couldn’t evaluate in this study8. In CD4 T cells, Yang et al. reported the presence of heterogeneity of PD-1-expressing CD4 T cells and identified a functionally antithetical population within CD4 T cells expressing PD-1 in follicular lymphoma28. In RCC patients, clinically important populations of CD4 T cells, except for Tregs, could not be identified in previous reports in which analysis was performed using 19 kinds of markers9. It has been suggested that CD4 T cells might play an important role as well as CD8 T cells in immunotherapy using IC inhibitors10–12 and further analysis is needed to identify the important population in CD4 T cells.
This study was also the first, to our knowledge, to show that WHO/ISUP grade could be a significant prognostic factor in RCC patients treated with anti-PD-1 antibody. Among RCC patients treated with anti PD-1 antibody monotherapy, the prognosis of the patients with a high PD-L1 expression was poorer than that in the patients with low PD-L1 expression1. Contrastingly, among RCC patients treated with combination therapy of anti PD-1 antibody and anti CTLA-4 antibody, the prognosis of the patients with high PD-L1 expression was better than that in the patients with low PD-L1 expression2. Moreover, the treatment efficacy of this combination was significantly superior to that of sunitinib in 112 patients with sarcomatoid tumour categorized as WHO/ISUP grade 4 regardless of PD-L1 expression according to the result of the combination of nivolumab plus ipilimumab in RCC patients (CheckMate 214)29. Previously, specific gene profiles correlating with sarcomatoid differentiation and T-cell infiltration were reported to predict the prognosis of metastatic RCC patients treated with sunitinib30. Interestingly, those genes included CXCL13 and PDCD1, which were upregulated in Fr. III and Fr. IV of CD8 T cells compared to Fr. I in the present study. So, the combination therapy of ipilimumab and nivolumab targeting both CD4 + PD-1lowTIM-3+ and CD8+ PD-1highTIM-3+ might be able to reverse the natural prognosis of RCC and be ideal for treating patients with higher tumour grade. Actually, this novel immune therapy was effective in metastatic RCC patients with intermediate and poor risk, but the superiority of nivolumab and ipilimumab could not be shown in metastatic RCC patients with good risk2. We speculated that one reason might be the presence of more patients with lower-grade RCC among the metastatic RCC patients with good risk, and combination therapy was unnecessary for these patients, although we could not verify the results of the clinical trial. The WHO/ISUP grading system is widely recognized to be useful for the prognostic prediction of clear cell and papillary RCC and is usually used to evaluate only these two histological types31. However, this system could be used to describe the morphological features of all histological types of RCC, and thus we should consider tumour grade across tumour subtypes when evaluating the efficacy of IC inhibitor treatments because it is more versatile than the evaluation of immunohistochemistry in general practice. In our cohort, only surgical specimens were used to define WHO/ISUP grade because the diagnostic accuracy of tumour grade between biopsy and surgical pathology was reported to be around 50–75%32,33. Recently, the importance and versatility of biopsy of the renal mass or metastatic lesions have rapidly increased, and the development of a means of accurate diagnosis using liquid biopsy is needed34.
This study has several limitations. First, we could not clarify a direct association between total dysfunction of TILs and tumour grade because our extraction method was difficult to analyse alive tumour cells, especially in RCC tissues. Second, this was a retrospective study, and the number of patients sampled and treated with nivolumab monotherapy was small. Moreover, the sample types were mostly extracted from untreated primary lesions. The lymphocyte subsets between primary and metastatic RCC are reported to be slightly different18, and thus we need to perform an expanded study using liquid biopsy to represent the total condition of the tumour.
In conclusion, this study showed that tumour grade correlated significantly with the overall dysfunction of CD4 and CD8 T cells and the efficacy of nivolumab monotherapy. These results suggested that we should revisit the importance of cell morphology along with genetic features because cancer has heterogeneous features that are difficult to overcome35. Also, ipilimumab in combination with nivolumab might be preferable to nivolumab alone in patients with high-grade tumour after second-line therapy for RCC although there is no supporting evidence for this suggestion at present.
Materials and Methods
Patient characteristics
Two independent cohorts were analysed for this study as shown in Supplementary Fig. 1. Ninety-seven tumour tissues and 48 adjacent normal tissues were collected from 97 RCC patients surgically treated from October 2014 to May 2017 without perioperative multidisciplinary therapies (Supplementary Fig. 6a). The initially diagnosed tumours were staged according to the 7th American Joint Committee on Cancer staging classification36. Pathological diagnosis was evaluated according to WHO/ISUP classification, grading, presence of coagulative necrosis and pathological staging of RCC31 by two specialized pathologists (N.S., M.E.). Patient characteristics including laboratory findings were evaluated as previously reported18.
Twenty patients treated with nivolumab monotherapy from November 2016 to December 2018 were retrospectively evaluated to clarify the relationship between tumour grade and their prognosis. Patients were evaluated at the time of nivolumab administration according to MSKCC risk groups for previously treated patients37, and tumour grade was diagnosed using surgical specimens, not biopsy specimens, obtained from primary or metastatic sites (Supplementary Fig. 6b).
This study was approved by the local ethics committee of the Graduate School of Medicine, Osaka University (approval nos. #708, #13397–2, #14069-3), and written informed consent was obtained from all patients. All experiments were performed in accordance with relevant guidelines and regulations.
Preparation of lymphocytes analysed by multicolour flow cytometry
PBMCs were extracted from 1 mL peripheral blood of RCC patients using BD Pharm Lyse lysing buffer (BD Biosciences, Tokyo, Japan). Ninety-seven TILs were extracted macroscopically from cancer tissues, and 48 NILs were extracted macroscopically from non-cancerous tissues just after surgery and all fresh samples were analysed by flowcytometry and RNA sequencing. In several cases where coagulative necrosis was difficult to distinguish macroscopically, extracted area was evaluated later by haematoxylin and eosin staining. Details of lymphocyte extraction, methods, and clones of the antibodies for staining cell surface molecules were as previously reported18.
T-cell sorting and RNA sequencing
Stained fresh T cells were sorted by FACSAria SOAP (BD Biosciences) and lysed by RLT buffer (Qiagen, Tokyo, Japan) supplemented with 2-mercaptoethanol. RNA was extracted using Agencourt RNAClean XP (Beckman Coulter, Tokyo, Japan) and reverse transcribed to cDNA with SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara Bio, Tokyo, Japan). Sequence libraries were prepared with KAPA Hyper Prep Kit (KAPA Biosystems, Wilmington, MA), and 2×100 paired-end sequencing was performed with a HiSeq. 4000 system (Illumina, Tokyo, Japan).
Transcriptomic analysis
Adaptor sequences were removed by Cutadapt38, and trimmed reads were mapped to human genome Grch38/hg3839,40 with HISAT2 version 2.041. Read alignments were sorted and converted by SAMtools version 1.3.142. The reads were normalized to fragments per kilobase of transcript per million mapped reads by Cufflinks version 2.2.143. For identification of differentially expressed genes, PCA, and normalization of reads, raw tag counts were generated by featureCounts44 and processed using the DESeq. 2 package in R45. Gene expression was visualized by heat mapping with the ComplexHeatmap package in R46. Gene Ontology (GO) enrichment analysis was performed by calculating overlaps with biological process gene sets in MSigDB47. GSEA48 was performed with ranked gene lists based on q-value. TCR clones were reconstructed from the reads obtained by the above RNA sequencing using MiXCR49. Estimation of TCR repertoire diversity was performed with the tcR50 and VDJ tools51.
Multi-cytokine production assay
TILs were suspended in AIM-V (Thermo Fisher Scientific, Tokyo, Japan) supplemented with 10% AB serum (Biowest, Nuaillé, France) and then treated with Golgi STOP (1:1000), PMA (50 ng/mL), and ionomycin (1 µM) for 4 h. Harvested cells were stained with Zombie Yellow Fixable Viability kit (BioLegend), anti-CD45-BV785, anti-CD45RA-APC-Cy7 [HI100], anti-PD-1-PE-Cy7, and anti-Tim3-PE. After fixing with Foxp3/transcription factor staining buffer set (Thermo Fisher Scientific), cells were stained with anti-CD3-BV421 [UCTH1], anti-CD4-APC [RPA-T4], CD8-BV711 [RPA-T8], anti-IL-2 [PerCP-eFluor710], anti-TNFα-BV510 [Mab11], and anti-IFNγ-FITC [4 S.B3].
Cytotoxicity assay and intracellular staining
Cell surfaces were stained with Zombie Yellow Fixable Viability kit, anti-CD45-BV785, anti-CD45RA-APC-Cy7, anti-PD-1-PE-Cy7, and anti-Tim3-APC. Fixation and permeabilization of cells were conducted with Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific). CD3-Alexa Fluor 700, CD8-BV711, anti-Granzyme B-BV421 [GB11], and anti-perforin-FITC [B-D48] were used for intracellular staining.
Patient follow-up
Patient follow-up generally included a history, physical examination, routine blood work, abdominopelvic computed tomography, and chest radiography. Details of the evaluation of therapeutic response were as previously reported18.
PFS time was retrospectively measured from the date of initiation of nivolumab treatment until disease progression, death from disease progression, or the date of the patient’s last follow-up visit. CSS time was retrospectively calculated from the date of initiation of nivolumab treatment until cancer-related death or the date of the patient’s last follow-up visit.
Statistics
Comparisons between the surface marker expressions and clinical features were evaluated by Kruskal-Wallis test and Bonferroni-corrected Mann-Whitney U test. Distributions of PFS and CSS times were estimated with the Kaplan-Meier method, and associations between PFS, CSS, and the immunological classifications were assessed with a log-rank test. A value of P < 0.05 was considered statistically significant. These statistical analyses were performed with SPSS software, version 20.0 (IBM SPSS Statistics, Tokyo, Japan) and R (The R Foundation for Statistical Computing, Vienna, Austria).
Supplementary information
Acknowledgements
This study was supported by the Osaka Kidney Bank (Osaka, Japan), the Public Trust Surgery Research Fund (Tokyo, Japan), the Osaka Cancer Society Grant (Osaka, Japan), and JSPS KAKENHI (Grant-in-Aid for Young Scientists (B), grant numbers JP15K20085 and JP17K16790). We also thank Ms. Yamami Nakamura for her important contribution to performance of the RNA sequencing.
Author contributions
Conception and design: A.K., T.K., H.W. and N.N. Development of methodology: T.K., M.H., Y.K., T.Y., M.H. and N.O. Acquisition of data: A.K., T.K., Y.K., T.Y., M.H., K.N., S.N. and E.M. Analysis and interpretation of data: A.K., T.K., Y.K. and N.O. Writing and review of the manuscript: A.K., T.K., Y.Y., T.K. and K.H. Administrative, technical or material support: T.K., T.U., A.N., K.F., A.M-O., K.I., M.U., R. I., N.O. and S.S. Study supervision: N.O., H.W. and N.N.
Competing interests
T. Kanazawa, Y. Kidani, T. Yoshida, and M. Hirata are employees of Shionogi & Co., Ltd. The other authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Atsunari Kawashima and Takayuki Kanazawa.
Supplementary information
is available for this paper at 10.1038/s41598-020-63060-1.
References
- 1.Motzer RJ, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motzer RJ, et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N Engl J Med. 2018;378:1277–1290. doi: 10.1056/NEJMoa1712126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thommen DS, et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med. 2018;24:994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamada T, et al. PD-1(+) regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc Natl Acad Sci USA. 2019;116:9999–10008. doi: 10.1073/pnas.1822001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seo H, et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8(+) T cell exhaustion. Proc Natl Acad Sci USA. 2019;116:12410–12415. doi: 10.1073/pnas.1905675116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng C, et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 2017;169:1342–1356 e1316. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 8.Jansen CS, et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature. 2019;576:465–470. doi: 10.1038/s41586-019-1836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevrier S, et al. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell. 2017;169:736–749 e718. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alspach E, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature. 2019;574:696–701. doi: 10.1038/s41586-019-1671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borst J, Ahrends T, Babala N, Melief CJM, Kastenmuller W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol. 2018;18:635–647. doi: 10.1038/s41577-018-0044-0. [DOI] [PubMed] [Google Scholar]
- 12.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackay LK, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol. 2013;14:1294–1301. doi: 10.1038/ni.2744. [DOI] [PubMed] [Google Scholar]
- 14.Wherry EJ, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Plitas G, et al. Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity. 2016;45:1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Simone M, et al. Transcriptional Landscape of Human Tissue Lymphocytes Unveils Uniqueness of Tumor-Infiltrating T Regulatory Cells. Immunity. 2016;45:1135–1147. doi: 10.1016/j.immuni.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonacci B, et al. Requirements for growth and IL-10 expression of highly purified human T regulatory cells. J Clin Immunol. 2012;32:1118–1128. doi: 10.1007/s10875-012-9701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawashima A, et al. Immunological classification of renal cell carcinoma patients based on phenotypic analysis of immune check-point molecules. Cancer Immunol Immunother. 2018;67:113–125. doi: 10.1007/s00262-017-2060-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motzer RJ, et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med. 2019;380:1103–1115. doi: 10.1056/NEJMoa1816047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rini BI, et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med. 2019;380:1116–1127. doi: 10.1056/NEJMoa1816714. [DOI] [PubMed] [Google Scholar]
- 21.Ljungberg B, et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2019 Update. Eur Urol (2019). [DOI] [PubMed]
- 22.Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 23.Hugo W, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2017;168:542. doi: 10.1016/j.cell.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 24.van Rooij N, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14:717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 26.Thompson RH, et al. Tumor B7-H1 is associated with poor prognosis in renal cell carcinoma patients with long-term follow-up. Cancer Res. 2006;66:3381–3385. doi: 10.1158/0008-5472.CAN-05-4303. [DOI] [PubMed] [Google Scholar]
- 27.Cai C, et al. Tim-3 expression represents dysfunctional tumor infiltrating T cells in renal cell carcinoma. World J Urol. 2016;34:561–567. doi: 10.1007/s00345-015-1656-7. [DOI] [PubMed] [Google Scholar]
- 28.Yang ZZ, et al. Mass Cytometry Analysis Reveals that Specific Intratumoral CD4(+) T Cell Subsets Correlate with Patient Survival in Follicular Lymphoma. Cell Rep. 2019;26:2178–2193 e2173. doi: 10.1016/j.celrep.2019.01.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Highlights in renal cell carcinoma from the seventeenth International Kidney Cancer Symposium. Clin Adv Hematol Oncol16Suppl 19, 1–16 (2018). [PubMed]
- 30.Beuselinck B, et al. Molecular subtypes of clear cell renal cell carcinoma are associated with sunitinib response in the metastatic setting. Clin Cancer Res. 2015;21:1329–1339. doi: 10.1158/1078-0432.CCR-14-1128. [DOI] [PubMed] [Google Scholar]
- 31.Warren AY, Harrison D. WHO/ISUP classification, grading and pathological staging of renal cell carcinoma: standards and controversies. World J Urol. 2018;36:1913–1926. doi: 10.1007/s00345-018-2447-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel HD, et al. Diagnostic Accuracy and Risks of Biopsy in the Diagnosis of a Renal Mass Suspicious for Localized Renal Cell Carcinoma: Systematic Review of the Literature. J Urol. 2016;195:1340–1347. doi: 10.1016/j.juro.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delahunt B, et al. The International Society of Urological Pathology (ISUP) grading system for renal cell carcinoma and other prognostic parameters. Am J Surg Pathol. 2013;37:1490–1504. doi: 10.1097/PAS.0b013e318299f0fb. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto Y, et al. Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci. 2019;110:617–628. doi: 10.1111/cas.13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edge, S. B. American Joint Committee on Cancer. AJCC cancer staging manual, 7th edn. Springer (2010). [DOI] [PubMed]
- 37.Motzer RJ, et al. Prognostic factors for survival in previously treated patients with metastatic renal cell carcinoma. J Clin Oncol. 2004;22:454–463. doi: 10.1200/JCO.2004.06.132. [DOI] [PubMed] [Google Scholar]
- 38.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011. 2011;17:3. [Google Scholar]
- 39.Haeussler M, et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47:D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 41.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 45.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 47.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolotin DA, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 50.Nazarov VI, et al. tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinformatics. 2015;16:175. doi: 10.1186/s12859-015-0613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shugay M, et al. VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Computational Biology. 2015;11:e1004503. doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.