Abstract
Superficial scald is one of the most serious postharvest physiological disorders that can affect apples after a prolonged cold storage period. This study investigated the impact of pre- and post-harvest climatic variations on superficial scald in a susceptible apple cultivar. Fruit batches with contrasting phenotypes for superficial scald incidence were identified among several years of “Granny Smith” fruit production. The “low scald” year pre-harvest climate was characterised by a warm period followed by a sudden decrease in temperature, playing the part of an in vivo acclimation to cold storage. This was associated with many abiotic stress responsive genes which were induced in fruit peel. In particular 48 Heat Shock Proteins (HSPs) and 5 Heat Shock transcription Factors (HSFs) were strongly induced at harvest when scald incidence was low. For “high scald” year, a post-harvest acclimation of 1 week was efficient in reducing scald incidence. Expression profiles of stress related genes were affected by the acclimation treatment and indicate fruit physiological adaptations to cold storage. The identified stress-responsive genes, and in particular HSPs, could be useful indicators of the fruit physiological status to predict the risk of scald occurrence as early as harvest.
Subject terms: Plant molecular biology, Abiotic
Introduction
Apple (Malus domestica Borkh.) is an economically important fruit crop, which is cultivated worldwide, with high nutritional value and a wide range of tastes. Apple fruit quality is affected by complex processes depending on genetic, environmental and agronomic factors, such as temperature, light, humidity or harvest stage1. To maximize their postharvest qualities, fruit are harvested early and stored at low temperature for several months. However, extended cold storage periods may lead to several physiological disorders such as superficial scald on ‘Granny Smith’, a sensitive variety. The appearance of brown patches on the epidermis and hypodermal cortical tissues during shelf life severely deteriorate fruit visual qualities, rendering the product unmarketable2.
The development of superficial scald symptoms is assumed to be an oxidative response due to cold stress during storage and the accumulation of oxidative products in apple peel3. Several studies have linked the acyclic sesquiterpene α-farnesene, a volatile compound accumulated in the peel under cold stress, and its auto-oxidation in conjugated trienols (Ctols) and ketone 6-methyl-5-hepten-2-one (MHO) with superficial scald symptoms2,4. Other studies have shown that superficial scald could also be induced even at low concentrations of α-farnesene5 and therefore suggested the involvement of other pathways. In particular, the oxidative stress from prolonged cold storage was considered to influence scald development2. New hypotheses arose from the study of the inhibitory effect of the ethylene inhibitor 1-MCP on scald development. It was shown that in addition to reduced ethylene and α-farnesene production, 1-MCP also reduced the accumulation of reactive oxygen species (ROS) in fruit peel, probably by promoting cell-membrane integrity6. Further studies revealed that 1-MCP stimulates the production of ROS scavengers, synthesis of fatty acids that could stabilize plastid and vacuole membranes against cold, and the accumulation of sorbitol that can act as a cryoprotectant7. In the proposed model, ROS production in response to cold stress plays a central role in scald development by disrupting membrane integrity and promoting the accumulation of phenolic compounds. The symptomatic peel browning would result from the enzymatic oxidation of the accumulated chlorogenic acid by the polyphenol oxidase (PPO), both initially in separate compartments8.
Although superficial scald was intensively studied for several years, its molecular or biochemical determinism was mainly analysed after long time cold storage during symptom development3,9, or under 1-MCP or DPA treatments4,7,8,10–12. Thus, early determinisms linked to the pre-harvest conditions, in particular to climate or fruit maturity stages remain poorly understood.
Maturity stage at harvest appears to be a key factor controlling scald development because immature apple fruit are more susceptible to develop symptoms. It has been speculated that the reason is a lower anti-oxidant to oxidant products ratio in immature apples13. There is also wide agreement that warm and dry growing periods contribute to increase susceptibility to superficial scald2. Therefore climate change could lead to an increase of scald incidence and severity through rising temperatures14. Although controversial and depending on several climatic factors, in certain locations the risk of superficial scald occurrence has been shown to be reduced when fruit were exposed to 150 hours below 10 °C during 4 or 6 weeks before harvest15. It was suggested that this ‘in vivo’ cold acclimation could modify the physiological status of the fruit at harvest, and thus preventing scald development during subsequent cold storage16. Based on the same idea, one experiment with step-wise cooling of harvest fruit has shown to reduce scald development for ‘Granny Smith’17.
Plant cold acclimation is associated with multiple physiological modifications participating in membrane stabilization and ROS reduction to maintain cellular homeostasis and prevent low temperature-induced oxidative injuries18. Studies have shown that pre-storage cold acclimation treatments can reduce chilling injury of harvested fruit for cucumber19, citrus20, zucchini21, avocado22, loquat23 and mango24. Production of protective compounds such as sorbitol could act as a cryoprotectant to promote directly membrane stabilization as demonstrated in transgenic Arabidopsis7. Gene expression and enzymatic activities of ROS scavengers such as superoxide dismutase (SOD), catalase (CAT) and ascorbate peroxidase (APX), as well as ascorbic acid and glutathione were also enhanced under cold acclimation as reported in cucumber19 and kiwi fruit25. Therefore, pre- or post-harvest cold acclimation could promote fruit resistance to oxidative stress triggered by cold storage and limit the development of scald symptoms.
The aim of this study was to investigate the effect of pre-harvest climate or post-harvest cold acclimation on superficial scald development in ‘Granny Smith’, and to identify its early molecular determinism. Fruit batches with contrasting phenotypes for superficial scald incidence and severity were identified among several years of fruit production. The “low” and “high” scald years were then analysed for their pre-harvest climatic variables, and transcriptomic analyses were set up between the corresponding fruit peel samples collected at harvest. The expression of identified genes and indicators of fruit physiological status were analysed in response to a post-harvest cold acclimation that was efficient in reducing scald incidence. The relevance of the identified genes is discussed with respect to their involvement in superficial scald determinism and prediction.
Materials and methods
Plant material and sampling strategies
‘Granny Smith’ apple fruit (Malus domestica Borkh.) were harvested in 2014, 2015 and 2017 from commercially run orchards at the Station Experimental de La Morinière (30 ha estate, Saint Epain, France). Each year, fruit from two different orchards were harvested at three different maturity stage based on starch index (SI): early (H1, SI = 3), optimal (H2, SI = 5) and late (H3, SI = 7) (Fig. 1a; harvest dates in Supplementary Table S1). Fruit were collected from orchards P24 and P31 in 2014, P24 and P36 in 2015, R06 and R11 in 2017. For each batch, 100 to 200 fruit were randomly collected on both side of the orchard ranks, at human height, from at least 15 trees. Fruit were then stored at the Station Experimental de La Morinière in controlled atmosphere cold rooms (0.5 °C, 2% O2, 2% CO2 in 2014, 0.5 °C, 1.5% O2, 1% CO2 in 2015 and 2017) for 5 to 6 months. Each year peel samples were collected immediately at harvest stage.
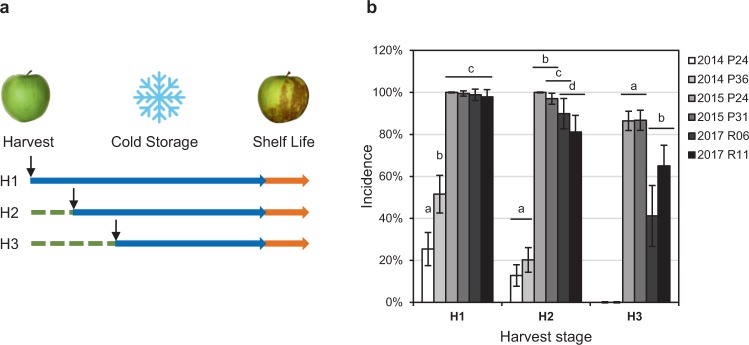
Figure 1.
Effect of year and fruit maturity on superficial scald incidence. (a) Schematic representation of the experimental design. Fruit were harvested at three different maturity stages, early (H1), optimal (H2) or late (H3), stored under cold controlled atmosphere (blue arrow) for 5 to 6 months, and phenotyped for scald incidence after 1 week of shelf-life at room temperature (orange arrow). (b) Annual incidence of superficial scald injuries according to maturity at harvest on fruit collected from two different orchards each year. Values are binomial proportions and confidence intervals for n = 100 to 200 and α = 0.05. Snowflake image unchanged according to https://commons.wikimedia.org/wiki/File:Emojione_2744.svg, (https://creativecommons.org/licenses/by-sa/4.0/deed.en).
An acclimation test was run on fruit collected in 2017 on R06 and R11 orchards. It consisted in 1 week storage at 8 °C (H1-acclim) before transfer to classic cold chambers at 2 °C up to 3, 4 and 5 months. For comparison, additional fruit batches harvested at H1 and H2 were immediately stored in classic cold chambers at 2 °C for 3, 4 and 5 months. Peel samples were collected after one day (1D) and one week (1 W) of acclimation or classic cold storage.
For each sample, peel was collected from both sun-exposed and shaded sides of 20 randomly selected fruit, immediately frozen, ground and homogenized in liquid nitrogen. Sample aliquots were stored at −80 °C before use.
Evaluation of superficial scald symptoms
Following cold storage, fruit were phenotyped for the development of superficial scald symptoms immediately and after 1 week of shelf life at room temperature (20 °C). Incidence (percentage of fruit with symptoms) and severity were assessed for 100 to 200 fruit per batch. Severity was recorded for each fruit according to the relative surface area affected by symptoms using a 0 to 4 scale as following: S0, no symptom; S1, > 0% to 25%; S2, > 25% to 50%; S3, > 50% to 75%; S4, > 75% of affected surface area (Supplementary Fig. S1). Binomial proportion confidence intervals were calculated using the Wald method with continuity correction and error rate α = 0.05.
Meteorological data analysis
Meteorological data were collected at the Station Experimental de La Morinière (Saint Epain, France) using an Agriscope weather station (http://www.agriscope.fr/). Temperature, radiation, relative humidity and pluviometry were recorded hourly. Daily data synthetized as average, minimum and maximum were analysed over the 60 d preceding the fruit harvest. PCA analyses were performed for data collected 60 d, 40 d or 20 d before harvest using the R package FactoMineR26 (Fig. 2).
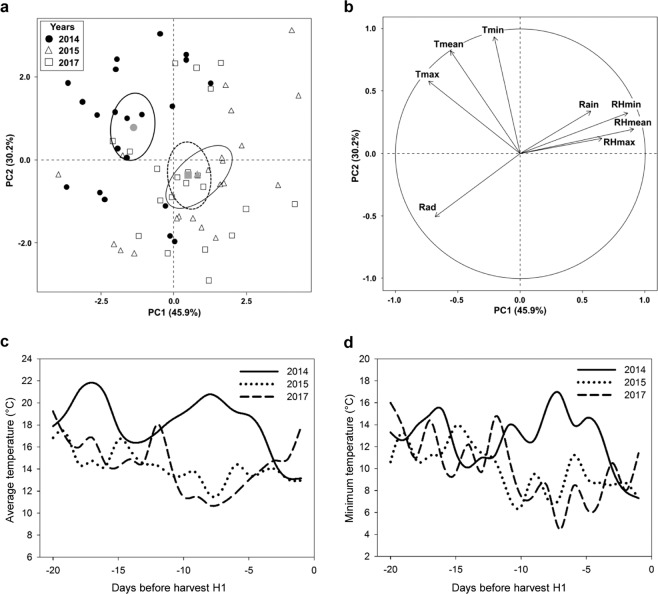
Figure 2.
Analysis of pre-harvest climatic variables. PCA analysis of daily temperature (T, °C), rain (mm), relative humidity (RH) and radiation (Rad, W m−2) variables collected in the orchard each year for 20 d before harvest. Individuals (a) and variables (b) are represented in the first two dimension of the PCA. Pre-harvest daily temperatures: (c) average (Tmean, °C) and (d) minimum (Tmin, °C).
RNA extraction, amplification and microarray hybridization
Total RNA was extracted from 3 × 10−3 kg FW of frozen fruit peel tissue finely ground in liquid nitrogen using two successive extraction procedures with CTAB and then SSTE buffers as described by Segonne et al.27. RNA quality indicator (RQI) was determined using the Experion TM RNA StdSens Analysis Kit (Bio-Rad, USA), only samples with RQI > 8 were kept for hybridizations and RT-qPCR experiments.
mRNAs were amplified from 200 ng of total RNA and labelled with either Cyanine-3 or Cyanine-5 fluorescent dye with the Agilent Low Input Quick Amp Labelling kit (Agilent, Foster City, CA, USA) according the manufacturer’s recommendations. Labelled samples were purified with Qiagen RNeasy kit (Qiagen, USA), combined as 20 pmol for each dye and co-hybridized to the Agilent microarray AryANE_v2 containing 60-mers oligonucleotide probes designed for each MDP gene from Velasco et al.28 Malus domestica genome v1, as described by Celton, Gaillard et al.29.
The experimental design included three comparisons between samples collected at harvest from fruit batches with low versus high incidence for superficial scald (Fig. 3a). Each comparison included a technical repetition with dye swap. Thus, fruit peel samples were compared as following: samples 2014-P24 vs 2015-P24 (C1), samples 2014- P36 vs 2015-P31 (C2), samples 2014-P24 vs 2017-R11 (C3).
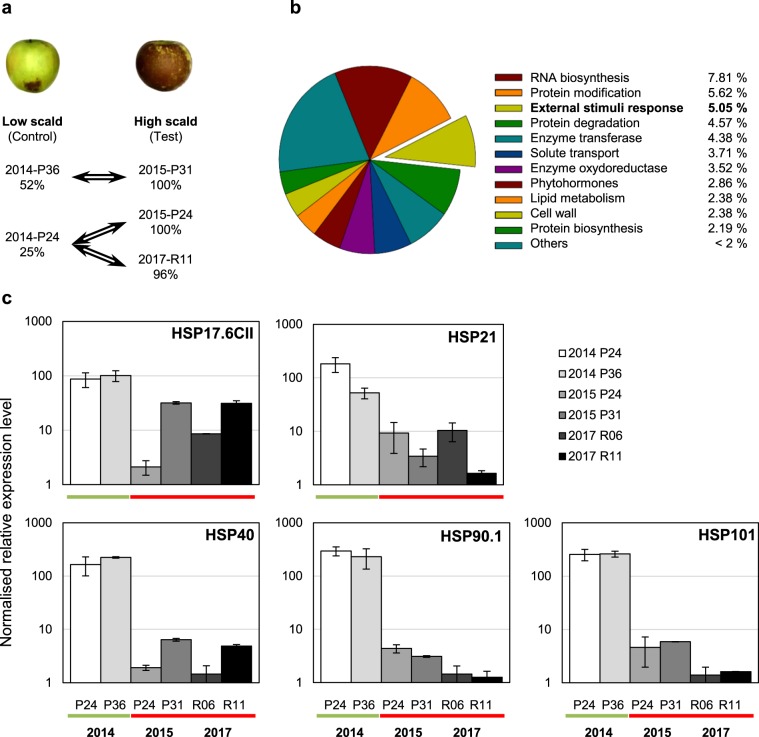
Figure 3.
Transcriptomic analysis at harvest for fruit batches with contrasted phenotype for superficial scald. (a) Three comparisons were setup in dye-swap between peel samples collected at early harvest from fruit batches with “low” versus “high” scald incidence after cold storage and shelf-life. The three comparisons were combined in one statistical analysis setting the “low” scald samples as “control”, and “high” scald samples as “test”. (b) DETs assignment to functional categories: 1050 DETs with BH < 0.05 were selected and 57% were assigned (based on Mapman ontology using the Mercator web tool). (c) Relative gene expression level (RT-qPCR) in fruit peel samples collected at harvest from “low” (2014) or “high” (2015 and 2017) scald years for HSP17.6CII (MD15G1053800), HSP21 (MD13G1108500), HSP40 (MD10G1289200), HSP90.1 (MD01G1208700) and HSP101 (MD06G1201600). Data are mean values ± SD of n = 3.
Microarrays analysis
The Agilent Feature Extraction 11.5 software was used to extract data files from the scanned images obtained using the MS 200 microarray scanner (Roche Nimblegen). All statistical analyses were conducted based on a dye swap approach as described by Celton, Gaillard et al.29 with the R software30. Briefly, for each comparison (C1 to C3) data were normalized with the lowess method, and differential expression analyses were carried out using the lmFit function and the Bayes moderated t test using the R package LIMMA31 from the Bioconductor project. To estimate gene expression levels, the normalized expression values were corrected from background. An additional statistical analysis, including a Benjamini–Hochberg procedure in order to controls the false discovery rate (FDR), was performed with combining the three initial comparisons (C1 to C3) considered as three biological repetitions.
Probes were considered reporting differentially expressed transcripts when their respective corrected P-value (BH) was equal or below 0.05. Only probes reporting sense transcription (96.3%) were then considered in this study. In order to take into account the improvements brought by the GDDH13 v1.1 assembly and annotation32, specific and 100% matching targets were search with blast for the selected AryANE_v1 probes. Best blast search results were reported in the Supplementary File S1 under the “spe_new” column. The “spe” code correspond to the identification of at least one target with 100% match among the GDDH13 v1.1 transcripts (76% of the selection). Only the identified set of MD GDDH13 v1.1 genes was then considered in the subsequent analyses and RT-qPCR experiments. Functional classification of DETs was based on Mapman ontology using the Mercator web tool (http://mapman.gabipd.org/web/guest/app/mercator)33. An enrichment analysis (Wilconxon rank sum test with Benjamin-Hochberg-correction) was performed with Mapman software 3.6.034. Arabidopsis genes GO annotations were retrieved from The Arabidopsis Information Resource (TAIR) on www.arabidopsis.org. The microarrays data are available under the accession number GSE135863 in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/).
Reverse transcription and quantitative real time PCR (RT-qPCR)
Total RNA samples used for the microarrays experiments and RNA extracted from the 2017 acclimation experiment peel samples were treated with 1 U of DNAse I (Promega, USA) and cDNAs were synthesized from 1 μg of DNA-free-RNA with oligo(dT) 15 and 200 U of MMLV-RT (Promega) according to Segonne et al.27. For each cDNA, qPCR experiments were carried out in triplicate according to Segonne et al.27 using the IQ SYBR Green Super Mix (Biorad) and specific forward and reverse primers at 0.3 µM. Amplifications were performed on a CFX MaestroTM Real-Time PCR instrument (Bio-Rad, USA) and data analysed with CFX Maestro™ Software (Bio-Rad, version 1.1).
Primer pairs were designed for short and specific amplification based on GDDH13 v1.1 genome sequence (Supplementary Table S2) using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) or retrieved from Vergne et al.35. Each primer pair was tested for its respective specificity and efficiency of the reactions: (1) amplicons were sequenced once and aligned to GDDH13 v1.1; (2) for each run, single product amplification was confirmed by melting curve analyses. Amplification efficiency was assessed using a dilution curve method over a 6 point dilution series. Only primer pairs with efficiencies higher than 85% were retained for further analysis.
Based on microarray results, three reference genes never differentially expressed in all comparisons were selected as reference genes to calculate a normalization factor (geometric mean). Relative expression levels were calculated using a formula derived from the 2–ΔΔCt method [ΔCt = (Ctgoi–Ctref)], where Ct is the threshold cycle, goi is the gene of interest, and ref is the reference gene36. Standard deviations were calculated from three replicates following the error-propagation rule formula.
Results
Superficial scald symptoms development
Superficial scald was evaluated for different maturity stages at harvest (H1 to H3) on 2 orchards from the same location over 3 years (Fig. 1a). Each year scald incidence decreased while fruit maturity at harvest increased. In particular in 2014 no symptom was detected on late H3 fruit, while 25% and 52% of the early H1 fruit, and 12% and 13% of the optimum H2 fruit were affected by scald (Fig. 1b). Scald incidence was significantly higher in 2015 and 2017 (Fig. 1b), with scald incidence reaching 98% to 100% at H1 and remaining above 80% at H2. 2015 and 2017 fruit were also more severely affected than in 2014 (Supplementary Fig. S1). In 2015 incidence only slightly decreased to 90% at the latest harvest stage H3 while, as observed in 2014, 2017 incidence and severity progressively decreased with higher maturity stage at harvest (Fig. 1b and Supplementary Fig. S1). However, 2017 incidence remained higher than in 2014. Therefore, 2014 was a “low scald” year, while 2015 and 2017 were “high scald” years.
Analysis of pre-harvest climate
Climatic variables were retrieved from the weather station located in the location of the orchards in order to analyse pre-harvest conditions each year. The total number of hours when hourly temperature was below 10 °C (NH10) was calculated each year from 60 d, 45 d, 30 d and 20 d before harvest (Table 1). Irrespective of the pre-harvest period considered, the NH10 was lower than 150 hours in all years. Contrary to expectation15, NH10 was the lowest for the “low scald” year 2014. Based on these results, principal component analysis (PCA) was performed for each previously cited period to identify climatic differences between years. The analysis performed for the period of 20 d before harvest showed the maximum difference between the years (Fig. 2a). The first two components of the PCA captured most of the variability accounting for respectively 46% and 30% of explained variance (Fig. 2b). These two components allow the clear separation of pre-harvest days between “low scald” year 2014 and “high scald” years 2015 and 2017. The 2014 pre-harvest days were negatively correlated with PC1 and positively correlated with PC2. PC1 was mainly defined by relative humidity (RH), in particular by the daily mean and minimum RH (RHmean correlation 0.91, P-value = 5 × 10−25; RHmin, correlation 0.86, P-value = 1.4 × 10−19). PC2 included variables linked to the daily temperature, in particular the minimum and the mean temperature of the day (Tmin, correlation 0.94, P-value 2.6 × 10−29; Tmean, correlation 0.82, P-value 1.5 × 10−16). Pre-harvest days were thus dryer and warmer in the “low scald” year of 2014 than in the “high scald” years of 2015 and 2017. Analysis of the daily average temperature confirms that the pre-harvest period was overall warmer in 2014 than in 2015 and 2017 (Fig. 2c). However three days before harvest the mean temperature dropped suddenly from 18 °C to 13 °C. This rapid decrease was especially marked for the daily minimum temperature that dropped from 15 °C to 7 °C (Fig. 2d). A drastic shift of temperature also occurred just before 2017 harvest, a “high-scald” year, but in the opposite way with a rapid increase of the mean and minimum daily temperatures. The low scald incidence observed only in 2014 could be explained by an “in vivo” acclimation to cold due to the pre-harvest warm period followed by a rapid decrease in temperature.
Table 1.
Number of pre-harvest hours with mean temperature below 10 °C (NH10).
| Numbers of days | NH10 | ||
|---|---|---|---|
| 2014 | 2015 | 2017 | |
| 60 | 40 | 99 | 96 |
| 45 | 40 | 99 | 96 |
| 30 | 18 | 99 | 95 |
| 20 | 18 | 72 | 82 |
Transcriptome analysis at harvest
In order to identify early determinants of scald development, transcriptomic analyses were performed at early harvest stage on the base of scald contrasted phenotypic data after cold storage. Expression profiles of apple peel sampled at harvest from low scald year 2014 were compared with the high scald years 2015 and 2017. Expression profiles of 2014 peel samples from P24 and P36 orchards were compared respectively with expression profiles of 2015 peel samples from P24 and P31 orchards. An additional comparison was set up between 2014-P24 and 2017-R11 peel samples (Fig. 3a). We hypothesized that genes involved in early events that prevent or induce scald development displayed similar differential expression patterns for each comparison. Therefore, we set up an additional t-test combining the three comparisons. 1491 differentially expressed transcripts (DETs) were identified with significant BH adjusted P-value (BH < 0.05). Thus, 1435 sense DETs were selected, of which 41.7% were up regulated and 58.3% were down regulated in high scald samples, when compared to low scald samples (Supplementary File S1).
A search for corresponding genes on the Malus domestica GDDH13 v1.1 genome version32 led to the identification of 1050 (73.2%) non-redundant genes. Based on MD gene annotation, 57.1% of these DETs were assigned to functional categories (Fig. 3b; Supplementary File S3). Among them, the most represented were “RNA biosynthesis” (7.81%), “Protein modification” (5.62%), “External stimuli response” (5.05%) and “Protein degradation” (4.57%). It is noteworthy that 42.9% of the DETs were not allocated to any category, and that 7.90% belonged to enzyme categories “oxidoreductase” or “transferase”. In order to validate data obtained from the microarray analysis, the relative transcript abundances of a subset of differentially expressed genes with contrasted expression profiles were tested by RT-qPCR using cDNA from low or high scald samples (Supplementary File S4). The results were consistent with those obtained from the microarray analysis (Pearson correlation coefficient = 0.83).
Candidate genes associated with later high scald symptoms
The most significantly over-represented category was “External stimuli response” (P-value = 8.61 × 10−14) and more specifically “response to temperature” (3.81% of DETs, P-value = 3.93 × 10−17) (Supplementary File S3). It included members from all known Heat Shock Protein families (HSP100, HSP90, HSP70, HSP60 and sHSP) (Table 2). All 48 DETs belonging to these families were down regulated in 2015 and 2017 when scald incidences were high. The 10 most differentially expressed transcripts of the analysis are potential HSPs, in addition 64% of the top 50 DETs are HSPs (Supplementary File S1). In particular, we verified that potential orthologous genes to Arabidopsis HSPs known to be induced in response to heat stress were down regulated in “high scald” samples: MD15G1053800 and MD13G1108500 respectively coding for small HSPs HSP17.6CII and HSP21, as well as MD01G1208700 and MD06G1201200 coding for HSP90.1 and HSP10137–40 (Fig. 2c). Furthermore, 74% of the DETs annotated as HSPs could be linked to a gene ontology (GO) annotation related to heat stress (Supplementary File S1). Most of them were also annotated as responding to light stimulus and/or oxidative stress. The subgroup of Heat shock transcription factors (HSFs) was also significantly over represented in the transcriptomic analysis. A potential HsfA2, activator of transcription for many HSPs in Arabidopsis41, and its potential targets were particularly repressed in “high scald” samples (Table 2). This transcription factor has been shown to respond to different environmental stresses, including the combination of heat and high light, as well as hydrogen peroxide (Supplementary File S1).
Table 2.
HSP and HSF transcripts down regulated in “high scald” fruit peel.
| Seq_id | LR | MD gene | TAIR | ||||
|---|---|---|---|---|---|---|---|
| evalue | name | Annotation | AtHSP | GO | |||
| Small HSP (sHSP) | |||||||
| MDP0000700383 | −3.26 | MD15G1053800 | 3 × 10−59 | AT5G12020 | 17.6 kDa class II heat shock protein | AtHsp17.6-CII | HLOx |
| MDP0000362505 | −2.43 | MD08G1068200 | 3 × 10−58 | AT5G12020 | 17.6 kDa class II heat shock protein | AtHsp17.6-CII | HLOx |
| MDP0000188935 | −3.00 | MD08G1068000 | 9 × 10−57 | AT5G12020 | 17.6 kDa class II heat shock protein | AtHsp17.6-CII | HLOx |
| MDP0000214382 | −3.79 | MD13G1108500 | 3 × 10−37 | AT4G27670 | 21 kDa class II heat shock protein | AtHsp25.4-P* | HLOx |
| MDP0000125300 | −2.68 | MD06G1060300 | 3 × 10−54 | AT4G25200 | 23.6 kDa class II heat shock protein | AtHsp23.6-M* | |
| MDP0000795157 | −2.62 | MD10G1289200 | 0 | AT2G20560 | DNAJ heat shock family protein | ||
| MDP0000290546 | −2.58 | MD05G1310300 | 6 × 10−179 | AT2G20560 | DNAJ heat shock family protein | ||
| MDP0000549793 | −1.95 | MD12G1172300 | 0 | AT3G08970 | DNAJ protein ERDJ3A | H | |
| MDP0000164489 | −2.70 | MD11G1089300 | 1 × 10−69 | AT1G07400 | HSP20-like | HOx | |
| MDP0000493154 | −2.94 | MD11G1087100 | 3 × 10−69 | AT1G07400 | HSP20-like | HOx | |
| MDP0000574524 | −4.56 | MD05G1240300 | 3 × 10−38 | AT1G07400 | HSP20-like | HOx | |
| MDP0000094857 | −3.22 | MD11G1089400 | 3 × 10−16 | AT1G07400 | HSP20-like | HOx | |
| MDP0000791550 | −1.74 | MD07G1210400 | 2 × 10−78 | AT1G53540 | HSP20-like | AtHsp17.6C-Cl* | HOx |
| MDP0000265157 | −2.59 | MD01G1144400 | 5 × 10−73 | AT1G53540 | HSP20-like | AtHsp17.6C-Cl* | HOx |
| MDP0000412799 | −2.08 | MD07G1210800 | 2 × 10−66 | AT1G53540 | HSP20-like | AtHsp17.6C-Cl* | HOx |
| MDP0000424976 | −1.64 | MD11G1087200 | 2 × 10−22 | AT1G53540 | HSP20-like | AtHsp17.6C-Cl* | HOx |
| MDP0000152564 | −2.45 | MD17G1151000 | 4 × 10−53 | AT1G54050 | HSP20-like | AtHsp17.4-CIII* | HLOx |
| MDP0000136609 | −3.02 | MD15G1443700 | 2 × 10−49 | AT4G10250 | HSP20-like | AtHsp22.0-ER* | HOx |
| MDP0000656080 | −2.97 | MD08G1249100 | 2 × 10−45 | AT4G10250 | HSP20-like | AtHsp22.0-ER* | HOx |
| MDP0000208958 | −1.46 | MD09G1271100 | 2 × 10−58 | AT5G37670 | HSP20-like | AtHsp15.7-Cl* | HOx |
| HSP60 - HSP70 | |||||||
| MDP0000752314 | −1.50 | MD10G1170700 | 0 | AT3G23990 | Heat shock protein 60 | H | |
| MDP0000859313 | −1.18 | MD05G1182500 | 0 | AT3G23990 | Heat shock protein 60 | H | |
| MDP0000867730 | −2.03 | MD07G1196600 | 3 × 10−39 | AT1G56410 | Heat shock protein 70 | H | |
| MDP0000620433 | −2.30 | MD07G1197200 | 0 | AT3G12580 | Heat shock protein 70 | AtHsp70-4* | HLOx |
| MDP0000122734 | −2.27 | MD17G1226000 | 0 | AT3G12580 | Heat shock protein 70 | AtHsp70-4* | HLOx |
| MDP0000220559 | −1.63 | MD01G1126500 | 0 | AT3G12580 | Heat shock protein 70 | AtHsp70-4* | HLOx |
| MDP0000311339 | −3.00 | MD15G1150500 | 0 | AT1G16030 | Heat shock protein 70B | AtHsp70-5* | H |
| MDP0000172536 | −2.16 | MD16G1192600 | 0 | AT2G32120 | Heat-shock protein 70T-2 | AtHsp70-8* | HLOx |
| MDP0000684170 | −1.83 | MD13G1191900 | 0 | AT2G32120 | Heat-shock protein 70T-2 | AtHsp70-8* | HLOx |
| HSP90 | |||||||
| MDP0000181929 | −1.06 | MD11G1037400 | 0 | AT5G56000 | Heat shock protein 81.4 | AtHsp90-4 | H |
| MDP0000254260 | −3.53 | MD01G1208700 | 0 | AT5G52640 | Heat shock protein 90.1 | AtHsp90-1 | H |
| MDP0000303430 | −3.00 | MD07G1279200 | 0 | AT5G52640 | Heat shock protein 90.1 | AtHsp90-1 | H |
| MDP0000948331 | −2.38 | MD00G1081900 | 6 × 10−88 | AT5G52640 | Heat shock protein 90.1 | AtHsp90-1 | H |
| HSP 100 | |||||||
| MDP0000217508 | −3.59 | MD06G1201600 | 0 | AT1G74310 | Heat shock protein 101 | AtHsp100-1* | HLOx |
| MDP0000303015 | −2.76 | MD14G1211000 | 0 | AT1G74310 | Heat shock protein 101 | AtHsp100-1* | HLOx |
| MDP0000308722 | −2.32 | MD06G1201200 | 0 | AT1G74310 | Heat shock protein 101 | AtHsp100-1* | HLOx |
| MDP0000755970 | −2.27 | MD14G1210600 | 0 | AT1G74310 | Heat shock protein 101 | AtHsp100-1* | HLOx |
| Other related chaperone | |||||||
| MDP0000197501 | −2.99 | MD16G1124100 | 1 × 10−21 | AT1G12060 | BAG chaperone regulator | ||
| MDP0000215062 | −1.79 | MD14G1054700 | 0 | AT3G12050 | Hsp90 binding protein | ||
| MDP0000932255 | −1.64 | MD12G1055300 | 0 | AT3G12050 | Hsp90 binding protein | ||
| MDP0000190008 | −1.17 | MD13G1024100 | 2 × 10−53 | AT1G23100 | Hsp10 Hsp60-co-chaperone | ||
| MDP0000422652 | −1.15 | MD04G1081900 | 0 | AT4G12400 | Hsp70-Hsp90 organizing protein | HLOx | |
| MDP0000161691 | −1.08 | MD06G1065400 | 0 | AT4G12400 | Hsp70-Hsp90 organizing protein | HLOx | |
| HSF | |||||||
| MDP0000527802 | −1.50 | MD02G1171800 | 2 × 10−92 | AT4G36990 | Heat shock factor 4 | HsfB1 | H |
| MDP0000925901 | −1.64 | MD03G1258300 | 2 × 10−124 | AT3G22830 | Heat shock transcription factor A6B | HsfA6b | H |
| MDP0000155667 | −1.05 | MD01G1198700 | 6 × 10−79 | AT5G62020 | Heat shock transcription factor B2A | HsfB2a | H |
| MDP0000243895 | −2.80 | MD15G1057700 | 8 × 10−126 | AT2G26150 | Heat shock transcription factor A2 | HsfA2* | HLOx |
| MDP0000489886 | −1.91 | MD08G1064100 | 5 × 10−120 | AT2G26150 | Heat shock transcription factor A2 | HsfA2* | HLOx |
Reference sequence for probes design (Seq_id), Log ratio (LR) of differential analysis, corresponding GDDH13 gene (MD gene), Arabidopsis gene annotation (TAIR e-value, gene name and annotation), Arabidopsis short name according to Swindell et al.77 (AtHSP), Arabidopsis GO annotation (response do heat (H), light (L) or ROS (Ox)). *Gene regulated by AtHSFA2 according to Nishizawa et al.41.
Other transcription factors involved in the light stress response were similarly down regulated in samples from “high scald” fruit batches (Table 3), like the potential orthologous genes of the Arabidopsis bZIP genes HY5 and HYH (MD08G1147100 and MD16G1132200)42. MD03G1297100 and MD11G1316800, two potential MYB12 regulators of the flavonoid pathways known to respond to high light43, were also down regulated. In agreement, a potential orthologous downstream regulated gene was also down regulated, namely a phenylalanine ammonia-lyase (PAL1, MD12G1116700). MD03G1099300, a potential ZAT12 gene C2H2 transcription factor also named RHL41 “responsive to high light 41”, was also down regulated. The Arabidopsis ZAT12 gene (AT5G59820) was shown to be also induced by heat and cold stress and to be involved in cold and photosynthetic acclimation44,45 (Table 3; Supplementary File S1).
Table 3.
Selected DETs associated with abiotic stress response.
| Seq_id | LR | MD gene | TAIR | ||||
|---|---|---|---|---|---|---|---|
| evalue | name | Short name | Annotation | GO | |||
| Response to heat | |||||||
| MDP0000175388 | −2.36 | MD04G1022400 | 0 | AT5G48570 | ROF2 | FKBP-type peptidyl-prolyl cis-trans isomerase | H |
| MDP0000275263 | −2.00 | MD09G1263000 | 5 × 10−112 | AT1G66080 | Protein of unknown function DUF775 | H | |
| MDP0000273764 | −1.06 | MD17G1258000 | 9 × 10−116 | AT1G66080 | Protein of unknown function DUF775 | H | |
| MDP0000917573 | −1.68 | MD02G1050200 | 5 × 10−70 | AT3G24500 | ATMBF1C | Multiprotein bridging factor 1 C | H |
| MDP0000129942 | −1.39 | MD15G1187400 | 2 × 10−69 | AT3G24500 | ATMBF1C | Multiprotein bridging factor 1C | H |
| MDP0000205111 | −1.40 | MD10G1156300 | 0 | AT3G25230 | ROF1 | Rotamase FKBP1 protein folding catalyst | H |
| MDP0000141863 | −1.04 | MD05G1166200 | 0 | AT3G25230 | ROF1 | Rotamase FKBP1 protein folding catalyst | H |
| Response to light | |||||||
| MDP0000586302 | −2.06 | MD08G1147100 | 1 × 10−66 | AT5G11260 | HY5 | bZIP transcription factor family protein | L |
| MDP0000388769 | −1.96 | MD12G1116700 | 0 | AT2G37040 | PAL1 | PHE ammonia lyase 1 | LOx |
| MDP0000268980 | −1.68 | MD11G1316800 | 3 × 10−73 | AT2G47460 | MYB12 | Myb domain protein 12 | L |
| MDP0000119725 | −1.67 | MD03G1297100 | 8 × 10−73 | AT2G47460 | MYB12 | Myb domain protein 12 | L |
| MDP0000281626 | −1.41 | MD10G1316100 | 0 | AT1G61800 | GPT2 | Glucose-6-phosphate/phosphate translocator 2 | L |
| MDP0000771031 | −1.31 | MD05G1305700 | 1 × 10−135 | AT2G29120 | ATGLR2.7 | Glutamate receptor 2.7 | L |
| MDP0000831937 | −1.21 | MD07G1285600 | 0 | AT5G24120 | SIGE | RNA polymerase sigma subunit E | L |
| MDP0000149332 | −1.17 | MD04G1144100 | 3 × 10−90 | AT2G37970 | ATHBP2 | SOUL heme-binding family protein | L |
| MDP0000159766 | −1.08 | MD17G1265700 | 0 | AT2G42690 | alpha/beta-Hydrolases superfamily protein | L | |
| Response to cold | |||||||
| MDP0000299872 | −1.15 | MD09G1043700 | 0 | AT5G40010 | AATP1 | AAA-ATPase 1 | C |
| MDP0000317816 | −1.13 | MD04G1164500 | 8 × 10−116 | AT5G01600 | ATFER1 | Ferretin 1 | COx |
| MDP0000203813 | −1.11 | MD07G1204100 | 0 | AT3G55580 | RCC1 | Regulator of chromosome condensation | C |
| Response to multiple stimuli | |||||||
| MDP0000739699 | −2.65 | MD14G1150400 | 3 × 10−50 | AT3G22840 | ELIP | Chlorophyll A-B binding family protein | HLC |
| MDP0000446914 | −1.87 | MD17G1280400 | 0 | AT2G47180 | ATGOLS1* | Galactinol synthase 1 | HLCOx |
| MDP0000595671 | −1.18 | MD03G1099300 | 3 × 10−37 | AT5G59820 | RHL41 | ZAT12 C2H2-type zinc finger family protein | HLCOx |
| MDP0000226817 | −1.27 | MD10G1110200 | 0 | AT1G17870 | EGY3* | Zinc metallopeptidase | HLOx |
| MDP0000238942 | −1.01 | MD01G1185500 | 0 | AT2G36530 | LOS2 | Enolase | LC |
| MDP0000755567 | −1.17 | MD03G1289900 | 1 × 10−21 | AT4G02380 | SAG21 | Senescence-associated gene 21 | LCOx |
| MDP0000286604 | −0.89 | MD17G1211100 | 4 × 10−15 | AT5G20230 | ATBCB | Blue-copper-binding protein | LCOx |
| MDP0000265874 | −0.87 | MD15G1253900 | 1 × 10−11 | AT1G20440 | COR47 | Cold-regulated 47 | HC |
| Response to oxidative stress/ROS processing | |||||||
| MDP0000166359 | −1.67 | MD06G1110300 | 2 × 10−98 | AT1G64500 | Glutaredoxin family protein | Ox | |
| MDP0000171695 | −1.87 | MD14G1014200 | 6 × 10−42 | AT3G10020 | Protein of unknown function | Ox | |
| MDP0000566567 | −2.42 | MD05G1211000 | 8 × 10−108 | AT1G78380 | ATGSTU19 | Glutathione S-transferase TAU 19 | Ox |
| MDP0000187493 | −1.76 | MD05G1211100 | 9 × 10−31 | AT1G78380 | ATGSTU19 | Glutathione S-transferase TAU 19 | Ox |
| MDP0000459010 | −1.46 | MD05G1210700 | 8 × 10−110 | AT1G78380 | ATGSTU19 | Glutathione S-transferase TAU 19 | Ox |
| MDP0000755113 | −1.03 | MD05G1210000 | 5 × 10−116 | AT1G78380 | ATGSTU19 | Glutathione S-transferase TAU 19 | Ox |
| MDP0000135807 | −1.15 | MD05G1309500 | 4 × 10−40 | AT1G28480 | Thioredoxin superfamily protein | Ox | |
| MDP0000179654 | −0.96 | MD10G1288100 | 2 × 10−41 | AT1G28480 | Thioredoxin superfamily protein | Ox | |
| MDP0000340109 | −1.26 | MD15G1395600 | 1 × 10−71 | AT1G22840 | CYTC-1 | CYTOCHROME C-1 | Ox |
| MDP0000205322 | −1.02 | MD08G1213100 | 7 × 10−72 | AT1G22840 | CYTC-1 | CYTOCHROME C-1 | Ox |
| MDP0000248822 | −1.15 | MD15G1085500 | 0 | AT2G26560 | PLP2 | Phospholipase A 2 A | Ox |
| MDP0000770103 | −1.17 | MD11G1004100 | 6 × 10−137 | AT5G05340 | PRX52 | Peroxidase superfamily protein | Ox |
| MDP0000642594 | −1.13 | MD04G1139400 | 3 × 10−80 | AT3G09270 | ATGSTU8 | Glutathione S-transferase TAU 8 | Ox |
| MDP0000320982 | −1.12 | MD04G1139500 | 4 × 10−83 | AT3G09270 | ATGSTU8 | Glutathione S-transferase TAU 8 | Ox |
| MDP0000126107 | −0.87 | MD04G1104900 | 1 × 10−154 | AT3G09640 | APX2* | Ascorbate peroxidase 2 | Ox |
| MDP0000326493 | 1.06 | MD02G1013800 | 9 × 10−33 | AT4G30380 | Barwin-related endoglucanase | Ox | |
| MDP0000126601 | 1.12 | MD17G1039200 | 4 × 10−27 | AT5G14920 | GASA14 | Gibberellin-regulated family protein | Ox |
Reference sequence for probes design (Seq_id), Log ratio (LR) of differential analysis, corresponding GDDH13 gene (MD gene), Arabidopsis gene annotation (TAIR e-value, gene name and annotation), Arabidopsis GO annotation (response do heat (H), light (L), cold (C) or ROS (Ox)). *Gene regulated by AtHSFA2 according to Nishizawa et al.41.
A few DETs responding to cold and/or involved in cold acclimation were identified (Table 3, Supplementary File S1). The most differentially expressed were down regulated in “high scald” samples, and as with ZAT12 they were also annotated as responding to heat and light stimuli. In particular, MD14G1150400, a potential orthologue for AT3G22840 coding for a chlorophyll binding protein ELIP1 involved in photoprotection was down regulated in “high scald” samples. The MD17G1280400 gene, potentially coding for an orthologue of the AtGOLS1 a galactinol synthase induced by heat, light and cold stimulus, was also down regulated.
Finally, among the DETs with GO for response to oxidative stress or probably involved in redox cellular homeostasis, several genes potentially coding for glutathione S-transferase were down regulated (MD05G1210700, MD05G1210000, MD05G1211000, MD05G1211100, MD04G1139400, MD04G1139500) as well as potential ascorbate peroxidase APX2 (MD04G1104900), glutaredoxin (MD06G1110300) and thioredoxin (MD05G1309500, MD10G1288100) (Table 3; Supplementary File S1).
Interestingly, only 1.9% of the genes were related to the biotic stress response while 12.5% of the DETs had GO related to the abiotic stress response. The whole transcriptome profile suggested that “low scald” fruit have been exposed to abiotic stresses before harvest, probably a combination of thermal and high light stresses.
Effect of post-harvest cold acclimation on scald
H1 fruit harvested in 2017 were submitted to a post-harvest cold period of 1 week at low temperature prior to classic cold storage (Fig. 4a). When fruit were cold acclimated (H1-acclim), scald incidence was significantly reduced for both fruit batches dropping from 82% to 45% for R06, and from 86% to 56% for R11 after 4 months cold storage (Fig. 4b). Scald symptoms were also less severe for H1-acclim fruit than for H1 fruit (Supplementary Fig. S3). Scald incidence for H2 fruit, harvested one week later and immediately stored in classic cold was also reduced when compared to H1 fruit, but less than for the H1 acclimated fruits. This intermediate phenotype for H2 fruit was not observed any more after 5 months. (Supplementary Fig. S3). Therefore, the observed reduced scald incidence for H1-acclim fruit was not only due to more mature fruit (1 week delay before classic cold storage), but principally due to the cold acclimation treatment.
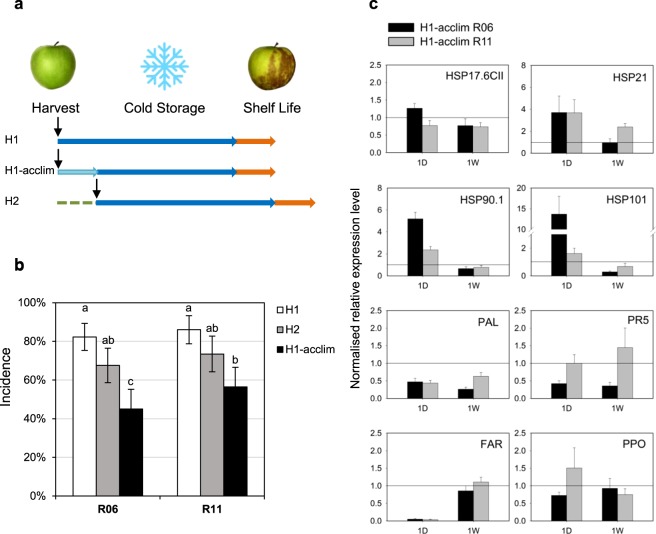
Figure 4.
Effect of cold acclimation on superficial scald incidence. (a) Schematic representation of the experimental design. Early harvested fruit were cold acclimated at 8 °C for 1 week (H1-acclim, light blue) and compared to fruit not acclimated (H1) or harvested 1 week later (H2). Fruit were stored in classic cold conditions (blue arrow) and phenotyped for scald incidence after one week of shelf-life at room temperature (orange arrow). (b) Incidence of superficial scald injuries after 4 months cold storage according to acclimation treatment and harvest stage on fruit collected from two different orchards (R06 and R11). Values are binomial proportions and confidence intervals for n = 100 and α = 0.05. (c) Effect of acclimation on relative gene expression in fruit peel samples after one day (1D) and one week (1W). Expression relatively to classic cold storage for HSP17.6CII (MD15G1053800), HSP21 (MD13G1108500), HSP90.1 (MD01G1208700), HSP101 (MD06G1201600), PAL (MD12G1116700), PR5 (MD04G1064200), FAR (MD10G1311000) and PPO (MD10G1299400). Data are mean values ± SD of n = 3 for fruit collected in both orchards R06 and R11. Snowflake image unchanged according to https://commons.wikimedia.org/wiki/File:Emojione_2744.svg, (https://creativecommons.org/licenses/by-sa/4.0/deed.en).
Effect of post-harvest cold acclimation on physiological stress indicators
As many HSPs responding to various stresses were found induced in 2014, the effect of cold acclimation on their expression was analysed. When compared with classic cold storage (H1 samples), the identified MD genes coding for HSP21, HSP90 and HSP101 were up-regulated after 1 day but not after 1 week of acclimation (Fig. 4c). Cold acclimation conditions tended to limit the major down regulation of HSP genes immediately observed after 1 day when fruit were stored in classic cold conditions (Supplementary Fig. S4). After one week of acclimation, all tested HSPs gene expression were similarly repressed as in classic cold conditions.
The effect of cold acclimation was further analysed using biotic and abiotic stress responsive gene expression46. The cold acclimation treatment had a repressive effect on MD12G1116700 gene coding for a phenyl ammonia lyase (PAL) (Fig. 4c). PAL was immediately induced by classic cold storage for 1 week, while the cold acclimation treatment limited its induction for the entire first week of storage (Supplementary Fig. S4). In contrast, for MD04G1064200, a PR5 Thaumatin coding gene, the acclimation had a clear repressive effect only for the R06 fruit batch (Fig. 4c). PR5 was progressively induced under classic cold storage, indicating an increasing fruit stress status (Supplementary Fig. S4).
The qPFD chip also included FAR (MD10G1311000), coding for a terpene synthase probably involved in the pathway leading to the biosynthesis of α-farnesene, and PPO (MD10G1299400), coding for a polyphenol oxidase. Both genes are reporters of enzymatic activities suspected of being involved in the appearance of scald symptoms. FAR and PPO were progressively induced by cold storage (Supplementary Fig. S4). The acclimation treatment transiently inhibited the strong induction of FAR on the first day but did not have a consistent effect on PPO (Fig. 4c) which was also induced in both conditions (Supplementary Fig. S4).
Discussion
Our study clearly demonstrated that pre-harvest ‘in vivo’ acclimation to cold is not necessarily due to the accumulation of cold temperature for 2 months before harvest as generally accepted, but also by a thermal variation due to a warm period followed by sudden cold temperatures three weeks before harvest. This thermal acclimation induced an increase in expression of HSP genes as well as other genes involved in abiotic stress responses. This transcriptomic response probably induced a physiological adjustment allowing the fruit to withstand cold storage and limiting further scald development.
The putative HSPs and HSFs apple orthologues highly expressed in low scald samples were probably induced by the high temperatures observed specifically through 2014 pre-harvest period. Indeed, the induction of HSP gene expression and proteins has already been observed in fruit flesh exposed to high daily temperatures in orchards47, and some HSF genes were shown to respond to heat stress in different apple tissues48. The sudden change of weather for cold temperatures three days before harvest may have also contributed to the induction of some HSPs. In particular, members of the HSP70 family were shown to be specifically induced by cold stress in spinach leaves and Arabidopsis49,50. Cold stress generates oxidative stress responses, and many AtHSPs were shown to be induced by ROS (Table 2). Therefore MdHSP expression could also have been indirectly induced by cold via ROS production.
The major negative effect of cold stress is that it induces severe membrane damage, alters the photosynthetic electron transfer machinery, and generates oxidative stress18. HSPs probably protect the cells in regulating the plant antioxidant system and ROS production51. Several chloroplastic sHSPs were shown to protect the Photosystem II (PSII) against formation of oxygen and photoinhibition in various stress conditions52,53. In particular, the tomato HSP21 and the sweet pepper CaHSP26 protect PSII from temperature dependent oxidative stress39,54. It was recently shown that the spinach SoHSC70 positively regulates thermotolerance by alleviating cell membrane damage, reducing ROS accumulation, and improving activities of antioxidant enzymes55. We can then hypothesize that among the identified apple HSFs and HSPs induced by pre-harvest conditions in 2014, several of them could have protected the fruit against cold stress induced by storage by reducing the oxidative stress and stabilizing the cell membranes (Fig. 5).
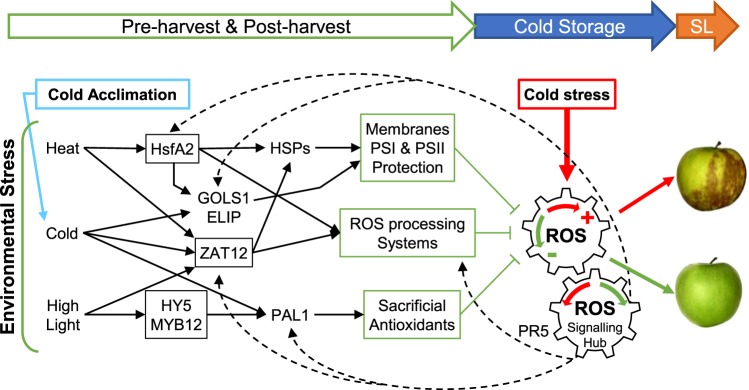
Figure 5.
Hypothetical model for pre- and post-harvest climatic effects on superficial scald incidence. Pre-harvest environmental stress and post-harvest cold acclimation activate interconnected regulation pathways (black arrows, transcription factors in black boxes). Fruit physiological status is therefore modified, in particular its redox status (physiological process in green boxes), which can counteract the oxidative stress due to cold storage (red arrow) and limit scald. The model involves the integration of the different signals at the ROS production level and feed-back through the ROS signalling hub76 (dash arrow). Sacrificial antioxidants include housekeeping compounds such as proline, as well as secondary compounds such as flavonoids56. The ROS processing system involves compounds and enzymes allowing repeated redox-cycling such as those involved in the glutathione-ascorbate cycle71. PR5 gene expression may be correlated with the increasing stress status of fruit during cold storage. PSI and PSII, photosystems I and II; ROS reactive oxygen species; SL, shelf life.
The protective effect of HSPs was also probably enhanced by the regulation network depending on HsfA2. Indeed, AtHsfA2 was identified as a key regulator in response to several types of environmental stress, inducing many HSPs but also genes involved in various cellular adaptation processes such as ascorbate peroxidase 2 (APX2) and galactinol synthase 1 (GolS1)41. In addition to HsfA2 and many HSPs, homologues of both APX2 and GolS1 genes were up regulated in the “low scald” fruit and could participate in the osmoprotection and ROS scavenging systems of the cells and prevent scald in response to cold stress (Table 3). Indeed, being involved in the response to a wide range of abiotic stresses, APX2 is a key enzyme of the redox homeostasis participating in the glutathione-ascorbate ROS scavenging cycle56, while GolS1 is involved in the synthesis of raffinose family oligosaccharides. In Arabidopsis galactinol and raffinose compounds can scavenge hydroxyl radicals to protect plant cells from oxidative damage caused by chilling57.
Protection against cold stress for “low scald fruit” was also probably enhanced by the expression of other genes involved in the maintenance of the cellular redox homeostasis. Apple genes coding for thioredoxin, glutaredoxin (GRX), glutathione-S-transferase (GST) and 2-oxoglutarate (2OG) or Fe(II)-dependent oxygenase were also relatively more expressed at harvest in “low scald” fruit. The Arabidopsis GRX gene AtGRXS17 was shown to confer chilling stress tolerance in tomato fruit by increasing antioxidant enzyme activities and reducing H2O2 accumulation58. GSTs are directly involved in reducing oxidative damage as well as in resistance and adaptation to various abiotic stresses in plants59. In the same way, over expression of bZIP gene HY5 and MYB transcription factors in “low scald” fruit could also participate in the maintenance of the cellular redox homeostasis through their role in anti-oxidant components synthesis and therefore promote cold acclimation of plants18,42,43.
The pre-storage acclimation treatment was sufficient in temperature and duration to reduce superficial scald, while preserving the fruit qualities (colour and firmness). A similar result was obtained by Moggia et al.17 with a step-wise cooling experiment. The acclimation treatment also delayed the repression of several HSP genes involved in abiotic stress response (Fig. 4c). This result indicates that HSP genes induction in 2014 was not specifically due to the progressive decrease in temperature three days before harvest but more probably to the previous warm period or by the combination of warm and cold periods (Fig. 3). This result supposes also that the increase in temperature in 2017 just before harvest was not long or intense enough to induce and maintain HSP gene expression until the end of the cold acclimation. Therefore, it could be interesting to test the post-harvest effect of a heat shock or a warm period followed by a cold acclimation on HSP gene expression and superficial scald development.
The expression profiles of FAR and PPO, coding respectively an α-farnesene synthase and a polyphenol oxidase often proposed to be involved in scald symptoms development, through the early phase of cold storage could not explain the efficiency of the acclimation treatment. In contrast, stress responsive genes expression profiles indicated differential fruit physiological status (Fig. 4c). PAL is the first enzyme of the polyphenol biosynthesis pathways leading to several groups of secondary metabolites such as lignins and flavonoids, including anthocyanins60. PAL regulation and ROS scavenging properties of the polyphenols led to the hypothesis that PAL was a key element in plant thermal stress acclimation61. PAL expression and activity has already been associated with chilling tolerance in several fruit species62,63. In particular, PAL induction of expression was observed in peel of citrus under cold stress and was attributed to a rapid adaptive response of the tissue to low temperature64. The rapid induction of PAL expression under classic cold or acclimation conditions is consistent with the observed increase in PAL enzyme when apple peel disc were chilled65. Probably because of milder temperatures, the acclimation treatment led to a lower induction of PAL and suggests a lower stress status for these fruit when compared with those subjected to classic cold conditions.
In addition to being a transcriptional biotic and abiotic stresses responsive marker, PR5 was shown to regulate physiological processes and interact with the ROS processing system66. PR5-osmotin helps in the accumulation of proline, an osmolyte which quenches reactive oxygen species and free radicals67. The overexpression of PR5-osmotin in chili pepper was shown to increase the activity of ascorbate peroxidase (APX) and superoxide dismutase (SOD) to detoxify the accumulated ROS68. Contrary to PAL, PR5 gene expression was not increased immediately but only after one week of cold storage (Supplementary Fig. S4). No consistent effect of acclimation on PR5 expression was observed at this stage. In potato under cold acclimation, the induction of osmotin-like proteins such as PR5 was progressive and increased 2 weeks after the treatment69. In this context, PR5 would be a marker of stress accumulation over the cold storage period. The benefit of acclimation on fruit stress status and PR5 differential expression should be observed over a longer time frame.
It is now well established that ROS are essential messengers involved in redox signalling to regulate a wide range of processes, including responses to abiotic stress70. Thus cold acclimation, with milder temperatures when compared with classic cold conditions, could have been sufficient to affect cellular redox homeostasis and stimulate the ROS processing system71. The generated signal may have entrained acclimation and improved apple stress tolerance by physiological adaptations (Fig. 5). The benefit of the acclimation in reduced scald symptoms would have been persistent until the end of the long cold storage period.
This hypothesis supports the idea that the physiological status of fruit and in particular the status of its ROS processing system before cold storage could be very critical for further superficial scald development during shelf-life. It would also explain the fruit maturity effect on scald incidence observed with H2 and H3 harvest irrespective of the year. Indeed, some studies showed that delayed harvests increased the accumulation of anti-oxidant metabolites such as α-tocopherol, carotenoids, anthocyanin in apple peel, and reduced superficial scald72. Levels of antioxidant products such as glutathione and ascorbate, and related enzyme activities, superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX) and glutathione peroxidase (GPX), were also found to increase during the ripening process of pear, peach and tomato73–75. This hypothesis is also in agreement with the theory that stress cross-tolerance is mediated through the ROS processing system and the associated regulation of gene expression through the redox signalling hub76 (Fig. 5). Therefore, it would be interesting to study the ROS processing system and its related gene network in response to pre-storage acclimation treatments and along the long cold storage of fruit.
Conclusion and Perspectives
In vivo acclimation by pre-harvest thermal variations, including a warm period of several days, as well as post-harvest acclimation by a mild-cold period significantly reduced superficial scald development. Post-harvest thermotherapy treatments, which are short intense heat shock, are currently under evaluation in order to reduce post-harvest fungal infections. The success of this strategy supposes a non-antagonistic effect on other post-harvest disorders such as superficial scald. The correlation between the induction of HSF and HSP gene expression and the limitation of scald incidence suggests that thermotherapy treatments could limit scald. A post-harvest procedure combining both thermotherapy and cold acclimation could be even more efficient in preventing scald disorders.
This study revealed that HSP gene expression could be a potential marker to predict as early as harvest the risk of scald development during subsequent cold storage. Additional stress responsive genes identified at harvest (Table 3) or during the early days of cold storage, as well as orchard temperatures would increase the reliability of such a prediction which could support decisions for post-harvest treatments and/or cold storage management.
Supplementary information
Acknowledgements
The authors thank the SFR QuaSAV and Muriel Bahut for access to the ANAN platform and the microarrays facilities, for technical assistance and advice; Dr. Marie-Noel Brisset (IRHS/UMR1345) and Dr. Matthieu Gaucher (IRHS/UMR1345) for access to qPFD primers; Pr Anis Limami and Pr Gerhard Buck-Sorlin for critical proof reading, Dr. Thomas K. Baldwin for English language editing. This research work was conducted in the framework of the “MPIA Predifruit” program supported by Angers Loire Métropole, the regional program “Objectif Végétal, Research, Education and Innovation in Pays de la Loire”, supported by the French Region Pays de la Loire, Angers Loire Métropole and the European Regional Development Fund. MM received a PhD grant from the Institut National de la Recherche Agronomique and the French Region Pays de la Loire.
Author contributions
M.O., M.D. and M.M., conceived and designed the experiments. M.M. performed experiments, with assistance of M.C., M.C.G. and A.S.P. and S.H. for fruit sampling and phenotyping. M.C. and S.P. contributed to the transcriptomic experiments and data processing. C.C. and C.T. ran the orchards and were involved in fruit sampling and phenotyping. L.F. and J.P.R. contributed to secure funding resources. M.O. and M.D. supervised MM to analyse and interpret data. M.O., M.D. and M.M. conceived and wrote the paper.
Data availability
The microarrays data are available from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE135863.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-63018-3.
References
- 1.Musacchi S, Serra S. Apple fruit quality: Overview on pre-harvest factors. Sci. Hortic. 2018;234:409–430. doi: 10.1016/j.scienta.2017.12.057. [DOI] [Google Scholar]
- 2.Lurie S, Watkins CB. Superficial scald, its etiology and control. Postharvest Biol. Technol. 2012;65:44–60. doi: 10.1016/j.postharvbio.2011.11.001. [DOI] [Google Scholar]
- 3.Whitaker, B. D. Genetic and biochimical bases of superficial scald storage disorder in apple and pear fruits. Acta Hortic. 47–60 10.17660/ActaHortic.2013.989.3 (2013).
- 4.Farneti B, et al. Untargeted metabolomics investigation of volatile compounds involved in the development of apple superficial scald by PTR-ToF–MS. Metabolomics. 2015;11:341–349. doi: 10.1007/s11306-014-0696-0. [DOI] [Google Scholar]
- 5.Whitaker BD, Nock JF, Watkins CB. Peel tissue α-farnesene and conjugated trienol concentrations during storage of ‘White Angel’x‘Rome Beauty’ hybrid apple selections susceptible and resistant to superficial scald. Postharvest Biol. Technol. 2000;20:231–241. doi: 10.1016/S0925-5214(00)00139-3. [DOI] [Google Scholar]
- 6.Sabban-Amin R, Feygenberg O, Belausov E, Pesis E. Low oxygen and 1-MCP pretreatments delay superficial scald development by reducing reactive oxygen species (ROS) accumulation in stored ‘Granny Smith’ apples. Postharvest Biol. Technol. 2011;62:295–304. doi: 10.1016/j.postharvbio.2011.06.016. [DOI] [Google Scholar]
- 7.Busatto N, et al. Apple fruit superficial scald resistance mediated by ethylene inhibition is associated with diverse metabolic processes. Plant J. 2018;93:270–285. doi: 10.1111/tpj.13774. [DOI] [PubMed] [Google Scholar]
- 8.Busatto N, et al. Target metabolite and gene transcription profiling during the development of superficial scald in apple (Malus x domestica Borkh) BMC Plant Biol. 2014;14:193. doi: 10.1186/s12870-014-0193-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karagiannis, E. et al. Ethylene –dependent and –independent superficial scald resistance mechanisms in ‘Granny Smith’ apple fruit. Sci. Rep. 8, (2018). [DOI] [PMC free article] [PubMed]
- 10.Du L, Song J, Campbell Palmer L, Fillmore S, Zhang Z. Quantitative proteomic changes in development of superficial scald disorder and its response to diphenylamine and 1-MCP treatments in apple fruit. Postharvest Biol. Technol. 2017;123:33–50. doi: 10.1016/j.postharvbio.2016.08.005. [DOI] [Google Scholar]
- 11.Gapper, N. E. et al. Delayed response to cold stress is characterized by successive metabolic shifts culminating in apple fruit peel necrosis. BMC Plant Biol. 17, (2017). [DOI] [PMC free article] [PubMed]
- 12.Leisso R, Buchanan D, Lee J, Mattheis J, Rudell D. Cell Wall, Cell Membrane, and Volatile Metabolism Are Altered by Antioxidant Treatment, Temperature Shifts, and Peel Necrosis during Apple Fruit Storage. J. Agric. Food Chem. 2013;61:1373–1387. doi: 10.1021/jf3046208. [DOI] [PubMed] [Google Scholar]
- 13.Emongor VE, Murr DP, Lougheed EC. Preharvest factors that predispose apples to superficial scald. Postharvest Biol. Technol. 1994;4:289–300. doi: 10.1016/0925-5214(94)90040-X. [DOI] [Google Scholar]
- 14.Rai, R. et al. Implications of changing climate on productivity of temperate fruit crops with special reference to apple. J. Hortic. 02, 1–6 (2015).
- 15.Thomai T, Sfakiotakis E, Diamantidis GR, Vasilakakis M. Effects of low preharvest temperature on scald susceptibility and biochemical changes in ‘Granny Smith’ apple peel. Sci. Hortic. 1998;76:1–15. doi: 10.1016/S0304-4238(98)00133-2. [DOI] [Google Scholar]
- 16.Barden CL, Bramlage WJ. Separating the effects of low temperature, ripening, and light on loss of scald susceptibility in apples before harvest. J. Am. Soc. Hortic. Sci. 1994;119:54–58. doi: 10.21273/JASHS.119.1.54. [DOI] [Google Scholar]
- 17.Moggia C, Hernández O, Pereira M, A. Lobos G, Yuri JA. Effect of the cooling system and 1-MCP on the incidence of superficial scald in ‘Granny Smith’ apples. Chil. J. Agric. Res. 2009;69:383–390. doi: 10.4067/S0718-58392009000300011. [DOI] [Google Scholar]
- 18.Janská A, Maršík P, Zelenková S, Ovesná J. Cold stress and acclimation - what is important for metabolic adjustment? Plant Biol. 2010;12:395–405. doi: 10.1111/j.1438-8677.2009.00299.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang B, Zhu S. Pre-storage cold acclimation maintained quality of cold-stored cucumber through differentially and orderly activating ROS scavengers. Postharvest Biol. Technol. 2017;129:1–8. doi: 10.1016/j.postharvbio.2017.03.001. [DOI] [Google Scholar]
- 20.Maul P, McCollum G, Guy CL, Porat R. Temperature conditioning alters transcript abundance of genes related to chilling stress in ‘Marsh’ grapefruit flavedo. Postharvest Biol. Technol. 2011;60:177–185. doi: 10.1016/j.postharvbio.2010.06.007. [DOI] [Google Scholar]
- 21.Carvajal F, Palma F, Jamilena M, Garrido D. Preconditioning treatment induces chilling tolerance in zucchini fruit improving different physiological mechanisms against cold injury: Preconditioning treatment induces chilling tolerance in zucchini fruit. Ann. Appl. Biol. 2015;166:340–354. doi: 10.1111/aab.12189. [DOI] [Google Scholar]
- 22.Mendieta B, Olaeta JA, Pedreschi R, Undurraga P. Reduction of cold damage during cold storage of Hass avocado by a combined use of pre-conditioning and waxing. Sci. Hortic. 2016;200:119–124. doi: 10.1016/j.scienta.2016.01.012. [DOI] [Google Scholar]
- 23.Jin P, et al. Low-Temperature Conditioning Alleviates Chilling Injury in Loquat Fruit and Regulates Glycine Betaine Content and Energy Status. J. Agric. Food Chem. 2015;63:3654–3659. doi: 10.1021/acs.jafc.5b00605. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, et al. Low-temperature conditioning induces chilling tolerance in stored mango fruit. Food Chem. 2017;219:76–84. doi: 10.1016/j.foodchem.2016.09.123. [DOI] [PubMed] [Google Scholar]
- 25.Yang Q, et al. Low-temperature conditioning induces chilling tolerance in ‘Hayward’ kiwifruit by enhancing antioxidant enzyme activity and regulating en-dogenous hormones levels: Low-temperature conditioning induces chilling tolerance in ‘Hayward’ kiwifruit. J. Sci. Food Agric. 2013;93:3691–3699. doi: 10.1002/jsfa.6195. [DOI] [PubMed] [Google Scholar]
- 26.Lê, S., Josse, J. & Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 25, (2008).
- 27.Segonne, S. M. et al. Multiscale investigation of mealiness in apple: an atypical role for a pectin methylesterase during fruit maturation. BMC Plant Biol. 14, (2014). [DOI] [PMC free article] [PubMed]
- 28.Velasco R, et al. The genome of the domesticated apple (Malus x domestica Borkh.) Nat. Genet. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 29.Celton J-M, et al. Widespread anti-sense transcription in apple is correlated with siRNA production and indicates a large potential for transcriptional and/or post-transcriptional control. New Phytol. 2014;203:287–299. doi: 10.1111/nph.12787. [DOI] [PubMed] [Google Scholar]
- 30.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. (2009).
- 31.Smyth, G. K. Limma: Linear Models for Microarray Data. in Bioinformatics and Computational Biology Solutions Using R and Bioconductor (eds. Gentleman, R., Carey, V. J., Huber, W., Irizarry, R. A. & Dudoit, S.) 397–420 (Springer-Verlag, 2005). 10.1007/0-387-29362-0_23.
- 32.Daccord N, et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet. 2017;49:1099–1106. doi: 10.1038/ng.3886. [DOI] [PubMed] [Google Scholar]
- 33.Lohse M, et al. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data: Mercator: sequence functional annotation server. Plant Cell Environ. 2014;37:1250–1258. doi: 10.1111/pce.12231. [DOI] [PubMed] [Google Scholar]
- 34.Usadel B, et al. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 2009;32:1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 35.Vergne E, et al. Membrane-Targeted HrpN Ea Can Modulate Apple Defense Gene Expression. Mol. Plant. Microbe Interact. 2014;27:125–135. doi: 10.1094/MPMI-10-13-0305-R. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Krishna P, Gloor G. The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones. 2001;6:238. doi: 10.1379/1466-1268(2001)006<0238:THFOPI>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G, Li J, Hao R, Guo Y. Activation of catalase activity by a peroxisome-localized small heat shock protein Hsp17.6CII. J. Genet. Genomics. 2017;44:395–404. doi: 10.1016/j.jgg.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Neta-Sharir I. Dual role for tomato heat shock protein 21: protecting photosystem II from oxidative stress and promoting color changes during fruit maturation. Plant Cell. 2005;17:1829–1838. doi: 10.1105/tpc.105.031914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Queitsch C, Hong S-W, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishizawa A, et al. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006;48:535–547. doi: 10.1111/j.1365-313X.2006.02889.x. [DOI] [PubMed] [Google Scholar]
- 42.An J-P, et al. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017;4:17023. doi: 10.1038/hortres.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang F, et al. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genomics. 2016;291:1545–1559. doi: 10.1007/s00438-016-1203-2. [DOI] [PubMed] [Google Scholar]
- 44.Davletova S. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in arabidopsis. PLANT Physiol. 2005;139:847–856. doi: 10.1104/pp.105.068254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis: Arabidopsis low temperature transcriptome. Plant J. 2004;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- 46.Brisset, M.-N. & Dugé De Bernonville, T. Device for determining or studying the state of stimulation of the natural defences of plants or portions of plants. (2011).
- 47.Ferguson I, Snelgar W, Lay-Yee M, Watkins C, Bowen J. Expression of heat shock protein genes in apple fruit in the field. Funct. Plant Biol. 1998;25:155–163. doi: 10.1071/PP97093. [DOI] [Google Scholar]
- 48.Giorno F, Guerriero G, Baric S, Mariani C. Heat shock transcriptional factors in Malus domestica: identification, classification and expression analysis. BMC Genomics. 2012;13:639. doi: 10.1186/1471-2164-13-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson JV, Li QB, Haskell DW, Guy CL. Structural organization of the spinach endoplasmic reticulum-luminal 70-kilodalton heat-shock cognate gene and expression of 70-kilodalton heat-shock genes during cold acclimation. Plant Physiol. 1994;104:1359. doi: 10.1104/pp.104.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sung DY, Vierling E, Guy CL. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001;126:789–800. doi: 10.1104/pp.126.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Timperio AM, Egidi MG, Zolla L. Proteomics applied on plant abiotic stresses: Role of heat shock proteins (HSP) J. Proteomics. 2008;71:391–411. doi: 10.1016/j.jprot.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 52.Al-Whaibi MH. Plant heat-shock proteins: A mini review. J. King Saud Univ. - Sci. 2011;23:139–150. doi: 10.1016/j.jksus.2010.06.022. [DOI] [Google Scholar]
- 53.Downs CA, Ryan SL, Heckathorn SA. The chloroplast small heat-shock protein: Evidence for a general role in protecting photosystem II against oxidative stress and photoinhibition. J. Plant Physiol. 1999;155:488–496. doi: 10.1016/S0176-1617(99)80043-1. [DOI] [Google Scholar]
- 54.Guo S-J, Zhou H-Y, Zhang X-S, Li X-G, Meng Q-W. Overexpression of CaHSP26 in transgenic tobacco alleviates photoinhibition of PSII and PSI during chilling stress under low irradiance. J. Plant Physiol. 2007;164:126–136. doi: 10.1016/j.jplph.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 55.Qi C, et al. SoHSC70 positively regulates thermotolerance by alleviating cell membrane damage, reducing ROS accumulation, and improving activities of antioxidant enzymes. Plant Sci. 2019;283:385–395. doi: 10.1016/j.plantsci.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Foyer CH, Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 2011;155:2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishizawa A, Yabuta Y, Shigeoka S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008;147:1251–1263. doi: 10.1104/pp.108.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu, Y. et al. Tomato expressing Arabidopsis glutaredoxin gene AtGRXS17 confers tolerance to chilling stress via modulating cold responsive components. Hortic. Res. 2, (2015). [DOI] [PMC free article] [PubMed]
- 59.Edwards R, Dixon DP, Walbot V. Plant glutathione S -transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000;5:193–198. doi: 10.1016/S1360-1385(00)01601-0. [DOI] [PubMed] [Google Scholar]
- 60.Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.2307/3870059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christie PJ, Alfenito MR, Walbot V. Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: Enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta. 1994;194:541–549. doi: 10.1007/BF00714468. [DOI] [Google Scholar]
- 62.Martifnez-Tellez M, Lafuente M. Chilling-induced changes in phenylalanine amminia-lyase, peroxidase, and polyphenol oxidase activities in citrus flavedo tissue. Acta Hortic. 1993;343:257–263. doi: 10.17660/ActaHortic.1993.343.58. [DOI] [Google Scholar]
- 63.Rivero RM, et al. Resistance to cold and heat stress: accumulation of phenolic compounds in tomato and watermelon plants. Plant Sci. 2001;160:315–321. doi: 10.1016/S0168-9452(00)00395-2. [DOI] [PubMed] [Google Scholar]
- 64.Sanchez-Ballesta MT, Lafuente MT, Zacarias L, Granell A. Involvement of phenylalanine ammonia-lyase in the response of Fortune mandarin fruits to cold temperature. Physiol. Plant. 2000;108:382–389. doi: 10.1034/j.1399-3054.2000.108004382.x. [DOI] [Google Scholar]
- 65.Tan S. Phenylalanine ammonia-lyase and the phenylalanine ammonia-lyase inactivating system: Effects of light, temperature and mineral deficiencies. Funct. Plant Biol. 1980;7:159–167. doi: 10.1071/PP9800159. [DOI] [Google Scholar]
- 66.Hakim, et al. Osmotin: A plant defense tool against biotic and abiotic stresses. Plant Physiol. Biochem. 2018;123:149–159. doi: 10.1016/j.plaphy.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 67.Hong, Z., Lakkineni, K., Zhang, Z. & Verma, D. P. S. Removal of feedback inhibition of 1-Pyrroline-5- Carboxylate Synthetase results in increased proline accumulation and protection of plants from osmotic stress. 122, 8 (2000). [DOI] [PMC free article] [PubMed]
- 68.Subramanyam K, Sailaja KV, Subramanyam K, Muralidhara Rao D, Lakshmidevi K. Ectopic expression of an osmotin gene leads to enhanced salt tolerance in transgenic chilli pepper (Capsicum annum L.) Plant Cell Tissue Organ Cult. PCTOC. 2011;105:181–192. doi: 10.1007/s11240-010-9850-1. [DOI] [Google Scholar]
- 69.Zhu B, Chen THH, Li PH. Activation of two osmotin-like protein genes by abiotic stimuli and fungal pathogen in transgenic potato plants. Plant Physiol. 1995;108:929. doi: 10.1104/pp.108.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baxter A, Mittler R, Suzuki N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014;65:1229–1240. doi: 10.1093/jxb/ert375. [DOI] [PubMed] [Google Scholar]
- 71.Noctor G, Reichheld J-P, Foyer CH. ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. 2018;80:3–12. doi: 10.1016/j.semcdb.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 72.Ju Z, Yuan Y, Liu C, Zhan S, Wang M. Relationships among simple phenol, flavonoid and anthocyanin in apple fruit peel at harvest and scald susceptibility. Postharvest Biol. Technol. 1996;8:83–93. doi: 10.1016/0925-5214(95)00062-3. [DOI] [Google Scholar]
- 73.Huan C, et al. Potential role of reactive oxygen species and antioxidant genes in the regulation of peach fruit development and ripening. Plant Physiol. Biochem. 2016;104:294–303. doi: 10.1016/j.plaphy.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 74.Jimenez A, et al. Changes in oxidative processes and components of the antioxidant system during tomato fruit ripening. Planta. 2002;214:751–758. doi: 10.1007/s004250100667. [DOI] [PubMed] [Google Scholar]
- 75.Lentheric I, Pinto E, Vendrell M, Larrigaudiere C. Harvest date affects the antioxidative systems in pear fruits. J. Hortic. Sci. Biotechnol. 1999;74:791–795. doi: 10.1080/14620316.1999.11511190. [DOI] [Google Scholar]
- 76.Bartoli CG, Casalongué CA, Simontacchi M, Marquez-Garcia B, Foyer CH. Interactions between hormone and redox signalling pathways in the control of growth and cross tolerance to stress. Environ. Exp. Bot. 2013;94:73–88. doi: 10.1016/j.envexpbot.2012.05.003. [DOI] [Google Scholar]
- 77.Swindell WR, Huebner M, Weber AP. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics. 2007;8:125–125. doi: 10.1186/1471-2164-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarrays data are available from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE135863.