Abstract
We investigated fosfomycin susceptibility in Escherichia coli clinical isolates from South Korea, including community-onset, hospital-onset, and long-term care facility (LTCF)-onset isolates. The resistance mechanisms and genotypes of fosfomycin-resistant isolates were also identified. Finally, the in vitro efficacy of combinations of fosfomycin with other antibiotics were examined in susceptible or extended spectrum β-lactamase (ESBL)-producing E. coli isolates. The fosfomycin resistance rate was 6.7% and was significantly higher in LTCF-onset isolates than community-onset and hospital-onset isolates. Twenty-one sequence types (STs) were identified among 19 fosfomycin-resistant E. coli isolates, showing diverse genotypes. fosA3 was found in only two isolates, and diverse genetic variations were identified in three genes associated with fosfomycin resistance, namely, GlpT, UhpT, and MurA. Some fosfomycin-resistant E. coli isolates carried no mutations. In vitro time-kill assays showed that fosfomycin alone did not exhibit an excellent killing activity, compared with ciprofloxacin in susceptible isolates and with ertapenem in ESBL producers. However, combining fosfomycin with cefixime or piperacillin-tazobactam eradicated susceptible or ESBL-producing isolates, respectively, even with 0.5× minimum inhibitory concentrations. Overall, we found a relatively high fosfomycin resistance rate in E. coli isolates from South Korea. Based on their genotypes and resistance mechanisms, most of the fosfomycin-resistant E. coli isolates might occur independently. Antibiotic combinations with fosfomycin could be a suitable therapeutic option for infections caused by E. coli isolates.
Keywords: fosfomycin, in vitro time-kill, cefixime, piperacillin-tazobactam
1. Introduction
Escherichia coli is one of the most common pathogens in community-acquired and nosocomial infections, including urinary tract infections (UTIs), biliary tract infections, and complicated intraabdominal infections. Indeed, one in every three adult women suffers from UTIs and about 50% of UTIs are caused by E. coli [1,2]. Although E. coli is intrinsically susceptible to many antimicrobial agents, antimicrobial resistance has been increasingly reported due to extended-spectrum β-lactamases (ESBLs), carbapenemases, plasmid-mediated quinolone resistance, and mcr genes causing colistin resistance [3]. The increase of antimicrobial resistance in E. coli leads to the reduction of usable therapeutic agents and prolongs the length of hospital stay due to the absence of effective oral antibiotics.
Fosfomycin, an old antimicrobial drug, has resurfaced as a therapeutic option for multidrug-resistant (MDR) gram-negative bacilli [4]. Its mechanism of action is to inhibit the formation of the peptidoglycan precursor UDP N-acetylmuramic acid (UDP-MurNAc) in the bacterial cell wall biosynthesis [5]. Since fosfomycin is structurally unrelated to any other antimicrobial agent, there is a small chance of cross-resistance [6]. Additionally, it has a broad-spectrum activity against both gram-negative and gram-positive bacteria, with limited side effects [7,8]. It is known that E. coli has two main nutrient transport systems essential for fosfomycin uptake: the glycerol-3-phosphate transporter (GlpT) and a hexose phosphate transporter, known as the glucose-6-phosphate transporter (UhpT) [9]. Thus, the key fosfomycin resistance mechanisms involve reduced permeability related to GlpT and UhpT, and target modification related to MurA. In addition, drug inactivation can be caused by the acquisition of fos genes mostly by plasmid, resulting in fosfomycin resistance [6,8,10].
The current resistance rate of E. coli to fosfomycin is estimated to be lower than 5%, and lower than 10% among extended spectrum β-lactamase (ESBL) producers worldwide [11,12,13]. In South Korea, the fosfomycin resistance rate has been reported to be up to 3% in clinical E. coli isolates, and up to 7% among ESBL producers [14,15,16,17]. However, only a few studies have explored the mechanisms of fosfomycin resistance and the genotypes of fosfomycin-resistant E. coli isolates in South Korea.
In this study, we investigated the antimicrobial susceptibility of different E. coli clinical isolates from South Korea, including community-onset, hospital-onset, and long-term care facility (LTCF)-onset isolates. We also identified the resistance mechanisms and genotypes of fosfomycin-resistant isolates. In addition, the in vitro efficacy of combinations of fosfomycin with other antibiotics were examined in E. coli isolates, including ESBL producers. Although combination therapy based on fosfomycin is not commonly used, we explored the possibilities of combination therapy, especially using oral antibiotics such as ciprofloxacin and cefixime.
2. Results
Among the 283 E. coli clinical isolates from South Korea, 19 isolates (6.7%) were resistant to fosfomycin based on the cut-off minimum inhibition concentration (MIC) (Table 1). The fosfomycin resistance rates were higher in LTCF-onset isolates (10.0%) than in community-onset and hospital-onset isolates (6.0% and 6.9%, respectively), but their difference was not statistically significant. The difference of fosfomycin resistant rates between hospitals was also not statistically significant, although it was significantly lower in urinary tract infection (UTI) isolates than in non-UTI isolates (P, 0.042) (Table 1).
Table 1.
Antibiotics susceptibilities of all E. coli isolates in this study.
| Antimicrobial Agents | Number of Resistant Isolates (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total (n = 283) |
Mode of Acquisition | Site of Infection c | Facility e | |||||||||
| Community-onset (n = 150) |
Hospital-onset (n = 102) |
LTCF b-onset (n = 31) |
P | UTI d (n = 129) |
Non-UTI (n = 154) |
P | SMC (n = 114) |
SCH (n = 107) |
KUAH (n = 62) |
P | ||
| Fosfomycin | 19 (6.7) | 9 (6.0) | 7 (6.9) | 3 (10.0) | 0.384 | 5 (3.9) | 14 (9.1) | 0.042 | 4 (3.5) | 9 (8.4) | 6 (9.7) | 0.054 |
| Ciprofloxacin | 181 (64.0) | 72 (48.0) | 70 (68.6) | 22 (71.0) | 0.285 | 70 (72.9) | 111 (59.4) | 0.023 | 67 (58.8) | 62 (57.9) | 52 (83.9) | 0.008 |
| Cefepime | 117 (41.3) | 48 (32.0) | 55 (53.9) | 14 (45.2) | 0.008 | 45 (34.9) | 72 (46.8) | 0.115 | 45 (39.5) | 40 (37.4) | 32 (51.6) | 0.022 |
| Cefixime | 131 (46.3) | 58 (38.7) | 57 (55.9) | 16 (51.6) | 0.033 | 54 (48.9) | 77 (50.0) | 0.363 | 48 (42.1) | 45 (42.1) | 38 (61.3) | 0.062 |
| P/T a | 88 (31.1) | 38 (25.4) | 42 (41.2) | 8 (25.8) | 0.025 | 42 (32.6) | 46 (29.9) | 0.813 | 49 (43.0) | 34 (31.8) | 5 (8.1) | <0.001 |
| Amikacin | 6 (2.1) | 0 | 4 (3.9) | 2 (6.5) | 0.017 | 4 (3.1) | 2 (1.3) | 0.534 | 5 (4.4) | 0 | 1 (1.6) | 0.116 |
| Ertapenem | 7 (2.5) | 1 (0.7) | 6 (5.9) | 0 | 0.012 | 4 (3.1) | 3 (1.9) | 0.880 | 1 (0.9) | 3 (2.8) | 3 (4.8) | 0.244 |
| Colistin | 30 (10.6) | 12 (8.0) | 15 (14.7) | 3 (9.7) | 0.219 | 18 (14.0) | 12 (7.8) | 0.121 | 20 (17.5) | 7 (6.5) | 3 (4.8) | 0.007 |
| Tigecycline | 3 (1.1) | 0 | 2 (2.0) | 1 (3.2) | 0.075 | 2 (1.5) | 1 (0.6) | 0.593 | 0 | 0 | 2 (3.2) | 0.117 |
a P/T, piperacillin/tazobactam. b LTCF, long-term care facility. c All patients accompanied by E. coli infection. d UTI, urinary tract infection. e SMC, Samsung Medical Center; SCH, Samsung Changwon Hospital; KUAH, Korea University Ansan Hospital.
The ciprofloxacin resistance rate was very high (64.0% in all isolates), especially in UTI isolates and isolates from KUAH (Table 1). Isolates resistant to cephalosporin (cefepime and cefixime) were found more frequently in hospital- and LTCF-onset isolates. The piperacillin–tazobactam resistance rate was higher in isolates from SMC and SCH, unlike fosfomycin, ciprofloxacin, and cefepime resistance rates. The amikacin resistance rate was very low (2.1%), and only seven ertapenem-resistant isolates were identified (2.5%). The colistin resistance rate was 10.6%. While the colistin resistance rate did not differ in the mode of acquisition and site of infection, it was significantly higher in isolates from SMC (17.5%) compared to those from the other two hospitals (6.5% in SCH and 4.8% in KUAH, respectively). Only three isolates were resistant to tigecycline. Multidrug resistance (MDR), defined as resistance to ≥3 antibiotic classes, was identified in 116 isolates (41.0%).
Using the MLST analysis, the genotypes of 19 fosfomycin-resistant E. coli isolates were identified (Table 2). A total of 14 STs were identified. Only three of these STs were found in multiple isolates: ST1193 in four isolates, while ST131 and ST1531-slv in two isolates each. Eight STs were newly identified, and they were defined as slv or dlv of the most closely related ST. The largest CCs were CC131 and CC14. All CC131 isolates except one (C072) were MDR. The fosfomycin resistance rates were similar between isolates from blood (16/243 isolates, 6.6%) and urine (3/40 isolates, 7.5%).
Table 2.
Characteristics of 29 fosfomycin-resistant E. coli isolates: genotype, clinical characteristics, and amino acid alterations in genes associated with fosfomycin resistance.
| Isolate No. | CC a | ST a | Allele no. b | Specimen | Site of Infection c |
Mode of Acquisition d | Amino Acid Alterations | |||
|---|---|---|---|---|---|---|---|---|---|---|
| fos | GlpT | UhpT | MurA | |||||||
| S020 | CC131 | ST131 | 53-40-47-13-36-28-29 | Blood | IAI | Hospital | ||||
| S074 | ST131 | 53-40-47-13-36-28-29 | Blood | UTI | Hospital | D220N | ||||
| C072 | ST131-slv1 | 53-40-193-13-36-28-29 | Blood | Cholangitis | Community | |||||
| C073 | ST131-slv2 | 53-40-47-200-36-28-29 | Blood | UTI | LTCF | |||||
| A011 | ST131-dlv1 | 53-35-47-13-36-5-29 | Blood | IAI | Hospital | |||||
| C025 | CC14 | ST1193 | 14-14-10-200-17-7-10 | Urine | UTI | Community | ||||
| C036 | ST1193 | 14-14-10-200-17-7-10 | Blood | Cholangitis | Community | G168R | ||||
| A049 | ST1193 | 14-14-10-200-17-7-10 | Blood | UTI | Community | M136K | ||||
| S019 | ST1193 | 14-14-10-200-17-7-10 | Blood | Prostatitis | LTCF | |||||
| C078 | ST1193-slv | 14-40-10-200-17-7-10 | Blood | UTI | Community | |||||
| C106 | CC69 | ST106 | 21-38-27-6-5-8-4 | Blood | UTI | Community | ||||
| A004 | ST106-dlv | 21-88-27-6-5-79-4 | Blood | UTI | Community | fosA3 | ||||
| A031 | CC95 | ST1531-slv | 37-35-19-37-17-5-26 | Blood | UTI | Community | A16T | |||
| A041 | ST1531-slv | 37-35-19-37-17-5-26 | Blood | UTI | LTCF | A16T | ||||
| C011 | CC155 | ST58 | 64-4-16-24-8-14 | Blood | Liver abscess | Community | Y60F | |||
| C045 | CC38 | ST38 | 4-26-2-25-5-5-19 | Urine | UTI | Hospital | Ins. of DG139 | |||
| C063 | CC10 | ST10 | 10-11-4-8-8-8-2 | Urine | UTI | Hospital | fosA3 | G168R | P99S | |
| A043 | CC398 | ST398-slv | 64-40-1-1-8-8-6 | Blood | Cholangitis | Hospital | A154T | |||
| S050 | CC95 | ST95-slv | 37-38-34-37-17-11-26 | Blood | NF | Hospital | A16T | |||
a CC, clonal complex; ST, sequence type. b adk-fumc-gyrB-icd-mdh-purA-recA, slv, single-locus variant; dlv, double-locus variant. The allele number different from the most closely related ST was underlined. c UTI, urinary tract infection; IAI, intraabdominal infection; NF, neutropenic fever. d LTCF, long-term care facility.
As for fosfomycin resistance mechanisms, fosA3 was identified in only two isolates (A004 and C063) (Table 2). Amino acid substitutions or insertions in GlpT, UhpT, and MurA were found in eight, one, and two fosfomycin-resistant isolates, respectively. Only one mutation, A16T in GlpT was identified in multiple fosfomycin-resistant E. coli isolates belonging to the same genotype (ST1531-slv). Only one isolate (C063) contained both fosA3 and amino acid alterations in GlpT and MurA. Neither fosA3 nor amino acid alterations in three genes were identified in eight fosfomycin-resistant isolates.
To investigate the antimicrobial effects of fosfomycin and other antibiotics with in vitro time-kill assays, we selected four E. coli isolates susceptible to fosfomycin, two susceptible to ciprofloxacin and cefixime, and two resistant to ciprofloxacin and cefixime and producing ESBLs (Table 3). These belonged to different STs, namely, ST216, ST144-slv, ST131, and ST1193. Two isolates producing ESBLs were identified (A038 and C047), producing CTX-M-15 and CTX-M-14, respectively.
Table 3.
Characteristics of E. coli isolates selected for time-kill assays.
| Isolate No. | ST a | ESBL Type b | MIC (mg/L) c | Site of Infection d | Mode of Acquisition | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOS | CIP | CFM | CFX | P/T | AMK | ETM | COL | TGC | |||||
| C093 | ST216 | - | 4 | 0.06 | 0.06 | 0.25 | 16/4 | 4 | 0.06 | 1 | 2 | Cholangitis | Community |
| S088 | ST144-slv | - | 8 | 0.06 | 0.06 | 0.06 | 1/4 | 4 | 0.06 | 0.5 | 1 | Cholangitis | Community |
| A038 | ST131 | CTX-M-15 | 8 | >64 | >64 | >64 | 16/4 | 4 | 0.06 | 1 | 4 | UTI | Community |
| C046 | ST1193 | CTX-M-14 | 16 | >64 | >64 | >64 | 4/4 | 4 | 0.06 | 1 | 1 | UTI | Community |
| Control Strains | |||||||||||||
| E. coli ATCC 25922 | 0.5 | 0.06 | 0.06 | 0.5 | 2/4 | 1 | 0.06 | 0.5 | 0.06 | ||||
| P. aeruginosa ATCC 27853 | 2 | 0.25 | 2 | NAe | 4/4 | 1 | 0.5 | 0.5 | NA | ||||
a ST, sequence type. b ESBL, extended-spectrum-β-lactamase. c MIC, minimum inhibitory concentration; FOS, fosfomycin; CIP, ciprofloxacin; CFM, cefepime; CFX, cefixime; P/T, piperacillin-tazobactam; AMK, amikacin; ETM, ertapenem; COL, colistin; TGC, tigecycline. d UTI, urinary tract infection. e NA, not available.
While 1× and 0.5× MICs of cefixime and fosfomycin did not eradicate both susceptible E. coli isolates (C093 and S088), 1× MIC of ciprofloxacin showed complete killing efficacy after 4 or 8 h (Figure 1A,B). Using 0.5× MIC of ciprofloxacin also killed the isolates, but the killing efficacy was lower than with 1× MIC. The combination of 0.5× MICs of fosfomycin and other antibiotics (ciprofloxacin or cefixime) eradicated the isolates (Figure 1C,D). Interestingly, the combination of 0.5× MICs of fosfomycin and cefixime showed a synergistic killing effect; while single regimens of each antibiotic did not kill the susceptible isolates, using a combination of the two completely eradicated the isolates.
Figure 1.
Time-kill curves for ciprofloxacin, cefixime, and fosfomycin against susceptible E. coli isolates, C093 (ST216) and S088 (ST144-slv). (A and B), the results of 1× MICs of single antibiotics, (C and D), the results of 0.5× MICs of single and combination of antibiotics. CIP, ciprofloxacin; CFX, cefixime; FOS, fosfomycin.
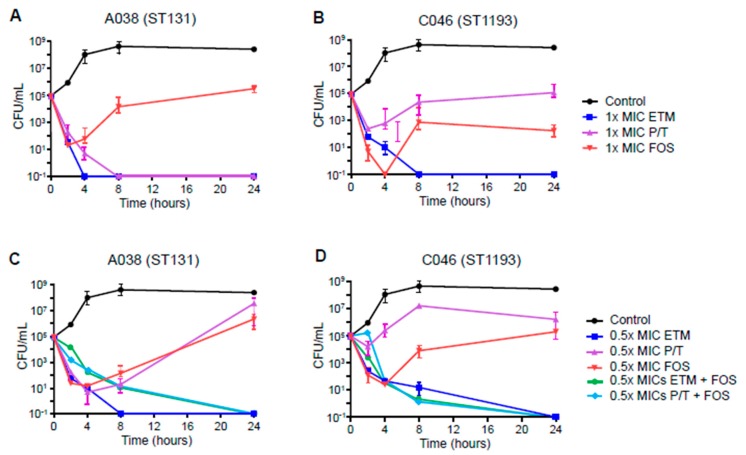
For the two ESBL-producing E. coli isolates, ertapenem and piperacillin-tazobactam were also tested in addition to fosfomycin (Figure 2). While 1× MIC of ertapenem completely killed both ESBL-producing isolates, 1× MIC of fosfomycin decreased the growth of both isolates within 2 or 4 h, after which they started growing again (Figure 2A,B). In contrast, 1× MIC of piperacillin-tazobactam produced different results between the two ESBL-producing isolates. Although the combinations of 0.5× MICs of fosfomycin with ertapenem or piperacillin-tazobactam completely eradicated both ESBL-producing isolates after 24 h, 0.5x MIC of ertapenem and fosfomycin also completely killed them after 4 h (Figure 2C,D). The combinations of 0.5× MICs of fosfomycin and ertapenem or piperacillin-tazobactam completely eradicated both ESBL-producing isolates after 24 h.
Figure 2.
Time-kill curves for ertapenem, piperacillin-tazobactam, and fosfomycin against ESBL-producing E. coli isolates, CTX-M-15-producing A038 (ST131) and CTX-M-14-producing C046 (ST1193). (A and B), the results of 1× MICs of single antibiotics, (C and D), the results of 0.5× MICs of single and combination of antibiotics. ETM, ertapenem; P/T, piperacillin-tazobactam; FOS, fosfomycin.
3. Discussion
In this study, the fosfomycin resistance rate was estimated to be 6.7%, which is somewhat higher than that previously reported in South Korea [14,15,16,17]. This study identified different genotypes among 19 fosfomycin-resistant E. coli isolates, suggesting that most of these isolates might emerge sporadically. The most prevalent clonal group in these isolates was CC131 and CC14. It is known that ST131 is tightly associated with the production of the CTX-M-type ESBL and with fluoroquinolone resistance, and is therefore commonly acknowledged as a significant threat to public health [18,19,20]. The fosfomycin-resistant E. coli CC131 isolates in this study were all MDR, except for one. Since the genetic alterations of fosfomycin resistance-associated genes were not identical among the CC131 isolates, it does not appear that all fosfomycin resistant strains belonging to CC131 have spread clonally. In addition, ST1193, found in four fosfomycin-resistant isolates, may not be spread clonally. Although it is still unclear whether fosfomycin resistance preferably occurs in certain genetic backgrounds, it is worth investigating whether ST131 or ST1193 increase the risk of developing fosfomycin resistance.
We investigated the presence of genetic mutations in several genes (fosA, fosC, glpT, uhpT, and murA) that are commonly associated with fosfomycin resistance [6,21]. Diverse genetic alterations in these genes were found, including the insertion of two amino acids in GlpT. This supports the idea that fosfomycin resistance in E. coli isolates might occur independently between each other. Additionally, the fosfomycin-modifying enzyme FosA3, a metalloenzyme acquired through plasmid-transfer [9], was identified in only two isolates, although the fosA gene has been reported to be prevalent in Asia [22]. This suggests that the horizontal transfer of fosfomycin resistance genes through mobile elements is not common in South Korea. However, no genetic alterations in GlpT, UhpT, and MurA were found in many isolates, particularly isolates belonging to CC131. Further fosfomycin resistance mutations reported in other studies, such as those in cyaA and ptsI resulting in lower cAMP levels and downregulation of fosfomycin transporters [23,24], may be associated to some fosfomycin-resistant E. coli isolates. Further unknown mechanisms might also mediate fosfomycin resistance in E. coli.
Some E. coli studies have shown a synergistic effect of fosfomycin in combination with other antibiotics, such as carbapenems, colistin, aztreonam, netilmicin, and tigecycline, against ESBL-producing strains [9]. In this study, the activity of antibiotic combinations based on fosfomycin were evaluated by in vitro time-kill assay using multiple E. coli isolates. Different antibiotic combinations were tested according to antibiotic susceptibility. For both antibiotic susceptible and ESBL-producing E. coli isolates, fosfomycin alone did not show lower killing activity than other antibiotics, such as ciprofloxacin against susceptible isolates and ertapenem against ESBL-producers. However, the combination of fosfomycin with other antibiotics, including cefixime and piperacillin-tazobactam, achieved complete bactericidal effects against susceptible isolates and ESBL-producing isolates, respectively, even using 0.5× MIC. This suggests that antibiotic combinations of fosfomycin and other antibiotics, even at low concentrations, could be a potential treatment option for infections caused by E. coli.
This study investigated fosfomycin resistance in clinical E. coli isolates from South Korea. The fosfomycin resistance rate was 6.7%, and the rates were significantly lower in UTI isolates. The fosfomycin-resistant isolates showed diverse genotypes and genetic variations in genes associated with fosfomycin resistance, indicating sporadic emergence of fosfomycin resistance in South Korea. Although fosfomycin monotherapy was not superior to other antibiotics in its killing activity against susceptible and ESBL-producing E. coli isolates, its combination with other antibiotics, even at low concentrations, resulted in a synergistic effect.
4. Materials and Methods
4.1. Bacterial Isolates
A total of 283 nonduplicated E. coli clinical isolates (243 from blood and 40 from urine) were collected from patients from January to June 2018 from three tertiary-care hospitals in South Korea: the Samsung Medical Center (SMC, 114 isolates), the Samsung Changwon Hospital (SCH, 107 isolates), and the Korea University Ansan Hospital (KUAH, 62 isolates). Species identification was performed using a VITEK-2 system (BioMérieux, Hazelwood, MO, USA).
4.2. Antimicrobial Susceptibility Testing
In vitro antimicrobial susceptibility testing was performed in all E. coli isolates according to the Clinical and Laboratory Standard Institute (CLSI) guidelines [25]. An agar dilution method with glucose-6-phophate was used for fosfomycin, and a broth microdilution method was applied for the other eight antimicrobial agents that were tested: ciprofloxacin, cefixime, cefepime, ertapenem, piperacillin-tazobactam, amikacin, colistin, and tigecycline. E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as control strains. The antimicrobial susceptibility testing was performed in duplicate.
4.3. Genotyping
Multilocus sequence typing (MLST) analysis was performed to determine the genotypes of fosfomycin-resistant E. coli isolates as previously described [26]. The sequence type (ST) and clonal complex (CC) were determined based on the database available at https://pubmlst.org/escherichia/, and those not matching exactly with STs assigned in the database were designated as single-, double-, and triple-locus variants (slv, dlv, and tlv) of the most closely related STs.
4.4. Fosfomycin Resistance Mechanisms
Polymerase chain reaction (PCR) and sequencing were performed for fosfomycin-resistant E. coli isolates to identify the following genes associated with fosfomycin resistance: murA, glpT, uhpT, fosA3, and fosC2. The primers used for amplification and sequencing were based on previous studies [13,27,28].
4.5. In vitro Time-Kill Assays
The activity of fosfomycin and other antibiotics was evaluated by time-kill assays using an inoculum of 1 × 106 CFU/mL of E. coli. Fosfomycin, ciprofloxacin, and cefixime were evaluated against two ESBL-nonproducing E. coli isolates (C093 and S088), while fosfomycin, ertapenem, and piperacillin-tazobactam were evaluated against ESBL-producing isolates (A038 and C046). ESBL production was identified using a double-disc synergy test and was confirmed by PCR for blaCTX-M [29]. Antibiotic dilutions were prepared in 10 ml of PBS adjusted to final concentrations of 0.5× and 1× the MIC of the test strains. Efficacy of the combination of fosfomycin and other antibiotics was evaluated using concentrations of 0.5× MICs. PBS without antibiotics served as a growth control; PBS without E. coli served as a negative control. Bacterial growth was quantified after 0, 2, 4, 8, and 24 h incubation at 37 °C by plating 10-fold dilutions on sheep blood agar. Colony forming unit (CFU) counts were determined for killing curves of antibiotics. Antimicrobials were considered bactericidal when a ≥ 3 log10 decrease in CFU/mL was reached compared with the initial inoculate.
4.6. Definition of Infection Onset
An infection occurring in the community or up to 48 h after hospital admission was defined as community-onset, whereas one occurring more than 48 h after admission was defined as hospital-onset. Patients who were referred from LTCF were considered as LTCF-onset. The site of infection was classified as urinary tract or non-urinary tract because of the heterogeneity of infection sites.
4.7. Statistical Analysis
To compare the two groups, Pearson χ² tests and Fisher’s exact tests were used for categorical variables, and student’s t-test and Mann–Whitney U tests were used for continuous variables where appropriate. All statistical tests were two-tailed, and P values ≤ 0.05 were considered statistically significant.
Acknowledgments
Some Escherichia coli isolates used in this study were obtained from the Asian Bacterial Bank (ABB) of the Asia Pacific Foundation for Infectious Diseases (APFID) (Seoul, South Korea).
Author Contributions
Conceptualization, H.S., K.R.P. and K.S.K.; methodology, J.Y.C. and K.S.K.; software, Y.M.W.; validation, Y.M.W., D.W.P. and K.R.P.; formal analysis, H.S. and J.Y.C.; investigation, H.S. and J.Y.C.; resources, H.S., Y.M.W. and D.W.P.; data curation, K.R.P. and K.S.K.; writing—original draft preparation, H.S.; writing—review and editing, K.R.P. and K.S.K.; visualization, H.S. and J.Y.C.; supervision, K.R.P. and K.S.K.; project administration, K.S.K.; funding acquisition, K.S.K. All authors have contributed equally to the writing and editing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (grant no. 2018R1D1A1B07049433).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bouchillon S.K., Badal R.E., Hoban D.J., Hawser S.P. Antimicrobial susceptibility of inpatient urinary tract isolates of gram-negative bacilli in the United States: Results from the study for monitoring antimicrobial resistance trends (SMART) program: 2009–2011. Clin. Ther. 2013;35:872–877. doi: 10.1016/j.clinthera.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 2.Foxman B. Epidemiology of urinary tract infections: Incidence, morbidity, and economic costs. Am. J. Med. 2002;113:5–13. doi: 10.1016/S0002-9343(02)01054-9. [DOI] [PubMed] [Google Scholar]
- 3.Vila J., Saez-Lopez E., Johnson J.R., Römling U., Dobrindt U., Cantón R., Giske C.G., Naas T., Carattoli A., Martínez-Medina M., et al. Escherichia coli: An old friend with new tidings. FEMS Microbiol. Rev. 2016;40:437–463. doi: 10.1093/femsre/fuw005. [DOI] [PubMed] [Google Scholar]
- 4.Giske C.G. Contemporary resistance trends and mechanisms for the old antibiotics colistin, temocillin, fosfomycin, mecillinam and nitrofurantoin. Clin. Microbiol. Infect. 2015;21:899–905. doi: 10.1016/j.cmi.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 5.Kahan F.M., Kahan J.S., Cassidy P.J., Kropp H. The mechanism of action of fosfomycin (phosphonomycin) Ann. N. Y. Acad. Sci. 1974;235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 6.Sastry S., Doi Y. Fosfomycin: Resurgence of an old companion. J. Infect. Chemother. 2016;22:273–280. doi: 10.1016/j.jiac.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falagas M.E., Giannopoulou K.P., Kokolakis G.N., Rafailidis P.I. Fosfomycin: Use beyond urinary tract and gastrointestinal infections. Clin. Infect. Dis. 2008;46:1069–1077. doi: 10.1086/527442. [DOI] [PubMed] [Google Scholar]
- 8.Castaneda-Garcia A., Blazquez J., Rodriguez-Rojas A. Molecular Mechanisms and Clinical Impact of Acquired and Intrinsic Fosfomycin Resistance. Antibiotics (Basel) 2013;2:217–236. doi: 10.3390/antibiotics2020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falagas M.E., Athanasaki F.A., Voulgaris G.L., Triarides N.A., Vardakas K.Z. Resistance to fosfomycin: Mechanisms, frequency and clinical consequences. Int. J. Antimicrob. Agents. 2019;53:22–28. doi: 10.1016/j.ijantimicag.2018.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Takahata S., Ida T., Hiraishi T., Sakakibara S., Maebashi K., Terada S., Muratani T., Matsumoto T., Nakahama C., Tomono K. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents. 2010;35:333–337. doi: 10.1016/j.ijantimicag.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Kresken M., Pfeifer Y., Hafner D., Wresch R., Korber-Irrgang B. Occurrence of multidrug resistance to oral antibiotics among Escherichia coli urine isolates from outpatient departments in Germany: Extended-spectrum beta-lactamases and the role of fosfomycin. Int. J. Antimicrob. Agents. 2014;44:295–300. doi: 10.1016/j.ijantimicag.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 12.Mendes A.C., Rodrigues C., Pires J., Amorim J., Ramos M.H., Novais Â., Peixe L. Importation of fosfomycin resistance fosA3 gene to Europe. Emerg. Infect. Dis. 2016;22:346–348. doi: 10.3201/eid2202.151301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y., Zheng B., Li Y., Zhu S., Xue F., Liu J. Antimicrobial susceptibility and molecular mechanisms of fosfomycin resistance in clinical Escherichia coli isolates in mainland China. PLoS ONE. 2015;10:e0135269. doi: 10.1371/journal.pone.0135269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko K.S., Suh J.Y., Peck K.R., Lee M.Y., Oh W.S., Kwon K.T., Jung D.S., Lee N.Y., Song J.H. In vitro activity of fosfomycin against ciprofloxacin-resistant or extended-spectrum beta-lactamase-producing Escherichia coli isolated from urine and blood. Diagn. Microbiol. Infect. Dis. 2007;58:111–115. doi: 10.1016/j.diagmicrobio.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Lee S.Y., Park Y.J., Yu J.K., Jung S., Kim Y., Jeong S.H., Arakawa Y. Prevalence of acquired fosfomycin resistance among extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J. Antimicrob. Chemother. 2012;67:2843–2847. doi: 10.1093/jac/dks319. [DOI] [PubMed] [Google Scholar]
- 16.Seo M.R., Kim S.J., Kim Y., Kim J., Choi T.Y., Kang J.O., Wie S.H., Ki M., Cho Y.K., Lim S.K., et al. Susceptibility of Escherichia coli from community-acquired urinary tract infection to fosfomycin, nitrofurantoin, and temocillin in Korea. J. Korean Med. Sci. 2014;29:1178–1181. doi: 10.3346/jkms.2014.29.8.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho Y.H., Jung S.I., Chung H.S., Yu H.S., Hwang E.C., Kim S.O., Kang T.W., Kwon D.D., Park K. Antimicrobial susceptibilities of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in health care-associated urinary tract infection: Focus on susceptibility to fosfomycin. Int. Urol. Nephrol. 2015;47:1059–1066. doi: 10.1007/s11255-015-1018-9. [DOI] [PubMed] [Google Scholar]
- 18.Johnson J.R., Johnston B., Clabots C., Kuskowski M.A., Castanheira M. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 2010;51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 19.Lee M.Y., Choi H.J., Choi J.Y., Song M., Song Y., Kim S.W., Chang H.H., Jung S.I., Kim Y.S., Ki H.K., et al. Dissemination of ST131 and ST393 community-onset, ciprofloxacin-resistant Escherichia coli clones causing urinary tract infections in Korea. J. Infect. 2010;60:146–153. doi: 10.1016/j.jinf.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Colpan A., Johnston B., Porter S., Clabots C., Anway R., Thao L., Kuskowski M.A., Tchesnokova V., Sokurenko E.V., Johnson J.R. VICTORY (Veterans Influence of Clonal Types on Resistance: Year 2011) Investigators. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin. Infect. Dis. 2013;57:1256–1265. doi: 10.1093/cid/cit503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silver L.L. Fosfomycin: Mechanism and resistance. Cold Spring Harb. Perspect. Med. 2017;7:a025262. doi: 10.1101/cshperspect.a025262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Díez-Aguilar M., Cantón R. New microbiological aspects of fosfomycin. Revista Española de Quimioterapia. 2019;32(Suppl 1):8–18. [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsson A.I., Berg O.G., Aspevall O., Kahlmeter G., Andersson D.I. Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob. Agents Chemother. 2003;47:2850–2858. doi: 10.1128/AAC.47.9.2850-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karageorgopoulos D.E., Wang R., Yu X.H., Falagas M.E. Fosfomycin: Evaluation of the published evidence on the emergence of antimicrobial resistance in Gram-negative pathogens. J. Antimicrob. Chemother. 2012;67:255–268. doi: 10.1093/jac/dkr466. [DOI] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing: Twenty-seventh Informational Supplement M100-S28. CLSI; Wayne, PA, USA: 2018. [Google Scholar]
- 26.Wirth T., Falush D., Colles F., Mensa P., Wieler L.H., Karch H., Reeves P.R., Maiden M.C., Ochman H., Achtman M. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu J.Y., Yang Y., Han H., Betzi S., Olesen S.H., Marsilio F., Schönbrunn E. Functional consequence of covalent reaction of phosphoenolpyruvate with UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA) J. Biol. Chem. 2012;287:12657–12667. doi: 10.1074/jbc.M112.342725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou J., Huang X., Deng Y., He L., Yang T., Zeng Z., Chen Z., Liu J.H. Dissemination of the fosfomycin resistance gene fosA3 with CTX-M beta-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob. Agents Chemother. 2012;56:2135–2138. doi: 10.1128/AAC.05104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J., Lim Y.M., Rheem I., Lee Y., Lee J.C., Seol S.Y., Lee Y.C., Cho D.T. CTX-M and SHV-12 beta-lactamases are the most common extended-spectrum enzymes in clinical isolates of Escherichia coli and Klebsiella pneumoniae collected from 3 university hospitals within Korea. FEMS Microbiol. Lett. 2005;245:93–98. doi: 10.1016/j.femsle.2005.02.029. [DOI] [PubMed] [Google Scholar]