Abstract
Stenotrophomonas maltophilia, an environmental Gram-negative bacterium, is an emerging nosocomial opportunistic pathogen that causes life-threatening infections in immunocompromised patients and chronic pulmonary infections in cystic fibrosis patients. Due to increasing resistance to multiple classes of antibiotics, S. maltophilia infections are difficult to treat successfully. This makes the search for new antimicrobial strategies mandatory. In this study, the antibacterial activity of the heterocyclic corticosteroid deflazacort and several of its synthetic precursors was tested against S. maltophilia. All compounds were not active against standard strain S. maltophilia K279a. The compound PYED-1 (pregnadiene-11-hydroxy-16α,17α-epoxy-3,20-dione-1) showed a weak effect against some S. maltophilia clinical isolates, but exhibited a synergistic effect with aminoglycosides. PYED-1 at sub-inhibitory concentrations decreased S. maltophilia biofilm formation. Quantitative real-time polymerase chain reaction (RT-qPCR) analysis demonstrated that the expression of biofilm- and virulence- associated genes (StmPr1, StmPr3, sphB, smeZ, bfmA, fsnR) was significantly suppressed after PYED-1 treatment. Interestingly, PYED-1 also repressed the expression of the genes aph (3′)-IIc, aac (6′)-Iz, and smeZ, involved in the resistance to aminoglycosides.
Keywords: antimicrobial activity; deflazacort (DFZ), S. maltophilia; quantitative real-time PCR; anti-virulence agent; checkerboard assay
1. Introduction
Stenotrophomonas maltophilia is an emerging opportunistic bacterium, which represents the third Gram-negative responsible for nosocomial infections [1,2,3]. S. maltophilia generally causes bacteremia and pneumonia, and infections are frequently associated with complications and death in immunosuppressed or immunocompromised patients [4,5]. S. maltophilia is frequently found in polymicrobial infections from the respiratory tract of cystic fibrosis (CF) patients [6]. In CF patients, S. maltophilia infections are associated with a severe lung disease and increased risk of the need for transplantation, or death [7,8]. Many virulence factors that may contribute to the pathogenicity of S. maltophilia have been reported [9,10,11,12,13,14,15]. The therapeutic treatment is hampered by intrinsic or acquired resistance to multiple antimicrobial agents [16,17]. S. maltophilia is intrinsically resistant to carbapenems, aminoglycosides, and tetracyclines [3,16] owing to its multidrug efflux pumps and overexpression of resistant determinants, such as carbapenemases and aminoglycoside-modifying enzymes [4,16,17]. Trimethoprim–sulfamethoxazole (STX) is the first-line antimicrobial combination for the treatment of S. maltophilia infections. However, the allergenic reaction, the intolerance, and the increasing rates of resistance limit the use of Trimethoprim–sulfamethoxazole [18,19]. S. maltophilia infections are also difficult to treat due to the ability of this bacterium to form highly structured and multilayered biofilms [20,21,22]. Because of the potential for resistance development, there is need of novel antimicrobials for S. maltophilia infection treatment.
Deflazacort (DFZ) is a heterocyclic corticosteroid derivative of prednisolone, used as an anti-inflammatory and immunosuppressant, and characterized by high efficacy and good tolerability [23,24]. Recently, the DFZ synthetic precursor PYED-1 (pregnadiene-11-hydroxy-16α,17α-epoxy-3,20-dione-1) showed a good antibacterial activity against Staphylococcus aureus ATCC 29213 and Acinetobacter baumannii ATCC 17978 without showing cytotoxicity [25].
The aim of this study was to evaluate the antimicrobial, antibiofilm, and anti-virulence activities of PYED-1 against S. maltophilia.
2. Results and Discussion
2.1. Antimicrobial Activity of a Panel of Steroid Derivatives
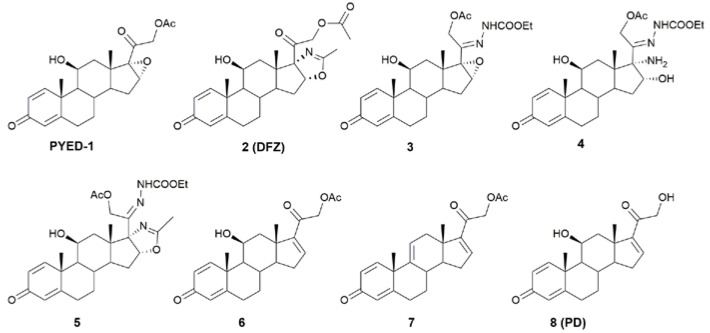
The antimicrobial activity of glucocorticoid DFZ and its synthetic precursors (Figure 1) against S. maltophilia K279a was assessed by broth microdilution assay (Table 1).
Figure 1.
Deflazacort (DFZ), prednisolone (PD), and DFZ synthetic precursors used in this study.
Table 1.
MIC (μg/mL) and MBC (μg/mL) values of PD, DFZ, and DFZ precursors against S. maltophilia K279a.
| Compounds | MIC | MBC |
|---|---|---|
| PYED-1 | 256 | 512 |
| 2 (DFZ) | >1000 | >1000 |
| 3 | 750 | >1000 |
| 4 | >1000 | >1000 |
| 5 | 512 | >1000 |
| 6 | >1000 | >1000 |
| 7 | 1000 | >1000 |
| 8 (PD) | >1000 | >1000 |
| STX | 1 | 1 |
All compounds were inactive against S. maltophilia K279a cells (Table 1). Instead, S. maltophilia K279a was susceptible to STX standard compound (Table 1). PYED-1, the compound that proved to be the most effective growth inhibitor against S. aureus ATCC 29213 and A. baumannii ATCC 17978 [25], showed MIC value at 256 μg/mL against S. maltophilia K279a, while MBC value at twofold upper MIC value (512 μg/mL). To examine whether the inhibition of bacterial growth could be related to the dimethyl sulfoxide (DMSO) used to dissolve the compounds tested, the growth of S. maltophilia K279a was measured in the presence of increasing concentrations of DMSO (ranging from 0.1% to 1%). Results indicated that there was no difference in S. maltophilia K279a growth in the presence of any of the DMSO concentrations used (data not shown). The antimicrobial activity of PYED-1 was also tested on a panel composed by eleven clinical isolates of S. maltophilia (Table 2). STX standard compound retained its activity against all S. maltophilia clinical isolates (data not shown).
Table 2.
MIC (μg/mL), MBC (μg/mL) values, and killing quotient (KQ) of PYED-1 against S. maltophilia K279a and S. maltophilia clinical isolates.
| Strain | MIC | MBC | KQ | Strain | MIC | MBC | KQ |
|---|---|---|---|---|---|---|---|
| K279a | 256 | 512 | 2 | Sm0707 | 256 | 512 | 2 |
| Sm0262 | 128 | 512 | 4 | Sm0916 | 256 | 512 | 2 |
| Sm0527 | 64 | 128 | 2 | Sm1053 | 512 | 2000 | 4 |
| Sm0528 | 128 | 256 | 2 | OBGTC3 | 64 | 128 | 2 |
| Sm0545 | 512 | 2000 | 4 | OBGTC9 | 256 | 512 | 2 |
| Sm0571 | 256 | 512 | 2 | OBGTC20 | 128 | 512 | 2 |
The MIC values ranging from 64 μg/mL to 512 μg/mL. MBC values were always at least twofold higher than MIC values (see Table 2). The killing quotient (KQ), which corresponds to the MBC/MIC ratio, indicated that the compound has bactericidal (≤4) or bacteriostatic (>4) activity [26]. As shown in Table 2, KQ was less than or equal to 4 for all strains, indicating that PYED-1 exhibited bactericidal activity.
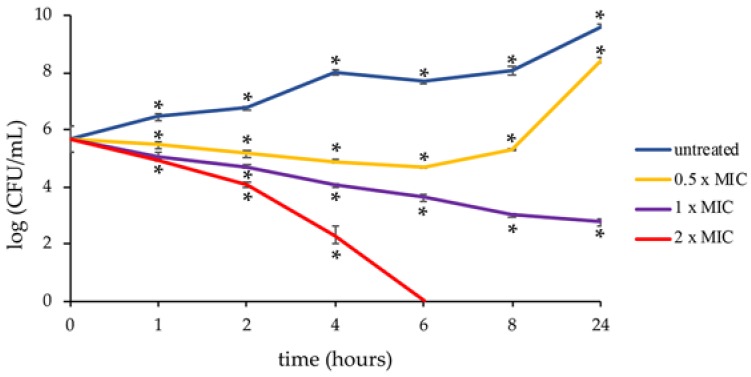
The time–kill assay showed that PYED-1 exhibited a significant bactericidal activity (Figure 2).
Figure 2.
Killing kinetics for S. maltophilia K279a following treatment with the PYED-1. Growth kinetics were monitored following exposure to PYED-1 at 0.5 × MIC (128 μg/mL), 1 × MIC (256 μg/mL), and 2 × MIC (512 μg/mL). Values are presented as mean ± SD. *P < 0.05.
At 0 h treatment, the colony forming unit (CFU) was approximately 6 × 105 /mL. After 2 h of incubation with 128 μg/mL, 256 μg/mL and 512 μg/mL, the bacterial load was reduced approximately five, ten, and fifty times, respectively. Following 4 h exposure with the indicated MIC, CFU declined to 8 × 104, 1 × 104, and 2 × 102, respectively. No viable S. maltophilia cells were recovered after six hours’ exposure at 2 × MIC. However, after 24 h the cell growth recovery was observed at 128 μg/mL. By definition, a compound that kills ≥3 log10 bacteria following 24 h incubation is considered bactericidal [27]. In this study, the PYED-1 MIC reduced the number of CFUs by 3.2 log10 following 24 h incubation, indicating that PYED-1 was bactericidal at this concentration.
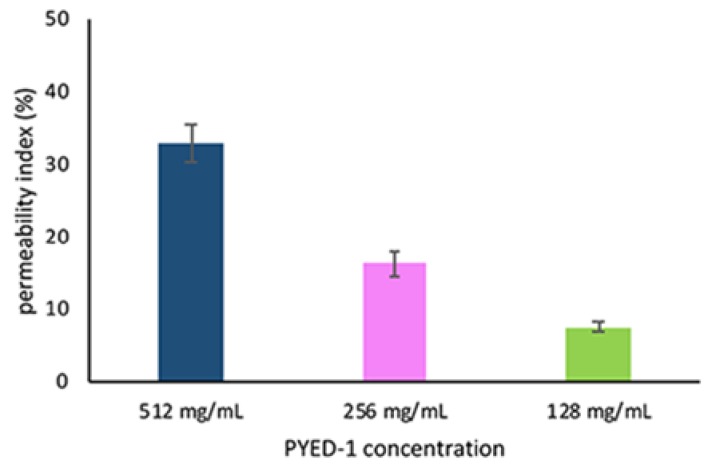
The bacterial uptake of the membrane impermeant dye propidium iodide (PI) was measured to assess the membrane permeability of S. maltophilia K279a following treatment with PYED-1. The increase of PI fluorescence indicated the loss of bacterial membrane integrity. As shown in Figure 3, PYED-1 increased PI uptake into S. maltophilia in a dose-dependent manner. PI-fluorescence intensity of S. maltophilia K279a cells increased by 33%, 16% and 7.5% after 4 h incubation with PYED at 2 × MIC, 1 × MIC, and 1/2 × MIC, respectively. Based on the above finding, we hypothesize that the permeabilization of the bacterial membrane might contribute to the bactericidal activity of PYED-1. Further experiments are necessary to validate this hypothesis.
Figure 3.
Effect of PYED-1 on S. maltophilia K279a membrane permeability. S. maltophilia K279a cells were treated with 2 × MIC (512 μg/mL), 1 × MIC (256 μg/mL) and 1/2 × MIC (128 μg/mL), and PI uptake was measured after 4 h of treatment. The error bars represent standard error of the mean from three independent experiments.
2.2. Checkerboard Assay
S. maltophilia is intrinsically resistant to aminoglycosides, a class of conventional antibiotics mostly used in the treatment of respiratory and urinary tract infections, blood, bone, and soft tissue infections caused both by Gram-negative and Gram-positive bacteria [16]. To find out whether PYED-1 combined with aminoglycosides may potentiate the antibacterial activity of these antibiotics against S. maltophilia, the activity of PYED-1 in combination with gentamycin and amikacin was determined using broth microdilution checkerboard assay against S. maltophilia K279a. The fractional inhibitory concentration (FIC) index for these combinations is reported in Table 3.
Table 3.
Synergistic effects of PYED-1 with antibiotics against S. maltophilia K279a.
| Bacterial strain | Combination | MICa (μg/mL) | MICc (μg/mL) | FIC index |
|---|---|---|---|---|
| S. maltophilia K279a | PYED-1 /gentamicin | 256/16 | 64/4 | 0.5 |
| PYED-1 /amikacin | 256/16 | 64/2 | 0.375 |
a, MIC of one sample alone; c, MIC of samples in combination; FIC index, fractional inhibitory concentration.
In combination with PYED-1, MIC was remarkably reduced by eightfold (from 16 to 2 μg/mL) and fourfold (from 16 to 4 μg/mL) for gentamicin and amikacin, respectively. These results demonstrated synergistic effect with aminoglycosides. The PYED-1 concentration of 64 μg/mL (1/4 MIC) was able to give the most synergistic effect with both aminoglycosides. The synergistic effect of PYED-1 and gentamicin and amikacin is an important finding because it allows the extension of the antimicrobial strategies against multidrug-resistant S. maltophilia [3,4,16,17].
2.3. Effects of PYED-1 on the Formation of S. maltophilia Biofilm
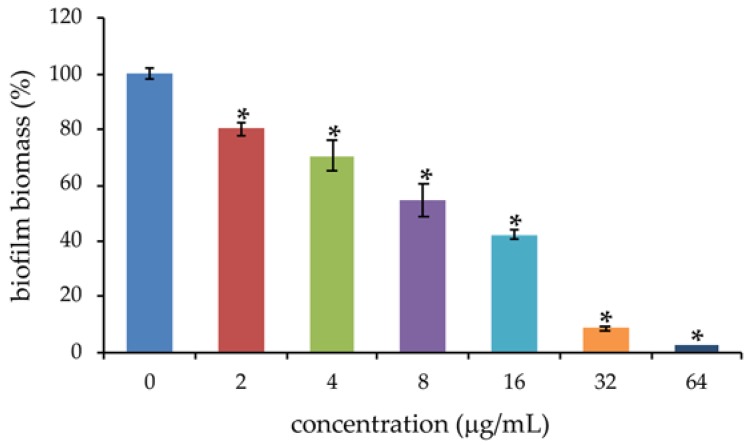
Most S. maltophilia strains form biofilms on several biotic and abiotic surfaces, and this greatly contributes to the pathogenicity of these bacteria [20,21,22]. Thus, the antibiofilm properties of PYED-1 against S. maltophilia K279a were investigated. We measured the biofilm biomass of S. maltophilia K279a cells treated with increasing concentrations of PYED-1 in static condition at 37 °C using abiotic crystal violet staining. Sub-inhibitory concentrations of PYED-1 were able to reduce biofilm formation of S. maltophilia K279a compared to the untreated control (Figure 4).
Figure 4.
S. maltophilia K279a biofilm formation following treatment with the PYED-1. Cells were incubated for 24 h in the presence of sub-inhibitory concentrations of PYED-1. Biofilms were quantified after crystal-violet staining. Values are presented as mean ± SD. * P < 0.05.
A reduction of 97%, 90%, and 57% was found at concentrations 64 μg/mL, 32 μg/mL, and 16 μg/mL, corresponding to 1/4 × MIC, 1/8 × MIC and 1/16 × MIC, respectively. To determine whether the inhibitory effect on biofilm formation was related to growth inhibition, planktonic growth was measured in same conditions used in biofilm assay. At the concentrations tested in biofilm assay, PYED-1 did not affect planktonic growth (data not shown). Several studies demonstrated that novel classes of antimicrobial compounds inhibit S. maltophilia biofilm formation [28,29,30,31,32,33]. In further support of this, our results showed that PYED-1 at 64 μg/mL significantly inhibited S. maltophilia biofilm formation on a polystyrene abiotic surface. Interestingly, the same concentration of PYED-1 has a synergistic effect with gentamicin or amikacin on bacterial growth inhibition. This is in agreement with other studies showing that S. maltophilia biofilm formation plays a notable role in the development of antibiotic resistance [20,21,22].
2.4. Transcriptional Changes Induced by PYED-1 in S. maltophilia K279a
A promising alternative strategy to treat infections caused by multidrug bacteria is anti-virulence therapy, based on the development of drugs able to specifically inhibit virulence factors [34,35]. To investigate the anti-virulence activity of PYED-1 the expression levels of a dozen known S. maltophilia virulence genes [9,10,11,12,13,14,15] were investigated by qRT-PCR. RNA was extracted from exponential S. maltophilia cells (3 × 108 CFU/mL) untreated and treated at sub-MIC concentration (128 μg/mL) of PYED-1 for 3 h. No growth differences between treated and untreated cells were observed. Also, 3 h’ PYED-1 treatment slightly increased the membrane permeability of S. maltophilia K279a (data not shown). In this type of experiments, genes with at least a twofold difference in relative transcript levels (with a P value of <0.05) are considered significant. As shown in Table 4, the expression of most of the tested genes was affected by PYED-1 treatment.
Table 4.
RT-PCR analysis of biofilm and virulence factors gene expression in S. maltophilia K279a in the presence of PYED-1.
| Gene | Fold Change ± SD | P Value | ||
|---|---|---|---|---|
| Smlt0648 | rmlA | glucose-1-phosphate thymidylyl transferase | −1.10 ± 0.045 * | 0.00002919 |
| Smlt0686 | StmPr1 | extracellular protease | −2.21 ± 0.108 * | <0.0000001 |
| Smlt0706 | smf-1 | fimbrial adhesin protein | −1.33± 0.144 * | 0.00003563 |
| Smlt1736 | hfq | host factor-I protein | −1.20± 0.241 | 0.07368922 |
| Smlt2120 | aph3’-IIc | aminoglycoside 3’-phosphotransferase II | −2.93 ± 0.193 * | <0.0000001 |
| Smlt2202 | smeZ | multidrug efflux pump | −3.97 ± 1.381 * | 0.00009759 |
| Smlt2299 | fsnR | response regulator protein | −2.03 ± 0.061 * | <0.0000001 |
| Smlt3524 | sphB | Serine-protease | −2.66 ± 0.067 * | <0.0000001 |
| Smlt3615 | aac6′-Iz | aminoglycoside 6’-N-acetyltransferase | −2.44± 0.486 * | 0.00000015 |
| Smlt3638 | transmembrane hemolysin protein | −1.23 ± 0.083 * | 0.00000143 | |
| Smlt4190 | sppA | protease IV | −1.63 ± 0.088 * | <0.0000001 |
| Smlt4209 | bfmA | two component response regulator | −2.36 ± 0.174 * | <0.0000001 |
| Smlt4395 | StmPr3 | Serine-protease | −2.90 ± 0.456 * | <0.0000001 |
* The asterisks indicate genes with a P value of <0.05.
PYED-1 significantly decreased the gene expression of fsnR, encoding the orphan response regulator FsnR, involved in motility and biofilm formation. S. maltophilia cell motility and biofilm formation can be inhibited by a reduction of fsnR gene expression [36]. Furthermore, the expression of bfmA gene, a transcription factor that stimulates the transcription of bfmAK operon involved in biofilm formation [37], was significantly downregulated by PYED-1. Based on our results, we postulate that inhibition of bfmA and fsnR gene expression by PYED-1 may contribute to the reduction of biofilm formation in S. maltophilia. Data are in agreement with a previous study showing that bfmA and fsnR gene expression is modulated by treatment of S. maltophilia K279a with celastrol [30].
In addition, PYED-1 significantly decreased the expression of sphB, StmPr1, and StmPr3 genes encoding for serine proteases that contribute to degradation of extracellular matrix proteins [38]. The ability of S. maltophilia to produce extracellular protease may contribute to S. maltophilia pathogenesis in the lungs of CF patients [39,40]. The major protease StmPr1 induces the death of A549 fibroblasts and IL-8 secretion by A549 cells [40]. PYED-1 treatment could reduce the release of extracellular proteases, and consequently tissue damage and inflammation in the host.
PYED-1 significantly reduced also the gene expression of smeZ, aph3′-IIc, and aac6′-Iz, three genes involved in the resistance to aminoglycosides. Interestingly, the RND-type efflux encoded by smeZ has been demonstrated in S. maltophilia to contribute to aminoglycosides resistance [41], and other virulence-related functions, such as swimming, protease secretion, and biofilm formation [42]. In further support of this, we hypothesize that PYED-1 decreases antibiotic resistance in S. maltophilia by inhibiting the biofilm formation and the expression of multidrug efflux pumps. Future experiments will be necessary to validate the hypothesis.
Overall, our data demonstrate that PYED-1 in combination with gentamicin or amikacin aminoglycosides shows antimicrobial activity against S. maltophilia. Also, PYED-1 acts as an antibiofilm drug and inhibits the expression of important biofilm and virulence genes in S. maltophilia, attenuating the virulence of this drug-resistant pathogen. These results make PYED-1 a promising candidate for clinical use against S. maltophilia infectious diseases. Although the pharmacokinetics of corticosteroids are well known [43], no information about the pharmacokinetics of the PYED-1 is available yet. Future studies will be necessary to establish the pharmacokinetics of PYED-1 and its derivatives and their potential clinical use.
3. Materials and Methods
3.1. Chemicals and Reagents
All chemicals and solvents were purchased with the highest degree of purity (Sigma-Aldrich, Alfa Aesar, VWR) and used without further purification. The reactions were monitored by TLC (precoated silica gel plate F254, Merck) and the products were detected by exposure to ultraviolet radiation, iodine vapor, and chromic mixture. The purity of the compounds was determined by CHNS analysis and was ≥ 95% in all cases. NMR spectra were recorded on NMR spectrometers operating at 400 MHz (Bruker DRX, Bruker AVANCE) using CDCl3 solutions. Coupling constant values (J) were reported in Hz. Chemical synthesis and structural characterization of compounds was realized as previously reported [25].
3.2. Antimicrobial Activity
Strains evaluated in this study included the S. maltophilia K279a reference strains and eleven S. maltophilia clinical isolates belonging to a bacterial collection previously established. Epidemiological features of strains were in accordance to previous publications [44,45]. No ethical approval was required for the study because there was no access to patients’ data. All strains were grown on blood agar plates (TSA). Minimum inhibitory concentration (MIC) values of steroidal compounds against planktonic bacteria were examined by a broth microdilution method previously described [46]. Briefly, stock solutions of all compounds at the concentration of 50 mg/mL were made by dissolving them in DMSO. Bacterial cell suspensions were prepared at 0.5 McFarland standard using a BD PhoenixSpec™ nephelometer and were subsequently diluted in cation-adjusted Mueller–Hinton broth (CA-MHB) to approximately 5 × 106 CFU/mL. One hundred microliter of bacteria (5 × 105 CFU) were then added to the microtiter plates containing 100 μL of serial dilutions of steroidal compounds. Only CA-MHB was added in negative control wells. Wells with no compounds were used on each plate as positive growth control. Plates were incubated at 37 °C for 18 h under shaking (300 rpm). The optical density at 595 nm was measured by using a microplate reader (Bio-Rad Laboratories S.r.l.). The effect of different concentrations of DMSO (ranging from 0.1% to 1%) on bacteria growth kinetics was separately tested. To calculate the minimum bactericidal concentration (MBC), bacterial suspensions from MIC assay microtiter wells were diluted in PBS and spot-plated on TSA plates, and the colonies were counted after incubation at 37 °C for 18 h. The MBC was determined as the lowest concentration of substance, which produced ≥99.9% killing (≥3 log10) after 24 h of incubation as compared to the colony count of the starting inoculum. All tests were performed in triplicate and repeated three times.
3.3. Time Killing Assay
The killing kinetics of PYED-1 at 0.5 ×, 1 × and 2 × MIC were determined against S. maltophilia K279a. Approximately 6 × 105 CFU/mL of S. maltophilia K279a strain was used to inoculate 3 mL of CA-MHB containing different concentrations of PYED-1 and incubated at 37 °C under shaking (300 rpm). A tube without PYED-1 was a growth control. Viable bacterial counts were performed after 0, 1, 2, 4, 6, 8, and 24 h incubation by plating serial tenfold dilutions of broth cultures onto TSA plates, and incubating at 37 °C for 24 h. All experiments were repeated three times.
3.4. Propidium Iodide Uptake Assay
Cell permeability was assessed as reported earlier [47]. Briefly, S. maltophilia cells were grown in CA-MHB up to the mid logarithmic phase, adjusted to 1 × 106 CFU/mL and incubated at 37 °C with PYED-1 (512 μg/mL, 256 μg/mL, and 128 μg/mL) for 4 h. After PYED-1 treatment, the cells were washed in PBS buffer and incubated with PI (10 μM) at 37 °C for 20 min in the dark. Bacterial cells permeabilized with 0.5% Triton X100 were used as a positive control. Untreated bacterial cells were used as a negative control. The PI fluorescence was measured at excitation and emission of 485 nm and 590 nm respectively, using the plate reader Synergy HT spectrofluorimeter (Biotek). Fluorescence was normalized by subtracting fluorescence of the untreated cells from that of the treated. Permeability index was expressed using the following equation: permeability index (%) = [(sample - negative control)/(positive control- negative control)] * 100.
3.5. Checkerboard Assay
The combination effects between PYED-1 and gentamicin or amikacin against S. maltophilia K279a cells were assessed by a microbroth checkerboard assay [48]. Serial twofold dilutions were prepared in CA-MHB to ranch the final concentration of gentamicin or amikacin ranging from 0.25 to 128 μg/mL, and PYED-1 ranging from 4 to 1000 μg/mL. Fifty microliters of antibiotic (gentamicin or amikacin) was added to the rows of a 96-well microtiter plate in decreasing concentrations, and 50 μl of PYED-1 was added to the column in decreasing concentrations. Microplates were inoculated with 100 μl of S. maltophilia K279a suspension with a final concentration of 105 × CFU/mL and incubated at 37 °C for 18 h. The optical density at 595 nm was measured by using a microplate reader (Bio-Rad Laboratories S.r.l.). The effect of the interactions of PYED-1 with each of the tested antibiotic was quantified by calculating the fractional inhibitory concentration (FIC) index as follows: FIC index = FIC of PYED-1 + FIC of antibiotic, where FIC of PYED-1 (or antibiotic) is the ratio of MIC of PYED-1 (or antibiotic) in combination and MIC of PYED-1 (or antibiotic) alone. The following intervals of FIC index were used to interpret the experimental outcome: ≤0.5, synergistic; >0.5 to ≤1.0, additive; >1.0 to ≤2.0, indifferent; and >2.0, antagonistic effects [48]. All experiments were repeated three times.
3.6. Biofilm Assay
Biofilm quantification assays were performed in 96-well microtiter plates using a crystal violet (CV) method as previously described [46]. Briefly, overnight cultures of S. maltophilia K279a were diluted with fresh trypticase soy broth (TSB) with 0.5 % glucose to obtain a bacterial suspension of 5 × 106 CFU/mL. One hundred of the bacterial suspension were added to 96-well sterile flat-bottom polystyrene plates in the presence of 100 μl of sub-MIC concentrations of PYED-1 ranging from 2 to 128 μg/mL. After 24 h incubation at 37 °C, the planktonic cells were gently aspirated, then the biofilms were washed twice times with sterile PBS (pH 7.2). Two hundred microliters of 0.1% crystal violet was added and incubated at room temperature for 15 minutes. Crystal violet was removed by pipetting, and wells were washed three times with 200 μl sterile PBS (pH 7.2). Plates were air-dried and 200 μL of ethanol was added. After 20 min, the biofilm biomass was quantified by measuring the optical density at 595 nm using a microplate reader (Bio-Rad Laboratories S.r.l.).
3.7. RNA
Total RNA was isolated from S. maltophilia K279a cells grown in in CA-MHB at 37 °C at 200 rpm to an OD600 of 0.4. Two mL of culture was subsequently treated with either PYED-1 at the concentration of 128 μg/mL or 0.016% DMSO and incubated at 37 °C at 200 rpm for 3 h. Two volumes (4 mL) of RNAprotect Bacteria Reagent was added to the cell suspensions and incubated for 5 min at room temperature. Next, the cell suspensions were centrifuged at 5000 × g for 10 min and the supernatant was decanted. RNA was purified according to the previously reported method [49]. RNA was quantified using a Nano-drop instrument (Thermo Fisher).
3.8. RT-PCR
Total RNA was reverse-transcribed into cDNA using QuantiTect Reverse Transcription Kit (Qiagen), according to the manufacturer’s protocol. The RT-PCR was performed as previously described [50], using a SYBR Green master mix (Applied Biosystems). The oligonucleotides used in PCR experiments are reported in Table 5.
Table 5.
Oligonucleotide sequences used in this study.
| Primer Name | Primer Sequence |
|---|---|
| aac6-Iz fw | TGTGGACTGATGCCGATG |
| aac6-Iz rv | GCACTTCAGCGAAACCAAC |
| aph3-IIc fw | CCGATCATGAAGACCTGGTG |
| aph3-IIc rv | GTCGATGAAACCGCTGAAAC |
| bfmA fw | AGTGAACTGCGCTTTTCTGG |
| bfmA rv | TGAATTCACCACGGCTGAG |
| fsnR fw | TCCTGATGGACCTGTCATTG |
| fsnR rv | TGCATGGTCATCATCACAAC |
| Hfq fw | TCTACAAGCACGCCATTTCC |
| Hfq rv | TACTCGTCTGCTTCATCACCTG |
| rmlA fw | TGCTGGGTGACAACATCTTC |
| rmlA rv | CCGGATCATTCACCCAATAG |
| rpoB fw | AGGAAATGCTGACGGTGAAG |
| rpoB rv | ACGAGCACGTTGAAGGATTC |
| smeZ fw | GCAGTGATGTACCTGTTTCTGC |
| smeZ rv | CAGCACATTGATCGAGAAGC |
| smf-1 fw | ACCGTGTCCAAGAACACTCTG |
| smf-1 rv | TGCACTTGGTCAGGTTGATG |
| Smlt3638 fw | GGTTGAAGGTATTCGACCACTG |
| Smlt3638 rv | ATCAGGGTGAACGGGGTATAG |
| sphB fw | CGCATCTTTCAGTCACCAAC |
| sphB rv | GTAATTGAAGTTGGCCAGCAC |
| sppA fw | AGTTTCTTCATCGGGCTGTG |
| sppA rv | ATGACGAACATCACCAGCAG |
| StmPr1 fw | GCCGAAGTCATCAACCTCTC |
| StmPr1 rv | ACACGTTGGTGTTGCTGTTG |
| StmPr3 fw | ATCGACAGCACCTGCAACTAC |
| StmPr3 rv | TTCACATCGCGATAGGACAG |
The rpoB gene was used as the housekeeping control to normalize the expressions of genes of interest. RNA samples not treated with reverse transcriptase were routinely included as no template controls. Changes in transcript levels were determined using the 2−ΔΔCT method [51]. RNA expression levels were determined by using three independent cultures, and all analyses were performed in triplicate.
3.9. Statistical Analysis
The calculation of arithmetic means and standard deviations was utilized to statistically analyze continuous variables. A t-test was used to determine statistical differences between treated and control groups for each dosage and each time point for the time–kill assay and for the biofilm assay. Statistical differences between treated and control samples expression values derived from RT-PCR were evaluated by using, for each gene, a Z-test on the null hypothesis that the average ΔΔCt values are equal to 1. All results were considered to be statistically significant at P < 0.05.
4. Conclusions
The results of the present study revealed that PYED-1 in combination with aminoglycosides, represent a significant tool to control S. maltophilia growth. Moreover, PYED-1 was identified as a promising agent for targeting biofilm and virulence of S. maltophilia. This might be a new strategy for the treatment of S. maltophilia biofilm-associated chronic infections.
Acknowledgments
We gratefully acknowledge Pierpaolo Di Nocera for comments and critical reading of the manuscript.
Author Contributions
Conceptualization, E.D.G. and A.G.; methodology, E.P.E., A.V., A.E., and D.D.; formal analysis, E.P.E., A.V. and E.D.G.; investigation, E.P.E., A.V., A.E., D.D.; data curation, A.G., R.Z. and E.D.G.; writing—original draft preparation, R.Z. and E.D.G.; supervision, A.G., R.Z. and E.D.G.; funding acquisition, R.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded in part by a grant from the Italian Ministry of Education, University and Research (MIUR): PRIN2017 (Grant 2017SFBFER to RZ).
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Fihman V., Le Monnier A., Corvec S., Jaureguy F., Tankovic J., Jacquier H., Carbonnelle E., Bille E., Illiaquer M., Cattoir V., et al. Stenotrophomonas maltophilia—the most worrisome threat among unusual non-fermentative gram-negative bacilli from hospitalized patients: A prospective multicenter study. J. Infect. 2012;64:391–398. doi: 10.1016/j.jinf.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Looney W.J., Narita M., Mühlemann K. Stenotrophomonas maltophilia: An emerging opportunist human pathogen. Lancet Infect. Dis. 2009;9:312–323. doi: 10.1016/S1473-3099(09)70083-0. [DOI] [PubMed] [Google Scholar]
- 3.Crispino M., Boccia M.C., Bagattini M., Villari P., Triassi M., Zarrilli R. Molecular epidemiology of Stenotrophomonas maltophilia in a university hospital. J. Hosp. Infect. 2002;52:88–92. doi: 10.1053/jhin.2002.1280. [DOI] [PubMed] [Google Scholar]
- 4.Brooke J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Rev. Clin. Microbiol. 2012;25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeon Y.D., Jeong W.Y., Kim M.H., Jung I.Y., Ahn M.Y., Ann H.W., Ahn J.Y., Han S.H., Choi J.Y., Song Y.G., et al. Risk factors for mortality in patients with Stenotrophomonas maltophilia bacteremia. Medicine. 2016;95:e4375. doi: 10.1097/MD.0000000000004375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frost F., Nazareth D., Shaw M., Walshaw M.J. Cystic fibrosis related diabetes is not independently associated with increased Stenotrophomonas maltophilia infection: Longitudinal data from the UK CF Registry. J. Cyst. Fibros. 2019;18:294–298. doi: 10.1016/j.jcf.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Barsky E.E., Williams K.A., Priebe G.P., Sawicki G.S. Incident Stenotrophomonas maltophilia infection and lung function decline in cystic fibrosis. Pediatr Pulmonol. 2017;52:1276–1282. doi: 10.1002/ppul.23781. [DOI] [PubMed] [Google Scholar]
- 8.Waters V., Atenafu E.G., Lu A., Yau Y., Tullis E., Ratjen F. Chronic Stenotrophomonas maltophilia infection and mortality or lung transplantation in cystic fibrosis patients. J. Cyst. Fibros. 2013;12:482–486. doi: 10.1016/j.jcf.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Adegoke A.A., Stenström T.A., Okoh A.I. Stenotrophomonas maltophilia as an Emerging Ubiquitous Pathogen: Looking Beyond Contemporary Antibiotic Therapy. Front. Microbiol. 2017;8:2276. doi: 10.3389/fmicb.2017.02276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayer-Santos E., Cenens W., Matsuyama B.Y., Oka G.U., Di Sessa G., Mininel I., Alves T., Farah C.S. The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing. PLoS Pathog. 2019;15:e1007651. doi: 10.1371/journal.ppat.1007651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Bonaventura G., Prosseda G., Del Chierico F., Cannavacciuolo S., Cipriani P., Petrucca A., Superti F., Ammendolia M.G., Concato C., Fiscarelli E., et al. Molecular characterization of virulence determinants of Stenotrophomonas maltophilia strains isolated from patients affected by cystic fibrosis. Int. J. Immunopathol. Pharmacol. 2007;20:529–537. doi: 10.1177/039463200702000311. [DOI] [PubMed] [Google Scholar]
- 12.Nas M.Y., White R.C., DuMont A.L., Lopez A.E., Cianciotto N.P. Stenotrophomonas maltophilia encodes a VirB/VirD4 Type IV Secretion System that modulates apoptosis in human cells and promotes competition against heterologous bacteria, including Pseudomonas aeruginosa. Infect. Immun. 2019;87:e00457-19. doi: 10.1128/IAI.00457-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicoletti M., Iacobino A., Prosseda G., Fiscarelli E., Zarrilli R., De Carolis E., Petrucca A., Nencioni L., Colonna B., Casalino M. Stenotrophomonas maltophilia strains from cystic fibrosis patients: Genomic variability and molecular characterization of some virulence determinants. Int. J. Med. Microbiol. 2011;301:34–43. doi: 10.1016/j.ijmm.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Pompilio A., Pomponio S., Crocetta V., Gherardi G., Verginelli F., Fiscarelli E., Dicuonzo G., Savini V., D’Antonio D., Di Bonaventura G. Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: Genome diversity, biofilm formation, and virulence. BMC Microbiol. 2011;11:159. doi: 10.1186/1471-2180-11-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trifonova A., Strateva T. Stenotrophomonas maltophilia—A low-grade pathogen with numerous virulence factors. Infect. Dis. 2019;51:168–178. doi: 10.1080/23744235.2018.1531145. [DOI] [PubMed] [Google Scholar]
- 16.Chang Y.T., Lin C.Y., Chen Y.H., Hsueh P.R. Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front. Microbiol. 2015;6:893. doi: 10.3389/fmicb.2015.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sánchez M.B. Antibiotic resistance in the opportunistic pathogen Stenotrophomonas maltophilia. Front. Microbiol. 2015;6:658. doi: 10.3389/fmicb.2015.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun E., Liang G., Wang L., Wei W., Lei M., Song S., Han R., Wang Y., Qi W. Antimicrobial susceptibility of hospital acquired Stenotrophomonas maltophilia isolate biofilms. Braz. J. Infect. Dis. 2016;20:365–373. doi: 10.1016/j.bjid.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toleman M.A., Bennett P.M., Bennett D.M., Jones R.N., Walsh T.R. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg. Infect. Dis. 2007;13:559–565. doi: 10.3201/eid1304.061378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alcaraz E., Garcıa C., Friedman L., Passerini de Rossi B. The rpf/DSF signalling system of Stenotrophomonas maltophilia positively regulates biofilm formation, production of virulence-associated factors and β-lactamase induction. FEMS Microbiol. Lett. 2019;366:fnz069. doi: 10.1093/femsle/fnz069. [DOI] [PubMed] [Google Scholar]
- 21.Flores-Treviño S., Bocanegra-Ibarias P., Camacho-Ortiz A., Morfín-Otero R., Salazar-Sesatty H.A., Garza-González E. Stenotrophomonas maltophilia biofilm: Its role in infectious diseases. Expert. Rev. Anti Infect. Ther. 2019;17:877–893. doi: 10.1080/14787210.2019.1685875. [DOI] [PubMed] [Google Scholar]
- 22.Pompilio A., Savini V., Fiscarelli E., Gherardi G., Di Bonaventura G. Clonal diversity, biofilm formation, and antimicrobial resistance among Stenotrophomonas maltophilia strains from cystic fibrosis and non-cystic fibrosis patients. Antibiotics (Basel) 2020;9:15. doi: 10.3390/antibiotics9010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez-Perez O., Luquin S., Garcia-Estrada J., Ramos-Remus C. Deflazacort: A glucocorticoid with few metabolic adverse effects but important immunosuppressive activity. Adv. Ther. 2007;24:1052–1060. doi: 10.1007/BF02877711. [DOI] [PubMed] [Google Scholar]
- 24.Parente L. Deflazacort: Therapeutic index, relative potency and equivalent doses versus other corticosteroids. BMC Pharmacol. Toxicol. 2017;18:1. doi: 10.1186/s40360-016-0111-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esposito A., De Gregorio E., De Fenza M., D’Alonzo D., Satawani A., Guaragna A. Expeditious synthesis and preliminary antimicrobial activity of deflazacort and its precursors. RSC Advances. 2019;9:21519–21524. doi: 10.1039/C9RA03673C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levison M.E. Pharmacodynamics of antimicrobial drugs. Infect. Dis Clin. North. Am. Rev. 2004;18:451–465. doi: 10.1016/j.idc.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing, 29th Informational CLSI Supplement M100. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2019. [Google Scholar]
- 28.Karunanidhi A., Thomas R., van Belkum A., Neela V. In vitro antibacterial and antibiofilm activities of chlorogenic acid against clinical isolates of Stenotrophomonas maltophilia including the trimethoprim/sulfamethoxazole resistant strain. Biomed. Res. Int. 2013;2013:392058. doi: 10.1155/2013/392058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karunanidhi A., Ghaznavi-Rad E., Hamat R.A., Pichika M.R., Lung L.T.T., Mohd Fauzi F., Chigurupati S., van Belkum A., Neela V. Antibacterial and Antibiofilm Activities of Nonpolar Extracts of Allium stipitatum Regel. against Multidrug Resistant Bacteria. Biomed. Res. Int. 2018;2018:9845075. doi: 10.1155/2018/9845075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim H.R., Lee D., Eom Y.B. Anti-biofilm and Anti-Virulence Efficacy of Celastrol against Stenotrophomonas maltophilia. Int. J. Med. Sci. 2018;15:617–627. doi: 10.7150/ijms.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollini S., Di Pilato V., Landini G., Di Maggio T., Cannatelli A., Sottotetti S., Cariani L., Aliberti S., Blasi F., Sergio F., et al. In vitro activity of N-acetylcysteine against Stenotrophomonas maltophilia and Burkholderia cepacia complex grown in planktonic phase and biofilm. PLoS ONE. 2018;13:e0203941. doi: 10.1371/journal.pone.0203941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pompilio A., Crocetta V., Scocchi M., Pomponio S., Di Vincenzo V., Mardirossian M., Gherardi G., Fiscarelli E., Dicuonzo G., Gennaro R., et al. Potential novel therapeutic strategies in cystic fibrosis: Antimicrobial and anti-biofilm activity of natural and designed α-helical peptides against Staphylococcus aureus, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia. BMC Microbiol. 2012;12:145. doi: 10.1186/1471-2180-12-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vidigal P.G., Müsken M., Becker K.A., Häussler S., Wingender J., Steinmann E., Kehrmann J., Gulbins E., Buer J., Rath P.M., et al. Effects of green tea compound epigallocatechin-3-gallate against Stenotrophomonas maltophilia infection and biofilm. PLoS ONE. 2014;9:e92876. doi: 10.1371/journal.pone.0092876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heras B., Scanlon M.J., Martin J.L. Targeting virulence not viability in the search for future antibacterials. Br. J. Clin. Pharmacol. 2015;79:208–215. doi: 10.1111/bcp.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Totsika M. Disarming pathogens: Benefits and challenges of antimicrobials that target bacterial virulence instead of growth and viability. Future Med. Chem. 2017;9:267–269. doi: 10.4155/fmc-2016-0227. [DOI] [PubMed] [Google Scholar]
- 36.Kang X.M., Wang F.F., Zhang H., Zhang Q., Qiana W. Genome-wide identification of genes necessary for biofilm formation by nosocomial pathogen Stenotrophomonas maltophilia reveals that orphan response regulator FsnR is a critical modulator. Appl. Environ. Microbiol. 2015;81:1200–1209. doi: 10.1128/AEM.03408-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhenga L., Wang F.F., Ren B.Z., Liu W., Liu Z., Qian W. Systematic mutational analysis of histidine kinase genes in the nosocomial pathogen Stenotrophomonas maltophilia identifies BfmAK systemcontrol of biofilm development. Appl. Environ. Microbiol. 2016;82:2444–2456. doi: 10.1128/AEM.03951-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molloy K., Smith S.G., Cagney G., Dillon E.T., Greene C.M., McElvaney N.G. Characterisation of the major extracellular proteases of Stenotrophomonas maltophilia and their effects on pulmonary antiproteases. Pathogens. 2019;8:92. doi: 10.3390/pathogens8030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DuMont A.L., Karaba S.M., Cianciotto N.P. Type II secretion-dependent degradative and cytotoxic activities mediated by Stenotrophomonas maltophilia serine proteases StmPr1 and StmPr2. Infect. Immun. 2015;83:3825–3837. doi: 10.1128/IAI.00672-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DuMont A.L., Cianciotto N.P. Stenotrophomonas maltophilia serine protease StmPr1 induces matrilysis, anoikis, and protease-activated receptor 2 activation in human lung epithelial cells. Infect. Immun. 2017;85:e00544-17. doi: 10.1128/IAI.00544-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu C.J., Huang Y.W., Lin Y.T., Ning H.C., Yang T.C. Inactivation of SmeSyRy two-component regulatory system inversely regulates the expression of SmeYZ and SmeDEF efflux pumps in Stenotrophomonas maltophilia. PLoS ONE. 2016;11:e0160943. doi: 10.1371/journal.pone.0160943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Y.T., Huang Y.W., Chen S.J., Chang C.W., Yang T.C. The SmeYZ efflux pump of Stenotrophomonas maltophilia contributes to drug resistance, virulence-related characteristics, and virulence in mice. Antimicrob. Agents and Chemother. 2015;59:4067–4073. doi: 10.1128/AAC.00372-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jusko W.J. Pharmacokinetics and receptor-mediated pharmacodynamics of corticosteroids. Toxicology. 1995;102:189–196. doi: 10.1016/0300-483X(95)03047-J. [DOI] [PubMed] [Google Scholar]
- 44.Di Nocera P.P., De Gregorio E., Rocco F. GTAG- and CGTC-tagged palindromic DNA repeats in prokaryotes. BMC genomics. 2013;14:522. doi: 10.1186/1471-2164-14-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rocco F., De Gregorio E., Di Nocera P.P. A giant family of short palindromic sequences in Stenotrophomonas maltophilia. FEMS Microbiol. Lett. 2010;308:185–192. doi: 10.1111/j.1574-6968.2010.02010.x. [DOI] [PubMed] [Google Scholar]
- 46.Pane K., Cafaro V., Avitabile A., Torres M.T., Vollaro A., De Gregorio E., Catania M.R., Di Maro A., Bosso A., Gallo G. Identification of novel cryptic multifunctional antimicrobial peptides from the human stomach enabled by a computational-experimental platform. ACS Synth. Biol. 2018;7:2105. doi: 10.1021/acssynbio.8b00084. [DOI] [PubMed] [Google Scholar]
- 47.Ayaz Ahmed K.B., Raman T., Veerappan A. Platinum nanoparticles inhibit bacteria proliferation and rescue zebrafish from bacterial infection. RSC Adv. 2016;50:44415–44424. doi: 10.1039/C6RA03732A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pillai S.K., Moellering R.C., Eliopoulos G.M. Antimicrobial combinations. In: Lorian V., editor. Antibiotics in Laboratory Medicine. 5th ed. The Lippincott Williams & Wilkins Co.; Philadelphia, PA, USA: 2005. pp. 365–440. [Google Scholar]
- 49.De Gregorio E., Esposito E.P., Zarrilli R., Di Nocera P.P. Contact-Dependent Growth Inhibition Proteins in Acinetobacter baylyi ADP1. Curr. Microbiol. 2018;75:1434–1440. doi: 10.1007/s00284-018-1540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinucci M., Roscetto E., Iula V.D., Votsi A., Catania M.R., De Gregorio E. Accurate identification of members of the Burkholderia cepacia complex in cystic fibrosis sputum. Lett. Appl. Microbiol. 2016;62:221–229. doi: 10.1111/lam.12537. [DOI] [PubMed] [Google Scholar]
- 51.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]