In 2007, researchers at Danisco interested in fortifying the lactic acid bacteria used to produce dairy products such as milk and cheese against attack by phage showed that CRISPR provided resistance (1). These phage infections were common in industrial dairy fermentation, and it was hugely significant that insertion of multiple spacers in the bacterial CRISPR system was shown to protect against them. Intriguingly, rare mutations in the phage regions from which these spacers were derived were also shown to give rise to lineages that could again reinfect the bacteria. Thus was born the study of the phage–bacteria immune system arms race. In PNAS, Bradde et al. (2) present a theory of the tradeoff between broad coverage of phage types and efficient use of a limited number of enzymes in each bacterium that leads to an optimal size of the CRISPR immune system.
Earlier bioinformatics studies had shown that the majority of spacers in bacterial CRISPR arrays matched phage or conjugative plasmids that naturally infect the bacteria containing the CRISPR array (3). These CRISPR spacers were found to be the most polymorphic sites in plague Yersinia pestis strains, and thus potentially useful for forensically tracing the origins of prokaryotic pathogens (4). In the same year, bioinformatic analysis showed that spacers from multiple strains and species of Streptococcus were homologous to phage and plasmid sequences (5). The sensitivity of Streptococcus thermophilus to phage was shown to correlate with the number of spacers in the CRISPR locus. A significant number of studies followed up on these observations that bacterial resistance to phage correlated with spacer diversity in the CRISPR array (6–8).
Later studies confirmed the ability of S. thermophilus to insert phage sequences into its CRISPR array and analyzed thousands of spacers from over a hundred strains (9). The data suggested that the activity of CRISPR was correlated with the spacer diversity. Metagenomic studies of Leptospirillum environmental biofilm community species confirmed the extensive diversity of the spacer loci, suggested it was the result of a population-level response to a changing population of phage (6), and emphasized the role of phage evolution in maintaining the diversity of relevant spacers (10).
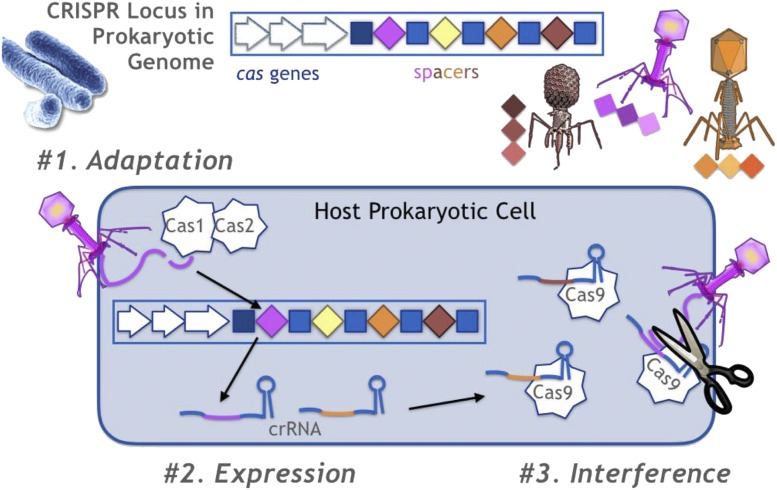
We now know that CRISPR is an adaptive immune system for bacteria that interacts with the diversity of the phage population (Fig. 1). Due to the insertion of spacers into the leader-proximal position, the CRISPR array provides a record of the phage challenges that the bacteria have faced. However, this record is shaped not only by the prevalence of phage but also by the selective advantages that the different spacers may confer. An early theoretical study showed that in environments with changing phage populations, leader-distal spacers are less diverse than leader-proximal spacers due to the selective pressure for targeting dominant phage genotypes (14), as had been observed in studies of environmental strains. A population dynamics model showed that a less diverse CRISPR spacer distribution that is focused on the most effective spacers evolved when phage had multiple protospacers that differ in their effectiveness, although a diverse spacer locus evolved when phage protospacers differed in their ease of acquisition (15). Moreover, large acquisition probabilities lead to a broader spacer distribution, whereas smaller acquisition probabilities lead to a spacer distribution focused on the most effective spacers. A spatially diverse model showed that the CRISPR array evolved to a length between 20 and 30 spacers (16).
Fig. 1.
CRISPR is an adaptive, heritable, genetic immune system for prokaryotes that operates in three phases of adaptation (1), expression (11), and interference (12). During adaptation, the CRISPR acquires spacers from the phages that the bacteria encounter in the environment. These spacers are inserted adjacent to the leader sequence at the start of the CRISPR array. During expression, small CRISPR RNAs (crRNAs) are produced from transcripts of the CRISPR array. During interference, the crRNAs guide Cas proteins to matching DNA sequences of invading phages, which are then cleaved by the Cas proteins. Reprinted from ref. 13, which is licensed under CC BY 3.0.
The article by Bradde et al. (2) complements these coevolutionary models to expand our understanding of how spacer diversity and CRISPR length relate to bacterial immunity against phage. By focusing on the limitations imposed due to an efficient use of a limited number of Cas proteins in a bacterium, Bradde et al. provide a theory that explains the driving forces that limit the size of the CRISPR immune system in bacteria. The theory quantifies the optimal size of the immune system as a balance between effectiveness of the CRISPR response against a specific phage strain and coverage of as many strains as possible.
In the typical limit of a high diversity of phage, the optimal length of the CRISPR array scales with the number of available Cas proteins, Np. What sets this scaling is the requirement of roughly dc Cas/crRNA complexes to suppress a given phage type within a single bacterium. If the array size is below the optimal value, fewer phage strains will be recognized, and this incurs a fitness penalty to the bacteria. This penalty explains the many observations of increased bacterial resistance to phage with increased spacer diversity (17). Conversely, if the size is above the optimal value, there are so many distinct potential spacers that the requirement of dc Cas/crRNA complexes per phage type is not able to be met. This penalty addresses the biochemical requirement for efficient use and low energy consumption of Cas proteins (13).
Interestingly, this argument holds even when all phage strains in the environment cannot be protected against. For typical phage diversities on the order of 103, the optimal CRISPR array may protect against 1 to 10% of the phage strains (2). The predicted spacer repertoire sizes of 10 to 100 are consistent with many empirical observations. The theory further predicts that a more effective CRISPR immune system, via either smaller dc or larger Np, leads to a longer optimal repertoire size and greater number of phage strains recognized. These predictions may explain why more active CRISPR loci tend to have a greater diversity of spacers (9). These results parallel previous theories of the adaptive immune system that predict a substantial coverage of rare antigens, rather than excess coverage of common antigens (18). Indeed, archaea seem to balance protection against low-abundance but persistent phages with protection against more abundant, but fleeting phage strains (19).
In PNAS, Bradde et al. present a theory of the tradeoff between broad coverage of phage types and efficient use of a limited number of enzymes in each bacterium that leads to an optimal size of the CRISPR immune system.
If the phage diversity is low, there is a different result for the optimal length of the CRISPR array. When multiple spacers can match any given phage type, there is no upper bound on the optimal array size in the theory. If the CRISPR is constrained to contain no more than a single spacer matching any given phage type, The CRISPR array scales as (Np K)1/2, where K is the phage diversity. If the array size is too small, the probability of recognizing any phage strain is too low. If the array size is too large, the assumption that only a single spacer in the array can match any given phage type implies that with high probability all of the Np Cas proteins are complexed with any of the L-1 spacers that are not specific for that phage type, i.e., that ineffective memory of prior phage strains inhibits an optimal response to current strains.
There are several biological implications of this theory. Since the optimal array size is limited by the number of Cas proteins, Cas protein expression is likely to be regulated. For example, a higher diversity of attacking phage is likely countered by greater Cas protein expression. Overexpression of Cas protein might even be desirable in engineered strains under these conditions. When expression of sufficient Cas protein imposes too high a fitness cost, other forms of immunity such as surface receptor modification likely become significant (20). While a larger activation threshold, dc, for CRISPR suppression might increase specificity and reduce off-target effects, it is predicted by the theory to reduce the optimal repertoire size, L ∼ Np/(2dc). If specificity is beneficial under conditions of high phage diversity, for example to avoid self-targeting, other mechanisms to increase specificity that do not involve the use of multiple Cas proteins might be expected to arise.
The development of the CRISPR field has relied heavily on bioinformatic methods (21). The paper by Bradde et al. (2) emphasizes that results from theory and simulation have a role to play in deepening our understanding of this fascinating bacterial immune system.
Footnotes
The author declares no competing interest.
See companion article, “The size of the immune repertoire of bacteria,” 10.1073/pnas.1903666117.
References
- 1.Barrangou R., et al. , CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Bradde S., Nourmohammad A., Goyal S., Balasubramanian V., The size of the immune repertoire of bacteria. Proc. Natl. Acad. Sci. U.S.A. 117, 5144–5151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mojica F. J. M., Díez-Villaseñor C., García-Martínez J., Soria E., Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Pourcel C., Salvignol G., Vergnaud G., CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Bolotin A., Quinquis B., Sorokin A., Ehrlich S. D., Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Tyson G. W., Banfield J. F., Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ. Microbiol. 10, 200–207 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Diez-Villasenor C., Almendros C., Garcia-Martinez J., Mojica F., Diversity of CRISPR loci in Escherichia coli. Microbiology 156, 1351–1361 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Weinberger A. D., et al. , Persisting viral sequences shape microbial CRISPR-based immunity. PLoS Comput. Biol. 8, e1002475 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath P., et al. , Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190, 1401–1412 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson A. F., Banfield J. F., Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320, 1047–1050 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Brouns S. J., et al. , Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garneau J. E., et al. , The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Bonomo M. E., Deem M. W., The physicist’s guide to one of biotechnology’s hottest new topics: CRISPR-Cas. Phys. Biol. 15, 041002 (2018). [DOI] [PubMed] [Google Scholar]
- 14.He J., Deem M. W., Heterogeneous diversity of spacers within CRISPR (clustered regularly interspaced short palindromic repeats). Phys. Rev. Lett. 105, 128102 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Bradde S., Vucelja M., Teşileanu T., Balasubramanian V., Dynamics of adaptive immunity against phage in bacterial populations. PLoS Comput. Biol. 13, e1005486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haerter J. O., Sneppen K., Spatial structure and Lamarckian adaptation explain extreme genetic diversity at CRISPR locus. MBio 3, e00126–e12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Houte S., et al. , The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer A., Balasubramanian V., Mora T., Walczak A. M., How a well-adapted immune system is organized. Proc. Natl. Acad. Sci. U.S.A. 112, 5950–5955 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emerson J. B., et al. , Virus-host and CRISPR dynamics in Archaea-dominated hypersaline Lake Tyrrell, Victoria, Australia. Archaea 2013, 370871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin B. R., Nasty viruses, costly plasmids, population dynamics, and the conditions for establishing and maintaining CRISPR-mediated adaptive immunity in bacteria. PLoS Genet. 6, e1001171 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lander E. S., The heroes of CRISPR. Cell 164, 18–28 (2016). [DOI] [PubMed] [Google Scholar]