Significance
The gut-originating pathogen Escherichia coli has been associated with a portion of cases of late-onset sepsis (LOS), a leading cause of neonatal mortality. While it remains unclear how E. coli may gain access to the systemic circulation from the intestine, breast milk may protect against bacterial translocation and reduces the risk of LOS. Here we show a mechanism whereby gut-residing E. coli gain systemic access and have developed an animal model replicating this mechanism to explore the protective effects of breast milk in LOS.
Keywords: goblet cells, breast milk, sepsis, neonate, bloodstream infections
Abstract
Late-onset sepsis (LOS) is a highly consequential complication of preterm birth and is defined by a positive blood culture obtained after 72 h of age. The causative bacteria can be found in patients’ intestinal tracts days before dissemination, and cohort studies suggest reduced LOS risk in breastfed preterm infants through unknown mechanisms. Reduced concentrations of epidermal growth factor (EGF) of maternal origin within the intestinal tract of mice correlated to the translocation of a gut-resident human pathogen Escherichia coli, which spreads systemically and caused a rapid, fatal disease in pups. Translocation of Escherichia coli was associated with the formation of colonic goblet cell-associated antigen passages (GAPs), which translocate enteric bacteria across the intestinal epithelium. Thus, maternally derived EGF, and potentially other EGFR ligands, prevents dissemination of a gut-resident pathogen by inhibiting goblet cell-mediated bacterial translocation. Through manipulation of maternally derived EGF and alteration of the earliest gut defenses, we have developed an animal model of pathogen dissemination which recapitulates gut-origin neonatal LOS.
Late-onset neonatal sepsis (LOS) can be caused by a variety of pathogens, occurs at least 72 h after birth, accounts for 26% of all deaths in preterm infants, and results in an increased risk of long-term neurocognitive problems (1, 2). LOS has remained an important challenge in preterm infant care (3) due to the continuing reduction in the age of viability of very low birth weight (VLBW, birth weight ≤ 1,500 g) infants, who are at greater risk for bloodstream infections (BSIs), with an incidence of 10% and a mortality rate between 30 to 50% (1). Recent clinical data indicate incidence of LOS may be decreasing, though it remains unclear which interventions have the most impact in reduction of neonatal BSIs and LOS (4). Great effort has been devoted to hygiene related to invasive procedures (5, 6), and there is increasing focus on breastfeeding (7, 8).
In a substantial portion of LOS cases, the causative pathogen can be found to reside in the gut before it disseminates (9–12). It is hypothesized that an immature intestinal barrier enables translocation of such resident gut bacteria, but the mechanisms allowing or inhibiting translocation of the gut microbiota in early life remain unknown. These infections occur during the first 60 d of life, a time in life when exclusive breastfeeding is recommended. Formula-fed preterm infants have greater intestinal permeability compared to breast milk-fed preterm infants (13), and in animal models, formula feeding enables bacterial translocation from the gut to the mesenteric lymph nodes (MLNs) and the liver (14–16). Epidermal growth factor (EGF) is a growth factor that is maternally supplied with high concentrations in colostrum and decreases in concentration in breast milk throughout lactation (17–19), and reduces bacterial translocation in formula-fed animals (20).
Here we disrupt EGF receptor (EGFR) signaling in neonatal mice and use asynchronously cross-fostered pups to reduce high luminal EGF concentrations originating from maternal milk perinatally, both of which result in bacterial translocation from the gut to the mesenteric lymph nodes and spleen. Following EGFR disruption or asynchronously cross-fostering, mice developed a rapid fatal response to oral challenge of Escherichia coli isolated from the bloodstreams of LOS patients, which were also of gut origin in those neonates. However, other gut-resident commensal E. coli from infants without disease were not lethal, despite their ability to translocate in this model. E. coli translocation was associated with goblet cell-associated antigen passages (GAPs) formation by colon goblet cells and inhibited by luminal EGF via EGFR activation in goblet cells. Thus maternal EGF, and potentially other EGFR ligands, inhibits translocation of a gut-residing pathogen in offspring by directly acting on goblet cells and inhibiting GAP formation in the offspring’s colon, thereby interdicting gut-resident pathogens from traversing the epithelium and gaining systemic access.
Results
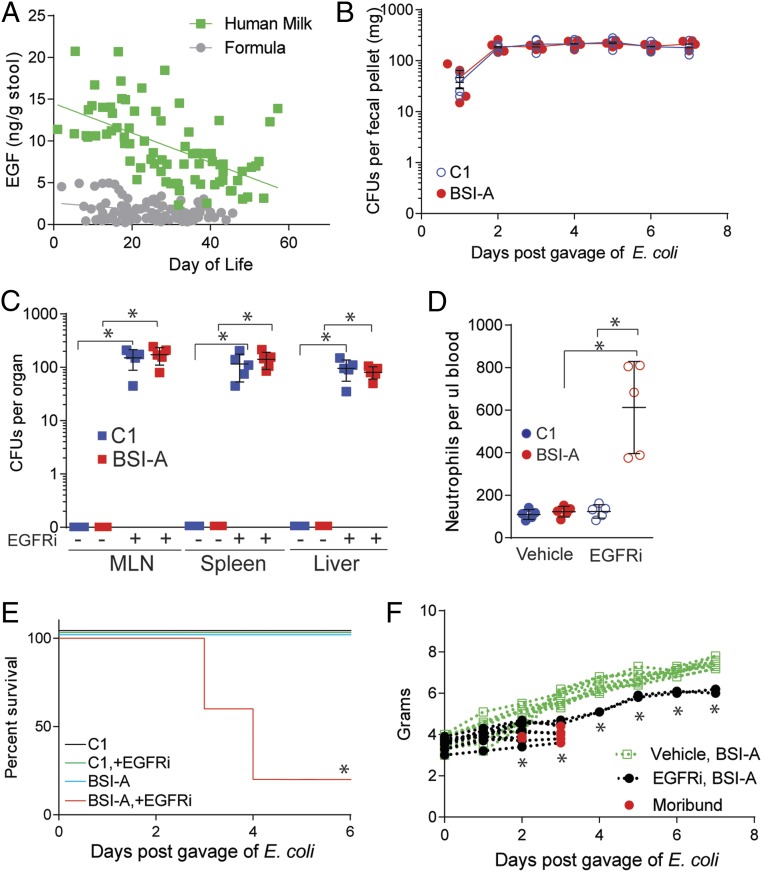
Infants born preterm and fed mother’s own milk (MOM) have a reduced risk of LOS (21, 22). EGF is abundant in breast milk after parturition, decreases throughout lactation, and has been observed to improve intestinal barrier function and reduce bacterial translocation in offspring (20). Therefore, we measured EGF in the stool of VLBW infants (SI Appendix, Table S1) that were fed MOM or formula over the first 60 d of life to assess the contribution of maternal breast milk to luminal EGF concentrations. Stools from the breastfed children contained significantly greater concentrations of EGF, and this concentration diminished over time, reflecting the reduction in concentration in breast milk of EGF (17–19) (Fig. 1A and SI Appendix, Fig. S1A). Similar temporal trends of EGFR ligands amphiregulin (AREG), transforming growth factor alpha (TGF-α), and heparin-binding EGF like growth factor (HB-EGF), which have been reported to be present in human breast milk (17, 23, 24), were not observed in the stool. Additionally, there were no differences in the fecal concentrations of AREG, TGF-α, and HB-EGF in the stools of MOM-fed or formula-fed children, which were overall less abundant than EGF in the stool, particularly in the first 4 wk of life (SI Appendix, Fig. S1 B–D, compare to Fig. 1A). EGF concentrations in murine milk also diminished over time, reflecting a gradient that could also be measured in the stool (SI Appendix, Fig. S2A) and lumen (25) of pups during early life, a time when neonatal mice immunologically resemble the preterm human infant gut (26, 27).
Fig. 1.
Inhibition of EGFR results in bacterial translocation of gut-residing E. coli. (A) Concentration of EGF as measured by ELISA from the stool of children fed mother’s own milk (green squares) or formula (gray circles). In B–F, conventionally reared mice were gavaged with 2 × 105 CFUs of E. coli and injected with EGFRi on DOL5. (B) CFUs in stool following gavage of 2 × 105 CFUs of E. coli C1nalR (blue) or BSI-AnalR (red) in conventionally reared mice on DOL5. (C) CFUs in mesenteric lymph node (MLN), spleen, and liver 3 d following gavage of 2 × 105 CFUs of E. coli C1nalR (blue) or BSI-AnalR (red) on DOL5 in conventionally reared mice injected with tyrphostin AG1478 (EGFRi) or vehicle. (D) Neutrophils in the blood 48 h following gavage of 2 × 105 CFUs of E. coli C1nalR or E. coli BSI-AnalR and injected with EGFRi as indicated or vehicle. (E) Survival of mice following vehicle or EGFRi injection and gavage of 2 × 105 CFUs of E. coli C1nalR or BSI-AnalR. (F) Weight of mice following gavage of 2 × 105 CFUs of E. coli BSI-AnalR and injected with EGFRi (black), or vehicle (green). Red dots denote moribund pups. n = 5 individuals in each group with multiple time points in A. n = 5 mice per group in B–D. n = 15 mice per group from 3 independent litters in E. n = 10 mice per group in F. Individual data points plotted in B–D with mean and SD plotted per group. Statistics used: Linear regression (A), Mann–Whitney (C), one-way ANOVA (D), two-way ANOVA (F), Kaplan–Meier (E), *denotes statistical significance, P < 0.05 or less.
We next asked if maternal EGF induces a response in the offspring that prevents translocation of gut bacteria via activation of the EGFR. Pups were colonized with E. coli by oral gavage of 2 × 105 colony-forming units (CFUs) of E. coli BSI-AnalR, a nalidixic acid-resistant mutant of an E. coli isolated on day of life 21 from the bloodstream of an infant born at 28 wk gestation (9) or colonized with a nalidixic acid-resistant mutant E. coli C1nalR (SI Appendix, Table S2), a commensal E. coli isolated from the intestinal tract of an infant without a bloodstream infection and injected intraperitoneally (i.p.) with vehicle or an inhibitor of EGFR activation, tyrphostin AG1478, on day of life (DOL)5. Both E. coli strains were found in the stool of the pups for at least 7 d following gavage confirming colonization (Fig. 1B); no nalidixic acid-resistant bacteria were found in the stool of the dam or pups prior to inoculation (SI Appendix, Fig. S2B). E. coli BSI-AnalR or C1nalR could be found in the stool of all mice 3 d after gavage, but only the MLNs, spleen, and liver of pups treated with epidermal growth factor receptor inhibitor (EGFRi) contained E. coli BSI-AnalR or C1nalR (Fig. 1C). EGFRi-treated mice developed neutrophilia (Fig. 1D) and died following colonization with E. coli BSI-AnalR but not C1nalR (Fig. 1E). Illness was characterized by a lack of weight gain (Fig. 1F), and lethargy. Mice injected i.p. with BSI-A, but not C1, succumbed rapidly, confirming the virulence of BSI-A as a pathogen when introduced directly into the body cavity of mice (SI Appendix, Fig. S3). The mean lethality rate per litter in EGFRi-treated mice exposed to BSI-A was 75% (SI Appendix, Fig. S4). Thus, following inhibition of EGFR in pups, E. coli resident in the gut disseminate and, depending on the microbe, can cause rapid and significant lethality.
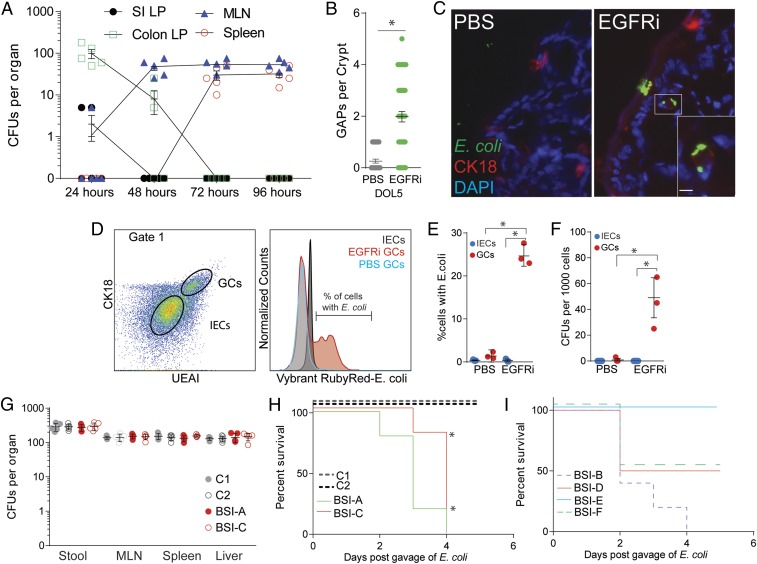
To identify where E. coli translocate following colonization, small intestine and colon lamina propria, mesenteric lymph nodes, and spleen were homogenized and plated on MacConkey agar plates containing nalidixic acid following oral challenge and EGFRi injection. At 24 h, E. coli BSI-AnalR were largely found in the lamina propria of the colon, but not small intestine. E. coli BSI-AnalR was recovered from the MLN after 48 h and from the spleen and liver after 72 h (Fig. 2A). These data suggest that E. coli BSI-AnalR translocate via the colon epithelium. Goblet cells, intestinal epithelial cells of the secretory lineage, have been observed to be a portal of entry for pathogens (28, 29) and are present and functional in the neonatal intestinal epithelium following birth (30). Specifically, colon goblet cells can deliver antigens and live bacteria from the intestinal lumen to the colon lamina propria and colon mesenteric lymph node, through the formation of goblet cell-associated antigen passages (GAPs) (25, 31). Luminal EGF inhibits colon GAP formation in neonates through EGFR activation specifically on goblet cells (25, 32), and accordingly GAP formation occurred in the colon of pups following treatment with EGFRi (Fig. 2B). Using immunohistochemistry and flow cytometry, we observed that fluorescently labeled E. coli BSI-A associated with cytokeratin (CK)-18+ goblet cells in the colon of mice following EGFRi treatment but not with intestinal epithelial cells (IECs) of EGFRi-treated mice or goblet cells of vehicle-treated mice (Fig. 2 C–E). Live BSI-AnalR could be isolated from fluorescence-activated cell (FACS)-sorted goblet cells (GCs), but not IECs, of EGFRi-treated mice (Fig. 2F), confirming that translocation of live E. coli occurs via GCs and correlates with GAP formation following EGFRi treatment.
Fig. 2.
Translocation of E. coli is associated with colon GAP formation. Conventionally reared mice were gavaged with 2 × 105 CFUs of E. coli and injected intraperitoneally with EGFRi on DOL5. (A) CFUs per organ (small intestine lamina propria [SI LP], colon lamina propria [Colon LP], mesenteric lymph node [MLN], and spleen), following gavage of 2 × 105 CFUs of BSI-ANalR in EGFRi-treated mice. (B) GAPs per colon crypt in conventionally reared mice treated with EGFRi or vehicle. (C) Immunohistochemistry image of colon epithelium from a vehicle (PBS, Left) or EGFRi-treated (Right) mouse following gavage of CFSE-labeled E. coli BSI-A (green) stained with CK-18 (red), and DAPI (blue), Inset of single CK-18+ goblet cell containing E. coli. (Scale bar: 10 μm.) (D) Gating strategy for colon CD45− epithelial cells showing UEAI+CK18+ goblet cells (GCs) and UEA-I−CK18− intestinal epithelial cells (IECs) containing Vybrant RubyRed-labeled E. coli BSI-A. (E) Percentage of GCs or IECs containing E. coli BSI-A. (F) CFUs of BSI-AnalR in IECs and GCs FACS-sorted based on the gating strategy in D. (G) CFUs in stool, mesenteric lymph node (MLN), spleen, and liver 3 d following gavage of 2 × 105 CFUs of E. coli C1nalR, 2nalR, BSI-AnalR, BSI-CnalR in EGFRi-treated mice. (H) Survival of EGFRi-treated mice following gavage of 2 × 105 CFUs of E. coli strains. Dotted lines denote commensal strains, not known to cause disease; solid lines denote E. coli isolated from patient’s bloodstream (SI Appendix, Table S2). (I) Survival of EGFRi-treated mice following gavage of 2 × 105 CFUs of E. coli strains isolated from patient’s bloodstream (SI Appendix, Table S2). n = 5 mice per group in A and G. n = 3 mice per group in B, E, and F. Representative image and flow plots of three independent mice shown in C and D. n = 5 mice per group in H. Individual data points plotted in A, B, and E–G with mean and SD plotted per group. Statistics used: Mann–Whitney (B), one-way ANOVA (E and F), Kaplan–Meier (H), *denotes statistical significance, P < 0.05 or less.
We expanded our testing to additional strains of E. coli isolated from preterm infants. The nalidixic acid-resistant mutants of E. coli BSI-C, a bloodstream isolate from a sepsis patient (9), and E. coli C2, a gut commensal not known to cause disease that was isolated from the stool of an infant, translocated similarly following colonization and EGFRi administration (Fig. 2G). Only BSI-AnalR and BSI-CnalR caused morbidity in pups that had been administered EGFRi (Fig. 2H). Furthermore, three of four other E. coli bloodstream isolates from sepsis patients were lethal in pups following EGFRi treatment (Fig. 2I and SI Appendix, Table S2), suggesting that E. coli from bloodstream infections result in systemic disease, but commensal E. coli do not, even though they both disseminate. Initial screen for recognized E. coli virulence factors using whole genome sequences from BSI-A, BSI-C, C1, and C2 revealed no common virulence factors present in both of the pathogenic strains that were not present in at least one of the nonpathogenic strains. Bloodstream isolates BSI-A and BSI-C and control isolates C1 and C2 have four, seven, four, and three putative virulence loci, respectively. Only four loci were shared by more than one isolate: fliC, which encodes flagellar filaments, hlyE, which encodes a pore-forming hemolysin, and iss and traT, which encode proteins that confer resistance to serum complement (SI Appendix, Table S3).
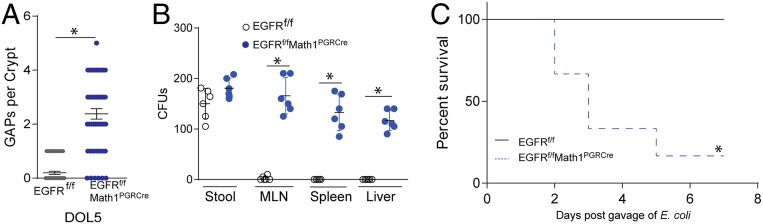
Deletion of EGFR from goblet cells using an inducible Math1-driven Cre recombinase resulted in GAP formation on DOL5 in EGFRf/fMath1PGRCre mice (Fig. 3A). E. coli BSI-AnalR and C1nalR could be found in the stool of all mice 3 d after gavage indicating colonization, but only disseminated to the MLN, spleen, and liver of EGFRf/fMath1PGRCre mice, replicating translocation of bacteria following EGFR inhibition (Fig. 3B). Again, mice quickly succumbed following infection with E. coli BSI-AnalR but not C1nalR following translocation (Fig. 3C). Thus, sensing by EGFR on goblet cells regulated bacterial translocation across the colon epithelium.
Fig. 3.
Goblet cell expression of EGFR is necessary for EGF-driven prevention of bacterial translocation. (A) Counts of GAPs per colon crypt in EGFRf/fMath1PGRCre or EGFRfl/fl littermates. (B) CFUs in stool, mesenteric lymph node (MLN), spleen, and liver 3 d after gavage of 2 × 105 CFUs of E. coli BSI-AnalR in conventionally reared EGFRf/fMath1PGRCre or EGFRfl/fl littermates. (C) Survival of EGFRf/fMath1PGRCre or EGFRfl/fl littermates following gavage of 2 × 105 CFUs of E. coli BSI-AnalR. n = 6 mice per group in A–C. Individual data points plotted in A and B with mean and SD plotted per group. Statistics used: Mann–Whitney (A and B) Kaplan–Meier (C), *denotes statistical significance.
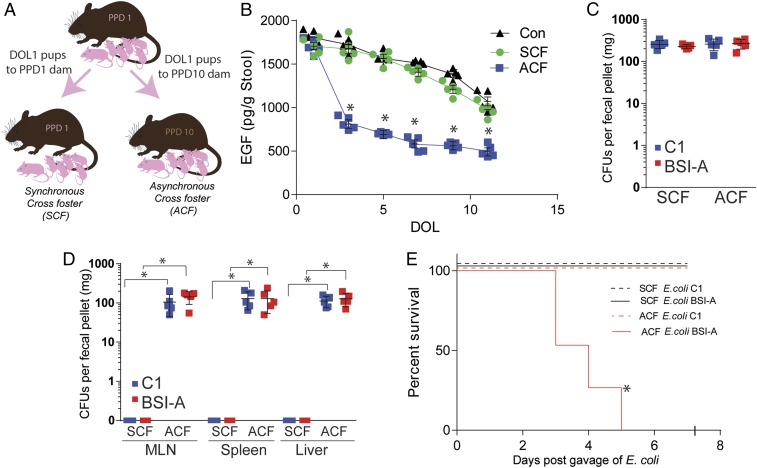
The concentration of EGF in murine milk decreases throughout lactation (SI Appendix, Fig. S2A). To model the exposure to low EGF concentrations seen in formula-fed children, we asynchronously cross-fostered mice by housing 1 d old pups with dams that delivered 10 d earlier (Fig. 4A). Asynchronously cross-fostered (ACF), but not synchronously cross-fostered (SCF) pups (1 d old pups cross-fostered to a different dam having delivered a litter on the same day, Fig. 4A), had reduced concentrations of EGF in their stool, suggesting that fecal EGF quantities in pups reflect the EGF concentration in their source of maternal milk (Fig. 4B).
Fig. 4.
Asynchronous cross-fostering results in translocation of gut-residing E. coli. (A) Schematic of asynchronous cross-fostering (ACF) protocol. (B) Concentration of EGF in stool of conventionally reared (Con, black triangle), SCF (green circles), or ACF (blue squares) pups. (C) CFUs in stool 3 d following gavage of 2 × 105 CFUs of E. coli C1nalR (blue) or BSI-AnalR (red) in SCF and ACF mice. (D) CFUs in mesenteric lymph node (MLN), spleen, and liver 3 d following gavage of 2 × 105 CFUs of E. coli C1nalR (blue) or BSI-AnalR (red) in SCF and ACF mice. (E) Survival of SCF and ACF mice following gavage of 2 × 105 CFUs of E. coli C1nalR or BSI-AnalR in SCF and ACF mice. n = 5 in B–D. n = 15 mice from three independent litters per group. Individual data points plotted in B–D with mean and SD plotted per group. Statistics used: two-way ANOVA (B), Mann–Whitney (D), Kaplan–Meier (E), *denotes statistical significance, P < 0.05 or less.
To evaluate if ACF enables bacterial translocation of gut-resident bacteria and potentiates sepsis, mice were ACF on DOL1 and colonized with C1nalR or BSI-AnalR on DOL5. Three days postinfection, E. coli BSI-AnalR and C1nalR were found in the stool of both SCF and ACF mice indicating all mice were colonized (Fig. 4C), but were only found in the MLN, spleen, and liver of ACF mice. This is similar to the results in the EGFRi-treated mice (Fig. 4D), and supports that ACF results in bacterial translocation and systemic dissemination. Additionally, ACF mice quickly succumbed following colonization with E. coli BSI-AnalR but not C1nalR (Fig. 4E). Both SCF and ACF mice died equally in response to i.p.-injected E. coli BSI-AnalR, suggesting maternal disruption of EGF in ACF mice specifically increases risk of sepsis due to enteric pathogens, but not systemically introduced pathogens (SI Appendix, Fig. S5).
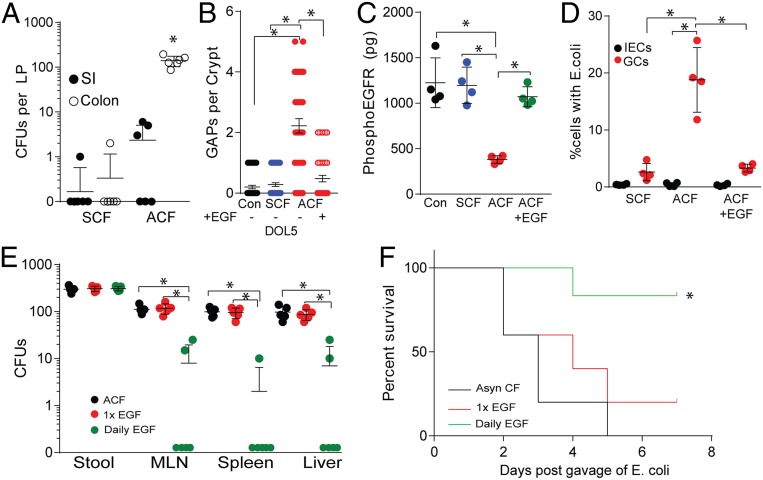
E. coli BSI-AnalR was recovered from the colon lamina propria and, to a significantly lesser extent, the small intestine lamina propria 24 h after gavage of ACF mice, (Fig. 5A) similar to the observations in EGFRi-treated mice (Fig. 2A). GAPs formed in the colons of ACF pups, but not SCF pups, and this formation was inhibited by luminal EGF (Fig. 5B). Additionally, the epithelium of ACF pups contained significantly decreased phosphorylated EGFR, indicating EGFR is less activated in ACF mice. Phosphorylation of EGFR on the epithelium was restored upon luminal EGF administration (Fig. 5C) suggesting reduced activation of epithelial EGFR is consistent with reduced luminal EGF in ACF mice. Additionally, E. coli BSI-AnalR was found within GCs, but not IECs, of the ACF mice, and the presence of E. coli BSI-AnalR in GCs was significantly diminished following EGF administration (Fig. 5D). To confirm that luminal EGF was sufficient to protect from gut-origin sepsis in ACF mice, ACF mice were infected with E. coli BSI-AnalR and gavaged with either a single dose of recombinant murine EGF at the time of infection, or daily administrations of EGF throughout the course of the infection model. ACF mice given daily EGF had reduced bacterial translocation to the MLN, spleen, and liver even though colonization was not affected (Fig. 5E). Finally, daily EGF significantly protected ACF mice following BSI-AnalR infection (Fig. 5F). In contrast, daily administration of EGF to EGFRi-treated mice or EGFRf/fMath1PGRCre mice failed to inhibit GAP formation, prevent bacterial translocation, or protect from disease (SI Appendix, Fig. S6), indicating the necessity of EGFR ligands to be actively sensed by the offspring to provide protection from translocating bacteria. Taken together, these data suggest disruption of maternally derived EGF, and potentially other EGFR ligands, reduces EGFR activation, GAP formation by GCs, and bacterial translocation via colon GAPs. Additionally, we found dissemination of pathogenic E. coli but not commensal E. coli resulted in a fatal sepsis-like disease in pups following EGFR ligand or EGFR disruption.
Fig. 5.
Luminal EGF protects against bacterial translocation and susceptibility to bloodstream infections. (A) CFUs in lamina propria of small intestine or colon following gavage of 2 × 105 CFUs of E. coli BSI-AnalR in ACF mice. (B) Counts of GAPs per colon crypt in SCF or ACF mice and gavaged with EGF (10 ng/mL) or vehicle. (C) Concentration of phosphorylated EGFR per colon epithelial fraction as measured by ELISA. GAPs (black), ACF given a single gavage of recombinant murine EGF at the time of infection (red), or ACF mice given daily gavages of EGF (green). (D) Percentage of GCs or IECs containing E. coli BSI-AnalR. (E) CFUs in stool, mesenteric lymph node (MLN), spleen, and liver in the days following gavage of 2 × 105 CFUs of E. coli BSI-AnalR in ACF mice treated with EGF. (F) Survival of ACF mice following gavage of 2 × 105 CFUs of E. coli BSI-AnalR in ACF mice (black), ACF gavaged with EGF (10 ng/mL) at the time of infection (red), or ACF mice given daily gavages of EGF (10 ng/mL) (green). n = 4 mice in A, n = 4 mice in B–D. n = 5 in E. n = 6 mice per group in F. Individual data points plotted in A–E with mean and SD plotted per group. Statistics used: one-way ANOVA (A–E), Kaplan–Meier (F), *denotes statistical significance, P < 0.05 or less.
Discussion
The many postulated beneficial effects of breast milk, particularly MOM, include reducing intestinal permeability (13) and preventing bacterial translocation (14–16). These effects plausibly contribute to the partial reduction of the risk of LOS among breastfed preterm infants (21, 22). EGF and other EGFR ligands, components of breast milk, can contribute to each of these functions through direct ligation of the EGFR on goblet cells in the colon of offspring. Breast milk contains EGF and other EGFR ligands, such as AREG, TGF-α, and HB-EGF (17, 23, 24), though at reduced concentrations compared to EGF (18, 19). We found no significant difference in the concentration of AREG, TGF-α, and HB-EGF in the stool between MOM-fed and formula-fed children, suggesting maternally derived EGF is the predominant EGFR ligand biologically available throughout the entirety of the GI tract, including the lower large intestine. We recognize that other EGFR ligands may also be affected following ACF and EGFR inhibition and that in some systems EGFR ligands can have complementary and overlapping roles (33). However, the relatively higher levels of EGF in breastmilk, and the ability of enteral recombinant EGF in mice to restore the intestinal barrier function, prevent bacterial translocation, and avert disseminated bacterial infection after enteral challenge with a pathogenic E. coli strongly suggest that EGF is a protective element in maternal milk.
Other breast milk constituents might confer protection from enteric pathogens including maternal immunoglobulin A (IgA) (34, 35) and oligosaccharides (36), both of which are also found in higher concentrations in breast milk soon after parturition compared to later in lactation and may also be disrupted by ACF. The individual contributions of these maternal factors can be explored in the ACF model as IgA may contribute to protection from invasive enteric pathogens through directly binding and preventing adherence and evasion (37, 38), and oligosaccharides support the maturation of the normal infant microbiota that may provide colonization resistance (39, 40).
Other models of enteric infections have demonstrated invasive organisms associate with goblet cells (28, 41). We extend this finding to GAP formation by goblet cells in the colon consistent with our previous work showing bacterial translocation through the colon occurs when bacteria pass transcellularly through goblet cells when forming GAPs (31). EGFR activation inhibits GAP formation in early life (25), suggesting a role for maternally derived EGFR ligands is to limit bacterial translocation and protect the offspring from potentially invasive enteric pathogens. EGF did not appear to have an effect on bacterial habitation in the lumen, as all pups were colonized similarly by evidence of E. coli measured in stool. Rather we find direct sensing of EGFR ligands by the EGFR on goblet cells in the colon of the offspring provides protection from bacterial translocation and bloodstream infection. The recent initiative to provide neonatal intensive care unit (NICU) patients with human milk through donor milk banks when MOM is unavailable is a laudable endeavor to replace formula with a more complete diet offering nutrition and protection to the neonate. However, our work raises the possibility that human milk from donors close to parturition may offer superior protection compared to human milk expressed later in lactation, due to temporal availability of bioactive factors present in human milk.
While all E. coli strains spread systemically, only those isolated from the bloodstream of LOS cases resulted in noticeable disease or death in the pups, despite similar translocation and bacterial load in extraintestinal tissue. We have previously shown gut-resident commensal bacteria translocating via GAPs circulate systemically without causing overt disease (31). Combined with the previous observation, the variation among E. coli isolates from LOS cases seen in this set of experiments suggest specific bacterial factors determine virulence following translocation from the intestine. Initial screens for virulence loci found no common factors present in both of the pathogenic strains that were not present in at least one of the nonpathogenic strains. Our identification of several virulence loci in the bloodstream isolates and in the controls is potentially interesting, but the assignment of a virulence locus in E. coli obligates large specimen sets from cases and controls and extensive in vitro and in vivo experimental data to infer mechanisms. There are many studies of extraintestinal pathogenic E. coli virulence loci, but bloodstream isolates from infants are underrepresented in these analyses. The association between strains and outcomes should be considered to be provisional, as one of five bloodstream isolates did not cause disease in mice. An intensive survey of pathogenicity of neonatal wild-type E. coli would be informative. Also, even though this study is limited to E. coli organisms, LOS can be caused by multiple different genera, including Enterococci, Streptococci, and other Enterobacteriaceae species (3). Though the generalizability of GAP translocation as a portal of entry of specific pathogens is not established by our data, we have previously shown both Gram-negative and Gram-positive bacteria can translocate via GAPs (31). Further work will determine the variety of bacteria that can utilize goblet cells to translocate in early life following EGFR disruption and should pursue additional and potentially complementary mechanisms, such as toll-like receptor (TLR) activation in the gut mucosa. Compared to models of neonatal sepsis utilizing i.p. injection of lipopolysaccharide (LPS), or cecal contents, ACF more faithfully recapitulates the enteric route of entry that has been associated with late-onset sepsis cases (9–12, 42). Combining the previous studies with the observation that the enteric route of entry has been shown to contribute to virulence and disease when compared to i.p. injection (43), ACF represents a physiologic route for bacterial translocation. ACF and colonization offer a minimally invasive model of dissemination without pharmacological manipulation or physical breach of the intestinal or skin barriers.
In summary, our model of enterally acquired bloodstream infections following translocation of a gut-residing pathogen demonstrates that maternally derived EGF, and potentially other EGFR ligands, can prevent this process. GAPs, which formed in response to decreased or disrupted EGFR signaling, were exploited by enteric bacteria. Depending on the bacteria, translocation resulted in the rapid sepsis-like death of offspring in this model of LOS. This model has profound relevance and application to the study of LOS and the development of therapies for the prevention and treatment of LOS unique to the neonatal phase of life.
Materials and Methods
Mice.
All mice were bred for 10 or more generations on the C57BL/6 background. Math1PGRCre mice (44) were purchased from The Jackson Laboratory and bred and maintained in-house. EGFRf/f mice (45) were a gift of Dr. David Threadgill, University of North Carolina, Chapel Hill, NC. To genetically deplete EGFR in GCs, mice were bred to generate EGFRf/fMath1PGRCre and EGFRf/f littermates. These mice were injected i.p. with mifepristone (Caymen Chemical Company) (10 mg/kg), which was dissolved at a concentration of 2 mg/mL in sunflower oil, on DOL2, 4, and 6. Cohoused littermates were used when possible as experimental groups and controls to minimize differences in the gut microbiota. Animal procedures and protocols were performed in accordance with the Institutional Animal Care and Use Committee at Washington University School of Medicine. In some experiments, mice were injected i.p. with 500 μg/kg tyrphostin AG1478 (EGFRi) (Sigma Aldrich) on DOL5 to inhibit EGFR activation. EGFRi was initially diluted in dimethyl sulfoxide (DMSO) to a concentration of 3.3 mg/mL, then further diluted in phosphate buffered saline (PBS) to a concentration of 33 μg/mL. Mice were injected with 16.6 μL, per gram bodyweight on DOL5 (25). In some experiments, mice were gavaged with 100 ng recombinant EGF (Shenandoah) in 50 μL PBS. Only litters from naïve mothers not previously exposed to E. coli isolates were used for experiments.
Human Studies.
Bloodstream and fecal isolates of E. coli, and infant stools, were obtained from a prospective case-cohort study of the role of gut microbial populations in necrotizing enterocolitis in infants hospitalized in the NICUs at St. Louis Children’s Hospital and the Children’s Hospital of Oklahoma University, and a study of the postdischarge microbiome in infants who had been hospitalized at St. Louis Children’s Hospital. Both site’s institutional review board approved this study; written informed consent was obtained from parents before enrollment. Details of the cohorts and methodology of stool collection are provided in the SI Appendix. Some of the E. coli strains used in this study have been previously published (9, 46). Pathogenic E. coli (E. coli BSI-A, BSI-B, BSI-C, BSI-D, BSI-E, BSI-F, SI Appendix, Table S2) were isolated from the bloodstream of NICU patients. Strains BSI-A and BSI-C were also identified in the patient’s stool prior to onset of clinical deterioration and positive blood culture diagnosing LOS (9), and we did not seek cognate E. coli in the stools from the other patients from whom we isolated bloodstream E. coli. Commensal E. coli species (E. coli, C1, C2, SI Appendix, Table S2) were isolated from the stool of discharged pediatric patients, and not known to cause disease. DNA was isolated from single colonies and sequenced following PCR amplification of the adk, fumC, gyrB, icd, mdh, purA, and recA genes for multilocus sequence type (MLST) identification (Genewiz) (SI Appendix, Table S2). Full genome sequences of BSI-A (9), BSI-C (9), C1 (46), and C2 (47) are publicly available. PRJNA/GenBank/Sequence Read Archive (SRA) run accession nos. BSI-A (ST69): 203059/ATNW00000000/SRR769039 and BSI-C (ST70): 203060/ATNV00000000/SRR769057; C1 (ST35): accession no. GCA_008081795.1; C2: PRJNA602838/SAMN13909231/JAADAA000000000.
Virulence Factor Screen.
Primers for extraintestinal pathogenic (ExPEC) virulence factors were retrieved from the literature and used for in silico PCR with the command line tool primer search from EMBOSS (48). Only assemblies with matches for both primers for each gene were reported. In addition, assemblies were analyzed against the VirulenceFinder database and hits against ExPEC virulence genes are reported (49).
Stool Samples.
Frozen stool previously collected was identified from patients 1) having either an exclusive human milk (MOM) or formula-fed diet during the first 60 d of life, 2) negative for necrotizing enterocolitis or sepsis, and 3) having at least 10 stool specimens available from birth through day of life 60. Five breastfed patients and five formula-fed patients were included in this study, with at least 10 time points per individual between DOL0 and DOL60. Patient metadata are available in SI Appendix, Table S1.
Quantification of EGFR Ligands from Stool.
Frozen stool specimens were resuspended in PBS, homogenized, and analyzed by enzyme-linked immunosorbent assay (ELISA) for human EGF (R&D Systems), human amphiregulin (AREG) (R&D Systems), human TGF-α (R&D Systems), and human heparin-binding EGF (HB-EGF) (R&D Systems), per the manufacturer’s protocol. For mice, stool was collected in PBS, homogenized, and analyzed by ELISA for murine EGF (R&D Systems), per manufacturer’s protocol.
Asynchronous Cross-Fostering.
At first sign of pregnancy, male breeders were removed from the cages of female breeders. On DOL1, pups were placed in a new cage with a dam that had delivered pups 10 d prior and was still actively nursing (asynchronous cross-fostering, ACF), or a dam that had delivered pups on the same birthdate as pups (synchronous cross-fostering SCF). All pups, bedding, and dam’s nostrils were wiped with imitation vanillin extract to increase litter acceptance by dam. Mice were monitored for litter acceptance and maternal care, and weighed daily.
E. coli Infection.
On DOL5, mice were gavaged with 20 µL PBS containing 107 CFUs/mL (equivalent to 2 × 105 CFU) E. coli. Stool was collected to monitor colonization. Mice were monitored for disease, including lack of weight gain, lethargy, pallor, lack of nursing, and death. In some experiments, moribund mice were euthanized to confirm translocation of enteric bacteria to extraintestinal organs and compare the MLST of recovered colonies to those of the challenge E. coli. Moribund was recognized as lethargic pups excluded from the nest that remained motionless when weighed on scale and failed to gain weight. In survival experiments, mice were monitored twice daily for signs of disease and lack of weight gain. In some mice, 24 h following gavage, small intestine and colon lamina propria were isolated to analyze bacterial translocation. In some mice, 3 d following gavage, spleen, MLN, and liver were isolated to analyze bacterial translocation. Litters of four or more were all colonized with the same E. coli strain to avoid cross-contamination and repeated at least three times to control for litter variability. In EGFRi experiments, litters of eight or more were all colonized with the same E. coli strain and then injected with EGFRi or PBS within the same litter to control for litter variability. EGFRf/fMath1PGRCre and EGFRf/f littermates within a single litter were all colonized with the same E. coli strain and treated with mifepristone.
Nalidixic Acid-Resistant E. coli Strains.
E. coli strains BSI-A, BSI-C, C1, and C2 were grown in 10 mL Luria-Bertani (LB) broth containing 4 μg/mL nalidixic acid (Acros Organics, Thermo Fisher Scientific). Isolates were plated on MacConkey agar plates containing 4 μg/mL nalidixic acid. The process was repeated using increasing amounts of nalidixic acid until E. coli strains grew in media containing 20 μg/mL nalidixic acid, and the MLST types were confirmed to remain unchanged. In all experiments, E. coli was grown by inoculating 10 mL of LB containing 4 μg/mL nalidixic acid with a frozen chip of E. coli glycerol stock (stock made from 109 CFUs in 1.5 mL PBS:glycerol in a 1:1 ratio). Cultures were grown shaking, at a 45° slant, at 37 °C to an optical density (OD) of 0.30 for a concentration of 108 CFUs/mL. Cultures were pelleted, washed in PBS, and resuspended to a concentration of 107 CFUs/mL in PBS for further use.
Blood Collection and Flow Cytometry.
On DOL5, mice were gavaged with 20 µL PBS containing 106 CFUs of E. coli and injected i.p. with EGFRi or vehicle. Forty-eight hours following gavage, whole blood was collected via cheek bleed. Two drops (10 μL) were collected directly into a microcentrifuge tube containing 90 μL of 50 mM ethylenediaminetetraacetic acid (EDTA). Neutrophils in blood were identified as CD45+ (monoclonal antibody 30-F11, eBioscience, Thermo Fisher Scientific) Ly6G/Ly6C+ (monoclonal antibody RB6-8C5, eBioscience). Flow cytometry was collected and analyzed on an Attune NxT flow cytometer (Invitrogen).
Bacterial Quantification in Organs and Stool.
Small intestines or colons were harvested, rinsed with PBS, and Peyer’s patches or colonic patches were removed and discarded. Epithelial cell populations were released by incubating the intestines for 15 min in a 37 °C rotating incubator in 20 mL Hank’s balanced salt solution (HBSS) media (BioWhittaker) containing 5 mM EDTA and gentamicin (50 μg/mL) as previously described (50). Following removal of epithelium, isolated lamina propria was cut into pieces. We recovered splenic, MLN, and lamina propria cells as previously described (50). Lamina propria pieces, spleen, MLN, liver, or stool were homogenized in 500 μL PBS with 200 mg 0.1 mm diameter zirconium silica beads (BioSpec), and homogenized Bullet Blender Tissue Homogenizer (Next Advance) vortexed on bead beater. Supernatant was plated on MacConkey agar containing 20 ng/mL nalidixic acid to identify nalidixic acid-resistant E. coli, and identity of E. coli strain was confirmed through MLST identification.
Extraction of Milk from Lactating Dams.
Milk was extracted from lactating dams every third day beginning on postpartum day 5 as described (51).
GAP Quantification, Goblet Cell Staining, and Bacteria Visualization.
With mice under anesthesia, tetramethylrodamine-labeled dextran 10,000 molecular weight (MW) (10 mg/mL) (Thermo Fisher Scientific) was injected intraluminally into the colon. Thirty minutes later, colons were removed and fixed in 10% buffered formalin for 30 min before blocking in optimal cutting temperature (OCT) compound and freezing as previously described (32, 52). Sections were cut and counterstained with DAPI. The number of GAPs per colon crypt was quantified by immunofluorescence microscopy as previously described (32, 52). Specimens were blinded for analysis, and GAPs were identified as dextran-filled columns measuring ∼20 µm (height) × 5 µm (diameter) traversing the epithelium and containing a DAPI+ nucleus. Colon GAPs were enumerated within the crypts as GAPs per crypt, which were identified as a ring of DAPI+ epithelial nuclei. Images were analyzed by fluorescent wide-field microscopy that was performed with an Axioskop 2 microscope using Axiovision software (Carl Zeiss). In some experiments, 106 CFUs of E. coli BSI-A were labeled with carboxyfluorescein succinimidyl ester (CFSE) as per the manufacturer’s directions (CellTrace CFSE, Thermo Fisher Scientific) and resuspended into a concentration of 106 CFUs/mL. One hundred microliters (105 CFUs) was intrarectally administered into the colon. Three hours later, colons were removed and fixed in 10% buffered formalin for 15 min before blocking in OCT and freezing as previously described (32, 52). Sections were cut and stained with CK-18 (Abcam) and DAPI. Immunohistochemistry was performed as previously described (52).
Flow Cytometry and Sorting of Goblet Cells.
CFUs (106) E. coli BSI-A were labeled with Vybrant RubyRed (Thermo Fisher Scientific) per the manufacturer’s directions and resuspended into a concentration of 106 CFUs/mL. CFUs (105; 100 μL) were intrarectally administered into the colon. Three hours later, colons were removed and the epithelial cells were isolated by shaking tissue in 20 mL of HBSS+5 mM EDTA with gentamicin (50 μg/mL) to kill extracellular bacteria. Goblet cells were identified as CK-18-fluorescein isothiocyanate (FITC)+ CD45-allophycocyanin (APC)lo while IECs were identified as CK-18-FITC− CD45-APC− (CK-18 antibody, monoclonal C-04, Abcam; CD45 antibody, monoclonal 30-F11, Thermo Fisher Scientific). Cells were either analyzed by flow cytometry on an Attune nxT (Invitrogen) or FACS isolated on a Synergy (Sony) sorter. FACS-isolated cells were homogenized and plated on MacConkey agar containing 20 ng/mL of nalidixic acid.
Statistical Analysis.
Data analysis using Kaplan–Meier survival, one-way ANOVA (nonparametric Kruskal–Wallis), two-way ANOVA (repeated measure two-way ANOVA), linear regression analysis, or Mann–Whitney U test was performed using GraphPad Prism (GraphPad Software Inc.).
Data Availability.
Whole genome sequences for BSI-A, BSI-C, C1, and C2 are publicly available: Bioproject accession PRJNA/GenBank/SRA run accession nos. BSI-A (ST69): 203059/ATNW00000000/SRR769039 and BSI-C (ST70): 203060/ATNV00000000/SRR769057; C1 (ST35): accession no. GCA_008081795.1; C2: PRJNA602838/SAMN13909231/JAADAA000000000. All materials, data, and associated protocols are available upon request from the corresponding author, K.A.K.
Supplementary Material
Acknowledgments
Supported by grants DK109006 (K.A.K.), AI144721 (K.A.K.), AI095542 MIST Scholars Award (K.A.K.), DK097317 (R.D.N.), AI112626 (R.D.N.), AI140755 (R.D.N.), AI131342 (R.D.N. and P.I.T.), UH3AI083265 (P.I.T.), MD-II-2009-201 (B.B.W.), MD-II-2018-725 (B.B.W.), Washington University Digestive Diseases Research Core Center (Biobank), and supported by grant P30 DK052574.
Footnotes
Competing interest statement: K.A.K. and R.D.N. are inventors on U.S. Nonprovisional Application Serial No. 15/880,658 Compositions And Methods For Modulation Of Dietary And Microbial Exposure. P.I.T. is a consultant to, holder of equity in, and a member of the Scientific Advisory Board of MediBeacon Inc, which is developing a noninvasive technology to measure intestinal permeability in humans. He also is an inventor on a patent that could generate royalty payments using this technology. He is also a consultant to Takeda Pharmaceuticals.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the GenBank database, https://www.ncbi.nlm.nih.gov/genbank/ (accession nos. 203059/ATNW00000000/SRR769039, 203060/ATNV00000000/SRR769057, GCA_008081795.1, PRJNA602838/SAMN13909231/JAADAA000000000).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912022117/-/DCSupplemental.
References
- 1.Cortese F., et al. , Early and late infections in newborns: Where do we stand? A review. Pediatr. Neonatol. 57, 265–273 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Stoll B. J., et al. ; National Institute of Child Health and Human Development Neonatal Research Network , Neurodevelopmental and growth impairment among extremely low-birth-weight infants with neonatal infection. JAMA 292, 2357–2365 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Bizzarro M. J., Raskind C., Baltimore R. S., Gallagher P. G., Seventy-five years of neonatal sepsis at Yale: 1928-2003. Pediatrics 116, 595–602 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Greenberg R. G., et al. ; Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network , Late-onset sepsis in extremely premature infants: 2000-2011. Pediatr. Infect. Dis. J. 36, 774–779 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplan H. C., Lannon C., Walsh M. C., Donovan E. F.; Ohio Perinatal Quality Collaborative , Ohio statewide quality-improvement collaborative to reduce late-onset sepsis in preterm infants. Pediatrics 127, 427–435 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Jones A. R., Kuschel C., Jacobs S., Doyle L. W., Reduction in late-onset sepsis on relocating a neonatal intensive care nursery. J. Paediatr. Child Health 48, 891–895 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Patel A. L., et al. , Impact of early human milk on sepsis and health-care costs in very low birth weight infants. J. Perinatol. 33, 514–519 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schüller S. S., et al. , Immunomodulation to prevent or treat neonatal sepsis: Past, present, and future. Front Pediatr. 6, 199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carl M. A., et al. , Sepsis from the gut: The enteric habitat of bacteria that cause late-onset neonatal bloodstream infections. Clin. Infect. Dis. 58, 1211–1218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almuneef M. A., Baltimore R. S., Farrel P. A., Reagan-Cirincione P., Dembry L. M., Molecular typing demonstrating transmission of gram-negative rods in a neonatal intensive care unit in the absence of a recognized epidemic. Clin. Infect. Dis. 32, 220–227 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Graham P. L. 3rd, Della-Latta P., Wu F., Zhou J., Saiman L., The gastrointestinal tract serves as the reservoir for Gram-negative pathogens in very low birth weight infants. Pediatr. Infect. Dis. J. 26, 1153–1156 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Smith A., et al. , Concordance of gastrointestinal tract colonization and subsequent bloodstream infections with Gram-negative Bacilli in very low birth weight infants in the neonatal intensive care unit. Pediatr. Infect. Dis. J. 29, 831–835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor S. N., Basile L. A., Ebeling M., Wagner C. L., Intestinal permeability in preterm infants by feeding type: Mother’s milk versus formula. Breastfeed. Med. 4, 11–15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Go L. L., Albanese C. T., Watkins S. C., Simmons R. L., Rowe M. I., Breast milk protects the neonate from bacterial translocation. J. Pediatr. Surg. 29, 1059–1063, discussion 1063–1064 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Yajima M., et al. , Bacterial translocation in neonatal rats: The relation between intestinal flora, translocated bacteria, and influence of milk. J. Pediatr. Gastroenterol. Nutr. 33, 592–601 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Nakayama M., Yajima M., Hatano S., Yajima T., Kuwata T., Intestinal adherent bacteria and bacterial translocation in breast-fed and formula-fed rats in relation to susceptibility to infection. Pediatr. Res. 54, 364–371 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Nojiri T., et al. , Clinical significance of amphiregulin and epidermal growth factor in colostrum. Arch. Gynecol. Obstet. 286, 643–647 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Matsuoka Y., Idota T., The concentration of epidermal growth factor in Japanese mother’s milk. J. Nutr. Sci. Vitaminol. 41, 241–251 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Kobata R., et al. , High levels of growth factors in human breast milk. Early Hum. Dev. 84, 67–69 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Okuyama H., Urao M., Lee D., Drongowski R. A., Coran A. G., The effect of epidermal growth factor on bacterial translocation in newborn rabbits. J. Pediatr. Surg. 33, 225–228 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Ashraf R. N., et al. , Breast feeding and protection against neonatal sepsis in a high risk population. Arch. Dis. Child. 66, 488–490 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J., et al. , A systematic review and meta-analysis of human milk feeding and morbidity in very low birth weight infants. Nutrients 10, E707 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michalsky M. P., Lara-Marquez M., Chun L., Besner G. E., Heparin-binding EGF-like growth factor is present in human amniotic fluid and breast milk. J. Pediatr. Surg. 37, 1–6 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Ballard O., Morrow A. L., Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. North Am. 60, 49–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knoop K. A., et al. , Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci. Immunol. 2, eaao1314 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu P., et al. , Animal models of gastrointestinal and liver diseases. Animal models of necrotizing enterocolitis: Pathophysiology, translational relevance, and challenges. Am. J. Physiol. Gastrointest. Liver Physiol. 306, G917–G928 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ares G. J., McElroy S. J., Hunter C. J., The science and necessity of using animal models in the study of necrotizing enterocolitis. Semin. Pediatr. Surg. 27, 29–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikitas G., et al. , Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J. Exp. Med. 208, 2263–2277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulkarni D. H., et al. , Goblet cell associated antigen passages are inhibited during Salmonella typhimurium infection to prevent pathogen dissemination and limit responses to dietary antigens. Mucosal Immunol. 11, 1103–1113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanai H., et al. , Intestinal stem cells contribute to the maturation of the neonatal small intestine and colon independently of digestive activity. Sci. Rep. 7, 9891 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knoop K. A., McDonald K. G., Kulkarni D. H., Newberry R. D., Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut 65, 1100–1109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knoop K. A., McDonald K. G., McCrate S., McDole J. R., Newberry R. D., Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol. 8, 198–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luetteke N. C., et al. , Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development 126, 2739–2750 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Goldsmith S. J., Dickson J. S., Barnhart H. M., Toledo R. T., Eiten-Miller R. R., IgA, IgG, IgM and lactoferrin contents of human milk during early lactation and the effect of processing and storage. J. Food Prot. 46, 4–7 (1983). [DOI] [PubMed] [Google Scholar]
- 35.Castellote C., et al. , Premature delivery influences the immunological composition of colostrum and transitional and mature human milk. J. Nutr. 141, 1181–1187 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Smilowitz J. T., Lebrilla C. B., Mills D. A., German J. B., Freeman S. L., Breast milk oligosaccharides: Structure-function relationships in the neonate. Annu. Rev. Nutr. 34, 143–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van de Perre P., Transfer of antibody via mother’s milk. Vaccine 21, 3374–3376 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Zheng W., et al. , Microbiota-targeted maternal antibodies protect neonates from enteric infection. Nature 577, 543–548 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanton L. V., et al. , Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351, aad3311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y.-G., et al. , Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 356, 315–319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birchenough G. M. H., et al. , Postnatal development of the small intestinal mucosa drives age-dependent, regio-selective susceptibility to Escherichia coli K1 infection. Sci. Rep. 7, 83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taft D. H., et al. , Center variation in intestinal microbiota prior to late-onset sepsis in preterm infants. PLoS One 10, e0130604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole B. K., et al. , Route of infection alters virulence of neonatal septicemia Escherichia coli clinical isolates. PLoS One 12, e0189032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose M. F., Ahmad K. A., Thaller C., Zoghbi H. Y., Excitatory neurons of the proprioceptive, interoceptive, and arousal hindbrain networks share a developmental requirement for Math1. Proc. Natl. Acad. Sci. U.S.A. 106, 22462–22467 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee T. C., Threadgill D. W., Generation and validation of mice carrying a conditional allele of the epidermal growth factor receptor. Genesis 47, 85–92 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gasparrini A. J., et al. , Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat. Microbiol. 4, 2285–2297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knoop K. A., Gasparrini A. J., Tarr P. I., C2 E.coli WGS. NCBI GenBank. https://www.ncbi.nlm.nih.gov/nuccore/JAADAA000000000. Deposited 24 January 2020.
- 48.Rice P., Longden I., Bleasby A., EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Joensen K. G., et al. , Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDonald K. G., et al. , Epithelial expression of the cytosolic retinoid chaperone cellular retinol binding protein II is essential for in vivo imprinting of local gut dendritic cells by lumenal retinoids. Am. J. Pathol. 180, 984–997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lanik W. E., et al. , Breast milk enhances growth of enteroids: An ex vivo model of cell proliferation. J. Vis. Exp., e56921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDole J. R., et al. , Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature 483, 345–349 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole genome sequences for BSI-A, BSI-C, C1, and C2 are publicly available: Bioproject accession PRJNA/GenBank/SRA run accession nos. BSI-A (ST69): 203059/ATNW00000000/SRR769039 and BSI-C (ST70): 203060/ATNV00000000/SRR769057; C1 (ST35): accession no. GCA_008081795.1; C2: PRJNA602838/SAMN13909231/JAADAA000000000. All materials, data, and associated protocols are available upon request from the corresponding author, K.A.K.