Significance
Heat shock protein 70 kDa (Hsp70) plays a central role in maintaining protein homeostasis. It cooperates with cochaperone Hsp40, which stimulates Hsp70 ATPase activity and presents protein substrates to Hsp70 to assist refolding. The mechanism by which Hsp40 regulates the intramolecular and intermolecular changes of Hsp70 is still largely unknown. Here, by bulk and single-molecule FRET, we report the conformational dynamics of Hsp70 and its regulation by Hsp40 as well as the kinetics of the multistep Hsp70–Hsp40 functional cycle. We show that Hsp40 modulates the conformations of ATP-bound Hsp70 to a domain-undocked ATPase-stimulated state, and facilitates the formation of a heterotetrameric Hsp70–Hsp40 complex. Our findings provide insights into the functional mechanism of this core chaperone machinery.
Keywords: single-molecule FRET, chaperone, Hsp70, kinetic analysis, conformational change
Abstract
Hsp70 is a conserved molecular chaperone that plays an indispensable role in regulating protein folding, translocation, and degradation. The conformational dynamics of Hsp70 and its regulation by cochaperones are vital to its function. Using bulk and single-molecule fluorescence resonance energy transfer (smFRET) techniques, we studied the interdomain conformational distribution of human stress-inducible Hsp70A1 and the kinetics of conformational changes induced by nucleotide and the Hsp40 cochaperone Hdj1. We found that the conformations between and within the nucleotide- and substrate-binding domains show heterogeneity. The conformational distribution in the ATP-bound state can be induced by Hdj1 to form an “ADP-like” undocked conformation, which is an ATPase-stimulated state. Kinetic measurements indicate that Hdj1 binds to monomeric Hsp70 as the first step, then induces undocking of the two domains and closing of the substrate-binding cleft. Dimeric Hdj1 then facilitates dimerization of Hsp70 and formation of a heterotetrameric Hsp70–Hsp40 complex. Our results provide a kinetic view of the conformational cycle of Hsp70 and reveal the importance of the dynamic nature of Hsp70 for its function.
The Hsp70 family of chaperones is involved in a variety of cellular processes, including protein folding, translocation, and degradation (1). Hsp70 homologs share a similar structure composed of an N-terminal 43-kDa nucleotide-binding domain (NBD) and a C-terminal 27-kDa substrate-binding domain (SBD) connected by a conserved flexible and hydrophobic linker. The SBD is further divided into two subunits including a two-layer β-sandwich subdomain (SBDβ) and an α-helical bundle lid subdomain (SBDα) that together form a hydrophobic cleft for substrate binding. The NBD of Hsp70 has weak ATPase activity, and the binding/hydrolysis of ATP to ADP allows allosteric regulation of the interdomain conformation and substrate binding of the SBD. In the ATP-bound state, Hsp70 has low affinity for substrate but with fast binding and release rates. After ATP hydrolysis to ADP, the substrate affinity dramatically increases and the association and dissociation rates also decrease (2).
High-resolution structures are available for full-length Hsp70 in ADP- and ATP-bound states (3–7), providing insights into the mechanism of Hsp70 interdomain allosteric communication. Based on these structures, in the nucleotide-free and ADP-bound states, the NBD and SBD tend to adopt an undocked conformation connected by the flexible linker, and the SBD cleft remains closed; substrate peptides are bound in the hydrophobic cleft enclosed by the SBDβ and SBDα (3). In the ATP-bound state, the SBD cleft tends to change to a wide-open conformation with the SBD docked onto the NBD (4, 5, 7). Although the dominant or snapshot structures of different nucleotide-bound states during the ATP–ADP functional cycle have been identified, detailed knowledge of the conformational dynamics is still under exploration. Growing evidence suggests that the individual domains of Hsp70 are highly dynamic (8, 9). Moreover, the application of sensitive techniques such as single-molecule fluorescence resonance energy transfer (smFRET), NMR, electron paramagnetic resonance, and ion mobility native mass spectroscopy suggests that full-length Hsp70 adopts multiple conformations (10–16). These studies not only provide supplemental dynamic information to the classical structural picture but also indicate the importance of the dynamic nature of Hsp70 in its allosteric regulation and functional cycle (17, 18).
Hsp70 usually functions together with Hsp40 cochaperones and nucleotide exchange factors (NEFs), which provides functional diversity and increases the efficiency of the ATP-hydrolysis cycle (19). Members of the cochaperone Hsp40 family (also called J proteins because of the conserved N-terminal J domain) can bind to unfolded and misfolded proteins and transfer them to Hsp70, enhance its ATPase activity, and facilitate substrate binding and refolding (20). The J domain of Hsp40 binds at the NBD–SBD interface of Hsp70, as indicated by structural studies and molecular simulations (21–23), and interacts with both domains of Hsp70 in a nucleotide-dependent manner (24–26). On the other hand, the peptide binding domain of Hsp40 has also been found to interact with the conserved C-terminal EEVD motif of Hsp70 (27, 28). To date, most of the knowledge about the Hsp70–Hsp40 interaction mechanism comes from analysis of the Escherichia coli homologs DnaK and DnaJ. Recently, the crystal structure of the J domain of DnaJ in complex with E. coli DnaK in the ATP-bound state was solved (21), revealing the key residues on both proteins that are crucial for ATPase stimulation. However, the kinetics of the intramolecular and intermolecular changes during the formation of the Hsp70–Hsp40 complex and how Hsp40 affects the conformational dynamics of Hsp70 are not clearly known.
Human Hsp70A1 (hHsp70) is stress-inducible and related to various human diseases such as cancer and neurodegenerative disease; thus, it is a potential target for drug design and development of therapies (29–31). Therefore, experimental characterization of the interdomain conformational dynamics of hHsp70 and its changes upon interaction with the cochaperone Hsp40 are of great importance in understanding the molecular mechanism of hHsp70 and development of effective allosteric inhibitors. Here, by ensemble FRET and smFRET, we investigated the conformational distribution and kinetics for hHsp70 and its modulation by the Hsp40 family member Hdj1. The results reveal the sequential movement of different domains of hHsp70 and also the Hdj1-induced intramolecular and intermolecular changes of hHsp70 in detail. Our study sheds light on the heterogeneous and dynamic nature of hHsp70 and its importance in the Hsp70 functional cycle.
Results
smFRET Measurements Reveal the Conformational Heterogeneity of hHsp70 in Different Nucleotide-Bound States.
In order to study the interdomain conformation of hHsp70, we constructed two FRET variants based on homology modeling of the structure of full-length hHsp70 (SI Appendix, Fig. S1 A and B). The hHsp70 variant E315C/I427C was designed to investigate the conformation of the NBD relative to the SBDβ, while I427C/K559C was designed to monitor movement of the SBDβ relative to the SBDα (Fig. 1 A and B and SI Appendix, Fig. S1 A and B). Since hHsp70 has five intrinsic cysteines, we mutated them either to alanine or serine (SI Appendix). These mutations did not affect the substrate binding ability or the substrate refolding activity of hHsp70 (SI Appendix, Fig. S1 C–F and Table S1). Moreover, fluorescence labeling of the introduced Cys residues with maleimide-functionalized dyes Cy3 and Cy5 did not affect the basal ATPase activity of hHsp70 or its stimulation by the cochaperone Hdj1 (SI Appendix, Fig. S1G and Table S2). Therefore, the FRET labeling variants we designed can be used to study the conformational changes of hHsp70 and its interaction with Hdj1. The fluorescence spectra obtained for the NBD–SBDβ labeling variant Cy3/Cy5–E315C/I427C showed a higher FRET efficiency in the presence of ATP than in the apo or ADP-bound states (Fig. 1A), indicating that the NBD and SBDβ are closer to each other in the ATP-bound state, while being prone to adopt an undocked conformation in the apo and ADP-bound states. In contrast, the fluorescence spectra for the SBDβ–SBDα labeling variant Cy3/Cy5–I427C/K559C showed a lower FRET efficiency in the ATP-bound state than in the apo or ADP-bound states (Fig. 1B), demonstrating that the SBDα and SBDβ domains adopt an open conformation in the ATP-bound state and move to a closed conformation in the apo and ADP-bound states. The above bulk fluorescence results are similar to the allosteric conformational changes observed by structural study of DnaK (3, 4).
Fig. 1.
Conformations of hHsp70 in different nucleotide-bound states measured by ensemble FRET and smFRET. (A and B) The FRET spectra of NBD–SBDβ labeling variant Cy3/Cy5–E315C/I427C (A) and SBDβ–SBDα labeling variant Cy3/Cy5–I427C/K559C (B) in apo, ADP-bound, and ATP-bound states. The fluorescence at the donor maximum was normalized to 1.0. The donor and acceptor fluorescence labeling sites are indicated by red and green stars in the cartoon. (C and D) The smFRET distribution histograms of NBD–SBDβ labeling variant Cy3/Cy5–E315C/I427C (C) and SBDβ–SBDα labeling variant Cy3/Cy5–I427C/K559C (D) in apo, ADP-bound, and ATP-bound states. The low FRET efficiency populations (zero peak) caused by inactive acceptor dye are shaded in gray. The red lines in the Left panels are the fits obtained by PDA, and the obtained distance distributions between the labeling sites are plotted on the Right. The results of PDA are listed in SI Appendix, Table S3.
To gain more detailed information about the conformations of hHsp70, confocal smFRET experiments were performed to analyze the conformational distributions in different nucleotide-bound states (Fig. 1 C and D). The smFRET histograms were analyzed using probability distribution analysis (PDA) to convert the data into distance distributions between the fluorescence labeling sites (32) (SI Appendix). The “zero peak” in the confocal smFRET histogram, which is generally caused by donor-only or acceptor-bleached molecules, has been shaded gray in each of the figures. The smFRET histogram of the NBD–SBDβ labeling variant Cy3/Cy5–E315C/I427C showed a relatively broad distribution in the apo and ADP-bound states with a major distance distribution between the labeling sites centered at 66 Å (Fig. 1C). This distance corresponds to a fully undocked conformation of the NBD and SBDβ, in agreement with the distance in the structure of ADP-bound DnaK (68 Å) (3). However, a minor subpopulation was also observed in the apo and ADP-bound states of hHsp70 that gave a distance of 50 Å between E315 and I427 (Fig. 1C), resembling the docked conformation in the structure of ATP-bound DnaK. For the ATP-bound state of hHsp70, a major peak at d = 50 Å was observed, which represents a completely docked conformation of the NBD–SBDβ, while a minor population centered at 66 Å resembles the undocked conformation. These results demonstrate the heterogeneous nature of hHsp70 in different nucleotide-bound states. These results are dramatically different from DnaK (10, 14, 16) but are generally consistent with the most recent findings on human cytosolic Hsp70 by NMR spectroscopy (16), highlighting that different Hsp70 homologs can show pronounced differences in structural and dynamic properties, despite the high degree of sequence similarity.
The smFRET histograms of I427C/K559C, which reflect the conformation of the SBD cleft, required at least two Gaussian populations to describe the smFRET distribution in the apo and ADP-bound states (in addition to the zero peak) (Fig. 1D). The major population showed a peak at 39 Å and indicates a fully closed conformation of the SBD cleft in the apo and ADP-bound states of hHsp70, similar to the distance in the structure of DnaK (34 Å) (3). We also observed a subpopulation, giving a distance of 52 to 54 Å, suggesting that in addition to the fully closed conformation, the SBD of hHsp70 also populates a partially open state. In the ATP-bound state, the major peak of the FRET efficiency distribution shifted below 0.3 and tended to overlap with the zero-peak background of the confocal smFRET measurement. Although we could not obtain a precise distance between the labeled sites in the ATP-bound state and its proportion by PDA, the separation between I427 and K559 is larger than 100 Å, indicating a completely open conformation. Additionally, in the ATP-bound state, there is a minor population showing FRET efficiency between 0.8 and 1.0, revealing the existence of a small population of the closed conformation of the SBD cleft in the ATP-bound state of hHsp70.
In order to distinguish whether the minor populations in the ATP-bound state can be attributed to conformational heterogeneity or whether they reflect the presence of the ATP-hydrolyzed ADP state, we tested two ATP analogs, AMP-PNP and ATPγS in place of ATP. However, we did not observe any interdomain conformational change in hHsp70 induced by either AMP-PNP or ATPγS in contrast to ATP, similar to what has been reported for DnaK (33–35). Therefore, we incorporated the T204A mutation into each of the two FRET mutants, which is equivalent to the E. coli DnaK–T199A and blocks the ATPase activity without disruption of the allosteric change (36). We used the T204A mutant (the residual ATPase activity was measured to be 15% of wild-type [WT]-hHsp70; SI Appendix, Table S2) to perform the smFRET experiments. The smFRET distributions of hHsp70 T204A in the ATP-bound state (SI Appendix, Fig. S2 A and B) were similar to unmutated hHsp70 (Fig. 1 C and D), indicating that the undocked conformation of the NBD–SBDβ and the closed conformation of the SBD cleft indeed exist in the ATP-bound state, and are not caused by ATP hydrolysis. In the apo and ADP-bound states, a slight difference was observed in the NBD–SBDβ labeling variant containing the T204A mutation compared to without the T204A mutation (SI Appendix, Fig. S2A and Fig. 1C), where the proportion of the docked conformation is higher in T204A variants (SI Appendix, Table S3). Therefore, subtle changes may occur in the T204A mutant, although it retains the predominant allosteric communication between domains. Overall, the above smFRET results demonstrate that hHsp70 shows conformational heterogeneity in both the ATP-bound and ADP-bounds states, with respect to both NBD–SBDβ docking/undocking and opening/closing of the SBDβ–SBDα cleft.
Kinetic Analysis of the ATP-Induced Conformational Changes Indicates Sequential Movements of hHsp70 Domains.
The kinetics of ATP-induced conformational changes has been studied for DnaK using intrinsic Trp fluorescence (33, 37) and fluorophore quenching (4). However, these measurements only reflect the change of the microenvironment around the fluorophore and cannot give accurate kinetic information about the long-range movements of specific domains. Using the FRET labeling variants, we monitored the conformational changes of hHsp70 at a series of ATP concentrations. The observed increase in FRET for the NBD–SBDβ labeling variant (Cy3/Cy5–E315C/I427C) and the decrease in FRET for the SBDβ–SBDα labeling variant (Cy3/Cy5–I427C/K559C) showed ATP concentration dependence (Fig. 2 A and B). None of the kinetic curves could be fitted to a simple single-exponential function, indicating that the observed kinetics represents more than one step. To further analyze this process, we used the following two-step reaction mechanism including both an ATP concentration-dependent step and a concentration-independent conformational conversion step:
| [1] |
where an intermediate state—the intermediate complex of hHsp70 and ATP—is included to account for the ATP concentration dependence of this process and the saturation effect observed when the ATP concentration is higher than 10 μM. In Eq. 1, and represent the forward and reverse rates of the reaction from apo hHsp70 to the intermediate hHsp70·ATP complex induced by ATP, and represents the conformational transition from the intermediate hHsp70·ATP complex to the final ATP-bound conformation. It should be noted that the above model is a mathematical abstraction of a single reaction pathway with multiple intermediate states. In Eq. 1, hHsp70·ATP represents an ensemble of various possible intermediate conformational states from apo state to ATP state, which readily interconvert with each other. As E315C/I427C detects the NBD–SBDβ docking process and I427C/K559C detects the SBDβ–SBDα opening process, different labeling modes may allow detection of different conformational intermediates on the reaction pathway.
Fig. 2.
Kinetics of ATP-induced conformational changes of hHsp70 measured by FRET. (A and B) Intramolecular FRET change of the NBD–SBDβ labeling variant Cy3/Cy5–E315C/I427C (A) and the SBDβ–SBDα labeling variant Cy3/Cy5–I427C/K559C (B) upon addition of different concentrations of ATP. The experimental data are shown as dots, while global fitting based on Eq. 1 is shown as dashed lines. Six ATP concentrations were measured: 1 μM (gray), 2 μM (blue), 5 μM (pink), 10 μM (green), 40 μM (red), and 100 μM (orange). (Insets) Detail of early time points. (C) The rate constants obtained from fitting of the data in A and B. (D and E) Intramolecular FRET change of NBD–SBDβ labeling variant Cy3/Cy5–E315C/I427C (D) and SBDβ–SBDα labeling variant Cy3/Cy5–I427C/K559C (E) at different temperatures. The experimental data are shown as dots, while parameterless global fitting based on the Arrhenius equation is shown as dashed lines. Six temperatures were measured: 25 °C (gray), 27 °C (blue), 30 °C (pink), 33 °C (green), 37 °C (red), and 42 °C (orange). The fluorescence labeling sites in hHsp70 are indicated by red and green stars in the cartoon. For each kinetic curve, at least three replicates were performed and averaged. (Insets) Detail of early time points. (F) Arrhenius plot of the reaction rates obtained from temperature-dependent conformational changes in D and E.
We performed global fitting of the kinetic data at different ATP concentrations for each FRET labeling variant using Eq. 1 (Fig. 2 A and B). The fitting results show that conversion from the intermediate state to the final ATP-bound conformation is the rate-limiting step for both the NBD–SBDβ and the SBDβ–SBDα labeling variants, with a rate of 0.12 ± 0.02 and 0.13 ± 0.01 s−1 independent of ATP concentration, which is the same order-of-magnitude as that measured for DnaK using intrinsic fluorescence (37). However, there is a dramatic difference in the observed rates for the ATP concentration-dependent process between the two labeling variants. The NBD–SBDβ labeling variant showed values of and of 0.61 ± 0.10 μM−1⋅s−1 and 0.43 ± 0.03 s−1, respectively, while the SBDβ–SBDα labeling variant has a slightly smaller of 0.39 ± 0.07 μM−1⋅s−1 and a negligible reverse reaction rate (Fig. 2C). The dramatic difference in indicates that the conformations of the hHsp70·ATP intermediate for the NBD–SBDβ and SBDβ–SBDα labeling variants are distinct from each other, which correspondingly can be denoted as hHsp70·ATP* and hHsp70·ATP**. Thus, the whole process of ATP-induced Hsp70 conformational changes can be classified into at least three substeps between four sequential states: 1) apo state, 2) the hHsp70·ATP* state (detected by E315C/I427C labeling), 3) the hHsp70·ATP** state (detected by I427C/K559C labeling), and 4) the ATP-bound state. Considering the fact that the SBDβ–SBDα labeling variant has a negligible reverse reaction rate, and in the absence of significant differences in forward rates, we can conclude that the hHsp70·ATP* state is significantly more reversible than the hHsp70·ATP** state. The higher degree of reversibility of the hHsp70·ATP* state indicates that NBD–SBDβ docking happens prior to opening of the SBDβ–SBDα cleft, suggesting transduction of the allosteric signal by a sequential mode from NBD to SBDβ and then to SBDα. Our results for hHsp70 are to some extent similar to the observations for E. coli DnaK, in which the relocation of the SBDα toward the NBD has been suggested to be slower than the docking of the SBDβ onto the NBD. However, the fluorescence quenching of dyes labeled at the interaction interface of DnaK only reflects the change near the fluorophores (4), and not the long-range domain movements, which were measured by FRET in this study.
We made further measurements to investigate the effect of temperature on the ATP-induced hHsp70 conformational changes to explore the energy barrier of the interdomain rearrangement. Since hHsp70 in the apo state is not stable at elevated temperature, we started the conformational cycle from ADP-bound hHsp70 and measured the conversion after exchange with excess ATP (1 mM) at a series of temperatures. The apparent reaction time increased compared to starting from the apo state (Fig. 2 A and B), indicating nucleotide exchange became the rate-limiting step. According to the Arrhenius equation, , we globally fitted the curves at different temperatures and obtained the activation energy barrier (Ea) for the whole process as 53.2 ± 3.0 kJ⋅mol−1 for the E315C/I427C labeling variant and 43.4 ± 1.2 kJ⋅mol−1 for the I427C/K559C labeling variant (Fig. 2 D–F). Considering the previous kinetic observation that NBD–SBDβ docking or formation of the hHsp70·ATP* state (detected by E315C/I427C labeling) occurs prior to opening of the SBDβ–SBDα cleft or formation of the hHsp70·ATP** state (detected by I427C/K559C labeling), the 10-kJ difference in Ea indicates that the transition from hHsp70·ATP* to hHsp70·ATP**, i.e., opening of the SBDβ–SBDα cleft, is likely to be a spontaneous process.
Hsp40 Induces Intramolecular Conformational Changes and Intermolecular Dimerization of hHsp70.
Structural studies indicate that the cochaperone Hsp40 interacts with the interface of the NBD and SBD (21). However, the kinetics of binding, the Hsp40-induced changes in Hsp70 during the ATPase cycle, and how ATP hydrolysis is stimulated are still under investigation. Based on the FRET variants we constructed, we investigated the influence of the human type II Hsp40 homolog, Hdj1, on the conformation of hHsp70 (Fig. 3 A and B). For the Cy3/Cy5–SBDβ–SBDα labeling variant, addition of Hdj1 led to an increase in the bulk FRET efficiency especially in the ATP-bound state (Fig. 3A), which is even higher than in the ADP-bound state. This result suggests that the FRET increase is not only caused by intramolecular conformational changes but that intermolecular interactions also occur. To further identify the origin of the FRET change, we premixed an excess of unlabeled hHsp70 (10-fold or 50-fold molar excess) with the Cy3/Cy5–SBDβ–SBDα labeling variant in the ATP-bound state, so that the inter-hHsp70 FRET signal would be suppressed if present. Under these conditions, the extent of the FRET increase induced by Hdj1 was much lower than that without unlabeled hHsp70 (SI Appendix, Fig. S3A), indicating the existence of an intermolecular interaction between hHsp70 molecules and an intramolecular conformational change of hHsp70, both of which are induced by Hdj1. We further verified this by directly detecting the intermolecular FRET between Cy3-labeled hHsp70 and Cy5-labeled hHsp70 upon addition of Hdj1 (SI Appendix, Fig. S3 B and C). The results show inter-hHsp70 FRET in the presence of Hdj1 in both ATP- and ADP-bound states. On the other hand, the FRET change for the Cy3/Cy5–NBD–SBDβ labeling variant E315C/I427C when interacting with Hdj1 was not as significant as that for the SBDβ–SBDα labeling variant in either the ATP- or ADP-bound states (Fig. 3B). This is possibly because the inter-hHsp70 FRET was offset by the intra-hHsp70 changes (see below). Size exclusion chromatography (SEC) and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis of the mixture of hHsp70 and Hdj1 shows that the molecular weight of the hHsp70–Hdj1 complex is around 200 kDa, and the resulting peak contains both hHsp70 and Hdj1, suggesting that the complex is composed of dimeric Hdj1 and two hHsp70 molecules (Fig. 3 C and D). Since full-length Hdj1 is a symmetric dimer, it is reasonable that Hdj1 induces the formation of a heterotetramer containing two hHsp70 molecules. The observation of similar amounts of the complex in the presence of ATP or ADP is consistent with the observation that the binding affinity between hHsp70 and Hdj1 in the ATP-bound state is similar or only slightly higher to that in the ADP-bound state as measured by FRET and biolayer interferometry (SI Appendix, Fig. S3 D–G).
Fig. 3.
Hdj1 induces intermolecular interactions and intramolecular conformational changes of hHsp70. (A and B) The FRET spectra of SBDβ–SBDα labeling variant Cy3/Cy5–I427C/K559C (A) and NBD–SBDβ labeling variant Cy3/Cy5–E315C/I427C (B) in the presence of Hdj1 in ATP- and ADP-bound states. The fluorescence at the donor maximum was normalized to 1.0. The J domain and substrate binding domain of Hdj1 are colored yellow and pink. The fluorescence labeling sites in hHsp70 are indicated by red and green stars in the cartoon. (C) SEC and SDS-PAGE analysis of the mixture of hHsp70 and Hdj1. A mixture of 2 μM hHsp70 and 10 μM Hdj1 in the ATP- or ADP-bound state was analyzed by SEC (red and blue lines). As a control, 2 μM hHsp70 and 10 μM Hdj1 were also run through the SEC column individually (black and green lines). The peak positions of standard samples including β-amylase (200 kDa, 11.27 mL), alcohol dehydrogenase (150 kDa, 12.19 mL), and bovine serum (66 kDa, 13.43 mL) are shown in the figure. (D) SDS-PAGE analysis of the eluted components of each sample in the SEC experiment, with a and b corresponding to the peak position indicated by arrows a (11.1 to 11.5 mL) and b (12.5 to 13.5 mL) in C.
The real-time kinetics of the Hdj1-induced intramolecular and intermolecular changes in hHsp70 was measured by stopped-flow FRET experiments. Since the hHsp70 protein has basal ATP hydrolysis activity, our measurements reflect the steady-state kinetics. Here, we used three different labeling modes to monitor: 1) the Hdj1-induced dimerization of hHsp70, 2) the intramolecular conformational change of hHsp70, and 3) binding between hHsp70 and Hdj1 (Fig. 4A). The dimerization of hHsp70 was monitored by inter-hHsp70 FRET between Cy3-labeled hHsp70 and Cy5-labeled hHsp70 when mixed with Hdj1 in the presence of ATP (Fig. 4B). An increase in inter-hHsp70 FRET was observed and was fitted to a single-exponential function giving an apparent rate constant of 0.0095 ± 0.0001 s−1 for the SBDβ–SBDα labeling variant and a similar rate of 0.0074 ± 0.0001 s−1 for the NBD–SBDβ labeling variant (Fig. 4B, green and blue curves). In order to measure the intramolecular conformational change of hHsp70 during the interaction with Hdj1, a 10-fold excess amount of unlabeled hHsp70 was premixed with the Cy3/Cy5–SBDβ–SBDα labeling variant to avoid inter-hHsp70 FRET caused by dimerization. The FRET increase of the sample upon addition of Hdj1 was then fitted to a single-exponential function giving a rate constant of 0.027 ± 0.001 s−1 (Fig. 4B, red curve), corresponding to the Hdj1-induced closing of the SBDβ–SBDα cleft, which is threefold faster than the dimerization process (SI Appendix, Table S4).
Fig. 4.
The kinetics of Hdj1-induced intermolecular interactions and intramolecular conformational changes of hHsp70. (A) The fluorescence labeling schemes for measuring the multistep kinetic parameters of the hHsp70–Hdj1 interactions. The J domain and substrate binding domain of Hdj1 are colored yellow and pink, respectively. The green and red stars in the cartoon indicate the Cy3 and Cy5 dyes labeled in hHsp70 and Hdj1. The schemes shown in A use the SBDβ–SBDα labeling variant hHsp70-I427C/K559C as an example. The NBD–SBDβ labeling variant hHsp70-E315C/I427C was also measured in the same manner. (B) The kinetics of the FRET change after addition of Hdj1 to the mixture of Cy3/Cy5-labeled I427C/K559C and 10-fold molar excess of unlabeled I427C/K559C (red curve), the mixture of Cy3-I427C/K559C and Cy5-I427C/K559C (green curve), and the mixture of Cy3-E315C/I427C and Cy5-E315C/I427C (blue curve) in the ATP-bound state. (C) The kinetics of the FRET changes after addition of Hdj1 to Cy3/Cy5-labeled E315C/I427C with or without 10-fold molar excess of unlabeled E315C/I427C in the ATP-bound state. The kinetic curves were fitted to either single-exponential or double-exponential functions (gray lines) as described in the text. (D) The kinetics of FRET changes of direct interaction between Cy3-labeled hHsp70-E315C/I427C (blue curve) or I427C/K559C (red curve) in the ATP-bound states and Cy5-labeled Hdj1 (red curve). (Insets) Early stage of the FRET change (0 to 20 s), which were fitted to a double-exponential function (gray lines) to obtain the apparent binding rate constants. (E) The observed rate constants (kobs) of direct binding between Cy3-labeled hHsp70-E315C/I427C and different concentrations of Cy5–Hdj1 measured by FRET. The kobs values were plotted against Hdj1 concentration and fitted linearly to obtain the second-order reaction rate from the slope. The fitting results of B–E are listed in SI Appendix, Table S4.
As shown in Fig. 3B, there is no significant FRET change for the NBD–SBDβ labeling variant E315C/I427C when interacting with Hdj1. However, in the ATP-bound state, we could observe an initial decrease and a subsequent increase in the FRET efficiency with high reproducibility (Fig. 4C). When 10-fold excess of unlabeled hHsp70 was premixed with the Cy3/Cy5–NBD–SBDβ labeling variant, only a FRET decrease was observed with a rate of 0.056 ± 0.001 s−1 after addition of Hdj1, indicating that the initial FRET decrease is caused by the intramolecular undocking of NBD–SBDβ induced by Hdj1. The rate constant of the following increase in FRET, which should represent intermolecular dimerization, is 0.0042 ± 0.0002 s−1, which is of the same order-of-magnitude as that obtained from direct measurement of dimerization of the NBD–SBDβ labeling variant (Fig. 4B).
We also measured the kinetics of the direct binding between Cy3-labeled hHsp70 and Cy5-labeled Hdj1 in the presence of ATP (Fig. 4D). The FRET change shows a fast initial binding phase on a subsecond timescale followed by a slow phase on a tens- to hundreds-of-seconds timescale, which may reflect the subsequent changes within the hHsp70–Hdj1 complex. The apparent rate constant of the fast binding phase obtained from the fit is 1.2 ± 0.1 s−1, which is about 60-fold faster than the conformational changes of the SBD and more than 100-fold faster than the inter-hHsp70 dimerization at the same initial concentration, demonstrating that hHsp70–Hdj1 binding occurs as the first step. Since the initial binding of hHsp70 and Hdj1 is a second-order reaction while the following intra-hHsp70 conformational change and dimerization of the two hHsp70 molecules within the complex can be considered to be first-order reactions, we performed further stopped-flow experiments on a short timescale (0 to 5 s) for different concentrations of Hdj1 to obtain the second-order binding rate, which was measured to be 0.35 ± 0.1 μM−1⋅s−1 (Fig. 4E). Considering that the concentration of Hsp70 in eukaryotic cells ranges from 5 to 50 μM and the concentration of Hdj1 is around 10-fold lower than Hsp70, ranging from 0.5 to 2.0 μM (38, 39), the apparent binding rate between hHsp70 and Hdj1 under physiological conditions should be similar to or even faster than that measured in our kinetic experiments.
There are two possibilities for the NBD–SBDβ undocking and SBDβ–SBDα closing induced by addition of Hdj1. One is that the binding of Hdj1 shifts the hHsp70 to an ATPase-stimulated conformation while still in the ATP-bound state. The other possibility is that the observed conformational change of hHsp70 is the result of accelerated hydrolysis of ATP to ADP. In order to distinguish between these two possibilities, we performed an ATPase assay to measure the ATP turnover rate in the presence of the same concentration of Hdj1 and at the same temperature used in the kinetic studies. The Hdj1-stimulated ATP hydrolysis rate was measured to be 0.0067 ± 0.0005 s−1 (SI Appendix, Table S2), much slower than the apparent rate constants of NBD–SBDβ undocking and SBDβ–SBDα closing. Therefore, the intramolecular conformational change of hHsp70 is due to interaction with Hdj1 and occurs before ATP hydrolysis. Notably, the ATP hydrolysis rate is close to the dimerization rate of hHsp70 induced by Hdj1, indicating the hydrolysis process could happen concurrently with Hdj1–hHsp70 heterotetramer formation, which may contain both ATP- and ADP-bound hHsp70.
Hsp40 Does Not Induce Intramolecular and Intermolecular Changes in T204A hHsp70.
We also checked the effect of Hdj1 on the FRET variants of hHsp70 containing the T204A mutation. We did not observe significant FRET changes after addition of Hdj1 to either the NBD–SBDβ or SBDβ–SBDα labeling variants containing the T204A mutation in the ATP-bound state (SI Appendix, Fig. S4 A and B), in contrast to what was observed when using the “WT” hHsp70 variants that have ATP hydrolysis ability (Fig. 3A). There is also no direct inter-hHsp70 FRET signal induced by Hdj1 (SI Appendix, Fig. S4 C and D). However, we could still observe direct binding between Hdj1 and hHsp70 containing the T204A mutation (SI Appendix, Fig. S4E) occurring with rate constants of 2.5 ± 0.1 and 1.5 ± 0.1 s−1 for the two labeling variants (SI Appendix, Table S4), which is of the same order-of-magnitude as the initial fast phase observed without the T204A mutation (Fig. 4D and SI Appendix, Table S4), further confirming that initial binding between Hdj1 and hHsp70 is a fast process, which occurs as the first step. To date, the only available Hsp70–Hsp40 complex structure is the truncated J domain of DnaJ in complex with DnaK in the ATP-bound state, revealing that the J domain interacts with the docked NBD and SBDβ of DnaK (21). In this structure, the ATPase-deficient mutant DnaK–T199A was used (equivalent to hHsp70–T204A), and the SBDα domain of DnaK was covalently fixed to the NBD by a disulfide bond in order to stabilize the SBD open conformation. Here, we show that although Hsp40 interacts with ATP–hHsp70 containing the T204A mutation (equivalent to DnaK–T199A) with fast kinetics, it does not lead to any of the following intramolecular conformational changes or intermolecular assembly. Additionally, flexibility of the NBD–SBD is necessary for activation of the ATPase, since cross-linking between the NBD and SBD dramatically impairs the ATPase stimulation and the interaction with Hsp40 (25, 40). Therefore, the available structure of the complex using the domain cross-linked DnaK–T199A probably provides a snapshot of the initial binding step during the Hsp70–Hsp40 functional cycle (21), while our study allows observation of the conformational changes required for normal functioning of Hsp70 and provides kinetic insights into the multistep reaction.
Based on our smFRET results, the T204A mutation itself does not have any significant influence on the conformational distribution of hHsp70 in the ATP-bound state (SI Appendix, Fig. S2 and Fig. 1 C and D). However, a recent structural study on the human endoplasmic reticulum Hsp70 homolog BiP showed that the ATPase-deficient T229A mutant of BiP (equivalent to T199A in DnaK and T204A in hHsp70) has an open L1,2 loop conformation in the SBDβ in contrast to the completely closed L1,2 loop in WT BiP in the ATP-bound state (6, 7). This result hints that subtle changes can be induced by the T204A mutation and transduced from the NBD through the linker and the NBD–SBD docking interface to the SBD. This kind of subtle change may also influence the conformational dynamics of hHsp70 especially between the NBD and SBDβ as well as its cooperation with Hsp40.
The Monomeric J+G/F-Rich Segment of Hdj1 Is Sufficient to Induce Intramolecular Conformational Changes in hHsp70 and Stimulate Its ATPase Activity.
The isolated J domain of E. coli DnaJ was found to be incapable of activating the ATP hydrolysis of DnaK and the following G/F-rich region is also required for ATPase stimulation (41, 42). We compared the ATPase activity of hHsp70 in the presence of full-length Hdj1 and the J+G/F-rich segment (JGF) of Hdj1 (residues 1–141, including the J domain and G/F-rich region), which is monomeric (SI Appendix, Fig. S5A) and has the same degree of ATPase stimulation above 2 μM as Hdj1 (SI Appendix, Fig. S1 G and H). We could also detect the direct binding between JGF and hHsp70 with an apparent rate constant of 0.7 ± 0.1 and 1.6 ± 0.1 s−1 for the two labeling variants of hHsp70 in the ATP-bound state (SI Appendix, Fig. S5B), which is of the same order-of-magnitude as the binding kinetics between full-length Hdj1 and hHsp70 in the ATP-bound state under the same conditions (SI Appendix, Table S4). However, when we measured the fluorescence spectra of Cy3-labeled hHsp70 and Cy5-labeled hHsp70 upon addition of JGF, there was no inter-hHsp70 FRET (SI Appendix, Fig. S5 C and D), indicating the absence of hHsp70 dimerization, in contrast to when using full-length dimeric Hdj1 (SI Appendix, Fig. S3 B and C). Therefore, we then proceeded to use an excess amount of monomeric JGF instead of full-length Hdj1 to study its effects on the intramolecular conformation of hHsp70, in order to avoid the inter-hHsp70 FRET contribution from hHsp70 dimer, which is induced by the presence of dimeric Hdj1. The FRET efficiency of the SBDβ–SBDα labeling variant increased after binding with JGF in the ATP state (Fig. 5A). The magnitude of the FRET increase was lower than in the presence of full-length dimeric Hdj1 because of the lack of the inter-hHsp70 FRET contribution from the hHsp70 dimer as we expected, similar to the results when a large excess of unlabeled hHsp70 was added to eliminate inter-hHsp70 FRET (Fig. 5A and SI Appendix, Fig. S3A). On the other hand, the FRET efficiency of the NBD–SBDβ labeling variant decreased when interacting with JGF in the presence of ATP (Fig. 5B), lacking the opposite contribution from the dimerization of hHsp70, which together leads to nearly unchanged FRET efficiency when adding full-length dimeric Hdj1 (Fig. 3B). These results indicate that the JGF segment can induce SBD closing and NBD–SBDβ undocking in the ATP-bound state. The real-time kinetics of the intramolecular conformational changes could be directly measured by stopped-flow because hHsp70 does not dimerize upon binding with the monomeric JGF. The kinetics of SBD closing was reflected by the FRET increase of the SBDβ–SBDα labeling variant, giving a rate constant of 0.018 ± 0.001 s−1 obtained by fitting to a single-exponential function (Fig. 5C). Meanwhile, the kinetics of domain undocking as reflected by the FRET decrease of the NBD–SBDβ labeling variant showed a dominant fast phase with a rate constant of 0.035 ± 0.001 s−1 (Fig. 5C). Both of the rates are close to the values derived from the kinetic measurements using full-length Hdj1 (Fig. 4 B and C and SI Appendix, Table S4). The kinetic measurements also confirmed that the detachment of the SBDβ from the NBD has a slightly faster rate than the SBD closing process, indicating that NBD–SBD detachment occurs prior to closing of the SBD.
Fig. 5.
JGF of Hdj1 induces intramolecular conformational changes in hHsp70. (A and B) The FRET spectra of SBDβ–SBDα labeling variant Cy3/Cy5–I427C/K559C (A) and NBD–SBDβ labeling variant Cy3/Cy5–E315C/I427C (B) in the presence of JGF of Hdj1 or full-length Hdj1 in the presence of ATP. The fluorescence at the donor maximum was normalized to 1.0. (C) The FRET changes after addition of JGF to SBDβ–SBDα labeling variant Cy3/Cy5–I427C/K559C (orange curve) or NBD–SBDβ labeling variant Cy3/Cy5–E315C/I427C (blue curve) in the presence of ATP. The kinetic curves were fitted to single-exponential or double-exponential functions (gray lines). The fitting results of C are listed in SI Appendix, Table S4. (D and E) The smFRET distribution histograms of SBDβ–SBDα labeling variant Cy3/Cy5–I427C/K559C (D) and NBD–SBDβ labeling variant Cy3/Cy5–E315C/I427C (E) in the ATP-bound state with or without the presence of JGF. The low FRET efficiency populations (zero peak) caused by inactive acceptor dye are shaded in gray. The red lines on the Left are the fits from PDA and the obtained distance distributions between the labeling sites are shown on the Right. The results of PDA are listed in SI Appendix, Table S3.
The above bulk FRET measurements show that JGF alone is sufficient to induce conformational changes in hHsp70 (Fig. 5 A and B). To identify whether the change in apparent ensemble FRET was caused by changing to an intermediate conformation between the “ATP-like” docked and “ADP-like” undocked states, or a shift in population between multiple conformational states in the ATP-bound state, we performed smFRET experiments. In the presence of an excess of unlabeled JGF, the proportion of the closed conformation of the SBDβ–SBDα and the proportion of the NBD–SBDβ undocked conformation both increased without any change in the peak positions of the distance distributions between the labeling sites (Fig. 5 D and E). Therefore, JGF can induce the interdomain reorganization of hHsp70 to achieve a higher proportion of the ADP-like undocked conformation in the ATP-bound state, which is probably an ATPase-stimulated conformational state. This is consistent with the observation in a cross-linking study that the same cross-linked fragments of Hsp40-induced hHsp70 dimer were found in the presence of ATP or ADP (43), and neither of these cross-linked sites could be accommodated in the crystal structure of ATP–DnaK dimer (5) but could be better matched to an antiparallel dimer of the DnaK ADP state (43), implying that the ATP-bound hHsp70 tends to populate the ADP-like conformation when interacting with Hsp40.
Taken together, the NBD–SBDβ undocking of hHsp70 caused by interaction with the J domain probably accounts for the ATPase stimulation, while the following dimerization of hHsp70 is induced by the dimeric nature of the full-length Hdj1. The ATPase stimulation of hHsp70 by JGF is lower than by the full-length Hdj1 in the substoichiometric range (SI Appendix, Fig. S1H), which indicates that the monomeric JGF cannot fully replace the full-length Hsp40 in ATPase stimulation. This is in accordance with the observation for the DnaK–DnaJ system that although the J+G/F-rich region is sufficient to stimulate the ATPase activity of DnaK, it shows lower apparent affinity to DnaK than full-length DnaJ (41, 42). So other domains of Hsp40 may also contribute to the interaction with and dimerization of Hsp70 to stabilize the ATPase-stimulated conformation of Hsp70 induced by the J domain, thus achieving maximum activation. Whether the formation of the Hsp70–Hsp40 heterotetramer identified in this study is functionally related to specific physiological processes such as substrate transfer and refolding as well as the synergistic cooperation between Hsp40 and substrate still needs further investigation.
Discussion
Although the Hsp70 family is highly conserved between organisms, differences exist in domain interactions and conformational dynamics among Hsp70 family members. For E. coli DnaK, its ATP-bound state is homogeneous, while in the ADP-bound state, the NBD–SBD is undocked with multiple conformations of the SBD pocket (10, 12). In the case of the yeast mitochondrial Ssc1, the ATP-bound state is also homogeneous, but the ADP-bound state of Ssc1 shows both docked and undocked populations of the NBD–SBD (10, 13). In higher eukaryotes, BiP shows population of both undocked and docked NBD–SBD conformations in the ATP-bound state (11), but the conformation of the NBD–SBD is homogenous in the ADP-bound state (11). In a recent NMR study, both the human stress-inducible Hsp70 and the constitutive cognate Hsc70 were shown to have docked and undocked populations in the ADP-bound state (16). In this study, using smFRET, we have revealed that in both the ATP- and ADP-bound states, there is not only conformational heterogeneity of docking/undocking between the NBD and SBD of hHsp70 but also conformational heterogeneity of opening/closing of the SBDβ–SBDα cleft. It is likely that the interdomain interaction of Hsp70 homologs increases in terms of dynamics and complexity from prokaryotes to eukaryotes, which may reflect adaption to more complex physiological environments and more specific cellular functions in higher organisms. For example, the conformational flexibility of the SBDα lid of Hsp70 might be beneficial for the accommodation of substrates with different sizes, especially for larger proteins and aggregates (11, 44, 45).
It is well established that Hsp40 can stimulate the ATPase activity of Hsp70, but how Hsp40 helps to reach an ATPase-activated state was not clearly known. In the ATP-bound state, Hsp70 has low ATPase activity because the NBD is docked onto the SBDβ and constrained in a conformation incompatible with ATP hydrolysis (46). The only complex structure currently available of DnaK–DnaJ is fixed in an NBD–SBD docked conformation by cross-linking (21), which is unlikely to be an ATPase-activated state. In this study, we directly observed the kinetic process whereby the binding of Hdj1 disrupts the docked NBD and SBDβ, which results in formation of an ADP-like domain-undocked conformation in the ATP-bound state, which then releases the constraints and directs Hsp70 into an optimal conformation for ATP hydrolysis (Fig. 6). Similarly, NBD–SBD undocking can also be induced by mutations and substrate binding in the ATP-bound state (14, 34, 46), leading to an ATPase-stimulated state. However, the ATPase stimulation effect of substrates is usually weaker than that of Hsp40, mainly because the interaction between substrates and the Hsp70 SBD is weak and transient in the ATP-bound state (34). Hsp40 has multiple binding sites for Hsp70 and directly interacts with the NBD–SBDβ interface of hHsp70, leading to higher binding affinity and a direct effect on the conformation of hHsp70. In another study of the interaction between bovine Hsc70 and a J domain protein, cross-linking of the NBD and SBD of Hsc70 dramatically diminishes the stimulation of the ATPase by the J domain, suggesting the docked conformation could generate steric clashes between the SBD and the J domain (25). Therefore, the interdomain conformational flexibility is crucial for the Hsp70–Hsp40 functional cycle. Interestingly, a recent study of human BiP by fluorescence quenching shows that the binding of Hsp40 ERdj3 to ATP-bound BiP can lead to changes in the conformation and dynamics of the β1–L1,2–β2 loop in the SBD to an ADP-like conformation (7). This observation to some extent supports our model, in which the conformational changes of Hsp70 are induced by binding of Hsp40 and occur prior to ATP hydrolysis. The allosteric signal upon interaction with Hsp40 is likely transduced from the linker and the domain interface to the SBD, which leads to the undocked conformation observed in our study and also regulates the conformation of the SBD as observed in the BiP/ERdj3 study (7).
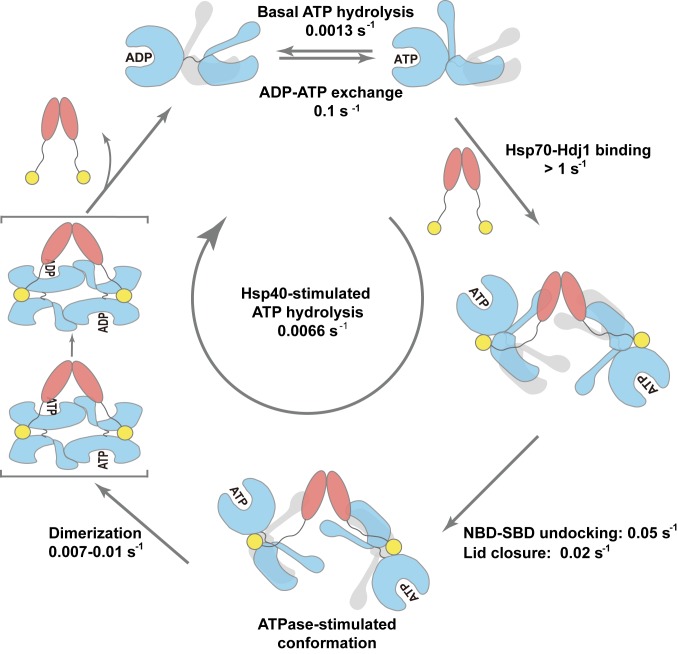
Fig. 6.
The kinetics of the Hsp70–Hdj1 functional cycle. The J domain and substrate binding domain of Hdj1 are colored yellow and pink, respectively. As the first step, dimeric Hdj1 binds two monomeric hHsp70 molecules either simultaneously or sequentially on a subsecond timescale. Next, the J domain of Hdj1 interacts with the NBD–SBD interface of hHsp70, causing the NBD and SBDβ to detach, and pushing the SBDα toward the SBDβ on a tens-of-seconds timescale, to form the ATPase-stimulated conformational state. Binding to dimeric Hdj1 and the consequent intramolecular conformational changes facilitates direct dimerization of hHsp70 and formation of the “two hHsp70 + Hdj1 dimer” tetrameric complex on a 100-s timescale, accompanying ATP hydrolysis. After ATP is hydrolyzed to ADP, Hdj1 dissociates from the complex and the hHsp70 dimer dissociates back to monomer in the ADP-bound state, which can then exchange with ATP to start a new cycle. The kinetic parameters shown in this figure are extracted from the kinetic measurements in Figs. 2 and 4. The ATPase activity shown in this figure was measured at the same temperature as the kinetic experiments, for comparison.
Recently, Hsp40 has been found by mass spectrometry and chemical cross-linking to facilitate the formation of an antiparallel dimer of monomeric Hsp70 in the ATP-bound state, showing the interaction between the NBD with the SBDα from the other Hsp70 molecule in the dimer (43). However, how this multistep process occurs and the intermediate states encountered were still unknown. Here, we directly observed the Hsp70–Hsp40 assembly process, which allows us to distinguish the sequence of the multistep reaction, showing that Hdj1 binds to monomeric hHsp70 in the ATP-bound state as the first step, then induces domain-undocking between the NBD and SBDβ and opening of the SBDα-SBDβ cleft, and finally causes dimerization of hHsp70 (Figs. 4 and 6). A crystal structure of DnaK in the dimeric ATP-bound state has been reported previously (5), and the dimeric form of ATP–DnaK was suggested to be required for efficient interaction with DnaJ as indicated by mutagenesis and biochemical studies (47). Although we also observed the formation of an hHsp70 dimer when interacting with Hsp40, we propose a different model for the Hsp70–Hsp40 functional cycle based on the real-time kinetic measurements of the intermolecular and intramolecular changes of Hsp70 modulated by Hsp40 (Fig. 6). First, it is not likely that hHsp70 maintains the same structure as the dimeric ATP–DnaK when assembled with Hdj1, because undocking of the NBD–SBDβ and closing of the SBDβ–SBDα induced by Hdj1 occur faster than the inter-hHsp70 dimerization as shown by the kinetic results. Another piece of direct evidence on this point is that cross-linking of the ATP–DnaK dimer, which fixes it in a “NBD–SBD docked and SBD open” conformation, greatly compromises rather than enhances its interaction with DnaJ (40). Second, the binding between hHsp70 and Hdj1 is over an order-of-magnitude faster than hHsp70 dimerization under both the in vitro experimental conditions and in vivo physiological concentrations (Fig. 4 B–E), indicating that the dimer formation of hHsp70 is induced by binding to symmetric Hsp40 dimers. Furthermore, the monomeric JGF of Hdj1 could also stimulate ATPase activity in the same way as full-length Hdj1 without formation of the Hsp70 dimer when present at saturating concentrations (Fig. 5 and SI Appendix, Figs. S1G and S5), which suggests that the dimeric form of Hsp70 is not indispensable at least for initial binding with Hsp40 and ATPase stimulation. Therefore, it is unlikely that only the preexisting, extremely low amount of dimeric ATP–Hsp70 could interact with Hsp40 and then move the equilibrium from Hsp70 monomer toward Hsp70 dimer. A possible explanation is that there may be some differences between eukaryotic Hsp70s such as hHsp70 and prokaryotic Hsp70s such as E. coli DnaK in the mechanism of assembly with Hsp40. However, as we observed that Hsp40 induces both intramolecular and intermolecular changes in Hsp70 upon interaction, the conformational flexibility of Hsp70 is clearly crucial for the normal function of the ATP hydrolysis cycle and its cooperation with Hsp40. Thus, any restriction on the conformation such as cross-linking or fixation may lead to abnormal behavior and loss of function.
In summary, based on our observations, we propose a scheme for the kinetics of the Hsp40-modulated Hsp70 chaperone cycle (Fig. 6): Hsp40 binds to ATP-bound Hsp70 on a subsecond timescale. The binding subsequently induces undocking of the NBD–SBDβ and closing of the SBD on a tens of seconds timescale to achieve a conformation resembling an ADP-like domain-undocked conformation. Binding with dimeric Hdj1 and the consequent intramolecular conformational changes facilitate direct dimerization of Hsp70 to form a “two Hsp70 + Hdj1 dimer” tetrameric complex on a 100-s timescale. Hsp70 itself has a low ATP hydrolysis rate due to the occasional undocking of the NBD and SBDβ, while Hsp40 modulates Hsp70 to adopt an ATPase-activated domain-undocked conformation. Therefore, the participation of Hsp40 increases the overall hydrolysis efficiency by nearly fivefold, although Hsp70 needs to go through multiple reaction steps. Our study provides insight into the dynamic and kinetic aspects of the Hsp70 functional cycle, which is of great importance for mechanistic understanding of the Hsp70–Hsp40 chaperone system as well as development of small-molecule inhibitors targeting Hsp70 and Hsp70-containing complexes to prevent human diseases (48, 49).
Materials and Methods
Ensemble Steady-State FRET.
Ensemble steady-state FRET measurements were performed at 25 °C on an RF-5301PC spectrofluorometer (Shimadzu) or an F7000 spectrofluorometer (Hitachi). Fluorescence-labeled hHsp70 was diluted to 500 nM with or without 1 mM ATP or ADP. Where used, the final concentration of Hdj1 or JGF of Hdj1 was 5 μM. The samples were excited at 532 nm, and the emission spectra were recorded from 540 to 800 nm. The purity of ATP and ADP (Sigma) used in this study was checked by high-performance liquid chromatography (SI Appendix).
Stopped-Flow Fluorescence Measurements.
The kinetics of the ATP- and Hdj1-induced conformational changes of hHsp70 reflected by changes in the FRET signal of hHsp70 were measured using a SX20 stopped-flow instrument (Applied Photophysics) with dual-channel fluorescence photomultiplier tube detectors. For the kinetic experiments on hHsp70 without Hdj1, the Cy3/Cy5-labeled hHsp70 and different concentrations of ATP were mixed at a 1:1 volume ratio to reach a final concentration of 100 nM for fluorescently labeled hHsp70. For the kinetic experiments related to Hdj1, the labeled hHsp70 in the presence of 2 mM ATP was mixed with Hdj1 (or its JGF) at a 1:1 volume ratio to reach final concentrations of 100 nM for hHsp70, 1 mM for ATP, and 5 μM for Hdj1 (or JGF). To prevent protein adsorption, 0.01% Tween 20 (Thermo) was added to the buffer. The samples were excited at 532 nm with a 10-nm slit width and the fluorescence signals for each of the two channels were recorded simultaneously using a 585 ± 20-nm filter for Cy3 and a 650-nm long-pass filter for Cy5. The FRET efficiencies were calculated as the ratio of Cy5 intensity to the sum of Cy3 and Cy5 intensity. The stopped-flow instrument was equipped with a circulating water bath, allowing precise control of the reaction temperature. All of the results were the average of four to five repeated runs. The analysis of the kinetic data in Fig. 2 is described in SI Appendix.
Confocal smFRET Experiment.
Confocal smFRET experiments were carried out using a home-built confocal microscope as described previously (50). The bin time for single-molecule data collection was 1 ms. A 30 photon/ms bin for the sum of donor channel and acceptor channel was set as the threshold for the selection of single-molecule bursts. The apparent FRET efficiency was calculated using the equation below:
| [2] |
where and are the fluorescence intensity of acceptor and donor, respectively. The statistical histograms of the FRET distribution were processed by PDA (SI Appendix) to obtain the distance distribution between the labeling sites. The calibration factors including the difference in quantum yield and detection efficiency of donor and acceptor, the donor fluorescence leakage to the acceptor channel, and the direct excitation of acceptor were measured and incorporated in the PDA (SI Appendix, SI text and Table S5).
Plasmid construction, protein expression, purification and labeling, SEC experiments, functional assays, and analysis of smFRET data and kinetic data are described in detail in SI Appendix.
Data Availability.
The code used for smFRET data analysis is available at GitHub: https://github.com/THUZCAM/PDA-method. All other methods and data needed to evaluate the conclusions are present in the main article and/or SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Xun Li (Nikon Instruments, Beijing Branch) for helpful discussions about single-molecule experiments. We thank Yuanyuan Chen, Zhenwei Yang, and Bingxue Zhou (Institute of Biophysics, Chinese Academy of Sciences) for technical help with BLI experiments. We acknowledge support from the Chinese Ministry of Science and Technology (Grant 2017YFA0504000), the National Natural Science Foundation of China (Grants 31570780, 21673278, 31770829, 31920103011, and 21877070), the National Laboratory of Biomacromolecules, and the Chinese Academy of Sciences Center of Excellence in Biomacromolecules.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The code used for single-molecule fluorescence resonance energy transfer data analysis is available at GitHub (https://github.com/THUZCAM/PDA-method).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914376117/-/DCSupplemental.
References
- 1.Bukau B., Weissman J., Horwich A., Molecular chaperones and protein quality control. Cell 125, 443–451 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Mayer M. P., et al. , Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol. 7, 586–593 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Bertelsen E. B., Chang L., Gestwicki J. E., Zuiderweg E. R., Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U.S.A. 106, 8471–8476 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kityk R., Kopp J., Sinning I., Mayer M. P., Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Qi R., et al. , Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat. Struct. Mol. Biol. 20, 900–907 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J., Nune M., Zong Y., Zhou L., Liu Q., Close and allosteric opening of the polypeptide-binding site in a human Hsp70 chaperone BiP. Structure 23, 2191–2203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J., et al. , Conformation transitions of the polypeptide-binding pocket support an active substrate release from Hsp70s. Nat. Commun. 8, 1201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhuravleva A., Gierasch L. M., Substrate-binding domain conformational dynamics mediate Hsp70 allostery. Proc. Natl. Acad. Sci. U.S.A. 112, E2865–E2873 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharya A., et al. , Allostery in Hsp70 chaperones is transduced by subdomain rotations. J. Mol. Biol. 388, 475–490 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mapa K., et al. , The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol. Cell 38, 89–100 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Marcinowski M., et al. , Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat. Struct. Mol. Biol. 18, 150–158 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Schlecht R., Erbse A. H., Bukau B., Mayer M. P., Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 18, 345–351 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Sikor M., Mapa K., von Voithenberg L. V., Mokranjac D., Lamb D. C., Real-time observation of the conformational dynamics of mitochondrial Hsp70 by spFRET. EMBO J. 32, 1639–1649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai A. L., et al. , Key features of an Hsp70 chaperone allosteric landscape revealed by ion-mobility native mass spectrometry and double electron-electron resonance. J. Biol. Chem. 292, 8773–8785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wieteska L., Shahidi S., Zhuravleva A., Allosteric fine-tuning of the conformational equilibrium poises the chaperone BiP for post-translational regulation. eLife 6, e29430 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng W., Clerico E. M., McArthur N., Gierasch L. M., Allosteric landscapes of eukaryotic cytoplasmic Hsp70s are shaped by evolutionary tuning of key interfaces. Proc. Natl. Acad. Sci. U.S.A. 115, 11970–11975 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer M. P., Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 38, 507–514 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Alderson T. R., Kim J. H., Markley J. L., Dynamical structures of Hsp70 and Hsp70-Hsp40 complexes. Structure 24, 1014–1030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young J. C., Mechanisms of the Hsp70 chaperone system. Biochem. Cell Biol. 88, 291–300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kampinga H. H., Craig E. A., The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kityk R., Kopp J., Mayer M. P., Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 chaperones. Mol. Cell 69, 227–237.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Malinverni D., Jost Lopez A., De Los Rios P., Hummer G., Barducci A., Modeling Hsp70/Hsp40 interaction by multi-scale molecular simulations and coevolutionary sequence analysis. eLife 6, e23471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmad A., et al. , Heat shock protein 70 kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proc. Natl. Acad. Sci. U.S.A. 108, 18966–18971 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suh W. C., et al. , Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc. Natl. Acad. Sci. U.S.A. 95, 15223–15228 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang J., et al. , Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell 28, 422–433 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J. H., Alderson T. R., Frederick R. O., Markley J. L., Nucleotide-dependent interactions within a specialized Hsp70/Hsp40 complex involved in Fe-S cluster biogenesis. J. Am. Chem. Soc. 136, 11586–11589 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Wu Y., Qian X., Sha B., Crystal structure of yeast Sis1 peptide-binding fragment and Hsp70 Ssa1 C-terminal complex. Biochem. J. 398, 353–360 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freeman B. C., Myers M. P., Schumacher R., Morimoto R. I., Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 14, 2281–2292 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R. E., Targeting heat shock proteins 70/90 and proteasome for cancer therapy. Curr. Med. Chem. 18, 4250–4264 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Broadley S. A., Hartl F. U., The role of molecular chaperones in human misfolding diseases. FEBS Lett. 583, 2647–2653 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Murphy M. E., The HSP70 family and cancer. Carcinogenesis 34, 1181–1188 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antonik M., Felekyan S., Gaiduk A., Seidel C. A., Separating structural heterogeneities from stochastic variations in fluorescence resonance energy transfer distributions via photon distribution analysis. J. Phys. Chem. B 110, 6970–6978 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Theyssen H., Schuster H. P., Packschies L., Bukau B., Reinstein J., The second step of ATP binding to DnaK induces peptide release. J. Mol. Biol. 263, 657–670 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Zhuravleva A., Clerico E. M., Gierasch L. M., An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151, 1296–1307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buchberger A., et al. , Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J. Biol. Chem. 270, 16903–16910 (1995). [DOI] [PubMed] [Google Scholar]
- 36.McCarty J. S., Walker G. C., DnaK as a thermometer: Threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc. Natl. Acad. Sci. U.S.A. 88, 9513–9517 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banecki B., Zylicz M., Real time kinetics of the DnaK/DnaJ/GrpE molecular chaperone machine action. J. Biol. Chem. 271, 6137–6143 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Nollen E. A., Brunsting J. F., Song J., Kampinga H. H., Morimoto R. I., Bag1 functions in vivo as a negative regulator of Hsp70 chaperone activity. Mol. Cell. Biol. 20, 1083–1088 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stankiewicz M., Nikolay R., Rybin V., Mayer M. P., CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. FEBS J. 277, 3353–3367 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Liu Q., et al. , A disulfide-bonded DnaK dimer is maintained in an ATP-bound state. Cell Stress Chaperones 22, 201–212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wall D., Zylicz M., Georgopoulos C., The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J. Biol. Chem. 269, 5446–5451 (1994). [PubMed] [Google Scholar]
- 42.Karzai A. W., McMacken R., A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J. Biol. Chem. 271, 11236–11246 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Morgner N., et al. , Hsp70 forms antiparallel dimers stabilized by post-translational modifications to position clients for transfer to Hsp90. Cell Rep. 11, 759–769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao X., et al. , Human Hsp70 disaggregase reverses Parkinson’s-linked α-synuclein amyloid fibrils. Mol. Cell 59, 781–793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nillegoda N. B., et al. , Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 524, 247–251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kityk R., Vogel M., Schlecht R., Bukau B., Mayer M. P., Pathways of allosteric regulation in Hsp70 chaperones. Nat. Commun. 6, 8308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarbeng E. B., et al. , A functional DnaK dimer is essential for the efficient interaction with Hsp40 heat shock protein. J. Biol. Chem. 290, 8849–8862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Assimon V. A., Gillies A. T., Rauch J. N., Gestwicki J. E., Hsp70 protein complexes as drug targets. Curr. Pharm. Des. 19, 404–417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rousaki A., et al. , Allosteric drugs: The interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J. Mol. Biol. 411, 614–632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lou F., Yang J., Wu S., Perrett S., A co-expression strategy to achieve labeling of individual subunits within a dimeric protein for single molecule analysis. Chem. Commun. (Camb.) 53, 7986–7989 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code used for smFRET data analysis is available at GitHub: https://github.com/THUZCAM/PDA-method. All other methods and data needed to evaluate the conclusions are present in the main article and/or SI Appendix.