Significance
Introgressive hybridization of pairs of species can affect the evolution of their populations in several important ways. More rarely, three species form an interbreeding complex (triad), but little is known of the consequences. A long-term study of interbreeding Darwin’s finches on Daphne Major island, Galápagos, shows that Geospiza fortis acts as a conduit for the passage of genes from Geospiza fuliginosa, a rare immigrant, to Geospiza scandens, a resident. Surprisingly, the species and the derived dihybrids and trihybrids had approximately equal fitness. Gene flow increased in frequency and apparently caused a morphological convergence of the resident species. These findings illustrate how evolutionary change is potentially increased by hybridization among three species and the assembly of novel combinations of genes.
Keywords: speciation, introgression, trihybrids, fitness, convergence
Abstract
Introgressive hybridization can affect the evolution of populations in several important ways. It may retard or reverse divergence of species, enable the development of novel traits, enhance the potential for future evolution by elevating levels of standing variation, create new species, and alleviate inbreeding depression in small populations. Most of what is known of contemporary hybridization in nature comes from the study of pairs of species, either coexisting in the same habitat or distributed parapatrically and separated by a hybrid zone. More rarely, three species form an interbreeding complex (triad), reported in vertebrates, insects, and plants. Often, one species acts as a genetic link or conduit for the passage of genes (alleles) between two others that rarely, if ever, hybridize. Demographic and genetic consequences are unknown. Here we report results of a long-term study of interbreeding Darwin’s finches on Daphne Major island, Galápagos. Geospiza fortis acted as a conduit for the passage of genes between two others that have never been observed to interbreed on Daphne: Geospiza fuliginosa, a rare immigrant, and Geospiza scandens, a resident. Microsatellite gene flow from G. fortis into G. scandens increased in frequency during 30 y of favorable ecological conditions, resulting in genetic and morphological convergence. G. fortis, G. scandens, and the derived dihybrids and trihybrids experienced approximately equal fitness. Especially relevant to young adaptive radiations, where species differ principally in ecology and behavior, these findings illustrate how new combinations of genes created by hybridization among three species can enhance the potential for evolutionary change.
In the early stages of speciation, nascent species are prone to hybridize and exchange genes through backcrossing and introgression (1–3). The circumstances and consequences of hybridization in animals have received much attention recently because gene exchange can affect the evolution of populations in several important ways: at the most extreme in facilitating the evolution of unisexuality in both vertebrates (4, 5) and invertebrates (6, 7). Hybridization may retard, arrest, or reverse divergence of species (8–11); enable the development of novel morphological and physiological traits (12–15); enhance the potential for future evolution by elevating levels of standing variation (16–20) and by strengthening or relaxing covariances (17, 21); alleviate inbreeding depression in small populations (22–24); and create new species (2, 25–27). Extensive hybridization in the past is known through inferences from phylogenetic reconstructions (3, 15, 28, 29). With regard to contemporary hybridization in nature, most of what is known comes from the study of pairs of species, either coexisting in the same habitat or distributed geographically with little overlap and separated by a hybrid zone (30, 31). In this article, we discuss an additional complexity, the occurrence of hybridization among triplets of species (triads), and consider its context and evolutionary significance.
Triad hybridization has been reported in fish (32–34), frogs (35), lizards (36), birds (17, 37, 38), mammals (39–41), snails (42), butterflies (43), mosquitoes (44), as well as several plant taxa (45, 46). One species acts as a genetic bridge (33, 47, 48) or conduit (41, 49) for the passage of genes (alleles) between two others that, in some cases, rarely if ever hybridize (33, 39, 41). Exchange of genes is generally not studied directly with breeding pairs but is inferred from the presence of individuals of mixed (trispecies) ancestry (33, 39, 41) or from the fact that hybridization occurs, or is enhanced, only in the presence of a third species (36, 50).
Triad hybridization, where one species acts as an intermediary for gene exchange between two other noninterbreeding species, raises questions of genome compatibility, selection, fitness, and the evolution of reproductive isolation. Evolutionary effects of bispecies hybridization may be either negated or amplified in trispecies hybridization. On the one hand, donated alleles may be gradually eliminated by selection and drift, resulting in no net change in genome compositions. In this case, hybridization is likely to be transitory and a prelude to the evolution of stronger reproductive isolation and a decline in gene exchange (51). On the other hand, introgressed alleles may be filtered in transit (52), with some being incorporated into the genomes of the bridging species and in the downstream recipient species, where they affect morphology or physiology and facilitate evolutionary divergence (53–55). Gene dynamics are potentially heterogeneous. Epistasis is expected to be complex because genes from one species function against a genetic background derived in part from two other species. Of interest is the question whether the recipient species receive alleles equally or unequally (filtered) via the intermediary conduit, and whether the conduit species itself donates more or less than it receives. Alternative outcomes depend on the frequency of interbreeding, the direction(s) of exchange, and fitness consequences. With rare exceptions (39, 43), all of these are unknown. Long-term demographic studies are needed to estimate the magnitude of these influences and to document and interpret the significance of triad hybridization.
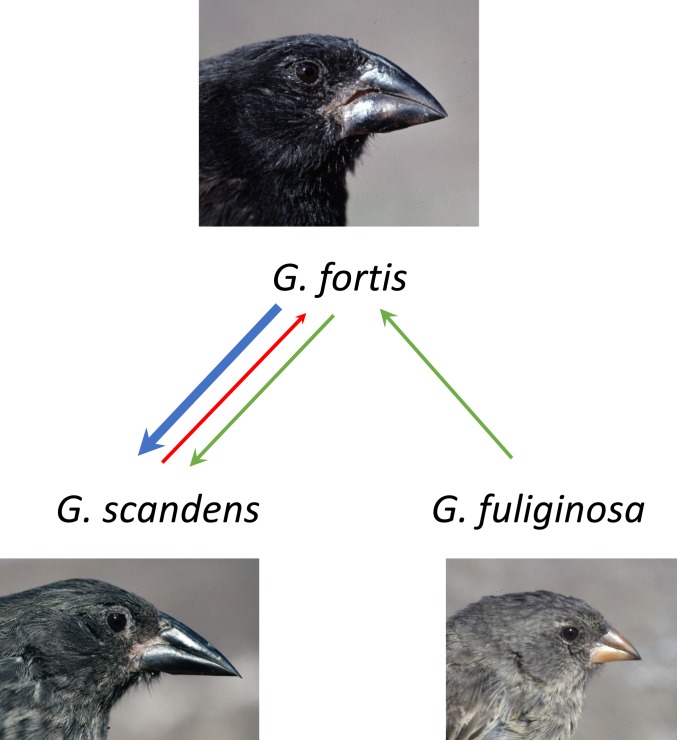
We report an example from a long-term demographic study of Darwin’s finches on the small Galápagos island of Daphne Major (0.34 km2), carried out from 1973 to 2012. On this island, the medium ground finch (Geospiza fortis, ∼17 g) occasionally breeds with the cactus finch (Geospiza scandens, ∼21 g), a resident species, and the small ground finch (Geospiza fuliginosa, ∼13 g), a rare immigrant and sister species to G. fortis (28, 56). In doing so, G. fortis acts as a conduit for the passage of genes between G. fuliginosa and G. scandens, two species that have never been observed to interbreed on Daphne (Fig. 1). Individuals were initially assigned to species on the basis of morphology (SI Appendix, section 1), and hybrids were identified from pedigree data (57). Mixed pairs, first observed in 1976 (57, 58), were generally at frequencies of one to three percent of the breeding populations (56). Offspring of known parents were uniquely marked with leg bands in the years 1976 to 1998, and breeding pairs were identified in every year up to 2012. Questions of paternity were resolved with microsatellite markers when a program of DNA sampling was initiated in 1988 (59). The microsatellite markers were used to identify hybrids throughout the whole study period (Methods and SI Appendix, sections 2 and 3). Known hybrids were observed to breed from 1983 onward when an El Niño event caused a major change to the environment (56, 60). Hybrids were found to backcross according to the same rules that govern intraspecific mate choice: finches choose mates on the basis of imprinting on paternal song and parental morphology, and occasionally hybridize when they learn the song of another species (61, 62). Frequencies of alleles at 16 unlinked polymorphic loci are used here to assign 3,165 individuals probabilistically (≥0.9) to the three species (63), and, if the 0.9 criterion is not met, they are assigned to hybrids (admixtures). We refer to them as primary admixtures when individuals are assigned to two species (dihybrids) and secondary admixtures when they are assigned to all three species (trihybrids).
Fig. 1.
Gene exchange among three species of Darwin’s finches. The green lines indicate an indirect transfer from G. fuliginosa to G. scandens via G. fortis acting as a conduit. G. fuliginosa is too rare to form an independent breeding population. The principal flow of genes is from G. fortis to G. scandens (blue line).
We present quantitative estimates of the frequencies of di- and trihybrids and introgression of neutral microsatellite markers. We show that, as expected, birds with genomic admixtures are intermediate in morphology between the means of the parental species that gave rise to them, and with elevated variances. The main finding is that di- and trihybrids survived surprisingly well, approximately as well as G. fortis and G. scandens over the first 6 to 10 y of life. High survival, even of trihybrids, may be characteristic of relatively young adaptive radiations in fluctuating environments.
Results
Frequencies of Primary Admixtures.
G. scandens × G. fortis (SF) and G. fortis × G. scandens (FS) hybrids were produced in every well-sampled year from 1978 onward (Table 1): the first letter denotes the species with majority assignment probability. G. fortis × G. fuliginosa (Ff) hybrids were produced from 1981 onward, and in greater numbers. A small number of admixed individuals (n = 27) were assigned to G. scandens and G. fuliginosa (Sf and fS) but not to G. fortis. Most (0.74) occurred after 1998 when the detailed breeding study ceased. They may have been the undetected product of interbreeding of the two species, which was never observed in 1976 to 1998 but may have occurred through extrapair mating (61). Alternatively, and perhaps more likely, they were the product of interbreeding of G. scandens and Ff, or some more complicated pairing such as SF × fF.
Table 1.
Numbers of sampled G. fuliginosa, G. fortis, G. scandens, and pairwise admixtures produced each year
| Year | fuliginosa | fortis | scandens | Ff + fF | FS | SF | Sf + fS |
| 1975 | — | — | 2 | — | — | — | — |
| 1978 | — | 7 | 3 | — | 1 | — | 1 |
| 1979 | — | 1 | 1 | — | — | 1 | — |
| 1981 | — | 9 | 3 | 2 | — | 1 | 1 |
| 1983 | 1 | 45 | 18 | 2 | 2 | 1 | 1 |
| 1984 | 1 | 24 | 2 | 3 | — | — | — |
| 1986 | 1 | — | — | — | — | — | |
| 1987 | 6 | 283 | 57 | 21 | 7 | 2 | 3 |
| 1990 | 27 | 4 | 3 | — | — | — | |
| 1991 | 5 | 648 | 127 | 41 | 11 | 4 | 2 |
| 1992 | 7 | 123 | 46 | 15 | 2 | 3 | 1 |
| 1993 | 7 | 126 | 102 | 5 | 5 | 5 | — |
| 1997 | — | 6 | 13 | — | — | 1 | — |
| 1998 | 1 | 140 | 98 | 8 | 6 | 6 | 3 |
| 2000 | — | — | 15 | 1 | 1 | 2 | 1 |
| 2001 | 1 | 53 | 4 | 5 | 1 | — | — |
| 2002 | 3 | 68 | 66 | 17 | 4 | 6 | 1 |
| 2005 | 7 | 93 | 27 | 10 | 5 | 6 | 1 |
| 2007 | 1 | 31 | 13 | 5 | 8 | 11 | 3 |
| 2008 | 3 | 75 | 82 | 4 | 3 | 14 | 3 |
| 2009 | 2 | 53 | 41 | 8 | 5 | 2 | 1 |
| 2010 | 8 | 74 | 59 | 6 | 3 | 9 | 3 |
| 2011 | 8 | 14 | 26 | 4 | 4 | 6 | 3 |
| Totals | 61 | 1,901 | 809 | 160 | 68 | 80 | 28 |
Abbreviations: letters designating hybrids refer to the species G. fuliginosa (f), G. fortis (F), and G. scandens (S). The first letter of admixtures denotes the species with majority assignment probability.
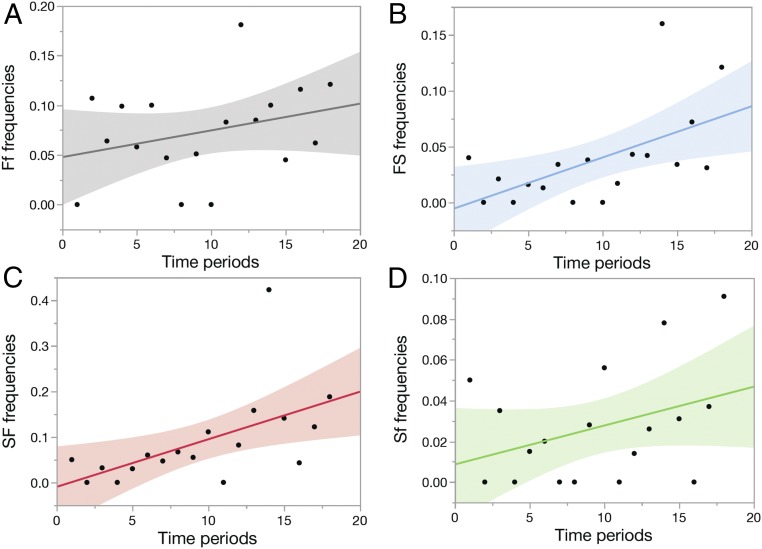
Hybrid production increased in frequency, especially after 2000 (Fig. 2). All slopes of regressions of numbers on year are positive. The Ff relationship is not significant, either with (F1,16 = 1.61, P = 0.2228) or without the outlier in 2002 (F1,15 = 1.36, P = 0.2617), and nor is the relationship for Sf and fS combined (F1,16 = 2.20, P = 0.1572). The FS relationship is significant with the two outliers in 2007 and 2011 included (F1,16 = 7.48, P = 0.0147) or excluded (F1,14 = 4.64, P = 0.0491), and the SF relationship is also significant without the strong outlier in 2007 (F1,15 = 15.00, P = 0.0015).
Fig. 2.
Frequencies of hybrids produced in each year of breeding. Hybrids are expressed as frequencies of the offspring produced in each of the G. fortis groups (G. fortis, Ff [A], FS [B]) and G. scandens groups (G. scandens, SF [C], Sf [D], fS). Regressions are significant for FS and SF frequencies (see Frequencies of Primary Admixtures).
To quantify the trends, we calculated means after deleting the conspicuous outliers. Since the remaining frequencies vary relatively little and are normally distributed, we did not transform them. Frequencies are much higher in Ff (0.073 ± 0.011 SE) and SF (0.089 ± 0.023) than FS (0.037 ± 0.010) and Sf (0.028 ± 0.007; n = 18 for each group). Sf frequencies are notably lower than Ff (P = 0.0160) and SF (P = 0.0327) frequencies by Kramer HSD tests. The inequalities imply that alleles were transferred twice as frequently from G. fuliginosa to G. fortis (Ff) and from G. fortis to G. scandens (SF) as in the reverse direction (FS).
Secondary Admixtures.
Secondary admixtures occur through interbreeding of G. scandens and Ff hybrids or backcrosses. Frequencies were uniformly low (Table 2), generally much lower than the frequencies of primary admixtures (Table 1). Allele transfer from G. fuliginosa to G. scandens via members of the G. fortis population (including hybrids) is indicated by the breeding of trispecific hybrids. Three of these hybrids with a majority assignment to G. fortis or G. fuliginosa produced offspring by breeding with G. scandens, and four others did so by breeding with G. fortis. In the four cases where fathers of the trihybrids were known (three G. fortis and one G. scandens), they all followed the mating rule of pairing according to paternal song type. As with the dihybrids, gene exchange involving trihybrids was bidirectional between G. fortis and G. scandens.
Table 2.
Numbers of trihybrids produced each year
| Year | FfS | fFS | FSf | fSF | SFf | SfF |
| 1987 | 3 | 1 | 4 | 1 | 1 | — |
| 1991 | 1 | 1 | 1 | — | — | — |
| 1992 | 1 | — | 1 | 1 | — | — |
| 1993 | 2 | — | — | 1 | — | — |
| 1997 | — | — | — | — | 1 | — |
| 1998 | — | — | 1 | — | 1 | — |
| 2002 | 1 | — | — | 1 | — | — |
| 2005 | 2 | — | 1 | — | 3 | 1 |
| 2007 | 1 | 2 | — | — | — | 1 |
| 2008 | 1 | — | 1 | — | — | 1 |
| 2009 | 1 | — | — | — | 1 | 1 |
| 2010 | 3 | — | 1 | 1 | 3 | 1 |
| Totals | 16 | 4 | 10 | 5 | 10 | 5 |
Abbreviations: letters designating hybrids refer to the species G. fuliginosa (f), G. fortis (F), and G. scandens (S), and the descending order of assignment probability to the three species.
Introgression.
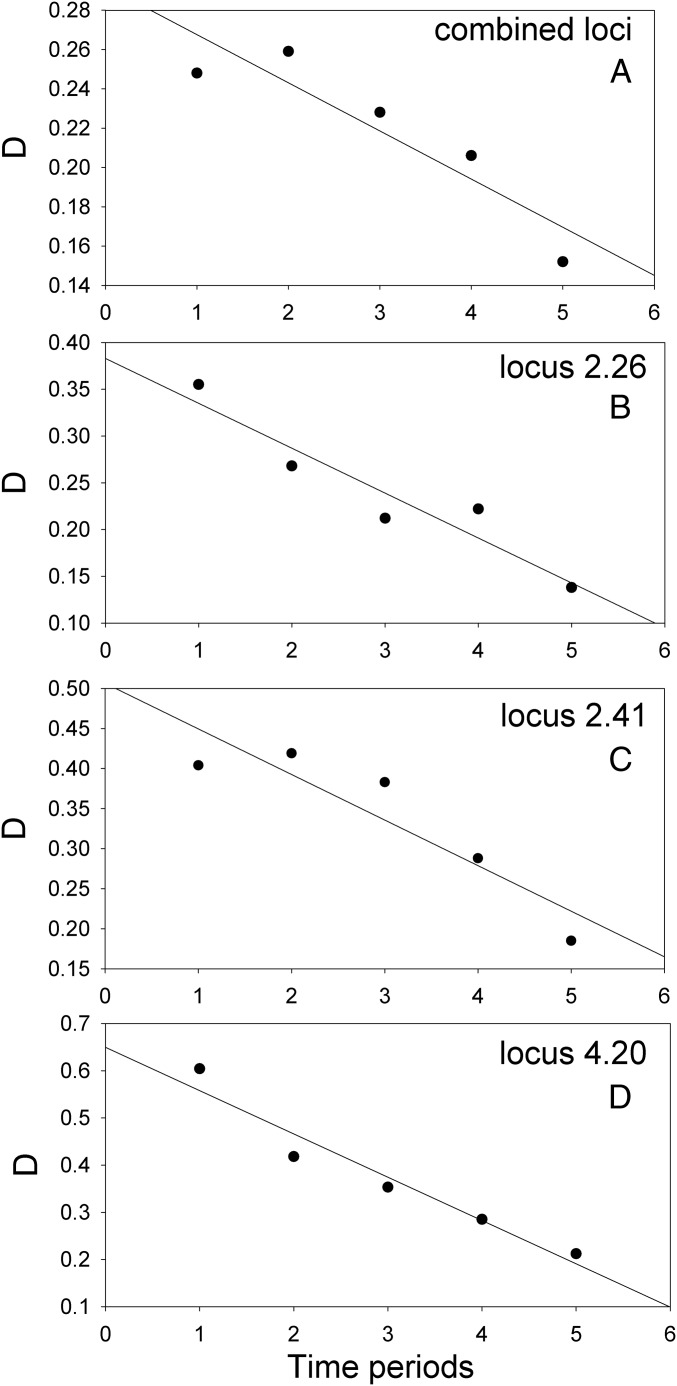
As a result of an exchange of alleles, G. scandens became progressively more similar genetically to G. fortis across five periods of breeding (Fig. 3). The pattern of genetic convergence has been reported before (56), but without statistical analysis. Fourteen of the 16 slopes for individual loci are negative, which is a significant departure from the null hypothesis expectation of eight (two-tailed binomial test, P = 0.004). Moreover, three of the slopes for individual loci are significant (Fig. 3).
Fig. 3.
Genetic convergence. (A) G. scandens became progressively more similar genetically to G. fortis across five periods (1975 to 1987, 1990 to 1993, 1997 to 1998, 2000 to 2002, 2005 to 2011) separated by years of little or no breeding. The regression relationship of annual mean DA values on time for the combined data are significant (F1,4 = 15.30, P = 0.0297, R2 adj = 0.78), and has a negative slope of −0.024 ± 0.006 (A). (B–D) Significant regression relationships at three of the individual loci: locus 2.26 (F = 27.92, P = 0.0132, b = −0.048 ± 0.009 SE, R2 adj = 0.87), locus 241 (F = 15.06, P = 0.0303, b = −0.057 ± 0.015, R2 adj = 0.78), and locus 420 (F = 47.13, P = 0.0063, b = −0.092 ± 0.013, R2 adj = 0.92).
We tested the null hypothesis that G. fortis and G. scandens contributed equally to genetic convergence. We did this by comparing the average Nei’s DA value, a measure of genetic distance, between the earliest G. scandens and latest G. fortis samples with the average DA value between the earliest G. fortis and latest G. scandens samples. They should be equal according to the null hypothesis. The power of the test for this type of comparison with small samples is low; nevertheless, the difference is close to being significant: paired t15 = 1.95, P = 0.07 with arc sign-transformed data. Late G. scandens are more similar to early G. fortis (average DA = 0.185) than late G. fortis are to early G. scandens (average DA = 0.241). Thus, G. scandens appear to have converged genetically more than G. fortis, consistent with the inference from the higher frequency of SF than FS, and consistent with the morphological trends (51). Furthermore, G. scandens gained 16 alleles present in the earliest G. fortis sample but not in the earliest G. scandens sample, whereas G. fortis gained only three alleles from G. scandens. However, the difference is partly explained by the fact that G. fortis had more alleles (n = 190) to donate than G. scandens (n = 159).
In contrast to these results, the genetic distance between G. fortis and G. fuliginosa remained unchanged across the five periods (F = 1.40, P = 0.3224), consistent with constant frequencies of Ff hybrids (as detailed earlier). We interpret the constancy as a steady-state gene flow, with input of G. fuliginosa alleles into G. fortis through repeated immigration and hybridization roughly balanced by output as the backcrosses to G. fortis across generations become increasingly indistinguishable from G. fortis itself. Input exceeds output in G. fortis and G. scandens. The net result is a predominant flow of genes from G. fuliginosa and G. fortis to G. scandens, with some reverse flow to G. fortis, as depicted in Fig. 1. Unequal exchange of genes has also been reported for two species of tree finches on Floreana island (Camarhynchus spp.), predominantly from a rare to a common species (64).
Phenotypic Consequences of Hybridization.
Morphological traits are highly heritable in G. fortis and G. scandens (65, 66). Therefore hybrids, both dihybrids and trihybrids, are predicted to have intermediate means and increased phenotypic variances in the absence of selection (17). We used principal components analysis (PCA; SI Appendix, section 4) to test these predictions.
Mean measurements match expectations. The hybrid groups are larger in size than smaller species (G. fuliginosa and G. fortis) and smaller than the larger species (G. scandens; Table 3). Beak shape means are similarly intermediate. For each of these traits, 14 comparisons were made between a group of hybrids and a species that contributed to them. Most differences in body size and beak shape are significant by Tukey’s HSD tests (Table 3).
Table 3.
Comparison of morphological means from PCA between species and hybrids by Tukey HSD tests, contrasting size and shape of species and hybrids
| Species | Hybrid | Body size | Beak size | Beak shape | |||
| P | Largest | P | Largest | P | Pointed | ||
| fuliginosa | Ff | 0.0001 | Hybrid | 0.0001 | Hybrid | 0.0001 | Species |
| fortis | Ff | 0.2723 | — | 0.0001 | Species | 0.0028 | Hybrid |
| fuliginosa | Sf | 0.0001 | Hybrid | 0.0001 | Hybrid | 0.4205 | — |
| fortis | FS | 0.9629 | — | 1.0000 | — | 0.0001 | Hybrid |
| fortis | SF | 0.0001 | Hybrid | 0.9995 | — | 0.0001 | Hybrid |
| scandens | FS | 0.0001 | Species | 0.0619 | — | 0.0001 | Species |
| scandens | SF | 0.0001 | Species | 0.0681 | — | 0.0001 | Species |
| scandens | Sf | 0.0002 | Species | 0.8883 | — | 0.0001 | Species |
| fortis | FfS+fFS | 0.0040 | Hybrid | 1.0000 | — | 0.0001 | Hybrid |
| fortis | FSf+fSF | 0.1922 | — | 0.9961 | — | 0.0003 | Hybrid |
| fortis | SfF+SfF | 0.0165 | Hybrid | 0.9998 | — | 0.0001 | Hybrid |
| scandens | FfS+fFS | 0.0001 | Species | 0.8100 | — | 0.0001 | Species |
| scandens | FSf+fSF | 0.0001 | Species | 0.1342 | — | 0.0001 | Species |
| scandens | SfF+SfF | 0.0001 | Species | 0.9138 | — | 0.0001 | Species |
The expectation of larger variances of the hybrid groups is realized in all 42 comparisons, and 35 of them are significant by one-tailed F tests (Table 4). Dihybrids differ from the species in variances of beak traits more than do the trihybrids, although not in variances of body size. Average ratios of variances for the dihybrids are 2.00 (body size), 3.24 (beak size), and 3.86 (beak shape), and, for the trihybrids, they are 2.65, 3.04, and 3.09 in the same sequence. These are notably large values.
Table 4.
Ratios of variances of hybrids to species
| Hybrid | Species | df | Body size | Beak size | Beak shape |
| Ff | fuliginosa | 112,17 | 2.76* | 8.45**** | 2.84* |
| Ff | fortis | 112,733 | 2.63**** | 2.63**** | 1.38*** |
| fS | fuliginosa | 22,17 | 2.80* | 5.81**** | 12.81**** |
| FS | fortis | 31,733 | 1.75* | 1.06 | 2.63**** |
| SF | fortis | 58,733 | 2.10**** | 1.12 | 2.80**** |
| FS | scandens | 31,200 | 1.06 | 1.83*** | 2.26**** |
| SF | scandens | 58,200 | 1.28 | 1.94**** | 1.75**** |
| Sf | scandens | 23,200 | 1.62* | 3.04**** | 3.44**** |
| FfS + fFS | fortis | 16,733 | 1.74* | 1.47 | 4.12**** |
| FSf + fSF | fortis | 12,733 | 4.85**** | 3.60**** | 3.48**** |
| SFf + SfF | fortis | 14,733 | 3.31**** | 1.60 | 3.83**** |
| FfS + fFS | scandens | 16,200 | 1.06 | 2.54**** | 2.57**** |
| FSf + fSF | scandens | 12,200 | 2.94**** | 6.23**** | 2.17* |
| SFf + SfF | scandens | 14,200 | 2.01* | 2.78**** | 2.38*** |
Transgressive Segregation.
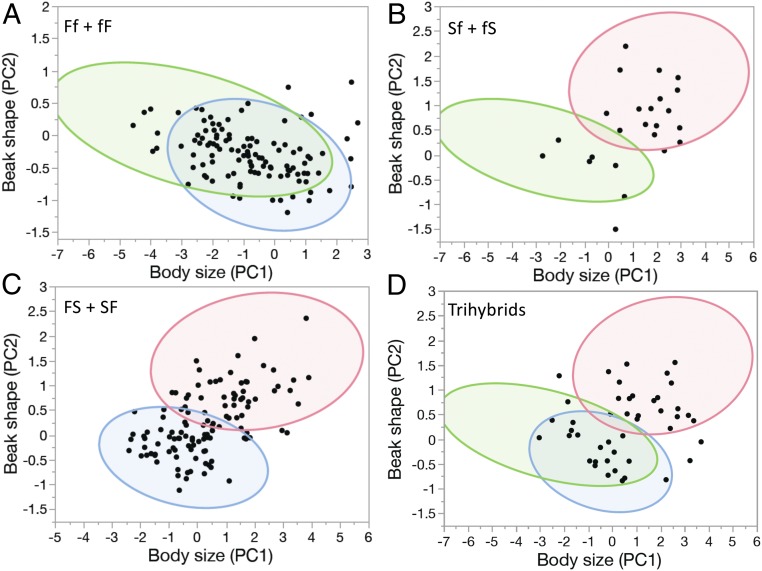
Some interbreeding populations of other species produce hybrids that are beyond the range of the combined phenotypic variation of the parental species, a phenomenon known as transgressive segregation and attributed to a variety of genetic and developmental causes (67–71). To detect transgressives, we compared admixed individuals (hybrids) with 99% ellipses around the joint beak shape–body size means of each of the three species, G. fortis, G. fuliginosa, and G. scandens (Fig. 4). The boundaries are conservative because some individuals assigned to a species by the ≥0.9 probability criterion may in fact be advanced-generation backcrosses, and, additionally, the sample size of one of the species, G. fuliginosa, is small. We found a single fS individual beyond the range of both parental species (Fig. 4B). However, this example is anomalous and should be discounted because admixture does not reflect ancestry: although they were not genotyped, both parent individuals were G. fortis morphologically. A few other individuals lay within the range of parental group phenotypes but just outside the ellipses. Noteworthy are three trihybrids that are smaller or larger than one of the parental species but with the same beak shape as the species to which they are most genetically affiliated. Six large Ff dihybrids have unusually pointed beaks for birds of their body size. Collectively, these few individuals should be considered allometric transgressives as they are displaced from the principal axes of parental group covariation.
Fig. 4.
Beak shape and body size of hybrids (dots) in relation to 99% ellipses around the distributions of G. fuliginosa (green), G. fortis (blue), and G. scandens (pink). (A) G. fuliginosa × G. fortis dihybrids. (B) G. fuliginosa × G. scandens dihybrids. (C) G. fortis × G. scandens dihybrids. (D) Trihybrids: beak pointedness increases from origin to the top, and body size increases from left to right. The smallest trihybrid outlier is fFS, and the other two are FfS.
Fitness.
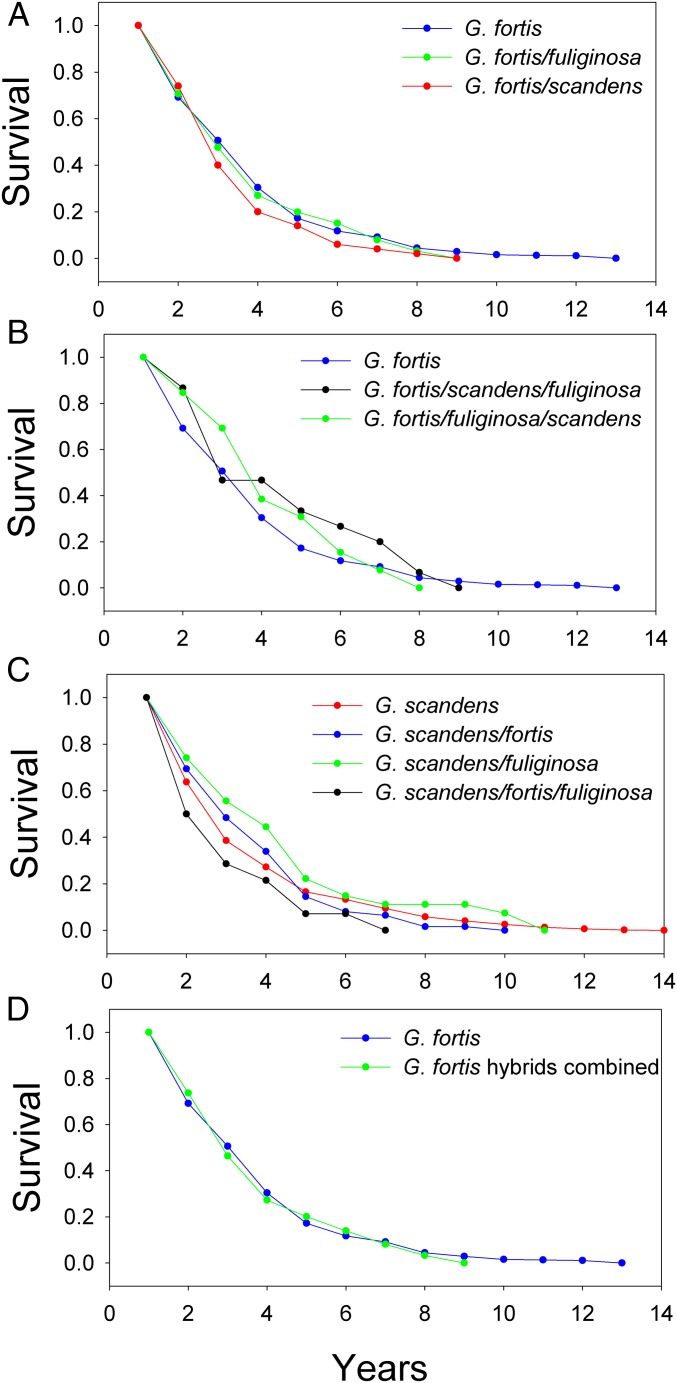
We measured fitness in terms of survival and, independently, the probability of becoming a recruit to the breeding population. Maximum longevities recorded on Daphne were 17 y for both G. fortis and G. scandens (56). Hybrid longevities are apparently shorter (Fig. 5), but this is likely to be an artifact of their smaller sample sizes. The oldest hybrids are not included in Fig. 5 because they hatched in years before genotyping began. They are approximately the same: 13 y (one Ff, one Sf), 12 y (one SF), and 11 y (one FS).
Fig. 5.
Survival of di- and trihybrids in relation to G. fortis and G. scandens from year 1 in the years 1987 to 2010. (A) Dihybrids Ff (green) and FS (red). (B) Trihybrids FSf + FSf (black) and FfS + fFS (green). (C) Di- and trihybrids SF (blue), Sf (green), and SFf + SfF (black). (D) G. fortis compared with dihybrids and trihybrids with predominant assignment to G. fortis (Ff, FS, FfS, fFS, FSf, fSF).
Overall, di- and trihybrids survived approximately as well as G. fortis and G. scandens over the first 6 to 10 y of life in the period 1987 to 2010 (Fig. 5). Ff hybrid survival was almost identical to G. fortis survival, FS survival was lower after year 2 by up to 10%, and SF survival was higher than G. scandens survival until year 5. Dihybrids with admixture of G. fuliginosa and G. scandens had the highest survival. Relative survival of the trihybrids is heterogeneous. The two G. fortis trihybrid groups, FfS and FSf, survived better than G. fortis, whereas SFf survival was inferior to that of G. scandens by about 10% at each age. Note each group of dihybrid and trihybrids deviate from their respective species in opposite directions and hence their effects tend to cancel each other, resulting in no, or reduced, net advantage or disadvantage of the combined hybrids relative to the species. This is illustrated with G. fortis and hybrids in Fig. 5. G. scandens combined with hybrids shows the same pattern (SI Appendix, section 5 and Fig. S2).
Recruitment success of the two cohorts with the most complete data did not differ between species and hybrids (Table 5). A possible exception is the low recruitment of FS in the 1991 cohort, but the numbers are small, without statistical evaluation, and, furthermore, this hybrid group of the 1987 cohort experienced the opposite, high success. Together, survival and recruitment of hybrids provide no indication of a fitness disadvantage.
Table 5.
Recruitment of hybrids and species from the two cohorts with most data
| Hybrid/species | Bred | Did not breed | Total | Proportion bred |
| 1987 | ||||
| G. fortis | 136 | 130 | 266 | 0.51 |
| Ff | 10 | 7 | 17 | 0.59 |
| FS | 5 | 2 | 7 | 0.71 |
| FfS | 5 | 3 | 8 | 0.62 |
| 1991 | ||||
| G. fortis | 204 | 428 | 632 | 0.32 |
| Ff | 16 | 24 | 40 | 0.40 |
| FS | 3 | 8 | 11 | 0.27 |
| FfS | 2 | 1 | 3 | 0.67 |
| G. scandens | 30 | 88 | 118 | 0.25 |
| SF | 2 | 2 | 4 | 0.50 |
Hybrid Fitness in Relation to Morphology.
Despite the high fitness of hybrids, those deviating most strongly in morphology from parental species may lie in a fitness valley between peaks occupied by the species. We tested the hypothesis with survival from year 1 posthatch to year 2 with the only samples sufficient for analysis from the period 2005 to 2012. The hypothesis was supported in the expected direction for the SF hybrids with PC data (n = 48, 0.62 survival) on body size (F = 6.85, P = 0.0119) and beak size (F = 10.13, P = 0.0026) but not beak shape (F = 0.02, P = 0.8936). The survival difference is not the consequence of males, the larger sex, surviving better than females, as it also occurs among males alone (n = 28, 0.75 survival) in body size (F = 11.19, P = 0.0025) and beak size (F = 15.34, P = 0.0006) but not beak shape (F = 1.72, P = 0.2006). There are no morphological differences between surviving and nonsurviving FS hybrids (n = 26, all P > 0.30, 0.50 survival) or Ff hybrids (n = 35, all P > 0.20, 0.54 survival).
Discussion
The main finding of the study is the role of G. fortis as a conduit species, which is demonstrated by the occurrence of two types of hybrids. The first group of hybrids are G. scandens × G. fuliginosa admixtures (dihybrids), since these two species are not known to interbreed on Daphne. G. scandens and G. fuliginosa differ strongly in the traits used in mate choice—beak size and shape, body size, and song (61, 62, 72)—whereas morphological difference between them are smaller on San Cristóbal island, where they do interbreed (personal observations, 1997 and 2018). The second group of hybrids are admixtures of three species (trihybrids).
We used neutral markers, microsatellites, to assign individuals to species or hybrids, using an arbitrary but often used and biologically justified criterion (36, 39, 43, 46) to separate them (SI Appendix, section 2). Assignments are subject to error (SI Appendix, section 2), and it is possible that the small number of loci results in an inflated number of erroneous assignments to hybrid classes. There are three reasons for believing this is not a source of strong bias. First, in a simulation study, most hybrids (0.85) were correctly identified (63). Second, each hybrid group has morphological features expected from its assigned genetic composition: intermediate means between the parental groups that are known or estimated to give rise to it, and increased variances. This would not be expected if many of the hybrids were not in fact hybrids. Third, there is no evidence of inflation in the samples from 1984 to 1998 when the pedigree allowed identification of first-generation backcrosses. Ten of 1,808 individuals with all four grandparents and both parents known have genomes with mixtures of G. fortis, G. scandens, and G. fuliginosa. Eight of the 1,808 individuals were assigned to trihybrids by microsatellites.
Existence of the two groups raises questions of genetics and fitness. How can a two-way or three-way mixture of genomes of different species function in individual organisms, what combinations of genes are compatible and which ones are not, how stable or transitory are their long-term dynamics, and what factors permit, promote, or hinder their existence? Answers to these questions can help to explain how barriers to interbreeding become strengthened in the process of speciation leading to complete reproductive isolation (1, 2, 15, 73, 74). Similar questions have been addressed in studies of interbreeding populations of Neanderthals, Denisovans, and ancestors of modern humans (75, 76), and the answers may portend future discoveries with finches and other organisms. Introgressive hybridization has been associated with a transfer of adaptive genes, for example those affecting skin color and immune function (75), deleterious genes affecting male fertility (76), and presumably an abundance of selectively neutral genes; in other words, a mixture, subject to repeated selective filtering.
To address these genetic questions, a preferable approach would be to use whole genomes. Nonetheless, microsatellites are sufficient to show that, in combination, dihybrids and trihybrids were at no disadvantage in terms of survival or recruitment when compared with the species that gave rise to them during a period of favorable food conditions following the change in ecology caused by the 1982 to 1983 El Niño event. High fitness is surprising, and indicates that selection against novel gene combinations in this group of finches must be weak at most. The only disadvantage we could detect was low survival of the smallest SF hybrids. This should slow the rate of convergence of G. scandens on G. fortis; however, we doubt if this happens because it is counteracted by relatively high survival of Sf hybrids (Fig. 5), which are also smaller in body size than G. scandens (Table 3).
We can identify two factors that help to explain how the genome of one species is apparently tolerant to invasion (77) by two others. The first is the phylogenetic youth of the group. G. fortis and G. fuliginosa are sister species according to a whole-genome phylogenetic analysis of autosomes (28). They shared a common ancestor ∼240,000 y ago, and they shared a common ancestor with G. scandens ∼260,000 y ago or earlier. These are minimum estimates because allele sharing makes them appear to be more similar and younger than would be revealed by their true history. Their youth means that relatively few genes have diverged in separate species; they mainly affect morphological traits related to feeding and breeding ecology, and they cause no incompatibility and have little or no transgressive effects upon phenotypes. The second factor is the genetic similarity of G. fuliginosa and G. scandens, as indicated by their microsatellites. Although G. fortis and G. fuliginosa are the most similar pair of species (Nei’s D = 0.28), G. scandens is more similar to G. fuliginosa (D = 0.46) than it is to G. fortis (D = 0.76). G. scandens is also more similar to G. fuliginosa in beak shape, although not in body size (Fig. 3D).
Triad hybridization, with one species acting as bridge or conduit between two noninterbreeding species, is valuable for what it reveals about the potential for the generation of novel phenotypes. Introgressive hybridization increases the potential by increasing additive genetic variances, altering genetic covariances, and constructing new combinations of interacting genes (12, 15). As expected, we found increased morphological variances in both dihybrids and trihybrids. A previous study provided evidence of relaxed genetic covariances in dihybrids (17), as has also been found in fish (21, 70). Multivariate effects should be greater through hybridization of three species than of two species. We found no clear case of extreme phenotypes beyond the range of variation of both of the contributing parental groups (transgressive segregation); however, three trihybrids displayed allometric transgression, lying outside the region of body size variation but not outside the range of beak shape variation of the parental groups. Some dihybrids displayed similar patterns. The evolutionary potential of allometric transgression has been demonstrated with the origin of a new lineage on Daphne Major. The lineage was initiated by hybridization of an immigrant Geospiza conirostris with a resident G. fortis, resulting in the formation of a reproductively isolated population that displayed allometric transgression, that is, a deviation from the two species in the relationship between beak size and body size (27, 78). Transgressive morphology is implicated in reproductive isolation because mate choice is based in part on beak and body size (61, 62, 72) and the relationship between the two (79).
Triad hybridization is likely to be more common than is currently recognized by the few cases that have been documented. For example, we would expect it in species-rich adaptive radiations that have diversified relatively recently. Foremost among them are the hundreds of cichlid fish species in the Great Lakes of Africa, for which genome data are becoming rapidly available (78–84), and the many species of Heliconius butterflies that participate in mimicry rings (3, 26, 43, 85). Other possible groups are numerous species complexes that are similar morphologically and genetically and sometimes difficult to resolve taxonomically, such as Cottoid fish in Lake Baikal (86), Anastrepha flies (87, 88), ant-nest beetles (89), some groups of mosquitoes (44) and, among plants, Andean lupins (90) and the Hawaiian Silversword Alliance (91).
Since triad hybridization occurs in contemporary time, it must have occurred in the past. Together with hybridization of one species with two others separated in time or in space (a pair of dyads) (92, 93), triad hybridization may be responsible for polytomies in phylogenetic trees. Polytomies occur when branching cannot be resolved into bifurcations because an ancestral species and two descendant species are genetically so similar (94–97). The power to measure divergence is likely to be reduced when the samples are affected by triad hybridization.
Hybridization on Daphne is relevant to a global outlook on the future. The frequency and success of hybridization on the island increased from 1983 onward as a result of a change in vegetation caused by abundant rain associated with an intense and prolonged El Niño event. This major perturbation is a natural analog of an anticipated unnatural, human-caused change in the global environment. Whether or not populations have sufficient genetic variation for adaptive responses is the subject of ongoing critical debate (98–101). One way that variation is enhanced is through interspecific gene exchange (19, 102). Hybridization is believed to be increasing in frequency as a result of anthropogenically caused change to climate and to habitat that reduces population sizes and brings together previously separated species (102–106). Hybridizing species may therefore be disproportionately successful in coping with a changing environment in the future, as in the past (9, 18, 107). Fur seals on Macquarie Island (39) provide a prime example of what might come. The original population became extinct as a result of overexploitation in the 19th century. The island has been recolonized by two interbreeding species, Arctocephalus gazella and Arctocephalus tropicalis. A ménage à trois was created by a single male of a third species, Arctocephalus forsteri, and it led to the production of offspring with genes from all three species. It remains to be seen in this case and more generally if multispecies hybridization has significant consequences in terms of evolution and conservation in a world of increasing habitat destruction and climate change.
Methods
Sampling Design.
Field methods have been extensively described in previous publications (56, 57, 63, 65). Birds were captured in mist nets every year from 1973 to 2012. All birds were measured as adults (63), defined as birds in the year following the year of hatching, except in 1983 and 1987, when some birds that hatched that year bred and so are included as adults. The proportion of adults captured, measured, and banded uniquely was ∼90% in 1979 and ∼98% in 1981 (108). By 1992, all breeding adults had been banded. A large number of nests were followed in the years 1976 to 1978, and, from 1979 to 1998, attempts were made to find all nests on the island and identify their owners (banded or not banded). In later years, the year of hatching was estimated by plumage of birds in the hand (56). Nestlings were banded at day 8. Blood samples were taken from birds captured in mist nets or from nestlings for microsatellite analyses. Sixteen unlinked polymorphic loci were used for parentage and hybrid assignment with the program STRUCTURE, as described previously (63). The Princeton University Animal Care Committee approved the research procedures.
Statistical Analyses.
Statistical analyses were performed in JMP (SAS Institute). All tests were two-tailed unless indicated otherwise.
Data Availability.
Supplementary Information.
Supplementary Material
Acknowledgments
We thank the Charles Darwin Research Station for logistical support, the many assistants who helped in the fieldwork, Erik Enbody for reading the manuscript, and two reviewers for their valuable comments. The research was supported by grants from the Natural Sciences and Engineering Research Council of Canada and the National Science Foundation, and was carried out with permission of the Galápagos National Parks Directorate.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000388117/-/DCSupplemental.
References
- 1.Abbott R., et al. , Hybridization and speciation. J. Evol. Biol. 26, 229–246 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Taylor S. A., Larson E. L., Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat. Ecol. Evol. 3, 170–177 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Edelman N. B., et al. , Genomic architecture and introgression shape a butterfly radiation. Science 366, 594–599 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mateos M., Vrijenhoek R. C., Ancient versus reticulate origin of a hemiclonal lineage. Evolution 56, 985–992 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Bogart J. P., Bi K., Fu J., Noble D. W., Niedzwiecki J., Unisexual salamanders (genus Ambystoma) present a new reproductive mode for eukaryotes. Genome 50, 119–136 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Ghiselli F., Milani L., Scali V., Passamonti M., The Leptynia hispanica species complex (Insecta Phasmida): Polyploidy, parthenogenesis, hybridization and more. Mol. Ecol. 16, 4256–4268 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Montelongo T., Gómez-Zurita J., Nonrandom patterns of genetic admixture expose the complex historical hybrid origin of unisexual leaf beetle species in the genus Calligrapha. Am. Nat. 185, 113–134 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Taylor E. B., et al. , Speciation in reverse: Morphological and genetic evidence of the collapse of a three-spined stickleback (Gasterosteus aculeatus) species pair. Mol. Ecol. 15, 343–355 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Seehausen O., Takimoto G., Roy D., Jokela J., Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol. Ecol. 17, 30–44 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Vonlanthen P., et al. , Eutrophication causes speciation reversal in whitefish adaptive radiations. Nature 482, 357–362 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Kleindorfer S., et al. , Species collapse via hybridization in Darwin’s tree finches. Am. Nat. 183, 325–341 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Stelkens R., Seehausen O., Genetic distance between species predicts novel trait expression in their hybrids. Evolution 63, 884–897 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Pereira R. J., Barreto F. S., Burton R. S., Ecological novelty by hybridization: Experimental evidence for increased thermal tolerance by transgressive segregation in Tigriopus californicus. Evolution 68, 204–215 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Brennan A. C., Hiscock S. J., Abbott R. J., Completing the hybridization triangle: The inheritance of genetic incompatibilities during homoploid hybrid speciation in ragworts (Senecio). AoB Plants 11, ply078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marques D. A., Meier J. I., Seehausen O., A combinatorial view on speciation and adaptive radiation. Trends Ecol. Evol. 34, 531–544 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Lewontin R. C., Birch L. C., Hybridization as a source of variation or adaptation to new environments. Evolution 20, 315–336 (1966). [DOI] [PubMed] [Google Scholar]
- 17.Grant P. R., Grant B. R., Phenotypic and genetic effects of hybridization in Darwin’s finches. Evolution 48, 297–316 (1994). [DOI] [PubMed] [Google Scholar]
- 18.Seehausen O., Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Hedrick P. W., Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 22, 4606–4618 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Arnold M. L., Kunte K., Adaptive genetic exchange: A tangled history of admixture and evolutionary innovation. Trends Ecol. Evol. 32, 601–611 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Selz O. M., Lucek K., Young K. A., Seehausen O., Relaxed trait covariance in interspecific cichlid hybrids predicts morphological diversity in adaptive radiations. J. Evol. Biol. 27, 11–24 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Arnold M. L., Bulger M. R., Burke J. M., Hempel A. L., Williams J. H., Natural hybridization: How long can you go and still be important? Ecology 80, 371–381 (1999). [Google Scholar]
- 23.Lopez G. A., Potts B. M., Tilyard P. A., F1 hybrid inviability in eucalyptus: The case of E. ovata × E. globulus. Heredity 85, 242–250 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Grant P. R., Grant B. R., Keller L. F., Markert J. A., Petren K., Inbreeding and interbreeding in Darwin’s finches. Evolution 57, 2911–2916 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Rieseberg L. H., Sinervo B., Linder C. R., Ungerer M. C., Arias D. M., Role of gene interactions in hybrid speciation: Evidence from ancient and experimental hybrids. Science 272, 741–745 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Jiggins C. D., Salazar C., Linares M., Mavarez J., Review. Hybrid trait speciation and Heliconius butterflies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 3047–3054 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamichhaney S., et al. , Rapid hybrid speciation in Darwin’s finches. Science 359, 224–228 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Lamichhaney S., et al. , Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518, 371–375 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Gopalakrishnan S., et al. , Interspecific gene flow shaped the evolution of the genus Canis. Curr. Biol. 28, 3441–3449.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barton N. H., Hewitt G. H., Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 16, 113–148 (1985). [Google Scholar]
- 31.Taylor S. A., Larson E. L., Harrison R. G., Hybrid zones: Windows on climate change. Trends Ecol. Evol. 30, 398–406 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crow K. D., et al. , Maintenance of species boundaries despite rampant hybridization between three species of reef fishes (Hexagrammidae): Implications for the role of selection. Biol. J. Linn. Soc. Lond. 91, 135–147 (2007). [Google Scholar]
- 33.McDonald D. B., Parchman T. L., Bower M. R., Hubert W. A., Rahel F. J., An introduced and a native vertebrate hybridize to form a genetic bridge to a second native species. Proc. Natl. Acad. Sci. U.S.A. 105, 10837–10842 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keck B. P., Near T. J., Geographic and temporal aspects of mitochondrial replacement in Nothonotus darters (Teleostei: Percidae: Etheostomatinae). Evolution 64, 1410–1428 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Drillon O., Dufresnes G., Perrin N., Crochet P.-A., Dufresnes C., Reaching the edge of the speciation continuum: Hybridization between three sympatric species of Hyla tree frogs. Biol. J. Linn. Soc. Lond. 126, 743–750 (2019). [Google Scholar]
- 36.Haines M. L., et al. , Geographic variation in hybridization and ecological differentiation between three syntopic, morphologically similar species of montane lizards. Mol. Ecol. 25, 2887–2903 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Toews D. P. L., Streby H. M., Burket L., Taylor S. A., A wood-warbler produced through both interspecific and intergeneric hybridization. Biol. Lett. 14, 20180557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweizer M., et al. , Parallel plumage colour evolution and introgressive hybridization in wheatears. J. Evol. Biol. 32, 100–110 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Lancaster M. L., Gemmell N. J., Negro S., Goldsworthy S., Sunnucks P., Ménage à trois on Macquarie island: Hybridization among three species of Fur seal (Arctocephalus spp.) following historical population extinction. Mol. Ecol. 15, 3681–3692 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Alves P. C., Melo-Ferreira J., Freitas H., Boursot P., The ubiquitous mountain hare mitochondria: Multiple introgressive hybridization in hares, genus Lepus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 2831–2839 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson P. J., Rutledge L. Y., Wheeldon T. J., Patterson B. R., White B. N., Y-chromosome evidence supports widespread signatures of three-species Canis hybridization in eastern North America. Ecol. Evol. 2, 2325–2332 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson M. S., Murray J., Clarke B., The ecological genetics and adaptive radiation of Partula on Moorea. Oxf. Surv. Evol. Biol. 9, 167–238 (1993). [Google Scholar]
- 43.Kronforst M. R., Young L. G., Blume L. M., Gilbert L. E., Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies. Evolution 60, 1254–1268 (2006). [PubMed] [Google Scholar]
- 44.Fontaine M. C., et al. , Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347, 1258524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hersch E. I., Roy B. A., Context-dependent pollinator behavior: An explanation for patterns of hybridization among three species of Indian paintbrush. Evolution 61, 111–124 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Fogelqvist J., et al. , Genetic and morphological evidence for introgression between three species of willows. BMC Evol. Biol. 15, 193 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leebens-Mack J., Milligan B., Pollination biology in hybridizing Baptisia (Fabaceae) populations. Am. J. Bot. 85, 500–507 (1998). [PubMed] [Google Scholar]
- 48.Broyles S. B., Hybrid bridges to gene flow: A case study in milkweeds (Asclepias). Evolution 56, 1943–1953 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Dowling T. E., Secor C. L., The role of hybridization and introgression in the diversification of animals. Annu. Rev. Ecol. Syst. 28, 593–619 (1997). [Google Scholar]
- 50.Comeault A. A., Matute D. R., Genetic divergence and the number of hybridizing species affect the path to homoploid hybrid speciation. Proc. Natl. Acad. Sci. U.S.A. 115, 9761–9766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grant P. R., Grant B. R., Hybridization increases population variation during adaptive radiation. Proc. Natl. Acad. Sci. U.S.A. 116, 23216–23224 (2019b). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinsen G. D., Whitham T. G., Turek R. J., Keim P., Hybrid populations selectively filter gene introgression between species. Evolution 55, 1325–1335 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Parchman T. L., et al. , The genomic consequences of adaptive divergence and reproductive isolation between species of manakins. Mol. Ecol. 22, 3304–3317 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Kingston S. E., Parchman T. L., Gompert Z., Buerkle C. A., Braun M. J., Heterogeneity and concordance in locus-specific differentiation and introgression between species of towhees. J. Evol. Biol. 30, 474–485 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Stryjewski K. F., Sorenson M. D., Mosaic genome evolution in a recent and rapid avian radiation. Nat. Ecol. Evol. 1, 1912–1922 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Grant P. R., Grant B. R., 40 Years of Evolution. Darwin’s Finches on Daphne Major Island (Princeton University Press, Princeton, 2014). [Google Scholar]
- 57.Boag P. T., Grant P. R., Darwin’s finches (Geospiza) on Isla Daphne Major, Galápagos: Breeding and feeding ecology in a climatically variable environment. Ecol. Monogr. 54, 463–489 (1984). [Google Scholar]
- 58.Grant P. R., Price T. D., Population variation in continuously varying traits as an ecological genetics problem. Am. Zool. 21, 795–811 (1981). [Google Scholar]
- 59.Petren K., Grant B. R., Grant P. R., Low extrapair paternity in the cactus finch (Geospiza scandens). Auk 116, 252–256 (1999). [Google Scholar]
- 60.Grant B. R., Grant P. R., Evolution of Darwin’s Finches caused by a rare climatic event. Proc. Biol. Sci. 251, 111–117 (1993). [Google Scholar]
- 61.Grant B. R., Grant P. R., Cultural inheritance of song and its role in the evolution of Darwin’s finches. Evolution 50, 2471–2487 (1996). [DOI] [PubMed] [Google Scholar]
- 62.Grant P. R., Grant B. R., Adult sex ratio influences mate choice in Darwin’s finches. Proc. Natl. Acad. Sci. U.S.A. 116, 12373–12382 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grant P. R., Grant B. R., Conspecific versus heterospecific gene exchange between populations of Darwin’s finches. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 1065–1076 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peters K. J., Myers S. A., Dudaniec R. Y., O’Connor J. A., Kleindorfer S., Females drive asymmetrical introgression from rare to common species in Darwin’s tree finches. J. Evol. Biol. 30, 1940–1952 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Grant P. R., Grant B. R., “Quantitative genetic variation in populations of Darwin’s finches” in Adaptive Genetic Variation in the Wild, Mousseau T. A., Sinervo B., Endler J., Eds. (Oxford Univ. Press, Oxford, 2000), pp. 3–41. [Google Scholar]
- 66.Keller L. F., Grant P. R., Grant B. R., Petren K., Heritability of morphological traits in Darwin’s finches: Misidentified paternity and maternal effects. Heredity 87, 325–336 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Rieseberg L. H., Archer M. A., Wayne R. K., Transgressive segregation, adaptation and speciation. Heredity 83, 363–372 (1999). [DOI] [PubMed] [Google Scholar]
- 68.Rieseberg L. H., Widmer A., Arntz A. M., Burke J. M., The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 1141–1147 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Albertson R. C., Kocher T. D., Genetic architecture sets limits on transgressive segregation in hybrid cichlid fishes. Evolution 59, 686–690 (2005). [PubMed] [Google Scholar]
- 70.Parsons K. J., Son Y. H., Albertson R. C., Hybridization promotes evolvability in African cichlids: Connections between transgressive segregation and phenotypic integration. Evol. Biol. 38, 306–315 (2011). [Google Scholar]
- 71.Kagawa K., Takimoto G., Hybridization can promote adaptive radiation by means of transgressive segregation. Ecol. Lett. 21, 264–274 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Grant P. R., Grant B. R., Role of sexual imprinting in assortative mating and premating isolation in Darwin’s finches. Proc. Natl. Acad. Sci. U.S.A. 115, E10879–E10887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Butlin R., et al. ; Marie Curie SPECIATION Network , What do we need to know about speciation? Trends Ecol. Evol. 27, 27–39 (2012). [DOI] [PubMed] [Google Scholar]
- 74.Bell G., Every inch a finch: A commentary on grant (1993). Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20140287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pääbo S., The diverse origins of the human gene pool. Nat. Rev. Genet. 16, 313–314 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Sankararaman S., Mallick S., Patterson N., Reich D., The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr. Biol. 26, 1241–1247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mallet J., Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Grant P. R., Grant B. R., The secondary contact phase of allopatric speciation in Darwin’s finches. Proc. Natl. Acad. Sci. U.S.A. 106, 20141–20148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ratcliffe L. M., Grant P. R., Species recognition in Darwin’s finches (Geospiza, Gould). I. Discrimination by morphological cues. Anim. Behav. 31, 1139–1153 (1983). [Google Scholar]
- 80.Meier J. I., et al. , Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8, 14363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Irisarri I., et al. , Phylogenomics uncovers early hybridization and adaptive loci shaping the radiation of Lake Tanganyika cichlid fishes. Nat. Commun. 9, 3159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Malinsky M., et al. , Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat. Ecol. Evol. 2, 1940–1955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Salzburger W., Understanding explosive diversification through cichlid fish genomics. Nat. Rev. Genet. 19, 705–717 (2018). [DOI] [PubMed] [Google Scholar]
- 84.Meier J. I., et al. , The coincidence of ecological opportunity with hybridization explains rapid adaptive radiation in Lake Mweru cichlid fishes. Nat. Commun. 10, 5391 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Enciso-Romero J., et al. , Evolution of novel mimicry rings facilitated by adaptive introgression in tropical butterflies. Mol. Ecol. 26, 5160–5172 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Kontula T., Kirilchik S. V., Väinölä R., Endemic diversification of the monophyletic cottoid fish species flock in Lake Baikal explored with mtDNA sequencing. Mol. Phylogenet. Evol. 27, 143–155 (2003). [DOI] [PubMed] [Google Scholar]
- 87.Segura D. F., Assortative mating among Anastrepha fraterculus (Diptera: Tephritidae) hybrids as a possible route to radiation of the fraterculus cryptic species complex. Biol. J. Linn. Soc. Lond. 102, 346–354 (2011). [Google Scholar]
- 88.Díaz F., et al. , Evidence for introgression among three species of the Anastrepha fraterculus group, a radiating species complex of fruit flies. Front. Genet. 9, 359 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moore W., Robertson J. A., Explosive adaptive radiation and extreme phenotypic diversity within ant-nest beetles. Curr. Biol. 24, 2435–2439 (2014). [DOI] [PubMed] [Google Scholar]
- 90.Hughes C., Eastwood R., Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proc. Natl. Acad. Sci. U.S.A. 103, 10334–10339 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baldwin B. G., Sanderson M. J., Age and rate of diversification of the Hawaiian silversword alliance (Compositae). Proc. Natl. Acad. Sci. U.S.A. 95, 9402–9406 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.vonHoldt B. M., et al. , A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids. Genome Res. 21, 1294–1305 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Seneviratne S. S., Davidson P., Martin K., Irwin D. I., Low levels of hybridization across two contact zones among three species of woodpeckers (Sphyrapicus sapsuckers). J. Avian Biol. 47, 887–898 (2016). [Google Scholar]
- 94.Kliman R. M., et al. , The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156, 1913–1931 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takahashi K., Terai Y., Nishida M., Okada N., Phylogenetic relationships and ancient incomplete lineage sorting among cichlid fishes in Lake Tanganyika as revealed by analysis of the insertion of retroposons. Mol. Biol. Evol. 18, 2057–2066 (2001). [DOI] [PubMed] [Google Scholar]
- 96.Rokas A., Carroll S. B., Bushes in the tree of life. PLoS Biol. 4, e352 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ottenburghs J., van Hooft P., van Wieren S. E., Ydenburgh R. C., Prins H. H. T., Birds in a bush: Toward an avian phylogenetic network. Auk 133, 577–582 (2016). [Google Scholar]
- 98.Bitter M. C. L., Kapsenberg L., Gattuso J.-P., Pfister C. A., Standing genetic variation fuels rapid adaptation to ocean acidification. Nat. Commun. 10, 5821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Catullo R. A., Llewelyn J., Phillips B. L., Moritz C. C., The potential for rapid evolution under anthropogenic climate change. Curr. Biol. 29, R996–R1007 (2019). [DOI] [PubMed] [Google Scholar]
- 100.Razgour O., et al. , Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. Proc. Natl. Acad. Sci. U.S.A. 116, 10418–10423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Radchuk V., et al. , Adaptive responses of animals to climate change are most likely insufficient. Nat. Commun. 10, 3109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chunco A. J., Hybridization in a warmer world. Ecol. Evol. 4, 2019–2031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Anderson E., Stebbins G. L., Hybridization as an evolutionary stimulus. Evolution 8, 378–388 (1954). [Google Scholar]
- 104.Hubbs C. L., Hybridization between fish species in nature. Syst. Zool. 4, 1–20 (1955). [Google Scholar]
- 105.Canestrelli D., et al. , Climate change promotes hybridisation between deeply divergent species. PeerJ. 5, e3072 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sánchez-Guillén R. A., Muñoz J., Rodríguez-Tapia G., Feria Arroyo T. P., Córdoba-Aguilar A., Climate-induced range shifts and possible hybridisation consequences in insects. PLoS One 8, e80531 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ivory S. J., et al. , Environmental change explains cichlid adaptive radiation at Lake Malawi over the past 1.2 million years. Proc. Natl. Acad. Sci. U.S.A. 113, 11895–11900 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Price T. D., Sexual selection on body size, territory and plumage variables in a population of Darwin’s finches. Evolution 38, 327–341 (1984). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary Information.