The phytohormone auxin regulates plant growth and development. Proper spatiotemporal distribution of active auxin (indole-3-acetic acid [IAA]), via formation of auxin gradients, ensures precise organ morphogenesis and physiology. Because plants are sessile organisms, they are exquisitely attuned to fluctuating environmental conditions, such as alterations in ambient temperature and light conditions. Response to environmental changes often incurs modulation of auxin gradients to coordinate appropriate growth responses (such as hypocotyl elongation in response to shade). Auxin’s role as a key hormonal regulator to translate environmental stimuli into an adaptive growth response requires optimal auxin levels and response throughout the plant. Thus, auxin is tightly regulated through multiple processes including metabolism (biosynthesis, conjugation, degradation), transport, and signaling. Despite significant progress toward understanding the mechanism and role of auxin biosynthesis in growth and environmental responses, unanswered questions regarding regulation of auxin levels by distinct inputs remain. In PNAS, Chen et al. (1) uncover an additional layer of complexity to the seemingly simple indole-3-pyruvic acid (IPyA) pathway of auxin biosynthesis.
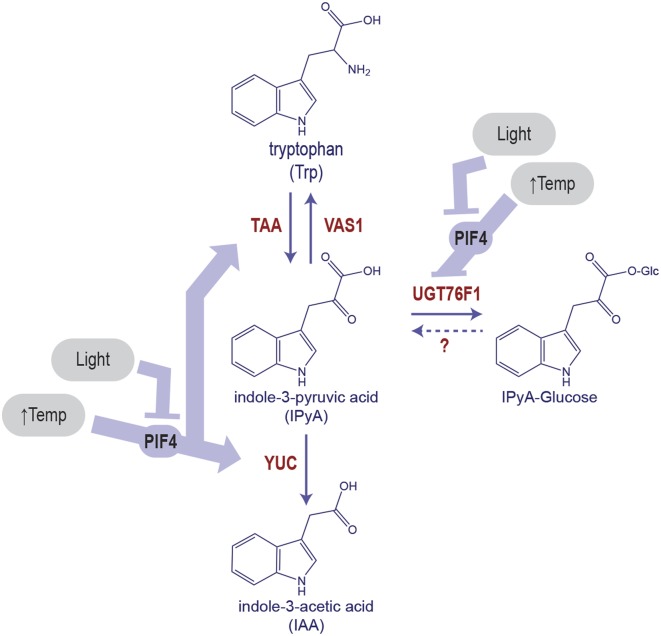
Although several parallel IAA biosynthesis pathways have been suggested (2–4), the IPyA pathway has emerged as the predominant source of auxin in the plant. This pathway consists of two steps: tryptophan (Trp)-to-IPyA conversion via the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA) family of Trp aminotransferases, followed by IPyA-to-IAA conversion via the YUCCA (YUC) family of flavin monooxygenases (Fig. 1) (5, 6). In Arabidopsis, the importance of IPyA pathway contributions is highlighted by significant reductions in endogenous levels of free IAA coupled with severe developmental defects found in mutants disrupted in this pathway (Table 1) (7, 8). Transgenic plants overexpressing TAA1 show little effect on free IAA levels, whereas plants overexpressing individual YUC genes hyperaccumulate IAA, suggesting the YUC family of flavin monooxygenases catalyzes the rate-limiting step in the IPyA pathway for IAA biosynthesis (6, 9, 10).
Fig. 1.
Fine-tuning the IPyA pathway in Arabidopsis. The two-step IPyA pathway is the predominant auxin biosynthesis pathway in Arabidopsis. Conversion of tryptophan to IAA (auxin) consists of two steps: TAA conversion of Trp to IPyA, followed by YUC conversion of IPyA to IAA. Fine-tuned IAA production in response to environmental stimuli, as seen in response to elevated temperature or light, is critical for developmental response to these stimuli. In the IPyA pathway for IAA biosynthesis, these environmental stimuli trigger PIF4-mediated transcriptional regulation of enzymes involved in modification or biosynthesis of IAA.
Table 1.
Arabidopsis components of IPyA pathway and their effect on free IAA
| Family | Role in pathway | Mutant | Effect on free IAA | Effect on IPyA |
| Synthesis | ||||
| TAA | TRP-to-IPyA conversion | wei8;tar2 | Decreased (7, 34) | Decreased (10) |
| YUC | IPyA-to-IAA conversion | yuc1;yuc2;yuc4;yuc6 | Unknown | Increased (10) |
| IPyA-to-IAA conversion | yuc1;yuc2;yuc6 | Unknown | Increased (6) | |
| IPyA-to-IAA conversion | yucQ | Decreased (8) | Unknown | |
| IPyA-to-IAA conversion | YUC6ox* | Increased (10) | Decreased (10) | |
| IPyA-to-IAA conversion | yuc1D (GOF) | No change (10) | Unknown | |
| IPyA-to-IAA conversion | YUC6ox; TAA1ox | Increased (10) | Unknown | |
| IPyA-to-IAA conversion | TAA1ox | No change (10) | Increased (10) | |
| IPyA-to-IAA conversion | TAA1ox; yuc1D | Increased (10) | Unknown | |
| VAS1 | IPyA-to-l-Trp conversion | vas1 | Increased (17) | Increased (17) |
| IPyA-to-l-Trp conversion | VAS1ox | Decreased (17) | Unknown | |
| IPyA-to-l-Trp conversion | vas1;sav3 | Increased (17) | Increased (17) | |
| Conjugation | ||||
| UGT76F1 | IPyA-to-IPyA–Glc conjugation | ugt76f1 | Increased (1) | Unknown |
| IPyA-to-IPyA–Glc conjugation | UGT76F1ox† | Reduced (1) | Unknown | |
Overexpression of any of the 11 YUC genes increases free IAA content.
Increased IPyA–Glc in UGT76F1ox and WT after yucasin treatment.
Because auxin gradients are critical to regulating plant growth and responses to environmental stimuli, it is not surprising that spatiotemporal expression of TAA and YUC genes are tightly regulated (reviewed in refs. 3 and 11). Furthermore, elevated temperature (12, 13) or shade (14–16) induces genes encoding IPyA pathway members through the PHYTOCHROME INTERACTING FACTOR (PIF) family of transcription factors. Thus, transcriptional regulation of pathway members plays key roles in plant development and environmental responses.
In addition to transcriptional regulation of TAA and YUC genes, recent studies suggest modulation of the amount of IPyA precursor available for conversion to IAA can regulate auxin homeostasis. In response to shade, the pyridoxal-phosphate–dependent aminotransferase REVERSAL OF SAV3 PHENOTYPE1 (VAS1) catalyzes the conversion of IPyA back to Trp to dampen shade-induced auxin biosynthesis to prevent plants from overcompensating (Fig. 1 and Table 1) (17). In PNAS, Chen et al. present compelling genetic, biochemical, and metabolic evidence for an additional layer of regulation of IPyA precursor via UGT76F1-catalyzed glucosylation of IPyA (1). Furthermore, this study shows UGT76F1-mediated glucosylation of IPyA is transcriptionally negatively regulated by PIF4 to alter temperature- and light-dependent hypocotyl growth (Fig. 1 and Table 1). Thus, IAA produced via the IPyA pathway is regulated through IPyA reversion to Trp and IPyA storage as a glucosylated form, further fine-tuning cellular auxin levels (18–20).
UGT76F1 serves a similar purpose as VAS1 by limiting the pool of available IPyA for IAA biosynthesis, thus raising the question as to why this primary auxin precursor is being so meticulously regulated. IPyA is known to be a highly unstable molecule capable of nonenzymatic breakdown into bioactive IAA (10, 21, 22). Given its instability, accumulation of IPyA poses a potential risk to the fine-tuned auxin homeostasis. Furthermore, IPyA is a chemically reactive molecule with potent hydroxyl radical scavenging properties (23, 24). In metazoans, IPyA oxidation (such as oxidative decarboxylation) can produce multiple reactive indole-derived by-products. Due to the instability and reactivity of IPyA, it is not unreasonable to postulate plants have evolved mechanisms mediated by VAS1 and UGT76F1 to prevent elevated IPyA levels not only to regulate auxin homeostasis but also to prevent damage from IPyA oxidation.
Of note in PNAS, Chen et al. show ugt76f1 mutants hyperaccumulate IAA, presumably due to increased availability of IPyA for conversion to IAA. The current dogma is that YUC-mediated conversion of IPyA to IAA is the rate-limiting step in the IPyA pathway. The elevated IAA levels found in ugt76f1 challenge this dogma by suggesting YUC is not a fully rate-limiting step in this instance (1). Follow-up genetic studies will lend insight into the links between UGT76F1- and VAS1-mediated IPyA modulation in response to environmental stimuli. Furthermore, studies aimed at elucidating whether IPyA-glucose can be hydrolyzed will be useful in determining whether UGT76F1 acts to store or catabolize IPyA.
Modified auxin forms have been identified, many of which involve conjugation of IAA to amino acids or sugars; however, little is known about integration of these modified auxins into the overall auxin biosynthesis and inactivation pathways (18–20, 25, 26). Some studies suggest the various conjugated forms of auxin are used by the plant as a method of temporal storage or detoxification when auxin is in excess, proposing the conjugated moiety itself may dictate the destiny of the bound auxin for transport, storage, or catabolism. In Arabidopsis, IAA-Asp and IAA-Glu amide conjugates are believed to function in IAA inactivation, whereas IAA-Ala and IAA-Leu are believed to function as inactive storage forms capable of being hydrolyzed into free IAA (26–29). IAA-Glc has also been observed in many plants, and likely contributes to active IAA formation (30–33), but there is not strong evidence for this in Arabidopsis. Interestingly, glucosylated forms of auxin precursors outside of the IPyA pathway, such as indole-3-butyric acid (IBA)-Glc, have also been observed (31).
Identification of two mechanisms (VAS1 and UGT76F1) to biochemically regulate IPyA contributions to auxin levels suggests that the two-step IPyA pathway for IAA biosynthesis is more complex than previously recognized. IPyA regulation appears to act as a hub between environmental stimuli and growth response. The auxin field is only just beginning to understand how plants regulate auxin levels and response on a cellular basis. In PNAS, Chen et al. (1) highlight the complexity of this regulation and provide compelling evidence that PIF4 acts as a major signaling hub between light and temperature signaling and auxin response through modulation of IPyA pools in auxin biosynthesis.
Acknowledgments
This research is supported by the National Institutes of Health (R01 GM112898), the National Science Foundation (NSF) (IOS-1453750), and the NSF Center for Engineering Mechanobiology (CMMI-1548571).
Footnotes
The authors declare no competing interest.
See companion article, “IPyA glucosylation mediates light and temperature signaling to regulate auxin-dependent hypocotyl elongation in Arabidopsis,” 10.1073/pnas.2000172117.
References
- 1.Chen L., et al. , IPyA glucosylation mediates light and temperature signaling to regulate auxin-dependent hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 117, 6910–6917 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casanova-Sáez R., Voß U., Auxin metabolism controls developmental decisions in land plants. Trends Plant Sci. 24, 741–754 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Kasahara H., Current aspects of auxin biosynthesis in plants. Biosci. Biotechnol. Biochem. 80, 34–42 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Ljung K., Auxin metabolism and homeostasis during plant development. Development 140, 943–950 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Stepanova A. N., et al. , The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23, 3961–3973 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Won C., et al. , Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18518–18523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stepanova A. N., et al. , TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Chen Q., et al. , Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55, 1072–1079 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y., et al. , A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Mashiguchi K., et al. , The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brumos J., Alonso J. M., Stepanova A. N., Genetic aspects of auxin biosynthesis and its regulation. Physiol. Plant. 151, 3–12 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Franklin K. A., et al. , Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. U.S.A. 108, 20231–20235 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun J., Qi L., Li Y., Chu J., Li C., PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 8, e1002594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y., et al. , Central clock components modulate plant shade avoidance by directly repressing transcriptional activation activity of PIF proteins. Proc. Natl. Acad. Sci. U.S.A. 117, 3261–3269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller-Moulé P., et al. , YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance. PeerJ 4, e2574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L., et al. , Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Z., et al. , Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat. Chem. Biol. 9, 244–246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig-Müller J., Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 62, 1757–1773 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Bajguz A., Piotrowska A., Conjugates of auxin and cytokinin. Phytochemistry 70, 957–969 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Korasick D. A., Enders T. A., Strader L. C., Auxin biosynthesis and storage forms. J. Exp. Bot. 64, 2541–2555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tam Y. Y., Normanly J., Determination of indole-3-pyruvic acid levels in Arabidopsis thaliana by gas chromatography-selected ion monitoring-mass spectrometry. J. Chromatogr. A 800, 101–108 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Suzuki M., et al. , Transcriptional feedback regulation of YUCCA genes in response to auxin levels in Arabidopsis. Plant Cell Rep. 34, 1343–1352 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Poeggeler B., et al. , Indole-3-propionate: A potent hydroxyl radical scavenger in rat brain. Brain Res. 815, 382–388 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Chowdhury G., et al. , Structural identification of diindole agonists of the aryl hydrocarbon receptor derived from degradation of indole-3-pyruvic acid. Chem. Res. Toxicol. 22, 1905–1912 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rampey R. A., et al. , A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135, 978–988 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam Y. Y., Epstein E., Normanly J., Characterization of auxin conjugates in Arabidopsis. Low steady-state levels of indole-3-acetyl-aspartate, indole-3-acetyl-glutamate, and indole-3-acetyl-glucose. Plant Physiol. 123, 589–596 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barlier I., et al. , The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. U.S.A. 97, 14819–14824 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowalczyk M., Sandberg G., Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol. 127, 1845–1853 (2001). [PMC free article] [PubMed] [Google Scholar]
- 29.Östin A., Kowalyczk M., Bhalerao R. P., Sandberg G., Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol. 118, 285–296 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campanella J. J., Olajide A. F., Magnus V., Ludwig-Müller J., A novel auxin conjugate hydrolase from wheat with substrate specificity for longer side-chain auxin amide conjugates. Plant Physiol. 135, 2230–2240 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tognetti V. B., et al. , Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22, 2660–2679 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szerszen J. B., Szczyglowski K., Bandurski R. S., iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 265, 1699–1701 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Ishimaru K., et al. , Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45, 707–711 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Tao Y., et al. , Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]