Significance
Spinocerebellar ataxia type 3 (SCA3) is a neurodegenerative disease with no effective treatments. SCA3 is etiologically linked to an abnormal polyglutamine (polyQ) tract at the C terminus of Ataxin-3 (ATXN3). How this polyQ stretch causes SCA3 pathology remains elusive. Here we provide evidence that wild-type ATXN3 plays an important role in error-free repair of DNA double-strand breaks in the transcribed genes. In contrast, mutant ATXN3 blocks the activity of a DNA end-processing enzyme, polynucleotide kinase 3′-phosphatase (PNKP), leading to progressive accumulation of double-strand breaks and abrogation of global transcription. Since PNKP overexpression in Drosophila rescued the SCA3 phenotype, this promising therapeutic avenue for SCA3 is worth exploring.
Keywords: ATXN3, DNA double-strand break repair, PNKP, RNA-templated TC-NHEJ, spinocerebellar ataxia type-3
Abstract
Spinocerebellar ataxia type 3 (SCA3) is a dominantly inherited neurodegenerative disease caused by CAG (encoding glutamine) repeat expansion in the Ataxin-3 (ATXN3) gene. We have shown previously that ATXN3-depleted or pathogenic ATXN3-expressing cells abrogate polynucleotide kinase 3′-phosphatase (PNKP) activity. Here, we report that ATXN3 associates with RNA polymerase II (RNAP II) and the classical nonhomologous end-joining (C-NHEJ) proteins, including PNKP, along with nascent RNAs under physiological conditions. Notably, ATXN3 depletion significantly decreased global transcription, repair of transcribed genes, and error-free double-strand break repair of a 3′-phosphate–containing terminally gapped, linearized reporter plasmid. The missing sequence at the terminal break site was restored in the recircularized plasmid in control cells by using the endogenous homologous transcript as a template, indicating ATXN3′s role in PNKP-mediated error-free C-NHEJ. Furthermore, brain extracts from SCA3 patients and mice show significantly lower PNKP activity, elevated p53BP1 level, more abundant strand-breaks in the transcribed genes, and degradation of RNAP II relative to controls. A similar RNAP II degradation is also evident in mutant ATXN3-expressing Drosophila larval brains and eyes. Importantly, SCA3 phenotype in Drosophila was completely amenable to PNKP complementation. Hence, salvaging PNKP’s activity can be a promising therapeutic strategy for SCA3.
Spinocerebellar ataxia type 3 (SCA3), also called Machado-Joseph disease, is an autosomal dominant neurodegenerative disorder characterized by progressive ataxia, spasticity, ocular movement abnormalities, and short life expectancy (1–5). The neurodegeneration is most prominent in the brainstem, spinal cord, and cerebellum (3). SCA3 is the most common inherited ataxia worldwide (6), and to date no curative therapy, except symptomatic treatments, are available (5, 7). The genetic basis of SCA3 is attributed to unstable CAG (encoding glutamine/Q) repeat expansion from 12 to 41 (in healthy normal) to 62 to 84 (diseased state) in the C-terminal region of the Ataxin-3 (ATXN3) gene (2, 3, 8), resulting in an abnormal polyQ tract in the ATXN3 polypeptide. However, the pathophysiologic mechanism of SCA3 still remains elusive (5, 9–11).
ATXN3 is a multifunctional protein with pleiotropic effect in a wide range of cellular activities. A number of studies (5, 12–17) reported ATXN3’s biological function as a deubiquitinase (DUB). ATXN3 contains multiple ubiquitin-interacting motifs and it binds to both canonical K48- and nonproteolytic K63-linked ubiquitin chains, indicating its diverse roles in ubiquitin signaling (14, 16–18). ATXN3 knockout mice showed an increase in total ubiquitinated protein levels (19). Consistently, knockdown of ATXN3 resulted in significant increase in the level of endogenous ubiquitinated proteins, whereas ectopic expression of WT ATXN3 decreased protein ubiquitination in HEK293 cells (14). ATXN3 also interacts with several E3 ubiquitin ligases, such as Parkin (15, 20), the neuroprotective cochaperone CHIP (21, 22), and Gp78 (23) that regulate protein quality control. Recently, several other proteins, such as α-5 integrin subunit (24, 25), p53 (26), MDC1 (27), and autophagy-initiator beclin 1 (28) have also been identified as substrates of ATXN3′s DUB activity. Moreover, the DUB activity of ATXN3 was found to be instrumental in maintaining a steady-state level of Chk1, promoting DNA repair and checkpoint signaling through the ATR-Chk1 pathway, and ATXN3 depletion led to a compromised DNA damage response and reduced cellular survival after replication stress (29). It has also been reported that pathological ATXN3 stimulates the generation of cellular reactive oxygen species that in turn induces DNA damage and activates the ataxia telangiectasia mutated (ATM) signaling pathway (30), indicating ATXN3’s role in DNA repair as well. We have recently shown that ATXN3 is a component of the polynucleotide kinase 3′-phosphatase (PNKP)-mediated single-strand break repair (SSBR) complex (31). PNKP is a major enzyme for processing both 3′ and 5′ termini at strand breaks in mammalian genomes (32, 33). It removes the 3′-phosphate (3′-P) group and catalyzes phosphorylation of the 5′-OH termini; thus, it is involved in both SSBR and double strand-break (DSB) repair. Several recent reports have documented the association of PNKP deficiency with neurological/developmental disorders (34–37). Our earlier study (31) demonstrated that PNKP’s 3′-phosphatase activity is potentiated by WT ATXN3, whereas the pathogenic form with polyQ expansion blocks PNKP’s activity, resulting in DNA single- and double-strand break accumulation and chronic activation of the DNA damage-response pathways (31, 38).
DSBs are repaired either via error-free homologous recombination (HR) (39, 40) or the nonhomologous end-joining (NHEJ) pathway (41–43), which is generally considered to be error-prone. The majority of neuronal cells in adult human brains are in the G0/G1 (nonproliferating) phase; thus, DSB repair cannot occur via the HR pathway and cells must utilize the NHEJ pathway instead (44). NHEJ is of two types: Classical or C-NHEJ, involving Ku, DNA-PK, 53BP1 (p53 binding protein 1), and DNA Lig IV-XRCC4; and alternative end-joining, or Alt-EJ, which involves PARP1, Lig IIIα/XRCC1, or Lig I. C-NHEJ is not only the major repair pathway in G0/G1 cells, but is the predominant repair pathway for the majority of DSBs, even in the G2 phase (42). Alt-EJ, inherently error-prone, is functionally relevant when the standard repair process fails, primarily as a backup for both C-NHEJ and HR (41, 45).
We recently reported that DSBs in the transcribed genome are repaired via the C-NHEJ pathway in postmitotic, noncycling cells like neurons, and showed that nascent homologous RNA provides the template for restoring the missing information, so this repair pathway is error-free (46). Furthermore, we have established the critical role of PNKP in such repair. Since ATXN3 interacts with PNKP and modulates its activity (31), we postulated that ATXN3 plays an important role in C-NHEJ–mediated DSB repair as well. This is supported by a recent study demonstrating ATXN3’s role in both HR- and NHEJ-mediated DSB repair (27). However, the mechanistic details of ATXN3’s role in C-NHEJ–mediated error-free repair of DSBs are still lacking. In the present study, we provide evidence that WT ATXN3 plays a critical role in error-free DSB repair of the transcribed genome via the C-NHEJ pathway, where the mammalian cells utilize nascent homologous transcripts as a template to restore the original genomic sequence at the break site. We also show that WT ATXN3 plays a role in maintaining RNA polymerase II (RNAP II) stability and consequently, the global transcription level. Furthermore, SCA3 mice and patients’ brain tissue showed decreased PNKP activity, leading to accumulation of DNA DSBs, particularly in the transcribed genome, and the RNAP II level is also significantly decreased in these tissues. A similar decrease in RNAP II level was evident in Drosophila eyes and larval brain motor neurons that express mutant ATXN3. All these data provide important mechanistic insight into SCA3 pathology. Finally, we demonstrate that PNKP complementation in a Drosophila SCA3 model significantly ameliorates mutant ATXN3-mediated neurotoxicity, opening up a promising avenue of therapeutic intervention for SCA3.
Results
ATXN3 Associates with RNAP II and Key C-NHEJ Proteins.
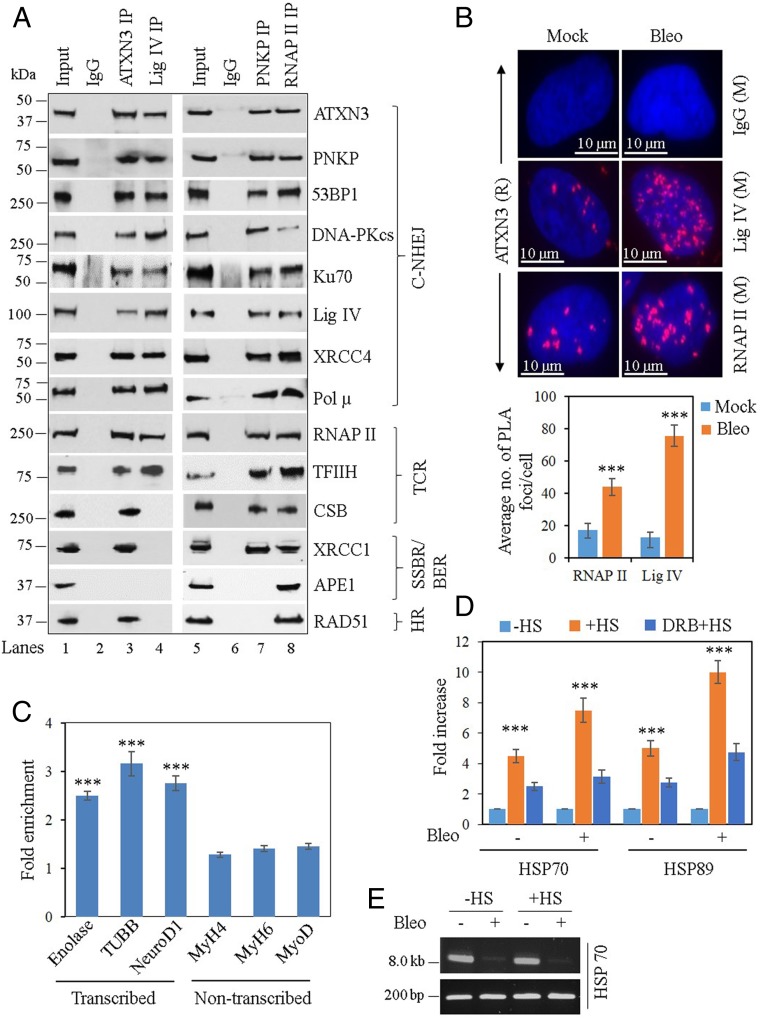
We demonstrated earlier that ATXN3 is a component of the PNKP-mediated DNA SSBR complex (31). Furthermore, we recently reported that PNKP is involved in preferential repair of DSBs in the transcribed genes via the C-NHEJ pathway, which we termed as transcription-coupled (TC) NHEJ (46). However, the involvement of ATXN3 in PNKP-mediated TC-NHEJ has not been characterized. To analyze ATXN3’s potential role in TC-NHEJ, we performed coimmunoprecipitation (co-IP) from nuclear extracts (NEs) of WT mouse cerebellum (an affected region of brain in SCA3) using an anti-ATXN3 antibody (Ab). It was found that ATXN3 does associate with the key C-NHEJ proteins (Fig. 1A, lane 3) (Ku70, DNA PKcs, 53BP1, PNKP, Pol µ, Lig IV, XRCC4) along with RNAP II. Importantly, the TC repair-related proteins TFIIH [subunit p89 (47)] and Cockayne syndrome B (48) were also present in the ATXN3 immunocomplex (IC). In reciprocal experiments, we analyzed the ICs of Lig IV, PNKP (two key C-NHEJ proteins), and RNAP II, and found ATXN3 along with C-NHEJ and TC repair proteins (Fig. 1A, lanes 4, 7, and 8, respectively). These data indicate a stable association of ATXN3 and TC-NHEJ components under physiological conditions. The absence of XRCC1 (an Alt-EJ/SSBR protein) (49) in the Lig IV IC (Fig. 1A, lane 4), APE1 [involved in OGG1/NTH1-mediated, PNKP-independent base excision repair (50, 51)] in the ATXN3/Lig IV/PNKP IC (Fig. 1A, lanes 3, 4, and 7, respectively) and RAD51 (a canonical HR protein) in Lig IV/PNKP IC (Fig. 1A, lanes 4 and 7, respectively) demonstrates the specificity of association. RAD51’s association with RNAP II (Fig. 1A, lane 8) is consistent with our earlier finding (46) and also supports the RNA-templated HR (52–54). ATXN3’s association with RAD51 (Fig. 1A, lane 3) is consistent with a recent report regarding ATXN3’s role in HR (27). We further found that ATXN3’s association with C-NHEJ proteins and RNAP II increased upon damage induction (SI Appendix, Fig. S1A) and ATXN3 associates with RNAP II and C-NHEJ proteins in a large multiprotein complex (SI Appendix, Fig. S1B; see also SI Appendix, SI Results). Supporting ATXN3’s role in TC-NHEJ, we found that siRNA-mediated depletion of ATXN3 in human induced pluripotent stem cells (iPSC)-derived neural stem cells (NSC) leads to widespread formation of γH2AX foci, a hallmark of DSB accumulation (SI Appendix, Fig. S2A). Moreover, ATXN3 depletion in postmitotic, noncycling SH-SY5Y cells (following differentiation by retinoic acid) leads to formation of p53BP1 foci (a C-NHEJ marker), whereas induction of HR marker RAD51 foci was insignificant in these cells (SI Appendix, Fig. S2B). This result suggests that C-NHEJ, although operative in all phases of the cell cycle, is the predominant DSB repair pathway in terminally differentiated neurons and ATXN3 plays a critical role therein.
Fig. 1.
Partial characterization of DSB repair complexes and ATXN3’s association with transcribed vs. nontranscribed genes. (A) Benzonase-treated NEs from WT mouse cerebellum were immunoprecipitated with anti-ATXN3 (lane 3), anti-Lig IV (lane 4), anti-PNKP (lane 7), anti-RNAP II (lane 8) Abs, or control IgG (mouse; lane 2 and rabbit; lane 6), and tested for the presence of associated proteins using specific Abs. One representative figure is shown (n = 3). BER, base excision repair; TCR, transcription-coupled repair. (B, Upper) Detection of ATXN3′s (anti-rabbit [R] Ab) association with Lig IV and RNAP II (both anti-mouse [M] Abs) in mock or Bleo-treated NSCs by proximity ligation assay (PLA). Nuclei were counterstained with DAPI (blue) and nonspecific Ab (IgG, mouse) was used as control. Scale bar: 10 µm. (Lower) Average number of proximity ligation assay foci per cell were calculated from 21 randomly selected cells in each sample. Error bars represent ± SD of the mean. The data were significant at ***P < 0.005 between mock and Bleo treatment (n = 3). (C) ChIP was performed from WT mouse cerebellum (n = 3) with anti-ATXN3 Ab and binding to the exonic regions of transcribed (Enolase, TUBB, and NeuroD1) vs. nontranscribed (MyH4, MyH6, and MyoD) genes was quantified (as fold-enrichment = percent input normalized over IgG) by qPCR from immunoprecipitated DNA. Error bars represent ± SD of the mean. ***P < 0.005, showing statistical significance between a particular transcribed gene and all of the nontranscribed genes. (D) SH-SY5Y cells were either mock-treated (−HS) or subjected to heat shock (+HS), or DRB treatment followed by HS (DRB + HS). The cells were further mock- (−) or Bleo- (+) treated and ChIP was performed with anti-ATXN3 Ab or control IgG. Binding to the HSP70 and HSP89 genes was quantified by qPCR from immunoprecipitated DNA. Percent input over IgG was calculated and represented as fold increase with −HS samples arbitrarily considered as unity. Error bars represent ± SD of the mean (n = 3). The data were significant at ***P < 0.005 between each set of −HS/+HS and +HS/DRB+HS. (E) LA-qPCR–mediated estimation of DNA damage in the HSP70 gene before (−HS) and after heat-shock (+HS). A short genomic fragment (∼200 bp) was amplified to normalize the LA-qPCR data.
To further confirm the in-cell association of ATXN3 with two key proteins, either involved in C-NHEJ (Lig IV) or transcription (RNAP II), we performed in situ proximity ligation assay in mock- or Bleo-treated iPSC-derived NSCs. Mock-treated cells had an average of 12 ± 3 and 17 ± 4 foci per cell, whereas Bleo-treated cells had 75 ± 6 and 43 ± 5 foci per cell for ATXN3–Lig IV and ATXN3–RNAP II interactions, respectively (Fig. 1B), indicating ∼6- and ∼2.5-fold increase in the association of ATXN3 with Lig IV and RNAP II, respectively, upon DSB induction. No fluorescence signals were detected when cells were stained with control IgG (Fig. 1B), indicating the specificity of the association. Collectively, these data suggest ATXN3’s role in the TC-NHEJ–mediated DSB repair of mammalian cells.
ATXN3 Preferentially Associates with the Transcribed Genes.
Since the C-NHEJ proteins preferentially associate with the transcribed genes (46), we examined the genomic association of ATXN3 by performing chromatin immunoprecipitation (ChIP) on soluble chromatin preparations of cerebellum from WT mice. Genomic DNA isolated after ChIP was subjected to qPCR using oligos for three neuron-specific transcribed genes [γ-enolase (Enolase) (55), tubulin β3 class III (TUBB) (56), and neuronal differentiation factor 1 (NeuroD1) (57)] and three nontranscribed genes (muscle specific myosin heavy chain 4 and 6 [MyH4, MyH6] and myogenic differentiation factor [MyoD]) (Fig. 1C). We confirmed cerebellum-specific expression of these genes by RT-PCR (SI Appendix, Fig. S2C). ATXN3 did indeed preferentially associate with the transcribed genes, providing in vivo evidence of ATXN3’s potential role in TC-NHEJ. This finding is also consistent with the data showing the physiological association of ATXN3 with RNAP II (Fig. 1A). However, how the proteins involved in the TC-NHEJ pathway specifically locate the DNA damage within the transcribed region is not clear. We have earlier shown that the recruitment of C-NHEJ proteins to DSB sites is transcription-dependent using a heat-shock–inducible system (46). Therefore, we tested the association of ATXN3 (in mock vs. Bleo-treated SH-SY5Y cells) with the heat-shock protein (HSP) genes HSP70 and HSP89 that are highly induced upon heat shock (+HS) (58). The association of ATXN3 with HSP genes was significantly increased (∼4- to 5-fold for mock-treated and ∼8- to 10-fold for Bleo-treated cells) only after heat-shock–induced transcription (Fig. 1D) (−HS vs. +HS), despite similar initial DSB levels after Bleo treatment (Fig. 1E). Notably, the transcription inhibitor DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazol) significantly reduced ATXN3’s association with these genes (Fig. 1D), further confirming the transcription-dependent recruitment of ATXN3 to DSB sites.
ATXN3 Is Involved in the Preferential Repair of Transcribed Genes.
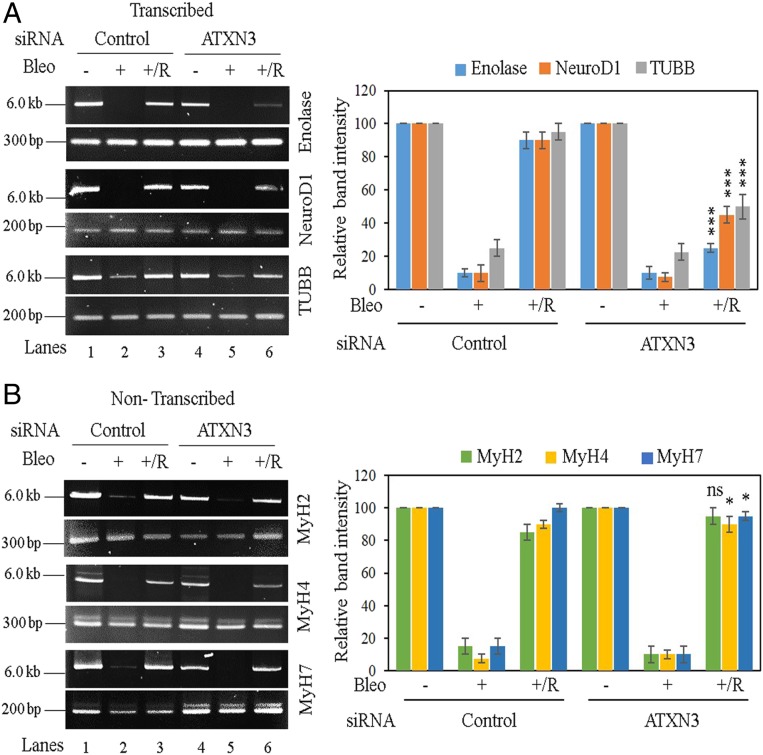
To investigate the role of ATXN3 in transcribed genome-specific repair, ATXN3-depleted SH-SY5Y cells (SI Appendix, Fig. S3A) were treated with Bleo to induce DSBs, and the strand-break accumulation was analyzed in transcribed (Enolase, NeuroD1, TUBB) vs. nontranscribed (MyH2, MyH4, and MyH7) genes (transcription profile confirmed by RT-PCR) (SI Appendix, Fig. S2D) using long-amplicon qPCR (LA-qPCR) (46, 59–62) after the cells were allowed to recover for 12 to 15 h after DNA damage induction. Despite comparable strand-break levels in both the transcribed and nontranscribed genes after Bleo treatment, depletion of ATXN3 significantly abrogated the recovery (R) of the transcribed (Fig. 2A, lane 6 vs. lane 4) but not the nontranscribed genes (Fig. 2B, lane 6 vs. lane 4). However, both transcribed and nontranscribed genes recovered equally well in control siRNA-transfected cells (Fig. 2, lane 3 vs. lane 1 in both panels). Interestingly, we found a significant decrease in the level of the largest subunit of RNAP II (RPB1) in ATXN3-depleted SH-SY5Y cells, while the PNKP level remained unaltered (SI Appendix, Fig. S3A).
Fig. 2.
Evaluation of genomic strand-break levels in transcribed vs. nontranscribed genes in ATXN3-depleted cells by LA-qPCR. SH-SY5Y cells were transfected with either control siRNA (lanes 1 to 3) or ATXN3 siRNA (lanes 4 to 6) and further mock- (−) or Bleo- (+) treated or kept for recovery (+/R) after Bleo treatment. (Left) Representative agarose gel images of each long (6 to 8 kb) and short (∼200 to 400 bp) amplicon of (A) the transcribed (Enolase, NeuroD1, and TUBB) genes and (B) the nontranscribed (MyH2, MyH4, MyH7) genes. (Right) The bar diagrams represent the normalized relative band intensity compared to the control siRNA/mock-treated sample (lane 1) arbitrarily set as 100. n ≥ 3 in each case. Error bars represent ± SD of the mean. The persistence of damage after recovery for each transcribed gene (A, lane 6) was significant (***P < 0.005) compared to corresponding mock-treated samples (A, lane 4); however, the damage was almost completely repaired in nontranscribed genes (B, lane 6 vs. lane 4; ns, nonsignificant [P > 0.05]; *P < 0.05).
We also performed similar studies in HEK293 cells and found that ATXN3 depletion (SI Appendix, Fig. S3B) caused impaired repair in the transcribed genes [HPRT and DNA polymerase β (POLB) (46)], while the nontranscribed genes [NANOG and OCT3/4 (46)] were repaired efficiently (SI Appendix, Fig. S3 C and D, respectively). These data indicate that ATXN3 plays an important role in preferential repair of DSBs in the transcribed genome.
ATXN3 Associates with the Nascent Transcripts.
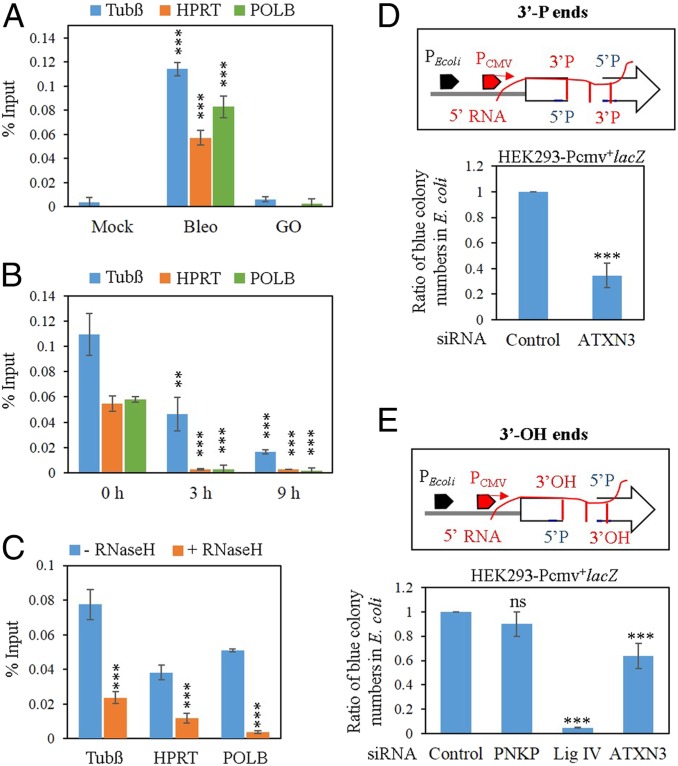
We demonstrated earlier that the nascent transcripts, associated with the C-NHEJ proteins, can be copied for error-free DSB repair of the transcribed genome (46). Our studies have shown that only PNKP and 53BP1, but neither the early (Ku70/DNA-PKcs) nor the late (Lig IV) proteins, recruit nascent RNA when the cells are treated with Bleo to induce DSBs. Since ATXN3 associates with the C-NHEJ proteins as well as RNAP II (Fig. 1A) and stably associates with PNKP (31), we investigated whether ATXN3 could also associate with nascent RNA (pre-mRNA) following DSB induction. Therefore, we performed RNA-ChIP using anti-ATXN3 Ab (Fig. 3A) from SH-SY5Y cells either mock-, Bleo-, or glucose oxidase- (GO, an SSB-inducing agent) treated and analyzed the immunoprecipitated RNA by qRT-PCR using intron-specific oligos for three randomly selected transcribed genes: Tubulin β (Tubß), HPRT, and POLB. We indeed observed an association of nascent RNA with ATXN3, but only in Bleo-treated cells, not in mock- or GO-treated cells. The association of ATXN3 with pre-mRNA was maximum immediately after DSB induction (recovery 0 h), started to decrease as early as 3 h into the recovery period, and became almost negligible at 9 h of recovery (Fig. 3B). This indicates that the pre-mRNA dissociates from the ATXN3 complex as the repair progresses and this is consistent with our previous report that shows almost complete DSB repair 9 to 15 h postdamage induction (46). In order to serve as a template, RNA should pair with DNA in an RNA–DNA hybrid, a pairing we confirmed in our previous study (46). Following a similar strategy, we treated SH-SY5Y cells with Bleo followed by incubation with RNase H, which specifically degrades RNA in an RNA–DNA hybrid, and then performed RNA-ChIP. We observed a significant decrease in the association of RNA with ATXN3 after RNase H treatment (Fig. 3C), suggesting the formation of an RNA–DNA hybrid at DSB sites. Control reactions were performed without reverse transcriptase (-RT) to rule out the possibility of any genomic DNA contamination during sample preparation (SI Appendix, Fig. S4 A–C). Thus, the above results clearly indicate the association of pre-mRNA with ATXN3.
Fig. 3.
Association of ATXN3 with nascent RNA. (A) SH-SY5Y cells were mock- or Bleo-, or GO-treated and their NEs subjected to RNA-ChIP using the anti-ATXN3 Ab. Real-time PCR was carried out for Tubβ, HPRT, and POLB genes using intron-specific primers. Data are presented as percent input; error bars show ± SD of the mean (n = 3, ***P < 0.005). (B) Time course analysis, where Bleo-treated SH-SY5Y cells were allowed to recover for the indicated times and NEs were subjected to RNA-ChIP using anti-ATXN3 Ab. Real-time PCR was performed using intron-specific primers and percent inputs were calculated using Ct values; error bars represent ± SD of the mean (n = 3) and the dissociation of pre-mRNA from the complex at 3 and 9 h was significant (***P < 0.005; **P < 0.01) compared to 0 h. (C) Bleo-treated cells were incubated with RNase H before RNA-ChIP using ATXN3 Ab to detect RNA-DNA hybrids and real-time qPCRs were done using intron-specific primers. Data are presented as percent input where error bars show ± SD of the mean (n = 3, ***P < 0.005). (D) Plasmid-based DSB repair (3′-P containing DSB ends) assay in control vs. ATXN3 depleted HEK293-Pcmv+lacZ stable cells. The number of E. coli blue colonies after transformation of the plasmids recovered from cells treated with control siRNA was arbitrarily set as 1. The data were the average of at least three independent experiments (n = 3), where ***P < 0.005. Error bars represent ± SD of the mean. (E) Similar plasmid-based assay with 3′-OH containing DSB ends, after Control or PNKP- or Lig IV- or ATXN3-specific siRNA-mediated depletion. The data are the average of at least three independent experiments, where ***P < 0.005; ns, nonsignificant (P > 0.05). D and E, Upper, show schematic representation of the plasmid DNA used for TC-NHEJ assay.
Plasmid-Based In-Cell DSB Repair Assay Shows ATXN3’s Role in RNA-Templated TC-NHEJ.
We described earlier the generation of an Escherichia coli lacZ- (under the control of both E. coli and viral promoters) expressing HEK293 stable cell line that provided the lacZ transcript as a template for repair of a transfected plasmid containing a DSB within the lacZ gene (46). This plasmid is replication-defective and contains DSB in such a way that a segment of DNA was deleted, resulting in inactivation of lacZ. We have shown earlier that this in-cell DSB repair takes place specifically via the C-NHEJ pathway since depletion of RAD51 (a canonical HR protein) had no effect, whereas depletion of C-NHEJ proteins significantly decreased the error-free repair (46). To examine the role of ATXN3 in such repair, ATXN3 was depleted (∼75%) in E. coli lacZ-expressing HEK293 cells using specific siRNA (SI Appendix, Fig. S4D). Additionally, we generated 3′-P or 3′-OH termini containing DSBs with deletion of several nucleotides in the plasmid, as shown schematically (Fig. 3 D and E, Upper) and then conducted the assay. Cells (+Pcmv; control vs. ATXN3 siRNA-transfected) were transfected with linearized plasmid with 3′-P ends and allowed to repair for 16 h. Plasmids were then recovered from transfected cells, introduced into rec– lacZ− E. coli, and blue colonies counted as a measure of error-free repair of lacZ gene. A significant decrease in E. coli blue colony number was observed (Fig. 3 D, Lower) in ATXN3-depleted cells (117 ± 54) compared to control siRNA-transfected cells (317 ± 75), confirming ATXN3’s role in DSB repair via the TC-NHEJ pathway. Since we found a decrease in RNAP II level concomitant with ATXN3 depletion (SI Appendix, Fig. S4D), we tested whether the repair deficiency of the reporter plasmid was due to the inadequate expression of the lacZ transcript itself. We performed lacZ-specific qRT-PCR from ATXN3-depleted cells vs. control and found no notable decrease in the lacZ transcript level following ATXN3 depletion (SI Appendix, Fig. S4E). We believe this result is due to constitutive expression of the lacZ gene under a strong viral pCMV promoter. This suggests that depletion of ATXN3 and not a transcriptional defect causes the low efficiency of repair of the reporter plasmid. To provide specificity of the assay, we monitored the repair of 3′-OH termini containing plasmid in PNKP- (SI Appendix, Fig. S4F) or Lig IV- (SI Appendix, Fig. S4G) or ATXN3-depleted Pcmv-lacZ–expressing cells. Lig IV (10 ± 1) or ATXN3 (92 ± 5) depletion significantly decreased the blue colony numbers (Fig. 3 E, Lower), compared to control (152 ± 6); however, PNKP depletion (139 ± 5) had no effect on error-free repair of 3′-OH–containing DSBs, as these plasmids did not require PNKP-mediated end processing for repair. Collectively, these data strongly suggest a role for ATXN3 in RNA-templated TC-NHEJ.
PNKP Activity Is Abrogated in SCA3 Mice and Patients’ Brain.
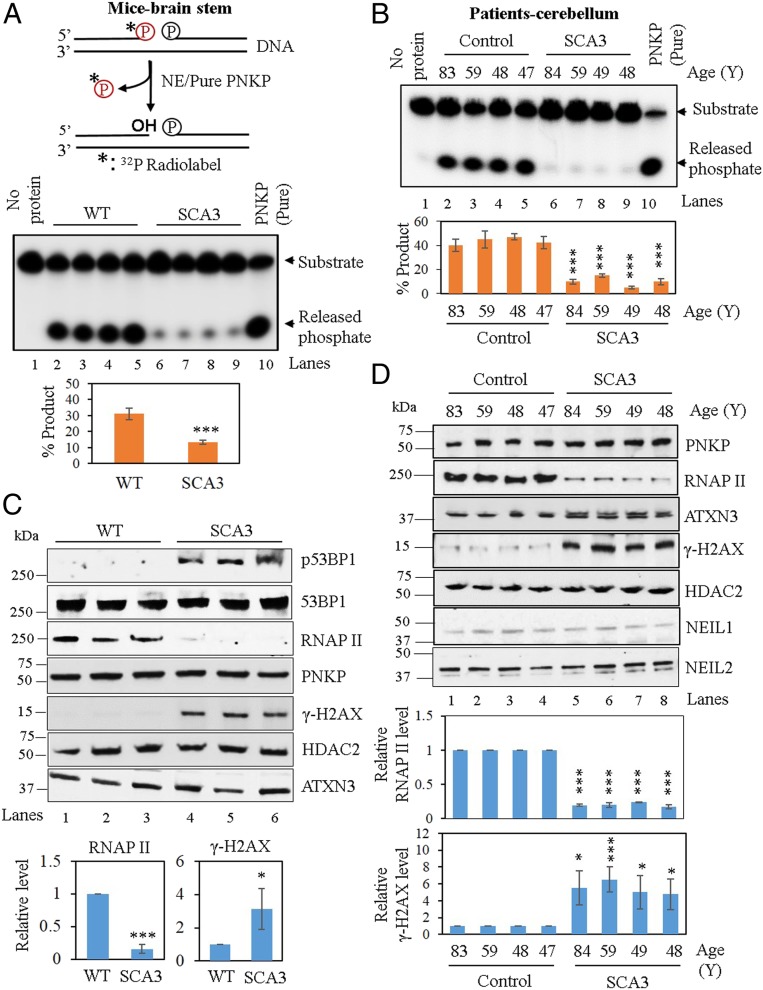
We have shown earlier that polyQ expanded ATXN3 blocks PNKP’s in vitro activity in deep cerebellar nuclei of SCA3 mice (31). We thus further assessed PNKP’s 3′-phosphatase activity (a schematic depiction of the assay is shown in Fig. 4 A, Upper) in the NEs prepared from the brainstem (one of the most affected regions in SCA3) of WT vs. SCA3 mice. We indeed found significant abrogation of PNKP’s activity (Fig. 4 A, Middle, lanes 6 to 9 vs. lanes 2 to 5, and Lower) whereas no significant change in the PNKP activity in the NEs of forebrain (unaffected region in SCA3) was observed (SI Appendix, Fig. S5 A, Upper, lanes 5 to 7 vs. 2 to 4, and Lower). Based on these findings, we postulated that mutant ATXN3 also abrogates PNKP activity in SCA3 patients. We thus assessed PNKP’s activity in the postmortem cerebellum tissues from four SCA3 patients and their age-matched controls (SI Appendix, Table S1). Activity of PNKP was indeed blocked significantly (∼90 to 95%) in the SCA3 patients’ cerebellum when compared to controls (Fig. 4 B, Upper , lanes 6 to 9 vs. lanes 2 to 5, and Lower). DNA repair is a multistep process. Therefore, we examined if mutant ATXN3 specifically blocks PNKP-mediated DNA end-processing (3′-phosphatase) activity or interferes with downstream DNA polymerase and DNA ligase activities. For this purpose, we used two nicked DNA duplexes: One with a 3′-OH end (requires DNA polymerase and ligase activities but not PNKP activity for repair) (SI Appendix, Fig. S6 A, Upper) and another with a 3′-phosphate end (requires PNKP-mediated 3′-P removal prior DNA polymerase and ligase activities for repair completion) (SI Appendix, Fig. S6 B, Upper). We observed that the NE from SCA3 patients’ cerebellum did not affect the repair of the duplex with the 3′ OH end (SI Appendix, Fig. S6 A, Middle, lanes 5 to 8 vs. lanes 1 to 4, and Lower). However, it did abrogate the repair of the duplex that requires PNKP-mediated end processing (SI Appendix, Fig. S6 B, Middle, lanes 5 to 8 vs. lanes 1 to 4, and Lower). This finding is consistent with our recent observation in another polyQ disease (Huntington’s disease), where we found similar specific inactivation of PNKP by the pathogenic form of huntingtin in both cell culture and an animal model (62).
Fig. 4.
PNKP activity in the WT vs. SCA3 mice and age-matched healthy normal vs. SCA3 patients’ brain. (A, Upper) A schematic representation of PNKP’s 3′ phosphatase assay. (Middle) 3′-phosphatase assay in the NEs of WT (lanes 2 to 5) vs. SCA3 mice (lanes 6 to 9) brainstem. Lane 1: No protein, substrate only. Lane 10: purified PNKP (25 fmol) as positive control. (Lower) Quantitation of the products (released phosphate) is represented in the bar diagram. Decrease in percent product was significant in SCA3 mice (n = 4, ***P < 0.005) compared to WT. Error bars represent ± SD of the mean. (B, Upper) 3′-phosphatase assay in the NE of age-matched control (lanes 2 to 5) vs. SCA3 patients’ (lanes 6 to 9) cerebellum as described above. Lane 1: No protein, substrate only. Lane 10: purified PNKP (25 fmol). (Lower) Quantitation of the products (released phosphate) is represented in the bar diagram. Decrease in percent product was significant in SCA3 patients (n = 3, ***P < 0.005) compared to age-matched healthy normal tissue. (C, Upper) Western blots showing the levels of proteins in the NE of WT vs. SCA3 mice brainstem. HDAC2: nuclear loading control. (Lower) Quantitation of RNAP II and γH2AX levels in the WT vs. SCA3 mice (n = 3). Error bars represent ± SD of the mean (*P < 0.05; ***P < 0.005). (D, Upper) Similar Western blots showing the levels of proteins in the NE of age-matched healthy normal vs. SCA3 patients’ cerebellum. (Lower) Quantitation of RNAP II and γH2AX levels in the SCA3 patients and the healthy controls from three independent gels. Error bars represent ± SD of the mean (*P < 0.05; ***P < 0.005).
We reported earlier that PNKP is also involved in Nei-like (NEIL) 1- and 2-mediated DNA base excision repair (50, 51). To provide further specificity of the mutant ATXN3-mediated abrogation of PNKP activity, we examined the activity of the NEILs in the NEs of WT vs. SCA3 mice and control vs. age-matched SCA3 patients and found no significant difference in the DNA glycosylase/AP lyase activity between the two groups (SI Appendix, Figs. S5B and S6C, respectively; Upper, lanes 6 to 9 vs. lanes 2 to 5, and Lower). Western analysis revealed no significant difference at the protein level of PNKP in WT vs. SCA3 mice (Fig. 4C) and in control vs. SCA3 patients (Fig. 4D). NEIL glycosylase (NEIL1 and NEIL2) levels were also similar in control vs. SCA3 patients (Fig. 4D). However, we found a dramatic decrease in the level of the largest subunit of RNAP II (80 to 90%) in all SCA3 mice (Fig. 4C, lanes 4 to 6 vs. lanes 1 to 3) and patients (Fig. 4D, lanes 5 to 8 vs. lanes 1 to 4). We also detected a significant increase in the γH2AX level in SCA3 mice (Fig. 4C, lanes 4 to 6 vs. lanes 1 to 3) and patients (Fig. 4D, lanes 5 to 8 vs. lanes 1 to 4, and SI Appendix, Fig. S6D, lanes 4 to 7 vs. lanes 1 to 3), indicating DSB accumulation. Notably, the brains of SCA3 mice and patients showed elevated level of p53BP1 (phospho-53BP1), whereas the total 53BP1 level remained unaltered (Fig. 4C and SI Appendix, Fig. S6D, respectively), indicating that the accumulation of DSB was due to impairment of the C-NHEJ pathway. It is thus apparent that mutant ATXN3-mediated specific abrogation of PNKP activity leads to DSB accumulation and consequent degradation of RNAP II, a plausible cause of SCA3 pathology.
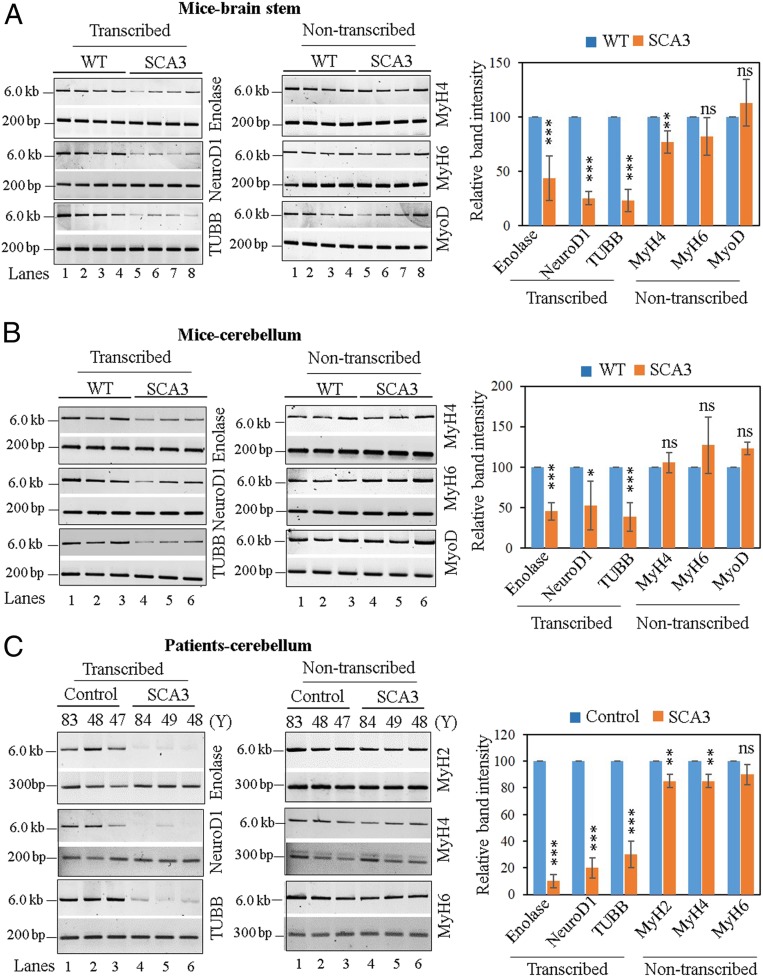
SCA3 Mice and Patients’ Brains Accumulate Higher Levels of DNA Strand-Breaks in the Transcribed Genes.
As PNKP activity is compromised in SCA3, we analyzed DNA damage by LA-qPCR in WT vs. SCA3 mouse brainstem and cerebella. We indeed detected higher levels of DNA strand-break accumulation in the transcribed (Enolase, NeuroD1, and TUBB) compared to the nontranscribed (MyH4, MyH6, and MyoD) genes (Fig. 5A, lanes 5 to 8 vs. lanes 1 to 4 and Fig. 5B, lanes 4 to 6 vs. lanes 1 to 3). As a control, we analyzed DNA damage accumulation in the transcribed genes (Enolase, NeuroD1, and TUBB) in WT vs. SCA3 mouse forebrain and we could not detect significant strand-break accumulation (SI Appendix, Fig. S5 C, Upper, lanes 4 to 6 vs. lanes 1 to 3, and Lower). This finding is consistent with our recent studies in the Huntington’s disease (HD) mouse model (62), where the HD mouse showed increased DNA strand breaks in the transcribed genes in the affected brain regions. Thereafter, we performed similar LA-qPCR analyses in the postmortem cerebella of SCA3 vs. control human subjects (three different age groups, as we could not recover the quality genomic DNA required to amplify the ∼6-kb region from one SCA3 patient sample). We found significantly higher strand-break accumulation in the actively transcribed genes than in the nontranscribed genes (Fig. 5C, lanes 4 to 6 vs. lanes 1 to 3 in each panel), consistent with our observation in cell culture and the SCA3 mouse model. These results also support our observation of significantly higher DNA strand-break accumulation in the transcribed genes in PNKP-depleted cells (46).
Fig. 5.
Evaluation of genomic strand-break levels in SCA3 mice and postmortem SCA3 patients’ brain. (Left) Representative agarose gel images of long (6 to 8 kb) and short (∼200 bp) amplicon of the corresponding transcribed (Enolase, NeuroD1, or TUBB) or nontranscribed (MyH2, MyH4, MyH6, or MyoD) genes from genomic DNA of (A) WT and SCA3 transgenic mice brainstem, (B) WT and SCA3 transgenic mice cerebellum, and (C) postmortem cerebellum of age-matched healthy normal (control) and SCA3 patients. (Right) The normalized relative band intensities are represented in the bar diagram with the WT/control sample arbitrarily set as 100 (n = 3, error bars represent ± SD of the mean). The damage accumulation for each transcribed gene in SCA3 mice/patients was significantly increased (***P < 0.005; *P < 0.05) compared to corresponding WT/control samples; however, there were far fewer DNA strand breaks in the nontranscribed genes (ns, nonsignificant [P > 0.05]; **P < 0.01).
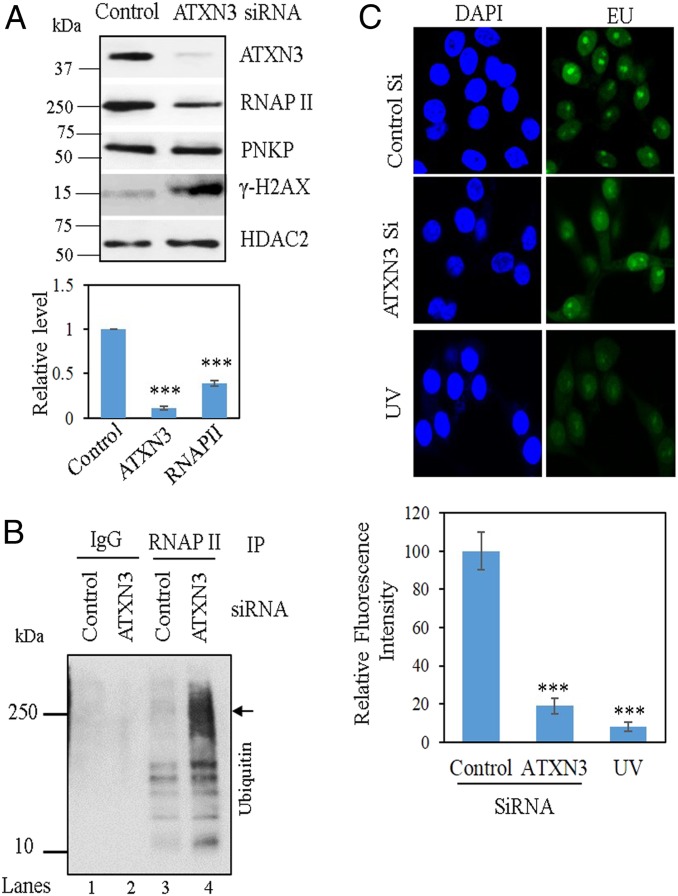
ATXN3 Depletion Facilitates RNAP II Degradation and Affects Global Transcription.
Our data so far indicate that ATXN3 plays a vital role in PNKP-mediated error-free DSB repair. Depletion of ATXN3 in multiple cell lines—as for example, SH-SY5Y cells (Fig. 6A and SI Appendix, Fig. S3A), NSCs (SI Appendix, Fig. S7A), HEK293 cells (SI Appendix, Fig. S4D), or human bronchial epithelial (BEAS-2B) cells (SI Appendix, Fig. S7B)—showed a concomitant decrease in the RNAP II level, but not that of PNKP or other proteins tested. This is consistent with our observation of decreased RNAP II level in SCA3 mice brainstem and patients’ cerebellum (Fig. 4 C and D). ATXN3, an important deubiquitinating enzyme in neuronal cells (63), associates with PNKP, RNAP II, and other C-NHEJ proteins (Fig. 1A and SI Appendix, Fig. S1 A and B), and is sequestered into the polyQ aggregates in SCA3 brains (64). We thus postulated that compromised ATXN3 activity will increase DNA strand-break levels, causing a prolonged stall of elongating RNAP II at the DSB sites. The enhanced ubiquitination of the stalled RNAP II may then lead to its subsequent degradation, adversely impacting both DNA repair and transcription. To explore this possibility, we performed IP using anti-RNAP II Ab (elongating form; pSer2; H5) from ATXN3-depleted (Fig. 6A) SH-SY5Y cells’ NEs. Western analyses with anti-Ubiquitin Ab showed enhanced ubiquitination of RNAP II in ATXN3-depleted cells compared to the control siRNA-transfected cells (Fig. 6B, lane 4 vs. lane 3). Similar results were found in ATXN3-depleted BEAS-2B cells (SI Appendix, Fig. S7C, lane 4 vs. lane 3) and in SCA3 mouse cerebellum (SI Appendix, Fig. S7D, lane 4 vs. lane 3). We have provided evidence that recruitment of C-NHEJ proteins (46), as well as ATXN3 to DSB sites within active genes, is transcription-dependent (Fig. 1D). Thus, degradation of RNAP II might explain why SCA3 mice and patients’ brains accumulate DNA strand breaks primarily on the transcribed genes.
Fig. 6.
Effect of ATXN3-depletion on transcription. (A, Upper) Western blots showing the levels of proteins in the NE of ATXN3-depleted SH-SY5Y cells. HDAC2, nuclear loading control. (Lower) The corresponding quantitation (n = 3, error bars represent ± SD of the mean, ***P < 0.005). (B) SH-SY5Y cells were transfected with control or ATXN3 siRNA and the NEs were immunoprecipitated with IgG (lanes 1, 2) or RNAP II Abs (elongating form, H5; lanes 3, 4) followed by Western blotting with anti-Ubiquitin Ab to assess the ubiquitination level of RNAP II. The arrow indicates the expected position of nonubiquitinated RNAP II. The positions of the protein markers are indicated on the left. (C) Fluorescent-labeled EU incorporation in the nascent RNA, captured by confocal microscopy (40× magnification), in control vs. ATXN3 siRNA-transfected SH-SY5Y cells. EU incorporation following UV exposure (20 J/m2) was used as control. The bar diagram shows the quantitation of the de novo transcript level from 50 randomly chosen cells. Fluorescence intensity of control siRNA-transfected cells was arbitrarily set as 100 (n = 3, error bars represent ±SD of the mean, ***P < 0.005).
Since degradation of RNAP II might impact de novo transcription, we evaluated the RNA synthesis in control vs. ATXN3 depleted SH-SY5Y (Fig. 6C) and BEAS-2B (SI Appendix, Fig. S7E) cells by measuring incorporation of 5-ethynyl-uridine (EU) in newly synthesized RNA and visualized by Click IT conjugation of Alexa Fluor 488. We indeed found significant reduction of EU incorporation in ATXN3-depleted cells. Fluorescence intensity (as a measure of EU incorporation) of 100 (arbitrary units) in control vs. ∼18 in ATXN3-depleted SH-SY5Y cells and ∼52 in ATXN3-depleted BEAS-2B cells indicated a global reduction in transcription following inactivation of ATXN3. UV-irradiation induces two of the most mutagenic and cytotoxic DNA lesions: Cyclobutane-pyrimidine dimers and 6-4 photoproducts (65). These bulky DNA adducts constitute a strong barrier for RNAP II translocation and result in polymerase stalling/arrest. Thus, de novo transcription is blocked and incorporation of EU in newly synthesized RNA is decreased. For this reason, UV-irradiated cells were used as positive control (65, 66) to provide specificity of the EU incorporation assay (Fig. 6C and SI Appendix, Fig. S7E). Collectively, these data suggest that ATXN3 can modulate RNAP II stability and the decreased RNAP II level in turn adversely affected global transcription.
Since our data indicate preferential accumulation of DNA strand breaks in the actively transcribed genes (Enolase, NeuroD1, and TUBB) in ATXN3-depleted SH-SY5Y cells (Fig. 2), SCA3 mice, and patients, and also decreased RNAP II levels therein, we examined the expression level of those genes by qRT-PCR. We indeed found a significant decrease in the expression of these genes in ATXN3-depleted (SI Appendix, Fig. S8B) SH-SY5Y cells (SI Appendix, Fig. S8A), SCA3 mouse brainstem and cerebellum (SI Appendix, Fig. S8 C and D, respectively), and postmortem SCA3 patients’ cerebellum (SI Appendix, Fig. S8E). These data are consistent with our observation of global decrease in transcription following ATXN3 depletion.
PNKP Complementation Rescues SCA3 Phenotype in Drosophila.
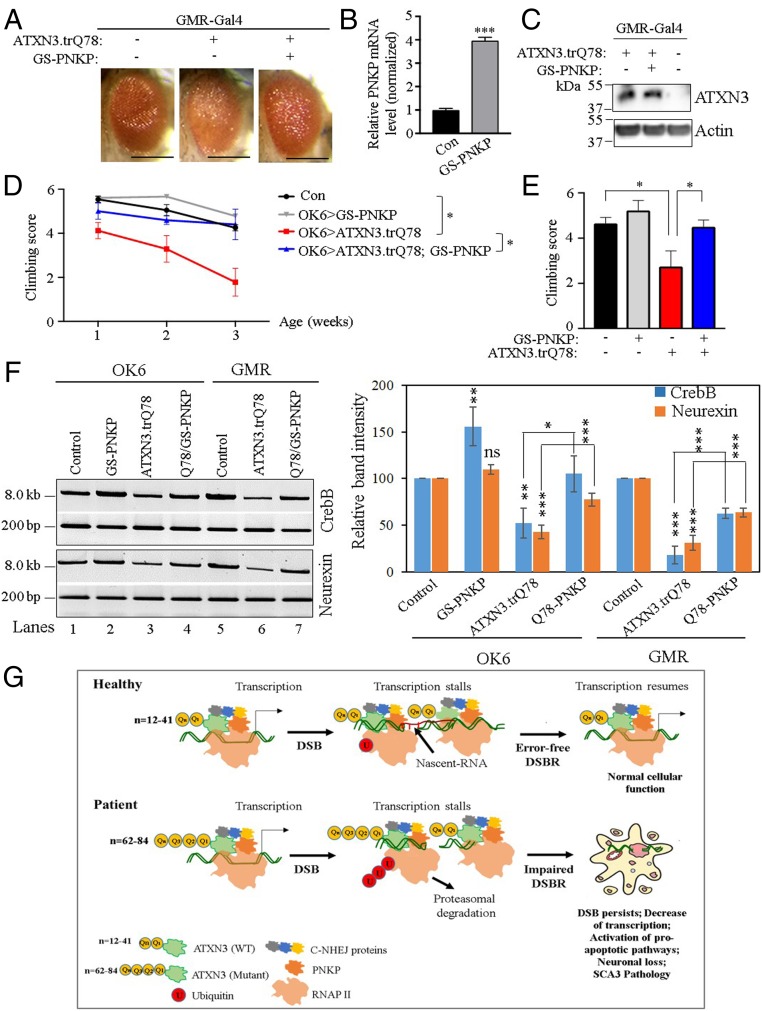
To test the functional outcome of increasing PNKP activity in vivo, we used a Drosophila model of SCA3. Expression of a truncated form of the human ATXN3 gene containing an expansion of the glutamine region (ATXN3.trQ78) in fly eyes by GMR-Gal4 resulted in progressive degeneration of photoreceptor cells, accompanied by eye depigmentation and roughness (67) (Fig. 7 A, Center) compared to control (Fig. 7 A, Left). We used the GS system to drive overexpression of the target gene, CG9601, a Drosophila homolog of PNKP (68). To assess the functionality of the gene, we expressed this gene in E. coli and purified the protein using affinity chromatography (SI Appendix, Fig. S9A). The protein product of this gene has 3′-phosphatase activity in vitro similar to that of human PNKP (SI Appendix, Fig. S9B). Driving a GS line located upstream of CG9601 (GS-PNKP) with Gal4 drivers resulted in an approximately fourfold increase of CG9601 mRNA levels, quantitated by qRT-PCR (Fig. 7B). Western blot revealed that the protein level of mutant ATXN3 was equivalent in GMR > ATXN3.trQ78 flies and GMR > ATXN3.trQ78;GS-PNKP flies (Fig. 7C). Coexpression of GS-PNKP with ATXN3.trQ78 in the eyes partially rescued the eye degeneration phenotype induced by ATXN3.trQ78 (Fig. 7 A, Right).
Fig. 7.
PNKP complementation rescues pathogenic ATXN3 (ATXN3.trQ78) -induced neurotoxicity in Drosophila. (A–C) Flies with eye-specific overexpression of ATXN3.trQ78 exhibit depigmentation and rough eye phenotype, which is mitigated by GS-PNKP coexpression. (A) Representative figures of a control eye (GMR-Gal4 only, Left), eyes expressing ATXN3.trQ78 (Center), and ATXN3.trQ78; GS-PNKP (Right). Scale bar: 100 µm. (B) Quantitation of mRNA levels of PNKP by qRT-PCR, normalized to housekeeping gene, RP49 and to control (Con). n = 3, cohorts, ***P < 0.005. (C) Representative Western blot of ATXN3 (monomer) in fly head lysates expressing eye-specific ATXN3.trQ78 with and without GS-PNKP coexpression. Actin: loading control. (D–E) Flies with motor neuron-specific overexpression of ATXN3.trQ78 exhibit progressive locomotor deficit, which is rescued by GS-PNKP coexpression. (D) Climbing score of flies of indicated genotype during 1 to 3 wk of aging. *P < 0.05, Mixed effect analysis, Tukey’s multiple comparison test. (E) Climbing score of flies at 3 wk old. *P < 0.05, one-way ANOVA, Sidak’s multiple comparison test. Genotypes: Con: OK6-Gal4 (motor neuron-specific driver). OK6 > GS-PNKP: OK6-Gal4/+; GS-PNKP/+. OK6 > ATXN3.trQ78: OK6-Gal4/UAS-ATXN3.trQ78. OK6 > ATXN3.trQ78; GS-PNKP: OK6-Gal4/UAS-ATXN3.trQ78; GS-PNKP/+. n = 4 cohorts each group. (F, Left) Representative agarose gel images of long (∼8 kb) and short (∼200 bp) amplicon of the CrebB and Neurexin genes from genomic DNA of OK6-control (lane 1), OK6 > GS-PNKP (lane 2), OK6 > ATXN3.trQ78 (lane 3), OK6 > ATXN3.trQ78;GS-PNKP (lane 4), GMR-control (lane 5), GMR-ATXN3.trQ78 (lane 6), and GMR-ATXN3.trQ78; GS-PNKP (lane 7) fly samples. (Right) The normalized relative band intensities were represented in the bar diagram with the control samples (OK6 and GMR) arbitrarily set as 100 (n = 3, error bars represent ± SD of the mean). The damage for each gene in SCA3 Drosophila was significant (**P < 0.01; ***P < 0.005) compared to the corresponding control samples. Also, the strand breaks were significantly repaired in PNKP coexpression samples compared to corresponding SCA3 samples (*P < 0.05; ***P < 0.005). (G) A model depicting the pathogenic mechanism of neurodegeneration in SCA3.
Next, we used a motor neuron model of SCA3 in Drosophila. Motor neuron-specific overexpression (driven by OK6-Gal4) of ATXN3.trQ78 led to an age-dependent reduction of locomotor capability, as shown by progressively reduced climbing score in negative geotaxis assay (Fig. 7D). GS-PNKP coexpression significantly enhanced the climbing ability of ATXN3.trQ78-expressing flies, which is most evident at 3 wk of age (Fig. 7 D and E). Consistent with our earlier observations in SCA3 mouse and patients, the RNAP II level was significantly decreased in both eyes (SI Appendix, Fig. S9C) and larval brains (motor neuron-specific expression of mutant ATXN3) (SI Appendix, Fig. S9D) of SCA3 Drosophila. In an attempt to test whether PNKP coexpression can ameliorate repair deficiency, we examined the DNA strand-break accumulation in Drosophila CrebB and Neurexin genes by LA-qPCR in adult head (for eye specific expression) and head with thorax (for motor neuron-specific expression) of flies expressing ATXN3.trQ78 alone or coexpressing ATXN3.trQ78 and GS-PNKP. We indeed observed higher strand-break accumulation following ATXN3.trQ78 expression and far fewer strand-breaks with GS-PNKP coexpression in both tissues (Fig. 7F). We also examined the larval brains of flies with motor neuron specific expression of ATXN3.trQ78 alone or coexpression of ATXN3.trQ78 and GS-PNKP for DSB accumulation by analyzing γH2A.v (the primary histone marker for DSBs in fly) foci. We observed a significant increase of γH2A.v foci in the neurons expressing ATXN3.trQ78 compared to OK6 control flies. With PNKP coexpression, these foci are largely resolved, indicating efficient DSB repair (SI Appendix, Fig. S10). Together, these results demonstrate that increasing PNKP activity rescued mutant ATXN3-induced DSB repair deficiency and neurotoxicity in vivo.
Discussion
Several reports (69–71) indicate that DNA repair deficiencies and genomic instability, causing persistent DSBs in the neuronal genome, contribute to various neuropathologies, including ataxia. The neurons in adults are postmitotic and are thought to incur frequent DNA lesions (43, 71) that might accumulate over a long period of time. Replication is a major source of endogenous DSBs in dividing neural progenitors (70); however, in postmitotic, mature neurons, reactive oxygen species that are frequently generated in the brain due to high oxygen consumption of this tissue remains one of the major source of endogenous DSBs (43, 71). Endogenous DSBs may also arise in the transcribing genome via topoisomerase-IIβ–dependent stimulation of transcription (71). Interestingly, a recent report showed that normal brain activity associated with exploration of a novel environment-caused transient DSB formation in young mice (72). Ongoing gene transcription is essential in maintaining cellular function. Hence, accumulation of DSBs in the transcribed genes represents a significant threat to neuronal homeostasis. More importantly, DSBs can often lead to deletion of sequence around the break point and such deletion, particularly in the transcribed sequences, could be lethal or mutagenic. Hence, a deficient DNA repair system, exaggerated by age and mutant protein-associated pathological processes, such as in SCA3, can lead to DNA damage accumulation. In our previous study (31, 38), we showed that persistent accumulation of DNA strand breaks may lead to subsequent chronic activation of the DNA damage-response ATM signaling pathway. This chronic activation of ATM triggers neuronal dysfunction and eventual neuronal degeneration that will enforce a point of no return, after which no drug or other type of intervention will work.
DSB repair is a complex, multistep process; many different proteins are likely to be involved to fine-tune the pathway, apart from the dedicated repair proteins. While screening for PNKP-associated proteins by two-dimensional gel electrophoresis and MS analysis, we identified ATXN3, along with several other proteins, in the PNKP IC (31). We thus tested ATXN3’s role in PNKP-mediated TC-NHEJ and its etiological linkage to SCA3. The present study revealed that ATXN3 is associated with the C-NHEJ proteins together with RNAP II. We also demonstrated that, in mammalian cells, ATXN3 forms a large megadalton complex along with Lig IV, PNKP, and RNAP II. Moreover, our data suggest that C-NHEJ is the major repair pathway in the postmitotic neurons and predominates over HR. ATXN3 plays a critical role in such neuronal DSB repair since depletion of WT ATXN3 leads to p53BP1 (C-NHEJ marker) foci formation in cultured neuronal cells, as well as expression of pathogenic ATXN3, leads to elevated levels of p53BP1 in SCA3 mice and patients’ brain tissues. Additionally, our RNA ChIP data and plasmid-based repair assay have conclusively shown that WT ATXN3 associates with the nascent RNA and facilitates RNA-templated error-free repair through RNA–DNA hybrid formation at the break sites in order to faithfully transfer the missing information of the original genomic sequence. Many recent studies have shown such transient RNA–DNA hybrid formation around DNA strand-break sites and established their role in efficient DNA DSB repair in yeast (73) and in mammalian cells (74). All of these observations are consistent with our model of RNA-templated repair of the transcriptionally active genome by TC-NHEJ where ATXN3 plays a significant role. Importantly, we demonstrated that pathogenic ATXN3 with polyQ expansion specifically impaired PNKP’s 3′-phosphatase activity, but not that of NEIL-initiated DNA glycosylase activity in SCA3 mice and patients. Persistent DSBs due to the inactivation of PNKP will stall transcription, interfering with the expression of a wide variety of genes that may contribute to the complexity of SCA3 pathology. Our data indeed revealed a global decrease in transcription in ATXN3-deficient cells. This global transcriptional dysregulation seems consistent with earlier reports of transcriptome analysis in cerebellums from SCA3 transgenic mice showing down-regulation of genes involved in glutamatergic neurotransmission, intracellular calcium signaling/mobilization of the MAP kinase pathway, GABAA/B receptor subunits, HSPs, and transcription factors regulating neuronal survival and differentiation (75).
There is increasing evidence for prolonged arrest of RNAP II at the site of strand-breaks causing poly-ubiquitination and proteasome-mediated degradation of the large subunit of RNAP II, which is conserved from yeast to humans (76–80). Interestingly, similar proteasome-mediated degradation of the RNAP II, stalled at the DSB sites, was evident in neuronal cells following ionizing radiation-induced DNA damage (44). Consistently, we observed enhanced ubiquitination and subsequent degradation of the RNAP II in ATXN3-depleted cells or mutant ATXN3 expressing SCA3 mice. However, we could not completely rule out the possibility of ubiquitination of RNAP II interactors, if any, in the RNAP II IC, despite maintaining stringent conditions in IP experiments. Nevertheless, the specific degradation of RNAP II suggests that ATXN3 modulates the ubiquitination level of temporarily stalled RNAP II at the DSB sites and thereby the level of functional RNAP II, which is solely responsible for transcription of the essential protein-coding genes. Collectively, these studies establish a molecular link between TC-NHEJ deficiency, transcriptional dysregulation, and SCA3 pathogenesis.
Based on these findings, we propose a model for SCA3 pathology (Fig. 7G) in which ATXN3 plays a bifunctional role in error-free DSB repair and protein quality control. We postulate that, during TC-NHEJ–mediated repair of DSBs, the transcription machinery pauses temporarily at the break site and the large subunit of RNAP II (RPB1) is monoubiquitinated for the functional assembly of the repair proteins (81) and subsequent strand invasion by a homologous nascent RNA to form an RNA–DNA hybrid. Thus, RNAP II stalled at a DNA strand break may act as a DNA damage sensor and provide a specific signal for activating DNA repair (82). Under normal conditions (in healthy individuals), WT ATXN3, being associated with TC-NHEJ components, activates PNKP to initiate repair by processing 3′-P–containing blocked DNA ends to generate 3′-OH ends that can be used by an RNA-dependent DNA polymerase to restore the missing sequence using nascent homologous RNA as a template (provided by the RNAP II–ATXN3–TC-NHEJ complex at the 5′ end of the DSB). After completion of repair, with the aid of other factors, RNAP II is deubiquitinated by ATXN3, and normal transcription resumes (Fig. 7 G, Upper). The potential deubiquitinating role of ATXN3 is supported by earlier reports showing ATXN3-mediated cleavage of the K63-linked ubiquitin chains that are mostly implicated in monoubiquitination-mediated signaling events (14). In the diseased state, mutant ATXN3 with polyQ expansion blocks the activity of PNKP, leading to impaired DSB repair in the transcribed genome. Persistent 3′-P due to PNKP inactivation or loss/sequestration of ATXN3 would cause transcriptional block, leading to prolonged arrest of RNAP II followed by its poly-ubiquitination and subsequent degradation (Fig. 7 G, Lower).
It is noteworthy that ubiquitination of RNAP II is a complex phenomenon. Several distinct ubiquitin ligases have been implicated in the ubiquitination and degradation of RNAP II (81). Identification and characterization of the ubiquitin ligase/deubiquitinase pair for RNAP II warrant further investigation. Nevertheless, our data provide compelling evidence that WT ATXN3 is a key regulatory component of the DSB repair complex and is pivotal in regulating both TC-NHEJ and transcription. We have also provided multiple lines of evidence strongly indicating that mutant ATXN3-mediated loss of DNA repair and transcriptional dysregulation may be critical proximal events that impair both TC-NHEJ–mediated DSB repair and transcription leading to SCA3 pathogenesis that can be rescued by the expression of PNKP in vivo. Thus, we hypothesize that PNKP overexpression in mutant ATXN3-expressing cells/tissues alone can circumvent both abrogated DSB repair and transcriptional block and eventually maintains a steady-state level of RNAP II to ensure essential cellular functions. In addition, our study provides a mechanistic link between transcriptional dysregulation and neurotoxicity in SCA3. This molecular understanding of 1) how or why neurons in SCA3 lack efficient DSB repair and 2) what is the role of ATXN3 in such repair is essential in determining the reversibility and treatment of such diseases. Recent studies show that PNKP plays a critical role in neurogenesis, and pnkp-null mice are embryonic-lethal (83). Our previous studies have shown that depletion of PNKP causes cell death, and ectopic PNKP expression rescues cells from mutant ATXN3-mediated toxicity (38). Our present study in a Drosophila model suggests that the SCA3 motor phenotype can be completely rescued by PNKP overexpression. Thus, molecular strategies that potentiate PNKP-mediated TC-NHEJ may help reduce neurotoxicity in SCA3, and therefore be instrumental in the development of mechanism-based treatment options for such disorders.
Materials and Methods
For detailed methods, see SI Appendix.
hNSC Culture.
To generate NSCs, a normal iPSC line KYOU-DXR0109B (201B7, ATCC) was grown in Cell Matrix basement membrane gel (Gibco) and pluripotent stem cell (PSC) SFM XF/FF media (Gibco) at 37 °C and 5% CO2. Derivation of NSCs was done using PSC neural induction medium (Gibco), as described previously (84, 85).
Coimmunoprecipitation.
Co-IP from NE was performed according to a previously described protocol (46, 61).
Proximity Ligation Assay.
In-cell protein–protein association was assessed by proximity ligation assay as per the manufacturer’s instructions (85).
DNA- and RNA-ChIP.
DNA/RNA-ChIP was performed as per protocol described previously (46, 61).
LA-qPCR.
LA-qPCR was performed following a standard protocol (46, 60–62). The oligos are listed in SI Appendix, Table S2.
DSB Repair Plasmid Assay.
Plasmid based DSB repair assay was performed in lacZ-expressing stable HEK293 cell line following a protocol standardized in our laboratory (46).
Human Tissue Samples.
Human postmortem cerebellum tissue from SCA3 patients and age-matched controls (SI Appendix, Table S1) were obtained from the Michigan Brain Bank.
3′-Phosphatase Activity of PNKP.
The 3′-phosphatase activity of PNKP in the NE was assessed following a protocol developed in our laboratory (31, 62).
RNA Synthesis Assay.
De novo RNA synthesis was measured by incorporation of 5-ethynyl-uridine visualized by Click chemistry according to the manufacturer’s protocol (Invitrogen, Cat# 10329).
Fly-Rearing Conditions.
All fly lines were maintained at 25 °C on standard fly food under a 12:12-h light–dark cycle. The adult fly climbing assay was performed by negative-geotaxis.
Animal Experiments.
All animal experiments were carried out as per protocols, DGAV 020317 of 2016-09-28 (approved by Direção Geral de Alimentação e Veterinária, for P.M.) and 0606029D (approved by UTMB Animal Care and Use Committee, for T.K.H.).
Data Availability Statement.
All data discussed in the text have been compiled in the main and supplementary figures.
Supplementary Material
Acknowledgments
We thank Sahn-ho Kim, Lawrence Berkeley National Laboratory, for providing critical help and necessary reagents for large-scale fractionations of HeLa-S3 cell nuclear extract by gel filtration; Dr. Sankar Mitra for critically reading the manuscript; and Dr. Sarah Toombs Smith, Editor in the Life Sciences, University of Texas Medical Branch (UTMB) for editing this manuscript. We thank the Michigan Brain Bank (5P30 AG053760 University of Michigan Alzheimer’s Disease Core Center) for providing us with the tissue of postmortem SCA3 patients and their age-matched controls. This work was supported by National Institute of Health Grants 2R01 NS073976 (to T.K.H.), R01 NS096305 (to P.S.S. and T.K.H.), and R01 NS088645 (to M.L.H.); University of California Tobacco Related Disease Research Program Grant 26IR-0017 (to A.H.S.) and P30 ES 06676 (to the National Institute on Environmental Health Sciences Center grant to Cell Biology and Molecular Genomics Core of the University of Texas Medical Branch). Research at the Life and Health Sciences Research Institute was supported by the European Regional Development Fund (FEDER), through the Competitiveness Internationalization Operational Programme (POCI), and by national funds, through the Foundation for Science and Technology, under the scope of the projects POCI-01-0145-FEDER-031987 and POCI-01-0145-FEDER-029056, and by the Northern Portugal Regional Operational Programme (NORTE 2020), under the Portugal 2020 Partnership Agreement (NORTE-01-0145-FEDER-000013).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917280117/-/DCSupplemental.
References
- 1.Coutinho P., Andrade C., Autosomal dominant system degeneration in Portuguese families of the Azores Islands. A new genetic disorder involving cerebellar, pyramidal, extrapyramidal and spinal cord motor functions. Neurology 28, 703–709 (1978). [DOI] [PubMed] [Google Scholar]
- 2.Kawaguchi Y., et al. , CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nat. Genet. 8, 221–228 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Zoghbi H. Y., Orr H. T., Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23, 217–247 (2000). [DOI] [PubMed] [Google Scholar]
- 4.McMurray C. T., Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 11, 786–799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matos C., Pereira de Almeida L., Nobrega C., Machado-Joseph disease/Spinocerebellar ataxia type 3: Lessons from disease pathogenesis and clues into therapy. J. Neurochem. 148, 8–28 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Ruano L., Melo C., Silva M. C., Coutinho P., The global epidemiology of hereditary ataxia and spastic paraplegia: A systematic review of prevalence studies. Neuroepidemiology 42, 174–183 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Schöls L., Bauer P., Schmidt T., Schulte T., Riess O., Autosomal dominant cerebellar ataxias: Clinical features, genetics, and pathogenesis. Lancet Neurol. 3, 291–304 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Maciel P., et al. , Improvement in the molecular diagnosis of Machado-Joseph disease. Arch. Neurol. 58, 1821–1827 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Costa M. d. C., Paulson H. L., Toward understanding Machado-Joseph disease. Prog. Neurobiol. 97, 239–257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X., Liu H., Fischhaber P. L., Tang T. S., Toward therapeutic targets for SCA3: Insight into the role of Machado-Joseph disease protein ataxin-3 in misfolded proteins clearance. Prog. Neurobiol. 132, 34–58 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Ward J. M., La Spada A. R., Ataxin-3, DNA damage repair, and SCA3 cerebellar degeneration: On the path to parsimony? PLoS Genet. 11, e1004937 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burnett B., Li F., Pittman R. N., The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 12, 3195–3205 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Mao Y., et al. , Deubiquitinating function of ataxin-3: Insights from the solution structure of the Josephin domain. Proc. Natl. Acad. Sci. U.S.A. 102, 12700–12705 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winborn B. J., et al. , The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 283, 26436–26443 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durcan T. M., et al. , The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum. Mol. Genet. 20, 141–154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Todi S. V., et al. , Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J. 28, 372–382 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Todi S. V., Paulson H. L., Balancing act: Deubiquitinating enzymes in the nervous system. Trends Neurosci. 34, 370–382 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carvalho A. L., Silva A., Macedo-Ribeiro S., Polyglutamine-independent features in ataxin-3 aggregation and pathogenesis of Machado-Joseph disease. Adv. Exp. Med. Biol. 1049, 275–288 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Schmitt I., et al. , Inactivation of the mouse Atxn3 (ataxin-3) gene increases protein ubiquitination. Biochem. Biophys. Res. Commun. 362, 734–739 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Durcan T. M., Fon E. A., Mutant ataxin-3 promotes the autophagic degradation of parkin. Autophagy 7, 233–234 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Jana N. R., et al. , Co-chaperone CHIP associates with expanded polyglutamine protein and promotes their degradation by proteasomes. J. Biol. Chem. 280, 11635–11640 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Scaglione K. M., et al. , Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol. Cell 43, 599–612 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ying Z., et al. , Gp78, an ER associated E3, promotes SOD1 and ataxin-3 degradation. Hum. Mol. Genet. 18, 4268–4281 (2009). [DOI] [PubMed] [Google Scholar]
- 24.do Carmo Costa M., et al. , Ataxin-3 plays a role in mouse myogenic differentiation through regulation of integrin subunit levels. PLoS One 5, e11728 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neves-Carvalho A., et al. , Dominant negative effect of polyglutamine expansion perturbs normal function of ataxin-3 in neuronal cells. Hum. Mol. Genet. 24, 100–117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H., et al. , The Machado-Joseph disease deubiquitinase ataxin-3 regulates the stability and apoptotic function of p53. PLoS Biol. 14, e2000733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeiffer A., et al. , Ataxin-3 consolidates the MDC1-dependent DNA double-strand break response by counteracting the SUMO-targeted ubiquitin ligase RNF4. EMBO J. 36, 1066–1083 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashkenazi A., et al. , Polyglutamine tracts regulate beclin 1-dependent autophagy. Nature 545, 108–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu Y., et al. , Ataxin-3 promotes genome integrity by stabilizing Chk1. Nucleic Acids Res. 45, 4532–4549 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertoni A., et al. , Early and late events induced by polyQ-expanded proteins: Identification of a common pathogenic property of polYQ-expanded proteins. J. Biol. Chem. 286, 4727–4741 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee A., et al. , The role of the mammalian DNA end-processing enzyme polynucleotide kinase 3′-phosphatase in spinocerebellar ataxia type 3 pathogenesis. PLoS Genet. 11, e1004749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karimi-Busheri F., et al. , Molecular characterization of a human DNA kinase. J. Biol. Chem. 274, 24187–24194 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Jilani A., et al. , Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem. 274, 24176–24186 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Shen J., et al. , Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat. Genet. 42, 245–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds J. J., Walker A. K., Gilmore E. C., Walsh C. A., Caldecott K. W., Impact of PNKP mutations associated with microcephaly, seizures and developmental delay on enzyme activity and DNA strand break repair. Nucleic Acids Res. 40, 6608–6619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Entezam M., Razipour M., Talebi S., Beiraghi Toosi M., Keramatipour M., Multi affected pedigree with congenital microcephaly: WES revealed PNKP gene mutation. Brain Dev. 41, 182–186 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Kalasova I., et al. , Novel PNKP mutations causing defective DNA strand break repair and PARP1 hyperactivity in MCSZ. Neurol. Genet. 5, e320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao R., et al. , Inactivation of PNKP by mutant ATXN3 triggers apoptosis by activating the DNA damage-response pathway in SCA3. PLoS Genet. 11, e1004834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.San Filippo J., Sung P., Klein H., Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Heyer W. D., Ehmsen K. T., Liu J., Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieber M. R., NHEJ and its backup pathways in chromosomal translocations. Nat. Struct. Mol. Biol. 17, 393–395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieber M. R., The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alt F. W., Schwer B., DNA double-strand breaks as drivers of neural genomic change, function, and disease. DNA Repair 71, 158–163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casafont I., et al. , Dynamic behavior of the RNA polymerase II and the ubiquitin proteasome system during the neuronal DNA damage response to ionizing radiation. Mol. Neurobiol. 53, 6799–6808 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Sallmyr A., Tomkinson A. E., Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J. Biol. Chem. 293, 10536–10546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakraborty A., et al. , Classical non-homologous end-joining pathway utilizes nascent RNA for error-free double-strand break repair of transcribed genes. Nat. Commun. 7, 13049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarker A. H., et al. , Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: Insights for transcription-coupled repair and Cockayne syndrome. Mol. Cell 20, 187–198 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Anindya R., et al. , A ubiquitin-binding domain in Cockayne syndrome B required for transcription-coupled nucleotide excision repair. Mol. Cell 38, 637–648 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Della-Maria J., et al. , The interaction between polynucleotide kinase phosphatase and the DNA repair protein XRCC1 is critical for repair of DNA alkylation damage and stable association at DNA damage sites. J. Biol. Chem. 287, 39233–39244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das A., et al. , NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells. DNA Repair 5, 1439–1448 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiederhold L., et al. , AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell 15, 209–220 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Storici F., Bebenek K., Kunkel T. A., Gordenin D. A., Resnick M. A., RNA-templated DNA repair. Nature 447, 338–341 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keskin H., et al. , Transcript-RNA-templated DNA recombination and repair. Nature 515, 436–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei L., et al. , DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 112, E3495–E3504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vizin T., Kos J., Gamma-enolase: A well-known tumour marker, with a less-known role in cancer. Radiol. Oncol. 49, 217–226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahly I., et al. , Prominent neuronal-specific tub gene expression in cellular targets of tubby mice mutation. Hum. Mol. Genet. 7, 1437–1447 (1998). [DOI] [PubMed] [Google Scholar]
- 57.Boutin C., et al. , NeuroD1 induces terminal neuronal differentiation in olfactory neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 1201–1206 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hunt C. R., et al. , Hyperthermia activates a subset of ataxia-telangiectasia mutated effectors independent of DNA strand breaks and heat shock protein 70 status. Cancer Res. 67, 3010–3017 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Ayala-Torres S., Chen Y., Svoboda T., Rosenblatt J., Van Houten B., Analysis of gene-specific DNA damage and repair using quantitative polymerase chain reaction. Methods 22, 135–147 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Santos J. H., Meyer J. N., Mandavilli B. S., Van Houten B., Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 314, 183–199 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Chakraborty A., et al. , Neil2-null mice accumulate oxidized DNA bases in the transcriptionally active sequences of the genome and are susceptible to innate inflammation. J. Biol. Chem. 290, 24636–24648 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao R., et al. , Mutant huntingtin impairs PNKP and ATXN3, disrupting DNA repair and transcription. eLife 8, e42988 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doss-Pepe E. W., Stenroos E. S., Johnson W. G., Madura K., Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol. Cell. Biol. 23, 6469–6483 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paulson H. L., et al. , Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 19, 333–344 (1997). [DOI] [PubMed] [Google Scholar]
- 65.Hanawalt P. C., Spivak G., Transcription-coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Tresini M., et al. , The core spliceosome as target and effector of non-canonical ATM signalling. Nature 523, 53–58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Warrick J. M., et al. , Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol. Cell 18, 37–48 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Barclay S. S., et al. , Systems biology analysis of Drosophila in vivo screen data elucidates core networks for DNA damage repair in SCA1. Hum. Mol. Genet. 23, 1345–1364 (2014). [DOI] [PubMed] [Google Scholar]
- 69.Rulten S. L., Caldecott K. W., DNA strand break repair and neurodegeneration. DNA Repair 12, 558–567 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Dumitrache L. C., McKinnon P. J., Polynucleotide kinase-phosphatase (PNKP) mutations and neurologic disease. Mech. Ageing Dev. 161, 121–129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McKinnon P. J., Genome integrity and disease prevention in the nervous system. Genes Dev. 31, 1180–1194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suberbielle E., et al. , Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci. 16, 613–621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohle C., et al. , Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell 167, 1001–1013.e7 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Lu W. T., et al. , Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat. Commun. 9, 532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chou A. H., et al. , Polyglutamine-expanded ataxin-3 causes cerebellar dysfunction of SCA3 transgenic mice by inducing transcriptional dysregulation. Neurobiol. Dis. 31, 89–101 (2008). [DOI] [PubMed] [Google Scholar]
- 76.Reid J., Svejstrup J. Q., DNA damage-induced Def1-RNA polymerase II interaction and Def1 requirement for polymerase ubiquitylation in vitro. J. Biol. Chem. 279, 29875–29878 (2004). [DOI] [PubMed] [Google Scholar]
- 77.Somesh B. P., et al. , Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121, 913–923 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Jung Y., Lippard S. J., RNA polymerase II blockage by cisplatin-damaged DNA. Stability and polyubiquitylation of stalled polymerase. J. Biol. Chem. 281, 1361–1370 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Heine G. F., Horwitz A. A., Parvin J. D., Multiple mechanisms contribute to inhibit transcription in response to DNA damage. J. Biol. Chem. 283, 9555–9561 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuehner J. N., Kaufman J. W., Moore C., Stimulation of RNA Polymerase II ubiquitination and degradation by yeast mRNA 3′-end processing factors is a conserved DNA damage response in eukaryotes. DNA Repair 57, 151–160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilson M. D., Harreman M., Svejstrup J. Q., Ubiquitylation and degradation of elongating RNA polymerase II: The last resort. Biochim. Biophys. Acta 1829, 151–157 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Lindsey-Boltz L. A., Sancar A., RNA polymerase: The most specific damage recognition protein in cellular responses to DNA damage? Proc. Natl. Acad. Sci. U.S.A. 104, 13213–13214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shimada M., Dumitrache L. C., Russell H. R., McKinnon P. J., Polynucleotide kinase-phosphatase enables neurogenesis via multiple DNA repair pathways to maintain genome stability. EMBO J. 34, 2465–2480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vasquez V., et al. , Chromatin-bound oxidized α-Synuclein causes strand breaks in neuronal genomes in in vitro models of Parkinson’s disease. J. Alzheimers Dis. 60 (suppl. 1), S133–S150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mitra J., et al. , Motor neuron disease-associated loss of nuclear TDP-43 is linked to DNA double-strand break repair defects. Proc. Natl. Acad. Sci. U.S.A. 116, 4696–4705 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the text have been compiled in the main and supplementary figures.