Significance
Intracellular pathogen infection in the nematode Caenorhabditis elegans induces a robust transcriptional response as the host copes with infection. This response program includes several ubiquitin ligase components that are predicted to function in protein quality control. In this study, we show that these infection-induced ubiquitin ligase components form a protein complex that promotes increased tolerance of acute heat stress, an indicator of improved protein homeostasis capacity. These findings show that maintaining protein homeostasis may be a critical component of a multifaceted approach allowing the host to deal with stress caused by intracellular infection.
Keywords: proteostasis, intracellular pathogen response, heat shock, C. elegans, cullin-RING ubiquitin ligase complex
Abstract
Intracellular pathogen infection leads to proteotoxic stress in host organisms. Previously we described a physiological program in the nematode Caenorhabditis elegans called the intracellular pathogen response (IPR), which promotes resistance to proteotoxic stress and appears to be distinct from canonical proteostasis pathways. The IPR is controlled by PALS-22 and PALS-25, proteins of unknown biochemical function, which regulate expression of genes induced by natural intracellular pathogens. We previously showed that PALS-22 and PALS-25 regulate the mRNA expression of the predicted ubiquitin ligase component cullin cul-6, which promotes thermotolerance in pals-22 mutants. However, it was unclear whether CUL-6 acted alone, or together with other cullin-ring ubiquitin ligase components, which comprise a greatly expanded gene family in C. elegans. Here we use coimmunoprecipitation studies paired with genetic analysis to define the cullin-RING ligase components that act together with CUL-6 to promote thermotolerance. First, we identify a previously uncharacterized RING domain protein in the TRIM family we named RCS-1, which acts as a core component with CUL-6 to promote thermotolerance. Next, we show that the Skp-related proteins SKR-3, SKR-4, and SKR-5 act redundantly to promote thermotolerance with CUL-6. Finally, we screened F-box proteins that coimmunoprecipitate with CUL-6 and find that FBXA-158 and FBXA-75 promote thermotolerance. In summary, we have defined the three core components and two F-box adaptors of a cullin-RING ligase complex that promotes thermotolerance as part of the IPR in C. elegans, which adds to our understanding of how organisms cope with proteotoxic stress.
Maintaining protein homeostasis (proteostasis) after exposure to environmental stressors is critical for organismal survival (1). Several signaling pathways have been identified that help organisms cope with stressors that perturb proteostasis. For example, elevated temperature triggers the conserved heat shock response (HSR) pathway, which helps organisms survive the toxic effects of heat (2). The HSR up-regulates expression of chaperones that help with refolding of misfolded proteins, to prevent the formation of protein aggregates and restore proteostasis (1). Disruptions of proteostasis and the formation of protein aggregates in humans are associated with severe neurodegenerative and age-related diseases, such as Alzheimer’s and Huntington’s diseases (1, 3–5).
Pathogen infection can perturb proteostasis and several studies in the nematode Caenorhabditis elegans have demonstrated intriguing connections between immune responses to extracellular pathogens and canonical proteostasis pathways (6–11). More recently, examining the C. elegans host response to intracellular pathogens has uncovered a novel stress response pathway that promotes proteostasis (12, 13). Microsporidia are intracellular, fungal-like pathogens that are the most common cause of infection of C. elegans in the wild, with Nematocida parisii being the most commonly found microsporidian species in C. elegans (14). N. parisii replicates inside the C. elegans intestine, and the infection is associated with hallmarks of perturbed proteostasis in the host, such as the formation of large ubiquitin aggregates in the intestine (12). Interestingly, the host transcriptional response to this infection is very similar to the host transcriptional response to another natural intracellular pathogen of the C. elegans intestine, the Orsay virus (12, 15, 16). These molecularly distinct pathogens induce a common mRNA expression pattern in C. elegans that we termed the “intracellular pathogen response” or IPR (13).
Functional insights into the IPR came from analysis of mutants that constitutively express IPR genes. Forward genetic screens identified two genes encoding proteins of unknown biochemical function called PALS-22 and PALS-25 that comprise an on/off switch for the IPR, with wild-type pals-25 acting as an activator of the IPR, which is repressed by wild-type pals-22 (13, 17). Constitutive up-regulation of IPR gene expression in pals-22 loss-of-function mutants is accompanied by a rewiring of C. elegans physiology, including increased resistance against natural pathogens like N. parisii and the Orsay virus, as well as slowed growth and shortened lifespan. pals-22 mutants also have increased proteostasis capacity characterized by improved thermotolerance and lowered levels of aggregated proteins (13). All of the pals-22 mutant phenotypes are reversed in pals-22 pals-25 loss-of-function double mutants (17). Interestingly, these phenotypes appear to be independent of canonical proteostasis factors (13), such as the transcription factors HSF-1 and DAF-16, which mediate the HSR (18), and the SKN-1/Nrf2 transcription factor, which mediates the proteasomal bounceback response (17, 19). In addition to genetic analysis, RNAseq has demonstrated that the genes induced by HSF-1 and SKN-1 are distinct from those up-regulated in pals-22 mutants. Instead, the proteostasis phenotypes of the pals-22 mutants require a cullin gene called cul-6, which is transcriptionally up-regulated in pals-22 mutants and by infection (13, 17). Cullins are components of multisubunit E3 ubiquitin ligases, which are enzymes that catalyze transfer of ubiquitin onto substrate proteins in order to alter their fate (20). Based on these findings we hypothesized that CUL-6-mediated ubiquitylation of target proteins may act as a protein quality control mechanism in the IPR to respond to proteotoxic stress.
CUL-6 belongs to the cullin-RING ligase (CRL) superfamily, which is found throughout eukaryotes (21). CRLs are multisubunit enzyme complexes, a subset of which are Skp, cullin, F-box (SCF) complexes consisting of four subunits: a RING-box/RBX protein, a Skp, a cullin, and an F-box protein, which serves as a substrate adaptor. Interestingly, the SCF class of ubiquitin ligases appears to have undergone a significant expansion in the evolutionary lineage that gave rise to the nematode C. elegans (22, 23). For example, there are around 520 F-box proteins in the C. elegans genome (22), in comparison to around 68 in humans (24), 22 in Drosophila, and 11 in Saccharomyces cerevisiae (25). In addition, the number of core SCF components has increased in nematodes, with C. elegans having 22 Skp-related proteins in comparison to 6 in Drosophila, just 1 in S. cerevisiae, and 1 in humans (26, 27). The SCF components up-regulated as part of the IPR include not only cul-6 as mentioned above, but also the Skp-related proteins skr-3, skr-4, and skr-5, and several F-box proteins (12). If cul-6 were functioning as part of a ubiquitin ligase complex to promote proteostasis as part of the IPR, it should be acting with skrs and other CRL components. Here we use a combination of biochemistry and genetics to describe how CUL-6 acts together with a RING domain protein, 3 Skp-related proteins and 2 F-box proteins to promote proteostasis in C. elegans.
Results
The Cullin CUL-6 Acts in the Intestine and Pharynx to Promote Thermotolerance.
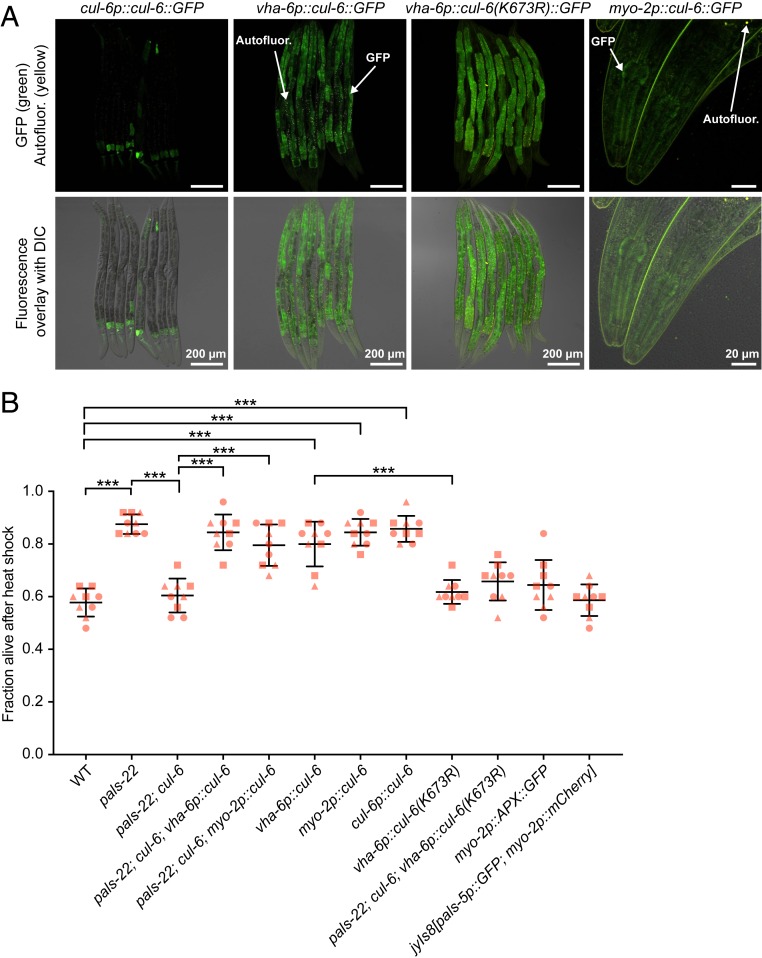
In comparison to wild-type animals, pals-22 loss-of-function mutants have increased thermotolerance, which is reduced to wild-type levels in pals-22; cul-6 double mutants (13). Our previous results indicated that pals-22 regulates thermotolerance in the intestine, where it also regulates cul-6 mRNA expression (13). Because cul-6 is expressed in both the intestine and the pharynx (Fig. 1A), we investigated where cul-6 acts to promote thermotolerance. We designed tissue-specific rescue constructs with cul-6 cDNA using the Mos1-mediated single copy insertion (MosSCI) system (28), to drive expression of GFP-tagged versions of cul-6 using intestinal (vha-6p) or pharyngeal promoters (myo-2p). Here we found that both the intestinal and pharyngeal strains expressed GFP::CUL-6 with the expected tissue distribution pattern (Fig. 1A). We then crossed these tissue-specific CUL-6 MosSCI transgenes into pals-22; cul-6 double mutants to test for rescue of thermotolerance. Here we found that expression of cul-6 in either the intestine or the pharynx was sufficient to increase the thermotolerance of pals-22; cul-6 double mutants (Fig. 1B).
Fig. 1.
CUL-6 expression in the intestine or in the pharynx promotes thermotolerance. (A) Confocal fluorescence images of L4 animals with cul-6::GFP transgenes driven either from the endogenous promoter and expressed from a multicopy in the case of cul-6p::cul-6::GFP (29), or by the vha-6 or myo-2 promoter and integrated with MosSCI. (B) Survival of animals after 2 h of 37.5 °C heat shock, followed by 24 h at 20 °C (hereafter referred to as “heat shock treatment”). Strains were tested in triplicate experiments, with three plates per experiment, 30 animals per plate. The genotypes myo-2p::APX::GFP and jyIs8[pals-5p::GFP; myo-2p::mCherry] were tested as controls for myo-2p-driven expression. Each dot represents a plate and different shapes represent the experimental replicates done on different days. Mean fraction alive of the nine replicates is indicated by the black bar with errors bars as SD. ***P < 0.001, one-way ANOVA with Tukey’s post hoc multiple comparisons test. DIC, differential interference contrast.
Previous studies had only found a functional role for CUL-6 in a pals-22 mutant background. Here we found that overexpression of CUL-6 from a multicopy array (CUL-6 tagged at the C terminus with GFP and 3×FLAG, surrounded by a ∼20-kb endogenous regulatory region) (29) increased thermotolerance in a wild-type animal background (Fig. 1 A and B). Furthermore, we found that either pharyngeal or intestinal expression of cul-6 cDNA promoted thermotolerance in a wild-type background (Fig. 1B). Importantly, transgenic strains with vha-6 and myo-2 promoters driving genes other than wild-type cul-6 did not have increased thermotolerance (see text below, and Fig. 1B). Thus, increased expression of cul-6 in a wild-type background under its own promoter, or only in the intestine, or only in the pharynx leads to increased thermotolerance.
Because ectopic expression of cul-6 can promote thermotolerance, we investigated whether it may lead to up-regulation of heat shock proteins (HSPs). Previous analyses indicated that IPR genes (those regulated by infection and pals-22/pals-25) did not include HSPs, supporting the model that the IPR is distinct from the HSR. Similarly, here we found that vha-6p::cul-6 and myo-2p::cul-6 strains did not have up-regulation of HSPs, consistent with the model that the increased thermotolerance of these strains is not because of HSR activation (SI Appendix, Fig. S1).
We also generated a myo-3p::cul-6 construct to determine whether expression in body wall muscle could promote thermotolerance. Here, we failed to recover myo-3p::cul-6 transgenic animals after several rounds of germline injections, suggesting expression of CUL-6 in muscles may be lethal. To quantify this effect, we injected either red fluorescent protein markers together with myo-3p::cul-6, or red fluorescent protein markers alone (see Materials and Methods). Here we found none of the eggs expressing red fluorescent protein markers hatched when coinjected with myo-3p::cul-6 (0/87), while more than half of the eggs hatched when injected with the red fluorescent protein markers alone (48/84). These results suggest that ectopic expression of CUL-6 in muscles is toxic.
The activity of cullin-RING ubiquitin ligases can be increased by neddylation, which is the process of conjugating the ubiquitin-like protein Nedd8 onto a cullin protein at a conserved lysine residue (30). To determine whether CUL-6 might be regulated by neddylation, we mutated the lysine residue that would likely be targeted for neddylation into an arginine, which would be predicted to disrupt neddylation (SI Appendix, Fig. S2A) (31). We used the MosSCI technique to generate a strain that contains this vha-6p::cul-6(K673R) transgene (expression visualized in Fig. 1A) and found that it could not rescue the thermotolerance of pals-22; cul-6 mutants (Fig. 1B). Furthermore, unlike vha-6p::cul-6(wild-type), the vha-6p::cul-6(K673R) transgene did not promote thermotolerance in a wild-type background. This K673R mutation appeared to have the expected effect on neddylation, as assessed by visualizing a slower migrating band using Western blotting (SI Appendix, Fig. S2B). Previous studies in C. elegans have similarly detected neddylated CUL-2 and CUL-4 proteins, which physically associate with the neddylation regulator CAND-1 (32). Of note, CAND-1 also associates with CUL-6, consistent with CUL-6 being neddylated. Altogether, these results suggest that CUL-6 requires neddylation to promote thermotolerance, which is consistent with CUL-6 acting as part of a ubiquitin ligase complex.
Coimmunoprecipitation/Mass Spectrometry Analysis Identifies CUL-6 Binding Partners.
Next we performed coimmunoprecipitation mass spectrometry (co-IP/MS) analysis to identify binding partners of CUL-6 (Dataset S1). Here we used the C. elegans strain with GFP::3×FLAG-tagged CUL-6, which is functional for thermotolerance (Fig. 1A). We also used similar GFP::3×FLAG-tagged strains for PALS-22 and PALS-25 (13, 17). Through analysis of binding partners for PALS-22 and PALS-25, we sought to obtain insight into their biochemical function, which is currently unknown. Two proteins, GFP::3×FLAG alone and an unrelated protein F42A10.5::GFP::3×FLAG, were added as controls for the co-IPs. For each strain used for co-IP/MS analysis we confirmed transgene expression using immunoblotting and microscopy. When we treated animals with the proteasome inhibitor bortezomib to up-regulate IPR gene expression, we saw an increase in CUL-6 protein by both Western and microscopy analysis, as expected from previous studies, demonstrating that bortezomib induces cul-6 mRNA expression (12, 17) (SI Appendix, Fig. S3 A and B). CUL-6 expression was seen most strongly in the pharynx and the anteriormost intestinal cells. PALS-22 and PALS-25 were broadly expressed throughout the animal and, consistent with previous studies, their expression was not affected by IPR activation (SI Appendix, Fig. S3 A and B) (17).
Co-IP/MS of PALS-22 identified 23 binding partners, including PALS-25 as one of the most highly enriched binding partners, as compared to co-IP/MS of control proteins (SI Appendix, Fig. S4A). PALS-22 also physically associated with PALS-23, which is a PALS protein of unknown function, as well as with F26F2.1, which is a protein of unknown function previously shown to be induced by intracellular infection (12). Co-IP/MS of PALS-25 identified 7 binding partners, with PALS-22 being the most highly enriched hit when compared with co-IP/MS of either control protein (SI Appendix, Fig. S4B). These reciprocal co-IP results suggest that PALS-22 and PALS-25 are in a physical complex together.
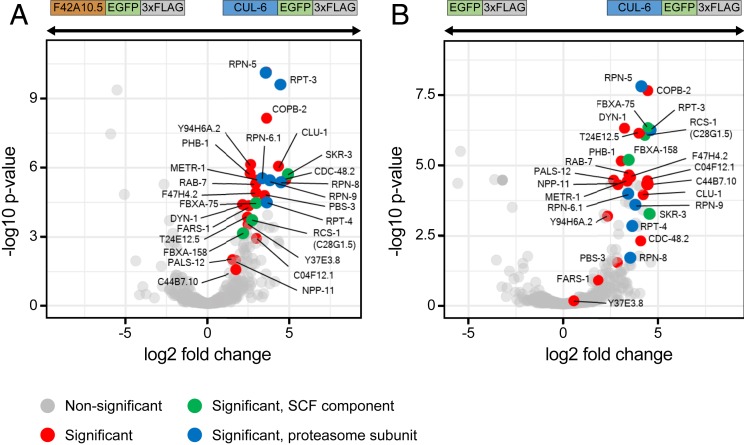
Co-IP/MS of CUL-6 identified 26 significant binding partners. These proteins included predicted SCF ubiquitin ligase components, such as the Skp-related protein SKR-3 and the F-box proteins FBXA-158 and FBXA-75 (Fig. 2 A and B). Additionally, 6 subunits of the 26S proteasome were identified (RPT-3 and -4 and RPN-5, -6.1, -8, and -9). Although not significant, we also saw an enrichment for the NED-8 protein, consistent with CUL-6 being neddylated (Dataset S1). An SCF ubiquitin ligase complex canonically contains an RBX RING box protein, which interacts with a cullin. C. elegans has two RBX proteins, RBX-1 and RBX-2, but neither of these proteins was identified as a significant binding partner for CUL-6 in the co-IP/MS. Instead, we identified a single RING domain protein, C28G1.5. Because of results described below, we renamed C28G1.5 as RING protein acting with cullin and Skr proteins (RCS-1). Interestingly, rcs-1 mRNA expression, like cul-6 mRNA expression, is higher in pals-22 mutants when compared to either wild-type animals (log2 fold change [FC] = 2.81, adjusted P value = 4.83E-08), or compared to pals-22 pals-25 mutants (log2 FC = 2.10, adjusted P value = 3.898E-07) (17).
Fig. 2.
Coimmunoprecipitation mass spectrometry analysis identifies binding partners for CUL-6. Volcano plot of proteins significantly enriched in CUL-6 IP compared to F42A10.5 IP (A) or to GFP IP (B). Proteins significantly more abundant compared to either of the control IPs (GFP alone control or F42A10.5 control, at adjusted P < 0.05 and log2 fold change >1) were considered interacting proteins (Dataset S1). Gray dots indicate nonsignificant proteins, red dots indicate significant proteins, green dots indicate significant SCF proteins, and blue dots indicate significant proteasome subunits.
RING Domain Protein RCS-1 Acts with CUL-6 to Promote Thermotolerance in pals-22 Mutants.
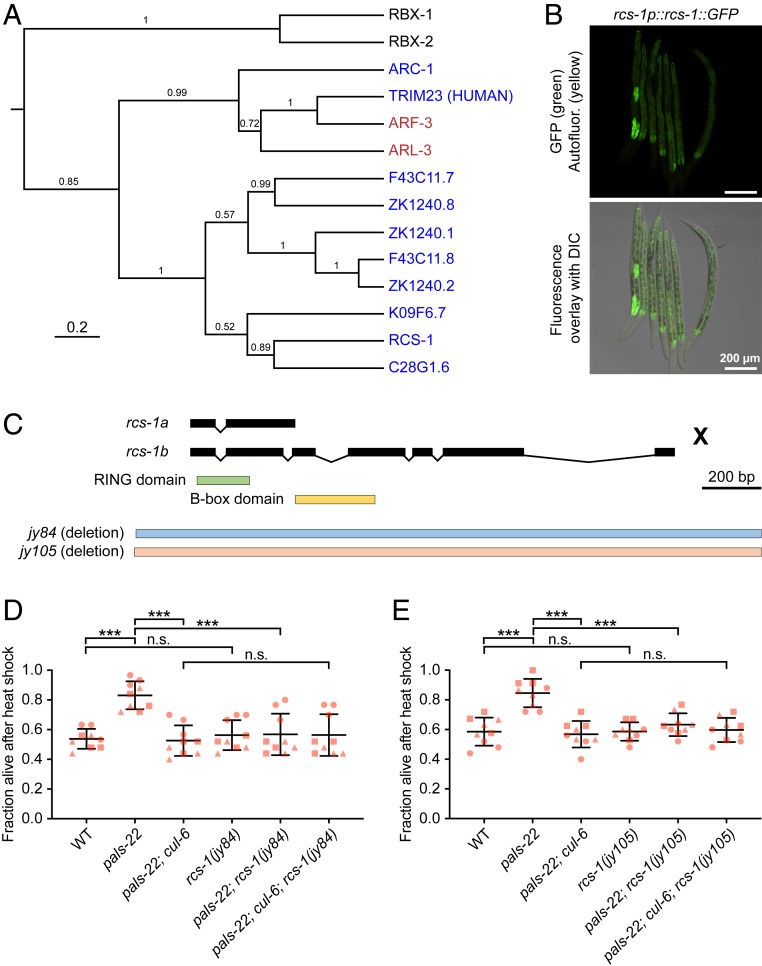
The rcs-1 gene had no previously described role in C. elegans and has two isoforms: rcs-1b has a RING domain and a B-box domain, and rcs-1a has the RING domain only. The closest potential homolog of rcs-1 in humans is the tripartite motif-containing protein 23 (TRIM23). The TRIM family is named for having three motifs (RING finger, B box domain, and coiled coil domain), and many TRIM proteins have E3 ubiquitin ligase activity (33). Phylogenetic analysis of the full-length RCS-1 (RCS-1B) indicated that it is part of the C. elegans TRIM protein family (Fig. 3A) (34), but several C. elegans proteins like the ADP-ribosylation factor related proteins (ARF-3 and ARF-6) are more closely related to TRIM23 than RCS-1. To determine the expression pattern of RCS-1 we generated transgenic C. elegans containing a 3×FLAG and GFP-tagged version of RCS-1 as a multicopy array under the control of its endogenous regulatory region (29). The resulting strain expressed GFP throughout the intestine of the worms, with particularly strong expression in the anteriormost intestinal cells (Fig. 3B), where CUL-6 is also expressed.
Fig. 3.
RING domain protein RCS-1 (C28G1.5) promotes thermotolerance in pals-22 mutants. (A) Phylogenetic relationships of RCS-1 protein with TRIM23 homolog proteins (in red), canonical RBX proteins (in black), and known TRIM proteins (in blue; all are C. elegans except noted human gene). The tree was built from a protein alignment using the Bayesian Markov chain Monte Carlo (MCMC) method. Posterior probabilities are indicated on the branches. (B) Confocal fluorescence images of L4 animals with rcs-1::GFP driven by the endogenous promoter and expressed from a multicopy array (29). (C) rcs-1 isoforms and exon/intron structures. Protein domains are colored in green and yellow. jy84 and jy105 are deletion alleles. (D and E) Survival of animals after heat shock treatment. Strains were tested in triplicate experiments, with three plates per experiment, 30 animals per plate. Each dot represents a plate, and different shapes represent the experimental replicates done on different days. Mean fraction alive of the nine replicates is indicated by the black bar with errors bars as SD. ***P < 0.001, n.s., not significant, P > 0.05, one-way ANOVA with Tukey’s post hoc multiple comparisons test. DIC, differential interference contrast.
Next we used CRISPR-Cas9 to generate two independent deletion alleles of rcs-1 (jy84 and jy105), which we crossed into pals-22 mutants (Fig. 3C). We found that, for both rcs-1 alleles, pals-22; rcs-1 double mutants had thermotolerance similar to wild-type animals, indicating that rcs-1 is required for the increased thermotolerance of pals-22 mutants (Fig. 3 D and E). Similar to cul-6, rcs-1 mutations had no effect in a wild-type background (Fig. 3 D and E). If RCS-1 were acting in a SCF together with CUL-6, then loss of rcs-1 would not further lower thermotolerance in a pals-22; cul-6 mutant. Indeed, we found that pals-22; cul-6; rcs-1 triple mutants had a similar level of thermotolerance to pals-22; cul-6 mutants, as well as to pals-22; rcs-1 mutants and wild-type animals. These findings, together with CUL-6 co-IP results and RCS-1::GFP expression pattern, are consistent with RCS-1 being the RING domain protein that acts with CUL-6 in a ubiquitin ligase complex.
SKP-Related Proteins SKR-3, SKR-4, and SKR-5 Act Redundantly to Promote Thermotolerance in pals-22 Mutants.
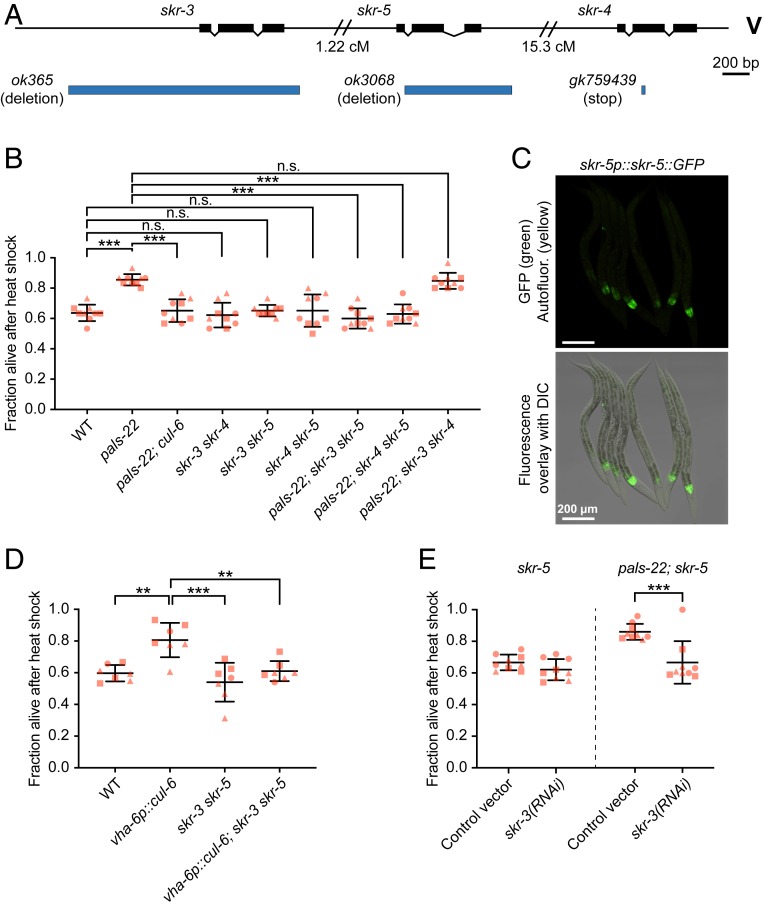
In addition to a cullin and a RING protein, SCF ubiquitin ligase complexes contain a Skp protein. Expression of three Skp-related genes (skr-3, skr-4, and skr-5) is up-regulated by both intracellular infection and mutation of pals-22, similar to cul-6 expression (12, 13). We previously found that mutation of either skr-3, skr-4, or skr-5 alone in a pals-22 mutant background had no effect on thermotolerance (Fig. 4A) (13). Therefore, we made all of the possible combinations of skr-3, skr-4, and skr-5 as double mutants and then crossed them into a pals-22 mutant background to determine whether they may act redundantly. Here we found that pals-22; skr-3 skr-5 and pals-22; skr-5 skr-4 triple mutants had a significant reduction in thermotolerance compared to pals-22 mutants, with levels similar to wild-type animals. In contrast, pals-22; skr-3 skr-4 mutants had thermotolerance similar to pals-22 mutants (Fig. 4B). These results indicate that either SKR-3, SKR-4, or SKR-5 can act together with CUL-6 in a SCF to promote thermotolerance, with SKR-5 being the most important. Consistent with the idea that SKR-5 acts together with CUL-6 and RCS-1, we found that SKR-5::GFP::3×FLAG under control of endogenous regulatory regions is strongly expressed in the anteriormost cells of the intestine (Fig. 4C). Other studies have indicated that SKR-3 has expression in the intestine and pharynx, while SKR-4 has expression in the intestine (27).
Fig. 4.
SKR-3, SKR-4, and SKR-5 act redundantly to promote thermotolerance in pals-22 mutants. (A) skr-3, skr-4, and skr-5 gene exon/intron structures. ok365 and ok3068 are deletion alleles, gk759439 is a premature stop mutation. (B) Survival of animals after heat shock treatment. ***P < 0.001, n.s., not significant, P > 0.05, one-way ANOVA with Tukey’s post hoc multiple comparisons test. n = 9 replicates per strain. (C) Confocal fluorescence images of L4 animals with skr-5::GFP driven by the endogenous promoter and expressed from a multicopy array (29). (D) Survival of animals after heat shock treatment. ***P < 0.001, **P < 0.01, one-way ANOVA with Tukey’s post hoc multiple comparisons test. n = 7 replicates per strain. (E) Survival of animals after heat shock treatment. skr-5 and pals-22; skr-5 mutants were fed on (R)OP50 expressing either L4440 (control vector) or skr-3 RNAi. ***P < 0.001, two-way ANOVA with Sidak’s multiple comparisons test. n = 9 replicates per condition. For B, D, and E strains were tested in triplicate experiments, 30 animals per plate. Mean fraction alive of the replicates is indicated by the black bar with errors bars as SD. Each dot represents a plate, and different shapes represent the experimental replicates done on different days. DIC, differential interference contrast.
Our results indicated that CUL-6 overexpression in the intestine was sufficient to increase thermotolerance in a wild-type background (Fig. 1B). To investigate whether CUL-6 acts together with SKR proteins in this context, we crossed the skr-3 skr-5 double mutant into the CUL-6 overexpressing strain (vha-6p::cul-6). As predicted, the resulting strain had a thermotolerance phenotype comparable to wild-type animals, consistent with CUL-6 acting together with SKR proteins (Fig. 4D).
Next we sought to use RNA interference (RNAi) to further validate the redundancy of SKR proteins acting with CUL-6. However, we found that pals-22 mutants do not have increased thermotolerance compared to wild-type animals when fed on the standard Escherichia coli bacteria used for feeding RNAi (HT115) (SI Appendix, Fig. S5A). This effect may be due to dietary differences between HT115 and the OP50 strain used for thermotolerance experiments described above (35). Therefore, we tested thermotolerance with an OP50 strain (R)OP50 that was modified to enable feeding RNAi studies (36). Here we found that pals-22 mutant animals fed on these RNAi-competent OP50 bacteria (transformed with empty vector L4440) have increased thermotolerance compared to wild-type animals, which is restored back to wild-type levels in pals-22; cul-6 mutants (SI Appendix, Fig. S5A). Using this system, we successfully knocked down expression of cul-6 with RNAi as assessed by lowered CUL-6::GFP::3×FLAG transgene expression (SI Appendix, Fig. S5 B and C). Here we found that cul-6 RNAi suppressed the enhanced thermotolerance of pals-22 mutants (SI Appendix, Fig. S5D). With this system, we then confirmed that skr-3 and skr-5 act redundantly to promote thermotolerance, as RNAi against skr-3 suppressed thermotolerance of pals-22; skr-5 double mutants but not in skr-5 mutants (Fig. 4E).
Analysis of CUL-6, SKR-3,4,5, and RCS-1 in Other Phenotypes Mediated by pals-22.
Previous analysis of IPR genes indicated that RNAi knockdown of cul-6, skr-3, or skr-5 increased susceptibility to intracellular infection in a sterile but otherwise wild-type background C. elegans strain (12). Here we investigated whether cul-6 mutants, skr-3 skr-5 or skr-4 skr-5 double mutants, or rcs-1 mutants could suppress the increased pathogen resistance of pals-22 mutants to microsporidia. In contrast to the complete suppression we found for increased thermotolerance of pals-22 mutants, we found only a minor suppression of pathogen resistance by these mutants in a pals-22 mutant background (SI Appendix, Fig. S6A). Similarly, cul-6 ectopic expression slightly increased pathogen resistance, to a lesser degree than a pals-22 mutation (SI Appendix, Fig. S6B). We also found that mutations in rcs-1, skr-3 skr-4, skr-3 skr-5, or skr-5 skr-4 did not suppress the slowed developmental rate in pals-22 mutants, similar to cul-6 (SI Appendix, Fig. S7). Therefore, cul-6, rcs-1, and skr-3,4,5 appear to be required for executing the thermotolerance phenotype of pals-22 mutants, but not other phenotypes.
FBXA-158 and FBXA-75 Act with CUL-6 to Promote Thermotolerance in pals-22 Mutants.
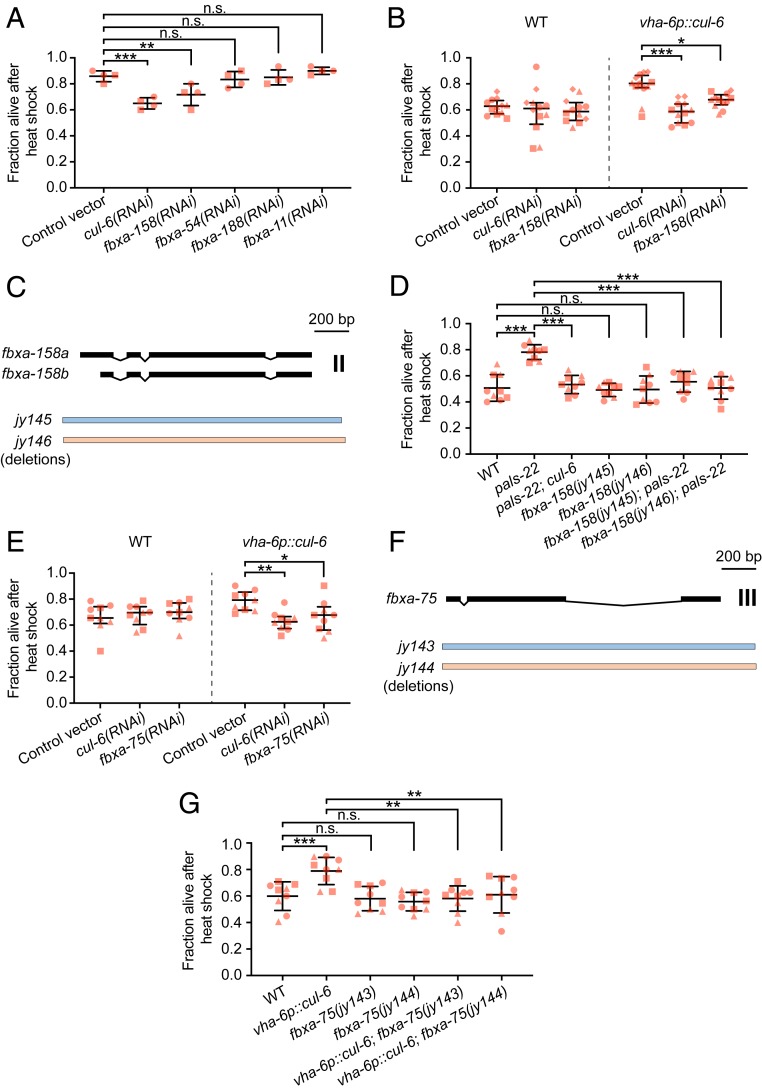
Next we sought to identify the F-box proteins that function as substrate adaptors with CUL-6, RCS-1, and SKR-3,4,5. Here we used RNAi to screen through the F-box proteins that were found to physically associate with CUL-6 from co-IP/MS. In addition to FBXA-158 and FBXA-75 as significant hits, there were other F-box proteins that we identified below the significance threshold, including FBXA-54, FBXA-188, and FBXA-11 (Dataset S1). Because there were RNAi clones available for fbxa-158, fbxa-54, fbxa-188, and fbxa-11, we first used the (R)OP50 RNAi system to screen these genes in a pals-22 mutant background. When treated with RNAi, only fbxa-158 showed significant suppression of the increased thermotolerance phenotype (Fig. 5A). To further validate the results from our initial screen, we also tested fbxa-158 RNAi in the CUL-6 overexpression strain vha-6p::cul-6. Here, fbxa-158 RNAi treatment similarly suppressed the vha-6p::cul-6 increased thermotolerance phenotype (Fig. 5B). Moreover, when treated with fbxa-158 RNAi, wild-type animals did not show further decreased thermotolerance, thus indicating that FBXA-158 is not acting independently from CUL-6 (Fig. 5B). To confirm our RNAi results, we used CRISPR-Cas9 to generate two independent deletion alleles of fbxa-158 (jy145 and jy146) and crossed them with pals-22 mutants (Fig. 5C). When tested for thermotolerance, both fbxa-158 deletion alleles suppressed the increased thermotolerance of pals-22 mutants, while in wild-type animals the survival rate was unchanged (Fig. 5D). We next investigated a role for FBXA-75, the other significant interacting F-box hit for CUL-6. Because there was not an existing clone available, we first made an RNAi clone for fbxa-75 and found that it suppressed the increased thermotolerance of vha-6p::cul-6 animals (Fig. 5E). We also made two independent CRISPR mutations in fbxa-75 (jy143 and jy144) and found that these mutations suppressed the increased thermotolerance of vha-6p::cul-6 animals (Fig. 5 F and G). Disruption of fbxa-75 function, either with RNAi or mutation, did not have an effect in a wild-type background.
Fig. 5.
FBXA-158 and FBXA-75 promote thermotolerance in pals-22 mutants. (A) Survival of animals after heat shock treatment. pals-22 mutants were fed on (R)OP50 expressing either L4440 (control vector) or RNAi for the indicated genes. pals-22 mutants were tested in duplicate experiments, with two plates per experiment, 30 animals per plate. ***P < 0.001, **P < 0.01, n.s., not significant, P > 0.05, one-way ANOVA with Tukey’s post hoc multiple comparisons test. (B) Survival of animals after heat shock treatment. Wild-type and vha-6p::cul-6 animals were fed on (R)OP50 expressing either L4440 (control vector), cul-6, or fbxa-158 RNAi. Strains were tested in quadruplicate experiments, with three plates per experiment, 30 animals per plate. ***P < 0.001, *P < 0.05, two-way ANOVA with Sidak’s multiple comparisons test. (C) fbxa-158 isoforms and exon/intron structures. jy145 and jy146 are deletion alleles. (D) Survival of animals after heat shock treatment. ***P < 0.001, n.s. P > 0.05, one-way ANOVA with Tukey’s post hoc multiple comparisons test. (E) Survival of animals after heat shock treatment. Wild-type and vha-6p::cul-6 animals were fed on (R)OP50 expressing either L4440 (control vector), cul-6, or fbxa-75 RNAi. Strains were tested in triplicate experiments, with three plates per experiment, 30 animals per plate. **P < 0.01, *P < 0.05, two-way ANOVA with Sidak’s multiple comparisons test. (F) fbxa-75 exon/intron structure. jy143 and jy144 are deletion alleles. (G) Survival of animals after heat shock treatment. ***P < 0.001, **P < 0.01, n.s. P > 0.05, one-way ANOVA with Tukey’s post hoc multiple comparisons test. For A, B, D, E, and G each dot represents a plate, and different shapes represent the experimental replicates done on different days. Mean fraction alive of the replicates is indicated by the black bar with errors bars as SD.
Similar to cul-6, rcs-1, and skr-3,4,5, the mRNA expression levels of both fbxa-158 and fbxa-75 are higher in pals-22 mutants when compared to wild-type animals (fbxa-75: log2 FC = 4.83, adjusted P value 0.0002; fbxa-158: log2 FC = 2.81, adjusted P value = 4.83E-08), and in pals-22 mutants compared to pals-22 pals-25 mutants (fbxa-75: log2 FC = 6.48, adjusted P value 6.86E-05; fbxa-158: log2 FC = 6.95, adjusted P value = 1.23E-06) (17). fbxa-158 expression is also enriched in the intestine (37). Together, these results indicate that both FBXA-75 and FBXA-158 are required as F-box adaptor proteins, likely acting with CUL-6/RCS-1/SKR-3,4,5 ubiquitin ligase complex components to promote thermotolerance in C.elegans.
Discussion
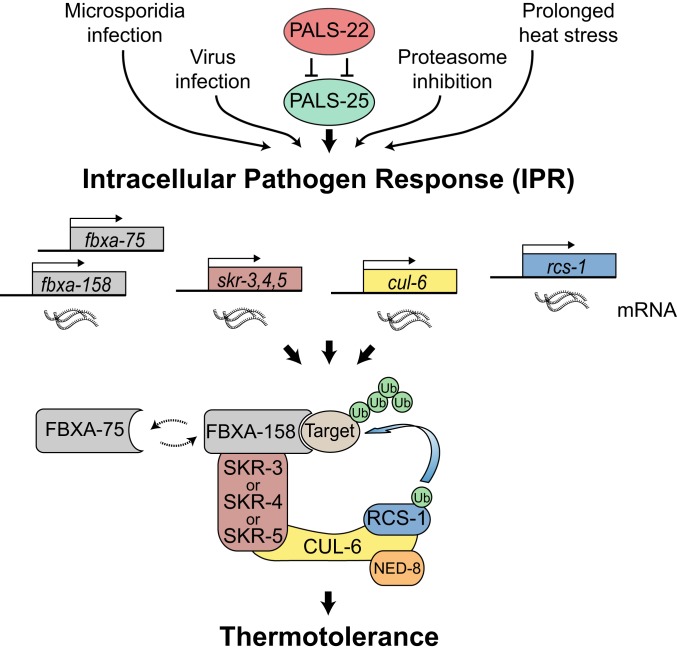
Thermal stress is one of many types of proteotoxic stress that can impair organismal health and survival. Here we used a combination of genetics and biochemistry to broaden our understanding of a recently identified proteostasis pathway called the IPR, which enables animals to survive exposure to thermal stress in a manner distinct from the canonical stress response pathways, including HSF-1/HSR and the SKN-1-mediated bounceback response (17). Specifically, we demonstrate that overexpression of CUL-6/cullin alone promotes thermotolerance, and it can act in either the intestine or the pharynx of C. elegans. Importantly, we found that CUL-6 acts together with other ubiquitin ligase components, including a previously uncharacterized RING protein we named RCS-1, as well as the Skp-related proteins SKR-3, SKR-4, and SKR-5 and the F-box proteins FBXA-158 and FBXA-75 (Fig. 6). We propose that these ubiquitin ligase complexes are able to target proteins for ubiquitin-mediated proteasomal degradation and that this activity is a critical part of the IPR program. Consistent with this model, our co-IP/MS identified several proteasomal subunits that interact with CUL-6 (Fig. 2).
Fig. 6.
Model for a RCS-1/CUL-6/SKR/FBXA-158 or FBXA-75 ubiquitin ligase that promotes proteostasis. Cullin-RING ligase components are transcriptionally up-regulated as part of the intracellular pathogen response. CUL-6 acts together with the RING domain protein RCS-1, functionally redundant Skp-related proteins SKR-3, SKR-4, and SKR-5, and both FBXA-75 and FBXA-158 to promote thermotolerance and improved proteostasis. CUL-6 also requires neddylation by NED-8. FBXA-158 and FBXA-75 may act sequentially to ubiquitinate the same target, or alternatively may have distinct targets.
We also investigated protein–protein interactions of the PALS-22 and PALS-25 proteins, which comprise an on/off switch in the IPR that regulates mRNA expression of cul-6, skr-3, skr-4, skr-5, rcs-1, fbxa-158, and fbxa-75. Previous studies indicated that pals-22 and pals-25 are in the same operon and interact genetically, and our co-IP/MS studies indicate they also interact biochemically. Notably, the pals gene family has expanded in the C. elegans genome, with 39 genes in C. elegans, in comparison to only 1 pals gene each in mouse and human (38). Although the divergent “pals” protein signature that defines PALS proteins is of unknown function, PALS-22 does have weak homology with F-box proteins (38), leading to the speculative idea that PALS proteins function as adaptor proteins in ubiquitin ligase complexes.
Like the pals gene family, C. elegans has a greatly expanded repertoire of SCF ligase components. This expansion has been suggested to reflect the results of an arms race against intracellular pathogens, as SCF components are among the most rapidly diversifying genes in the C. elegans genome (23). The most dramatically expanded class of SCF components includes ∼520 F-box adaptor proteins in C. elegans (22). C. elegans also has 22 SKR proteins, in comparison to only 1 Skp in humans, indicating there has been an expansion of core SCF components as well. Here we identified SKR-3 as a binding partner for CUL-6, which is consistent with previous two-hybrid results (26). Interestingly, we found redundancy in the role of SKRs at the phenotypic level: either SKR-3, SKR-4, or SKR-5 appears capable of acting with CUL-6 to promote thermotolerance, with SKR-5 being the most important. In contrast to the redundancy in SKRs, we found nonredundant roles for 2 different F-box proteins, FBXA-75 and FBXA-158; both are required to promote thermotolerance. Together, with the Skp-related proteins, the F-box proteins are responsible for substrate specificity of the SCF (39). FBXA-75 and FBXA-158 may ubiquitylate distinct targets, or perhaps different sites on the same target that are both required to promote thermotolerance. It is also possible that they are in a CRL that forms a dimer with 2 F-box proteins (40). The identification of FBXA-75, FBXA-158, and SKR-3,4,5 as members of the complex may help in future studies to identify what specific proteins are ubiquitylated by a CUL-6 SCF.
Canonical CRLs contain an RBX protein as the RING domain protein, which interacts with both a cullin and an E2 ubiquitin ligase (21). However, our co-IP/MS studies with CUL-6 did not identify RBX-1 or RBX-2 as interacting partners for CUL-6, but rather identified RCS-1. Given our genetic and biochemical results, we propose that RCS-1 plays the same role as an RBX protein would in a canonical SCF complex (Fig. 6). RCS-1 does not have a clear human ortholog, but the closest human protein is TRIM23 (41). Interestingly, the TRIM family contains 68 genes in humans, many of which encode single subunit E3 ubiquitin ligases, including those that restrict viral infection and regulate inflammatory signaling (33). C. elegans has 18 TRIM proteins, and they appear to have a simpler structure than human TRIM proteins, given the absence of additional motifs in the C-terminal domains normally found in the majority of mammalian TRIM proteins (34). If these other C. elegans TRIM proteins can act in SCF ligases like RCS-1, it suggests there may also be an expansion of the RING core SCF components in C. elegans, in addition to the previously described expansion of SKRs and adaptor proteins.
The role of CUL-6 in promoting thermotolerance was first demonstrated in pals-22 mutants, where there is an up-regulation of cul-6 mRNA as well as several other SCF components, including skr-3, skr-4, skr-5, rcs-1, fbxa-158, and fbxa-75 (13, 17). However, here we found that animals overexpressing only cul-6, without overexpression of the other components of the SCF, have increased thermotolerance. One explanation for this result is that CUL-6 is the limiting factor in a SCF that promotes thermotolerance in the IPR. Consistent with this idea, other components of the complex like SKR-3,4,5 are functionally redundant for thermotolerance, so even basal expression level might be sufficient to build a functional SCF, once CUL-6 expression increases past a certain threshold level.
What substrate(s) is targeted by the RCS-1/CUL-6/SKR-3,4,5/FBXA-158 and FBXA-75 ubiquitin ligase complex? It is possible that the effects of this complex are mediated through targeting a single regulatory protein for ubiquitylation and degradation. For example, ubiquitylation of the DAF-2 insulin receptor by the ubiquitin ligase CHIP can alter DAF-2 trafficking, and it appears that ubiquitylation of just this one target has significant effects on proteostasis in C. elegans (42). In contrast, CRL2 and CRL4 complexes in humans have been shown to target the C termini of a large number of proteins for degradation as part of a newly identified protein quality control system (43, 44), Given the modular nature of the SCF ligase family, the large number of F-box substrate adaptor proteins in C. elegans, and the redundancy we found in the SKR-3,4,5 proteins, it seems possible that there are multiple substrates and adaptors used in the IPR. An exciting possibility is that the IPR involves distinct CRLs that ubiquitylate several different targets to improve proteostasis and tolerance against environmental stressors, including infections. Identifying these factors will be the subject of future studies.
Materials and Methods
Cloning and Generation of cul-6 Tissue-Specific Rescue Strains.
A full list of strains used in this study is in SI Appendix, Table S1. A full list of constructs used in this study is in SI Appendix, Table S2. To generate the vha-6p::cul-6 transgene (pET499), vha-6p::SBP::3xFLAG, cul-6 cDNA, and the unc-54 3′ UTR were assembled in pCFJ150 using Gateway LR. To generate myo-2p::cul-6 (pET686) and myo-3p::cul-6 (pET687) constructs, the promoters myo-2p, myo-3p, and the pET499 linearized backbone without the vha-6p promoter were amplified by PCR from pCFJ90, pCFJ104, and pET499, respectively, and assembled by Gibson recombination (45). To generate the vha-6p::cul-6(K673R) construct (pET688) a 86-bp single-strand oligonucleotide and a linearized pET499 backbone that had been amplified by PCR were assembled by Gibson recombination. To generate the spp-5p::3xFLAG::GFP (pET555) transgene, spp-5p::3xFLAG::GFP and let-858 3′ UTR were assembled in pCFJ150 using Gateway LR. MosSCI was performed as described previously (28). Briefly, the plasmid of interest (25 ng/µL) was injected with pCFJ601 (50 ng/µL), pMA122 (10 ng/µL), pGH8 (10 ng/µL), pCFJ90 (2.5 ng/µL), and pCFJ104 (5 ng/µL) into the EG6699 strain. Injected animals were incubated at 25 °C for 7 d and then subjected to heat shock for 2 h at 34 °C. After 5 h, non-Unc animals were selected and the presence of the insertion was verified by PCR and sequencing. Transgenic strains with TransgeneOme fosmids (SI Appendix, Table S2) were generated as extrachromosomal arrays (29) by injecting into the EG6699 strain and then selecting non-Unc worms.
Lethality Scoring of myo-3p::cul-6 Tissue-Specific Rescue Strains.
To score the lethality of myo-3p::cul-6 expression, worms were injected with a complete MosSCI mix (see above) containing either myo-3p::cul-6 as the plasmid of interest, or water. Injected animals were incubated at 25 °C and after 1 d, eggs expressing red fluorescence were transferred onto new plates at 25 °C for 24 h. The hatching ratio of transferred eggs was then scored for both conditions. The assay was repeated two independent times.
Thermotolerance Assays.
Thermotolerance assays were performed as described (13, 17), and were based on recommendations from ref. 46. Animals were grown on standard nematode growth medium (NGM) plates at 20 °C. L4 stage animals were selected based on observation of the “half-moon” crescent dot in the vulva and were transferred onto fresh NGM plates seeded with OP50-1 and then subjected to heat shock for 2 h at 37.5 °C in an incubator. The plates were then placed in a single layer on a benchtop at room temperature for 30 min and then transferred to a 20 °C incubator. Then, 24 h later the survival was scored in a blinded manner. Worms not responding to touch were scored as dead, and 30 worms were scored per plate. Three replicate plates were scored for each strain per experiment, and each experiment was performed at least three independent times. Statistical significance was tested using one-way ANOVA with Tukey’s HSD (honestly significant difference) for post hoc multiple comparisons, two-way ANOVA with Sidak’s multiple comparisons, or Student’s t test where indicated.
Synchronization of Animals.
To obtain synchronized populations of L1s, gravid adults were washed from NGM plates with M9 into a 15-mL tube, centrifuged, and supernatant was removed leaving 2 mL of M9. Added were 800 µL of 5.65 to 6% sodium hypochlorite solution and 200 µL of 5 M NaOH. Eggs released after bleaching were washed five times with 15 mL of M9 and resuspended in 5 mL of M9. Eggs were put into a 20 °C incubator for 16 to 20 h with rotation to hatch L1s.
Coimmunoprecipitation.
Each sample for co-IP/MS was prepared in three independent experimental replicates. For each sample, 200,000 synchronized L1 animals were transferred onto NGM plates and grown for 48 h at 20 °C. Bortezomib was added to reach a final concentration of 22 µM or the equivalent volume of dimethyl sulfoxide (DMSO) for the control plates. After 6 h at 20 °C, worms were washed off of the plates with M9, washed twice with M9, resuspended in 500 µL of ice-cold lysis buffer (50 mM Hepes, pH 7.4, 1 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), 1 mM MgCl2, 100 mM KCl, 1% glycerol, 0.05% Nonidet P-40, 0.5 mM dithiothreitol (DTT), 1× protease inhibitor tablet) and immediately frozen dropwise in liquid N2. Frozen pellets were ground into powder with a prechilled mortar and pestle. Protein extracts were spun for 15 min 21,000 × g at 4 °C and supernatants were filtered on 0.45-µm filters (Whatman). Protein concentration was determined using Pierce 660-nm protein assay and adjusted to 1 µg/µL with fresh lysis buffer. One milligram of each sample was mixed with 25 µL of ANTI-FLAG M2 Affinity Gel (Sigma) and incubated at 4 °C with rotation (12 rpm) for 1 h. The resin was washed twice with 1 mL lysis buffer for 5 min, twice with 1 mL wash buffer for 5 min (50 mM Hepes, pH 7.4, 1 mM MgCl2, 100 mM KCl) and 20 min with 1 mL wash buffer with rotation. The liquid was removed and the beads were then stored at −80 °C. The strains used were expressing FLAG and GFP-tagged versions of our proteins of interest from extrachromosomal arrays generated with constructs from the “TransgeneOme” project, which contain ∼20 kb genomic DNA surrounding the gene of interest. The strains ERT365, ERT465, ERT422, and ERT488 were used for the proteins PALS-22, PALS-25, CUL-6, and the control protein F42A10.5, respectively. Additionally, the GFP only control IP was done using the strain ERT413 expressing GFP under the intestinal promoter spp-5 as a single integrated copy (SI Appendix, Table S1).
Trypsin Digestion.
The immunoprecipitated proteins bound to the beads were digested overnight in 400 ng trypsin (Promega, V511A) in 25 mM ammonium bicarbonate (Sigma) at 37 °C. Samples were then reduced with 1 mM final concentration of DTT (Acros Organics) for 30 min and alkylated with 5 mM final concentration of iodoacetamide (MP Biomedicals, LLC) for 30 min in dark. The peptides were extracted from the beads by adding 50 µL of 5% formic acid (Sigma). The extraction was repeated one more time and the eluted peptides were combined. Digested peptides were desalted using the Stage-Tip, C18 peptide cleanup method. The eluates were vacuum dried and peptides were reconstituted in 15 µL of 5% formic acid, 5% acetonitrile solution for LC-MS/MS analysis.
LC-MS/MS Analysis.
Samples were analyzed in triplicate by LC-MS/MS using an EASY-nLC 1000 high performance liquid chromatography (HPLC) (Thermo Scientific) and Q-Exactive mass spectrometer (Thermo Scientific) as described previously (47) with the following modifications. The peptides were eluted using a 60-min acetonitrile (ACN) gradient (45-min 2 to 30% ACN gradient, followed by a 5-min, 30 to 60% ACN gradient, a 2-min 60 to 95% ACN gradient, and a final 8-min isocratic column equilibration step at 0% ACN) at 250-nL/min flow rate. All of the gradient mobile phases contained 0.1% formic acid. Mass spectrometry data was collected using a top 10 data-dependent analysis (DDA) method in positive ion mode. Data was collected over a scan range of 400 to 1,800 m/z (mass/charge) at 70,000 resolution, and an AGC (automatic gain control) target of 3e6. A dynamic exclusion time of 20 s was implemented and unassigned, singly charged, and charge states above 6 were excluded for the data-dependent MS/MS scans. The MS2 scans were triggered with a minimum AGC target threshold of 1e5 and with maximum injection time of 60 ms. The peptides were fragmented using the normalized collision energy (NCE) setting of 25. Apex trigger and peptide match settings were disabled.
Peptide and Protein Identification and Quantification.
The RAW files obtained from the MS instrument were converted into mzXML format. The SEQUEST search algorithm was used to search MS/MS spectra against the concatenated target decoy database comprised of forward and reverse, reviewed C. elegans FASTA sequences from Uniprot (downloaded on 6/8/2015) along with GFP and E. coli sequences appended in the same file. The search parameters used were as follows: 20 ppm peptide mass tolerance; 0.01 Da fragment ion tolerance; trypsin (1 1 KR P) was set as the enzyme; maximum of two missed cleavages were allowed; oxidation on methionine (15.99491 Da) and N-terminal acetylation (42.01056 Da) were set as differential modifications; static modification (57.02146 Da) was set on cysteine for alkyl modification. Peptide matches were initially filtered to a peptide false discovery rate (FDR) of 2% using the linear discriminant analysis. Peptides were assembled into proteins and again filtered to a 2% FDR. This protein level filtering resulted in a final peptide FDR of less than 1%. The protein level matches were filtered at 2% FDR using the protein sieve analysis. The spectral counts from the triplicates were then summed and used for the data analysis.
Analysis of Mass Spectrometry Data.
Peptide spectral counts were used to calculate fold change ratio, P value, and adjusted P value between sample IPs and control IPs (GFP and F42A10.5) using the DEP package in R (48). Briefly the data were filtered to keep only the peptides present in at least two replicates in one condition. Filtered data were normalized using variance stabilizing normalization. Missing values were imputed using the MiniProb method from the DEP package by randomly selecting values from a Gaussian distribution centered on a minimal value of the dataset. Fold change ratio and adjusted P values were calculated. Proteins with adjusted P < 0.05 and log2 fold change >1 in comparison with at least one of the controls were considered as significant. Any protein with a negative log2 fold change with one control or the other (i.e., more affinity to the control protein than the tested bait) was considered as nonsignificant.
Phylogenic Analysis of RCS-1.
Amino acid sequences of 15 proteins were aligned using MUSCLE (version 3.7) and trimmed with trimAI (version 1.3) (49) using the phylemon2 online platform (50). Bayesian Markov chain Monte Carlo inference (LG + I + G + F) was performed using BEAST (version 1.10.4) (51). Analysis was run using a Yule model tree prior, an uncorrelated relaxed clock (lognormal distribution), and a chain length of 10 million states sampled every 1,000 iterations. Results were assessed with Tracer (version 1.7.1); a maximum clade credibility tree was built after a 25% burn-in. Posterior probability values greater than 0.5 are marked on branch labels.
CRISPR Deletions of rcs-1, fbxa-158, and fbxa-75.
To generate deletions of rcs-1, fbxa-158, and fbxa-75 a co-CRISPR strategy was used, adapted from the Integrated DNA Technologies (IDT) proposed method for C. elegans. To generate the ∼2.18-kb deletion alleles of rcs-1 we designed two crRNAs encompassing the whole rcs-1 locus (crRNA1: 5′-GTTTGTTGAAGGAAATGCACAGG-3′, crRNA2: 5′-GGTTTCCTATAGCTGTGACACGG-3′). These two oligonucleotides were synthetized by IDT and used with a crRNA targeting the dpy-10 gene (dpy-10 crRNA3: 5′-GCTACCATAGGCACCACGAG-3′) and assembled with commercial tracrRNA (50 µM crRNA1 and crRNA2, 25 µM dpy-10 crRNA, and 40 µM tracrRNA). After an annealing step for 5 min at 95 °C, the resulting guide RNA mixture was added to CAS9-NLS protein (27 µM final, ordered from QB3 Berkeley) and microinjected into N2 worms. F1 Dpy progeny were screened by PCR, confirmed by sequencing for a deletion, and positive lines were backcrossed four times to the N2 background before testing. The same co-CRISPR strategy was used to generate the ∼1.65-kb deletion alleles of fbxa-158 and the ∼2-kb deletion alleles of fbxa-75. Two crRNAs encompassing the whole fbxa-158 locus (crRNA1: 5′-ATAGTCGGGTACAAAACAAATGG-3′, crRNA2: CTACTCCATCTTTAAGAACACGG-3′) or of the fbxa-75 locus (crRNA1: 5′-AATACATTTCAACCAGCAAGAGG-3′, crRNA2: 5′-GAGCATTTTAATCGGATTGGTGG-3′) were used with the same dpy-10 crRNA and assembled as described above. Deletion positive lines were backcrossed two times to the N2 background before testing.
RNA Interference Assays.
RNA interference assays were performed using the feeding method. Overnight cultures of HT115 or OP50 strain (R)OP50 modified to enable RNAi (36) (a gift from M. Wang’s laboratory, Baylor College of Medicine, Houston, TX) were plated on RNAi plates (NGM plates supplemented with 5 mM isopropyl β-d-1-thiogalactopyranoside [IPTG], 1 mM carbenicillin) and incubated at 20 °C for 3 d. Gravid adults were transferred to these plates, and their F1 progeny (L4 stage) were transferred onto new RNAi plates before being tested for thermotolerance as previously described.
Western Blot Analysis.
For each strain, 1,500 synchronized L1 worms were placed on NGM plates seeded with OP50 bacteria and incubated at 20 °C for 48 h. These animals were then treated with bortezomib (22 µM final) or control DMSO for 6 h and washed off the plate with M9. Proteins were extracted in lysis buffer (50 mM Hepes, pH 7.4, 1 mM EGTA, 1 mM MgCl2, 100 mM KCl, 1% glycerol, 0.05% Nonidet P-40, 0.5 mM DTT, 1× protease inhibitor tablet) as previously described (13). Protein levels were determined using the Pierce 660-nm assay. Equal amounts of protein (5 µg) were boiled in protein loading buffer, separated on a 4 to 20% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS/PAGE) precast gel (Bio-Rad), and transferred onto polyvinylidene fluoride or polyvinylidene difluoride (PVDF) membrane. Nonspecific binding was blocked using 5% nonfat dry milk in phosphate-buffered saline (PBS)-Tween (0.1%) for 1 h at room temperature. The membranes were incubated with primary antibodies overnight at 4 °C (mouse anti-FLAG diluted 1:1,000 and mouse anti-tubulin diluted 1:7,500), washed five times in PBS-Tween, and blotted in horseradish peroxidase-conjugated secondary antibodies at room temperature for 2 h (goat anti-mouse diluted 1:10,000). Membranes were then washed five times in PBS-Tween, treated with enhanced chemiluminescence (ECL) reagent (Amersham), and imaged using a Chemidoc XRS+ with Image Lab software (Bio-Rad). To resolve CUL-6 neddylation by gel shift, 1,500 synchronized L1 worms were incubated for 48 h as described above and boiled in 6× SDS/PAGE loading buffer (375 mM Tris⋅HCl pH 6.8, 600 mM DTT, 12% SDS, 0.06% bromophenol blue, and 60% glycerol). Protein samples were separated on a 7.5% continuous SDS/PAGE precast gel (Bio-Rad) at 100 V for ∼6.5 h until the 100-kDa loading marker was run off the gel. Separately, equivalent amounts of each protein sample were run on another 7.5% gel at 100 V for 2 h for the tubulin loading control. Proteins were transferred to PVDF membranes, blocked, and incubated with anti-FLAG and anti-tubulin antibodies as described above.
Fluorescence Microscopy.
For cul-6, rcs-1, and skr-5 tissue-specific expression lines shown in Figs. 1, 3, and 4, respectively, images were taken using a Zeiss LSM700 confocal microscope with 10× and 40× objectives. For the expression analysis of GFP::3×FLAG-tagged proteins PALS-22, PALS-25, CUL-6, and F42A10.5 in SI Appendix, Fig. S3, images were taken with a Zeiss LSM700 confocal microscope with 10× objective. For RNAi knockdown in SI Appendix, Fig. S5, L4-stage F1 progeny of ERT422 worms fed with OP50 expressing cul-6 RNAi were anesthetized using 10 µM levamisole in M9 buffer and mounted on 2% agarose pads for imaging with a Zeiss LSM700 confocal microscope. Using ImageJ software (version 1.52e), GFP signal in the pharynx and the first intestinal cell ring were measured, as well as three adjacent background regions. The total corrected fluorescence (TCF) was calculated as TCF = integrated density − (area of selected cell × mean fluorescence of background readings). For each condition, 30 worms were imaged. Significance was assessed with a Student’s t test.
Pathogen Load.
N. parisii spores were prepared as previously described (52). Synchronized L1 worms were prepared as described above and 1,200 animals were plated on 6-cm NGM plates, in triplicate, with a mixture of OP50 bacteria and 1 × 105 N. parisii spores. The plates were incubated at 25 °C for 30 h before worms were collected off plates in M9, pelleted, and fixed in acetone. Fixed worms were stained with fluorescence in situ hybridization (FISH) probes conjugated to CAL Fluor 610 dye (Biosearch Technologies) targeting N. parisii ribosomal RNA. N. parisii pathogen load was measured with a COPAS Biosort machine (Union Biometrica). For SI Appendix, Fig. S6B, F42A10.5::GFP was used as a negative control for comparison to cul-6p::cul-6::GFP because both strains express TransgeneOme transgenes from extrachromosomal arrays in the unc-119(ed3) background.
qRT-PCR.
Approximately 6,000 synchronized L1 worms were prepared as described above and grown on 10-cm NGM plates at 20 °C until the L4 stage. Worms were washed off plates with M9, then concentrated to <100 µL. RNA was isolated with TRI reagent and 1-bromo-3-chloropropne (BCP) (Molecular Research Center, Inc.) and resuspended in nuclease-free H2O. cDNA was prepared from 100 ng/µL total RNA using iScript cDNA synthesis kit (Bio-Rad). qPCR was performed using iQ SYBR Green Supermix with a final concentration of 5 ng/µL cDNA (Bio-Rad). Each experimental replicate was measured in technical duplicate, and all gene expression was normalized to snb-1 expression. qPCR primers are listed in SI Appendix, Table S3.
Measurement of Developmental Rate.
A total of 40 to 50 gravid adults were transferred onto standard 10-cm NGM plates and incubated at 20 °C to lay eggs for 2 h before being removed. These plates were incubated at 20 °C and the proportion of eggs that hatched and developed into L4s was scored at 48 h, 64 h, and 72 h by scoring 100 animals each replicate.
Data Availability.
RAW files for co-IP/MS performed in this study have been uploaded to the Mass Spectrometry Interactive Virtual Environment (MassIVE) public repository (accession no. MSV000084936) and will be made freely available upon publication.
Statistical analysis of spectral counts from this dataset, as described above, are summarized in Dataset S1.
Supplementary Material
Acknowledgments
We thank Vladimir Lazetic, Ivana Sfarcic, Jessica Sowa, and Eillen Tecle for comments on the manuscript. This work was supported by NIH under R01 AG052622 (to E.R.T. and E.J.B.) and GM114139 grants and a Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases fellowship (to E.R.T.). S.S.G. was supported by NIH/National Institute of General Medical Sciences (NIGMS) Institutional Research and Academic Career Development Awards (IRACDA) K12 grant GM068524.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: RAW files for co-IP/MS performed in this study have been uploaded to the Mass Spectrometry Interactive Virtual Environment (MassIVE) public repository (accession no. MSV000084936).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1918417117/-/DCSupplemental.
References
- 1.Labbadia J., Morimoto R. I., The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomez-Pastor R., Burchfiel E. T., Thiele D. J., Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 4–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klaips C. L., Jayaraj G. G., Hartl F. U., Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 217, 51–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor R. C., Dillin A., Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 3, a004440 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kourtis N., Tavernarakis N., Cellular stress response pathways and ageing: Intricate molecular relationships. EMBO J. 30, 2520–2531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tillman E. J., et al. , Endoplasmic reticulum homeostasis is modulated by the forkhead transcription factor FKH-9 during infection of Caenorhabditis elegans. Genetics 210, 1329–1337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh V., Aballay A., Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc. Natl. Acad. Sci. U.S.A. 103, 13092–13097 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellegrino M. W., et al. , Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature 516, 414–417 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohri-Shiomi A., Garsin D. A., Insulin signaling and the heat shock response modulate protein homeostasis in the Caenorhabditis elegans intestine during infection. J. Biol. Chem. 283, 194–201 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Miles J., Scherz-Shouval R., van Oosten-Hawle P., Expanding the organismal proteostasis network: Linking systemic stress signaling with the innate immune response. Trends Biochem. Sci. 44, 927–942 (2019). [DOI] [PubMed] [Google Scholar]
- 11.O’Brien D., et al. , A PQM-1-mediated response triggers transcellular chaperone signaling and regulates organismal proteostasis. Cell Rep. 23, 3905–3919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bakowski M. A., et al. , Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog. 10, e1004200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reddy K. C., et al. , An intracellular pathogen response pathway promotes proteostasis in C. elegans. Curr. Biol. 27, 3544–3553.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G., et al. , A large collection of novel nematode-infecting microsporidia and their diverse interactions with Caenorhabditis elegans and other related nematodes. PLoS Pathog. 12, e1006093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkies P., Ashe A., Le Pen J., McKie M. A., Miska E. A., Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome Res. 23, 1258–1270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen K., Franz C. J., Jiang H., Jiang Y., Wang D., An evolutionarily conserved transcriptional response to viral infection in Caenorhabditis nematodes. BMC Genomics 18, 303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy K. C., et al. , Antagonistic paralogs control a switch between growth and pathogen resistance in C. elegans. PLoS Pathog. 15, e1007528 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu A.-L., Murphy C. T., Kenyon C., Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300, 1142–1145 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Li X., et al. , Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 7, e1002119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng N., Shabek N., Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Petroski M. D., Deshaies R. J., Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Thomas J. H., Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 16, 1017–1030 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kipreos E. T., “Ubiquitin-mediated pathways in C. elegans” in WormBook (The C. elegans Research Community, WormBook, 2005). [DOI] [PMC free article] [PubMed]
- 24.Jin J., et al. , Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18, 2573–2580 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kipreos E. T., Pagano M., The F-box protein family. Genome Biol. 1, reviews3002.1 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nayak S., et al. , The Caenorhabditis elegans Skp1-related gene family: Diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol. 12, 277–287 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Yamanaka A., et al. , Multiple Skp1-related proteins in Caenorhabditis elegans: Diverse patterns of interaction with Cullins and F-box proteins. Curr. Biol. 12, 267–275 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Frøkjær-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9, 117–118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarov M., et al. , A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150, 855–866 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duda D. M., et al. , Structural insights into NEDD8 activation of cullin-RING ligases: Conformational control of conjugation. Cell 134, 995–1006 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu K., Chen A., Pan Z.-Q., Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 275, 32317–32324 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Bosu D. R., et al. , C. elegans CAND-1 regulates cullin neddylation, cell proliferation and morphogenesis in specific tissues. Dev. Biol. 346, 113–126 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatakeyama S., TRIM family proteins: Roles in autophagy, immunity, and carcinogenesis. Trends Biochem. Sci. 42, 297–311 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Sardiello M., Cairo S., Fontanella B., Ballabio A., Meroni G., Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol. Biol. 8, 225 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pang S., Curran S. P., Adaptive capacity to bacterial diet modulates aging in C. elegans. Cell Metab. 19, 221–231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynn D. A., et al. , Omega-3 and -6 fatty acids allocate somatic and germline lipids to ensure fitness during nutrient and oxidative stress in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 112, 15378–15383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lingala S. M., Mhs M. G. M., Ghany, Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development 25, 289–313 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leyva-Díaz E., et al. , Silencing of repetitive DNA is controlled by a member of an unusual Caenorhabditis elegans gene family. Genetics 207, 529–545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skaar J. R., Pagan J. K., Pagano M., Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14, 369–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosu D. R., Kipreos E. T., Cullin-RING ubiquitin ligases: Global regulation and activation cycles. Cell Div. 3, 7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dawidziak D. M., Sanchez J. G., Wagner J. M., Ganser-Pornillos B. K., Pornillos O., Structure and catalytic activation of the TRIM23 RING E3 ubiquitin ligase. Proteins 85, 1957–1961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tawo R., et al. , The ubiquitin ligase CHIP integrates proteostasis and aging by regulation of insulin receptor turnover. Cell 169, 470–482.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koren I., et al. , The eukaryotic proteome is shaped by E3 Ubiquitin ligases targeting C-terminal degrons. Cell 173, 1622–1635.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin H.-C., et al. , C-terminal end-directed protein elimination by CRL2 Ubiquitin ligases. Mol. Cell 70, 602–613.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson D. G., et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Zevian S. C., Yanowitz J. L., Methodological considerations for heat shock of the nematode Caenorhabditis elegans. Methods 68, 450–457 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gendron J. M., et al. , Using the Ubiquitin-modified proteome to monitor distinct and spatially restricted protein homeostasis dysfunction. Mol. Cell. Proteomics 15, 2576–2593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X., et al. , Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 13, 530–550 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sánchez R., et al. , Phylemon 2.0: A suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res. 39, W470-4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suchard M. A., et al. , Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Estes K. A., Szumowski S. C., Troemel E. R., Non-lytic, actin-based exit of intracellular parasites from C. elegans intestinal cells. PLoS Pathog. 7, e1002227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RAW files for co-IP/MS performed in this study have been uploaded to the Mass Spectrometry Interactive Virtual Environment (MassIVE) public repository (accession no. MSV000084936) and will be made freely available upon publication.
Statistical analysis of spectral counts from this dataset, as described above, are summarized in Dataset S1.