Significance
We performed a systematic analysis of transposable elements (TEs) in enhancers associated with immune functions. In CD8+ T cells, specific TE subfamilies are enriched in enhancers and provide transcription factor recognition motifs and architectural sequences to central and peripheral domains, respectively. Enhancers unique to immune cell types are more prone to putative regulatory TE cooption as compared to enhancers unique to other tissues. Genomic neighborhoods of immune-related genes harbor more TEs compared to other gene-bearing regions, likely reflecting reduced local purifying selection. We hypothesize that TE abundance in immune-associated genomic regions facilitated active selection and functionalization of TE motifs beneficial for immune functions. This may have favored accelerated evolution of enhancers in immune cells adapting to changing infectious challenges.
Keywords: transposable element, enhancer, T lymphocyte, immune tissue
Abstract
Transposable elements (TEs) compose nearly half of mammalian genomes and provide building blocks for cis-regulatory elements. Using high-throughput sequencing, we show that 84 TE subfamilies are overrepresented, and distributed in a lineage-specific fashion in core and boundary domains of CD8+ T cell enhancers. Endogenous retroviruses are most significantly enriched in core domains with accessible chromatin, and bear recognition motifs for immune-related transcription factors. In contrast, short interspersed elements (SINEs) are preferentially overrepresented in nucleosome-containing boundaries. A substantial proportion of these SINEs harbor a high density of the enhancer-specific histone mark H3K4me1 and carry sequences that match enhancer boundary nucleotide composition. Motifs with regulatory features are better preserved within enhancer-enriched TE copies compared to their subfamily equivalents located in gene deserts. TE-rich and TE-poor enhancers associate with both shared and unique gene groups and are enriched in overlapping functions related to lymphocyte and leukocyte biology. The majority of T cell enhancers are shared with other immune lineages and are accessible in common hematopoietic progenitors. A higher proportion of immune tissue-specific enhancers are TE-rich compared to enhancers specific to other tissues, correlating with higher TE occurrence in immune gene-associated genomic regions. Our results suggest that during evolution, TEs abundant in these regions and carrying motifs potentially beneficial for enhancer architecture and immune functions were particularly frequently incorporated by evolving enhancers. Their putative selection and regulatory cooption may have accelerated the evolution of immune regulatory networks.
Upon encountering cognate antigens, resting naïve CD8+ T cells undergo activation and functional differentiation into cytotoxic effectors, which fight infections. Mechanistically, adaptive immune responses rely on a profound reorganization of enhancer networks that orchestrate transcriptional outputs (1, 2). Mutations in enhancers have been associated with autoimmune and inflammatory disorders, such as asthma and rheumatoid arthritis (3, 4). Epigenetic events accompanying enhancer remodeling during CD8+ T cell differentiation have been comprehensively documented (1, 2). Genomic features constituting CD8+ T cell enhancers and their role in regulation, however, have not yet been explored.
Enhancers are distantly operating modular elements that stimulate gene transcription by looping with target promoters (5), acting individually or jointly on one or multiple genes. The structural framework of an active enhancer includes an accessible DNA core, surrounded by more condensed chromatin harboring short arrays of nucleosomes (6, 7). These flanking nucleosomes are decorated with H3K4me1, which broadly marks both active and poised regulatory regions (8), and eventually with H3K27ac, which correlates with enhancer activity (9). Enhancers evolving from background DNA include transposable elements (TEs) (10), which are abundant in mammalian genomes (up to 50% of total DNA). TEs are selfish genetic elements that disseminate throughout genomes via autonomous self-replication and reinsertion. Based on distinct mobilization machinery, TEs broadly divide into retrotransposons and DNA transposons (11). Retrotransposons are by far more abundant and split into long-terminal repeat (LTR) and non-LTR groups. LTRs include endogenous retroviruses (ERVs), while non-LTR TEs subdivide into long-interspersed (LINEs) and short-interspersed elements (SINEs), nonautonomous transposons mobilized by the LINE integration machinery. These lineages are composed of phylogenetically related families, further branching out into multiple subfamilies, each originating from one precursor copy. With time, the accumulation of mutations introduced divergence in the consensus sequence within members of each subfamily.
A growing body of evidence suggests that specific TE subfamilies have been coopted for transcriptional regulation in a number of biological contexts (12–22). In particular, TE-associated enhancers contributed to the evolution of early development (17), placentation (18), mammalian pregnancy (14), and innate immune responses (21). TEs with regulatory potential are thought to accelerate evolution. Identical regulatory copies spread across the genome can rapidly rewire novel transcriptional patterns, resulting in functional novelty or an increase in regulatory complexity (23, 24). This mechanism could be particularly relevant for the evolution of the immune system, which needs to adapt rapidly to effectively respond to a variety of mutable stimuli.
Here, we use a combination of genome-wide approaches to characterize enhancers enriched in TEs in differentiating mouse CD8+ T lymphocytes. We map the topology of TE-lineages across enhancer central and boundary domains and show that core-enriched TEs carry ancestral transcription factor (TF) recognition motifs, better preserved compared to nonenhancer TE equivalents. Flanking TEs harbor high densities of H3K4me1 histone mark and provide dA:dT-rich sequences that match enhancer boundary nucleotide composition. TE-rich enhancers are linked to important T cell functions and are part of a regulatory network with a potentially high degree of functional redundancy. Enhancers selectively active in CD8+ T cells and other immune tissues are richer with TEs, as compared to enhancers active in other tissues. Similarly, TEs are more abundant in genomic neighborhoods of immune-related genes, as compared to regions bearing genes silent in immune tissues. These results reveal the differential contribution of distinct TE lineages to enhancer domains and highlight the putative role of TEs in the evolution of immune functions.
Results
Enhancer Network Reorganizes Early on during CD8+ T Cell Differentiation.
To study the role of TEs in cis-regulation in T lymphocytes, we isolated naïve (D0) C57BL/6J TCR transgenic (OT-I) mouse CD8+ T cells (expressing transgenic T cell receptors specific for ovalbumin). OT-I cells were activated in vitro for 24 h (D1, activated) or 3 d (D3, cycling cells) using antibodies directed against CD3 and CD28. To identify active enhancers, we first performed genome-wide mapping of open chromatin (accessible to nuclease digestion), using an assay for transposase-accessible chromatin coupled with high-throughput sequencing (ATAC-seq) (25). After peak calling, we identify a total of 42,019 high-fidelity ATAC sites (see mapping and reproducibility details in SI Appendix, Fig. S1 A and B).
To ascertain possible bias induced by in vitro differentiation, we compared our dataset to the published ATAC-seq carried out on OT-I cells, naïve and in vivo-differentiated in response to bacterial infection (GSE95237 series) (1). Of ATAC peaks from our dataset, 97% in naïve and 87% in D3 cells are also present in naïve and differentiated cells from the GSE95237 series, respectively, yielding highly similar profiles in the genome browser (SI Appendix, Fig. S1C). This not only demonstrates the reliability of our ATAC regions but also reveals that the majority of peaks found in D3 are independent of the type of stimulus. Importantly, we further used the in-house dataset because the sequencing conditions (longer paired-end reads of 100 bp) allowed less ambiguous mapping of ATAC signals on genomic repeats and transposons, as compared to single-end sequencing of shorter reads (26). Higher resolution of this strategy allows mapping TEs to ATAC peak summits, as well as slopes to better define boundaries of the accessible chromatin.
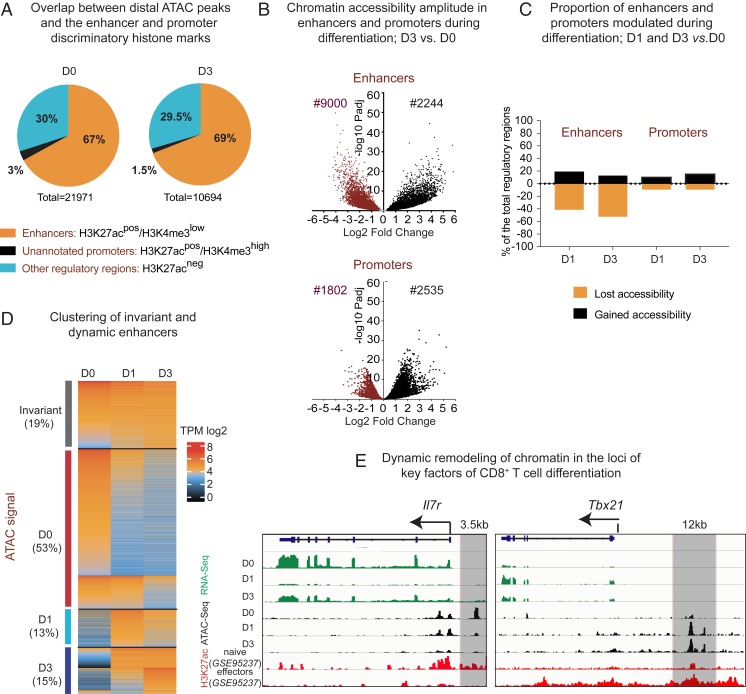
To distinguish putative enhancers, we split ATAC peaks into two groups: Promoters and the distal peaks (see the full pipeline in SI Appendix, Fig. S2A). Regions situated within ±2 kb from annotated transcription start sites (TSS) were considered as holding a promoter activity. More distal peaks were considered as cis-regulatory regions. To select distal ATAC peaks with the signature of active enhancers, we performed an overlap with the hallmark histone modification H3K27ac using chromatin immunoprecipitation-sequencing (ChIP-seq) datasets for naïve and differentiated cells from the above-mentioned GSE95237 series. As these H3K27ac+ distal ATAC peaks may contain a small portion of not yet unannotated promoters, we intersected them with the H3K4me3 peaks (in-house ChIP-seq), typically higher in promoters than in enhancers (27) (cut-off is shown in SI Appendix, Fig. S2B). After filtering out unannotated promoters, we observe an expected proportion of distal ATAC peaks with features of active enhancers (9), 67% in D0 and 69% in D3 (Fig. 1A). Finally, we obtained a total of 16,807 putative active enhancers and 12,526 of active promoters (all stages pooled).
Fig. 1.
Remodeling of enhancers during CD8+ T cell differentiation. (A) Pie charts depicting overlap between distal ATAC-peaks in D0 and D3 and the enhancer- and promoter-distinguishing histone marks (H3K27ac, GSE95237 series, and H3K4me3, in-house). (B) Volcano plots showing differential accessibility of ATAC regions during differentiation (log2 fold-change of ATAC signal modulation) in promoters and active enhancers during CD8+ T cell differentiation (shown is D3 vs. D0). (C) Bar plot showing proportion of promoters and enhancers significantly modulated during D0–D1 and D0–D3 transition (false-discovery rate 0.05, fold-change 1.5). Shown are proportions of the total promoters or enhancers. (D) Supervised heatmap of the transcripts per million (TPM)-normalized ATAC signal in enhancers, clustered by the significant or not modulation of accessibility during the D0–D3 transition. Each dynamic cluster is further subclustered into early (modulated at D1) and late responders. (E) Parallels between normalized RNA-seq, ATAC-seq, and CHIP-seq tracks on the IL7ra gene and its previously validated enhancer, and the Tbx21 gene and a putative enhancer in the vicinity (shadowed in gray).
To identify regions with significant alterations in accessibility during D0–D3 transition, we performed differential quantification of ATAC signals in enhancers found in different conditions. Enhancers were assigned to a differentiation stage only if they were either more accessible or uniquely found in this stage compared to another. D1 mostly served to follow the kinetics of enhancers’ chromatin accessibility between D0 and D3. As shown in Fig. 1 B, Upper, the amplitude (fold-change) of enhancer chromatin accessibility during differentiation in most cases exceeds twofold. Promoters are modulated to a somewhat lesser extent (Fig. 1 B, Lower). The vast majority of enhancers (nearly 80%) undergo statistically significant modulation of chromatin accessibility during differentiation, in contrast to 34% of modulated promoters (see numbers in Fig. 1B and proportions in Fig. 1C). Together, these data highlight the paramount importance of enhancers in the process of CD8+ T cell differentiation.
Based on differential quantification, we generated a supervised heatmap of ATAC signals clustered by their dynamic profile during differentiation (Fig. 1D). We observed a broader D0-specific enhancer cluster (50% of the total enhancers), as compared to D3. The majority of D0-specific enhancers shrink early on, at D1 (Fig. 1D). Less than 20% either gain accessibility by D3 or remain stable across differentiation (Fig. 1D). A small proportion of enhancers is temporarily more accessible at D1. Higher numbers of the D0-specific enhancers cannot be explained by higher numbers of genes overexpressed at D0, as compared to D3. The differential analysis of the stage-matched RNA sequencing (RNA-seq; in-house) reveals comparable numbers of genes selectively up-regulated at D0 or D3 (SI Appendix, Fig. S3). This result suggests a higher enhancer/gene ratio in naïve, as compared to activated T cells.
Loci of the key determinants of CD8+ T cell differentiation (Il7ra coding for a major marker of naïve cells, and Tbx21 coding for T-bet, the master regulator of effectors), show the expected modulation of chromatin accessibility, including the experimentally validated enhancer that regulates Il7ra gene expression in naïve T cells (28) and a putative enhancer in the proximity of Tbx1 gene (Fig. 1E).
Altogether, these results show that during CD8+ T cell differentiation chromatin remodeling is initiated early on (before the first cell division) and that naïve cells embody broader enhancer accessibility and potentially higher enhancer redundancy, as compared to activated cells.
Selected TE Species Are Differentially Enriched in Central and Peripheral Enhancer Domains.
To colocalize TEs with active enhancers, we first defined enhancer boundaries using hallmark histone modifications: H3K27ac (GSE95237 series) and H3K4me1 (GSE95237 series), as well as H3K4me3 (in-house), present at a low level in active enhancers. We observed a comodulation of H3K4me1 and ATAC signals in D0- and D3-specific developmental enhancers (SI Appendix, Fig. S4). Aggregation of histone marks around ATAC peak centers reveals a classic peak–valley–peak pattern (29), with the ATAC peak nearly perfectly coinciding with the valley (Fig. 2A). The highest densities of H3K27ac and H3K4me3 occur within ATAC-adjacent regions (roughly equal to the size of one nucleosome in deep sequencing; 170 bp), whereas H3K4me1 spans 500-bp boundaries of the size-averaged ATAC-peak (Fig. 2A).
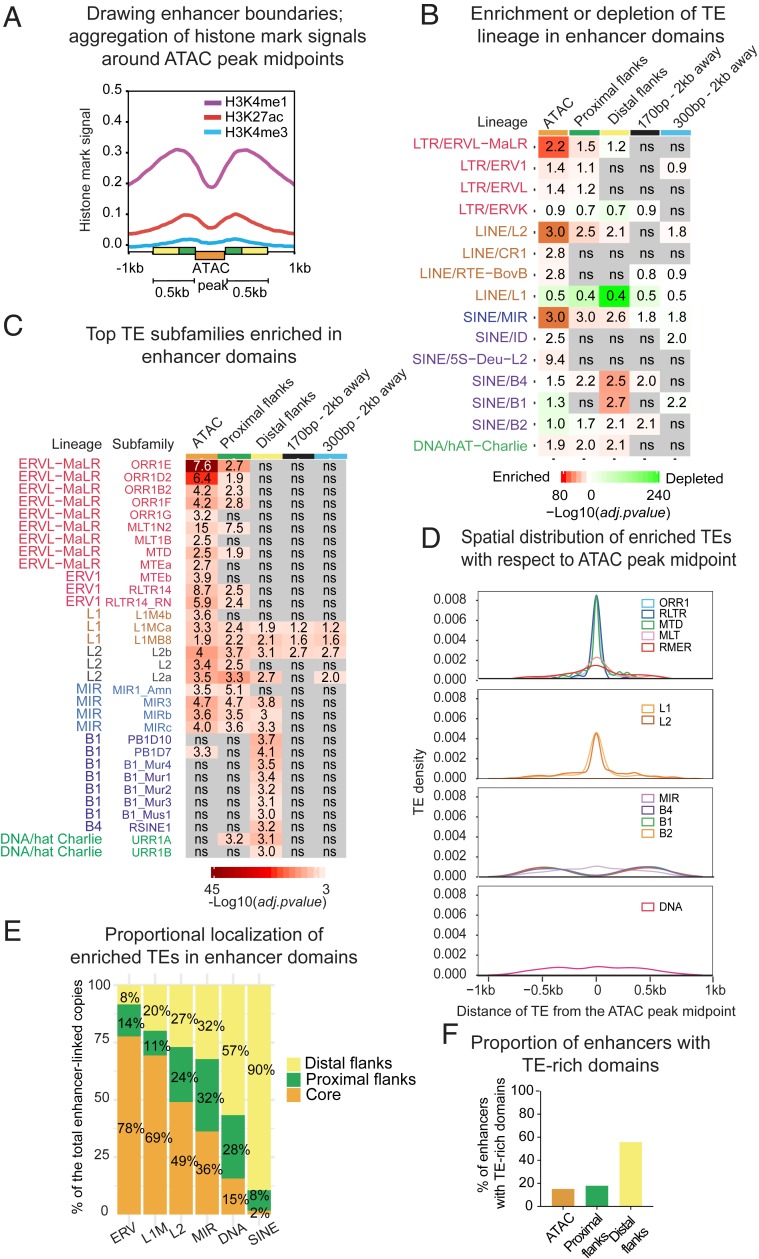
Fig. 2.
Specific lineages of transposable elements are differentially enriched and distributed within CD8+ T cell enhancers. (A) Signal aggregation for histone marks H3K27ac/H3K4me1 (GSE95237 series), and H3K4me3 (in-house) around ATAC peak midpoints in naïve (D0) cells. Enhancer domains are represented by colored boxes; size-averaged ATAC peak (300 bp), proximal flanks (170 bp), and more distal flanks (330 bp). (B) Heatmap showing significance of enrichment (red) or depletion (green) vs. genome of TE family according to the binomial two-tailed test inferred on TE counts in enhancer domains and in control size-matched regions located 2 kb away from the enhancer. Labels represent fold-change ratio in-set vs. in-genome (by length). ns = not significant. (C) Heatmap showing significance of enrichment vs. genome of TE subfamilies in enhancer domains inferred on the number of counts using standard hypergeometric test. Labels represent fold-change ratio in-set vs. in-genome (by length). ns = not significant. (D) Distribution of distance between enhancer-enriched TEs and the ATAC peak midpoint (related subfamilies are combined into groups). (E) Proportional localization of enhancer-enriched TEs within distinct enhancer domains (related subfamilies are combined into groups). (F) Proportion of enhancers enriched for TE subfamilies in ATAC-peaks, proximal, and distal flanks. Shown are proportions from the total number of identified enhancers.
These observations suggest that enhancers are composed of the three distinct domains: An accessible core (ATAC peak, orange label in Fig. 2A), proximal flanks (170 bp, green labels in Fig. 2A), and distal flanks (330 bp, yellow labels in Fig. 2A). Although this separation is somewhat arbitrary, it fits enhancer biology, with most accessible chromatin—including proximal nucleosomes—serving as the epicenter for TF binding (25). Least-accessible flanks harbor enhancer signature histone marks and may contribute to TF binding by providing favorable sequence environment and DNA shape (30–33), as well as host sites of attachment to nuclear scaffold/matrix (S/MARs) carrying a variety of regulatory codes (34, 35).
To investigate the contribution of TEs to different domains, we intersected TE annotations from the RepeatMasker with enhancer coordinates. To cut through pervasive association and to reveal potentially functional TEs, we compared proportions of the major TE lineages in enhancer domains with their genomic abundance. In parallel, we analyzed local genomic neighborhood, represented by 300-bp and 170-bp regions 2 kb away from enhancers that roughly match by size the ATAC peak (300 bp on average) and distal flanks (330 bp), as well as proximal flanks (170 bp). This analysis showed that TEs are overrepresented in enhancer domains in a lineage-specific way. Accessible cores are significantly enriched in ERVs, in particular in the mammalian apparent LTR retrotransposons (MaLRs), LINEs L2, and ancient SINEs belonging to the mammalian interspersed repeats (MIR) (Fig. 2B). Both their significance and fold-enrichment over background decrease while moving away from the enhancer core. In contrast, other SINE families, in particular the most numerous rodent-specific B1s, are depleted from cores but enriched in distal flanks. LINEs L1s, although being most abundant in the mouse genome, are depleted from enhancers altogether. Certain lineages, such as MalRs, L2s, MIRs, and hat-Charlies, are recurrently linked to human and mouse enhancers, likely reflecting their regulatory propensity in a variety of biological contexts (13–15, 36).
For a better resolution within large families, we performed similar tests for subfamilies, applying statistics to their enrichment in enhancer domains vs. genome by both length and copy number. To ensure that enriched subfamilies are associated with enhancers specifically, we sampled size-matching regions (170 bp and 300 bp) 10,000 times genome-wide. No significant association of the enhancer-enriched subfamilies with random regions was observed, except for a few L1M species. After removing them, we obtained 84 subfamilies (among 1,200 in total) enriched in enhancer domains both by length and by copy number (see enriched subfamilies in Dataset S1 and random sampling results in Dataset S2). The most enriched are subfamilies belonging to ORR1, MTE, and MTD species of ERVs, L2s, and MIRs, as well as B1, B2, and B4 SINEs (see Fig. 2C for the most numerically abundant subfamilies). Most L1s are depleted from enhancers, but 11 (of 300) L1M subfamilies are significantly enriched. Recently, the link between immunity and ORR1Es, MTEa and -b, and MIR3s has been highlighted by their enrichment in enhancers of dendritic cells (37).
Subfamilies demonstrate lineage-specific enrichment preferences, with ERVs enriched in cores and B1, B2, and B4 SINEs, in distal flanks (Fig. 2C). Because MIR species enrich all enhancer domains, in contrast to other SINEs, mostly found in distal flanks, we separated MIRs from other SINEs in the subsequent analyses. The majority of overrepresented subfamilies are not enriched in the local genomic environment (Fig. 2C), except for longer LINEs, which nevertheless lose enrichment while moving away from the core. In concordance with the enrichment, TEs are distributed within enhancers in a lineage-dependent way. Distinct groups of ERVs, LINEs, and MIRs are positioned toward the most accessible chromatin, while other SINE species favor flanks (Fig. 2D). More precisely, nearly 80% of ERVs are located in most accessible regions, while up to 90% of SINEs are in distal flanks (Fig. 2E).
For further analyses, we defined enhancers as “TE-rich” only if they contain at least one copy of a TE belonging to a subfamily enriched in one or more enhancer domains. All other enhancers were qualified as “TE-poor,” even if they overlapped with TEs (from nonenriched subfamilies). Finally, 60% of enhancers were defined as TE-rich, including 15% TE-rich in cores, and 56% in flanks (Fig. 2F). While SINE-rich enhancers numerically dominate, 10% include TEs from different lineages (SI Appendix, Fig. S5A).
Enrichment tests applied to the developmental clusters reveal a stronger association of TEs with the D0-specific enhancers, as compared to the D3-specific and invariant enhancers (SI Appendix, Fig. S5B). TE-rich enhancers in naïve cells seem to be more responsive to stimulation, as higher proportions of ERV- and LINE-containing enhancers lose accessibility during differentiation, as compared to TE-poor enhancers (SI Appendix, Fig. S5C).
Altogether, these results demonstrate an enrichment of specific TE subfamilies in CD8+ T cell enhancers and their particular topology. They also suggest that some species might have been coopted to specifically contribute to the maintenance of naïve T cell homeostasis.
TE Species Enriched in Enhancers Differentially Associate with Domain-Specific Chromatin Features.
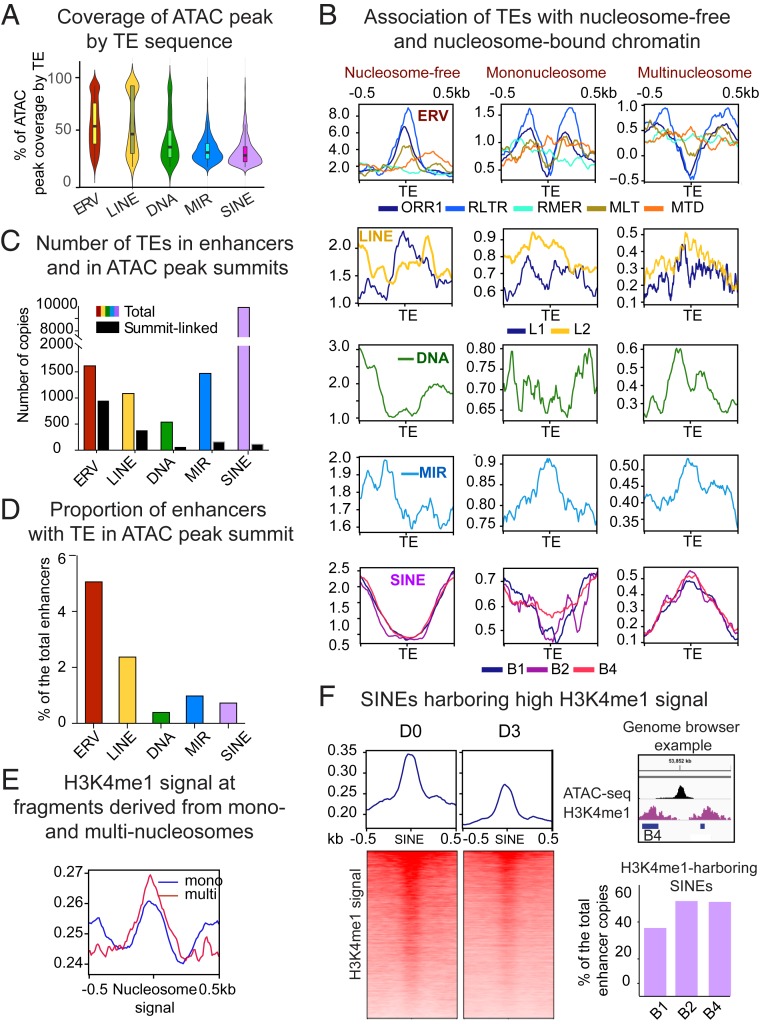
To investigate the possible contribution of TEs to enhancer core domains, we next analyzed copies directly overlapping ATAC peaks. Since related TE species show a similar distribution patterns, we combined them into higher hierarchical groups. As shown in Fig. 3A, only a small proportion of TEs can fully cover the ATAC peak. ERVs and LINEs on average cover up to 50% of the ATAC peak, while other species, either because they are smaller or further away, contribute less. Thus, most accessible chromatin domains are composed of chimeric TE/non-TE DNA. As TEs can span both open and nucleosome-occupied regions, we quantified their association with ATAC sequencing fragments derived from shorter nucleosome-free regions (harboring the highest signal), and longer fragments containing mononucleosomes (less accessible chromatin) and multinucleosomes (condensed chromatin). Typically, mononucleosomes are closed to the open chromatin, while nucleosomes from longer fragments are further away (25). This analysis shows that ERVs, in particular the most numerous ORR1 species, avoid nucleosomes and mostly overlap with nucleosome-free reads (Fig. 3B), except for MTD and RMER species that may span nucleosomes. LINEs overlap with all types of fragments by virtue of their length. MIRs are associated with both mononucleosomes and multinucleosome fragments, again contrasting other SINE species, mostly associated with multinucleosome reads (Fig. 3B).
Fig. 3.
TEs enriched in CD8+ T cell enhancers differentially contribute to chromatin states. Related TE lineages are hierarchically grouped to produce. (A) Violin plot showing distribution of ATAC-peak coverage by TE sequence (percent of ATAC peak length composed of TE). Median values are indicated. (B) TE-centered coverage plots showing association of enhancer-enriched TEs with ATAC-seq fragments derived from nucleosome-free, mono-, or multinucleosomes (di- and trinucleosomal fragments are combined). (C) Bar plot showing total number of enriched TEs in enhancers (colored bars) and TEs overlapping with ATAC peak summits (black bars). (D) Bar plot showing proportion of enhancers with TE in ATAC-peak summit. (E) Coverage plot showing intensity of H3K4me1 signal (GSE95237 series) in ATAC sequencing fragments derived from mono- or multinucleosomes. (F, Left) Heatmaps representing bins per million mapped reads-normalized H3K4me1 signal plotted over enhancer-linked SINEs harboring higher reads, as compared to adjacent regions. (Upper Right) A genome browser example of the H3K4me1-loaded B4 SINE. Upper bar plot shows proportion of the most numerically abundant enhancer-enriched SINE species targeted by H3K4me1.
Association of core TEs with nucleosome-free sequencing reads suggests that they may provide origins of chromatin accessibility. For each enriched TE lineage, we quantified the overlap with the ATAC peak summit. Over half of ERV copies underlie summits, while this proportion decreases for other TE species (Fig. 3C). As a result, ERVs are present in summits of the higher proportion of enhancers, as compared to other TE species (Fig. 3D). Overall, 10% of enhancer chromatin accessibility seems to be TE-originated (Fig. 3D).
We next studied the contribution of flanking TEs to boundary epigenetics. H3K4me1 signal is stronger at multinucleosome fragments, as compared to single nucleosomes (Fig. 3E). As SINEs also colocalize with these fragments, we quantified their association with the H3K4me1. We reasoned that a positive ratio between the H3K4me1 signal on SINE elements and adjacent nucleosome-sized flanks would indicate a direct deposition of this histone mark on SINE-bound nucleosomes (higher H3K4me1 signal in SINEs compared to surrounding regions is illustrated in Fig. 3 F, Left and Upper Right). As shown in Fig. 3 F, Lower Right, for nearly half of the copies of the most abundant SINE species, this ratio is positive. In total, 25% of enhancers are enriched for these SINEs. This suggests that flanking SINEs may contribute to nucleation and spreading of the H3K4me1, which has been shown to bind important chromatin modifiers (38). H3K4me1 signal at these SINEs decreases during activation (Fig. 3 F, Left), suggesting their putative developmental role. Altogether, these results highlight substantial contribution of enhancer-enriched TEs to building open chromatin and boundary regions.
TEs Enriched in Enhancers Carry Ancestral Regulatory Motifs.
As the majority of genomic transposons are truncated and diverged from consensus sequence during evolution, we identified constituent parts of the enhancer-enriched TE subfamilies that overlap with enhancer domains. To compare them with inaccessible TEs, we analyzed in parallel the equivalent subfamilies located in gene-poor regions (over 200 kb from the annotated TSS) with lower enhancer content and less accessible chromatin. Similar to genomic ERVs, enhancer-linked counterparts are reduced to solo LTRs (viral promoters) (SI Appendix, Fig. S6A). They, however, systematically show higher preservation of the consensus sequence (SI Appendix, Fig. S6A) and longer length (by 70 bp on average) (SI Appendix, Fig. S6B). As predicted, LINEs are mostly truncated to 3′UTRs and strongly diverged from the consensus. Only a small proportion of younger L1Ms carry promoter parts (5′UTRs). Enhancer-linked copies, however, are strikingly longer (by hundreds of base pairs) than their counterparts from gene deserts. Relatively young B1, B2s, and B4s show high degrees of conservation and almost no difference from their genomic equivalents (SI Appendix, Fig. S6 A and B).
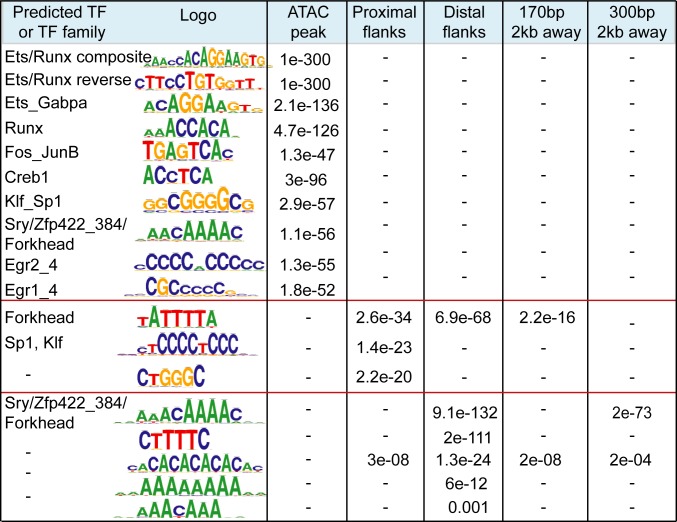
Since transposons are proficient in binding multiple TFs (16–20, 39) and some of them have preserved original promoters that may contain ancestral regulatory elements, we performed a search for TF-recognition motifs. We first analyzed domains of all CD8+ T cell enhancers (pooled from distinct stages of differentiation) and then enhancer-enriched TEs. The Regulatory Sequence Analysis Tool (RSAT) (40) helped to reveal that core domains specifically harbor the majority of recognition motifs for various TFs important for T cell biology (Fig. 4). Sequences recognized by ETS and RUNX domain-binding proteins are the most abundant. ETS1 and GAPBA (ETS domain) and Runx1 (RUNX domain) are linked to the key T cell functions (41–44). Neither enhancer flanks nor the size-matched control regions from the local genomic neighborhood are enriched for these motifs (Fig. 4). The sequence predicted to bind the pleiotropic transcriptional activators Sp1 and Sp1-like Klf (Kruppel-like Zinc finger proteins) is overrepresented in proximal flanks, together with AT-rich motifs (in both flanking domains), with putative affinity to Forkhead family proteins that play various context-dependent functions in immune cells (45), as well as A-favoring Zinc finger proteins. The main overall flanking feature is the abundance in A- and T-rich repeats. Some of these repeats (ATTTA or AACAAA) are enriched in S/MARs (46), suggesting that boundaries of CD8+ T cell enhancers may foster sites of the attachment to nuclear matrix.
Fig. 4.
Distinct sequence composition in enhancer cores and flanks. Sequence enrichment analysis using RSAT for enhancer domains and size- and number-matching control regions 2 kb away from enhancer. Shown are logos of the most enriched sequences, predicted TF or TF family, and e-values of enrichment. Recognition motifs are embedded within enriched sequences in direct or reverse orientation. Dash indicates that the motif is not found.
Overall, CpG dinucleotide frequency in flanks is lower than in cores (0.08 in core vs. 0.05 in distal flanks), consistent with a recent report showing that the low-flanking CG content is a hallmark of human enhancers associated with immune functions (47). dA:dT-rich environments carry regulatory properties and yield specific DNA shapes sensed by certain types of TFs (48), as well as dictate nucleosome positioning (49). This particular flanking composition seems to be enhancer-specific since the majority of sequences are not enriched in enhancer vicinity. The cross-comparison of similar numbers of the D0- and D3-specific enhancers (using counterpart as a background) reveals that the D0-specific cluster is enriched in binding motif for Tcf7, a critical TF for naïve and memory T cell-related functions (50) and AT-rich flanking sequences of various lengths and An:Tn periodicity, which are positionally biased toward flanks (SI Appendix, Fig. S6C). In contrast, the D3-specific cluster outnumbers by activation-linked Fos-Jun motifs and various CG-rich sequences that include Sp1/Klf motifs, all positioned toward enhancer cores (SI Appendix, Fig. S6C).
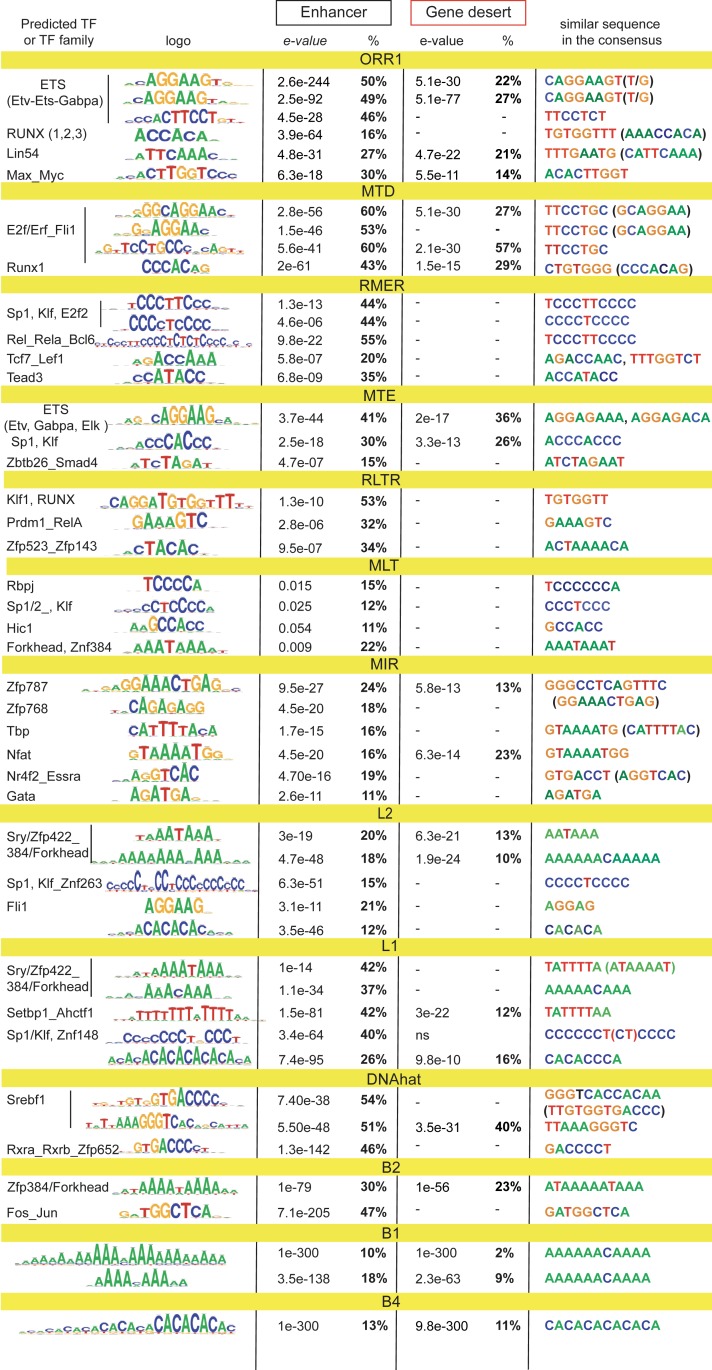
To study the putative contribution of TEs to TF-binding, we performed sequence analysis of the enhancer-enriched TEs and their subfamily equivalents from gene deserts, pooling them into phylogenetically related groups. This reveals that a high proportion of core TEs, in particular ERVs, carry recognition motifs for various TFs, including those for ETS and Runx domain proteins (Fig. 5). Both L1s and L2s bear Sp1-Klf and other Zinc finger protein motifs. MIRs also carry several TF recognition motifs, in contrast to flanking SINEs that are almost uniquely enriched for dA:dT-rich sequences (including CA repeats), which are predicted to be favored by A-prone proteins. Similar dA:dT-rich sequences are overrepresented in LINEs that span flanks. These sequences almost perfectly match enhancer dA:dT flanking lexicon. Nearly all motifs enriched in enhancer-linked TEs are present in the corresponding consensus, including A-rich tails of SINEs (51). The Fos-Jun binding motif found in B2s has been presumably evolved together with enhancers because it is not present in B2 consensus sequences. The majority of enriched motifs are absent or proportionally decreased from TE counterparts from gene deserts, suggesting a putative selective advantage for T cell cis-regulation.
Fig. 5.
Enriched TEs carry regulatory motifs. Sequence analysis (RSAT) performed on enhancer-enriched TE subfamilies and their equivalents from gene-poor regions (>200 kb away from the TSS). Related subfamilies combined into bigger groups. Shown are logos of the sequences most enriched vs. background, predicted TF, e-values of enrichment, proportion of TEs carrying at least one sequence, and the same or highly similar motifs in the TE consensus from the Dfam database. In case of opposite orientation, reverse-complement of the sequence is shown in brackets. Dash indicates that the motif is not found (in any orientation).
Since the A-rich Forkhead-like motifs are inherent to many TEs, we examined their capacity to bind Forkhead protein Foxo1, a major regulator of T cell homeostasis (45). To this end, we exploited the public dataset of ChIP-seq for Foxo1 performed on naïve CD4+ T cells (52), whose transcriptome is fundamentally similar to the one of naïve CD8+ T cells (53). We find a remarkable overlap of Foxo1 peaks with the promoter- and active enhancer-linked ATAC peaks in naïve (D0) cells (SI Appendix, Fig. S7A), confirming the biological relevance of the dataset. Our quantification shows that various TE species colocalize with Foxo1 peak summits in enhancers (SI Appendix, Fig. S7B), providing altogether 20% of binding sites. This suggests substantial participation of TEs in the maintenance of naïve cell homeostasis.
In sum, we show that enhancer core-linked TEs carry a rich array of ancestral motifs recognized by TFs relevant to T cell biology. These motifs are better preserved in enhancer-core TEs compared to nonaccessible TE equivalents. Flanking TEs carry sequences matching the inherent enhancer flanking lexicon, particularly featured in naïve cells, and may putatively contribute to sequence-based boundary regulation.
The Link Between TEs and Immune Functions.
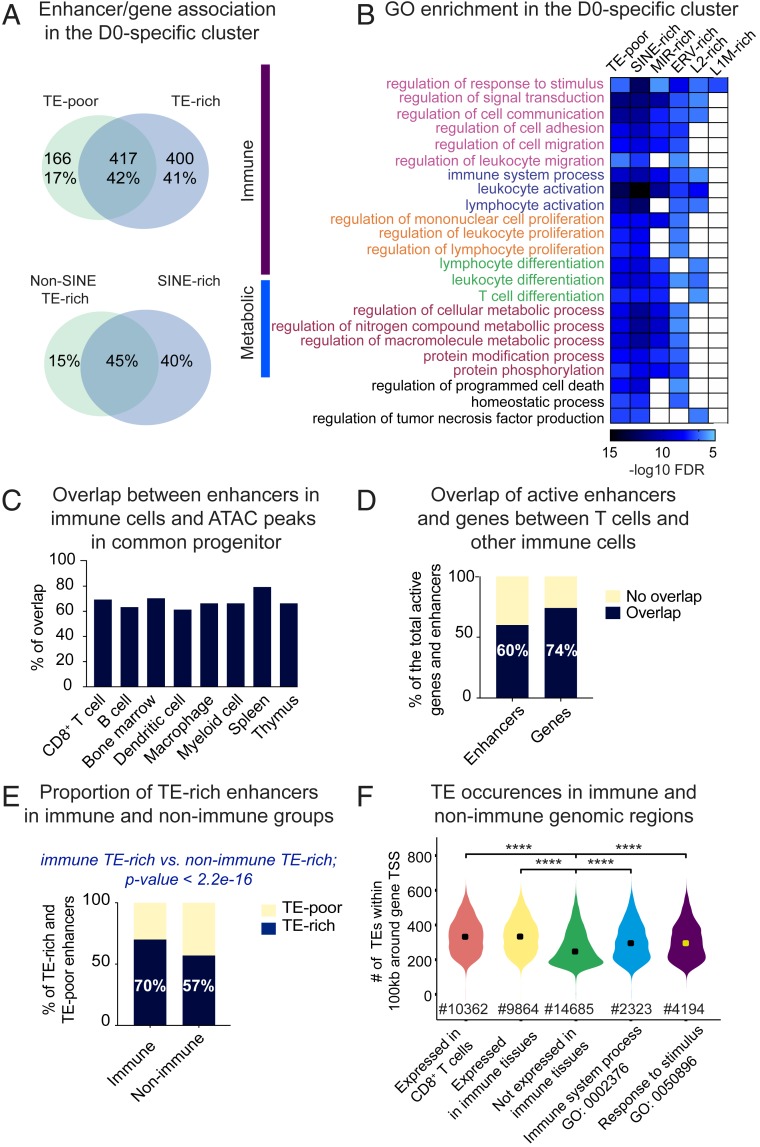
To identify transcriptional programs putatively rewired by TEs, we predicted enhancers’ gene targets via the Genomic Regions Enrichment of Annotations Tool (GREAT) (54). Using matching RNA-seq, we kept only developmentally comodulated enhancer/gene pairs (modulation of enhancer accessibility [by ATAC-seq] and change in gene transcription [by RNA-seq] are in concordance). This strategy allowed coupling of nearly 1,000 D0-specific genes to developmentally cobehaving enhancers (Dataset S3). Over 40% of the gene targets are shared between TE-rich and TE-poor enhancers, suggesting a high degree of putative enhancer redundancy (Fig. 6 A, Upper). A substantial proportion is uniquely assigned to TE-rich enhancers. Despite SINEs outnumbering other TE families, the majority of genes are shared between enhancers rich in SINEs and rich in other TE types (Fig. 6 A, Lower). Nearly one-third of the predicted enhancer/gene pairs in the D0-specific cluster has been previously identified by Hi-C (2) performed on naïve CD8+ T cells, likely reflecting a direct physical contact (Dataset S4). SI Appendix, Fig. S8A illustrates the loci of Irf1 and FbxI5 genes and corresponding TE-rich enhancers confirmed by Hi-C.
Fig. 6.
Link between TEs and immune functions. (A, Upper) Venn diagram depicting overlap between genes assigned to TE-poor and TE-rich enhancers in the D0-specific cluster. Shown are gene numbers and proportion of the total linked to enhancers in the D0-specific cluster. (Lower) Venn diagram depicting proportional overlap between genes assigned to enhancers enriched in numerically dominant flanking SINEs (without MIRs) and other TE species (pooled). (B) Heatmap showing enrichment of GO biological processes associated with TE-poor and distinct categories of TE-rich enhancers (split into lineages). GO terms are divided into two major functional classes, immune and metabolic. Same color is given to the hierarchically related processes, which are ranked by descending order starting with the parental term. (C) Bar plot showing proportion of enhancers in distinct immune lineages overlapping with ATAC-seq peaks in HSC (ENCFF190DGJ). (D) Bar plot showing overlap between enhancers and genes (fragments per kilobase of transcript per million mapped reads 0.5) active in CD8+ T cells (in-house datasets) and enhancers (Enhancer Atlas 2.0) and genes [Lara-Astiaso et al. (55)] active in other immune cells. (E) Barplot showing proportion of TE-rich among enhancers unique to immune tissue and unique to nonimmune tissues. Results of the Fisher’s exact test is shown to illustrate significantly higher proportion of TE-rich enhancers in the “immune” population, as compared to the “nonimmune” population. (F) Violin plot showing number of TE instances around genes (±100 kb from the TSS) expressed or not in CD8+ T cells and all other immune cells, as well as genes enriched in GO “immune system process” and its higher hierarchy, “response to stimulus.” Significance was assessed using the pairwise t test. ****P 0.001.
Gene ontology (GO) enrichment analysis performed on genes linked to TE-rich and TE-poor enhancers (split by TE lineage) showed that overrepresented functional terms mostly divide into two functional classes: Immune and metabolic (Fig. 6B). All of the enhancer categories are enriched for the “regulation of response to stimuli” biological process. The majority of them associate with various semiredundant processes linked to leukocyte and lymphocyte biology, such as migration, adhesion, and differentiation. LINE-linked enhancers themselves are restricted to a few processes, perhaps due to their lower number. ERV-, MIR-, and SINE-rich enhancers functionally overlap with each other and with TE-poor enhancers. Despite a high proportion of genes uniquely linked to TE-rich enhancers, functional patterns highly overlap between the distinct enhancer categories. This suggests that TE-rich enhancers may have complemented regulatory branches to each process instead of rewiring innovative functions. Similarly, D3-specific TE-rich and TE-poor enhancers show high functional overlap in leukocyte and immune effector functions (T cell activation, cytokine production) (SI Appendix, Fig. S8B). In the invariant cluster, ERV- and LINE-rich enhancers alone are not associated with any particular term, while MIR and other SINE-type–rich enhancers are linked to metabolic functions related to protein, DNA, and RNA turnover (SI Appendix, Fig. S8C). Several processes—for example, catabolic and “chromosome localization”—are uniquely assigned to SINE-bearing enhancers, suggesting their possible contribution to the evolution of specific housekeeping processes.
Association with leukocyte functions suggests that CD8+ T cell enhancers may originate from precursor lineages. To address this, we exploited the public dataset of ATAC-seq performed on hematopoietic stem cells (HSC, a common precursor of all immune lineages) (Encode ENCFF190DGJ). We observed a high proportion of CD8+ T cell enhancers and enhancers found in other immune cell types (extracted from the Enhancer Atlas 2.0; http://www.enhanceratlas.org/index.php) premarked in progenitors (Fig. 6C). This suggests that HCS-accessible chromatin is a major source of enhancers for immune tissues. Besides, CD8+ T cell enhancers overlap by 60% with enhancers pooled from other immune lineages (51,000 in total) (Fig. 6D). Moreover, a high proportion (74%) of genes expressed in CD8+ T cells are shared with genes expressed in other immune lineages (Fig. 6D) [data obtained from the publicly available RNA-seq analysis of all immune lineages (55)]. Despite differential enhancer activation and gene expression during lineage commitments (55), altogether this suggests that a high proportion of genomic regions evolved to broadly support immune functions.
To investigate whether these regions are different from other genomic regions, we extracted enhancer coordinates from 14 nonimmune tissues (listed in SI Appendix, Fig. S9A) from the Enhancer Atlas database 2.0. Cross-tissue comparison revealed 25,000 enhancers unique to immune tissues (immune enhancers) and 70,544 unique to nonimmune tissues (nonimmune enhancers). The majority of genes collectively expressed by immune cells (60%) colocalize with immune enhancers (within 100 kb around the TSS). To address putative TE cooption, we applied TE subfamily enrichment tests. Because it was impossible to precisely map centers and define domains, we trimmed public enhancer coordinates down to ±750 bp from the midpoint, obtaining uniform 1,500-bp regions. Significance of TE enrichment was inferred on TE copies located within loosely defined enhancer cores (400 bp around midpoint) and the full enhancer. This test revealed a significantly higher number of immune enhancers as TE-rich, as compared to the nonimmune enhancers (70% and 58%, respectively) (Fig. 6E). Specific subfamilies of ORR1s, RLTR19s, L1Ms, and B1Mus are particularly enriched in the population of immune enhancers, as compared to the nonimmune enhancers (SI Appendix, Fig. S9B). As these TE species are also overrepresented in CD8+ T cell enhancers, we compared occurrences of all enriched TE subfamilies in genomic regions containing genes expressed or silent in CD8+ T cells (within 100 kb around the TSS). There was a higher number of TEs around expressed genes and the “immune system process” genes (GO: 0002376), as compared to regions hosting genes expressed weakly or not expressed in CD8+ T cells (SI Appendix, Fig. S9C). Moreover, the total number of all TEs around genes expressed in CD8+ T and other immune cells (including genes of the “response to stimulus” and the “immune system” processes) is higher, as compared to “silent-gene” regions (Fig. 6F).
These results show that TE-rich enhancers in CD8+ T cells putatively rewire complementary branches of biological processes related to leukocyte and lymphocyte biology and that they are originated from the pool of enhancers supplied by HSC to distinct immune lineages. Enhancers unique to immune tissue encompass more TEs, as compared to enhancers unique to other tissues, and this correlates with higher TE occurrences around genes expressed in immune cells, as compared to unexpressed genes.
Discussion
Evolution of enhancers from the genomic background is assumed to be accompanied by the functionalization of regulatory sequences derived from transposons. TEs have supplied mammalian enhancers with multiple built-in regulatory motifs, contributing substantially to the shaping of cis-regulation in a variety of human and mouse tissues (10). While unique TE-derived regulatory cues have not yet been reported to our knowledge, functional TE cooption is assumed to accelerate the evolution of regulatory networks (10, 23, 24). Widespread TE copies carrying similar regulatory cues have the potential to rapidly rewire remotely located genes into novel coordinated transcriptional patterns. Specific TEs have participated in the emergence of functional innovations, such as mammalian pregnancy (14) and inflammatory innate immune response (21).
Immune cells, permanently adapting to novel stimuli, may have drawn a particular benefit from rapid TE-based adaptive strategies. We have performed a genome-wide analysis of the contribution of TEs to immune-related enhancers. Our findings suggest that enhancers specific to immune tissues are more prone to a putative TE cooption, as compared to enhancers specific to other tissue types. We show that different hematopoietic lineages have overlapping transcriptomes and employ a largely shared enhancer repertoire. A major source of these enhancers is a common hematopoietic progenitor. Even if the levels of gene expression and the enhancer engagement are variables between immune lineages (55), this shared regulatory pool suggests that during evolution, specific genomic regions have been tailored to serve immune functions. These regions differ from other gene-rich regions in several ways. First, genes responding to stimuli, including immune genes, evolve at a fast pace (56, 57). Second, we observed higher TE occurrences in the local environment of genes expressed in immune cells, as compared to silent genes. The forces that drive such biased TE representation in immune-associated genomic regions remain unclear. Reduced purifying selection is likely to be the primary determinant for the local accumulation of TE insertions. The efficiency of purifying selection may be weakened by low recombination rate, which is known to positively associate with a regional accumulation of transposons (58). It is also possible that immune genes have evolved under strong diversifying selection, resulting in nearby bystander mutations propagated through genetic linkage. Or else, TEs—in particular LINEs and SINEs—may have accumulated because of the immune-linked gene maintenance. Our observations suggest that reduced purifying selection alone is unable to explain higher proportion of TE-rich immune enhancers compared to nonimmune. We suggest that transposons carrying regulatory sequences beneficial for immune-related functions have been actively selected postintegration. Indeed, ancestral regulatory motifs are better preserved in enhancer-enriched TEs compared to their counterparts located in gene-poor regions. Regional abundance could have facilitated selection and cooption of regulatory transposons for the rewiring of immune transcriptional networks.
In CD8+ T cells, specific TE subfamilies are overrepresented either in enhancer cores or boundaries and contribute with sequences typical to these domains (TF-recognition and architectural motifs, respectively). TEs are mostly included in enhancers in an additive manner, forming TE/non-TE chimeras, while directly originating open chromatin in only 10% of enhancers. Nonrandom topology of enhancer-enriched TE lineages (ERVs favoring most accessible, while SINEs favor the least accessible chromatin) suggests distinct functional tasks, with ERVs putatively participating in TF binding, while SINEs contributing to “hardware” enhancer features. Indeed, A-tails of SINEs might be beneficial for enhancer flanks, as they match an overall AT-rich flanking lexicon. Such a sequence composition is able to physically yield particular DNA conformation (59), disfavoring local nucleosomal occlusion (49) and attracting specific TFs (30–33). It has been shown that B1-like human Alu SINEs can influence nucleosome positioning (60). Furthermore, enhancer-linked SINEs are enriched in sequences previously linked to S/MARs, which host versatile regulatory signals (46, 61) and where TEs have already been observed (62). It is possible that “well-positioned” SINEs passively participate in sequence-specific functions, although their enrichment within enhancers may argue for active selection. We show that SINEs harbor the highest H3K4me1 signals at one-quarter of CD8+ T cell enhancers. Because this histone mark binds important chromatin modifiers (38), flanking SINEs may provide platforms for chromatin rearrangement not only via physical but also biochemical properties of their sequences.
By contributing with ready-made regulatory motifs, TEs could have accelerated the formation of immune regulatory networks. In CD8+ T cells, we observe a high overlap between regulation computationally assigned to TE-rich and TE-poor enhancers. TE-rich enhancers share a cohort of putative gene targets with TE-poor enhancers. This likely reflects enhancer redundancy described as a widespread feature of mammalian genomes (63, 64). It is assumed to provide a sort of “regulatory buffer” to protect from the loss of individual enhancers (63, 64). TEs may also contribute to secondary enhancers, shown to protect phenotypic integrity only in extreme conditions (64). Besides sharing genes with TE-poor, TE-rich enhancers associate with selected gene group. Hypothetical contribution of TEs to rewiring these genes did not result in functional innovation. Rather, it nurtured biological processes similar to those linked to TE-poor enhancers. It is tempting to speculate that such regulatory arrangement may embrace high functional redundancy (different elements with similar functions organized into buffering network) described as another major trait of robust biological systems, protecting from environmental fluctuations and yielding diversity of a response via competitive exclusion and cooperative facilitation (65). Thus, TEs (at least in CD8+ T cells), by providing ready-made regulatory sequences, may have accelerated acquisition of functional robustness.
Robustness is likely critical to immune regulatory circuits. It may prevent hasty responses and ectopic losses of homeostasis, which in the case of naïve T cells can lead to ineffective or harmful immune responses. Additionally, robustness may support the diversity of immune responses. We propose a model in which a high density of TEs in immune-associated genomic regions favored selection and regulatory cooption of functional TE-sequences. This may have accelerated evolution of immune enhancers and acquisition of robustness of immune functions.
Materials and Methods
Isolation and Differentiation of Naïve CD8+ T Cells.
C57BL/6J TCR transgenic mice (OT-I) were purchased from the Jackson Laboratory and housed in Curie Institute’s specific pathogen-free animal facility. Live animal experiments were performed according to the guidelines of the European Veterinary Department (Project Authorization N:02465.02). CD8+ T cells were obtained from lymph nodes and spleens of OT-I mice and enriched using the naïve CD8+ isolation kit (StemCell). Cell labeling was performed in PBS supplemented with 0.5% BSA and 2 mM EDTA, using antibodies against CD62L, CD44, and CD25 (BD Biosciences). Polyclonal naïve, CD62Lhigh CD44low CD25− CD8+ cell subset was FACS sorted (FACSVantage Diva, FACSAria flow cytometers; BD Biosciences) to 99% purity. To activate cells in vitro, plates were coated with 10 μg/mL of CD3ε antibody (eBioscience) and naïve cells were seeded and cultured in the RPMI medium supplemented with 10% of FCS, rhIL-2 (40 UmL Proleukin; Novartis), 2-mercaptoethanol, Pen-Strep, l-glutamine, nonessential amino acids, and 1 μg/mL of anti-CD28 antibodies (eBioscience).
ATAC-seq and ChIP-seq.
ATAC-seq was performed as in ref. 25. ChIP-seq was performed as in ref. 66. Libraries were sequenced using 100-bp paired-end reads setting. Properly paired reads were aligned to the unique locations of the mm10 genome using Bowtie2 v2.1.0. For ATAC-seq, ATACseqQC Bioconductor package (67) with default parameters was used to identify nucleosome-free and nucleosome-containing inserts. Peaks were called using MACS2 v2.0.10 with default parameters. Modulation of the enhancer chromatin accessibility across differentiation was quantified using HTseq v0.6.1 followed by DESeq2 (v1.18.0). For ChIP-Seq, peaks were called using SICER v1.1 (window size 500 bp, gap size 1,500 bp, and 5% false-discovery rate threshold). See SI Appendix for detailed information.
RNA-seq.
Total RNA was purified using the TriZol reagent (Thermo Fisher Scientific) following the manufacturer’s instructions and isolated using RNAesy Mini kit (Qiagen). Libraries were sequenced using 100-bp paired-end reads. Properly paired reads were aligned to the unique locations of the mm10 genome using Tophat v2.0.6. Gene-expression values were quantified using HTseq v0.6.1, and the differential analysis was performed using DEseq2 (v1.18.0). See SI Appendix for detailed information.
Identification of Active-Enhancers.
To reveal active enhancers, ATAC peaks were split into two groups, the promoter (±2 kb around the gene TSS) and the distal ATAC peaks (>2 kb from the TSS). Distal ATAC peaks were intersected with H3K27ac peaks (GSE95237 series). Unannotated promoters with high level of H3K4me3 (in-house dataset) were filtered out (See SI Appendix, Fig. S2B for cut-off details). H3K27ac+ distal ATAC peaks with low H3K4me3 levels were defined as putative active enhancers. See SI Appendix for detailed information
Identification of TE-Rich Enhancers.
First, the effective length of active enhancers was determined using H2K27ac (GSE95237 series), H3K4me1 (GSE95237 series), and H3K4me3 (in-house). Enhancer was defined as a region centered upon ATAC-peak, carrying the highest densities of the three histone marks and comprised of three domains: ATAC peak, proximal (170 bp, roughly equal to the size of one nucleosome in deep sequencing), and distal flanks (around 330 bp). Enhancer coordinates were intersected with TE coordinates from the RepeatMasker (v4.0.9 produced for the mm10 assembly, version GRCm38.p6) parsing by “one code to find them all” tool (68) and using BEDTools (69). The ratio between TE family or subfamily abundance (by counts or length) in-set versus in-genome was analyzed using perl script of the TE-analysis_pipeline.pl as in Lynch et al. (14). The significance of enrichment was estimated using standard binomial and hypergeometric tests. Fold-change in figures was generated on length, as better representing the contribution of a given TE. The specificity of TE subfamily association with enhancer domains was accessed by randomly sampling size-matched regions 10,000 times parsing with TE-analysis_pipeline.pl. See SI Appendix for detailed information.
Sequence Analysis.
Perl script dfamscan.pl from the Dfam database (3.0) (70) was used to identify constituent parts of the enhancer-enriched TEs. Prediction of regulatory motifs within enhancer domains or TEs was performed using the peak-motif pipeline of the RSAT (40). Overrepresented motifs were screened by TF motif databases (JASPAR core nonredundant vertebrates, 2018) and cis-BP mouse (2019-06_v2.00) to predict TF binding. Most frequent oligonucleotides in TE-sets were searched within TE consensus sequences from the Dfam database (https://dfam.org/home). See SI Appendix for detailed information.
Predicting Enhancer/Gene Pairs and Enriched Biological Processes.
Enhancers were assigned to genes using GREAT (http://great.stanford.edu/public/html/) with default parameters. Only enhancer/gene pairs comodulated during differentiation were selected (ATAC signal is comodulated with the change in gene expression). Enhancer/gene pairs in naïve cells were compared with those predicted by Hi-C from He et al. (2). Functional enrichment analysis was performed using the Gene Ontology Resource (http://geneontology.org/). See SI Appendix for detailed information.
Comparison of Enhancer Repertoires and Transcriptomes Between Tissues.
Enhancer coordinates for different mouse tissues were retrieved from the Enhancer Atlas 2.0 database. To estimate TE enrichment, coordinates were trimmed down to 750 bp from the midpoint. TE enrichment was inferred on counts in loosely defined cores (400 bp) and full enhancer regions. To normalize for dataset size differences, the bootstrap sampling strategy was applied. Genes expressed in all major hematopoietic lineages were identified using the public dataset of RNA-seq (55). See SI Appendix for detailed information.
Source of the Public Data Used in This Study.
The sources of the public data used in this study are as follows: GSE95237 series: TN_H3K27ac (GSM2357745/46), TE_H3K27ac (GSM2357747/48), TN H3K4me1 (GSM2357729/30), TE_H3K4me1 (GSM2357731/32), and Inputs for TN (GSM2357753/54) and for TE (GSM2357755/56); GSE46525 dataset: Foxo1 ChIP-Seq (GSM1131775) and Input (GSM1131776) from the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/); and ATAC-seq on HSC (ENCFF190DGJ from the Encode; https://www.encodeproject.org).
Data Availability.
Raw and processed data of ATAC-seq, ChIP-seq, and RNA-seq have been uploaded to the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/) and are available under serial accession number GSE142151.
Supplementary Material
Acknowledgments
We thank George Kassiotis and Jean-Christophe Andrau for critical reading of the manuscript and fruitful discussions; Morgane Thomas-Chollier and Leandro Quadrena for fruitful discussions; and Fiorella Kotsias for critical reading of the manuscript. This work is supported by Institut Curie, Inserm Paris Sciences et Lettres (PSL) University, and Grants ANR-11-LABX-0043 and ANR-10-IDEX-0001-02 PSL, ANR-14-CE16, INCA 2017-1-PL BIO-03-ICR-1, INCA 2018-1-RT-02-ICR-1, and HERVimmune PRT-K-17-072. High-throughput sequencing has been performed by the Institut Curie Genomix of Excellence Next-Generation Sequencing platform of the Institut Curie supported by Grants ANR-10-EQPX-03 (Equipex) and ANR-10-INBS-09-08 (France Génomique Consortium) from the Agence Nationale de la Recherche (“Investissements d’Avenir” program), by the Canceropole Ile-de-France and by the Integrated Cancer Research Site (SiRIC)-Curie program/SiRIC Grant “INCa-DGOS- 4654”.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE142151).
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912008117/-/DCSupplemental.
References
- 1.Yu B., et al. , Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat. Immunol. 18, 573–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He B., et al. , CD8+ T cells utilize highly dynamic enhancer repertoires and regulatory circuitry in response to infections. Immunity 45, 1341–1354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seumois G., et al. , Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat. Immunol. 15, 777–788 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vahedi G., et al. , Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520, 558–562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babu D., Fullwood M. J., 3D genome organization in health and disease: Emerging opportunities in cancer translational medicine. Nucleus 6, 382–393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gross D. S., Garrard W. T., Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57, 159–197 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Schones D. E., et al. , Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heintzman N. D., et al. , Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Creyghton M. P., et al. , Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuong E. B., Elde N. C., Feschotte C., Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 18, 71–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finnegan D. J., Eukaryotic transposable elements and genome evolution. Trends Genet. 5, 103–107 (1989). [DOI] [PubMed] [Google Scholar]
- 12.Thompson P. J., Macfarlan T. S., Lorincz M. C., Long terminal repeats: From parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol. Cell 62, 766–776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huda A., et al. , Prediction of transposable element derived enhancers using chromatin modification profiles. PLoS One 6, e27513 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch V. J., et al. , Ancient transposable elements transformed the uterine regulatory landscape and transcriptome during the evolution of mammalian pregnancy. Cell Rep. 10, 551–561 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jjingo D., et al. , Mammalian-wide interspersed repeat (MIR)-derived enhancers and the regulation of human gene expression. Mob. DNA 5, 14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacques P. É., Jeyakani J., Bourque G., The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 9, e1003504 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunarso G., et al. , Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 42, 631–634 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Chuong E. B., Rumi M. A. K., Soares M. J., Baker J. C., Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 45, 325–329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sundaram V., et al. , Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 24, 1963–1976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourque G., et al. , Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 18, 1752–1762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuong E. B., Elde N. C., Feschotte C., Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083–1087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trizzino M., et al. , Transposable element exaptation is the primary source of novelty in the primate gene regulatory landscape. Genome Res. 27, 1623–1633 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Britten R. J., Davidson E. H., Gene regulation for higher cells: A theory. Science 165, 349–357 (1969). [DOI] [PubMed] [Google Scholar]
- 24.Davidson E. H., Britten R. J., Regulation of gene expression: Possible role of repetitive sequences. Science 204, 1052–1059 (1979). [DOI] [PubMed] [Google Scholar]
- 25.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sexton C. E., Han M. V., Paired-end mappability of transposable elements in the human genome. Mob. DNA 10, 29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heintzman N. D., et al. , Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Abe A., et al. , An enhancer of the IL-7 receptor α-chain locus controls IL-7 receptor expression and maintenance of peripheral T cells. J. Immunol. 195, 3129–3138 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Pundhir S., Bagger F. O., Lauridsen F. B., Rapin N., Porse B. T., Peak-valley-peak pattern of histone modifications delineates active regulatory elements and their directionality. Nucleic Acids Res. 44, 4037–4051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dror I., Golan T., Levy C., Rohs R., Mandel-Gutfreund Y., A widespread role of the motif environment in transcription factor binding across diverse protein families. Genome Res. 25, 1268–1280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jolma A., et al. , DNA-binding specificities of human transcription factors. Cell 152, 327–339 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Slattery M., et al. , Absence of a simple code: How transcription factors read the genome. Trends Biochem. Sci. 39, 381–399 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levo M., et al. , Unraveling determinants of transcription factor binding outside the core binding site. Genome Res. 25, 1018–1029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gasser S. M., Laemmli U. K., Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell 46, 521–530 (1986). [DOI] [PubMed] [Google Scholar]
- 35.Jenuwein T., et al. , Extension of chromatin accessibility by nuclear matrix attachment regions. Nature 385, 269–272 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Cao Y., et al. , Widespread roles of enhancer-like transposable elements in cell identity and long-range genomic interactions. Genome Res. 29, 40–52 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donnard E., et al. , Comparative analysis of immune cells reveals a conserved regulatory lexicon. Cell Syst. 6, 381–394.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Local A., et al. , Identification of H3K4me1-associated proteins at mammalian enhancers. Nat. Genet. 50, 73–82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T., et al. , Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl. Acad. Sci. U.S.A. 104, 18613–18618 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas-Chollier M., et al. , A complete workflow for the analysis of full-size ChIP-seq (and similar) data sets using peak-motifs. Nat. Protoc. 7, 1551–1568 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Zhao J. Y., Osipovich O., Koues O. I., Majumder K., Oltz E. M., Activation of mouse Tcrb: Uncoupling RUNX1 function from its cooperative binding with ETS1. J. Immunol. 199, 1131–1141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uchino R., Domain analyses of the Runx1 transcription factor responsible for modulating T-cell receptor-β/CD4 and interleukin-4/interferon-γ expression in CD4(+) peripheral T lymphocytes. Immunology 128, 16–24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grenningloh R., et al. , Ets-1 maintains IL-7 receptor expression in peripheral T cells. J. Immunol. 186, 969–976 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo C. T., et al. , Ets transcription factor GABP controls T cell homeostasis and immunity. Nat. Commun. 8, 1062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouyang W., Beckett O., Flavell R. A., Li M. O., An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity 30, 358–371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liebich I., Bode J., Reuter I., Wingender E., Evaluation of sequence motifs found in scaffold/matrix-attached regions (S/MARs). Nucleic Acids Res. 30, 3433–3442 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lecellier C., Wasserman W. W., Mathelier A., Human enhancers harboring specific sequence composition, activity, and genome organization are linked to the immune response. Genetics 209, 1055–1071 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohs R., et al. , The role of DNA shape in protein-DNA recognition. Nature 461, 1248–1253 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Field Y., et al. , Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLOS Comput. Biol. 4, e1000216 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xing S., et al. , Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nat. Immunol. 17, 695–703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy-Engel A. M., A tale of an A-tail: The lifeline of a SINE. Mob. Genet. Elements 2, 282–286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stone E. L., et al. , ICOS coreceptor signaling inactivates the transcription factor FOXO1 to promote Tfh cell differentiation. Immunity 42, 239–251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Godec J., et al. , Compendium of immune signatures identifies conserved and species-specific biology in response to inflammation. Immunity 44, 194–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLean C. Y., et al. , GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lara-Astiaso D., et al. , Immunogenetics. Chromatin state dynamics during blood formation. Science 345, 943–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Obbard D. J., Welch J. J., Kim K. W., Jiggins F. M., Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 5, e1000698 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rech G. E., et al. , Stress response, behavior, and development are shaped by transposable element-induced mutations in Drosophila. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kent T. V., Uzunović J., Wright S. I., Coevolution between transposable elements and recombination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160458 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harteis S., Schneider S., Making the bend: DNA tertiary structure and protein-DNA interactions. Int. J. Mol. Sci. 15, 12335–12363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka Y., Yamashita R., Suzuki Y., Nakai K., Effects of Alu elements on global nucleosome positioning in the human genome. BMC Genom. 11, 309 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liebich I., Bode J., Frisch M., Wingender E., S/MARt DB: A database on scaffold/matrix attached regions. Nucleic Acids Res. 30, 372–374 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Sternberg R., Shapiro J. A., How repeated retroelements format genome function. Cytogenet. Genome Res. 110, 108–116 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Osterwalder M., et al. , Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frankel N., et al. , Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitacre J. M., Biological robustness: Paradigms, mechanisms, and systems principles. Front. Genet. 3, 67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allan R. S., et al. , An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature 487, 249–253 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Ou J., et al. , ATACseqQC: A bioconductor package for post-alignment quality assessment of ATAC-seq data. BMC Genom. 19, 169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bailly-Bechet M., Haudry A., Lerat E., “One code to find them all”: A perl tool to conveniently parse RepeatMasker output files. Mob. DNA 5, 13 (2014). [Google Scholar]
- 69.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hubley R., et al. , The Dfam database of repetitive DNA families. Nucleic Acids Res. 44, D81–D89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data of ATAC-seq, ChIP-seq, and RNA-seq have been uploaded to the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/) and are available under serial accession number GSE142151.