Significance
The cell surface proteome (surfaceome) mediates interactions between the cell and the extracellular environment and is a major target for immunotherapy in cancer. Here, we compared how six neighboring proliferative oncogenes cause large and bidirectional change in expression of some 700 surface proteins. These large changes converge to common functional consequences that are reversed by small-molecule inhibition of the MAPK pathway. We further complemented the surfaceome analysis with bottom-up glycoproteomics enabled by activated ion electron transfer dissociation and found a dynamic regulation of the glycoproteome. This large-scale comparative study provides important insights for how oncogenes remodel isogenic cells in a cell autologous fashion and suggests opportunities for antibody drug discovery in cancer.
Keywords: oncogenes, glycoproteomics, surfaceome, MAPK signaling pathway
Abstract
The cell surface proteome, the surfaceome, is the interface for engaging the extracellular space in normal and cancer cells. Here we apply quantitative proteomics of N-linked glycoproteins to reveal how a collection of some 700 surface proteins is dramatically remodeled in an isogenic breast epithelial cell line stably expressing any of six of the most prominent proliferative oncogenes, including the receptor tyrosine kinases, EGFR and HER2, and downstream signaling partners such as KRAS, BRAF, MEK, and AKT. We find that each oncogene has somewhat different surfaceomes, but the functions of these proteins are harmonized by common biological themes including up-regulation of nutrient transporters, down-regulation of adhesion molecules and tumor suppressing phosphatases, and alteration in immune modulators. Addition of a potent MEK inhibitor that blocks MAPK signaling brings each oncogene-induced surfaceome back to a common state reflecting the strong dependence of the oncogene on the MAPK pathway to propagate signaling. Cell surface protein capture is mediated by covalent tagging of surface glycans, yet current methods do not afford sequencing of intact glycopeptides. Thus, we complement the surfaceome data with whole cell glycoproteomics enabled by a recently developed technique called activated ion electron transfer dissociation (AI-ETD). We found massive oncogene-induced changes to the glycoproteome and differential increases in complex hybrid glycans, especially for KRAS and HER2 oncogenes. Overall, these studies provide a broad systems-level view of how specific driver oncogenes remodel the surfaceome and the glycoproteome in a cell autologous fashion, and suggest possible surface targets, and combinations thereof, for drug and biomarker discovery.
The cell surface proteome, or surfaceome, is the main interface for cellular signaling, nutrient homeostasis, and cellular adhesion, and defines immunologic identity. To survive, cancer cells adjust to promote increased nutrient import, progrowth signaling, and evasion of immunological surveillance, among others (1). There are some 4,000 different membrane proteins encoded in the human genome (2, 3), yet antibodies to only about two dozen cell surface targets have been approved for therapeutic intervention, prompting the need to discover novel tumor specific antigens (4, 5).
Recent surfaceome studies in an isogenic MCF10A breast epithelial cell line transformed with oncogenic KRAS have identified more than two dozen up-regulated surface proteins that function in a cell autologous fashion to promote increased cell proliferation, metastasis, metabolic activity, and immunologic suppression (6–8). Many of the most powerful oncogenes are linked to KRAS and the MAPK pathway including overactivation of receptor tyrosine kinases, such as EGFR and HER2, or mutations in BRAF or RAS (Fig. 1A). It is well known that these neighboring oncogenes are mutually exclusive in human tumors. For example, lung cancer patients with oncogenic EGFR mutations seldom harbor oncogenic KRAS or BRAF mutations and vice versa (9). Also, recent evidence shows that oncogene coexpression can induce synthetic lethality or oncogene-induced senescence, further reinforcing that activation of one oncogene without the others is preferable in cancer (10, 11).
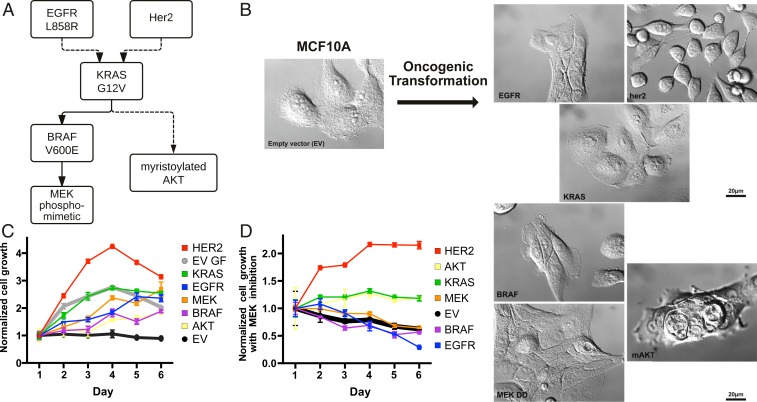
Fig. 1.
Growth rates and morphologies for MCF10A cells transformed with neighboring proliferative oncogenes in the MAPK pathway. (A) Simplified signaling schematic relationship of six proliferative oncogenes studied, , HER2 overexpression, , , , and . (B) Oncogenic transformation of MCF10A induces diverse cellular morphologies. Note that images are presented in the same order as the schematic in A. (C) MCF10A cells stably transformed with lentivirus with the different oncogenes grow independent of growth factor. Gray and black lines indicate cellular growth of MCF10A EV control with and without growth factors, respectively. Cell growth (n = 3) was measured each day for 6 d by CellTiter-Glo luminescent cell viability assay and normalized to viability on day 1. (D) Suppression of growth for all cell lines by treatment with 100 nM MEK inhibitor (PD0325901) in the absence of growth factors. MCF10A cells transformed with HER2 appears to be least sensitive to MEKi, followed by and .
There is also substantial evidence that cell surface glycosylation is altered in cancer. Incomplete or truncated synthesis, extended branching, core fucosylation, and sialylation of cell surface glycans are hallmarks of tumor cells, and alter physiological mechanisms of cell–cell adhesion, communication, and immune system recognition (12–18). Over the past decade, chemical glycoproteomics has revealed specific examples of altered glycosylation and heterogeneity of glycans on particular proteins (19). However, we do not know how expression of different oncogenes globally alters glycosylation on the individual proteins at a proteome-wide scale. Very recently, hybrid-type electron transfer dissociation (ETD) methods, such as activated ion ETD (AI-ETD) and ETD with supplemental activation (EThcD), have emerged as forerunners for glycoproteomic profiling, due to their ability to generate sequence-informative tandem mass spectra that link both the peptide backbone and corresponding glycan modification (20–22). These techniques can provide granularity to the altered glycosylation states of cell surface proteins upon oncogenic transformation.
Here we address how neighboring driver oncogenes (, HER2 overexpression, , , phosphomimetic , and myristoylated AKT) stably expressed in a common noncancerous isogenic epithelial cell alter the surfaceome and glycoproteome. Using cell surface capture (CSC) (23, 24) and AI-ETD glycoproteomics (21), we found that each oncogene induced common and unique sets of up- and down-regulated surface proteins and associated glycans. These sets of protein revealed common biological themes, including increased expression of nutrient transporters and decreased expression of adhesion molecules. These effects were massively reversed upon inhibition of the MAPK pathway, emphasizing its central importance in remodeling the surfaceome. These oncogene-induced surface proteins highlight targets or combinations to consider for immunotherapy.
Results
Phenotypic Analysis of Oncogene-Transformed MCF10A Cells.
Tumor biology is highly complex and varies depending on the cell type of origin, stage, epigenome, stroma, vascularization, the immune system, and metabolism. We deliberately took a simplistic reductionist approach to compare how specific proliferative oncogenes alter the cell surface proteome in a cell autologous fashion by using isogenic cells stably transformed with different oncogenes. No single cell type, culturing condition, or context is representative of all or even one cancer type. For practical reasons, we chose the spontaneously immortalized breast epithelial MCF10A as our parental cell line, because it is often used as a neutral starting point for oncogene studies (25–28). Although MCF10A certainly does not recapitulate the diversity of mammary cell biology, it is of epithelial origin like most common tumors; it is nontumorigenic, requires growth factors for survival, and does not harbor gene amplifications or other chromosomal aberrations typical of advanced cancer cell lines. Our intent is to compare how neighboring driving oncogenes remodel their surfaceomes and glycoproteomes in isogenic cells in a cell autologous fashion.
Lentivirus was used to stably transform MCF10A cells with six prevalent oncogenes that are neighbors in proliferative signaling: HER2 overexpression, , , , the phosphomimetic (), and myristoylated AKT () (Fig. 1A and SI Appendix, Fig. S1). Remarkably, the morphologies of each of the transformed cells varied from each other when cultured in the absence of growth factors, indicative of differences in proteomic landscape (Fig. 1B). In the absence of growth factors, cells transformed with HER2 and grew to confluence, while cells harboring , , and did not reach confluency, indicative of contact-dependent growth inhibition. Cells transformed with , which signals through a parallel pathway relative to the other five oncogenes, had the most dramatic morphology change, displaying vertically stacked clusters of cells. Unlike the parental cell line, all of the MCF10A cells stably transformed with any of the six oncogenes proliferated in the absence of growth factors to various degrees (Fig. 1C). The HER2- and -transformed cells proliferated most rapidly in the absence of growth factors, and were even comparable to or faster than the untransformed MCF10A cultured in the presence of growth factors. The HER2 and cells also lifted off the plates much more readily than the others, suggesting reduced adhesion phenotype.
These oncogenes can drive multiple branched pathways, yet it was previously shown that inhibition of the MAPK pathway with the potent and selective MEK inhibitor (PD032590, MEKi) significantly reverses the surfaceome changes of MCF10A cells transformed with (6). Indeed, MEKi substantially hampered growth for all cell lines either in the absence or presence of growth factors (Fig. 1D and SI Appendix, Fig. S2). Overexpression of HER2 was most resistant to MEKi, followed by and , whereas cells containing , , or were most sensitive to MEKi.
Differential Expression of Oncogene-Induced Surfaceomes in MCF10A Cells.
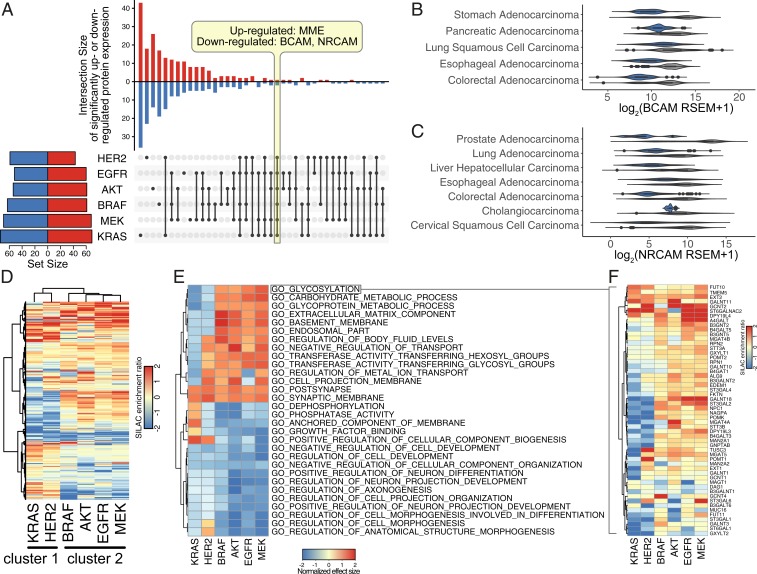
We next probed how the cell surfaceome is altered in the oncogene-transformed cells compared to the empty vector (EV) control. N-glycosylation is present on >85% of cell surface proteins (29) and can be exploited to capture the N-glycosylated proteins using a biotin hydrazide enrichment method (CSC) (24, 30). Here, we utilized a CSC protocol coupled with stable isotope labeling by amino acids in cell culture (SILAC) to compare the surfaceomes from the oncogene-transformed MCF10A cells to the EV control (SI Appendix, Figs. S3 and S4) (31). We identified and quantified a total of 654 cell surface proteins across the six oncogenic cell lines (Fig. 2A). Remarkably, the expression for 43% of the aggregate surface proteins (280 of 654) was altered by at least twofold for the oncogene-transformed cell lines relative to EV control, reflecting significant remodeling of the surfaceomes. In each of the six datasets, we observed at least twofold changes for 100 to 150 different surface proteins (Fig. 2A and SI Appendix, Fig. S5 A–F); these changes were evenly split between up- and down-regulated sets, reflecting bidirectional remodeling (SI Appendix, Fig. S6). Many of the differentially expressed proteins overlapped, but each cell line had a substantial number of uniquely differentially regulated proteins, presumably resulting from slight differences in signaling between each oncogene. Although it is well known that correlations between protein and RNA levels are not often strongly correlated (6, 32, 33), we were prompted to determine whether the most common changes were observed in previous studies. Down-regulation of BCAM and NRCAM transcription, in particular, was found in patient samples harboring the same oncogenic signatures in a number of cancers types of epithelial origin (Fig. 2 B and C). Using the 17 TCGA provisional dataset from The Cancer Genome Atlas (TCGA) with carcinoma (epithelial origin) annotation (34), activating oncogenic signature was defined as G12 or Q61 mutation in KRAS, L858 or amplification of EGFR, V600E mutation in BRAF, or amplification of HER2. Transcriptional up-regulation of MME, however, was not found in any of the dataset searched, suggesting regulation at the translational level or additional factors at play.
Fig. 2.
Proliferative oncogenes cause large changes in the surfaceome that are diverse in detail but have common functional themes. (A) Many differentially regulated proteins are unique to each cell line, and only three proteins were commonly up- or down-regulated among all six oncogene-transformed cell lines when compared to the EV control. In the vertical bar graph, up-regulated proteins (red) are indicated by the upward bars, and down-regulated proteins (blue) are indicated by the downward bars. The specific overlapping groups are indicated by the black solid dots below each bar. The total number of differentially regulated proteins for each cell line are indicated by the horizontal bar graph. Up-regulated (red) and down-regulated (blue) proteins are defined as (fold change) > 1 and P value < 0.05. (B) BCAM and (C) NRCAM are significantly down-regulated across various carcinoma with activating oncogenic signature in KRAS, HER2, BRAF, and EGFR (blue) compared to no mutations in these genes (gray). Activating oncogenic signature was defined as G12 or Q61 mutation in KRAS, L858 mutation or amplification of EGFR, V600E mutation in BRAF, or amplification of HER2. Genomic and expression data were obtained from the 17 TCGA provisional dataset with carcinoma (epithelial origin) annotation. (D) Hierarchal clustering of surfaceome changes revealed similarities between HER2- and KRAS-transformed cells. (E) Top 30 enriched gene sets identified by GSEA of the proteomics dataset using Gene Ontology terms show clustering between HER2- and KRAS-transformed cells. Positive normalized effect size (up-regulation) is shown in red, and negative (down-regulated) normalized effect size is shown in blue. Proteins were preranked by median SILAC peptide ratio, and GSEA was performed using MySigDB C5 GO gene set collection. (F) Proteins involved in glycosylation (GO:0070085) are down-regulated in the KRAS and HER2 cluster (blue), while the same proteins are up-regulated in the other oncogenes cluster. Heatmap for each gene set identified is also available at https://wellslab.ucsf.edu/oncogene_surfaceome.
At a global level, there were greater similarities between particular oncogenes. For example, cells harboring and HER2 clustered more closely together (cluster 1), and those containing , , , and clustered together (cluster 2) as seen either in the upset plot (Fig. 2A) or a heatmap with hierarchical clustering (Fig. 2D). The Pearson correlation coefficients were also higher within rather than across the two clusters (SI Appendix, Fig. S7A). One of the striking findings was the up-regulation of HER2 expression in the -transformed cells but not in cluster 2 transformed cells (SI Appendix, Fig. S8), which may help to explain the stronger similarity in oncogene-induced surfaceomes between the HER2 and cell lines. This same analysis also showed striking compensating regulation, where HER2 is down-regulated in the EGFR oncogene-expressing cell line.
Despite detailed differences at the individual target level, these harmonized into common biological processes when viewed by Gene Set Enrichment Analysis (GSEA) (Fig. 2E). For example, glycosylation and carbohydrate metabolism were similarly altered features for all of the oncogenes (Fig. 2F), consistent with numerous reports that altered glycosylation correlates with the development and progression of cancer (35–37). In addition, we see strong down-regulation of proteins involved in differentiation and adhesion, reflecting cell attachment and migration (SI Appendix, Fig. S9A). There were also large changes in some cell surface phosphatases involved in down-regulation of receptor tyrosine kinases (SI Appendix, Fig. S9B). These specific heatmaps reinforce the general division between cluster 1 and cluster 2. Due to the complexity of this data type, a data browser was made to view each individual gene set identified (https://wellslab.ucsf.edu/oncogene_surfaceome).
MEK Inhibition Induces Common Surfaceome Changes.
The MAPK pathway is a central driver of cell proliferation and has been a major therapeutic target in cancer. Indeed, MEKi significantly impedes the growth of all of the oncogene-transformed cells either in the absence (Fig. 1D) or presence of growth factors (SI Appendix, Fig. S2). To determine how MEKi alters the surfaceome of the oncogene-transformed cells, we compared the proteomic landscape of each oncogenic cell line in the presence and absence of MEKi. The proteomics dataset identified and quantified a total of 772 proteins, including an intersection of 492 proteins with the EV dataset discussed above, for a total of 934 proteins quantified between the two experiments. (Fig. 3 A–C).
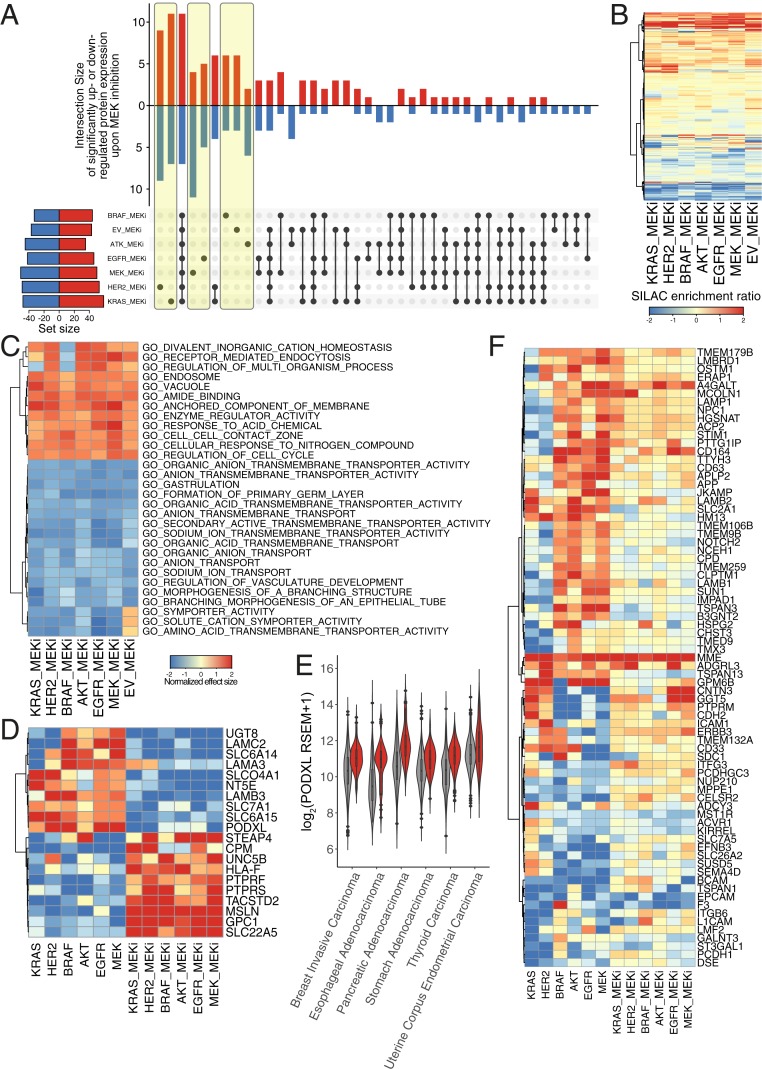
Fig. 3.
Inhibition of MEK led to similar perturbations of the surfaceome regardless of oncogenic transformation. (A) The majority of differentially regulated surface proteins were either completely unique (highlighted in yellow) or completely common (bar 3) to all seven cell lines upon MEK inhibitor treatment compared to control treatment. In the vertical bar graph, up-regulated proteins (red) are indicated by the upward bars, and down-regulated proteins (blue) are indicated by the downward bars. The specific overlapping groups are indicated by the black solid dots below each bar. The total number of differentially regulated proteins for each cell line are indicated by the horizontal bar graph. Up-regulated (red) and down-regulated (blue) proteins are defined as (fold change [FC]) > 1 and P value < 0.05. (B) Hierarchal clustering of surfaceome changes shows MEK inhibition treatment affected each cell line similarly. (C) Top 30 enriched gene sets identified by GSEA of the proteomics dataset using Gene Ontology terms show similarities between all cell lines, indicating that unique changes could be functionally redundant. Positive normalized effect size (up-regulation) is shown in red, and negative (down-regulated) normalized effect size is shown in blue. Proteins were preranked by median SILAC ratio, and GSEA was performed using MySigDB C5 GO gene set collection. Heatmap for each gene set identified is also available at https://wellslab.ucsf.edu/oncogene_surfaceome. (D) MEK-dependent regulation of protein expression induced by oncogenic transformation. Proteins shown were differentially regulated by oncogene (either (FC) > 1 or (FC) < −1 in oncogene vs. EV) and were then reversed (flipped (FC) < −1 and (FC) > 1 in MEK inhibition vs. respective cell line) in at least three cell lines. (E) PODXL is significantly up-regulated across various carcinoma with activating oncogenic signature in KRAS, HER2, BRAF, and EGFR (red) compared to no mutations in these genes (gray). Activating oncogenic signature was defined as G12 or Q61 mutation in KRAS, L858 or amplification of EGFR, V600E mutation in BRAF, or amplification of HER2. Genomic and expression data were obtained from the 17 TCGA provisional dataset with carcinoma (epithelial origin) annotation. (F) MEK-independent regulation of protein expression induced by oncogenic transformation. Proteins shown were differentially regulated by oncogene (either (FC) > 1 or (FC) < −1 in onco vs. EV) and were then reverted to a median level but not reversed (flipped (FC) < −1 and (FC) > 1 in MEK inhibition vs. respective cell line) in at least three cell lines.
In large measure, MEKi reversed the effects of the oncogenes and remarkably induced a more common state between all of the cell lines, including the EV control. For example, in contrast to the uninhibited datasets where 43% of proteins were differentially regulated by more than twofold in the oncogene-expressing cells, only 17% of the proteins (129 of 772) were altered by more than twofold in the presence of MEKi (Fig. 3A and SI Appendix, Fig. S6A). These changes were mostly bidirectional, except for BRAF and EGFR cell lines where MEK inhibition has skewness >1 or <−1 (SI Appendix, Fig. S6B). Of the proteins detected, 18 proteins were commonly changed across all oncogenes, as opposed to 3 in the absence of MEK inhibition (Fig. 3A and SI Appendix, Fig. S5 G–M). The similarity can be seen by hierarchical clustering of the MEKi datasets showing common changes across all cell lines (Fig. 3B). The Pearson correlation between the six oncogenes and the untransformed control were also higher, in general (SI Appendix, Fig. S7B). MEKi in and HER2 are still most closely correlated. GSEA of the MEKi data indicated a general common phenotypic reversal with down-regulation of membrane transporters, metabolism, and up-regulation of cell adhesion proteins consistent with a decrease in cancer-associated phenotypes such as cellular proliferation and metastasis (Fig. 3C and SI Appendix, Fig. S9 C and D).
Integration of the oncogenic transformation and MEKi datasets show 20 protein targets symmetrically flip from being significantly up- to down-regulated or visa versa in at least three of the cell lines, suggesting that the expression of these proteins is strongly dependent on the MAPK signaling pathway (Fig. 3D). One such target, PODXL, appears to be stringently regulated by the MAPK pathway. Using the same informatics approaches described above, transcription of PODXL was also found to be strongly up-regulated in several cancer types of epithelial origin (Fig. 3E). Additionally, 75 targets that were markedly up- or down-regulated revert to a median level of expression upon treatment with MEKi in at least three cell lines (Fig. 3F). Interestingly, there were a handful of oncogene-induced targets that are further up-regulated upon MEKi, such as MME, suggesting they are maintained by circuitous pathways outside of MAPK (MEKi independent). Overall, the six oncogenes cause profound changes to the surfaceome that alter common biological processes, which can be largely blunted by inhibition of the MAPK pathway.
Oncogenes Induce Large Changes to the Glycoproteome.
Glycosylation has long been known as a biomarker for cancer (35, 38–40), and our GSEA data show systematic up-regulation of proteins involved in glycosylation, especially in cluster 2 (Fig. 2E). We thus sought to identify the N-glycosylation modifications on specific membrane proteins for the six oncogene-transformed cell lines to compare among themselves and with the EV control. For maximal coverage, we enriched the glycoproteomes using the lectin ConA and hydrophilic interaction liquid chromatography (HILIC) that provide a complementary means for capturing high-mannose-type and complex-type glycans, respectively (41, 42). We processed the N-glycoproteomes in biological triplicate using liquid chromatography - tandem mass spectrometry (LC-MS/MS) and coupled with AI-ETD (SI Appendix, Fig. S10) (43). AI-ETD fragmentation combines radical-driven dissociation and vibrational activation and was recently shown to afford robust fragmentation of intact glycopeptides (21). Spectral assignments were made using the Byonic search engine (44); glycopeptides that were not identified across each of the three biological replicates for each cell line were removed (SI Appendix, Fig. S11).
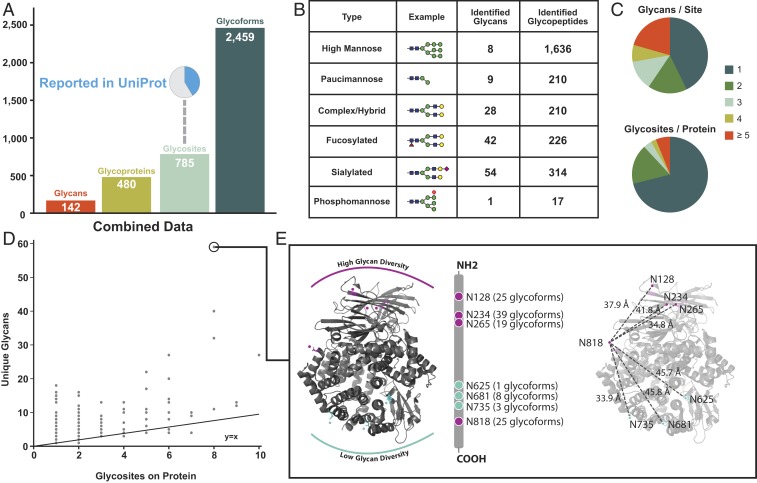
Combining ConA and HILIC enrichments allowed for sampling of different glycan classes, and the aggregated results of intact N-glycoproteomic analyses identified a total of 2,648 unique N-glycopeptides that passed conservative filtering described in Materials and Methods (Fig. 4 and SI Appendix, Fig. S12). Of this total, 189 were redundant glycan–glycosite pairs, that is, glycoforms that resulted from miscleaved or differentially oxidized peptides, leaving a total of 2,459 unique glycoforms (Fig. 4A). These glycoforms map to 785 N-glycosites found on a total of 480 glycoproteins. Thus, on average, each glycoprotein had 1.6 N-linked glycosites, and each site had, on average, 3.1 different glycans, reflecting significant heterogeneity in glycosylation. Similar yet unique glycoforms indicated that the heterogeneity of protein N-glycosylation is driven largely by the biology and not artifacts of in-source fragmentation. Of the 785 experimentally detected glycosites, only 324 were previously known or predicted to be glycoyslated in UniPort (45).
Fig. 4.
A global overview of the MCF10a glycoproteome identified from all cell lines. (A) Unique glycans, glycoproteins, glycosites, and glycoforms identified across all six cell lines. A pie chart represents a high percentage of glycosites not previously reported in uniport. (B) High-mannose glycan type is the most common glycan identified. A total of six common glycan types are used to categorize glycans identified. (C) The varying degrees of microheterogeneity (Top) and macroheterogeneity (Bottom) of protein N-glycosylation. (D) A scatter plot of unique glycans versus number of glycosites for each protein provides an additional view of heterogeneity in protein N-glycosylation. The protein with the most heterogenous glycoform was ANPEP (UniProtKB accession no. P15144). (E) Mapping glycan diversity on structure of ANPEP (Protein Data Bank ID 4FYR) reveals spatial regulation of glycosylation patterns. Asparagine residues with high (magenta) and low (teal) number of unique glycans are highlighted. The most C-terminus glyan, N818, has high glycan diversity distance and has similar distance to all other asparagine residues.
In total, we identified 142 different glycan structures. The glycans can be categorized into six structural classes based on their maturation state as they transition from ER to Golgi and then split off to either lysosomal, granular, or cell surface destinations (SI Appendix, Fig. S13) (46). The six glycan categories represent a gradation of maturation from the least mature high mannose, to paucimannose, phosphomannose, complex/hybrid, and finally fucosylated and sialylated (Fig. 4B). We identified 1,636 mannose glycopeptides containing any of 8 high-mannose-type glycans and 210 were found trimmed to contain any of 9 different paucimannose structures. The remaining 767 N-glycopeptides had more complex glycans including: 210 complex/hybrid glycopeptides with 28 different structures, 314 of the sialylated class with 54 different structures, and 226 of the fucosylated class with 42 different structures. We found only 17 phosphomannose containing glycopeptides each having the same structural type that are on proteins typically found in lysosomes (47–49). Thus, over 60% of the N-glycopeptides identified contained less mature high mannose or paucimannose structures originating from processing in the ER or early Golgi. It is not surprising to find these high-mannose modifications associated with cell surface proteins as others have seen that high-mannose N-glycosylation is abundant at the cell surface and especially associated with oncogene transformation (50–52). Furthermore, we have validated the expression of high-mannose glycans on the cell surface by means of ConA lectin staining (SI Appendix, Fig. S14A). Since CSC techniques label sialic acid, we have also shown that sialic acid is not differentially regulated at the global level shown by SNA lectin staining (SI Appendix, Fig. S14B).
We found significant heterogeneity in the number of different glycan structures at any given site (Fig. 4C). Approximately 45% of the sites were observed with only one glycan structure, but the range of glycans on these sites was broad. Some glycosites, such as position N-234 on aminopeptidase N (ANPEP), had up to 39 different glycans detected. Additionally, the number of glycosites per protein varies considerably; about 70% of the glycoproteins have only one site of N-glycosylation, but about 10% have over five sites. There is a general trend between the number of glycosites identified on a given protein and the number of unique glycans it has; however, some proteins show significantly increased heterogeneity of glycans relative to their number of total glycosites (Fig. 4D). The most extreme glycan diversity was observed on ANPEP, on which 59 unique glycans were identified across eight glycosites (Fig. 4E). Interestingly, the glycosites with higher glycan diversity mapped to specific regions on the three-dimensional structure of ANPEP. One face of the protein containing a cluster of four asparagines (at positions 128, 234, 265, and 818) had high glycan diversity ranging from 19 to 39 different glycan structures per glycosite, while the opposite face containing three clustered asparagines (at positions 625, 681, and 735) had much lower diversity, ranging between 1 and 8 different glycan structures per site. This suggests topological bias of glycosylation on the folded protein structure.
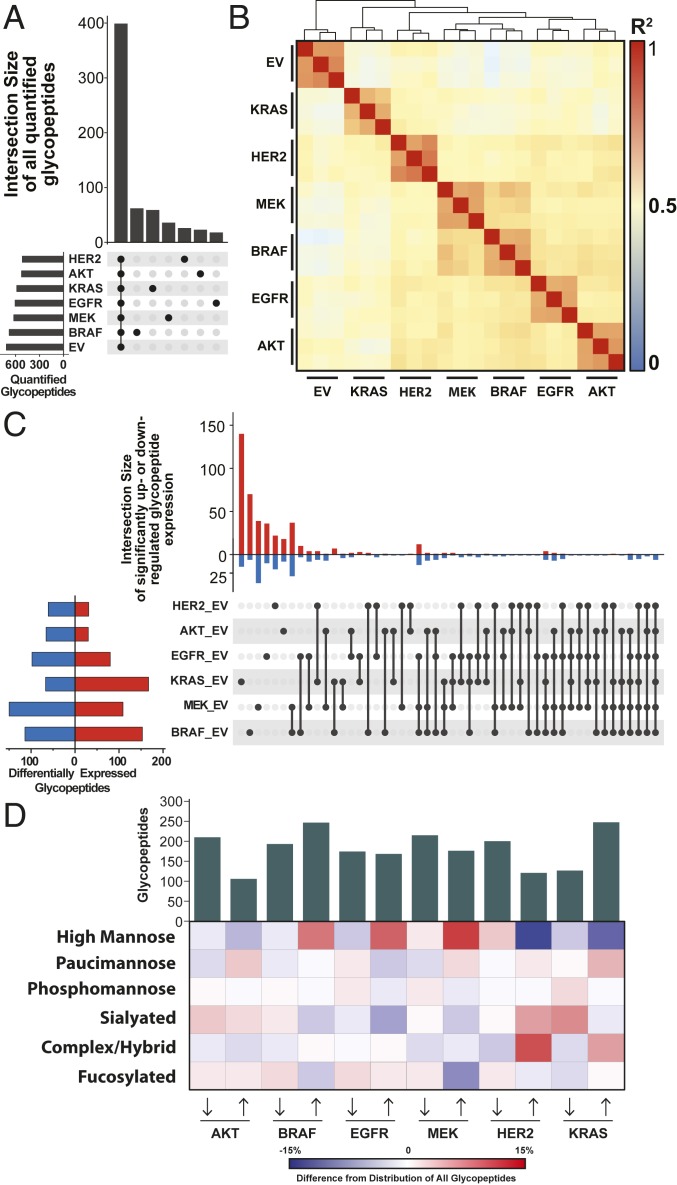
We next applied quantitative analysis to compare the different oncogene-transformed cell lines in terms of their glycoproteome landscapes and changes between them and the EV control. We observed hundreds of glycopeptides that were significantly and differentially expressed (q < 0.05) upon oncogenic transformation relative to the EV control (SI Appendix, Fig. S15). In general, the volcano plots were symmetric, reflecting bidirectional changes, and the fold changes ranged from 50-fold down-regulated to 50-fold up-regulated, representing significant remodeling of the glycoproteome. Changes in glycosylation were greater than changes in surface protein expression, which range from twofold to eightfold (SI Appendix, Fig. S5). We quantified about 600 glycopeptides in each dataset, of which two-thirds are shared in all datasets (Fig. 5A). There was high quantitative reproducibility of the glycopeptide measurements based on the close hierarchical clustering of the three biological replicates (Fig. 5B), tight clustering by principal component analysis, and low (<30%) median CV from the EV control cell line (SI Appendix, Fig. S16). and had the greatest glycoproteome similarity, while EV was the farthest removed from all of the oncogenes.
Fig. 5.
Quantitative glycopeptide measurements across mutant cell lines. (A) An UpSet plot shows glycopeptide identifications that are unique to or shared between datasets. (B) Pairwise Pearson correlations from all replicate analyses illustrate clustering between and glycoproteome. (C) An UpSet plot displays the shared and unique glycopeptides that are significantly differentially expressed upon oncogenic transformation (q < 0.05) as assessed by linear models for microarray data (LIMMA) with Bonferroni-adjusted P values. (D) A heatmap displays the differences in glycan type distribution for up- and down-regulated glycopeptides across cell lines.
The UpSet plot in Fig. 5C displays significant glycopeptide differential expression that is shared and unique to each cell line. MCF10A transformed with the oncogene resulted in the largest set of uniquely changing glycopeptides; 154 of the 234 differentially expressed glycopeptides in the cell line were unique to transformation. Some of these were highly protein specific. For example, 28 of the 154 glycopeptides uniquely differentially expressed by were identified from ANPEP, and all were up-regulated upon oncogenic transformation, as was the protein itself (SI Appendix, Fig. 17). Other glycopeptides from ANPEP were differentially expressed in different sets of cell lines. In fact, 51 of 69 glycopeptides from ANPEP were differentially expressed in at least one cell line; 12 ANPEP glycopeptides were significantly up-regulated in HER2, and 5 were shared between HER2 and .
, , and shared the most overlap of significantly changing glycopeptides between any group of three cell lines (Fig. 5C); 24 glycopeptides belonged to this intersection: 12 were overexpressed and 12 were underexpressed upon oncogenic transformation. Laminin subunit alpha-3 (LAMA3) and N-acetylglucosamine-6-sulfatase were most represented in the overexpressed and underexpressed group, respectively. Four glycopeptides from each protein were identified and significantly differentially expressed in these three cell lines. Six glycopeptides were significantly differentially expressed across all six cell lines, three of which were identified from galectin-3 binding protein (Gal-3BP), and all were down-regulated glycopeptides modified with high-mannose glycans (SI Appendix, Fig. S18). To explore this dataset, a data browser was made to view each individual glycopeptide identified (https://wellslab.ucsf.edu/oncogene_surfaceome).
We explored the glycan composition of differentially expressed glycopeptides to capture a broader view of differential glycosylation in the oncogene-transformed cell lines and to see whether general trends emerged. The heatmap in Fig. 5D displays the differential glycome composition of glycopeptides changing more than twofold upon oncogenic transformation compared to EV control. We, again, observe greatest similarity between , , and cell lines, which have an increased proportion of high-mannose glycans in up-regulated glycopeptides. In contrast, HER2 and expressed fewer up-regulated high-mannose-modified glycopeptides and showed an increased proportion of complex/hybrid-type glycopeptides. Further inspection revealed that nearly all of the up-regulated glycopeptides with a complex/hybrid glycan from the cell lines harboring HER2 (12 of 12) and (13 of 18) mapped to ANPEP. This protein was also up-regulated on the surfaceome (6), displayed the highest degree of glycan heterogeneity within the glycoproteomic data, and has previously been implicated in tumorigenesis (53, 54).
Discussion
Oncogenesis is a complex phenomenon that involves aberrant changes in multiple biological processes to promote cancer cell survival (1). Here we study how the surfaceome remodels in a simplified cell autologous model by six prevalent and neighboring oncogenes that drive proliferation through the MAPK signaling node. Genetic studies have shown that these oncogenes typically exhibit mutual exclusivity in tumors from cancer patients (9). The surfaceome is a terminal manifestation of these signaling pathways. We find significant differences in detailed expression patterns, consistent with previously reported differences in feedback loops and collateral signaling pathways between these oncogenes (55). However, we find that these oncogene-induced surfaceome differences harmonize in similar functional outcomes overall, and consistent with observed mutual exclusivity.
We took a reductionist approach, starting from an immortalized epithelial cell line stably transformed with each of the six different oncogenes to determine how the surfaceome is remodeled in a cell autologous fashion. This system is clearly an approximation and does not recapitulate the complexity of tumors that vary in cellular context, genetic variation, heterogeneity, oncogene expression, the presence of host immune, and metabolism. Nonetheless, cell culture models are a practical reality allowing isogenic comparison between oncogenes, renewable access to materials that permits studies to be readily reproduced (56–58). We chose the spontaneously immortalized breast epithelial cell line, MCF10A, where the functional loss of p16INK4a allows cells to be cultured with a myriad of oncogenes without oncogene-induced senescence. We picked this over cancer cell lines which are already transformed or other artificially immortalized cell lines, because they often contain genetic lesions that may cause genomic instability, leading to a more idiosyncratic background. Overall, we find that independent expression of each of these six oncogenes induced profound changes in the surfaceome, both in the proteins expressed and the glycans that decorate them.
Common Phenotypes and Biological Themes Induced by the Six Oncogenes.
Although the six oncogenes we studied (HER2 overexpression, , , , , and ) have many variants that could have different phenotypes, we chose well-known representatives, to begin to understand the similarities and differences between them at a coarse-grain level. Transformation with each of these oncogenes led to rapid growth, to varying degrees, in the absence of growth factors and produced somewhat different cell morphologies characteristic of a cancer cell transformed phenotype. Each oncogene caused large changes in the surfaceome, where about 40% of the detected N-glycoproteins had altered expression level by more than twofold, evenly divided between up- and down-regulated proteins, reflecting bidirectional cell surface remodeling.
There were important differences between the oncogenes, and they clustered into two groups based on growth rates, surfaceomes and associated glycans. Cluster 1, containing HER2 overexpression and , was most aggressive in proliferation and reduced adhesion. Cluster 2 was less aggressive, and included , , , and . We believe much of the similarity between HER2 and KRAS derives from KRAS-induced expression of HER2, whereas EGFR suppresses expression of HER2, probably reflecting their signaling redundancy. MCF10A is derived from normal breast epithelial cells, and, among these oncogenes, HER2 overexpression is most commonly seen in breast cancer patients, at 13% compared to 2% for KRAS, 2.8% for EGFR, 5% for AKT1, 1.7% for BRAF, and 0.6% for MEK1 (SI Appendix, Fig. S19). Each of these also shows remarkable mutual exclusivity relative to the others in breast cancer patients, reflecting oncogene functional redundancies. Further in silico comparison of oncogenic mutational occurrence across all cancer types (59, 60) indicates a strong mutual exclusivity between BRAF and KRAS, and a slight mutual exclusivity with EGFR (SI Appendix, Fig. S20).
Alterations in Transporter Expression.
One of the most pronounced oncogene-induced changes we observed is to proteins involved in solute transport that reverse upon MEK inhibition, including up-regulation of SLC2A1, SLC6A15, SLC7A1, SLCO4A1, and MF12/melanotransferrin, and down-regulation of SLC22A5 and STEAP4 (Fig. 3D). SLC6A15 acts as a preferential methionine amino acid transporter and shows dramatic up-regulation in both KRAS- and HER2-transformed cells. This is consistent with recent studies showing that KRAS-transformed cells exhibit an extreme sensitivity to methionine deprivation (61). The Warburg Effect preferentially shifts cancer cells toward glycolysis, thereby promoting accelerated growth and division (62). We find that SLC2A1, also known as glucose transporter-1, is significantly up-regulated in all oncogene-transformed cells, which would facilitate anaerobic glycolysis and increased growth for these cells. Our data show up-regulation of SLCO4A1, a part of the organic anion transporter family that assists in transporting hormones such as prostaglandins and vasopressin. Heightened expression of this hormone transporter has been previously seen in metastatic colorectal cancer (63). SLC7A1 has been shown to be up-regulated across many cancer types, including colorectal cancer (64). Others have shown that SLC7A1 messenger RNA expression level decreases upon MAPK pathway inhibition (65), which is consistent with our proteomics data.
General Down-Regulation of Receptor Tyrosine Phosphatases and Tumor Suppressors.
Another important theme found within our data is a general down-regulation of surface proteins involved in receptor tyrosine phosphatases and other tumor suppressors such as PTPRF, PTPRS, UNC5B, and BCAM (Figs. 2A and 3D). The expression of PTPRF and PTPRS, in particular, has been associated with decreased metastasis through the inactivation of EGFR signaling, emphasizing their importance as tumor suppressor proteins in numerous cancer contexts (66, 67). Both of these proteins show a marked decrease in expression across the majority of the oncogene-transformed cells, perhaps as a means to promote growth and metastasis. Similarly, UNC5B has recently been shown to halt tumor progression in an in vivo model of bladder cancer, through inducing cell cycle arrest in the G2/M phase (68). Lastly, BCAM has been previously shown to be a tumor suppressor in a model of hepatocellular carcinoma (69), and decreased expression in all oncogene-transformed cells may signify the removal of tumor-suppressive signaling.
Alteration in Adhesion Molecules.
Metastasis is the leading cause of death for patients with cancer, and tumor cells acquire the ability to penetrate the surrounding tissues, thus leading to invasion (70). Many of these acquired functionalities are through changes in adhesion molecules on the cell surface, which play the role of mediating cell–cell interactions. Within our dataset, we identified five targets, LAMC2, LAMA3, LAMB3, PODXL, and MME, that are known to play important roles in mediating metastasis in various tumor types and we find to be up-regulated across the majority of oncogenic mutants (Figs. 2A and 3D). LAMC2 has previously been shown to promote epithelial–mesenchymal transition [EMT] and invasion in an in vivo model of lung adenocarcinoma (71). It has also been shown that colorectal cancer with high MAPK activity expresses heightened levels of LAMC2, supporting our proteomic results that show reversal upon MEK inhibition (72). PODXL had increased expression in all six mutant cell lines, and has been implicated in increasing the aggressiveness of breast and prostate cancer through the induction of both MAPK and PI3K signaling (73). PODXL is also a glycoprotein with extensive mucin-type O-glycosylation, the overexpression of which can lead to increased metastatic potential through increased cell cycle progression via the PI3K and MAPK axes (74–76). MME, known as neprilysin, was found up-regulated in all six oncogenic mutants compared to the EV control. While the function of MME has not been studied in great detail, it is considered to be an adverse prognostic marker for patients with lung adenocarcinoma (77) and has been found to play an important role in the growth and progression of colorectal cancers (78). MME was also found significantly up-regulated in surfaceomics studies of isogenic cells expressing the tubular sclerosis gene associated with bladder cancer (79). Interestingly, MME is up-regulated across all oncogenes in both the oncogene and MEKi datasets, suggesting that MME expression is not regulated through the MAPK or PI3K axis. This heightened expression of MME in all oncogenic contexts presents an opportunity for future research into synthetic lethality studies with MME inhibition in combination with MEKi or other MAPK targets.
Immune Modulation.
Another prominent cause of cancer progression is the evasion of immune surveillance. This can be achieved by overexpressing proteins that have a net immunosuppressive effect or by down-regulating proteins that increase immune activation. Here, we identified three differentially regulated proteins, NT5E, HLA-F, and DSE, that play important roles in immune functions (Fig. 3D). NT5E, which was up-regulated in all the proliferative oncogene cell lines, promotes immunosuppression through the production of adenosine from AMP, which can decrease the capacity of natural killer cells to produce immune-activating IFN molecules and prevents the clonal expansion of cytotoxic T cells in the surrounding tumor tissue (80). Conversely, HLA-F and DSE (also known as dermatan sulfate epimerase) were down-regulated in all oncogene-expressing cell lines, and both play a role in immune system activation and tumor rejection, through activation of NK cells or cytotoxic T cells, respectively, in the tumor microenvironment (81, 82). Both NT5E and HLA-F are down-regulated upon MEKi (Fig. 3D).
Down-Regulation by Shedding.
Proteolytic cleavage of proteins at the cell surface leads to ectodomain shedding and the existence of neo N termini. The loss of ectodomains would lead to decreased peptide detection by LC-MS/MS. We identified four surface proteins reduced in detection, MSLN, TACSTD2, NRCAM, and GPC1, that are known to undergo proteolysis to enhance the growth and metastasis of cancer (83–85). In particular, NRCAM, which had lower surface levels in all six oncogenic mutants, is a cell–cell adhesion molecule; it is known to be shed by proteolysis, and the shed form stimulates cell proliferation via the AKT pathway (86). Perhaps unsurprisingly, NRCAM did not exhibit a renormalization of expression levels after MEK inhibition, due to its exclusive function through the AKT pathway.
Changes in Glycosylation Machinery and Protein N-glycosylation.
Alterations in glycosylation are common features of cancer cells (35) which can result from a variety of factors, including changes in expression of glycosyltransferases, availability of sugar nucleotide substrates, alteration in expression of substrate proteins, or changes to the tertiary structure of the protein substrate that disrupt transfer of the glycan. We find that glycosyl transferases FUT10, EXT2, GALNT11, GCTN2, and ST6GALNAC2 are highly up-regulated across all oncogenic cell lines. FUT10 is an -1,3-focusyltransferase, and FUTs are critical for the production of LewisX and Sialyl LewisX antigens which are hallmarks of invasion (87). In fact, the most aggressive of our oncogene cell lines (HER2 and KRAS) are significantly up-regulated in complex hybrid-type glycans for which these are members, and especially noteworthy for ANPEP. EXT2 forms a heterodimer with EXT1 in the Golgi functioning as a glycosyl transferase involved in catalyzing the formation of heparin, a substrate that contributes to structural stability of the extracellular matrix, thereby mediating processes such as adhesion, immune infiltration, and signaling (88). Unsurprisingly, alterations in heparan sulfate formation have been shown to have emerging roles in oncogenesis, of which EXT2 is a primary facilitator (89). GALNT11 is a protein involved in the initiation of mucin-type O-glycosylation, a well-known marker that is overexpressed in cancer (90–92). MUC-1 is also a very important drug target for immunotherapy (93–96). Interestingly, a common substrate for this family of enzymes, PODXL, is also up-regulated across all oncogenic cell lines (see above), suggesting a possible mechanism by which increased GALNT11 promotes progrowth phenotypes in these cell lines. GCTN2 is a glycosyltransferase (97), and specifically implicated in the transition from naïve to germinal center B cell (98).
ST6GalNAc2 is a sialyltransferase that catalyzes attachment of GalNAc to core glycans and is paradoxically up-regulated in all of the oncogene cell lines. A genome-wide short hairpin RNA screen in mice for genes that suppress metastasis identified and validated ST6GalNAc2 as the strongest metastasis suppressor. ST6GalNAc2 is strongly down-regulated in estrogen receptor-negative breast cancer tumors. Silencing of ST6GalNAc2 promotes binding of Galectin-3 and retention of tumor cells to sites of metastasis. Galectin-3 is a member of S-type lectins and a known modulator of tumor progression (99) by enhancing aggregation of tumor cells during metastasis and preventing anoikis (100–102). Galectin-3 binds to terminal fucose on glycans of Gal-3BP which ST6GalNAc2 blocks via sialylation (102). We found that all glycopeptides from the eight glycosites Gal-3BP were down-regulated up to 50-fold in all cell lines (SI Appendix, Fig. S18).
Our glycoproteomic analyses show that protein N-glycosylation is dramatically changed upon oncogenic transformation, and that distinct genetic drivers of oncogenesis promote unique changes to the glycoproteome. The majority of differentially expressed glycopeptides were unique to individual cell lines. We observed greatest similarity between , , and , which has been consistent throughout different analyses of these cell lines (Fig. 5B). There were significant differences between the oncogene cluster 1 (/HER2) and cluster 2 (///). Specifically, we found differential increase in complex hybrid glycans and decrease in high mannose in cluster 1 and just the opposite for cluster 2.
There is abundant evidence to suggest that high-mannose-type glycans are present at the cell surface and, furthermore, that cancerous cells display increased abundance of high-mannose glycans at their cell surface (52, 103). A similar observation was reported in the glycomic comparison of transformed versus human embryonic stem cells (hESCs), where high-mannose glycans were observed at significantly higher abundance on plasma membranes of hESCs (50). Together, these observations help to explain the similarities between cancerous cells and hESCs, such as the regulation of tumor suppressor genes and the ability to self-renew (104). This is consistent with our studies, although we cannot exclusively rule out that some of the high-mannose glycans we observe are coming from proteins in route to the membrane.
Our data suggest glycosylation is quite heterogeneous, possibly reflecting incomplete maturation. The most stunning example in our data of heterogeneity in glycosylation was for ANPEP, an amino peptidase also up-regulated at the protein level across most cell lines. The most extreme example was site N234 of ANPEP, where we found 29 different glycan structures. When these are arranged in order of glycan maturation, we observed a close representation of its steady-state progression through the secretion pathway (SI Appendix, Fig. S13). ANPEP also showed the greatest representation within complex/hybrid glycopeptides that were up-regulated in HER2 and cell lines (Fig. 5D). ANPEP was most up-regulated in in surfaceomic analyses, and its expression was not dramatically influenced by MEK inhibition. ANPEP expression in our surfaceomic and glycoproteomic data represents one way in which diverges from other members of the MAPK pathway. These data, combined with ANPEP’s previously identified role in angiogenesis (54), underscore its importance in -positive tumors. ANPEP glycosylation may serve as a very sensitive biomarker of glycan metabolism in cells, given its highly diverse and heterogeneous patterns.
Conclusions
We provide a large-scale comparative study of how six neighboring proliferative oncogenes cause large-cell autologous remodeling in the surfaceome and glycoproteome. While many of the changes are specific to given oncogenes at both the protein and glycopeptide level, we observe common biological themes suggesting functional redundancy of the specific cellular expression. This is consistent with observations that common tumors can express these different oncogenes, although not together. We commonly observe up-regulation of surface proteins involved in metabolite transport, glycosylation, and immune suppressors and down-regulation of adhesion proteins and tumor suppressors, which is consistent with increased cell growth and invasion that are well-known properties of cancer cells. These studies were deliberately conducted in isogenic cell lines to isolate the oncogene-induced changes. Even in a simplified autologous cell model, we recapitulate many of the hallmarks of transformed cells driven by complex but functionally redundant changes to the surfaceome. We believe the work provides important insights into the similarities and differences among the neighboring oncogenes and provides opportunities to pursue antibody tools to follow in more complex tumor settings.
Data Archival.
All proteomics dataset has been deposited to ProteomeXchange Consortium (proteomecentral.proteomexchange.org) via the PRIDE partner repository accessible (identifier PXD017039). Interactive illustrations of several figures are also available in a data browser (https://wellslab.ucsf.edu/oncogene_surfaceome).
SI Appendix.
SI Appendix, Supplemental Materials and Methods, Figs. S1–S20, and Description of Datasets S1 to S25 is included as a single PDF.
Materials and Methods
All MCF10A cells were cultured according to established protocols unless otherwise stated. Surfaceome of each oncogene-transformed cell line was compared to EV control using SILAC-based quantification. For inhibition study, each oncogene-transformed cell line was treated with 100 nM MEK inhibitor PD032590 or dimethyl sulfoxide control for 3 d, and surfaceome changes were compared in each respective cell line using SILAC-based quantification. Glycoproteome of each oncogene-transformed cell line was quantified using label-free quantification. Detailed materials and methods are included in SI Appendix, and supplementary data are available as Datasets S1–25.
Supplementary Material
Acknowledgments
We thank Professor Sourav Bandyopadhyay at University of California, San Francisco (UCSF) for some of the lenti constructs and cell lines, and Michael Westphall, Evgenia Shishkova, and Jean Lodge at University of Wisconsin for instrumentation and helpful discussions. J.A.W. is grateful for funding from NIH Grant R35GM122451 and Celgene Corporation. K.K.L. thanks Canadian Institutes of Health Research for postdoctoral funding. L.L.K. is indebted to Chemistry and Chemical Biology Training Grant T32 GM064337 at UCSF for funding. J.J.C. wishes to thank NIH for generous funding from P41 Grant GM108538 (The National Center for Quantitative Biology of Complex Systems). G.M.W. was supported in part by the Biotechnology Training Grant T32 GM008349. N.M.R. gratefully acknowledges support from NIH Grant K00CA21245403.
Footnotes
Competing interest statement: K.K.L., L.L.K, and J.A.W. received research funding from Celgene Corporation but no personal financial gain or equity.
This article is a PNAS Direct Submission.
Data deposition: All of the proteomics datasets have been deposited to ProteomeXchange Consortium (proteomecentral.proteomexchange.org) via the PRIDE partner repository (identifier PXD017039). Interactive illustrations of several figures are also available in a data browser (https://wellslab.ucsf.edu/oncogene_surfaceome).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917947117/-/DCSupplemental.
References
- 1.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez R., et al. , Screening the mammalian extracellular proteome for regulators of embryonic human stem cell pluripotency. Proc. Natl. Acad. Sci. U.S.A. 107, 3552–3557 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallin E., von Heijne G., Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 7, 1029–1038 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter P. J., Lazar G. A., Next generation antibody drugs: Pursuit of the ’high-hanging fruit.’ Nat. Rev. Drug Discov. 17, 197–223 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Chames P., Van Regenmortel M., Weiss E., Baty D., Therapeutic antibodies: Successes, limitations and hopes for the future. Br. J. Pharmacol. 157, 220–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinko A. J., et al. , Targeting RAS driven human cancer cells with antibodies to upregulated and essential cell-surface proteins. eLife 7, e31098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye X., et al. , Comparative proteomics of a model MCF10A-KRasG12V cell line reveals a distinct molecular signature of the KRasG12V cell surface. Oncotarget 7, 86948–86971 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ying H., et al. , Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network , Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cisowski J., Sayin V. I., Liu M., Karlsson C., Bergo M. O., Oncogene-induced senescence underlies the mutual exclusive nature of oncogenic KRAS and BRAF. Oncogene 35, 1328–1333 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Unni A. M., Lockwood W. W., Zejnullahu K., Lee-Lin S. Q., Varmus H., Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. eLife 4, 1–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandes B., Sagman U., Auger M., Demetrio M., Dennis J. W., Beta 1-6 branched oligosaccharides as a marker of tumor progression in human breast and colon neoplasia. Canc. Res. 51, 718–723 (1991). [PubMed] [Google Scholar]
- 13.Hakomori S., Tumor malignancy defined by aberrant glycosylation and sphingo(glyco)lipid metabolism. Canc. Res. 56, 5309–5318 (1996). [PubMed] [Google Scholar]
- 14.Hudak J. E., Canham S. M., Bertozzi C. R., Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat. Chem. Biol. 10, 69–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia L., Schrump D. S., Gildersleeve J. C., Whole-cell cancer vaccines induce large antibody responses to carbohydrates and glycoproteins. Cell Chem. Biol. 23, 1515–1525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao H., Woods E. C., Vukojicic P., Bertozzi C. R., Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 113, 10304–10309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kannagi R., et al. , Altered expression of glycan genes in cancers induced by epigenetic silencing and tumor hypoxia: Clues in the ongoing search for new tumor markers. Canc. Sci. 101, 586–593 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varki A., Gagneux P., Multifarious roles of sialic acids in immunity. Ann. N. Y. Acad. Sci. 1253, 16–36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palaniappan K. K., Bertozzi C. R., Chemical glycoproteomics. Chem. Rev. 116, 14277–14306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu M. Q., et al. , pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat. Commun. 8, 438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riley N. M., Hebert A. S., Westphall M. S., Coon J. J., Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nat. Commun. 10, 1311 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y., Franc V., Heck A. J. R., Glycoproteomics: A balance between high-throughput and in-depth analysis. Trends Biotechnol. 35, 598–609 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Bausch-Fluck D., et al. , A mass spectrometric-derived cell surface protein Atlas. PLoS One 10, e0121314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wollscheid B., et al. , Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 27, 378–386 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen-Petersen B. L., Carter C. J., Ohm A. M., Reyland M. E., Protein kinase C is required for ErbB2-driven mammary gland tumorigenesis and negatively correlates with prognosis in human breast cancer. Oncogene 33, 1306–1315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martins M. M., et al. , Linking tumor mutations to drug responses via a quantitative chemical-genetic interaction map. Canc. Discov. 5, 154–167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu Y., et al. , Evaluation of MCF10A as a reliable model for normal human mammary epithelial cells. PLoS One 10, e0131285 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soule H. D., et al. , Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Canc. Res. 50, 6075–6086 (1990). [PubMed] [Google Scholar]
- 29.Apweiler R., Hermjakob H., Sharon N., On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Bausch-Fluck D., et al. , The in silico human surfaceome. Proc. Natl. Acad. Sci. U.S.A. 115, E10988–E10997 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung K. K., et al. , Multiomics of azacitidine-treated AML cells reveals variable and convergent targets that remodel the cell-surface proteome. Proc. Natl. Acad. Sci. U.S.A. 116, 695–700 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lundberg E., et al. , Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 6, 450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwanhäusser B., et al. , Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network et al. , The Cancer Genome Atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinho S. S., Reis C. A., Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Canc. 15, 540–555 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Varki A., Kannagi R., Toole B., Stanley P., “Glycosylation changes in cancer” in Essentials of Glycobiology, Varki A., et al., Eds. (Cold Spring Harbor Laboratory Press, 2015), pp. 597–609. [Google Scholar]
- 37.Möckl L., et al. , Quantitative super-resolution microscopy of the mammalian glycocalyx. Dev. Cell 50, 57–72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adamczyk B., Tharmalingam T., Rudd P. M., Glycans as cancer biomarkers. Biochim. Biophys. Acta 1820, 1347–1353 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Mechref Y., Hu Y., Garcia A., Hussein A., Identifying cancer biomarkers by mass spectrometry-based glycomics. Electrophoresis 33, 1755–1767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peracaula R., Barrabés S., Sarrats A., Rudd P. M., de Llorens R., Altered glycosylation in tumours focused to cancer diagnosis. Dis. Markers 25, 207–218 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Totten S. M., Feasley C. L., Bermudez A., Pitteri S. J., Parallel comparison of N-linked glycopeptide enrichment techniques reveals extensive glycoproteomic analysis of plasma enabled by SAX-ERLIC. J. Proteome Res. 16, 1249–1260 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Zhang C., et al. , Evaluation of different N-glycopeptide enrichment methods for N-glycosylation sites mapping in mouse brain. J. Proteome Res. 15, 2960–2968 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Riley N. M., Westphall M. S., Hebert A. S., Coon J. J., Implementation of activated ion electron transfer dissociation on a quadrupole-orbitrap-linear ion trap hybrid mass spectrometer. Anal. Chem. 89, 6358–6366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bern M., Kil Y. J., Becker C., Byonic: Advanced peptide and protein identification software. Curr. Protoc. Bioinformatics 2012, 1–22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.UniProt Consortium , The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 38, 142–148 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yarema K. J., Bertozzi C. R., Characterizing glycosylation pathways. Genome Biol. 2, reviews0004.1 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kornfeld S., Mellman I., The biogenesis of lysosomes. Annu. Rev. Cell Biol. 5, 483–525 (1989). [DOI] [PubMed] [Google Scholar]
- 48.Rohn W. M., Rouillé Y., Waguri S., Hoflack B., Bi-directional trafficking between the trans-Golgi network and the endosomal/lysosomal system. J. Cell Sci. 113, 2093–2101 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Rouillé Y., Rohn W., Hoflack B., Targeting of lysosomal proteins. Semin. Cell Dev. Biol. 11, 165–171 (2000). [DOI] [PubMed] [Google Scholar]
- 50.An H. J., et al. , Extensive determination of glycan heterogeneity reveals an unusual abundance of high mannose glycans in enriched plasma membranes of human embryonic stem cells. Mol. Cell. Proteomics 11, M111.010660 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balog C. I. A., et al. , N-glycosylation of colorectal cancer tissues: A liquid chromatography and mass spectrometry-based investigation. Mol. Cell. Proteomics 11, 571–585 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holst S., et al. , N-glycosylation profiling of colorectal cancer cell lines reveals association of fucosylation with differentiation and caudal type homebox 1 (CDX1)/Villin mRNA expression. Mol. Cell. Proteomics 15, 124–140 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong X., et al. , Modification of the amino terminus of a class II epitope confers resistance to degradation by CD13 on dendritic cells and enhances presentation to T cells. J. Immunol. 164, 129–135 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Pasqualini R., et al. , Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Canc. Res. 60, 722–727 (2000). [PMC free article] [PubMed] [Google Scholar]
- 55.Vaseva A. V., et al. , KRAS suppression-induced degradation of MYC is antagonized by a MEK5-ERK5 compensatory mechanism. Canc. Cell 34, 807–822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Domcke S., Sinha R., Levine D. A., Sander C., Schultz N., Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 4, 2126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rockwell S., In vivo-in vitro tumour cell lines: Characteristics and limitations as models for human cancer. Br. J. Canc. Suppl. 4, 118–122 (1980). [PMC free article] [PubMed] [Google Scholar]
- 58.Wilding J. L., Bodmer W. F., Cancer cell lines for drug discovery and development. Canc. Res. 74, 2377–2384 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Canc. Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Sanctis G., Spinelli M., Vanoni M., Sacco E., K-Ras activation induces differential sensitivity to sulfur amino acid limitation and deprivation and to oxidative and anti-oxidative stress in mouse fibroblasts. PLoS One 11, e0163790 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liberti M. V., Locasale J. W., The Warburg Effect: How does it benefit cancer cells? Trends Biochem. Sci. 41, 211–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buxhofer-Ausch V., et al. , Tumor-specific expression of organic anion-transporting polypeptides: Transporters as novel targets for cancer therapy. J. Drug Delivery 2013, 863539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu Y., et al. , Overexpression of arginine transporter CAT-1 is associated with accumulation of L-arginine and cell growth in human colorectal cancer tissue. PLoS One 8, e73866 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Calder M. D., Watson P. H., Watson A. J., Culture medium, gas atmosphere and MAPK inhibition affect regulation of RNA-binding protein targets during mouse preimplantation development. Reproduction 142, 689–698 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Davis T. B., et al. , PTPRS regulates colorectal cancer RAS pathway activity by inactivating Erk and preventing its nuclear translocation. Sci. Rep. 8, 9296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tian X., Yang C., Yang L., Sun Q., Liu N., PTPRF as a novel tumor suppressor through deactivation of ERK1/2 signaling in gastric adenocarcinoma. OncoTargets Ther. 11, 7795–7803 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kong C., et al. , Overexpression of UNC5B in bladder cancer cells inhibits proliferation and reduces the volume of transplantation tumors in nude mice. BMC Canc. 16, 892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Akiyama H., et al. , The FBI1/Akirin2 target gene, BCAM, acts as a suppressive oncogene. PLoS One 8, e78716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin T. A., Ye L., Sanders A. J., Lane J., Jiang W. G., “Cancer invasion and metastasis: Molecular and cellular perspective” in Metastatic Cancer: Clinical and Biological Perspectives, Jandial R, Ed (Landes Bioscience, 2013). [Google Scholar]
- 71.Moon Y. W., et al. , LAMC2 enhances the metastatic potential of lung adenocarcinoma. Cell Death Differ. 22, 1341–1352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blaj C., et al. , Oncogenic effects of high MAPK activity in colorectal cancer mark progenitor cells and persist irrespective of RAS mutations. Canc. Res. 77, 1763–1774 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Sizemore S., Cicek M., Sizemore N., Ng K. P., Casey G., Podocalyxin increases the aggressive phenotype of breast and prostate cancer cells in vitro through its interaction with ezrin. Canc. Res. 67, 6183–6191 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Nielsen J. S., McNagny K. M., The role of podocalyxin in health and disease. J. Am. Soc. Nephrol. 20, 1669–1676 (2009). [DOI] [PubMed] [Google Scholar]
- 75.Paszek M. J., et al. , The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature 511, 319–325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Woods E. C., et al. , A bulky glycocalyx fosters metastasis formation by promoting G1 cell cycle progression. eLife 6, e25752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leithner K., et al. , Hypoxia increases membrane metallo-endopeptidase expression in a novel lung cancer ex vivo model—Role of tumor stroma cells. BMC Canc. 14, 40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jang T. J., Park J. B., Lee J. I., The expression of CD10 and CD15 is progressively increased during colorectal cancer development. Korean J. Pathol. 47, 340–347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei J., et al. , Profiling the surfaceome identifies therapeutic targets for cells with hyperactive mTORC1 signaling. Mol. Cell. Proteomics 19, 297– 307 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao Zw., Dong K., Zhang H.-z., The roles of CD73 in cancer. BioMed Res. Int. 2014, 460654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dulberger C. L., et al. , Human leukocyte antigen F presents peptides and regulates immunity through interactions with NK cell receptors. Immunity 46, 1018–1029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liao W. C., et al. , DSE regulates the malignant characters of hepatocellular carcinoma cells by modulating CCL5/CCR1 axis. Am. J. Cancer Res. 9, 347–362 (2019). [PMC free article] [PubMed] [Google Scholar]
- 83.Kawahara R., et al. , Mass spectrometry-based proteomics revealed Glypican-1 as a novel ADAM17 substrate. J. Proteomics 151, 53–65 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Morello A., Sadelain M., Adusumilli P. S., Mesothelin-targeted CARs: Driving T cells to solid tumors. Canc. Discov. 6, 133–146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stoyanova T., et al. , Regulated proteolysis of Trop2 drives epithelial hyperplasia and stem cell self-renewal via -catenin signaling. Genes Dev. 26, 2271–2285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Conacci-Sorrell M., et al. , The shed ectodomain of Nr-CAM stimulates cell proliferation and motility, and confers cell transformation. Canc. Res. 65, 11605–11612 (2005). [DOI] [PubMed] [Google Scholar]
- 87.Blanas A., Sahasrabudhe N. M., Rodríguez E., van Kooyk Y., van Vliet S. J., Fucosylated antigens in cancer: An alliance toward tumor progression, metastasis, and resistance to chemotherapy. Front. Oncol. 8, 39 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nagarajan A., Malvi P., Wajapeyee N., Heparan sulfate and heparan sulfate proteoglycans in cancer initiation and progression. Front. Endocrinol. 9, 483 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Knelson E. H., Nee J. C., Blobe G. C., Heparan sulfate signaling in cancer. Trends Biochem. Sci. 39, 277–288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Villacis R. A. R., et al. , Contribution of rare germline copy number variations and common susceptibility loci in Lynch syndrome patients negative for mutations in the mismatch repair genes. Int. J. Canc. 138, 1928–1935 (2016). [DOI] [PubMed] [Google Scholar]
- 91.Libisch M. G., et al. , GALNT11 as a new molecular marker in chronic lymphocytic leukemia. Gene 533, 270–279 (2014). [DOI] [PubMed] [Google Scholar]
- 92.Hussain M. R. M., Hoessli D. C., Fang M., N-acetylgalactosaminyltransferases in cancer. Oncotarget 7, 54067–54081 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bafna S., Kaur S., Batra S. K., Membrane-bound mucins: The mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene 29, 2893–2904 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Posey A. D., et al. , Engineered CAR T cells targeting the cancer-associated tn-glycoform of the membrane mucin MUC1 control adenocarcinoma. Immunity 44, 1444–1454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rivalland G., Loveland B., Mitchell P., Update on mucin-1 immunotherapy in cancer: A clinical perspective. Expert Opin. Biol. Ther. 15, 1773–1787 (2015). [DOI] [PubMed] [Google Scholar]
- 96.Vassilaros S., et al. , Up to 15-year clinical follow-up of a pilot phase III immunotherapy study in stage II breast cancer patients using oxidized mannan-MUC1. Immunotherapy 5, 1177–1182 (2013). [DOI] [PubMed] [Google Scholar]
- 97.Bierhuizen M. F., Mattei M. G., Fukuda M., Expression of the developmental I antigen by a cloned human cDNA encoding a member of a beta-1,6-N-acetylglucosaminyltransferase gene family. Genes Dev. 7, 468–478 (1993). [DOI] [PubMed] [Google Scholar]
- 98.Meyer S. J., Linder A. T., Brandl C., Nitschke L., B cell siglecs—News on signaling and its interplay with ligand binding. Front. Immunol. 9, 2820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu F. T., Rabinovich G. A., Galectins as modulators of tumour progression. Nat. Rev. Canc. 5, 29–41 (2005). [DOI] [PubMed] [Google Scholar]
- 100.Bair E. L., Nagle R. B., Ulmer T. A., Laferté S., Bowden G. T., 90K/Mac-2 binding protein is expressed in prostate cancer and induces promatrilysin expression. Prostate 66, 283–293 (2006). [DOI] [PubMed] [Google Scholar]
- 101.Grassadonia A., et al. , 90K (Mac-2 BP) and galectins in tumor progression and metastasis. Glycoconj. J. 19, 551–556 (2002). [DOI] [PubMed] [Google Scholar]
- 102.Lin T. W., et al. , Galectin-3 binding protein and galectin-1 interaction in breast cancer cell aggregation and metastasis. J. Am. Chem. Soc. 137, 9685–9693 (2015). [DOI] [PubMed] [Google Scholar]
- 103.Hua S., et al. , Differentiation of cancer cell origin and molecular subtype by plasma membrane N-glycan profiling. J. Proteome Res. 13, 961– 968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shackleton M., Normal stem cells and cancer stem cells: Similar and different. Semin. Canc. Biol. 20, 85–92 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.