Abstract
Here, we report the identification of Histoplasma causing an unexplained disease cluster in Matthews Ridge, Guyana. In March 2019, 14 employees of Chongqing Bosai Mining Company, China, working in a manganese mining of Guyana, had unexplained fever, and two of them died. We obtained lung and brain tissues as well as the blood samples from the two deceased cases (patient No. 1 and 2), and bronchoscopy lavages and cerebrospinal fluid samples from one severe case (patient No. 3), respectively. All samples were tested by pathological examination, high-throughput sequencing, and real-time PCR. Pathological detection showed the presence of spore-like structures in the lung tissue of patient No. 1, indicating a fungal infection in this patient. Nanopore sequencing identified the existing of H. capsulatum in the lung tissue sample within 13 h. Next-generation sequencing identified specific fragments of H. capsulatum in all of the samples tested (lung, brain and blood serum from the deceased cases, and plasma from the severe case). Real-time PCR assays did not reveal any viral infection related to transmission from bat feces. We conclude that H. capsulatum was the causative pathogen of this disease cluster based on epidemiologic, clinical, pathological and nucleic acid evidence.
Keywords: Histoplasma, Fever, Histochemical examination, Nanopore high-throughput sequencing, Real-time PCR
Highlights
-
•
Scientific question
This study reported the identification of Histoplasma as the cause of an unexplained disease cluster in Matthews Ridge, Guyana.
-
•
Evidence before this study
In March 2019, 14 Chinese employees from Chongqing Bosai Mining Company, China, were engaged in manganese mining in Guyana and presented with unexplained fever. Two of them died. After preliminary examination by the local hospital, some potential infectious pathogens were excluded, including Leptospira, HIV, influenza H1N1, Zika virus, Chikungunya virus, Dengue virus, and Influenza A and B viruses. Histoplasmosis is a fungal disease caused by members of the genus Histoplasma and is mainly prevalent in the American continents. Histoplasma is capable of survival in moist soils and can often be isolated from soils containing decaying feces of bats and birds. Human activities in the surface soil produce aerosols, which in turn are inhaled to cause infection.
-
•
New findings
In response to the unexplained disease cluster, pathological examination, high through-put sequencing and real-time PCR were implemented. A TGS platform found Histoplasma within 13 hours. NGS was also successfully applied in response to this event. Compared with NGS, the main features of nanopore sequencing are long sequencing ability, simplicity of use, the fastest turn-around time, high portability and real-time analysis of sequencing data. Though NGS had a longer turnaround time (24 hours), it worked well with different sample types (lung tissue, brain tissue and serum from deceased cases and plasma from a severe case) and was more sensitive than nanopore sequencing. Real-time PCR assays did not reveal any infection by viruses related to bat feces transmission. Pathological detection results showed the presence of spore-like structures, indicating fungus infection in this patient. All the results were consistent with the NGS analysis, supporting the fungus infection.
-
•
Significance of the study
We concluded that H. capsulatum is the causative pathogen for this disease cluster based on epidemiologic, clinical, pathological and nucleic acid supportive evidence.
1. Introduction
Histoplasmosis is a fungal disease caused by the family of genus Histoplasma, which exists prevalently in the Americas, and rarely reported in China. Histoplasma can survive at temperature 22–29 °C in moist soils, with high acidity and nitrogen content. They can often be isolated from soils containing decaying bat and bird feces, but rarely isolated from fresh feces. Human activities cause the formation of fungal aerosols from surface soil, which can cause infection once inhaled [[1], [2], [3]].
In March 2019, 14 employees from Chongqing Bosai Mining Company, China, working in a manganese mining of Guyana, suffered from unexplained fever. Ten of them were treated in the Georgetown Public Hospital (GPHC) of Guyana, one in the mine hospital, and one was isolated in their hotel. Two patients died. Ten patients and the brain and lung tissues of these deceased individuals were transported to China by a Chinese Red Cross rescue aircraft on April 5th. The autopsy was performed by local doctors in the GPHC. After preliminary examination of the 10 patients by GPHC, potential infectious pathogens like Leptospira, HIV, Influenza H1N1, Zika virus, Chikungunya virus, Dengue virus, and Influenza A and B viruses were excluded.
We report here the identification of Histoplasma as the pathogen of this unexplained disease cluster in Matthews Ridge Guyana. Pathological examination, high-throughput sequencing, and real-time PCR were applied, and the strategy and workflow of the identification were summarized in Figure 1 .
Figure 1.
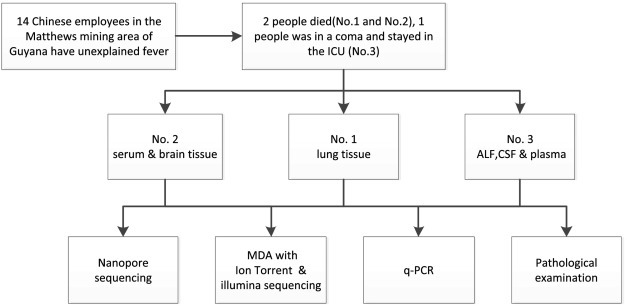
Strategy and workflow of pathogen identification.
2. Materials and methods
2.1. Sample preparation and nucleic acid extraction
Basic information of the 12 patients (two deaths, one severe case, and nine mild cases) and sample types were listed in Table 1 . All patients were male at ages from 30 to 56, and in general good health condition before infected. The 10 patients hospitalized in the GPHC showed all patients were male and their median age was 46 years. Tissue samples from the two deceased subjects (No. 1 and No. 2) were cut into 5 mm3 pieces, then put into freeze-storage tubes containing 500 μL sterile phosphate-buffered saline and repeatedly homogenized using an electric tissue grinder for 30 s with 1 min intervals until the tissues were completely dispersed. Nucleic acids were then extracted from 200 μL of homogenized samples using a QIAamp MinElute Virus Spin Kit (Qiagen, Germany) following the manufacturer's instructions. All operations were performed in a BSL-3 laboratory.
Table 1.
Basic demographic data of the patients involved in this epidemic.
| Patient No. | Age | Sample types |
|---|---|---|
| 1 | 46 | Lung tissue (autopsy) |
| 2 | 44 | Brain tissue (autopsy), serum |
| 3 | 42 | Plasma, bronchoscopy lavages, cerebrospinal fluid (severe, but survived) |
| 4 | 50 | Blood, nasopharyngeal swabs |
| 5 | 32 | Blood, nasopharyngeal swabs |
| 6 | 43 | Blood, nasopharyngeal swabs |
| 7 | 50 | Blood, nasopharyngeal swabs |
| 8 | 48 | Blood, nasopharyngeal swabs |
| 9 | 56 | Blood, nasopharyngeal swabs |
| 10 | 30 | Blood, nasopharyngeal swabs |
| 11 | 40 | Blood, nasopharyngeal swabs |
| 12 | 47 | Blood, nasopharyngeal swabs |
2.2. Pathological examination
2.2.1. Histochemical staining
Tissue specimens obtained from the autopsy of multiple tissue locations were embedded in paraffin blocks, then stained with hematoxylin-eosin (HE) and periodic acid-Schiff (PAS). For liquid specimens, including bronchoalveolar lavage fluid, anticoagulated blood, plasma, and cerebrospinal fluid (CSF) were made into smears; Giemsa staining and PAS staining were used respectively for histochemical examination.
2.2.2. Nanopore sequencing
For nanopore sequencing, the Ligation Sequencing Kit 1D R9 (ONT, SQK-LSK108) was applied to prepare libraries. End-repair was performed using NEBNext Ultra II End-repair/dA-tailing Module (New England BioLabs). Ligation was performed using a NEBNext Quick Ligation Module (New England BioLabs). The library was then ready for sequencing on the MinION system. During library incubation, the MinION was assembled with the Flow Cell Mk 1 Spot-ON Pk.1 (ONT, FLO-MIN 106 R9) and was connected to a computer (Windows 10; two Xeon E5-2640 v4; 128 GBDDR4 of 2133 MHz; SSD1TB) via a USB 3.0 hub. The sequencing library was loaded into the flow cell using Library Loading Bead Kit R9 (ONT, EXP-LLB001) according to the manufacturer's instructions. A 48-h sequencing plus base-caller protocol was selected in MinKNOW software and the sequencing process was stopped according to experimental needs [4]. Data were obtained by MinKNOW software and analyzed using the Albacore pipeline.
2.2.3. Next-generation sequencing (NGS)
NGS was independently performed by the National Institute for Viral Disease Control and Prevention (IVDC); Beijing University of Chemical Technology (BUCT); and Beijing Institute of Genome Research-Chinese Academy of Sciences and BGI Group (BGI). NGS is used by IVDC in response to emergency situations [[5], [6], [7]]. In the present case, the NGS processes in IVDC were conducted on an Ion Torrent PGM with Hi-Q View Kit and 318 v2 chip, and on an Illumina MiniSeq system with Nextera XT Kit and MiniSeq High-output sequencing Kit, and on an MGISEQ 2000. The data were downloaded and analyzed with our in-house Virus Identification Pipeline (VIP). Reads from NGS platforms were analyzed by an advanced version of the VIP [8]. Based on the results of third-generation sequencing (TGS, specifically nanopore sequencing in this study), we used the genome sequence of H. capsulatum as a reference for VIP. Generally, raw NGS reads were first preprocessed by the removal of adapter, low-quality, and low-complexity sequences. Next, clean reads were classified at the genus level. This step performed both nucleotide and amino acid homology alignments against a custom database which included the host genome (hg38) and a comprehensive collection of bacteria, fungi, parasite and virus sequences (derived from NT collection, May 2018) by Centrifuge [9] and Kaiju [10]. In addition, all classified reads under a genus were subject to de novo assembly and phylogenetic analysis [11]. The selected high-confidence reads and contigs were rechecked on web BLAST.
2.2.4. Real-time PCR
Real-time PCR was performed on all samples from the 12 patients to screen for Influenza A virus (in-house developed), Coronavirus (in-house developed), Black creek canal virus (in-house developed), Andes virus (in-house developed), Guanarito virus (in-house developed), Sabia virus (in-house developed), Nipah virus (in-house developed), Flavivirus (in-house developed), Leptospira [12,13], and Histoplasma [14,15], according to the cited publications or in-house protocols.
3. Results
3.1. Clinical symptoms and epidemiologic investigation
Most patients started with fever, headache, myalgia, loss of appetite, and hemorrhagic eruption on the skin. Two patients had intestinal symptoms. No human-to-human transmission was identified as the interpreter and patient roommates remained healthy. Six patients were admitted to the hospital on March 30th, and four on April 1st, 2019. The median time from onset to admission was 4 days (range 1–8 days).
3.2. Pathological analysis
Monocytes and neutrophils were examined in the plasma sample from patient No. 2 (deceased), and no abnormalities were observed. Bacteria were not observed in the plasma sample from patient No. 2, or in the CSF sample from patient No. 3 (severe case but survived). In the Giemsa-stained bronchoalveolar lavage fluid smear from patient No. 3, a large number of bacteria and scattered neutrophils were visible (Figure 2 ). PAS staining of the paraffin sections of the lung tissue sample from deceased patient No. 1 showed spore-like structures (Figure 3 ).
Figure 2.
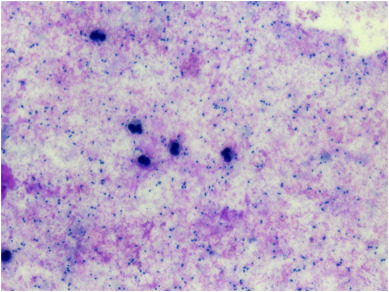
Giemsa stain of bronchoalveolar lavage fluid from patient No. 3 (severe but survived).
Figure 3.
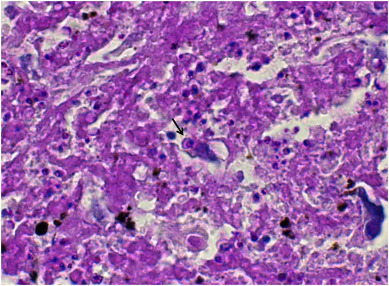
Periodic acid-Schiff staining of the lung tissue from patient No. 1 (deceased). The arrow indicates a spore-like structure.
Figure 4.
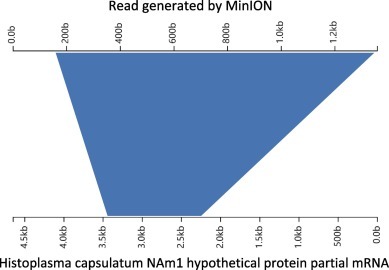
Aligned result of a typical nanopore sequencing read obtained from the lung tissue of patient No. 1.
3.3. High-throughput sequencing for samples from the two deceased cases and one severe case
After nucleic acid extraction was completed, NGS and TGS were simultaneously implemented by IVDC. Brain tissue and serum from patient No. 2, and lung tissue from patient No. 1 were detected sequentially using the MinION system. The rate of data generation was monitored, and sequencing was stopped when the rate of data generation became very slow or the number of active nanopores was significantly lower. The sequencing times for the serum, brain tissue and lung tissue were approximately 10, 5, and 13 h, respectively, resulting in 252,000 reads, 96,261 reads, and 1,553,164 reads, respectively. No significant evidence was found in data from the brain tissue or serum sample obtained by MinION, but the reads of Histoplasma were identified in the lung tissue sample. A read of 280 bp was generated during the sequencing and matched with the NCBI (nt/nr) database sequence of Histoplasma NAm1 predicted protein partial mRNA (accession no. XM_001538005.1) with an identity of 93%. Through analysis of all data from the lung tissue sample, a read of 1358 bp was found to have an identity of 90.88% with Histoplasma NAm1 hypothetical protein partial mRNA (accession no. XM_001541862.1) (Figure 4 ) [16].
Figure 5.
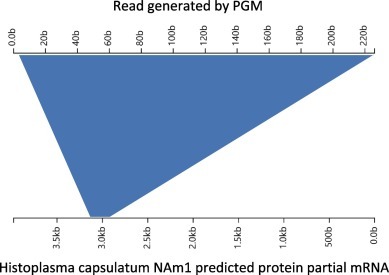
Aligned result of typical PGM sequencing read obtained from plasma from patient No. 3.
The serum sample from patient No. 2 and mixed tissue samples from patients No. 1 and 2 were also tested sequentially using the Ion Torrent PGM system, while a bronchoalveolar lavage fluid (BALF) sample, plasma and CSF from patient No. 3 (severe case were tested by IVDC using the Illumina MiniSeq apparatus). The time for Ion Torrent sequencing of the deceased patients was around 40 h; a total of 9,810,285 reads were obtained from the serum, and a total of 11,284,703 reads were obtained from the mixed tissue samples. The time for Illumina sequencing of samples from the severe case was around 20 h; a total of 5,829,534 reads, 8,739,522 reads, and 6,100,936 reads were obtained from the BALF, plasma and CSF sample respectively. The data generated by the Ion Torrent platform were aligned with the genomic database of Histoplasma, and specific reads were found in the serum and mixed brain and lung tissue from the deceased patients and in the plasma from the severe case (Figure 5 ) [16].
3.4. Real-time PCR
Regardless of the sample type (whole blood, serum, CSF, or nasopharyngeal swabs) or severity of disease of these patients, real-time PCR results for all the samples were virus-negative (data not shown).
4. Discussion
Generally, Histoplasma can be diagnosed by microscopic identification of H. capsulatum from the microbial culture of tissues or body fluids or from biopsy tissue samples of patients H. Capsulatum can be found in pathological sections by PAS staining, Gomori hexamine silver staining, and Wright-Giemsa staining, but not by HE staining. The fungus needs to be distinguished from other pathogens such as Penicillium marneffei and Leishmania, which to a great degree depends on the experience of the expert in pathobiology. The positive rate of clinical specimen culture is related to the symptom severity of the disease and the quality of the sample. Detection of Histoplasma antigens targeting the Histoplasma polysaccharide antigen in urine and serum samples can be also used for the diagnosis of this disease, and usually, urinary detection is more sensitive than serum detection. Unfortunately, commercial kits to detect Histoplasma antigens based on enzyme immunoassay are still not available in China. PCR, nested PCR and real-time PCR using Histoplasma specific primers were reported to distinguish cultured Histoplasma from other pathogenic fungi [14]. There are few reports on the detection of capsular Histoplasma by high-throughput sequencing.
Human activities such as excavation, demolition, and cave exploration in areas where Histoplasma is endemic result in the risk of infection. Therefore, construction workers, demolition workers, farmers, gardeners, air-conditioning maintenance personnel, historic building reformers, geologists, cave explorers, and so on are at high risk of infection. In the event described here, all patients were exposed to bat excretion during work in four abandoned mining tunnels in Matthews Ridge, Guyana. The presenting clinical symptoms of the patients and the mortality rate (10%–20%) were in accordance with those expected for Histoplasma infection [17,18].
Different testing methods were implemented in response to this unexplained disease cluster. A TGS platform (nanopore) was the first to provide evidence that Histoplasma caused this outbreak. In comparison with NGS, the advantages of nanopore sequencing are its ability to record long sequences. In addition, the system is simple to use, fast, high portability, thus, allows it to be used on real-time analysis of sequencing. MinION has been increasingly applied in the fields of virus detection, viral whole-genome sequencing, and viral evolution [[19], [20], [21], [22]]. NGS was also successfully applied in response to the event described in this paper. Though NGS using MiniSeq and MGI2000 systems worked slower (24 h) than nanopore sequencing (13 h), it fitted well with different sample types and was more sensitive. The sensitivity is closely related to the read numbers by NGS. We obtained read numbers of 228,453,930, 189,759,510 and 236,103,930 from the three samples (BALF, CSF and plasma) obtained from the severe case by, and all of them tested positive for H. capsulatum. In this case, NGS also identified specific fragments of Histoplasma in all tested samples (lung, brain and blood serum) from the deceased patients, which consolidated the identification of Histoplasma as the causative pathogen, indicating higher detection sensitivity by NGS than nanopore sequencing.
Real-time PCR assays did not reveal any infection with viruses related to bat feces transmission in any samples we tested, indicating viral agents were likely, not involved in this epidemic. Pathological detection results showed the presence of spore-like structures in the lung tissue from deceased patient No. 1, indicating a fungal infection in this patient.
Additionally, however, NGS and TGS technology showed many reads from Acinetobacter baumannii and herpes simplex virus 1 (HSV1) in the bronchoscopy lavage samples from the severe case (patient No. 3). These results indicated that the severe case was originally infected with capsular Histoplasma, and then infected with A. baumannii, and latent HSV1. These conclusions provide important clues for further study of the pathogenesis of Histoplasma.
The presence of Histoplasma nucleic acids in tissue, serum, and plasma from postmortem and severely ill patients were cross-validated by three independent laboratories (China CDC, Beijing University of Chemical Technology and BGI) and Histoplasma antigen was detected in five patients by the CARPHA Laboratory, Guyana, using enzyme immunoassay on April 12th, 2019. We conclude that Histoplasma was the causative pathogen of this disease cluster based on epidemiologic, clinical, pathological and nucleic acid evidence.
Ethics statement
All aspects of this study were performed in accordance with national ethics regulations and approved by the Institutional Review Boards of the National Institute for Viral Disease Control and Prevention, Center for Disease Control and Prevention of China. Written consent was obtained from the patients for using their data for analysis and to improve patient care activities. Their identities remain confidential.
Acknowledgments
Acknowledgments
We are grateful to the BGI team for helpful advice. We appreciate physicians providing clinical data to the Chinese Center for Disease Control and Prevention, and all the staff members who were responsible for specimen collection and shipment in provincial Centers for Disease Control and Prevention. This work was supported by grants from the China Mega-Projects for Infectious Disease (2018ZX10713-002, 2018ZX10711001, 2017ZX10104001, and 2017ZX10302301-004-002).
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Author contributions
G. F. Gao, X. Ma, and G. Wu designed the study; J. Wang, Y. Zhang, W. Zhou, H. Ling, R. Zhang, R. Wang, J. Li, Y. Zhang, J. Song, W. J. Liu, W. Zhen, K. Cai, S. Zhu, D. Wang, J. Xiao, Y. Tong, W. Liu, L. Song, W. Wu, Y. Liu, X. Zhao, R. Lu, B. Huang, and F. Ye performed the experiments; X. Dong, WenboXu, W. Lei, R. Gao, S. Ye, J. Wang, Q. Shi, C. Chen, and J. Han analyzed and interpreted the data; X. Ma, J. Wang, and Y. Zhang wrote the paper. All authors provided a critical review and approved the final manuscript.
Contributor Information
George F. Gao, Email: gaofu@chinacdc.cn.
Xuejun Ma, Email: maxj@ivdc.chinacdc.cn.
Guizhen Wu, Email: wgzcdc@hotmail.com.
References
- 1.Sepúlveda V.E. Genome sequences reveal cryptic speciation in the human pathogen Histoplasma capsulatum. mBio. 2017;8(6):e01339–17. doi: 10.1128/mBio.01339-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mok W.Y., Luiz O R.C., Silva M.D.S., Barreto D. Isolation of fungi from bats of the Amazon basin. Appl. Environ. Microbiol. 1982;44(3):570–575. doi: 10.1128/aem.44.3.570-575.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan W., Dermatology D.O. The epidemic characteristics and prevention and treatment of histoplasmosis in China. Dermatology Bulletin. 2017;5:101–110. [Google Scholar]
- 4.Wang J. Rapid and accurate sequencing of enterovirus genomes using MinION nanopore sequencer. Biomed. Environ. Sci. 2017;30(10):718–726. doi: 10.3967/bes2017.097. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z. A fatal yellow fever virus infection in China: description and lessons. Emerg. Microbes Infect. 2016;5(7):e69. doi: 10.1038/emi.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C.H. An improved barcoded oligonucleotide primers-based next-generation sequencing approach for direct identification of viral pathogens in clinical specimens. Biomed. Environ. Sci. 2017;30(1):22–34. doi: 10.3967/bes2017.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y. Next-generation sequencing study of pathogens in serum from patients with febrile jaundice in Sierra Leone. Biomed. Environ. Sci. 2019;32(5):363–370. doi: 10.3967/bes2019.048. [DOI] [PubMed] [Google Scholar]
- 8.Li Y. VIP: an integrated pipeline for metagenomics of virus identification and discovery. Sci. Rep. 2016;6:23774. doi: 10.1038/srep23774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim D. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26(12):1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menzel P., Ng K.L., Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016;7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camacho C. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteves L.M. Diagnosis of human leptospirosis in a clinical setting: real-time PCR high resolution melting analysis for detection of Leptospira at the onset of disease. Sci. Rep. 2018;8(1):9213. doi: 10.1038/s41598-018-27555-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riediger I.N. Rapid, actionable diagnosis of urban epidemic leptospirosis using a pathogenic Leptospira lipL32-based real-time PCR assay. PLoS Negl. Trop. Dis. 2017;11(9):e0005940. doi: 10.1371/journal.pntd.0005940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muraosa Y. Detection of Histoplasma capsulatum from clinical specimens by cycling probe-based real-time PCR and nested real-time PCR. Med. Mycol. 2016;54(4):433–438. doi: 10.1093/mmy/myv106. [DOI] [PubMed] [Google Scholar]
- 15.Koepsell S.A., Hinrichs S.H., Iwen P.C. Applying a real-time PCR assay for Histoplasma capsulatum to clinically relevant formalin-fixed paraffin-embedded human tissue. J. Clin. Microbiol. 2012;50(10):3395–3397. doi: 10.1128/JCM.01705-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wintersinger J.A., Wasmuth J.D. Kablammo: an interactive, web-based BLAST results visualizer. Bioinformatics. 2014;31(8):1305–1306. doi: 10.1093/bioinformatics/btu808. [DOI] [PubMed] [Google Scholar]
- 17.Wheat L.J. Histoplasmosis. Infect Dis Clin North Am. 2016;30(1):207–227. doi: 10.1016/j.idc.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Azar M.M., Hage C.A. Clinical perspectives in the diagnosis and management of histoplasmosis. Clin. Chest Med. 2017;38(3):403–415. doi: 10.1016/j.ccm.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Warwick-Dugdale J. Long-read viral metagenomics captures abundant and microdiverse viral populations and their niche-defining genomic islands. PeerJ. 2019;7:e6800. doi: 10.7717/peerj.6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oude Munnink B.B. Towards high quality real-time whole genome sequencing during outbreaks using Usutu virus as example. Infect. Genet. Evol. 2019;73:49–54. doi: 10.1016/j.meegid.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Naveca F.G. Genomic, epidemiological and digital surveillance of Chikungunya virus in the Brazilian Amazon. PLoS Negl. Trop. Dis. 2019;13(3):e0007065. doi: 10.1371/journal.pntd.0007065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchida Y. A case of genotype-3b hepatitis C virus in which the whole genome was successfully analyzed using third-generation nanopore sequencing. Hepatol. Res. 2019;49(9):1083–1087. doi: 10.1111/hepr.13339. [DOI] [PubMed] [Google Scholar]


