Key points
-
•
Syndromic and point-of-care molecular diagnostic testing methods have revolutionized infectious disease testing by permitting the simultaneous detection of multiple pathogens and drug resistance mechanisms directly from clinical specimens and positive blood cultures.
-
•
Syndromic testing panels currently include those designed for the detection of pathogens responsible for bloodstream, central nervous system, gastrointestinal tract, and respiratory tract infections as well as some biothreat agents.
-
•
Point-of-care molecular diagnostic systems afford providers the ability to generate laboratory-quality results without the need for confirmatory testing of negative specimens.
-
•
Implementation of syndromic and point-of-care molecular infectious disease testing methods has curbed the unnecessary use of antibiotics, strengthening antimicrobial stewardship practices.
-
•
Development of methods for the detection of infectious diseases beyond those that are detectable by current technologies is an area of investigation.
Introduction
For decades, the clinical laboratory diagnosis of many infectious diseases relied solely on time-consuming and often labor-intensive manual cultivation-based, microscopic, and immunoserologic methods that required experienced technical personnel to perform and interpret. The introduction of semiautomated and fully automated microbial phenotyping systems in the later part of the 20th century vastly improved the processes of bacterial and yeast isolate workup by decreasing identification and antimicrobial susceptibility testing turnaround times (TATs); however, the detection of many viruses and parasites still required traditional techniques. Subsequent improvements to automated platforms, including the refinement of the automated expert systems used by these devices to generate and interpret data, led to further decreases in TATs and a concomitant increase in culture throughput.
In the 1990s, the era of molecular diagnostics was ushered in with the introduction of nucleic acid analysis methods, including the hybridization protection assay (eg, AccuProbe, Salem, MA) for the identification of isolates and polymerase chain reaction (PCR) for the detection of pathogens directly from patient specimens [1], [2], [3], [4], [5], [6], [7]. Later, the implementation of real-time nucleic acid amplification chemistries permitted both faster pathogen detection and the enablement of nucleic acid quantitation [8]. These methods quickly migrated from research laboratories into clinical laboratories, first as laboratory-developed tests or so-called home brew tests. This revolution enabled laboratorians to more precisely identify the causes of infectious diseases by detecting pathogen-specific nucleotide sequences in cultured isolates and clinical specimens. Many of these methods proved to be far superior in terms of accuracy and result TAT compared with cultivation-dependent approaches, especially for the identification of viruses. Over time, these methods also underwent refinements that included the adaptation of several assays to automated platforms, allowing users to minimize the manual handling of specimens and reaction components.
One recent advancement in pathogen detection is the syndromic approach in which groups of pathogens are tested for simultaneously in a single reaction vessel. These assays incorporate components of older methods, including real-time PCR; however, rather than ordering separate tests for various pathogens, the syndromic tests allow simultaneous detection of a variety of agents that are associated with a specific disease syndrome [9].
The newest systems are those that allow users to perform laboratory-quality molecular testing at the point of patient care, a major advancement that has moved molecular pathology to the forefront of modern diagnostics. Many Clinical Laboratory Improvement Amendments (CLIA)-waived point-of-care (POC) systems are now available and permit rapid result reporting, enabling prescription of targeted treatment at the time of clinic visit. The current assays on these platforms largely target infections diagnosed in the ambulatory setting such as influenza and streptococcal pharyngitis.
Significance
Syndromic and POC molecular testing methods have revolutionized the diagnosis of infectious diseases by increasing the accuracy of microbial detection, substantially decreasing the time needed to generate clinically useful laboratory test results, and enabling the performance of laboratory-quality testing at or near the point of care by nonlaboratorians [9]. In return, patients are able to receive appropriate treatment sooner, avoiding prolonged exposure to unnecessary antimicrobial drugs, thereby avoiding the selection of drug-resistant pathogens and strengthening antimicrobial stewardship practices [9], [10], [11]. In addition, many of these methods permit the rapid detection of pathogens that pose significant infection control and public health hazards, including high-consequence and travel-related pathogens (eg, Bacillus anthracis, Ebola virus, and Plasmodium spp.) that can be associated with either naturally acquired infections or infections resulting from the deliberate release of these agents. As a consequence of the rapid TATs of these tests, containment and epidemiologic interventions can be instituted very soon after specimen acquisition.
In addition to the syndromic and POC molecular testing solutions available for aiding in the identification of pathogens, many test panels are also capable of detecting antimicrobial resistance mechanisms [9], [12], [13], the rapid detection of which affords clinicians and infection control practitioners the ability to quickly implement appropriate therapies and infection control precautions. The diagnostic power of these technologies coupled with their user friendliness have made them highly attractive alternatives to traditional methods that rely on the procurement of isolates. As a consequence, methods such as viral culture and direct immunofluorescence have largely disappeared from modern clinical microbiology laboratories. With these methods removed, many laboratories have streamlined the workup of clinical specimens by using one syndromic testing platform for the analysis of a variety of specimen types for a large array of pathogens.
Present relevance and future avenues to consider/investigate
Currently, numerous diagnostic product manufacturers market infectious disease syndromic panels and POC molecular tests that are available in a variety of formats, including customizable panels, single-analyte tests, and CLIA-waived and moderate complexity systems. These tests were designed to provide all of the advantages that traditional molecular diagnostic tests offer plus the benefits of rapid result delivery, portability, and ease of use. The ability of many of these test platforms to be successfully used by nonlaboratorians have made them amenable to deployment in patient care facilities such as clinics and hospital emergency departments, which are traditionally not staffed by medical laboratory scientists.
Several syndromic and POC testing systems that provide qualitative results are described herein. Please note that not all available systems are mentioned, but those with a visible market presence in the United States are discussed.
Currently Available Syndromic Panel Platforms
BD MAX system
The BD MAX System (BD Diagnostics, Quebec, Canada; Fig. 1 ) is an automated real-time PCR platform using TaqMan hydrolysis probes for detection. In addition to syndromic panels (see Table 1 and Table 3), the system is designed to run singleplex and user-defined assays. To perform testing, specimens in sample buffer, reagents, extraction wells, pipette tips, and a real-time PCR microfluidic cartridge is placed on board the BD MAX instrument, which automates all sample handling and real-time PCR steps (see Fig. 1). Up to 24 assays can be run simultaneously and results are available within 3 hours. Panel performance characteristics are reviewed in refs. [14], [15], [16], [17].
Fig. 1.

The BD MAX System, which is comprised of (A) a rack for holding specimens (blue-capped tubes in front) and reagent strip, (B) the PCR cartridge, and (C) the BD MAX instrument.
(Courtesy of BD, Sparks, MD; with permission.)
Table 1.
Overview of Analytes Detected by the BD MAX Women’s Health Panels, CT/GC/TV, and Vaginal Panel
| Analyte | FDA Cleared? | Atopobium vaginae | Bacterial Vaginosis-Associated Bacteria-2 | Candida spp.a | Chlamydia trachomatis | Gardnerella vaginalis | Lactobacillus spp.b | Megasphaera-1 | Neisseria gonorrhoeae | Trichomonas vaginalis |
|---|---|---|---|---|---|---|---|---|---|---|
| BD MAX CT/GC/TV | + | − | − | − | + | − | − | − | + | + |
| BD MAX Vaginal Panelc | + | + | + | + | − | + | + | + | − | + |
Abbreviations: CT/GC/VT, Chlamydia trachomatis/gonococcus (Neisseria gonorrhoeae)/Trichomonas vaginalis; FDA, US Food and Drug Administration.
Symbols: +, yes or present on panel; -, no or absent from panel.
The BD MAX vaginal panel detects Candida albicans, C dubliniensis, C glabrata, C krusei, C parapsilosis, and C tropicalis but reports them as “Candida group” for all except C glabrata and C krusei, which it reports individually.
The BD MAX Vaginal Panel detects Lactobacillus crispatus and L jensenii, which are grouped as Lactobacillus species.
Detection of 1 or more bacterial analytes are reported as positive or negative for bacterial vaginosis.
Table 3.
Syndromic Panels that Are Currently Available in the United States for the Detection of Gastrointestinal Tract Pathogens
| Analyte | FDA Cleared? | Adenovirus F 40/41 | Astrovirus | Campylobacter spp. | Clostridium difficile (Toxin A/B) | Cryptosporidium spp. | Cyclospora cayetanensis | Entamoeba histolytica | Escherichia coli O157 | E coli (EAEC) | E coli (EPEC) | E coli (EIEC) | E coli (ETEC) | E coli (STEC) or stx1 and stx2 | Giardia intestinalis (G lamblia) | Norovirus | Plesiomonas shigelloides | Rotavirus | Salmonella spp. | Sapovirus | Shigella spp. | Vibrio cholerae | Vibrio parahaemolyticus | Vibrio vulnificus | Yersinia enterocolitica |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BD MAX Enteric Bacterial Panel | + | − | − | + | − | − | − | − | + | − | − | + | − | + | − | − | − | − | + | − | + | − | − | − | − |
| BD MAX Extended Enteric Bacterial Panel | + | − | − | + | − | − | − | − | + | − | − | − | + | + | − | − | + | − | + | − | + | + | + | + | + |
| BD MAX Enteric Parasite Panel | + | − | − | − | − | + | − | + | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − |
| FilmArray GI Panela | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| VERIGENE EP Testb | + | − | − | + | − | − | − | − | − | − | − | − | − | + | − | + | − | + | + | − | + | + | + | − | + |
| xTAG GPPc | + | + | − | + | + | + | − | + | + | − | − | − | + | + | + | + | − | + | + | − | + | + | − | − | − |
Abbreviations: EIEC, Shigella/enteroinvasive E coli; EAEC, Enteroaggregative E coli; EPEC, Enteropathogenic E coli; ETEC, Enterotoxigenic E coli; STEC, Shiga Toxin-producing E coli; FDA, US Food and Drug Administration; GI, gastrointestinal; GPP, gastrointestinal pathogen panel.
Symbols: +, yes or present on panel; -, no or absent from panel.
The FilmArray GI panel detects Campylobacter jejuni, C coli, and C upsaliensis and reports them as “Campylobacter (jejuni, coli and upsaliensis).” Shigella spp. are reported along with E coli (EIEC) as “Shigella/enteroinvasive E coli (EIEC).” V cholerae, V parahaemolyticus, and V vulnificus are detected and are reported as “Vibrio (parahaemolyticus, vulnificus and cholerae); however, V cholerae is also reported independently if it alone is detected. Norovirus genogroups I and II are detected, and astrovirus genotypes I, II, IV, and V are detected.
The VERIGENE EP test detects C jejuni, C coli, and C lari and reports them as Campylobacter group. Shigella boydii, S dysenteriae, S flexneri, and S sonnei are detected and reported as Shigella spp. V cholerae and V parahaemolyticus are detected and reported as Vibrio group. Norovirus genogroups I and II are detected.
The xTAG GPP detects C jejuni, C coli, and C lari and reports them as Campylobacter; Cryptosporidium parvum and C hominis, and reports them as Cryptosporidium; and S boydii, S dysenteriae, S flexneri, and S sonnei, and reports them as Shigella.
ePlex, ePlex NP, and eSensor XT-8 systems
The ePlex, ePlex NP, and XT-8 systems (GenMark Diagnostics, Inc., Carlsbad, CA; Fig. 2 ) use patented eSensor technology to detect a variety of pathogens directly from patient specimens (ePlex and ePlex NP systems) and amplified nucleic acid mixtures (eSensor XT-8 system).
Fig. 2.
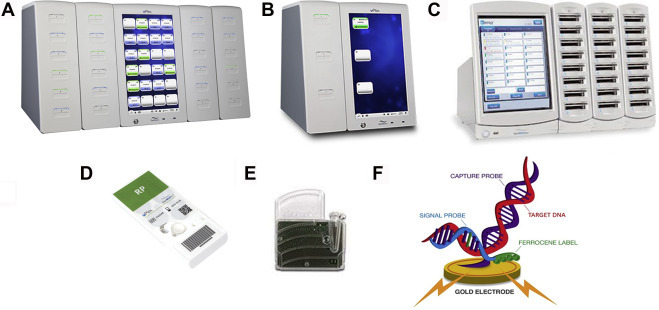
The ePlex (A), ePlex NP (B), and eSensor XT-8 (C) systems available from GenMark Diagnostics. Also shown are examples of ePlex (D) and XT-8 (E) panels. The nucleic acid hybridization complex described in the text is shown in (F).
(Courtesy of GenMark Diagnostics, Carlsbad, CA; with permission.)
For the ePlex system, the specimen in a buffer solution is added to the ePlex panel in which all liquid handling steps are performed by digital microfluidics technology (electrowetting) [14]. Electrochemical detection of ferrocene-labeled PCR amplicons occurs via capture probes that have been immobilized on gold-plated electrodes [15]. Results are generally available within 90 minutes, and anywhere from 3 to 24 assays can be ran simultaneously, depending on the instrument configuration (the ePlex NP performs a maximum of 3 tests at once). In contrast, the eSensor XT-8 system, the predecessor of the ePlex system, is designed to simultaneously interrogate up to 24 samples that have undergone offline nucleic acid amplification; results are available within 30 minutes [15]. Analytes detectable by the current panels are listed in Table 2 and performance characteristics and impacts of these systems are discussed in refs. [18], [19], [20], [21].
Table 2.
Syndromic Panels that Are Currently Available in the United States for the Detection of Upper Respiratory Tract Pathogens
| Analyte | FDA Cleared? | Adenovirus | Adenovirus Subtyping | Bordetella bronchiseptica | Bordetella holmesii | Bordetella pertussis/B. parapertussis | Chlamydophila pneumoniae | Coronavirus HKU1 | Coronavirus NL63 | Coronavirus 229E | Coronavirus OC43 | Enterovirus/Rhinovirus | Human Bocavirus | Human Metapneumovirus | Human Metapneumovirus Subtyping | Human Parainfluenza Virus 1 | Human Parainfluenza Virus 2 | Human Parainfluenza Virus 3 | Human Parainfluenza Virus 4 | Influenza A | Influenza A Subtyping | Influenza B | Legionella pneumophila | Mycoplasma pneumoniae | Respiratory Syncytial Virus | Respiratory Syncytial Virus Subtyping |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ePlex RP | + | + | − | − | − | −/− | + | + | + | + | + | + | − | + | − | + | + | + | + | + | + | + | − | + | + | + |
| eSensor RVP | + | + | + | − | − | − | − | − | − | − | − | +c | − | + | − | + | + | + | − | + | + | + | − | − | + | + |
| FilmArray RP Panel | + | + | − | − | − | +/− | + | + | + | + | + | + | − | + | − | + | + | + | + | + | + | + | − | + | + | − |
| FilmArray RP2 Panel | + | + | − | − | − | +/+ | + | + | + | + | + | + | − | + | − | + | + | + | + | + | + | + | − | + | + | − |
| FilmArray RP EZa | + | + | − | − | − | +/− | + | + | + | + | + | +/+ | − | + | − | + | + | + | + | + | + | + | − | + | + | − |
| NxTAG RPPb | + | + | − | − | − | −/− | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | − | − | + | + |
| VERIGENE RP Flexb | + | + | − | + | + | +/+ | − | − | − | − | − | +c | − | + | − | + | + | + | + | + | + | + | − | − | + | + |
Abbreviations: FDA, US Food and Drug Administration; RPP, respiratory pathogen panel.
Symbols: +, yes or present on panel; -, no or absent from panel.
The FilmArray RP EZ assay tests for human coronaviruses HKU1, NL63, 229E, and OC43 and human parainfluenza viruses 1, 2, 3, and 4, but it reports those analytes as coronavirus and parainfluenza virus, respectively.
The NxTAG and VERIGENE RP flex assays allow users to selectively report targets.
Rhinovirus alone is reported by the eSensor RVP, VERIGENE RP flex, US versions of the NxTAG RVP assays.
FilmArray system
The FilmArray system (BioFire Diagnostics and BioFire Defense, LLC, Salt Lake City, UT; Fig. 3 ) family of syndromic panels includes several assays that are cleared by the US Food and Drug Administration (FDA) and several research use only assays (Box 1 , see Table 2; Tables 3 and 4 ). Of note, BioFire Defense offers research-use-only reagents panels that are designed for detection of high-consequence and emerging pathogens whose detection could signal possible bioterrorism events, and for pathogens associated with travel to areas of the world where certain infectious diseases that are rare in the United States are endemic (Table 5 ).
Fig. 3.

The FilmArray system. The FilmArray pouch (A) and (B) the FilmArray 2.0 and (C) Torch instruments.
(Courtesy of BioFire Diagnostics, Salt Lake City, UT; with permission.)
Box 1. Central Nervous System Pathogens Detected by the FilmArray ME Panel.
Bacteria
Escherichia coli K1
Haemophilus influenzae
Listeria monocytogenes
Neisseria meningitidis
Streptococcus agalactiae
Streptococcus pneumoniae
Fungia
Cryptococcus neoformans/C gattii
Viruses
Cytomegalovirus
Enterovirus
Herpes simplex virus 1
Herpes simplex virus 2
Human herpes virus 6
Human parechovirus
Varicella-zoster virus
a The FilmArray ME Panel detects C neoformans and C gattii but reports them together as C neoformans/C gattii.
Table 4.
Syndromic Panels that Are Currently Available in the United States for the Detection of Bloodstream Infection Pathogens
| Analyte | FDA Cleared? | Acinetobacter baumannii or Acinetobacter spp. | Candida spp. | Citrobacter spp. | Enterobacteriaceae | Enterobacter spp. | Enterococcus spp. | Escherichia coli | Haemophilus influenzae | Klebsiella oxytoca | Klebsiella pneumoniae | Listeria spp. | Micrococcus spp. | Neisseria meningitidis | Proteus spp. | Pseudomonas aeruginosa | Serratia spp. | Staphylococcus spp. | Staphylococcus aureus | Streptococcus spp. | Streptococcus agalactiae | Streptococcus pneumoniae | Streptococcus pyogenes | Carbapenemase Genes | Extended-Spectrum β-Lactamase Genes | mecA | vanA/vanB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FilmArray BCID Panel | + | + | + | − | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | − | + | + |
| VERIGENE GN BC Test | + | + | X | + | − | − | X | + | − | − | + | X | X | − | + | + | + | X | X | X | X | X | X | + | + | X | X |
| VERIGENE GP BC Test | + | X | X | X | X | X | + | X | X | X | X | X | + | X | X | X | X | + | + | + | + | + | + | X | X | + | + |
Abbreviation: FDA, US Food and Drug Administration.
Symbols: +, yes or present on panel; -, no or absent from panel.
Table 5.
Multiplex Nucleic Acid Amplification Panels Available for the Detection of High-Consequence and Travel-Associated Pathogens
| Analyte | FDA Cleared? | Bacillus anthracis | Brucella melitensis | Burkholderia mallei /B pseudomallei | Chikungunya Virus | Clostridium botulinum | Coxiella burnettii | Crimean-Congo Hemorrhagic Fever Virus | Dengue Virus | Ebola Virus (Zaire ebolavirus) | EEE Virus | Francisella tularensis | Lassa Virus | Leptospira spp. | Leishmania spp. | Marburg Virus | Orthopox Virus | Plasmodium spp. | Ricinus communis | Rickettsia prowazekii | Salmonella enterica Serovar. Paratyphi A | S enterica Serovar. Typhi | Variola Virus | VEE Virus | WEE Virus | Yellow Fever Virus | Yersinia pestis | West Nile Virus | Zika Virus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FilmArray BioThreat Panela | − | + | + | + | − | + | + | − | − | + | + | + | − | − | − | + | + | − | + | + | − | − | + | + | + | − | + | − | − |
| FilmArray BioThreat−E Testb | −c | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| FilmArray Global Fever Paneld | − | + | − | − | + | − | − | + | + | + | − | + | + | + | + | + | − | +e | − | − | + | + | − | − | − | + | − | + | + |
Abbreviations: EEE virus, eastern equine encephalitis virus; FDA, US Food and Drug Administration; VEE virus, Venezuelan equine encephalitis virus; WEE, western equine encephalitis virus.
Symbols: +, yes or present on panel/test; -, no or absent from panel/test.
The FilmArray BioThreat Panel is designed to detect analytes from swab, liquid, culture, and powder samples.
The FilmArray BioThreat-E Test is designed to detect Ebola virus (Zaire ebolavirus) in blood and urine samples.
The FilmArray BioThreat-E Test was granted FDA EUA status in 2014 and it is only authorized for use by CLIA moderate and high complexity laboratories throughout the duration of the EUA. This test is intended to detect Ebola virus (Zaire ebolavirus) from blood and urine specimens.
The FilmArray Global Fever Panel is designed to detect analytes in whole blood.
The FilmArray Global Fever Panel detects and reports Plasmodium spp. and it also distinguishes Plasmodium falciparum from Plasmodium ovale/Plasmodium vivax and reports them as such.
The FilmArray System incorporates lyophilized reagents and assay reaction vessels on a small plastic film pouch topped by a solid plastic reagent housing (Fig. 3A). After reagent rehydration, the sample in buffer is added to a pouch that is then loaded into a FilmArray instrument for automated nucleic acid extraction, real-time PCR, detection, and high-resolution melt steps. The hands-on time of this system is minimal (approximately 2 minutes) and results are generally available in approximately 60 minutes [18]. Like the ePlex system, the FilmArray 2.0 and Torch instruments are scalable. With the exception of the FilmArray BCID Panel, which is meant for testing blood culture broths, all other panels are amenable to the direct testing of clinical specimens. The FilmArray BioThreat Panel, BioThreat-E Test, and Global Fever Panel are designed to test a variety of sample types, which are listed in the notes in Box 1. Numerous studies describing the performance characteristics and benefits of the FilmArray System have been published; examples can be found in refs. [13], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31].
Unyvero system
The Unyvero Lower Respiratory Tract Panel (Curetis USA Inc., San Diego, CA; Fig. 4 ), known as the Pneumonia Panel in Europe, is the first multiplex lower respiratory tract infection testing system to receive FDA clearance as of April 2018. This system uses PCR to detect an array of bacterial pathogens and antimicrobial resistance genes (Table 6 ) directly from tracheal aspirates in 4 to 5 hours. The Unyvero system is composed of 3 hardware components: a sample lysis device (Unyvero L4 Lysator), a panel analyzer (Unyvero A50 Analyzer), and a touchscreen computer interface and barcode scanner (Unyvero C8 Cockpit). Performance characteristics are described in refs. [32], [33].
Fig. 4.

The Curetis Unyvero system, composed of the Unyvero L4 Lysator (A, left), the Unyvero C8 Cockpit (A, middle), and the Unyvero A50 Analyzer (A, right), and the Unyvero LRT panel (B).
(Courtesy of Curetis, San Diego, CA; with permission.)
Table 6.
Curetis Unyvero Lower Respiratory Tract Panel for the Diagnosis of Community-Acquired and Health Care-Associated Pneumonia
| Microbial Targets | Antimicrobial Resistance Genes |
|---|---|
|
Acinetobacter spp. Chlamydophila pneumoniae Citrobacter freundii Enterobacter cloacae complex Escherichia coli Haemophilus influenzae Klebsiella oxytoca Klebsiella pneumoniae Klebsiella variicola Legionella pneumophila Moraxella catarrhalis Morganella morganii Mycoplasma pneumoniae Proteus spp. Pseudomonas aeruginosa Serratia marcescens Staphylococcus aureus Stenotrophomonas maltophilia Streptococcus pneumoniae |
blaTEM blaSHV blaVIM blaNDM blaKPC blaCTX-M blaOXA-23 blaOXA-24 blaOXA-48 blaOXA-58 gyrA83 mutation (of Escherichia coli) gyrA87 mutation (of E coli) gyrA83 mutation (of Pseudomonas aeruginosa) gyrA87 mutation (of P aeruginosa) mecA |
VERIGENE system
The VERIGENE system (Luminex Corporation. Austin, TX; Fig. 5 ) consists of VERIGENE Processor SP modules and the VERIGENE Reader, designed around a family of FDA-cleared syndromic panels. Multiple Processor SP units can be combined with a single Reader to accommodate the simultaneous testing of multiple samples. The extraction tray containing the sample, test cartridge (see Tables 2 and 3 for cartridges and analytes), tip holder assembly, and utility tray are loaded into the Processor SP for automated nucleic acid extraction, purification, target amplification (if required by the specific assay), and hybridization of the target molecules to a glass detection array in the test cartridge. NanoGrid Technology is used to capture, detect, and identify target molecules. After the processing step, the test cartridge array is placed into the VERIGENE Reader to obtain results, which may be selectively reported. Total hands-on, automated processing, and test interpretation time is less than 3 hours. Performance characteristics are described in refs. [34], [35].
Fig. 5.

The Luminex VERIGENE system, composed of (A, left) the VERIGENE Reader, (A, right) VERIGENE Processor SP (A, right), and (B) a VERIGENE Test Cartridge.
(Courtesy of Luminex, Austin, TX; with permission.)
xTAG technology
In addition to the VERIGENE system, Luminex offers FDA-cleared bead hybridization-based (xTAG) assays that require offline nucleic acid extraction followed by multiplex PCR and bead hybridization in 96-well plates. Analysis of beads is carried out by the MAGPIX instrument (Fig. 6 ). The MAGPIX instrument interrogates beads in each well of the reaction plate to detect fluorescent reporters that are linked to bead-hybridized target molecules. The total time required to perform a run is approximately 5 hours, so batch testing of samples is required. The NxTAG next-generation Respiratory Pathogen Panel allows users to selectively report analytes (see Table 2) whereas the xTAG Gastrointestinal Pathogen Panel (see Table 3) does not. See refs. [36], [37], [38] for performance characteristics.
Fig. 6.

The Luminex MAGPIX instrument and computer for NxTAG and xTAG assays.
(Courtesy of Luminex, Austin, TX; with permission.)
Brief Description of Currently Available Syndromic Panels
Blood culture
Accurate diagnosis and early, appropriate treatment of sepsis is a life-saving event. Syndromic panels are designed to use the exponential growth of organisms in broth-based blood culture systems to detect the most common causes of bacteremia and key resistance mechanisms that would alter therapeutic management (eg, methicillin resistance). The performance of these systems has been thoroughly reviewed elsewhere [39]. Of note, FilmArray and VERIGENE correctly detect more than 95% of identifiable organisms in monomicrobial cultures when compared with conventional methods. The Achilles heel of blood culture syndromic panels is miscalls, which are associated with polymicrobial cultures and, more specifically, those errors associated with antimicrobial resistance.
Central nervous system
Infectious meningitis and encephalitis are often medical emergencies requiring prompt and accurate diagnosis and intervention for favorable outcomes. Culture is suboptimal for detecting many etiologies; however, most laboratories lack the infrastructure to perform laboratory-developed molecular tests and, therefore, rely on reference laboratories for viral detection in the cerebrospinal fluid or when the patient is on antimicrobials. Widespread early adoption has been plagued by concerns around performance in a setting where misdiagnosis could be catastrophic. As with the respiratory panel, the correlation of results without a sensitive and specific gold standard is challenging. A multicenter evaluation established a high percent agreement (>99%) with comparator testing; however, the percent agreement for positive results was only 84.4% [40]. Furthermore, reports of false-negative (eg, potential suboptimal detection of herpesviruses and Cryptococcus) and false-positive results (eg, Streptococcus pneumoniae and herpes simplex virus-1) have resulted in delayed or missed meningitis diagnoses [41].
Gastrointestinal
Acute gastroenteritis presents a clinical and public health dilemma because the symptoms of infectious and noninfectious causes of diarrhea overlap. Traditional diagnostics (eg, culture, microscopy, antigen detection) are time consuming and may require multiple specimens for optimal sensitivity. Studies have reproducibly shown that 2 to 3 times more pathogens are detected when a molecular assay is used, compared with traditional methods [39]. This is due in part to an enhanced range of the targets, an increase in sensitivity of the assays over traditional methods, and the recognition of co-infections that were previously unrecognized. Sensitivity and specificity of these assays, overall, is high, with few exceptions. Notably, detection of rotavirus, Campylobacter spp., and Salmonella spp. may be problematic with VERIGENE; one publication noted false positivity with norovirus using BioFire. Overall, these platforms allow timely, accurate diagnosis of diarrhea and can assist with pathogen-directed therapy [42].
Respiratory
Owing to a relative paucity of accurate and rapid diagnostics, pathogen determination in acute respiratory illness had traditionally fallen to clinical presentation, despite its poor prognostic value. Rapid, accurate diagnosis is critical for antibiotic stewardship, informing the clinician if antivirals are warranted, and infection control initiatives (eg, isolation, cohorting). Unsurprisingly, respiratory illness has been a key target for syndromic testing (see Table 2). Performance characteristics have been reviewed in depth elsewhere [39], with key attributes highlighted herein. All commercially available respiratory panels significantly outperform traditional methods of detection and have expanded our recognition of coinfections (approximately 5% of samples tested). At present, no gold standard method for comparison of these assays has hampered exact determination of performance; however, the generally accepted overall agreement between molecular methods is between 85% and 99% for each target, with some exceptions. Accurate detection of adenovirus is problematic in many first-generation assays; however, a multicenter analysis of the FilmArray RP2 [43] indicates increased sensitivity with this assay.
Sexually transmitted infections/women’s health
Sexually transmitted infections, bacterial vaginosis, and vaginal yeast infections are associated with significant morbidity and can have long-term consequences, including infertility. The diagnosis of these highly prevalent infectious diseases once required the use of multiple testing methods such as microscopy, culture, and nucleic acid amplification testing; however, the advent of syndromic testing systems for these pathogens (eg, BD MAX CT/GC/TV and BD MAX Vaginal Panel; see Table 1) has streamlined their detection. Overall, both of the assays listed in Table 1 outperform traditional methods, including Chlamydia trachomatis culture and microscopic screening for bacterial vaginosis, candidiasis, and trichomoniasis. In one study that evaluated the performance of the BD MAX CT/GC/TV assay, the sensitivities for detection of all three analytes were 91.5% or greater and the specificities were 98.6% or greater [14]. According to clinical trial data of the BD MAX Vaginal Panel, the sensitivities for analytes ranged from 75.9% (analyte [collection method]: Candida glabrata [clinician-collected specimens]) to 100% (C glabrata [simulated specimens] and Candida krusei [simulated specimens]) and specificities ranged from 84.5% (bacterial vaginosis [self-collected specimens]) to 100% (C glabrata [simulated specimens] and C krusei [self-collected and simulated specimens]) [44]. Overall, sexually transmitted infection/women’s health syndromic panels outperform traditional methods in terms of sensitivity, specificity, and TATs.
Currently Available Point-of-Care Molecular Methods
The push to get faster diagnostic answers to guide admission or discharge strategies and therapeutics is likely to propel multiplex testing into the realm of POC testing. At present, issues with contamination, quality control performance and monitoring, interpretation of results, and overall good laboratory practices as technologies migrate into a less controlled setting is an obvious concern. Current CLIA-waived platforms are discussed briefly, because these systems may be readily adaptable to multiplex syndromic testing in the POC setting.
Alere i
The Alere i (Alere Scarborough, Inc., Scarborough, ME; Fig. 7 ) system uses nicking enzyme amplification reaction technology, an isothermal amplification method, to detect nucleic acids of target pathogens, including influenza A and B viruses, respiratory syncytial virus, and group A Streptococcus, either directly from swabs or from swab eluates in transport media. The consumable is a 3-component system that consists of a reagent base, elution buffer container, and transfer cartridge that is assembled on the Alere i instrument. Positive results are available within 8 to 15 minutes (depending on the assay type). Performance characteristics and additional details are described in refs. [45], [46], [47], [48], [49], [50].
Fig. 7.

The Alere i system, including the 3-part test cartridge (left) and the Alere i instrument (right).
(Courtesy of Alere, Waltham, MA; with permission.)
cobas Liat system
The cobas Liat System (Roche Diagnostics, Indianapolis, IN; Fig. 8 ) integrates all reagents necessary for nucleic acid purification and amplification (by real-time PCR) into a segmented soft plastic tube housed within a rigid plastic frame. After specimen collection (eg, nasopharyngeal swab) and elution into a suitable specimen transport medium (eg, viral transport medium), a small aliquot of the specimen is pipetted into the tube, the tube is capped, and the entire tube-frame assembly is loaded into the cobas Liat instrument, which automates all reaction and amplicon detection steps. Results for influenza detection are available within 20 minutes, and those for group A Streptococcus are available within 15 minutes of assay initiation. Currently, three FDA-cleared, CLIA-waived assays are available for the detection of influenza A/B viruses alone, influenza A/B viruses and respiratory syncytial virus, and group A Streptococcus. Performance characteristics are discussed in refs. [45], [51], [52], [53].
Fig. 8.

The cobas Liat system from Roche Diagnostics includes the test cartridge (foreground) and the Liat instrument (background).
(Courtesy of Roche Diagnostics, Indianapolis, IN; with permission.)
Solana
The Solana system (Quidel Corporation, San Diego, CA; Fig. 9 ) uses isothermal, helicase-dependent amplification and fluorescent-probe detection of target nucleic acids. Currently, FDA-approved assays are available for the detection of Clostridium difficile, groups A and B streptococci, herpes simplex viruses, varicella-zoster virus, influenza viruses, respiratory syncytial virus, human metapneumovirus, and Trichomonas vaginalis.
Fig. 9.

The Quidel Solana instrument.
(Courtesy of Quidel, San Diego, CA; with permission.)
To perform testing, a patient specimen (eg, nasal swab in viral transport medium) in Process Buffer is heated before transfer to a reaction tube. The reaction tube contains all reagents needed for amplification and detection in a lyophilized form. The reaction tube is next inserted into the Solana instrument where amplification and detection are performed. Results are available in 30 minutes or less. Up to 12 Solana diagnostic assays can be analyzed simultaneously. Relevant performance characteristics are described in ref. [54] and in assay package inserts available on the Quidel website (https://www.quidel.com).
The Future of Syndromic Testing and Point-of-Care Molecular Diagnostics
Each of these systems, plus those that have not been described herein, are constantly undergoing refinement to ensure their continued relevance in the molecular diagnostics marketplace. Test systems are continuously challenged with new strains of pathogens and antimicrobial resistance mechanisms to evaluate their inclusivity and/or exclusivity, and panels are frequently updated to include additional analytes. Also, new assays that enable detection and quantitation of pathogens associated with disease processes besides those mentioned previously are in development. The goal of most manufacturers is to offer infectious disease diagnostic testing solutions that allow identification of the greatest breadth of pathogens associated with a specific disease process or syndrome. To that end, future applications of these technologies include the enhancement of currently available systems, the development of additional panels that are amenable to the diagnosis other infectious processes, and the development of tests that enable users to comprehensively profile the antimicrobial susceptibility of pathogens.
Summary
The use of multiplex panels for pathogen detection provides several advantages over traditional laboratory diagnostic approach. Providers tout multiplex assays as a means to simplify the ordering process, decrease the number of required specimens, detect pathogens that may not be part of the initial diagnostic differential owing to rarity of the agent or inaccessibility of an alternative in-house test method, and provide results in a clinically actionable timeframe. The incorporation of molecular multiplex panels is of financial and logistical benefit to the laboratory. Specifically, one can decrease the number of methods needed to detect the same diversity of infectious agents and decrease technologist hands-on time, which results in a simplified workflow for the purposes of training, competency, use, and often cost. For the patient, these assays provide superior accuracy over traditional approaches and a greater breadth of targeted pathogens, which may decrease the number of medical visits, interventions, and durations of diagnosis. Certain multiplex assays provide benefits beyond direct patient care, such as early recognition of drug-resistant pathogens for swift implementation of infection control measures or improved detection of gastrointestinal pathogens that may impact outbreak investigations. Finally, rapid pathogen detection and the potential for improved, early intervention has been shown to positively impact associated costs by aiding in selective test use and reduced length of stay [39].
Taken together, the benefits of molecular syndromic panels and POC tests vastly outweigh any disadvantages, which is centered around cost to the health care system, namely, to the hospital or patient. The claim that this so-called shotgun approach adds unnecessary cost to the patient is an oversimplification. Although molecular multiplex assays are often more costly in terms of reagents and patient billing compared with a single culture- or serology-based assay, when multiple diagnostic tests are ordered, the multiplex assay is often less expensive than the full battery of traditional tests. Anecdotally, physicians desire to provide the patient with a definitive diagnosis and treatment plan, which may inadvertently lead to the overuse of medical resources such as injudicious antibiotic use and additional diagnostic tests (eg, imaging) to help establish a diagnosis when the causative pathogen is not easily identified. Therefore, the ability to rapidly and accurately provide a cause for the patient’s illness may improve both physician and patient satisfaction.
At present, FDA-cleared syndromic and POC molecular assays center around a few syndromes; however, this menu is likely to expand in the coming years as laboratories rapidly adopt this approach to testing. When combined with consultation (eg, antimicrobial stewardship intervention), the implementation of these assays has been shown to decrease the time to appropriate therapy, improve patient survival, and decrease overall health care-associated costs [9]. Care must be taken in the selection and appropriate use of multiplex panels. Specifically, one must balance the potential for increased laboratory reagent and equipment costs against the potential for reduced labor costs and increased revenue. To provide the highest quality of care while limiting unnecessary expense, laboratories must develop algorithms that assist providers in test ordering and use.
References
- 1.Daly J.A., Clifton N.L., Seskin K.C., et al. Use of rapid, nonradioactive DNA probes in culture confirmation tests to detect Streptococcus agalactiae, Haemophilus influenza, and Enterococcus spp. from pediatric patients with significant infections. J Clin Microbiol. 1991;29:80–82. doi: 10.1128/jcm.29.1.80-82.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis T.E., Fuller D.D. Direct identification of bacterial isolates in blood cultures by using a DNA probe. J Clin Microbiol. 1991;29:2193–2196. doi: 10.1128/jcm.29.10.2193-2196.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lumb R., Lanser J.A., Lim I.S. Rapid identification of mycobacteria by the Gen-Probe Accuprobe system. Pathology. 1993;25:313–315. doi: 10.3109/00313029309066597. [DOI] [PubMed] [Google Scholar]
- 4.Padhye A.A., Smith G., Standard P.G., et al. Comparative evaluation of chemilluminescent DNA probe assays and exoantigen tests for rapid identification of Blastomyces dermatitidis and Coccidioides immitis. J Clin Microbiol. 1994;32:867–870. doi: 10.1128/jcm.32.4.867-870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sninsky J.J. The polymerase chain reaction (PCR): a valuable method for retroviral detection. Lymphology. 1990;23:92–97. [PubMed] [Google Scholar]
- 6.Persing D.H., Mathiesen D., Marshall W.F., et al. Detection of Babesia microti by polymerase chain reaction. J Clin Microbiol. 1992;30:2097–2103. doi: 10.1128/jcm.30.8.2097-2103.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karron R.A., Froehlich J.L., Bobo L., et al. Rapid detection of parainfluenza virus type 3 RNA in respiratory specimens: use of reverse-transcription-PCR-enzyme immunoassay. J Clin Microbiol. 1994;32:484–488. doi: 10.1128/jcm.32.2.484-488.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orlando C., Pinzani P., Pazzagli M. Developments in quantitative PCR. Clin Chem Lab Med. 1998;36:255–269. doi: 10.1515/CCLM.1998.045. [DOI] [PubMed] [Google Scholar]
- 9.Abbott A.N., Fang F.C. Clinical impact of multiplex syndromic panels in the diagnosis of bloodstream, gastrointestinal, respiratory, and central nervous system infections. Clin Microbiol Newsl. 2017;39:133–142. [Google Scholar]
- 10.Messacar K., Hurst A.L., Child J., et al. Clinical impact and provider acceptability of real-time antimicrobial stewardship decision support for rapid diagnostics in children with positive blood culture results. J Pediatric Infect Dis Soc. 2016;6:267–274. doi: 10.1093/jpids/piw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rappo U., Schuetz A.N., Jenkins S.G., et al. Impact of early detection of respiratory viruses by multiplex PCR assay on clinical outcomes in adult patients. J Clin Microbiol. 2016;54:2096–2103. doi: 10.1128/JCM.00549-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ward C., Stocker K., Begum J., et al. Performance evaluation of the Verigene® (Nanosphere) and FilmArray® (BioFire®) molecular assays for identification of causative organisms in bacterial bloodstream infections. Eur J Clin Microbiol Infect Dis. 2015;34:487–496. doi: 10.1007/s10096-014-2252-2. [DOI] [PubMed] [Google Scholar]
- 13.Salimnia H., Fairfax M.R., Lephart P.R., et al. Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol. 2016;54:687–698. doi: 10.1128/JCM.01679-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Der Pol B. Profile of the triplex assay for detection of chlamydia, gonorrhea, and trichomonas using the BD MAX system. Expert Rev Mol Diagn. 2017;17:539–547. doi: 10.1080/14737159.2017.1321988. [DOI] [PubMed] [Google Scholar]
- 15.Madison-Antenucci S., Relich R.F., Doyle L., et al. Multicenter evaluation of the BD MAX enteric parasite real-time PCR assay for detection of Giardia duodenalis, Cryptosporidium hominis, Cryptosporidium parvum, and Entamoeba histolytica. J Clin Microbiol. 2016;54:2681–2688. doi: 10.1128/JCM.00765-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knabl L., Grutsch I., Orth-Höller D. Comparison of the BD MAX enteric pathogen panel with conventional diagnostic procedures in diarrheal stool samples. Eur J Clin Microbiol Infect Dis. 2016;35:131–136. doi: 10.1007/s10096-015-2517-4. [DOI] [PubMed] [Google Scholar]
- 17.Simner P.J., Oethinger M., Stellrecht K.A., et al. Multisite evaluation of the BD MAX extended enteric bacterial panel for detection of Yersinia enterocolitica, enterotoxigenic Escherichia coli, Vibrio, and Plesiomonas shigelloides from stool specimens. J Clin Microbiol. 2017;55:3258–3266. doi: 10.1128/JCM.00911-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nijhuis R.H.T., Guerendiain D., Claas E.C.J. Comparison of the ePlex respiratory pathogen panel with laboratory-developed real-time PCR assays for detection of respiratory pathogens. J Clin Microbiol. 2017;55:1938–1945. doi: 10.1128/JCM.00221-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierce V.M., Hodinka R.L. Comparison of the GenMark diagnostics eSensor respiratory viral panel to real-time PCR for detection of respiratory viruses in children. J Clin Microbiol. 2012;50:3458–3465. doi: 10.1128/JCM.01384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Babady N.E., England M.R., Jurcic Smith K.L., et al. Multicenter evaluation of the ePlex respiratory pathogen panel for the detection of viral and bacterial respiratory tract pathogens in nasopharyngeal swabs. J Clin Microbiol. 2018;56 doi: 10.1128/JCM.01658-17. [pii:e01658-17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Rijn A.L., Nijhuis R.H.T., Bekker V., et al. Clinical implications of rapid ePlex respiratory pathogen panel testing compared to laboratory-developed real-time PCR. Eur J Clin Microbiol Infect Dis. 2018;37:571–577. doi: 10.1007/s10096-017-3151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCoy M.H., Relich R.F., Davis T.E., et al. Performance of the FilmArray blood culture identification panel utilized by non-expert staff compared to conventional microbial identification and antimicrobial resistance gene detection from positive blood cultures. J Med Microbiol. 2016;65:619–625. doi: 10.1099/jmm.0.000277. [DOI] [PubMed] [Google Scholar]
- 23.Rogers B.B., Shankar P., Jerris R.C., et al. Impact of a rapid respiratory panel test on patient outcomes. Arch Pathol Lab Med. 2015;139:636–641. doi: 10.5858/arpa.2014-0257-OA. [DOI] [PubMed] [Google Scholar]
- 24.Kanack K.J. Rapid respiratory panel testing influences patient management and clinical outcomes. MLO Med Lab Obs. 2014;46:16. [PubMed] [Google Scholar]
- 25.MacVane S.H., Nolte F.S. Benefits of adding a rapid PCR-based blood culture identification panel to an established antimicrobial stewardship program. J Clin Microbiol. 2016;54:2455–2463. doi: 10.1128/JCM.00996-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Southern T.R., Van Schooneveld T.C., Bannister D.L., et al. Implementation and performance of the BioFire FilmArray blood culture identification panel with antimicrobial treatment recommendations for bloodstream infections at a midwestern academic tertiary hospital. Diagn Microbiol Infect Dis. 2015;81:96–101. doi: 10.1016/j.diagmicrobio.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Buss S.N., Leber A., Chapin K., et al. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol. 2015;53:915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prakash V.P., LeBlanc L., Alexander-Scott N.E., et al. Use of a culture-independent gastrointestinal multiplex PCR panel during a shigellosis outbreak: considerations for clinical laboratories and public health. J Clin Microbiol. 2015;53:1048–1049. doi: 10.1128/JCM.03374-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanson K.E., Slechta E.S., Killpack J.A., et al. Preclinical assessment of a fully automated multiplex PCR panel for detection of central nervous system pathogens. J Clin Microbiol. 2016;54:785–787. doi: 10.1128/JCM.02850-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duff S., Hasbun R., Ginocchio C.C., et al. Economic analysis of rapid multiplex polymerase chain reaction testing for meningitis/encephalitis in pediatric patients. Future Microbiol. 2018;13:617–629. doi: 10.2217/fmb-2017-0238. [DOI] [PubMed] [Google Scholar]
- 31.Gay-Andrieu F., Magassouba N., Picto V., et al. Clinical evaluation of the BioFire FilmArray BioThreat-E test for the diagnosis of Ebola virus disease in Guinea. J Clin Virol. 2017;92:20–24. doi: 10.1016/j.jcv.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Papan C., Meyer-Buehn M., Laniado G., et al. Assessment of the multiplex PCR-based assay Unyvero pneumonia application for detection of bacterial pathogens and antibiotic resistance genes in children and neonates. Infection. 2018;46:189–196. doi: 10.1007/s15010-017-1088-y. [DOI] [PubMed] [Google Scholar]
- 33.Personne Y., Ozongwu C., Platt G., et al. ‘Sample-in, answer-out’? Evaluation and comprehensive analysis of the Unyvero P50 pneumonia assay. Diagn Microbiol Infect Dis. 2016;86:5–10. doi: 10.1016/j.diagmicrobio.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Ledeboer N.A., Lopansri B.K., Dhiman N., et al. Identification of Gram-negative bacteria and genetic resistance determinants from positive blood culture broths by use of the Verigene Gram-negative blood culture multiplex microarray-based molecular assay. J Clin Microbiol. 2015;53:2460–2472. doi: 10.1128/JCM.00581-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buchan B.W., Ginocchio C.C., Manii R., et al. Multiplex identification of Gram-positive bacteria and resistance determinants directly from positive blood culture broths: evaluation of an automated microarray-based nucleic acid test. PLoS Med. 2013;10:e1001478. doi: 10.1371/journal.pmed.1001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esposito S., Principi N. The role of the NxTAG respiratory pathogen panel assay and other multiplex platforms in clinical practice. Expert Rev Mol Diagn. 2017;17:9–17. doi: 10.1080/14737159.2017.1266260. [DOI] [PubMed] [Google Scholar]
- 37.Tang Y.W., Gonsalves S., Sun J.Y., et al. Clinical evaluation of the Luminex NxTAG respiratory pathogen panel. J Clin Microbiol. 2016;54:1912–1914. doi: 10.1128/JCM.00482-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang R.S., Johnson C.L., Pritchard L., et al. Performance of the Verigene enteric pathogens test, BioFire FilmArray gastrointestinal panel and Luminex xTAG gastrointestinal pathogen panel for detection of common enteric pathogens. Diagn Microbiol Infect Dis. 2016;86:336–339. doi: 10.1016/j.diagmicrobio.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 39.Ramanan P., Bryson A.L., Binnicker M.J., et al. Syndromic panel-based testing in clinical microbiology. Clin Microbiol Rev. 2017;31 doi: 10.1128/CMR.00024-17. [pii:e00024-17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leber A.L., Everhart K., Balada-Llasat J.M., et al. Multicenter evaluation of BioFire FilmArray Meningitis/Encephalitis Panel for detection of bacteria, viruses, and yeast in cerebrospinal fluid specimens. J Clin Microbiol. 2016;54(9):2251–2261. doi: 10.1128/JCM.00730-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dien Bard J., Alby K. Point-counterpoint: meningitis/encephalitis syndromic testing in the clinical laboratory. J Clin Microbiol. 2018;56(4) doi: 10.1128/JCM.00018-18. [pii:e00018-18] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cybulski R.J., Jr., Bateman A.C., Bourassa L., et al. Clinical impact of a multiplex gastrointestinal PCR panel in patients with acute gastroenteritis. Clin Infect Dis. 2018 doi: 10.1093/cid/ciy357. [DOI] [PubMed] [Google Scholar]
- 43.Leber A.L., Everhart K., Daly J.A., et al. Multicenter evaluation of BioFire FilmArray Respiratory Panel 2 for detection of viruses and bacteria in nasopharyngeal swab samples. J Clin Microbiol. 2018;25(6):56. doi: 10.1128/JCM.01945-17. [pii:e01945-17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawa D, Paradis S, Yu JH, et al. Evaluating the standard of care for women’s health: the BD MAX Vaginal Panel and management of vaginal infections (white paper). 2017. Available at: http://moleculardiagnostics.bd.com/wp-content/uploads/2017/08/MAX-Vaginal-Panel-Whitepaper.pdf. Accessed June 25, 2018.
- 45.Young S., Illescas P., Nicasio J., et al. Diagnostic accuracy of the real-time PCR cobas Liat influenza A/B assay and the Alere i influenza A&B NEAR isothermal nucleic acid amplification assay for the detection of influenza using adult nasopharyngeal specimens. J Clin Virol. 2017;94:86–90. doi: 10.1016/j.jcv.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 46.Davis S., Allen A.J., O’Leary R., et al. Diagnostic accuracy and cost analysis of the Alere i influenza A&B near-patient test using throat swabs. J Hosp Infect. 2017;97:301–309. doi: 10.1016/j.jhin.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 47.Hassan F., Hays L.M., Bonner A., et al. Multicenter clinical evaluation of the Alere i respiratory syncytial virus isothermal nucleic acid amplification assay. J Clin Microbiol. 2018;56 doi: 10.1128/JCM.01777-17. [pii:e01777-17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schnee S.V., Pfeil J., Ihling C.M., et al. Performance of the Alere i RSV assay for point-of-care detection of respiratory syncytial virus in children. BMC Infect Dis. 2017;17:767. doi: 10.1186/s12879-017-2855-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cohen D.M., Russo M.E., Jaggi P., et al. Multicenter clinical evaluation of the novel Alere i Strep A isothermal nucleic acid amplification test. J Clin Microbiol. 2015;53:2258–2261. doi: 10.1128/JCM.00490-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berry G.J., Miller C.R., Prats M.M., et al. Comparison of the Alere i Strep A test and the BD Veritor system in the detection of group A Streptococcus and the hypothetical impact of results on antibiotic utilization. J Clin Microbiol. 2018;56 doi: 10.1128/JCM.01310-17. [pii:e01310-17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melchers W.J.G., Kuijpers J., Sickler J.J., et al. Lab-in-a-tube: real-time molecular point-of-care diagnostics for influenza A and B using the cobas Liat system. J Med Virol. 2017;89:1382–1386. doi: 10.1002/jmv.24796. [DOI] [PubMed] [Google Scholar]
- 52.Gibson J., Schechter-Perkins E.M., Mitchell P., et al. Multi-center evaluation of the cobas Liat Influenza A/B & RSV assay for rapid point of care diagnosis. J Clin Virol. 2017;95:5–9. doi: 10.1016/j.jcv.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 53.Ling L., Kaplan S.E., Lopez J.C., et al. Parallel validation of three molecular devices for simultaneous detection and identification of influenza A and B and respiratory syncytial viruses. J Clin Microbiol. 2018;56 doi: 10.1128/JCM.01691-17. [pii:e01691-17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaydos C.A., Schwebke J., Dombrowski J., et al. Clinical performance of the Solana point-of-care trichomonas assay from clinician-collected vaginal swabs and urine specimens from symptomatic and asymptomatic women. Expert Rev Mol Diagn. 2017;17:303–306. doi: 10.1080/14737159.2017.1282823. [DOI] [PMC free article] [PubMed] [Google Scholar]


