Abstract
Programmed ribosomal frameshifting in viral messenger RNA occurs in response to neighboring sequence elements consisting of: a frameshift site, a spacer, and a downstream enhancer sequence. In human immunodeficiency virus type 1 (HIV-1) mRNA, this sequence element has a potential to form either a stem-loop or a pseudoknot structure. Based on many mutational studies, the stem-loop structure has been proposed for the downstream enhancer region of the HIV-1 mRNA. This stimulatory stem-loop structure is separated from the shift site by a spacer of seven nucleotides. In contrast, a recent report has proposed an alternative model in which the bases in the spacer sequence form a pseudoknot structure as the downstream enhancer sequence [Du et al., Biochemistry 35 (1996) 4187–4198.]. Using UV melting and enzymatic mapping analyses, we have investigated the conformation of the sequence region involved in ribosomal frameshifting in HIV-1. Our S1, V1, and T1 endonuclease mappings, together with UV melting analysis, clearly indicate that this sequence element of the HIV-1 mRNA frameshift site forms a stem-loop structure, not a pseudoknot structure. This finding further supports the stem-loop structure proposed by many mutational studies for the downstream enhancer sequence of the HIV-1 mRNA.
Keywords: Frameshifting, HIV, Hairpin, Pseudoknot, RNA
1. Introduction
Human immunodeficiency virus (HIV) is a retrovirus that employs −1 ribosomal frameshifting to synthesize essential viral enzymes, including reverse transcriptase, integrase, and protease, as fusion proteins with gag proteins 1, 2, 3. The signals for this frameshifting process consist of a heptanucleotide shift site and a downstream enhancer sequence in the viral RNA. These two important signals are separated by a spacer with a nearly constant number of nucleotides.
The mechanism of ribosomal frameshifting is not well understood, although the simultaneous slippage model is the most plausible. The pre-translocation slippage model proposes that when the ribosome enters the frameshift region, the peptidyl (P) and aminoacyl (A) sites of the ribosome slip backward simultaneously into the −1 frame at the shift site, and that translation continues in the new reading frame 4, 5. In addition to this pre-translocation slippage model, a report by Horsfield et al. [6]proposes an E/P site post-translocation slippage model in E. coli containing HIV-1 gag-pol frameshift sequence. The enhancer secondary RNA structure, either a stem-loop or a pseudoknot, downstream of the shift site induces pausing of the ribosome and stimulates slippage at the shift sequence 4, 5, 6, 7.
Enhancement of ribosomal frameshifting by RNA secondary structures has been studied by many mutational analyses. An RNA pseudoknot is a common enhancer for highly efficient frameshifting ([8]and references therein). However, in HIV-1 and human T-cell leukaemia virus type II (HTLV-II), frameshifting occurs at a frequency of 1–5% both in vivo and in vitro, and a simple stem-loop structure has been implicated for the enhancer region 9, 10. This downstream enhancer structure is separated from the shift site by a spacer sequence of typical length. The importance of the proper distance between this two signals was evidenced by a mutational study [11]. However, recently Du et al. [12]proposed an alternative model involving a pseudoknot for the frameshifting region of the HIV-1 mRNA. In this pseudoknot model, the bases in the spacer region are part of stem 1 of the pseudoknot, thus leaving no nucleotides between the shift site and the downstream enhancer structure. Direct probing of the RNA secondary structure by single-strand and double-strand specific enzymes can distinguish between these two models.
Here, we determine the secondary structure of the HIV-1 mRNA frameshifting sequence elements including the shift site, the spacer, and the downstream enhancer sequence by using ultraviolet melting and enzymatic mapping analysis. The RNA sequence element has a potential to form either a stem-loop or a pseudoknot structure, as illustrated in Fig. 1 . We show that the downstream enhancer sequence adopts a simple stem-loop structure. The spacer region is not involved in the formation of the stem of a pseudoknot as predicted by Du et al. [12], but is in a single-stranded conformation. Our results agree well with the mutational studies showing a simple stem-loop structure for the downstream enhancer sequence of the frameshifting region of the HIV-1 mRNA.
Fig. 1.
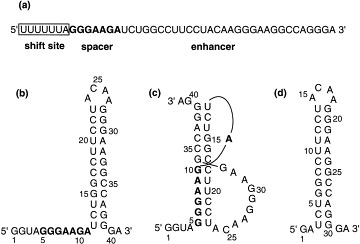
(a) The sequence of the frameshifting signals in HIV-1 RNA consists of a shift site (boxed), a spacer (bold), and an enhancer. This sequence has the potential to form either, (b) a stem-loop structure or, (c) a pseudoknot structure as proposed by Du et al. [12]. A truncated RNA sequence without a spacer can form (d) only a stem-loop structure.
2. Materials and methods
2.1. RNA synthesis and purification
Oligoribonucleotides were synthesized in vitro from DNA templates by using T7 RNA polymerase 13, 14. DNA templates were synthesized on an Applied Biosystem 381A and purified by 20% polyacrylamide gel electrophoresis under denaturing conditions (7 M urea). RNA oligonucleotides were purified by denaturing 20% polyacrylamide gel electrophoresis. The RNA transcripts were visualized by UV shadowing, were cut from the gel, were eluted from the gel by electroelution (Schleicher and Schuell), and were precipitated with ethanol. The purified RNA oligonucleotides were dialyzed first against 10 mM sodium phosphate, pH 7.0, 100 mM NaCl, 5 mM EDTA, next against 10 mM sodium phosphate, pH 6.4, 100 mM NaCl, and finally against 10 mM sodium phosphate, pH 6.4, 50 mM NaCl, 5 mM MgCl2.
2.2. UV absorbance melting experiments
All RNA melting experiments were done with a Gilford model 250 spectrophotometer interfaced to a personal computer. RNA samples were in 10 mM sodium phosphate, pH 6.4, 50 mM NaCl, 5 mM MgCl2. Melting curves were obtained at RNA concentrations from 2 μM to 1 mM. RNA samples were heated to 90°C for 1 min, slowly cooled, and equilibrated to 0°C for 20 min. Data were recorded at either 260 or 280 nm with the temperature increased from 0 to 95°C at a rate of 0.5°C min−1 by a Gilford model 2527 thermoprogrammer.
2.3. Enzymatic mapping of oligoribonucleotides
RNA oligonucleotides were dephosphorylated using calf intestinal phosphatase and were 5′-end labeled using γ-32P-ATP and bacteriophage T4 polynucleotide kinase (USB). Labeled RNA was purified by denaturing 20% polyacrylamide gel electrophoresis. The secondary structures of 5′-32P-labeled RNA were probed by using nuclease S1 and RNase V1 [15]. RNase T1 digestions were performed in 10 mM Tris–HCl, pH 7.0, 100 mM KCl, 10 mM MgCl2 on ice for 10 min. The reactions were stopped by adding 9 M urea, 0.05% (w/v) xylene cyanol in TBE (50 mM Tris–HCl, pH 8.1, 50 mM boric acid, 1 mM EDTA). The digestion products of each reaction were analyzed on the denaturing 20% polyacrylamide gel.
3. Results and discussion
3.1. UV melting indicates that the frameshifting sequence element of HIV-1 RNA adopts a stem-loop structure
Optical melting studies were done for two RNA fragments (Fig. 1, b and d) in 10 mM sodium phosphate, pH 6.4, 50 mM NaCl, 5 mM MgCl2. The UV absorbance profiles and their first derivatives as a function of temperature are presented in Fig. 2 . As expected for a unimolecular melting, the melting temperature was found to be independent of the RNA concentration from 2 μM to 1 mM. After heating the RNA to 95°C, the RNA oligonucleotides were refolded by cooling to 0°C, and the melting experiments were repeated. The same melting profiles and melting temperatures were recorded, indicating that the melting profiles were not perturbed by any hydrolysis of the RNA. The melting temperatures of the HIV-1 RNA frameshifting-enhancer sequence (Fig. 2a) and of the truncated RNA with the spacer region deleted (Fig. 2b) are identical. This indicates that the spacer region is not involved in the secondary structure of the sequence.
Fig. 2.
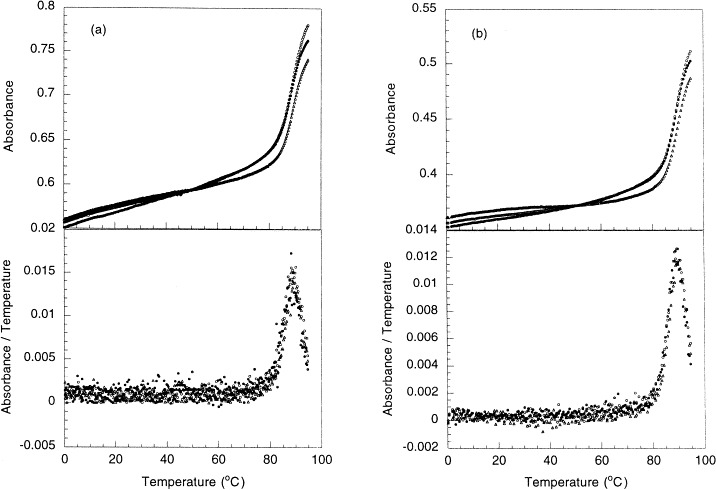
(a) Absorbance (260 nm) vs. temperature profiles for the RNA oligonucleotide containing both the spacer and the enhancer (Fig. 1b,c). (b) Absorbance (260 nm) vs. temperature profiles for the truncated RNA without the spacer sequence (Fig. 1d). Both RNA oligonucleotides from 2 μM to 1 mM have nearly identical melting profiles, and their first derivatives give the same melting temperature.
It is possible that the stem-loop and the pseudoknot shown in Fig. 1 have a same melting temperature. However, considering the truncated RNA oligonucleotide without the spacer region can form only a stem-loop conformation, the nearly identical UV melting profiles and melting temperatures for the two RNA oligonucleotides indicate indirectly that the RNA containing the spacer and the enhancer sequences adopts a stem-loop structure, but not a pseudoknot structure.
3.2. S1, V1, and T1 mapping confirm that the frameshifting sequence elements of HIV-1 RNA form a stem-loop structure
In order to obtain structural information for the frameshifting sequence elements, enzymatic mapping of the RNA was done. Before probing the secondary structure of the RNA oligonucleotides using enzymatic mapping analysis, we tested for the presence of a single conformation in the samples by a native polyacrylamide gel electrophoresis at 4°C. Only one band was observed on the native gel (data not shown), indicating that the RNA oligonucleotides in 10 mM sodium phosphate, pH 6.4, 50 mM NaCl, 5 mM MgCl2 adopt a single conformation.
Fig. 3 shows the nuclease S1, RNase V1, and RNase T1 cleavage patterns of the frameshifting sequence elements consisting of the spacer and the enhancer of HIV-1 mRNA (The sequence shown in Fig. 1a,b). Nucleotides from G1 to G5 were strongly cleaved by single-strand specific nuclease S1, but were protected from cleavage by double-strand specific RNase V1. However, at a high enzyme concentration, some of the nucleotides in the region of G1 to G10 was cleaved by RNase V1, suggesting that residues 1 to 10 are partially stacked. Guanine residues G5–G7, and G9, which are located in the stem region of the proposed pseudoknot structure (Fig. 1c), were strongly attacked by RNase T1 that cleaves the G in single-stranded region. These results indicate that residues 1–10 are in the single-stranded conformation. Nucleotides A24–A27 were cleaved by nuclease S1, indicating these residues are in a single-stranded conformation. Residues C13–C22 and G29–G38 were cleaved by RNase V1. No cleavage were observed for the nucleotides G28, G29, G30, and G33 by RNase T1, indicating these residues are in a double-stranded region.
Fig. 3.
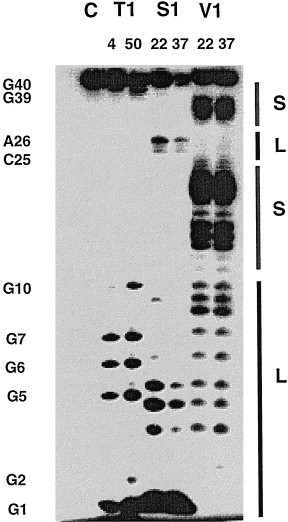
Enzymatic mappings of the RNA oligonucleotides containing the spacer and the enhancer sequence as shown in Fig. 1b,c. The control lane (C) shows no hydrolytic cleavage of the RNA under the reaction conditions. The ribonuclease T1 reaction was done at both 4 and 50°C; the ribonuclease V1 and S1 cleavage reactions were done at 22 and 37°C. The stem regions (S) and loop or single-strand regions (L) based on a stem-loop secondary structure are labeled. The cleavage patterns for S1 and T1 (specific for single-strands), and for V1 (specific for double-strands and stacked bases) at a high concentration are consistent with a stem-loop.
This cleavage pattern, as summarized in Fig. 4 , indicates that in the context of the frameshifting sequence elements of the HIV-1 RNA consisting of the spacer and the enhancer, the enhancer sequence adopts a stem-loop structure separated from the shift site by a spacer sequence. It is not consistent with the formation of a pseudoknot. This stem-loop structure agrees with the absorbance melting results and is consistent with the calculated secondary structure from the Zuker algorithm. The spacer and the enhancer is solely folded into the stem-loop structure in Fig. 1b with a free energy of −21.1 kcal mol−1. The same free energy of folding is also calculated for the stem-loop of the truncated RNA with the spacer region deleted (Fig. 1d).
Fig. 4.
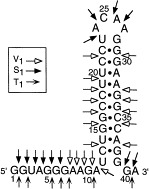
Summary of the V1, S1, and T1 mapping analysis. Nuclease S1 and ribonuclease T1 at 4°C strongly cleave the nucleotides in the spacer sequence, indicating that the spacer sequence is in a single-stranded conformation.
3.3. Mutational studies indicate a stem-loop enhancer and a spacer region
The requirement of a downstream RNA structure to enhance ribosomal frameshifting in HIV has been verified by many mutational studies. By expressing the HIV-1 gag-pol domain in cultured vertebrate cells, Parkin et al. [9]showed that a stem-loop structure downstream of the HIV-1 shift site is important for wild-type levels of frameshifting. In a cell culture analysis for the sequences and distance between two cis-acting signals in HIV-1 and HTLV-2, Kollmus et al. [11]observed that the slippery sequence alone is sufficient for basal level of frameshifting, but that the frameshifting efficiency is enhanced by a downstream stem-loop structure. A similar stimulating role of the stem-loop sequence in HIV-1 mRNA to enhance frameshifting was also observed in mammalian cell culture [16]. Honda et al. [17]used an in vitro luciferase assay system for HIV-1 translation frameshifting and noted that the stem-loop is not essential for basal-level frameshifting, but it does influence the efficiency of ribosomal frameshifting.
The simultaneous slippage model of ribosomal frameshifting proposes that an RNA secondary structure downstream of the shift site may slow or stall ribosomes during translation, thus increase the probability of the ribosome to slip backward into the −1 frame at the slippery site. Such a model predicts that the correct spacing must be maintained between the shift site and the downstream RNA structure. In analysis of the RNA sequences responsible for frameshifting in the avian corona virus infectious bronchitis virus (IBV), Brierley et al. [18]observed that alteration of the relative position of the shift sequence by as little as three nucleotides with the respect to the downstream pseudoknot causes a dramatic reduction in frameshifting efficiency. Morikawa and Bishop [19]also demonstrated for the gag-pol ribosomal frameshift site of feline immunodeficiency virus (FIV) that mutations which alter the distance between the frameshift signal and the downstream pseudoknot structure reduces frameshifting efficiency. In a systematic analysis of the effect of the sequence and the distance between two signals on frameshifting efficiency in HTLV-II, Kollmus et al. [11]demonstrated that the base composition 3′ to the slippery sequence does not have a significant influence on frameshifting, but that the spacing between the slippery sequence and the 5′ end of the stem-loop is stringently restricted to an optimal distance of seven nucleotides.
All of these mutational results imply that the downstream RNA structure should be located at a precise position relative to the shift site to enhance frameshifting. In the viruses reported so far, the relative spacing distance between the stimulator and the slippery sequence varies between five and eight nucleotides [8]. Of course it is possible that the distance from the more efficient slippery sequence to the downstream RNA structure needs not be so tightly constrained. Since HIV-1 contains the highly slippery sequence UUUUUUA, the distance between two signals needs not be so stringently maintained, and the spacing may vary from three to nine nucleotides to promote different level of frameshifting; the maximum frameshifting efficiency is observed in the context of relative spacing between two signals separated by seven nucleotides [11]. Although a pseudoknot is the usual enhancer structure in retroviral frameshifting, there is no support for the proposed pseudoknot structure for HIV-1 by Du et al. [12]. In a recent paper, Du and Hoffman [20]mentioned that the possible pseudoknot structure for HIV-1 is speculative, since it lacks a spacing sequence between the frameshift site and the pseudoknot. The mutation data and our direct structural studies all agree that a stem-loop structure separated from the slippery sequence by about seven nucleotides is necessary for efficient ribosomal frameshifting in HIV-1.
Acknowledgements
I thank professor Ignacio Tinoco, Jr. at the Department of Chemistry, University of California, Berkeley for the helpful advice and discussion, and Mr. David Koh for synthesizing DNA templates. The UV melting experiments were done at professor I. Tinoco's laboratory.
Footnotes
Kumho Life and Environmental Science Laboratory Publication No. 8.
References
- 1.Jacks T., Power M.D., Masiarz F.R., Luciw P.A., Barr P.J., Varmus H.E. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 2.Atkins J.F., Weiss R.B., Gesteland R.F. Cell. 1990;62:413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacks T. Curr. Top. Microbiol. Immunol. 1990;157:93–124. doi: 10.1007/978-3-642-75218-6_4. [DOI] [PubMed] [Google Scholar]
- 4.Tu C., Tzeng T.-H., Bruenn J.A. Proc. Natl. Acad. Sci. U.S.A. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Somogyi P., Jenner A.J., Brierley I., Inglis S.C. Mol. Cell. Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horsfield J.A., Wilson D.N., Mannering S.A., Adamski F.M., Tate W.P. Nucleic Acids Res. 1995;23:1487–1494. doi: 10.1093/nar/23.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schimmel P. Cell. 1989;58:9–12. doi: 10.1016/0092-8674(89)90395-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brierley I. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 9.Parkin N.T., Chamorro M., Varmus H.E. J. Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falk H., Mador N., Udi R., Panet A., Honigman A. J. Virol. 1993;67:6273–6277. doi: 10.1128/jvi.67.10.6273-6277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kollmus H., Honigman A., Panet A., Hauser H. J. Virol. 1994;68:6087–6091. doi: 10.1128/jvi.68.9.6087-6091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du Z., Giedroc D.P., Hoffman D.W. Biochemistry. 1996;35:4187–4198. doi: 10.1021/bi9527350. [DOI] [PubMed] [Google Scholar]
- 13.Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyatt J.R., Chastain M., Puglisi J.D. BioTechniques. 1991;11:764–769. [PubMed] [Google Scholar]
- 15.Wyatt J.R., Puglisi J.D., Tinoco I., Jr. J. Mol. Biol. 1990;214:455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- 16.Reil H., Kollmus H., Weidle U.H., Hauser H. J. Virol. 1993;67:5579–5584. doi: 10.1128/jvi.67.9.5579-5584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda A., Nakamura T., Nishimura S. Biochem. Biophys. Res. Commun. 1995;213:575–582. doi: 10.1006/bbrc.1995.2170. [DOI] [PubMed] [Google Scholar]
- 18.Brierley I., Digard P., Inglis S.C. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morikawa S., Bishop D.H.L. Virology. 1992;186:389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du Z., Hoffman D.W. Nucleic Acids Res. 1997;25:1130–1135. doi: 10.1093/nar/25.6.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]


