Significance
Every cell produces thousands of lipid species, but studying the function of individual lipids in living cells is almost impossible with existing methodologies. Addressing this experimental bottleneck, we developed a strategy to quantify dissociation constants for lipid–protein interactions and transmembrane flip-flop rates of native lipids in live-cell experiments. Using a combination of plasma membrane-specific photochemical probes and mathematical modeling, we demonstrate that, for diacylglycerols as a model lipid class, the inherent lipid structural diversity caused by variations in acyl chain composition determines lipid protein affinities and transbilayer kinetics. In fact, subtle chemical differences change these values by orders of magnitude. Our approach represents a generally applicable method for elucidating the biological function of single lipid species on subcellular scales.
Keywords: signaling lipids, diacylglycerol, protein kinase C, mathematical modeling, caged lipid probes
Abstract
Every cell produces thousands of distinct lipid species, but insight into how lipid chemical diversity contributes to biological signaling is lacking, particularly because of a scarcity of methods for quantitatively studying lipid function in living cells. Using the example of diacylglycerols, prominent second messengers, we here investigate whether lipid chemical diversity can provide a basis for cellular signal specification. We generated photo-caged lipid probes, which allow acute manipulation of distinct diacylglycerol species in the plasma membrane. Combining uncaging experiments with mathematical modeling, we were able to determine binding constants for diacylglycerol–protein interactions, and kinetic parameters for diacylglycerol transbilayer movement and turnover in quantitative live-cell experiments. Strikingly, we find that affinities and kinetics vary by orders of magnitude due to diacylglycerol side-chain composition. These differences are sufficient to explain differential recruitment of diacylglycerol binding proteins and, thus, differing downstream phosphorylation patterns. Our approach represents a generally applicable method for elucidating the biological function of single lipid species on subcellular scales in quantitative live-cell experiments.
Membrane lipids play a central role in cellular signal transduction. As receptor ligands, enzyme cofactors, and allosteric modulators, they control cellular excitability (1), immune responses (2), cell migration (3, 4), and stem cell differentiation (5, 6). In line with their fundamental importance, dysregulation of signaling lipids has been firmly established as a hallmark of severe diseases such as cancer (7) and diabetes (8). Lipids are grouped into classes characterized by common chemical features, such as their headgroup. Each of these classes comprises many molecularly distinct lipid species that differ in subtle chemical details, e.g., number of double bonds, ether or ester linkages, as well as fatty acid chain length and positioning, ultimately suggesting the presence of thousands of individual lipid species in mammalian cells (9, 10). While the heterogeneity of the cellular lipidome in general and of signaling lipids in particular is well established, it is much less clear whether this heterogeneity has causal relations to cellular function (11, 12).
Intriguingly, a growing body of evidence suggests that changes in the levels of individual lipid species rather than entire lipid classes determine cellular signaling outcome. For instance, early studies reported that activation of individual cell surface receptors leads to the formation and degradation of distinct patterns of diacylglycerol (DAG) species during signal transduction (13–15) on minute timescales. This suggests that crucial information could be encoded in the molecular spectrum of generated signaling lipids. Supporting this notion, drastically altered levels of distinct lipid species were correlated with cellular processes, e.g., the increase of a phosphatidic acid ether lipid during cytokinesis (16) or the reciprocal regulation of ceramide species during Toll-like receptor signaling in innate immunity (17). DAGs appear to be prime targets to study the importance of lipid heterogeneity in cell signaling, as they act as second messengers at the plasma membrane and function in many cellular processes, including insulin signaling, ion channel regulation, and neurotransmitter release (18, 19). Many of these processes involve effector proteins such as protein kinase C (PKC) isoforms, which are recruited to cellular membranes by DAG binding to their C1 domains (20). Faithful process initiation thus requires the activation of a subset of DAG effector proteins in the presence of others as observed during the formation of the immunological synapse (21). However, the molecular mechanisms of such specific recruitment events are not well understood. Here, specificity could be provided by differential activation of effectors by structurally distinct DAG species, which recruit specific DAG binding proteins due to differences in lipid-protein affinities, local lipid densities, and lifetimes. Determining these parameters requires quantitative experimental strategies that allow perturbing and monitoring levels of native lipid species and lipid–protein complexes in specific membranes of living cells. However, such methods are not yet available (22).
Closing this methodological gap, we developed chemical probes for rapid, leaflet-specific ultraviolet (UV) uncaging of individual DAG species at the plasma membrane of living cells. This allowed temporally well-defined increases of native DAG species in a quantitative and dose-dependent fashion as a prerequisite for kinetic analysis. By combining DAG uncaging and live-cell fluorescence imaging of DAG-binding proteins with mathematical modeling, we demonstrate that 1) structural differences between DAG species are sufficient to trigger different recruitment patterns of various PKC isoforms and lead to differential phosphorylation of downstream signaling targets; 2) Kd values of DAG–C1–domain interactions as well as transbilayer movement and turnover rates differ by orders of magnitude between DAG species; 3) the affinity of the lipid–protein interaction primarily influences the magnitude of DAG signaling events (recruitment of a specific effector protein); whereas 4) the kinetics of DAG signaling events are largely determined by lipid transbilayer movement and turnover rates. Overall, our data demonstrate that subtle differences in DAG structure affect lipid-protein affinities and the kinetics of transbilayer movement and lipid turnover. This results in preferential recruitment of DAG-binding proteins, which may serve as a mechanism to encode information during cellular signaling events.
Results
Photoactivation Allows Acute DAG Density Increases at the Plasma Membrane.
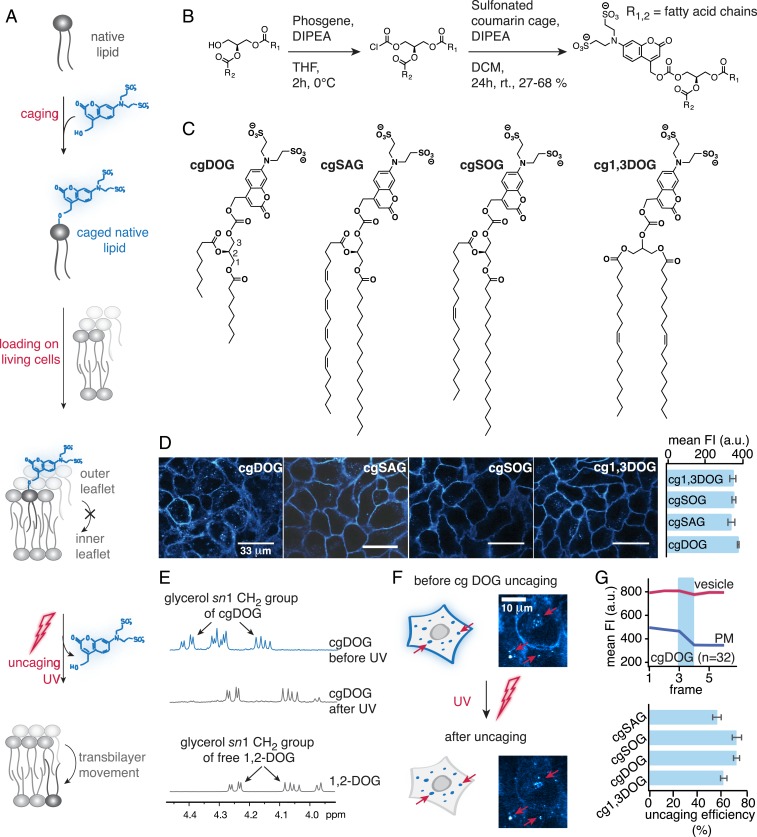
Photoliberation of native lipid species from caged lipids constitutes the most straightforward experimental approach to induce well-defined, temporally controlled density increases of a single lipid species in individual membrane leaflets (23–25), which is essential for kinetic analysis (Fig. 1A). In order to study the influence of DAG chemical heterogeneity on cellular signaling, we prepared four caged DAGs: One variant with short acyl chains (dioctanoylglycerol, cgDOG) and two typical, naturally occurring DAGs with long acyl chains, featuring one and four double bonds, respectively (stearoyl-arachidonylglycerol, cgSAG and stearoyl-oleoylglycerol, cgSOG) (Fig. 1 B and C). As a negative control, we prepared a caged regioisomer of the native species, 1,3-dioleoylglycerol (cg1,3DOG), which does not recruit DAG effector proteins to cellular membranes (26). To ensure plasma membrane-specific DAG photorelease, we used a sulfonated coumarin photo-caging group (27), which allows lipid side chains to incorporate selectively into the outer plasma membrane leaflet but completely blocks transbilayer movement (flip-flop) due to two negative charges (Fig. 1A). The absorption and emission spectra of coumarin derivatives cgDOG, cgSAG, cgSOG, and cg1,3DOG were very similar (SI Appendix, Fig. S1-1 B and C).
Fig. 1.
Caged diacylglycerols for acute lipid density increases at the outer plasma membrane leaflet. (A) Schematic description of the experimental approach. (B) Synthesis of the cgDAGs. (C) Structures of synthesized cgDAGs. (D, Left) Cellular localization of cgDAGs visualized by coumarin fluorescence. (Scale bars, 33 μm.) (D, Right) Quantification of FI after cellular uptake. (E–G) Quantification of cgDAG incorporation and photoreaction efficiency in vitro in solution and in living cells. (E) NMR spectra of cgDOG before and after irradiation using a 345-nm high-pass filter and DOG as reference compound. Photocleavage was monitored by the distinct shift of sn1 glycerol protons signal from 4.39 ppm (cgDAG) to 4.24 ppm (free DAG). (F) Whole field of view uncaging of cgDAGs after allowing for membrane turnover causes loss of fluorescence at the plasma membrane but not in endosomes (red arrows), in line with exclusive initial localization of the caged lipid at the outer plasma membrane leaflet. Both images are displayed at the same magnification. (G, Upper) Quantification of fluorescence intensity changes at the plasma membrane and in endosomes upon photoactivation. The blue bar indicates the uncaging event. (G, Lower) Uncaging efficiency for the different DAG species at 40% laser power calculated from FI loss at the plasma membrane. All live-cell experiments were carried out in HeLa Kyoto cells, n, numbers represent cell numbers, in a typical experiment 5 to 10 cells were imaged simultaneously. Data are mean; error bars represent SEM.
Brief loading followed by extensive washing ensured sufficient and comparable plasma membrane incorporation of all caged DAGs into HeLa Kyoto cells (Fig. 1D and SI Appendix, Extended Experimental Procedures) and membrane localization was confirmed by confocal microscopy using the intrinsic fluorescence of the coumarin caging group (Fig. 1D). Endocytosis of caged DAGs was slow (SI Appendix, Fig. S1-2A), resulting in a period of 40–60 min suitable for uncaging experiments. The uncaging reaction was confirmed in vitro in solution by NMR spectroscopy, found to be similarly efficient for all probes (Fig. 1E and SI Appendix, Fig. S1-1C) and comparable to previously reported probes bearing traditional caging groups (SI Appendix, Fig. S1-1D) (26).
To assess the uncaging efficiency in living cells, whole-field-of-view UV irradiation was combined with monitoring coumarin fluorescence at low intensity illumination. The coumarin fluorescence decreased at the plasma membrane, consistent with uncaging and subsequent dissipation of the cleaved coumarin alcohol, whereas the fluorescence signal remained unchanged in endosomes (where the coumarin alcohol is trapped in the lumen) (Fig. 1F and Movie S1). By quantifying the observed fluorescence decreases at the plasma membrane and correcting for baseline fluorescence levels, we found very similar (on average 66 ± 4%) uncaging efficiencies for all caged DAGs (Fig. 1G and SI Appendix, Fig. S1-2 B–D and Extended Experimental Procedures). Since these experiments were done with a laser scanning microscope in a single plane, we also assessed whether the uncaging changed throughout the cell in the z direction. To test this, we acquired z-stacks of coumarin fluorescence before and after uncaging at low laser intensity and found that the observed uncaging fluorescence loss was very similar throughout the z-stack suggesting that out of plane uncaging and/or rapid lateral diffusion of caged DAGs contribute to near-homogenous DAG photo release (SI Appendix, Fig. S1-3). These settings were used for all uncaging experiments in this study unless stated otherwise (SI Appendix, Extended Experimental Procedures). Taken together, our approach enables acute DAG density increases of different DAG species at the outer leaflet of the plasma membrane.
DAG Fatty Acid Composition Determines Selective Recruitment of PKC Isoforms.
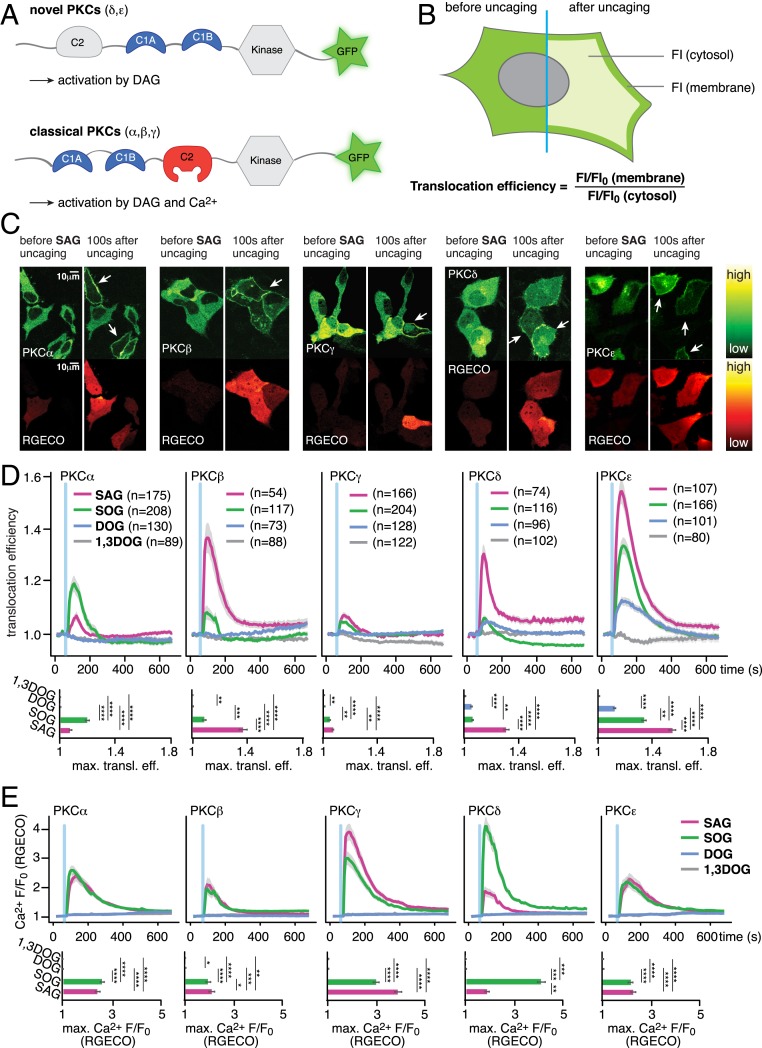
Varying DAG species patterns generated after receptor activation (13–15) can only encode information during signal transduction if chemically distinct DAG species differentially recruit DAG-binding proteins and, ultimately, cause different phosphorylation patterns of downstream effectors. We thus first tested whether uncaging of molecularly distinct DAG species resulted in specific recruitment patterns of individual EGFP-tagged PKC isoforms (Fig. 2A) in HeLa Kyoto cells. The extent of PKC recruitment was measured as “translocation efficiency,” the ratio between normalized fluorescence intensities (FI) at the plasma membrane and in the cytosol (28, 29) (Fig. 2B). Simultaneously, we monitored accompanying Ca2+ signaling events using RGECO, a red fluorescent, intensiometric Ca2+ indicator (30), as elevated DAG levels can directly induce Ca2+ signaling via activation of transient-receptor potential (TRP) channels (31).
Fig. 2.
Cellular responses after DAG uncaging. (A) Domain architecture of novel and classical PKCs. (B) Schematic illustration of the image analysis approach FI/FI0 are normalized fluorescence intensities at each time point in the time series. (C) Time-lapse montage of representative PKC (Upper) and RGECO (Lower) responses to uncaging of SAG. (D) Quantification of the recruitment of PKC isoforms PKCα, PKCβ, PKCγ, PKCδ, and PKCε to uncaging of different DAG species. The blue bar indicates the uncaging event. (Upper) Mean translocation efficiency traces. (Lower) Bar graphs show maximal translocation efficiency. Significance was tested using ANOVA followed by Dunn’s post hoc test and is represented by * (multiplicity adjusted P value, **P < 0.01; ***P < 0.001; ****P < 0.0001). Error bars represent SEM, data are mean. n, numbers represent cell numbers. (E) Quantification of Ca2+-signaling events in response to DAG uncaging. (Upper) Mean normalized fluorescence intensity traces for RGECO. (Lower) Bar graphs show maximal normalized fluorescence intensity. Significance was tested using the Dunn’s post hoc test and is represented by * (multiplicity adjusted P value, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). Shaded areas indicate SEM; blue bars indicate the uncaging event.
Upon uncaging of DAGs, we observed a unique temporal recruitment profile in response to the photorelease of individual 1,2-DAG species, while no recruitment was observed for the negative control 1,3-DOG. (Fig. 2 C and D and SI Appendix, Figs. S2-3 B–K and S2-4 B–F). The PKC isoforms (PKCδ and PKCε) were recruited to the plasma membrane in a staggered manner after uncaging of all three active 1,2-DAGs (Fig. 2D, PKCδ and PKCε), with SAG typically eliciting the strongest responses and DOG the weakest. PKCε recruitment was more pronounced than PKCδ recruitment for all DAGs, suggesting a higher affinity of PKCε for DAG. All PKCε translocation events observed after DAG uncaging were less intense than endogenous DAG production after ionomycin treatment, which should trigger a maximal, PLC-mediated response (SI Appendix, Fig. S2-1), indicating that the DAG concentration increases caused by DAG uncaging are within the concentration range of endogenous DAG generation events.
Both long-chain DAGs elicited Ca2+ transients, both in the presence and absence of PKC expression (Fig. 2E and SI Appendix, Fig. S2-2A), whereas no Ca2+ transients were observed for uncaging of the short-chain variant 1,2-DOG or the negative control lipid 1,3-DOG. We also found that the SAG- and SOG-induced Ca2+ transients appeared rather variable between cells and experiments (and in some cases were even absent). This could potentially be due to varying expression levels of DAG-sensitive Ca2+ channels (e.g., TRPC3/6). Thus, we drew comparative conclusions only from uncaging datasets acquired strictly in parallel under identical conditions. To assess whether the uncaging-triggered DAG signaling events could be obscured by Ca2+-induced production of endogenous DAG (via Ca2+-induced, PLC-mediated cleavage of PIP2), we performed an analysis of single-cell traces of PKCε translocation events. This isoform should only react to DAG levels, irrespective of the DAG source. We used the inherent variability of the Ca2+ transients to sort these cells into ones with large Ca2+ transients (larger than 50% RGECO fluorescence increase) and ones with lower Ca2+ transients and compared whether strong Ca2+ transients affected the observed PKC translocation events. This was not the case, and the resulting PKC recruitment profiles were very similar, even in cases where Ca2+ transients were not visible (SI Appendix, Fig. S2-2 B and C). Furthermore, analysis of the peak fluorescence ratios in the PKC and RGECO responses revealed no correlation between the two observables (SI Appendix, Fig. S2-2 B and C). Consequently, Ca2+-induced production of endogenous DAG in the uncaging experiments had no major effects on the observed PKC recruitment patterns.
The three classical, Ca2+-dependent PKC isoforms (PKCα, PKCβ, PKCγ) were only recruited by the long-chain DAGs (SAG and SOG) and did not respond to photoactivation of the short-chain probe cgDOG (Fig. 2D and SI Appendix, Figs. S2-3 B–K and S2-4 B–F). This is likely due to the fact that DOG never triggered elevated cytosolic Ca2+ levels (Fig. 2E), consistent with the requirement on coincident DAG and Ca2+ binding of these classical PKCs. The average PKCα translocation event was larger and longer-lasting for SOG uncaging than for SAG uncaging, whereas the opposite effect was observed for PKCβ (Fig. 2D). This particular difference appeared to depend exclusively on PKC–DAG interactions and not on Ca2+ levels, as similar Ca2+ transients were observed for both species (Fig. 2E). PKCγ was the only isoform similarly recruited by both native DAGs (SAG and SOG) (Fig. 2D). Interestingly, uniform elevation of plasma membrane DAG levels by photoactivation often led to the formation of localized patches of PKC recruitment or triggered global responses, which transformed into localized patches over time. Both effects were most pronounced for cgSAG (Fig. 2C and SI Appendix, Figs. S2-3 B–K and S2-4 B–F and H and Movies S2–S6), suggesting a varying capacity of individual DAG species to stabilize or form lipid gradients in the plasma membrane.
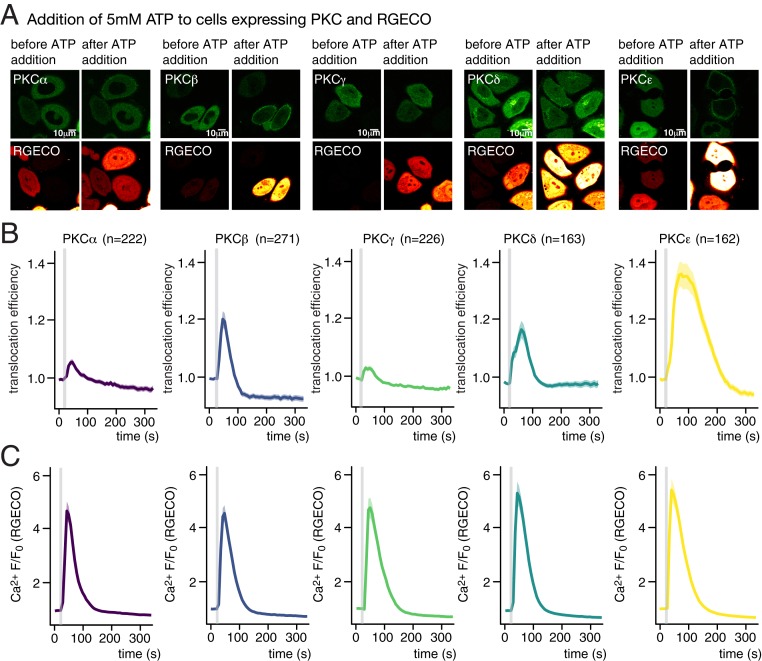
To compare PKC recruitment patterns after DAG uncaging with responses to a physiological stimulus, we monitored Ca2+ transients and PKC recruitment after ATP addition at different concentrations (1 and 5 mM), which should lead to the generation of endogenous DAG by PLCβ-mediated cleavage of PIP2, as well as IP3-induced Ca2+ transients. We observed that PKCε was recruited to the plasma membrane after ATP addition for both concentrations, featuring uniform, strong translocation events, whereas PKCδ-expressing cells exhibited less pronounced events when treated with 5 mM ATP (Fig. 3), in line with the pattern observed for all DAGs (compare Fig. 2). In contrast to the treatment with 5 mM ATP, PKCδ translocation events after adding 1 mM ATP were seldom observed (SI Appendix, Fig. S3). Moreover, translocation of the classical PKC isoforms was almost never observed when adding 1 mM ATP (SI Appendix, Fig. S3) despite the fact that pronounced Ca2+ transients were observed in these cases (SI Appendix, Fig. S3). The most likely explanation is that the necessary combined thresholds for DAG and Ca2+ concentrations were not reached. When adding 5 mM ATP, we observed pronounced translocation of PKCβ, whereas translocation was rare in cells expressing either PKCα or PKCγ. Importantly, the relative recruitment patterns for the five analyzed PKC isoforms after adding 5 mM ATP matched the ones observed for SAG uncaging (Fig. 2 C and D). Since PLC-mediated cleavage of PIP2 typically results in the generation of highly unsaturated DAG species such as SAG (32), this finding suggests that physiological DAG signaling events can be faithfully replicated by DAG uncaging with regard to effector protein recruitment patterns.
Fig. 3.
Cellular responses after ATP addition. (A) Time-lapse montage of representative PKC (Upper) and RGECO (Lower) responses following addition of 5 mM ATP. (Scale bars, 10 μM.) (B) Quantification of the recruitment of PKC isoforms PKCα, PKCβ, PKCγ, PKCδ, and PKCε to addition of 5 mM ATP. The gray bar indicates the ATP addition. (C) Quantification of Ca2+ signaling events in response to addition of 5 mM ATP as mean normalized fluorescence intensity traces for RGECO. Shaded areas represent SEM.
Next, we investigated whether uncaging of distinct DAGs gave rise to different phosphorylation patterns of bona fide PKC targets (C-Raf and GSK3β) and key players in cellular signaling cascades (MAPK, MEK1/2, and HDAC7). Cells were loaded with the caged DAGs (cgDAGs), and uncaging was performed on the whole dish. Cells were then collected, the cell lysate captured, and phosphorylation levels monitored with Western blot analysis (SI Appendix, Fig. S2-4 G and Extended Experimental Procedures). As expected, the negative control 1,3DOG did not significantly increase phosphorylation of any of the proteins studied. Significantly increased phosphorylation was observed in C-Raf at S298/296 after stimulation with SAG and in MEK1/2 and MAPK both after SAG and SOG uncaging, while DOG uncaging only significantly increased the phosphorylation of p-MAPK. This analysis confirmed that distinct phosphorylation profiles were triggered by the respective DAGs. Taken together, our data indicate that chemical differences between individual DAG species are sufficient to modulate DAG effector protein recruitment as well as downstream phosphorylation patterns.
Live-Cell Quantification of DAG Dynamics and DAG-Protein Affinities.
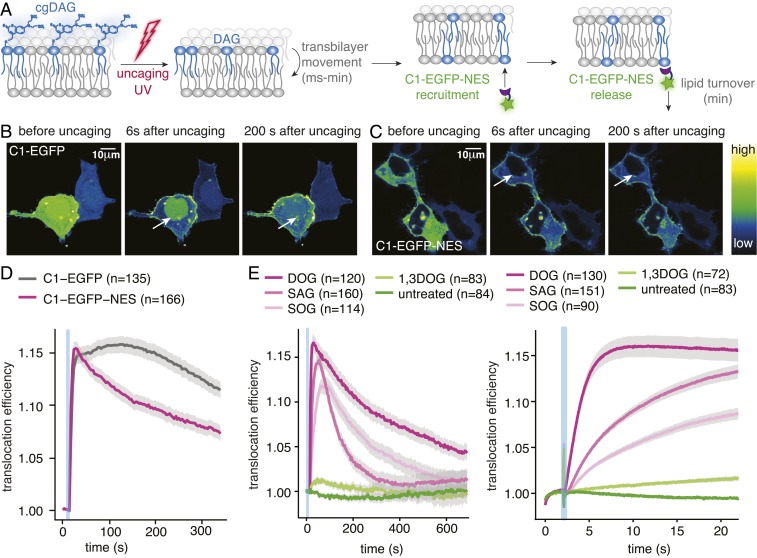
While our data suggest that DAG chemical diversity may provide a basis for specific recruitment of effector proteins, it is unclear how the chemical differences between individual lipid species are mechanistically translated into the different recruitment profiles. We decided to address this question using a minimal DAG-binding protein to isolate and quantify the lipid-driven protein recruitment process (Fig. 4A). We chose to use the known GFP fusion protein of the C1a domain of PKCγ, which is commonly used as a DAG biosensor (33) rather than screening for the most responsive C1 domains as this rendered our data as comparable as possible to our published data (26, 28) and the dynamic range of this sensor was assumed to be sufficient for kinetic analysis. However, we noticed that a significant part of the protein was retained in the nucleus and only exported when the cytosolic fraction was recruited to the plasma membrane upon DAG uncaging (Fig. 4B and Movie S7). This leads to an intrinsic delay of the sensor (Fig. 4D) and could hinder a kinetic analysis of the actual recruitment process. To avoid this, we equipped the protein with a nuclear export sequence (NES) to have the protein expressed solely in the cytosol (Fig. 4C and Movie S8). This suppressed the distortion of translocation kinetics by nuclear export compared to the original C1-EGFP construct (Fig. 4D).
Fig. 4.
An improved DAG biosensor allows precise analysis of membrane association kinetics and reveals differences between DAG species. (A) Schematic representation of the uncaging experiment. (B) Time-lapse montage of the C1-EGFP response to cgDOG uncaging in HeLa Kyoto cells. Note the presence of a nuclear protein pool indicated by the white arrows. (C) Time-lapse montage of the C1-EGFP-NES response to cgDOG uncaging in HeLa Kyoto cells. Note the absence of a nuclear protein pool indicated by the white arrows. (D) Translocation efficiency traces of C1-EGFP and C1-EGFP-NES after cgDOG uncaging. (E, Left) C1-EGFP-NES release kinetics after DAG uncaging in HeLa Kyoto cells. (E, Right) C1-EGFP-NES recruitment kinetics after DAG uncaging in HeLa Kyoto cells shown in more detail during the initial response. Uncaging experiments were carried out on a spinning disk microscope (SI Appendix, Additional Experimental Procedures). In all images, the blue bar indicates the uncaging event. n, numbers represent cell numbers; in a typical experiment, 5–10 cells were imaged simultaneously. Shaded regions surrounding mean traces in D and E represent SEM.
Uncaging of cgDOG, cgSAG, and cgSOG using the above described conditions triggered C1-EGFP-NES translocation to the plasma membrane, whereas neither cg1,3DOG uncaging nor illumination of unloaded cells caused translocation (Fig. 4E and SI Appendix, Fig. S4 A–D). A thorough characterization of C1-EGFP-NES in comparison with the C1-EGFP construct revealed no significant differences regarding the response rates to cgDAG uncaging (SI Appendix, Fig. S4 E–G). Importantly, striking differences between the individual lipid species were again observed (Fig. 4E): C1-EGFP-NES was recruited much faster to the plasma membrane after cgDOG uncaging compared to cgSOG or cgSAG uncaging (Fig. 4 E, Right), whereas the subsequent release of the sensor from the plasma membrane appeared to be fastest for SAG (Fig. 4 E, Left).
Quantitative Live-Cell Imaging Combined with Mathematical Modeling Reveals Specific Differences in Kinetics and Protein Affinities Among DAG Species.
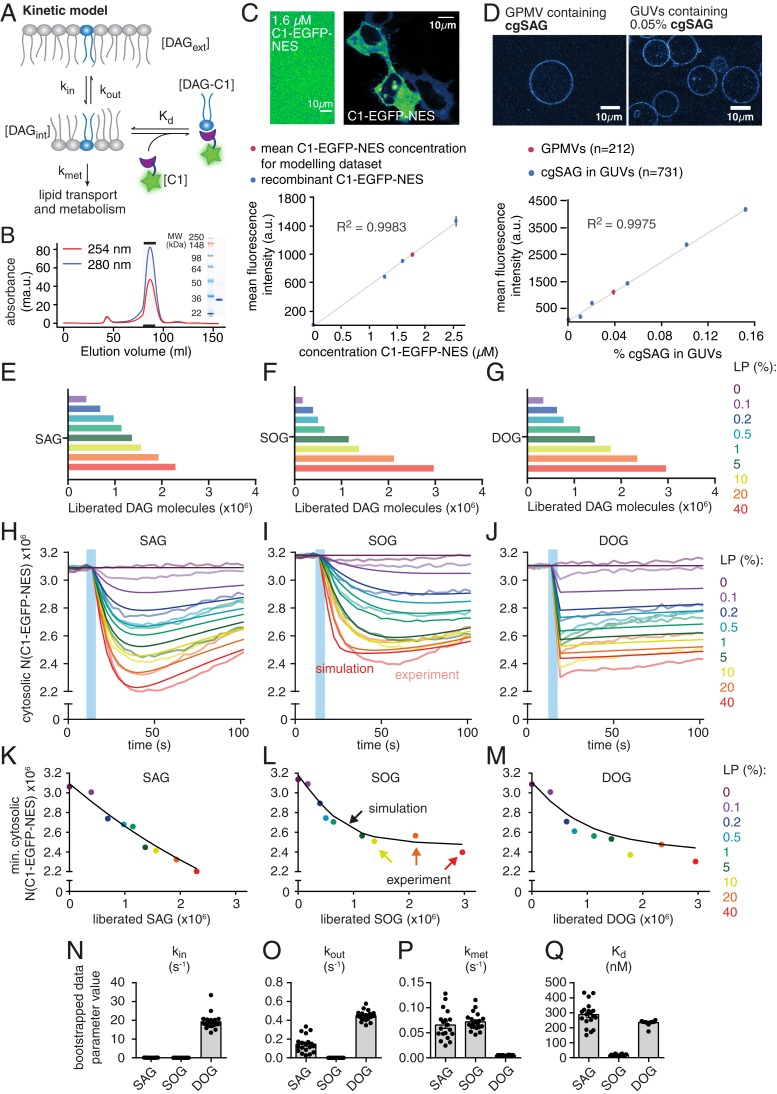
We hypothesized that the observed differences in protein recruitment between DAGs might be caused by distinct temporal DAG density profiles in the inner leaflet and, therefore, sought to characterize the kinetics of lipid transbilayer movement, lipid turnover, and lipid-protein affinities. For this, we developed a minimal kinetic model that could then be compared to the experimentally obtained temporal C1-EGFP-NES fluorescence profiles (Fig. 5A). This required quantitative knowledge of the number of C1-EGFP-NES protein copies and of the number of photoliberated DAG molecules. The number of free C1-EGFP-NES molecules of the resting cell was calculated from the cytosolic fluorescence intensity using a calibration curve generated from recombinantly produced C1-EGFP-NES at known concentrations (Fig. 5 B and C) and by estimating the cellular volume (SI Appendix, Extended Experimental Procedures). We determined the number of cgDAG in the outer leaflet of the plasma membrane before UV uncaging and the number of liberated DAG upon uncaging using the coumarin fluorescence. For this, the intensity of giant plasma membrane unilamellar vesicles (GPMVs) derived from cgDAG-loaded cells was quantified and compared to a calibration curve obtained from giant unilamellar vesicles (GUVs) containing defined mole percentages of caged DAG (Fig. 5D) (quantitative incorporation into GUVs was confirmed by mass spectrometry [SI Appendix, Extended Experimental Procedures and Table S1]). The uncaging efficiencies were determined for different laser powers, and this number was multiplied with the total amount of caged DAG to obtain the absolute number of liberated DAG molecules (Fig. 5 E–G). We found that uncaging of merely 1–2 × 10−3 mol% of an individual DAG species sufficed to induce the recruitment of C1-EGFP-NES to the plasma membrane, resulting in a sizable reduction of free C1-EGFP-NES in the cytosol (Fig. 5 H–J and SI Appendix, Extended Experimental Procedures). This observation allows the conclusion that the molecular composition of plasma membrane appears to be tuned in a way that enables the efficient recruitment of DAG-binding proteins in response to remarkably small elevations of DAG levels.
Fig. 5.
Kinetic model and determination of kinetic rate constants and Kd values for DAGs. (A) Kinetic scheme of relevant processes for DAG-induced plasma membrane recruitment of C1-EGFP-NES. (B) Size exclusion chromatography profile and corresponding SDS/PAGE analysis for purified C1-EGFP-NES. (C) Quantification of C1-EGFP-NES concentration in HeLa Kyoto cells. (Upper) Comparison of C1-EGFP-NES fluorescence intensity in HeLa Kyoto cells with a C1-EGFP-NES solution with a defined concentration of 1.6 µM. (Lower) Calibration curve with purified C1-EGFP-NES and comparison to average intracellular protein concentration. (D) Quantification of cgDAG density in HeLa Kyoto cells. (Upper) Comparison of GPMVs (giant plasma membrane vesicles) derived from cgSAG loaded cells with GUVs containing 0.05% cgSAG. (Lower) Quantification of fluorescence intensity of GPMVs (red) and GUVs featuring varying cgSAG content (blue). Error bars represent SEM. (E–G) Number of liberated DAG molecules for the different laser powers used to titrate C1-EGFP-NES responses. (H–J) Experimentally determined number of free C1-EGFP-NES proteins (mean number shown by faint lines) in the cytosol as a function of time for the three DAG species. Upon UV uncaging using the indicated nine different laser powers (0–40%, time of uncaging indicated by blue bar), the number of free proteins drops due to the recruitment of C1-EGFP-NES molecules to the plasma membrane upon DAG binding there. The measured/calculated mean amount of the free C1-EGFP-NES in the cytosol (faint lines) is shown together with predictions of this behavior by the model with its best fit parameters (solid lines). (K–M) For each of the nine laser powers, the minimal number of free C1-EGFP molecules in the cytosol that were observed upon uncaging (i.e., at the time point of maximal recruitment of the sensor to the plasma membrane) is plotted as a function of the amount of liberated DAG for the three DAG species. Colored dots represent experimental mean values, black lines show simulated results. (N–Q) Estimation of kinetic parameters and their respective variability by bootstrapping (one extreme outlier of SAG is not shown for scaling reasons and because it likely represents a nonfeasible local minimum). Data are shown as mean ± SEM and respective n numbers are given in SI Appendix, Fig. S5-1K.
To study how varying the amount of liberated DAG affected the translocation of C1-EGFP-NES, we used different laser powers for uncaging. This revealed an unexpected saturation of responses even when sizable fluorescence was still onserved in the cytosol (Fig. 5 H–M). This was accounted for in the model (which cannot by itself produce such a behavior due to its minimal design) by subtracting the experimentally observed nonresponsive fraction (determined from the experiment, not a model parameter). There are a few possible explanations for this phenomenon. Most likely, not all fluorescence in the cytosol corresponds to C1-EGFP-NES or the number of binding sites at the plasma membrane might be limiting.
Our minimal kinetic model was designed to describe the observed C1-EGFP-NES responses to DAG uncaging and featured DAG transbilayer movement, metabolism, and association to C1-EGFP-NES in equilibrium (Fig. 5A and SI Appendix, Fig. S5-1). Although the model only contained four free parameters, parameter optimization to all 510 experimental data points led to a very good agreement between model and experiment over the full range of laser powers (0–40%) used to liberate increasing amounts of DAG (compare experimental traces, faint lines, with model predictions, solid lines in Fig. 5 H–J). Specifically, the rate constant for outside-in movement across the plasma membrane (kin), the rate constant for inside-out movement (kout), and the rate constant for DAG turnover (kmet), and the affinity for binding C1-EGFP-NES (Kd) could be estimated by a parameter optimization sampling a large region of the parameter space (see SI Appendix, Extended Experimental Procedures for details). To investigate how variability in the experimental data might affect parameter estimates, we performed a bootstrapping analysis where the model parameters were repeatedly estimated from random subsets of the experimental traces (see SI Appendix, Extended Experimental Procedures for details). This lead to largely similar parameter estimates close to the ones obtained from the full dataset, in line with reliable and reproducible parameter estimates. We also performed a sensitivity analysis by investigating the agreement between model and data for various parameter combinations (SI Appendix, Fig. S5-2). This revealed a global minimum for SAG and for SOG, but the situation was less clear for DOG where multiple solutions for kin and kout produced similarly good agreement of the model with the data (SI Appendix, Fig. S5-2 C and F). This aligns with the experimental observation, where the recruitment of C1-EGFP-NES is already maximal with the first acquired data point (Fig. 5J), meaning that the sampling rate in the experiment is too low to define the speed of outside-in movement and only the ratio between kin and kout should be considered (the estimate of kmet was not affected by this, Fig. 5P). This was different for SAG and SOG, where multiple measurement points were acquired before the maximal C1-EGFP-NES recruitment was reached, which allowed for a robust estimate of kin, which was much lower (SAG: 0.065 s−1, SOG: 0.036 s−1, DOG: 17.02 s−1; for all parameters, see SI Appendix, Table S5). The estimated rate constant for inside-out transbilayer movement (kout) was much larger for SAG than for SOG (Fig. 5O). In fact, kout was essentially zero in the case of SOG and, indeed, a model without any inside-out transbilayer movement was preferred for SOG according to Akaike’s information criterion (34) (while the full model was preferred for SAG and DOG, although the major species-specific differences were overt in both models, see SI Appendix, Extended Experimental Procedures and Fig. S5-3). While the DAG turnover rate constant kmet was very similar for SAG and SOG, its value was much lower in the case of DOG (Fig. 5P), indicating that this nonnatural variant cannot be metabolized via the same pathways. Importantly, the DAG:C1-EGFP-NES affinities (1/Kd) of the two natural lipids SAG and SOG differed by one order of magnitude (in both models, Fig. 5Q and SI Appendix, Fig. S5-3L), indicating a clear side-chain specificity of the DAG-binding domain. This finding demonstrates that species-specific lipid–protein interactions can occur within biological membranes, a hypothesis that has been frequently put forward (12), but only rarely experimentally tested (35).
Simulating DAG Signaling Highlights the Importance of Lipid Dynamics.
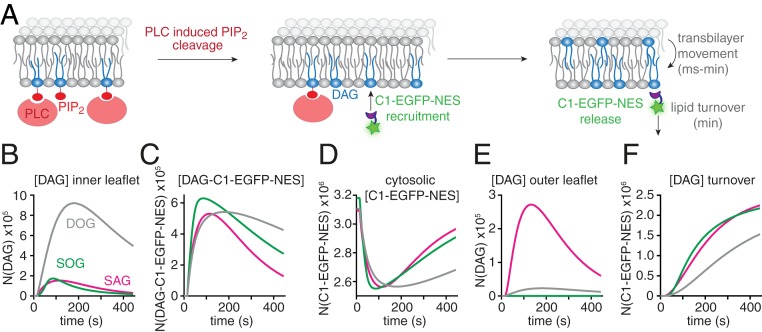
Knowledge of the species-specific kinetic parameters and affinities allowed us to simulate the relevant lipid and protein pools during physiological signaling events, where DAG is generated at the inner leaflet by PLC-mediated PIP2 cleavage (36) (Fig. 6A). We investigated how the temporal profile of DAG-mediated C1-EGFP-NES recruitment would deviate assuming (in theory) that either SAG, SOG, or DOG would exclusively be generated. Although we assumed that either species would be generated at the same rate, the resulting temporal profiles deviated significantly, owing to the species-specific properties: The model predicted much higher levels of DOG in the inner leaflet as compared to both SAG and SOG (Fig. 6B). Conversely, the respective maximal amounts of DAG-bound C1-EGFP-NES were much more similar (Fig. 6 C and D). The differences between DOG and SOG are a consequence of deviating lipid-protein affinities and turnover rates (Fig. 5 P and Q). However, lower SAG levels on the inner leaflet were also caused by differences in transbilayer movement (Fig. 5O), as SAG was the only species that accumulated to a significant amount on the outer leaflet of the plasma membrane (Fig. 6E). Physiologically, this would constitute a nonmetabolizable SAG buffer on the outer plasma membrane leaflet, prolonging the duration of SAG-mediated signaling events at the cost of an attenuated amplitude.
Fig. 6.
Simulation of physiological DAG signaling events. (A) Scheme depicting the in silico experiment of DAG generation in the inner leaflet of the plasma membrane by stimulation of PLC-mediated cleavage of 2.5 × 106 PIP2 molecules generating different DAG species with a time constant of τ = 100 s, in line with observations made in ATP stimulation experiments (SI Appendix, Fig. S3). (B–F) Temporal development of SAG (magenta), SOG (green), and DOG (gray) molecule numbers on the inner leaflet (B), the number of membranous DAG–C1–EGFP–NES complexes (C), the number of cytosolic C1-EGFP-NES molecules (D), the number of DAG molecule on the outer leaflet (E), the number of DAG molecule removed from the plasma membrane due to lipid turnover (F). Note that SAG is the only species exhibiting a sizable outer-leaflet pool.
Discussion
Quantitative Lipid Biochemistry in Living Cells.
In this study, we report a conceptual strategy to analyze the dynamics and molecular interactions of native lipid species in quantitative live-cell experiments. As biological membranes—unlike model membranes—feature an asymmetric lipid distribution (37) and highly complex lipid composition (12), such experiments are needed to understand the biological function of distinct lipid species. We developed a generation of caged DAG probes equipped with a sulfonated caging group designed for triggering rapid and temporally defined, quantitative increases of DAG levels specifically at the outer plasma membrane leaflet. Our experimental strategy enables quantitative kinetic analysis of DAG signaling events as it provides a defined starting point for both the amount of liberated DAG and signal initiation, which is not possible for receptor-induced DAG production, where the exact amount and time course of DAG production is experimentally not accessible.
Importantly, the induced changes appear to be comparable with alterations of DAG levels observed during physiological signaling events, as PKC recruitment patterns after stimulation of endogenous DAG production by ATP were replicated by SAG uncaging-induced recruitment (Figs. 2 and 3). While our approach does not capture effects during physiological signaling events that might be caused by localized DAG production in preorganized signaling clusters below the diffraction limit, it allows testing the hypothesis that lipid diversity provides a mechanistic basis for lipid function (12, 22). Importantly, a recent report suggests a high degree of species selectivity for a major family of DAG metabolizing enzymes, diacylglycerol kinases (38). The authors report distinct specificities for individual isoforms, a pattern that is reminiscent of the differences in recruitment patterns that we observe for PKC isoforms. Both findings strongly suggest that individual DAG species play distinct roles in lipid signaling networks.
Species-Specific DAG Dynamics and DAG–Protein Interactions Regulate Signaling.
We show that comparable increases of individual DAG species result in strikingly different PKC recruitment and downstream phosphorylation patterns, consistent with a functional role of DAG fatty acid composition in cellular signaling events. To understand the mechanistic basis for the observed differences, we devised a minimal mathematical model and optimized its parameters to fit the temporal recruitment profiles of the DAG sensor C1-EGFP-NES. We determined Kd values for DAG–C1–domain interactions as well as rate constants for transbilayer movement and turnover of individual DAG species using dose-dependent DAG photorelease.
We found that subtle variations in DAG acyl chain length and unsaturation degree result in markedly different parameter values. For instance, although structurally very similar, SAG and SOG deviate by an order of magnitude in their affinity for binding DAG sensing proteins (Fig. 5Q). Moreover, while our parameter estimates implied no movement of SOG from the inner to the outer leaflet of the plasma membrane, SAG was predicted to accumulate on the outer leaflet of the plasma membrane during physiological signaling, which could markedly affect the temporal signaling profile, because SAG on the outer leaflet would constitute a nonmetabolizable buffer, thus resulting in attenuated but temporally extended elevation of SAG levels.
We show that combining quantitative live imaging with mathematical modeling enables a quantitative comparison of specific molecular properties related to the chemical structure of diverse DAG species, which are not directly experimentally accessible. As with all modeling approaches, these parameter values can only be judged in the context of the model itself, which will always be an approximation of reality. Yet, such approximations are particularly useful to make predictions and to generate key hypotheses in membrane biology, which can then be addressed experimentally. Future work will identify the limitations and further requirements of the model and, together with experimental refinements, iteratively lead to improvements of the model’s predictive value.
Taken together, the combination of quantitative lipid biochemistry in living cells and mathematical modeling allowed us to study the mechanistic basis of cellular lipid signaling events in unprecedented molecular detail down to the elementary reactions that govern the behavior of distinct lipid species. We anticipate that the described experimental strategy could be expanded to other lipid classes, as the fundamental design principle of the utilized caged lipids is universal, and recently developed screening approaches have streamlined the discovery and characterization of lipid-binding domains (39). As more and more individual lipid species are linked to specific cellular processes by lipidomic screens, the need for experimental strategies to validate their involvement in live-cell assays and to investigate the underlying mechanisms will only increase.
Materials and Methods
The materials and methods used in this study are described in detail in SI Appendix, Additional Experimental Procedures. Information includes general synthetic procedures, photophysical characterization of compounds, description of plasmids and cloning, virus production, protein production and purification, Western blot analysis, preparation of giant unilamellar vesicles, mass spectrometric analysis of vesicle lipid content, cell culture and cDNA transfection protocols, life cell imaging, loading procedures for caged lipids, uncaging experiments, quantification of uncaging reactions, image analysis and data processing, and the kinetic model and fitting procedures. Data availability statement: All relevant datasets are included as figure items in the main text or SI Appendix. All software codes used for analysis will be made available to readers via the institute repository of the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG) (https://dx.doi.org/21.11101/0000-0007-DF4A-C) and upon request from the corresponding authors.
Supplementary Material
Acknowledgments
We thank the following services and facilities at MPI-CBG Dresden for their support: Protein Expression Facility, Mass Spectrometry Facility, and the Light Microscopy Facility. We thank Jan Peychl, Britta Schroth-Diez, and Sebastian Bundschuh for the outstanding support and expert advice. A.N. acknowledges funding by the European Research Council under the European Union’s Horizon 2020 research and innovation program Grant Agreement GA758334 ASYMMEM, the Max Planck Lipid Research Center, and the Deutsche Forschungsgemeinschaft (DFG) as a member of the TRR83 consortium. A.M.W. was funded by the DFG from Emmy Noether Grant and TRR186, Project 278001972, and A.T.G. was further part of the international program Medical Neurosciences. Ü.C. acknowledges financial support from the DFG as a member of the TRR83 and FOR2682 consortia and the German Federal Ministry of Education and Research (BMBF) grant to the German Center for Diabetes Research. A.S. acknowledges financial support from the DFG as a member of the TRR83 and FOR2682 consortia and BMBF Grant “Lipidomics for Life Sciences.”
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All software codes used for analysis have been deposited on the Max Planck Institute of Molecular Cell Biology and Genetics repository, https://dx.doi.org/21.11101/0000-0007-DF4A-C.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912684117/-/DCSupplemental.
References
- 1.Baukrowitz T., Fakler B., K(ATP) channels: Linker between phospholipid metabolism and excitability. Biochem. Pharmacol. 60, 735–740 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Tall A. R., Yvan-Charvet L., Cholesterol, inflammation and innate immunity. Nat. Rev. Immunol. 15, 104–116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai F.-C., et al. , A polarized Ca2+, diacylglycerol and STIM1 signalling system regulates directed cell migration. Nat. Cell Biol. 16, 133–144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krause M., Gautreau A., Steering cell migration: Lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 15, 577–590 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Mancini R., et al. , Metabolic features of cancer stem cells: The emerging role of lipid metabolism. Oncogene 37, 2367–2378 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Wang B., et al. , Phospholipid remodeling and cholesterol availability regulate intestinal stemness and tumorigenesis. Cell Stem Cell 22, 206–220.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beloribi-Djefaflia S., Vasseur S., Guillaumond F., Lipid metabolic reprogramming in cancer cells. Oncogenesis 5, e189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perry R. J., Samuel V. T., Petersen K. F., Shulman G. I., The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature 510, 84–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coskun U., Simons K., Cell membranes: The lipid perspective. Structure 19, 1543–1548 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Wenk M. R., Lipidomics: New tools and applications. Cell 143, 888–895 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Shevchenko A., Simons K., Lipidomics: Coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 11, 593–598 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Harayama T., Riezman H., Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 19, 281–296 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Lee C., Fisher S. K., Agranoff B. W., Hajra A. K., Quantitative analysis of molecular species of diacylglycerol and phosphatidate formed upon muscarinic receptor activation of human SK-N-SH neuroblastoma cells. J. Biol. Chem. 266, 22837–22846 (1991). [PubMed] [Google Scholar]
- 14.Pessin M. S., Baldassare J. J., Raben D. M., Molecular species analysis of mitogen-stimulated 1,2-diglycerides in fibroblasts. Comparison of alpha-thrombin, epidermal growth factor, and platelet-derived growth factor. J. Biol. Chem. 265, 7959–7966 (1990). [PubMed] [Google Scholar]
- 15.Peter-Riesch B., Fathi M., Schlegel W., Wollheim C. B., Glucose and carbachol generate 1,2-diacylglycerols by different mechanisms in pancreatic islets. J. Clin. Invest. 81, 1154–1161 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atilla-Gokcumen G. E., et al. , Dividing cells regulate their lipid composition and localization. Cell 156, 428–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Köberlin M. S., et al. , A conserved circular network of coregulated lipids modulates innate immune responses. Cell 162, 170–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma C., Su L., Seven A. B., Xu Y., Rizo J., Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339, 421–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yaney G. C., Corkey B. E., Fatty acid metabolism and insulin secretion in pancreatic beta cells. Diabetologia 46, 1297–1312 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Newton A. C., Protein kinase C: Poised to signal. Am. J. Physiol. Endocrinol. Metab. 298, E395–E402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quann E. J., Liu X., Altan-Bonnet G., Huse M., A cascade of protein kinase C isozymes promotes cytoskeletal polarization in T cells. Nat. Immunol. 12, 647–654 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muro E., Atilla-Gokcumen G. E., Eggert U. S., Lipids in cell biology: How can we understand them better? Mol. Biol. Cell 25, 1819–1823 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honigmann A., Nadler A., The next frontier: Quantitative Biochemistry in living cells. Biochemistry 57, 47–55 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Wagner N., Stephan M., Höglinger D., Nadler A., A click cage: Organelle-specific uncaging of lipid messengers. Angew. Chem. Int. Ed. Engl. 57, 13339–13343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng S., et al. , Mitochondria-specific photoactivation to monitor local sphingosine metabolism and function. eLife 7, e34555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadler A., et al. , The fatty acid composition of diacylglycerols determines local signaling patterns. Angew. Chem. Int. Ed. Engl. 52, 6330–6334 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Nadler A., et al. , Exclusive photorelease of signalling lipids at the plasma membrane. Nat. Commun. 6, 10056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Höglinger D., et al. , Trifunctional lipid probes for comprehensive studies of single lipid species in living cells. Proc. Natl. Acad. Sci. U.S.A. 114, 1566–1571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein F., Kress M., Reither S., Piljić A., Schultz C., FluoQ: A tool for rapid analysis of multiparameter fluorescence imaging data applied to oscillatory events. ACS Chem. Biol. 8, 1862–1868 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y., et al. , An expanded palette of genetically encoded Ca2+ indicators. Science 333, 1888–1891 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofmann T., et al. , Direct activation of human TRPC6 and TRPC3 channels by diacylglycerol. Nature 397, 259–263 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Bozelli J. C. Jr, Epand R. M., Specificity of acyl chain composition of phosphatidylinositols. Proteomics 19, e1900138 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Oancea E., Teruel M. N., Quest A. F., Meyer T., Green fluorescent protein (GFP)-tagged cysteine-rich domains from protein kinase C as fluorescent indicators for diacylglycerol signaling in living cells. J. Cell Biol. 140, 485–498 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akaike H., A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19, 716–723 (1974). [Google Scholar]
- 35.Björkholm P., et al. , Identification of novel sphingolipid-binding motifs in mammalian membrane proteins. Biochim. Biophys. Acta 1838, 2066–2070 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Carrasco S., Mérida I., Diacylglycerol, when simplicity becomes complex. Trends Biochem. Sci. 32, 27–36 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi T., Menon A. K., Transbilayer lipid asymmetry. Curr. Biol. 28, R386–R391 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Ware T. B., et al. , Reprogramming fatty acyl specificity of lipid kinases via C1 domain engineering. Nat. Chem. Biol. 16, 170–178 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saliba A.-E., et al. , A quantitative liposome microarray to systematically characterize protein-lipid interactions. Nat. Methods 11, 47–50 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.