Our recent article reports the important suppressing role of MRTFB (myocardin-related transcription factor B) in colorectal cancer (CRC) and reveals several target genes of MRTFB’s transcription activity, including SPDL1 and MCAM (1). We initiated the study because Mrtfb was identified as a candidate tumor suppressor gene in a forward genetic screen using the Sleeping Beauty (SB) transposon system (2). We do not investigate whether single-nucleotide polymorphism (SNP) contributes to MRTFB’s function in CRC, as the SB transposon system does not generate point mutations (3).
Zhao et al. (4) propose a new target gene, NOMO1 (NODAL Modulator 1), for MRTFB’s transcription activity. They find three SNPs in two human colon tissue samples and analyze their potential effects on NOMO1’s expression using the GTEx Consortium’s Genotype-Tissue Expression (GTEx) expression quantitative trait locus (eQTL) dataset (5). This finding carries potential significance, as it extends the list of MRTFB’s target genes in CRC development. We must point out that NOMO1 was not identified as a candidate target gene of MRTFB by our RNA sequencing (RNA-seq) analysis (1). We would also suggest the inclusion of a much larger number of colon samples for SNP analysis, as only two tissues were used by Zhao et al. (4).
The function of NOMO1 in CRC remains unknown, and the only relevant published study is from Perea et al. (6), which reports that NOMO1 deletion might serve as a molecular marker for early-onset CRC. Detailed functional assays are needed for us to appreciate the function of NOMO1 in CRC development, and the newly established tissue-specific Nomo1 knockout mice will serve as a valuable tool for this purpose (7).
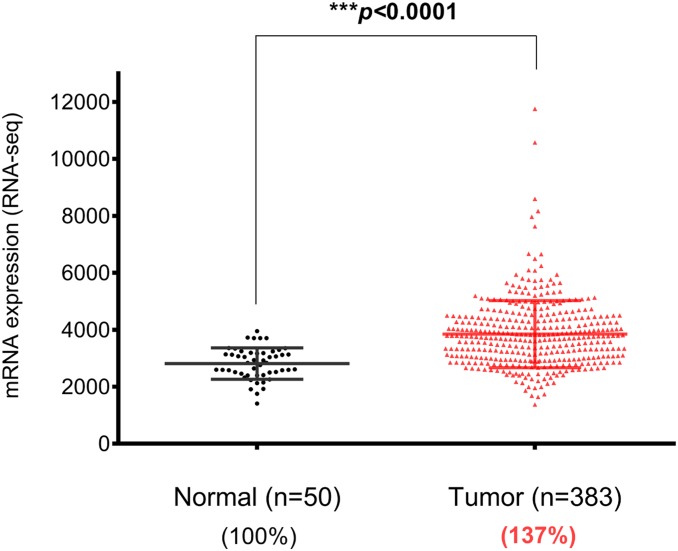
Another important question about NOMO1 is whether it plays a suppressing role in CRC. NOMO1 is expected to be a tumor suppressor if it is a direct target of MRTFB, a transcription activator and a tumor suppressor (1). This would be consistent with the observation of NOMO1 deletion in early-onset CRC (6). However, NOMO1’s expression level seems to be up-regulated in human CRC, as shown by the analysis results of TCGA’s RNA-seq data (Fig. 1) (http://firebrowse.org/?cohort=COADREAD) (8). This does not support the prediction that NOMO1 suppresses CRC development.
Fig. 1.
NOMO1 expression is significantly elevated in human CRC patients. TCGA RNA-seq data for CRC were downloaded from FireBrowse (http://firebrowse.org/). The samples included 50 normal tissue samples and 383 CRC samples; mRNA, messenger RNA.
In summary, NOMO1 might be a target of MRTFB’s transcription activity, as suggested by Zhao et al. (4). However, it is simply too early to conclude that NOMO1 plays any roles in CRC development. Detailed functional analyses, as done in our study (1), are required to reveal whether NOMO1 and its regulation by MRTFB are important for CRC development.
Footnotes
The authors declare no competing interest.
References
- 1.Kodama T., et al. , MRTFB suppresses colorectal cancer development through regulating SPDL1 and MCAM. Proc. Natl. Acad. Sci. U.S.A. 116, 23625–23635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeda H., et al. , Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat. Genet. 47, 142–150 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Dupuy A. J., et al. , A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 69, 8150–8156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao T., Hu Y., Zang T., Cheng L., MRTFB regulates the expression of NOMO1 in colon. Proc. Natl. Acad. Sci. U.S.A. 117, 7568–7569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.GTEx Consortium , Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perea J., et al. , NOMO-1 gene is deleted in early-onset colorectal cancer. Oncotarget 8, 24429–24436 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-Tuñón I., et al. , Establishment of a conditional Nomo1 mouse model by CRISPR/Cas9 technology. Mol. Biol. Rep. 47, 1381–1391 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Network , Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]