Significance
The cell cycle is a biological process requiring the interaction of multiple components to produce a new cell. How these processes synchronize with cell cycle progression, however, is currently unknown. This work performs a quantitative multiomic analysis of the eukaryal cell division cycle, to map which processes change at each level of regulation (transcriptome, proteome, phosphoproteome, and metabolome). This study provides a comprehensive resource for identifying possible targets for therapeutic intervention when cell cycle dysregulation occurs.
Keywords: multiomics, cell cycle, Saccharomyces cerevisiae, absolute quantitation, metabolism
Abstract
For cells to replicate, a sufficient supply of biosynthetic precursors is needed, necessitating the concerted action of metabolism and protein synthesis during progressive phases of cell division. A global understanding of which biosynthetic processes are involved and how they are temporally regulated during replication is, however, currently lacking. Here, quantitative multiomics analysis is used to generate a holistic view of the eukaryal cell cycle, using the budding yeast Saccharomyces cerevisiae. Protein synthesis and central carbon pathways such as glycolysis and amino acid metabolism are shown to synchronize their respective abundance profiles with division, with pathway-specific changes in metabolite abundance also being reflected by a relative increase in mitochondrial volume, as shown by quantitative fluorescence microscopy. These results show biosynthetic precursor production to be temporally regulated to meet phase-specific demands of eukaryal cell division.
The eukaryal cell division cycle is an essential process in most complex living organisms. It is defined by unidirectional steps in growth, DNA synthesis, and mitosis, with the involvement of essential cell division cycle genes (CDC) in multiple checkpoints during replication, underscoring the importance for fidelity in this process (1). Indeed, when the demands of division are unmet or components malfunction, this can lead to cell death and/or uncontrolled proliferation. One primary checkpoint is whether or not metabolic reserves exist in adequate supply before replication is committed to. This phenomenon of gating by metabolite availability implies that the metabolic status of the cell is of critical importance for cells to complete division. For example, before DNA synthesis can occur, a sufficient supply of nucleotide precursors and redox cofactors needs to be generated to allow genetic material to be copied and at the same time protected from oxidation by reactive oxygen species. Similarly, energy-rich storage carbohydrates such as trehalose have shown to be important to stockpile before replication can occur to fuel cell biosynthesis, with mutants in related metabolic genes affecting the completion time of cell division (2). This would suggest that timely provision of biosynthetic precursors is of fundamental importance for cell cycle progression. A global understanding of what biosynthetic processes are involved as well as how they act in synchrony with division is, however, currently lacking.
Emerging technologies such as RNA sequencing (RNA Seq) and mass spectrometry-based proteomics have begun to enable the characterization of cell systems at the absolute quantitative level, providing insight into resource allocation and copy number abundances of messenger RNA (mRNA) and proteins of interest (3). Furthermore, phosphoproteomics and metabolomics offer the opportunity to identify cell components that may regulate processes that would otherwise be challenging to trace from gene expression alone. Here, we perform quantitative multiomic analysis on the eukaryal cell cycle model organism, budding yeast (Saccharomyces cerevisiae), to create a global and quantitative perspective on how metabolism synchronizes with cell division, including what processes are involved, when they are active, and by how much.
Systems biology studies of the eukaryal model organisms, budding and fission yeast, during their cell cycle has previously afforded insight into the relationship between the transcriptome and proteome (4, 5), and the proteome and phosphoproteome (6, 7), adding to related work that has focused on individual omic studies of the metabolome (2) and the transcriptome (8, 9). Recent relative quantification of the proteome, transcriptome, and metabolome of budding yeast (10) has also added to this growing body of work for understanding cell division in eukaryal organisms. Here we performed absolute quantitative measurements of the transcriptome and proteome, alongside semiquantitative analysis of the phosphoproteome and metabolome, to identify and clarify key changes during cell division, shedding light on how different processes may be regulated from a multiomic perspective.
First, we compared changes at each omic layer, and found the most widespread changes to be at the transcriptomic level. Changes in protein abundance occurred predominantly via transcriptional regulation, with dynamic protein phosphorylation occurring mainly for proteins related to cell cycle regulation. On analysis of phase-specific processes, we find that protein synthesis predominantly occurs in G1, while the majority of metabolic pathways were active after cells committed to division in S and G2/M. Complementary analysis of the mitochondria, the energy powerhouse of the cell, also showed that this organelle’s volume increases disproportionately to total cell volume after the cell cycle was committed to, suggesting a relative up-regulation of metabolite and energy generation for sustaining cell biosynthesis. An untargeted analysis of metabolite abundance showed that cells modulate precursor abundance according to their role in metabolism, with amino acid abundance peaking in G1, nucleotides increasing in S phase alongside redox cofactors and lipids, and central carbon metabolism products peaking at the end of the cell cycle in G2/M. By identifying what biosynthetic processes synchronize with the cell cycle, these results could have important therapeutic implications for the development of metabolic targets in anticancer strategies.
Results
A Quantitative Multiomic Perspective of the Eukaryal Cell Cycle.
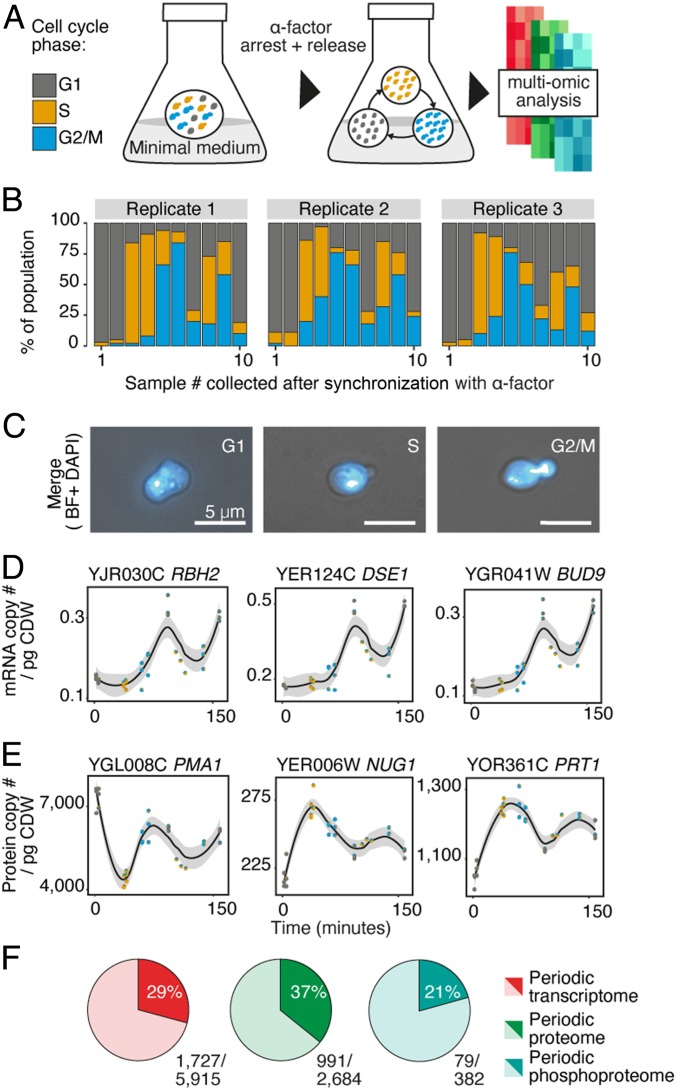
In this work, a quantitative multiomic analysis of the yeast cell cycle was generated, incorporating information from the transcriptome, proteome, phosphoproteome, and metabolome. Prototrophic yeast cells were synchronized in batch culture by addition of alpha (α) factor (Fig. 1A), producing close to 100% homogeneous cell populations for each cell cycle stage, allowing sample collection for up to three cell cycles (Fig. 1B). Although the cell cycle is divided into four separate cell cycle phases, in our study, G2 and mitosis (M) were coupled together, as these are inherently difficult to discriminate using fluorescence microscopy, resulting in samples being identified as either unbudded (G1), in the early stage of division with bud emergence (S phase), or at the final stage of the cell cycle prior to cytokinesis (G2/M) (Fig. 1C). At each time point, replicates were collected and submitted to transcriptome, proteome, phosphoproteome, and metabolome analysis. Biological triplicates and, for the first cycle, technical duplicates were collected, allowing an unprecedented level of coverage from one culture condition. Multiple replicates also enabled almost 100% coverage of the genome to be acquired for the transcriptome, alongside almost 70% of the expressed proteome, over 1,000 protein phosphorylation sites, and almost 300 metabolites for each time point.
Fig. 1.
A quantitative multiomic perspective of the eukaryal cell cycle. (A) Experimental workflow of yeast synchronization with alpha (α)-factor mating pheromone. Samples were taken for up to three cell cycles in batch culture and submitted for absolute quantitative transcriptomics, proteomics, and semiabsolute quantitative phosphoproteomics and metabolomics. (B) Population composition for each sample following synchronization with α-factor. (C) Cells in G1: unbudded, S: budded (without DNA in daughter cell), and G2/M: mitotic cell cycle stages, identified using fluorescence microscopy and the DNA stain, 4′,6-diamidino-2-phenylindole. (Scale bar: 5 μm.) (D and E) Top-ranked periodic genes for the (D) transcriptome and (E) proteome identified by limma modeling. All biological replicates are plotted for each time point, with population composition for each sample represented in each point as a pie chart. Span for fitting each local regression, as part of the loess smoother (locally estimated scatterplot smoothing), is 0.6. CI for regression line is 95%. pg CDW, picogram of cell dry weight. (F) Fraction of each omic level identified by limma modeling to be periodic during the eukaryal cell cycle.
RNA Seq/nCounter and stable isotope labeling with amino acids in cell culture/iBAQ tandem mass spectrometry (MS/MS) methods, respectively, were used to generate absolute quantitative mRNA and protein abundances (expressed in copy number per picogram of cell dry weight) for each sample. The abundance of most mRNA in the cell (>99%) varied between two orders of magnitude, while their translated protein demonstrated considerable amplification in abundance, spanning approximately six orders of magnitude, correlating positively with mRNA abundance (r2 = 0.61; SI Appendix, Fig. S1A), adding to the emerging consensus that protein levels are largely determined by transcript abundance (3). Most gene products were amplified during translation by ∼200-fold (SI Appendix, Fig. S1B), with proteins related to translational and metabolic processes being the most amplified, while proteins involved in transport, posttranslational modifications, and the cell cycle were the least amplified (SI Appendix, Fig. S1C). This process-dependent difference in amplification may explain why the proteome is disproportionately composed, by abundance, of proteins with a translational and metabolic function (SI Appendix, Fig. S1D).
To identify components that were periodic in abundance across the different omic levels, we used linear modeling based on differential expression analysis (limma) (Fig. 1 D and E). This approach was selected due to its ability to incorporate additional information about time points, such as sample heterogeneity, and to mitigate possible confounders, such as pheromone treatment. For example, in the linear model, we fit different coefficients for the pheromone effect and the effect of cells being in G1 phase of the cell cycle. We assume that only the samples in the first time point are affected by the pheromone, while the other G1 samples (in the second and third cycle) are not. Therefore, the G1 phase and the pheromone effect are not confounded, and it is possible to distinguish between them.
When compared to other methods such as Fourier transform (11) or rank-based methods (12), this approach performed favorably on the benchmark set of 113 genes known to have periodic expression (13), despite using relatively few time points, with 87% being detected in our dataset (98/113) (SI Appendix, Fig. S2). Phase-specific up-regulation of genes was also cross-compared between our data and results from previously curated data, where we found many genes up-regulated to have previously been identified as periodically peaking in the same phase, confirming the accuracy of our model (SI Appendix, Fig. S2). Across all omic levels and their abundance ranges (Dataset S1), at least 20% of components were identified as periodic (Dataset S2), with most changes observed at the transcriptomic level (Fig. 1F and SI Appendix, Fig. S3). Additionally, when comparing the largest differences in abundance between samples for individual mRNA and proteins, the majority (>94%) of both background (nonperiodic) and oscillating components increased within a twofold abundance from the least to the highest abundant value measured (SI Appendix, Fig. S4).
The acquisition of multiomic data also allowed the tracking of periodic genes from mRNA abundance to protein phosphorylation state (SI Appendix, Fig. S5). Here, it was clear the overlap between genes was not complete (SI Appendix, Fig. S5A). This may be, in part, due to the phosphoproteome coverage, which only included 382 proteins (14%) of the measured proteome (n = 2,684). Despite the difference in coverage, several genes were still found to have periodic abundance at all levels, including the protein of unknown function MTC1 that is synthetically lethal with cdc13-1 mutants, suggesting it may play a key role during division (SI Appendix, Fig. S5B).
Not all periodic transcripts gave rise to an associated periodic protein (SI Appendix, Fig. S5C). This result cannot be explained entirely by the lower coverage for the proteomic MS, as more than half (60%, n = 1,033/1,727) of proteins, which had periodic mRNA, were detected, such as SEC14, BCY1, and MSF1. Similarly, some proteins with changing abundance had nonperiodic mRNA, such as TRP5, LSM7, and CAR1 (SI Appendix, Fig. S5C). In a recent study by Kelliher et al. (14), for some cell cycle-related genes, protein abundance also did not reflect periodic mRNA behavior. This included SWI6, which here is also found to have significantly periodic mRNA (P ≤ 0.001) and, as previously found, could not be identified as periodic at the protein level (P = 0.297), nor were any periodic changes found for its protein phosphorylation state either. A lack of correlation between mRNA and protein could be attributed to mRNA localization and its effect on protein translation, which was found to be true for mammalian cells and tissues where some newly synthesized mRNA are retained in the nucleus to buffer against stochastic mRNA synthesis (3, 15, 16). Protein levels that don’t synchronize with mRNA abundance, aside from the possible influence of posttranslational modifications, may also be affected by translation machinery that buffers against changes in mRNA abundance via varying protein translation or protein degradation rates, through differences in ribosome distribution on mRNA (3).
One feature we discovered that coincides with periodic mRNA is that genes with periodic expression have different intergene distances (SI Appendix, Fig. S5D). Using a method previously described in Kristell et al. (17), the distance between all possible gene pairs was tested. Here, we find that genes identified as periodic and peaking in S phase locate closer to each other on the genome than expected by chance (SI Appendix, Fig. S5D). This suggests cells may regulate gene expression by spatially arranging genes to organize them into similar euchromatic or heterochromatic states, enabling them to undergo similarly timed transcriptional activation or repression.
We next used our list of periodic mRNA and protein to identify how many of these genes either had an essential function or were conserved in human genes or both. First, using the list of genome-wide essential genes, compiled by Liu et al. (18) (n = 1,074, Dataset S3), our results show almost half of genes with an essential function to also be periodic during the cell cycle, with 40% and 47% of the measured essential transcriptome (411/1,027) and proteome (308/659), respectively, being periodic, equating to 24% and 31% of all periodic mRNA and protein. Of the 175 yeast genes which could be replaced by human orthologs in the study by Kachroo et al. (19) (Dataset S4), 46% (80/175) and 45% (64/142) were periodic at the mRNA and protein levels, respectively. We next extracted genes from both the essential and conserved lists to see how many were periodic at both the mRNA and protein levels and which overlapped, to determine which proteins that were transcriptionally regulated were both conserved in humans and had an essential function. Remarkably, >90% (n = 32) of the 35 conserved genes that were periodic at both the mRNA and protein levels in human orthologs were also found to be essential. Of these 32 genes, the main processes were found to relate to protein synthesis (ribosomal RNA [rRNA] processing, and large and small ribosome subunit biogenesis) as well as metabolism with ∼30% (n = 10) having a metabolic function (CDC48, CDC21, KRS1, PCM1, OST1, UGP1, ERG12, KRE33, GLN4, DPM1).
The Periodic Multiome Reflects the Demand of Protein Synthesis during Division.
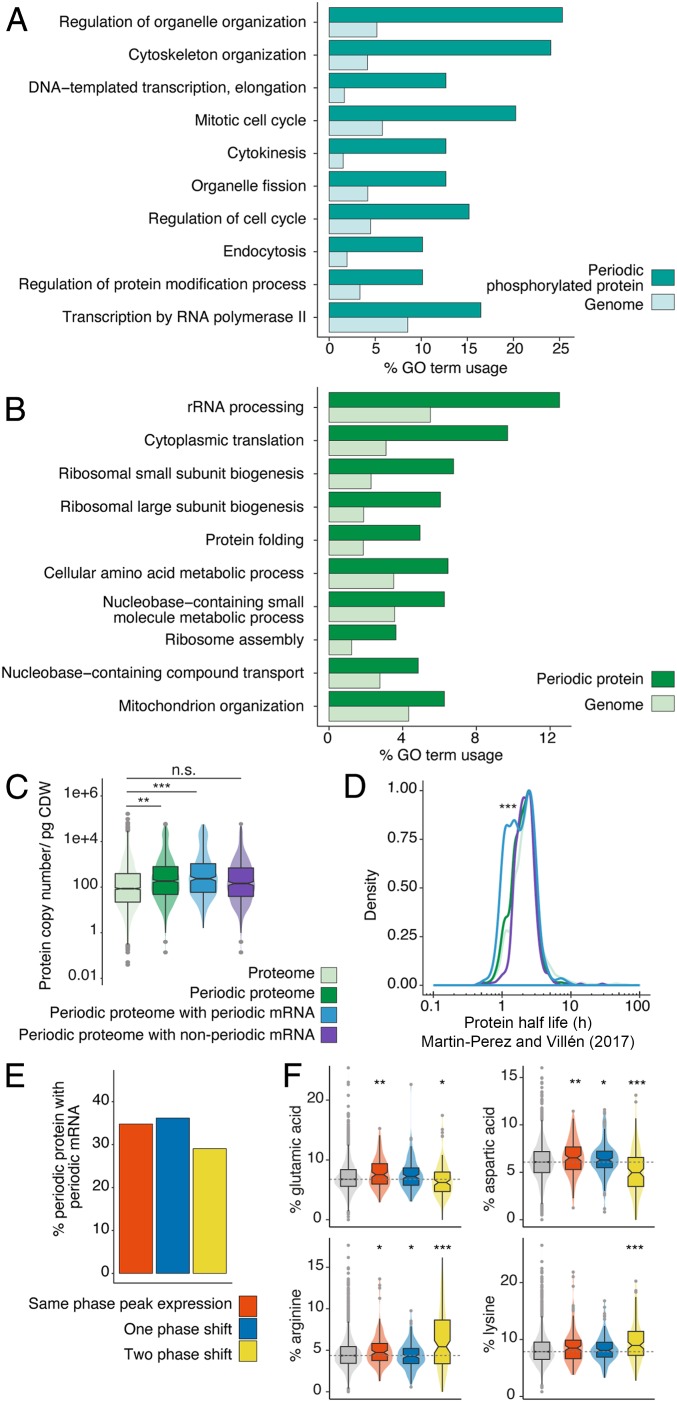
After our initial analysis of periodic genes, we next analyzed which biological processes were enriched at each omic level (Fig. 2 and SI Appendix, Fig. S6). Periodic protein phosphorylation affected organelle and cytoskeleton organization as well as several processes related to the cell cycle (mitosis, cytokinesis, and cell cycle regulation; Fig. 2A). The periodic proteome, however, showed enrichments for processes mainly related to biosynthetic processes, principally translation (rRNA processing and small and large ribosome subunit biogenesis) alongside several metabolic processes related to amino acid and nucleobase metabolism (Fig. 2B). Protein synthesis-related processes were also enriched in the periodic transcriptome (SI Appendix, Fig. S6) alongside processes principally related to the cell cycle (mitosis, cell cycle regulation, DNA replication, and chromosome segregation), with the absence of an enrichment for these processes at the periodic proteome possibly being due to their lower relative detection by MS (SI Appendix, Fig. S6).
Fig. 2.
The periodic multiome reflects the demand of protein synthesis during division. (A and B) Biological process GO terms ranked by enrichment in the periodic (A) phosphoproteome and (B) proteome relative to the entire genome (all GO terms were mapped via Saccharomyces Genome Database GO Slim Term Mapper function [https://www.yeastgenome.org/goSlimMapper]). (C) Median abundance of proteins with or without periodic abundance, and, for periodic protein, those which also have or do not have periodic mRNA abundance. P values: *** ≤ 0.001; ** ≤ 0.01; n.s., not significant. pg CDW, picogram of cell dry weight. (D) Densities for the distribution of turnover values for protein groups in C, using half-life values from Martin-Perez and Villén (22), with density estimate scaled to a maximum of 1. P value: *** ≤ 0.001. (E) Percentage of periodic protein which has periodic mRNA binned according to the number of phase shifts between its expression peak and that of its mRNA. (F) Percentage of negative (Glu and Asp) and positive (Arg and Lys) amino acids in each protein for protein groups in E based on protein sequences taken from UniProt (https://www.uniprot.org). Percentage abundance is also shown in gray for all S. cerevisiae protein sequences available in UniProt (n = 6,721). Median percentage of amino acid abundances (Glu, Asp, Arg, or Lys) across all proteins is also highlighted by a horizontal dashed line. P values: *** ≤ 0.001; ** ≤ 0.01; * ≤ 0.05.
Of the 79 proteins found to be periodic in their phosphorylation state during the cell cycle (across 134 sites), we analyzed how this correlated with their protein abundance (SI Appendix, Fig. S7). Here, most proteins that had multiple sites phosphorylated tended to correlate either only negatively or only positively, with only three proteins having both types of correlation between protein and phosphorylation abundance (SSD1, PAH1, and YLR257W). As almost half of all proteins negatively correlated in abundance with their protein phosphorylation state (46%, 36/79), this indicated a possible role of phosphorylation in degradation (SI Appendix, Fig. S7). Protein phosphorylation, as well as altering activity and localization of a protein, often serves as a marker to trigger ubiquitination, leading to eventual protein degradation (20). We therefore mined data from a previous study by Swaney et al. (21) that isolated ubiquitinated protein that had also been phosphorylated. Of the proteins that positively correlated in abundance with their phosphorylation state, 17% were previously identified as having cooccurring ubiquitination and phosphorylation (8/46), whereas, for proteins that negatively correlated with their phosphorylation abundance, 31% were known to have cooccurring ubiquitination (11/36). These results suggest that one function of phosphorylation in the cell cycle may include marking periodic protein for degradation.
We next explored the differences in protein abundance between protein groups that have periodic mRNA and those that do not (Fig. 2C). Of the total periodic proteome, 44% had periodic mRNA (434/991). However, when the total abundance of these protein groups was considered, this group of transcriptionally regulated proteins comprised >60% of the total periodic proteome. This would suggest that the majority of cell cycle-affected proteins, by abundance, are transcriptionally regulated. Using recently published protein half-life data (22), transcriptionally regulated periodic protein was also found to have a significantly (P ≤ 0.001) higher number of shorter-living proteins relative to the other protein groups analyzed (Fig. 2D). These results suggest that proteins that cycle in abundance are mostly involved in translation and metabolism, transcriptionally regulated, and shorter living.
On analysis of the timing of expression for transcriptionally regulated protein, we find most protein either to have peak expression at the same time as the mRNA it was translated from (for example, both having peak expression in G1) or to be within one phase of separation (Fig. 2E). Recently, Riba et al. (23) found that the rate of protein translation can be impacted by amino acid composition of synthesized proteins as much as codon and transfer RNA adaptation. In particular, they show that negatively charged proteins are translated faster than positively charged protein. To investigate whether or not the amino acid composition of our protein groups may explain the difference in timing for peak expression between periodic mRNA and its respective protein, we analyzed the percentage of both negative and positive amino acids in these three protein groups that had either the same phase for peak expression of mRNA and protein, a one-phase shift, or a two-phase shift (Fig. 2F). Using all S. cerevisiae protein sequences in UniProt as a control group, we find that proteins that are expressed in the same phase as their mRNA have a significantly higher proportion of negative amino acids, glutamic acid, and aspartic acid, and a significantly lower proportion of these amino acids in proteins with peak expression two phases after their mRNA’s phase for peak abundance. Moreover, we find that this protein group, with a two-phase delay between peak mRNA and peak protein expression, contains a significantly higher proportion of positive amino acids arginine and lysine (Fig. 2F). Overall, these results suggest protein abundance during the cell cycle may be broadly regulated by the proteins’ transcript abundance, while their expression timing is fine-tuned by additional properties of the proteins themselves, in particular, their amino acid composition.
Biosynthetic Processes Act in Synchrony with Cell Cycle Phases.
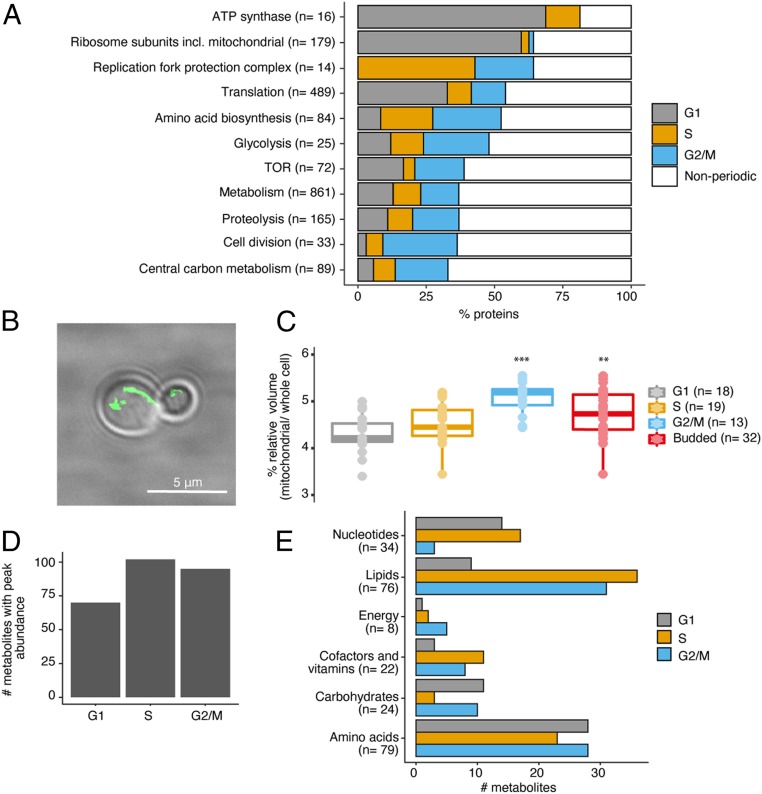
We next analyzed how periodic proteins peaked in abundance in different phases of the cell cycle, to determine when processes synchronized with cell cycle progression (Fig. 3A). Here, we found that, for biosynthetic processes related to protein synthesis, including energy generation via adenosine 5′-triphosphate (ATP) synthase, translation, and ribosome subunit biogenesis, there was a peak in activity in G1, the growth phase of the cell cycle (Fig. 3A). Absolute quantitation of these complexes revealed ribosome subunit and ATP synthase subunit proteins to both increase by ∼8% in abundance in this phase (SI Appendix, Fig. S8). S-phase proteins which peaked in activity included the replication fork protection complex, which is involved in coordinating leading- and lagging-strand DNA synthesis and in replication checkpoint signaling, while the majority of metabolic processes had peak activity at the end of the cell cycle in G2/M (Fig. 3A). For several metabolic processes, activity was spread across more than one phase, such as amino acid biosynthesis and TOR-related proteins which had peak expression mainly in S and G2/M (amino acids), and G1 and G2/M (TOR). The relatively low activity in S phase for TOR would suggest that this kinase regulates growth and metabolism, instead, during G1 when there is peak protein synthesis and during G2/M when there is peak protein activity in central carbon metabolism, respectively.
Fig. 3.
Biosynthetic processes act in synchrony with cell cycle phases. (A) Examples of biological processes with periodic protein that synchronize in peak abundance based on cell cycle phase. (B) Fluorescent-labeled mitochondrial network, using MitoLoc mitochondrial marker. (Scale bar: 5 μm.) (C) Mitochondrial volume relative to whole cell volume during each cell cycle phase. P values: *** ≤ 0.001; ** ≤ 0.01. (D) Number of metabolites with peak average abundance in each cell cycle phase (averages are based on replicates in G1, n = 12; S phase, n = 9; G2/M, n = 9). (E) Same as D with metabolites binned by superpathway. Annotation by Metabolon, Inc. was used as a reference for superpathway classification.
To cross-verify whether metabolic activity was synchronizing with a particular phase of the cell cycle, we performed an independent analysis on the mitochondria, the major metabolic and energetic hub in the cell (24). This investigation was also driven by our GO term enrichment results for periodic protein, which showed mitochondrial organization to be affected by the cell cycle (Fig. 2B). Using a genomically integrated fluorescent marker which localizes to the mitochondria, MitoLoc, and absolute quantitative fluorescence microscopy (Fig. 3B), we found that mitochondrial volume, relative to the whole cell, significantly increased (P < 0.01) as cells committed to the cell cycle (Fig. 3C). This relative increase in mitochondrial volume between budded and nonbudded cells, by ∼12%, suggests cells indeed are elevating their metabolic activity to sustain cell biosynthesis.
We next analyzed whether or not this increase in metabolic activity could also be seen at the metabolite level. To achieve this, we grouped the metabolites we had measured (n = 267) into cell cycle phase groups depending on which phase had the highest abundance of each metabolite (for example, if the maximum abundance of a metabolite occurred in G1, this phase would increase the number of metabolites it had in its group by one). This revealed that a higher proportion of metabolites peak in expression after cells committed to dividing in S and G2/M phases (Fig. 3D), reflecting what we had observed for protein activity (Fig. 3A) and mitochondrial dynamics (Fig. 3C). On inspection of how metabolites peaked in each cell cycle phase, this was shown to depend on what superpathway they belonged to (Fig. 3E). Nucleotides peaked in their synthesis in G1 and S phase, while lipids increased in S and G2/M, mirroring results for cofactors and vitamins. For carbohydrates and amino acids, their biosynthesis increased in G1 and G2/M with a relative drop in S phase (Fig. 3E). This strongly suggests that different metabolic precursors are generated by the cell, depending on the type of biosynthesis that is in demand during each respective phase of the cell cycle.
Biosynthetic Precursors Are Synthesized on Demand during Cell Division.
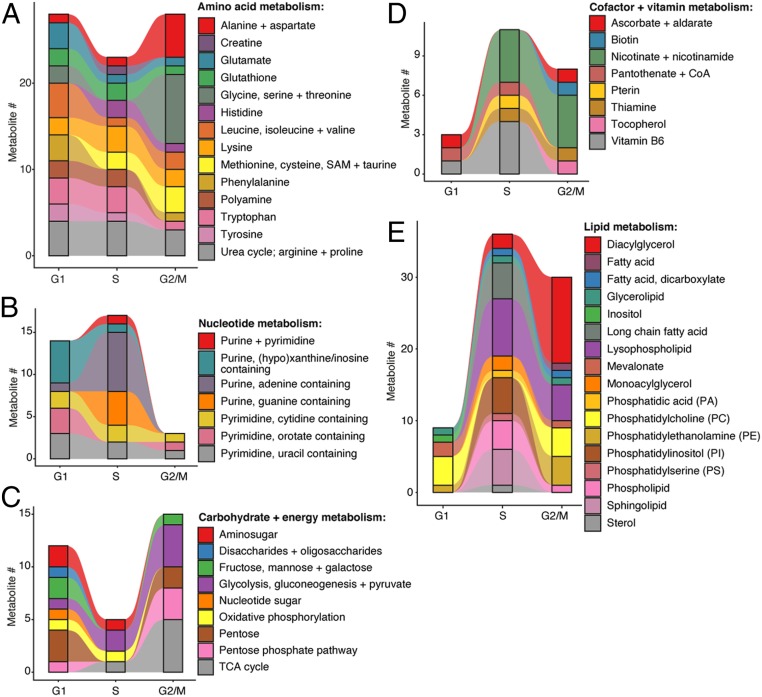
To understand further how metabolism responds to the biosynthetic demands of cell division, we used our metabolite data to dissect which specific pathways peaked in abundance in the different cell cycle phases (Fig. 4). On analysis of metabolites belonging to amino acid (Fig. 4A), nucleotide (Fig. 4B), energy (Fig. 4C), cofactor (Fig. 4D), and lipid metabolism (Fig. 4E), it was clear that some pathways demonstrated different trends to others within the same superpathway. For example, within the superpathway of amino acid metabolism, biosynthetic precursors of phenylalanine metabolism increased in G1, while methionine and cysteine precursors increased in S and G2/M (Fig. 4A). For nucleotides, adenine containing purine metabolites increased in S phase, while inosine-containing pyrimidine metabolites were mostly generated in G1 (Fig. 4B). For cofactors and vitamins, vitamin B6 metabolites were mostly abundant in G1 and S phase, while redox cofactors involved in nicotinate and nicotinamide metabolism had peak abundances in S phase and G2/M (Fig. 4D). The latter result supports the theory that DNA is highly sensitive to redox changes during replication in S phase, subsequently necessitating a relative increase in the supply of redox cofactors. Finally, for lipid metabolism, most pathways were shown to peak in S phase, including long-chain fatty acids, and precursors of lysophospholipid, phosphatidylinositols, phospholipid, and sphingolipid metabolism. However, for some pathways, this was true, instead, for G2/M, including metabolites related to diacylglycerol, phosphatidylcholine, and phosphatidylethanolamine metabolism (Fig. 4E). Taken together, these results suggest metabolite precursor supply is highly subject to different phase requirements of the cell cycle.
Fig. 4.
Biosynthetic precursors are synthesized on demand during cell division. Frequency of metabolites across five main superpathways: (A) amino acid metabolism, (B) nucleotide metabolism, (C) carbohydrate and energy metabolism, (D) cofactor and vitamin metabolism, and (E) lipid metabolism. Metabolites were binned according to which cell cycle phase they have peak abundance in, and which submetabolic pathway they belong to. Abundance values were calculated by averaging replicate abundance in each cell cycle phase (G1, n = 12; S phase, n = 9; G2/M, n = 9), with the cell cycle phase that contained the maximum abundance value for a given metabolite increasing in metabolite frequency in each bar plot. Annotation by Metabolon, Inc. was used as a reference for superpathway and subpathway classification.
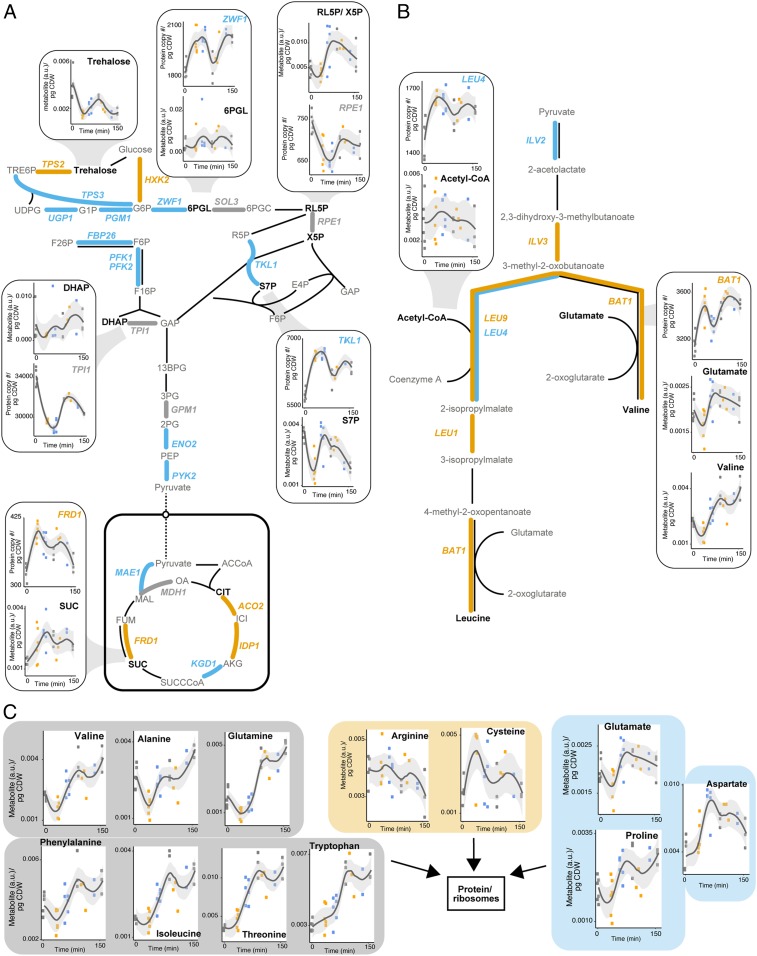
Using both proteomic and metabolomic insight, we next sought to see how these two omic levels synchronized in activity during the cell cycle to see whether any general trends could be observed (Fig. 5). By mapping enzyme abundance onto central carbon metabolic pathways, it was clear they demonstrated differences in activity depending on the cell cycle phase. For example, while glycolysis showed peak enzyme abundance for the majority of periodic proteins in G2/M (Fig. 5A), branched chain amino acid metabolism showed that proteins synchronize their peak abundance in S phase (Fig. 5B). Not all metabolites reflected enzyme abundance changes with, for example, the storage carbohydrate trehalose peaking in G1, whilst associated pathway enzymes mostly showed an increase in S and G2/M (Fig. 5A). Similarly, we found that, for the synthesis of valine, its biosynthetic enzyme, BAT1, increased in S phase, with its substrate glutamate concomitantly decreasing, implying its consumption for fueling valine synthesis; however, nonintuitively, this amino acid was found to increase instead during G1, reflecting that additional enzyme activity may be affecting this amino acid’s abundance. Indeed, G1 phase appears to be the predominant phase for most amino acids to peak in abundance (Fig. 5C). This would suggest that the increase in protein synthesis activity, shown by the periodic proteome GO enrichment for translation-related processes (Fig. 2B), is met by an increase in amino acid supply during this growth phase.
Fig. 5.
Synchronization of central carbon enzyme and metabolite abundance with cell cycle phases. (A) Glycolysis, TCA, and the pentose phosphate pathway with periodic enzyme abundance represented by colored connections between metabolites according to cell cycle phase they demonstrate peak abundance in (G1, gray; S, orange; G2/M, blue). Examples of periodic metabolites with neighboring periodic enzymes are also shown in subpanels. Points reflect individual replicates, with line representing a loess curve fitted to replicate abundance values for each time point, with span for fitting each local regression = 0.6. CI for regression line is 95%. (B) As in A but for the branched chain amino acid biosynthetic pathway. (C) Amino acid abundance grouped according to the cell cycle phase where each amino acid demonstrates peak abundance. a.u., arbitrary units; pg CDW, picogram of cell dry weight.
Discussion
In this study, a data-driven and quantitative multiomic approach was used to characterize the budding yeast cell cycle. Absolute quantitative omic analysis of mRNA and protein abundance has previously been performed by Marguerat et al. (4) for another eukaryal cell cycle model organism, fission yeast (Schizosaccharomyces pombe). Considering their data, several insights can be gained with respect to genes identified as periodic over the cell cycle as well as the overall abundance landscape for mRNA and protein in both of these eukaryotes. In the study by Marguerat et al., for example, they highlight several genes (H2Aα, H2Aβ, H2B, H3.1, H3.2, H3.4, H4.1, H4.2, H4.3, H2A.Z, mik1, mde6, and mei2) that they find to contain a switch-like pattern in transcription, proposing these genes to be confined to a specific cell cycle phase, as well as suggesting they may have deleterious effects if expressed at the wrong time (4). On analysis of the S. cerevisiae orthologs for these genes, we find their mRNA abundance to also be periodic (SI Appendix, Fig. S9). Eight of the histone gene orthologs (HTA2, HTB1, HTB2, HHT1, HHT2, HHF1, HHF2, and HTZ1) have significant (P < 0.05) mRNA expression in S phase, with SWE1, the ortholog of S. pombe’s mik1, and an inhibitor of mitosis, shown to have peak expression in G1 (SI Appendix, Fig. S9). Therefore, despite these two yeast models having an evolutionary distance of ∼350 million years, conservation in periodic behavior for these related genes is still shown to exist (25).
Our quantitative measurements also confirm that protein abundance greatly exceeds mRNA in number and dynamic range. On average, we find the total proteome to be >2,500 times more abundant than mRNA, a similar estimate as found for fission yeast (∼1,850 times) (4). Moreover, this amplification step is shown to be determined by protein function, with proteins involved in translation and metabolism being disproportionately amplified, leading to them representing the majority of the proteome.
Correlations between mRNA and protein copy number in this work additionally add to the growing consensus that protein levels are, to a large extent, reflected by their mRNA abundance (3, 4, 26, 27). In our analysis of cell cycle-related changes in mRNA and protein abundance, the similarities between the periodic proteome and periodic transcriptome also highlight that most protein changes in abundance are transcriptionally regulated. Moreover, we find properties of the protein themselves to impact their abundance changes over time. Amino acid composition of periodic proteins, in particular, amino acid charge, is shown to have a possible role in regulating the timing of peak protein expression, with proteins that have relatively more positive amino acids being translated slower than those which are relatively more negative.
For proteins which demonstrate an absence of periodic activity, this does not necessarily imply these are not active during the cell cycle. Our phosphoproteomic data, indeed, show that several processes, including those directly involved in the cell cycle, as well as other processes, such as cytoskeleton and organelle organization, change in their protein phosphorylation state in synchrony with cell division, changes which were less clear at the transcriptome or proteome. Using both proteomic and phosphoproteomic datasets together, it was also possible to identify a possible role of phosphorylation in degradation, when proteins and their phosphorylation states are anticorrelated. Additional posttranslational data would no doubt shed further light on how different processes are regulated in this way.
Proteomic data also unveiled an increase in protein synthesis activity before cell division, in G1, which could be supported by the increase in amino acids that was observed in the same phase. Litsios et al. (28), using microfluidics and time-lapse microscopy in single cells of S. cerevisiae, found that cells similarly demonstrate a pulse in protein production rate during G1. We also find that central carbon metabolism, as shown by an increase in protein abundance, increased after cell division was committed to, most notably with glycolytic pathway enzymes increasing in relative abundance in G2/M. Both metabolite abundance data and morphological changes in the mitochondria similarly highlight that metabolic activity increases as cells begin to replicate. In particular, phase-dependent differences in biosynthetic precursors, seen both at the enzyme level and, less intuitively, at the metabolite level, highlight a complex coordination between metabolism and the cell cycle (2, 7).
Recent work by Blank et al. (10), using an alternative approach of elutriation to study the yeast cell cycle, also found notable dynamic changes in metabolism through changes in enzyme abundance. Despite different experimental approaches used, several findings are shown to be reflected in both our studies. For example, Blank et al. find that ergosterol enzymes peak during mitosis, with our results similarly finding that several ERG genes peak in G2/M, with mRNA and protein increasing in this phase for ERG27, ERG12, and ERG7. We see that ergosterol itself peaks in S phase, again demonstrating a nonintuitive association between enzyme and metabolite abundance. Similar to their study, we also find that thiamine diphosphate-dependent enzymes peak in G2/M (in our case, TKL1 alongside KGD1), both reflecting a general peak in activity for glycolysis and the tricarboxylic acid cycle (TCA) after cell cycle commitment, that is demonstrated at both the enzyme and metabolite levels for these pathways.
Finally, using our list of genes identified as being periodic in their expression products, we show that almost half of human orthologs in yeast are periodic, and that almost all of the genes in this subset had an essential function. One-third of these genes contained a metabolic function, suggesting these could be targeted for modulating cell cycle progression, with relevance for human diseases such as cancer.
Materials and Methods
All details on materials and methods associated with omics sample collection, measurement, and analysis can be found in SI Appendix, SI Materials and Methods. This includes cell culture synchronization conditions, transcriptomic, proteomic and metabolomic workflows, the limma modeling method, omics visualization information, intergene distance analysis, and mitochondrial volume quantification by fluorescence microscopy.
Data and Materials Availability.
Correspondence and requests for materials should be addressed to nielsenj@chalmers.se. All omic information may be viewed interactively at https://www.sysbio.se/tools/cellcycle/ with instructions for use available in SI Appendix, SI Materials and Methods. RNA-seq data have been deposited in the ArrayExpress database (29) at EMBL-EBI (https://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-8565. The mass spectrometry proteomics and phosphoproteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (30) partner repository (https://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD016519.
Supplementary Material
Acknowledgments
We thank Rui Pereira and Xin Chen (Chalmers University of Technology, Sweden) for their support with sample collection, Jianye Xia (East China University of Science and Technology, China) for donation of heavy lysine-labeled yeast, Anna Koza (Technical University of Denmark, Denmark) for help with RNA sequencing, and Rudy van Eijsden and Erroll Rueckert (NanoString Technologies) and Annika Eriksson and Malin Alvehus (Karolinska University Hospital, Sweden) for their technical assistance with absolute mRNA quantification. Thanks also go to Rosemary Yu, Eduard Kerkhoven, and Quanli Liu (Chalmers University of Technology, Sweden) for providing feedback during manuscript preparation. This project has received funding from the Novo Nordisk Foundation (Grant NNF10CC1016517) and the Knut and Alice Wallenberg Foundation (J.N.). J.W. is financially supported by the Knut and Alice Wallenberg Foundation as part of the National Bioinformatics Infrastructure Sweden at SciLifeLab.
Footnotes
The authors declare no competing interest.
Data deposition: RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (https://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-8565. The mass spectrometry proteomics and phosphoproteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD016519. All omic information from Table S1 may be viewed interactively in SysBio at https://www.sysbio.se/tools/cellcycle/.
See QnAs on page 7548.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919535117/-/DCSupplemental.
References
- 1.Hartwell L. H., Culotti J., Pringle J. R., Reid B. J., Genetic control of the cell division cycle in yeast. Science 183, 46–51 (1974). [DOI] [PubMed] [Google Scholar]
- 2.Ewald J. C., Kuehne A., Zamboni N., Skotheim J. M., The yeast cyclin-dependent kinase routes carbon fluxes to fuel cell cycle progression. Mol. Cell 62, 532–545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y., Beyer A., Aebersold R., On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Marguerat S., et al. , Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151, 671–683 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flory M. R., et al. , Quantitative proteomic analysis of the budding yeast cell cycle using acid-cleavable isotope-coded affinity tag reagents. Proteomics 6, 6146–6157 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Carpy A., et al. , Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (Fission yeast). Mol. Cell. Proteomics 13, 1925–1936 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L., et al. , Multiple layers of phospho-regulation coordinate metabolism and the cell cycle in budding yeast. Front. Cell Dev. Biol. 7, 338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spellman P. T., et al. , Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9, 3273–3297 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho R. J., et al. , A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2, 65–73 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Blank H. M., et al. , Abundances of transcripts, proteins, and metabolites in the cell cycle of budding yeast reveals coordinate control of lipid metabolism. bioRxiv:2019.12.17.880252 (18 December 2019). [DOI] [PMC free article] [PubMed]
- 11.Futschik M. E., Herzel H., Are we overestimating the number of cell-cycling genes? The impact of background models on time-series analysis. Bioinformatics 24, 1063–1069 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Ahdesmäki M., Lähdesmäki H., Gracey A., Shmulevich L., Yli-Harja O., Robust regression for periodicity detection in non-uniformly sampled time-course gene expression data. BMC Bioinf. 8, 233 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lichtenberg U., et al. , Comparison of computational methods for the identification of cell cycle-regulated genes. Bioinformatics 21, 1164–1171 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Kelliher C. M., et al. , Layers of regulation of cell-cycle gene expression in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 29, 2644–2655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battich N., Stoeger T., Pelkmans L., Control of transcript variability in single mammalian cells. Cell 163, 1596–1610 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Bahar Halpern K., et al. , Nuclear retention of mRNA in mammalian tissues. Cell Rep. 13, 2653–2662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kristell C., et al. , Nitrogen depletion in the fission yeast Schizosaccharomyces pombe causes nucleosome loss in both promoters and coding regions of activated genes. Genome Res. 20, 361–371 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu G., et al. , Gene essentiality is a quantitative property linked to cellular evolvability. Cell 163, 1388–1399 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Kachroo A. H., et al. , Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348, 921–925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen L. K., Kolch W., Kholodenko B. N., When ubiquitination meets phosphorylation: A systems biology perspective of EGFR/MAPK signalling. Cell Commun. Signal. 11, 52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swaney D. L., et al. , Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 10, 676–682 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Perez M., Villén J., Determinants and regulation of protein turnover in yeast. Cell Syst. 5, 283–294.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riba A., et al. , Protein synthesis rates and ribosome occupancies reveal determinants of translation elongation rates. Proc. Natl. Acad. Sci. U.S.A. 116, 15023–15032 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spinelli J. B., Haigis M. C., The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 20, 745–754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman C. S., Wood V., Fantes P. A., An ancient yeast for young geneticists: A primer on the Schizosaccharomyces pombe model system. Genetics 201, 403–423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blevins W. R., et al. , Extensive post-transcriptional buffering of gene expression in the response to severe oxidative stress in baker’s yeast. Sci. Rep. 9, 11005 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee M. V., et al. , A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 7, 514 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litsios A., et al. , Differential scaling between G1 protein production and cell size dynamics promotes commitment to the cell division cycle in budding yeast. Nat. Cell Biol. 21, 1382–1392 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Athar A., et al. , ArrayExpress update—From bulk to single-cell expression data. Nucleic Acids Res. 47, D711–D715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.