Significance
The human brain is organized into complex networks whose interaction explains individual variability in cognitive abilities as well as symptoms of neurological and psychiatric disorders. Even though networks’ activity is canonically measured by recording spontaneous brain activity, recent evidence suggests individual differences in brain patterns are better captured in response to external perturbations. Here we used individualized brain stimulation to induce controlled perturbations in two brain networks, and observed the propagation of neural activity across local and distal regions with millisecond resolution. We show unique propagation patterns across stimulated networks, predicting individual differences in cognitive performance otherwise not observable through canonical resting-state recordings. Results promote perturbation-based neuroimaging for the identification of novel biomarkers of cognition as well as clinical conditions.
Keywords: TMS-EEG, fMRI, resting-state networks, cognition
Abstract
Large-scale brain networks are often described using resting-state functional magnetic resonance imaging (fMRI). However, the blood oxygenation level-dependent (BOLD) signal provides an indirect measure of neuronal firing and reflects slow-evolving hemodynamic activity that fails to capture the faster timescale of normal physiological function. Here we used fMRI-guided transcranial magnetic stimulation (TMS) and simultaneous electroencephalography (EEG) to characterize individual brain dynamics within discrete brain networks at high temporal resolution. TMS was used to induce controlled perturbations to individually defined nodes of the default mode network (DMN) and the dorsal attention network (DAN). Source-level EEG propagation patterns were network-specific and highly reproducible across sessions 1 month apart. Additionally, individual differences in high-order cognitive abilities were significantly correlated with the specificity of TMS propagation patterns across DAN and DMN, but not with resting-state EEG dynamics. Findings illustrate the potential of TMS-EEG perturbation-based biomarkers to characterize network-level individual brain dynamics at high temporal resolution, and potentially provide further insight on their behavioral significance.
The characterization of the functional role of spontaneous brain activity in cognition and behavior has led to a fundamental paradigm shift in the study of human brain function. Numerous functional MRI (fMRI) studies have revealed that spatially distant brain regions exhibit temporally coherent fluctuations in their spontaneous blood oxygenation level-dependent (BOLD) signal, and form a distinct set of functional networks in the resting brain (resting-state networks [RSNs]). These RSNs closely overlap with networks activated during task performance, providing a backbone for information transfer even in the absence of external stimuli. RSNs (1–4) are believed to reflect the underlying functional architecture of both healthy and abnormal brain activity (5–7). Thus, a growing body of clinical and translational neuroscience research has focused on fMRI-based connectivity dynamics within and across distributed brain networks as a promising approach for the diagnosis—as well as the identification of treatment targets—of various neurological and psychiatric disorders (8).
However, after two decades of research, our understanding of the functional relevance of large-scale distributed brain networks is still limited. The BOLD signal provides an indirect measure of neuronal activity, reflecting slow-evolving hemodynamic responses that typically fail to capture the rich temporal dynamics of ongoing oscillations across RSNs at the timescale of normal physiological function (9, 10). Furthermore, fMRI measures of resting-state connectivity identify bivariate correlations in brain activity, but the causal significance of identified network relationships remains unclear (11, 12).
The combination of transcranial magnetic stimulation (TMS) with simultaneous electroencephalography (TMS-EEG) may help address these limitations. TMS provides a unique method to perturb individual nodes of RSNs and examine both local and distributed network-level responses, characterizing so-called perturbation-based connectome. Unlike task-evoked brain activations, which likely involve various intermediate cortical/subcortical processes and are highly dependent on subjects’ attendance to/cognitive strategy for a given task, TMS can directly perturb specific cortical regions with good spatial resolution while requiring minimal involvement from a subject/patient. Simultaneous EEG recordings enable assessment of TMS-evoked effects and their propagation across functionally connected cortical regions (13, 14). Accumulating evidence supports the validity of such TMS-evoked potentials (TEPs) in characterizing the healthy and pathological brain (15–17). Furthermore, TMS-EEG might uncover subtle alterations in brain electrophysiology that are not apparent with more conventional resting-state EEG analyses. For example, TMS-EEG has been able to identify abnormalities in cerebral reactivity even when conventional scalp EEG features fail to reveal disease specific alterations, distinguishing patients who are minimally conscious from those in persistent vegetative state (18). However, evidence is lacking about the possibility of using such TMS-EEG measures to index network-level brain dynamics. A growing body of literature suggests the importance of looking at alterations of brain networks as well as network-to-network interactions as potential biomarkers of both pathological states and cognitive function (6, 18). For instance, alteration of the same RSNs (i.e., default mode network [DMN] and anterior salience network [ASN]) has been documented in both Alzheimer’s disease and frontotemporal dementia patients using fMRI; however, while the former group of patients shows increased DMN−AS functional connectivity, the latter display the opposite pattern, even though both conditions shared a significant neuropathological substrate (19). Whether TMS-EEG can index such finely grained network-level dynamics remains an open question, as well as the possibility for such technique to significantly increase the resolution of canonical fMRI/EEG-based biomarkers.
Here we use fMRI-guided TMS-EEG (see Fig. 1 A and B for details of network targeting) to selectively perturb two neighboring parietal lobe nodes of the DMN or the dorsal attention network (DAN) (Fig. 1B), which are thought to underlie internally and externally directed attention processes, respectively. In fMRI, the temporal relationship between these networks is characterized by a negative correlation pattern that is dynamically modulated between the “resting” and “task-related” brain states (20). Specifically, the “task-negative” DMN remains relatively silent when attention is paid to an external stimulus or a task, but exhibits increased activity during unconstrained resting state. In contrast, the “task-positive” DAN shows an opposite activation pattern, with increased activity during attention-demanding tasks but decreased activity at rest (1). Furthermore, disruptions or abnormalities in the functional connectivity pattern between these two networks have been associated with individual variability in several cognitive functions, motor behaviors, and symptomatology of various neurological and psychiatric disorders (21–25). However, because of the correlational nature of fMRI-based approaches, it remains unclear whether the observed reciprocal spatial−temporal dynamics of these two networks is a causal phenomenon. By employing a noninvasive and individualized network perturbation approach along with simultaneous whole-brain neuroimaging using EEG, the reciprocal dynamics between these two functionally opposing networks can be characterized with high temporal resolution. This also provides an opportunity to study the behavioral significance of such fast-evolving network dynamics.
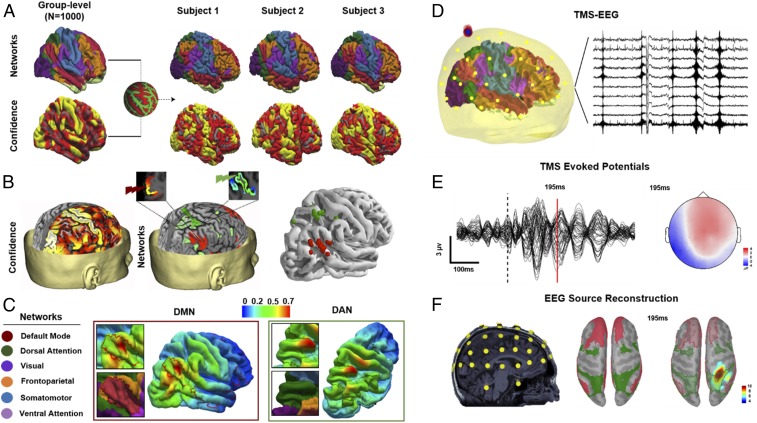
Fig. 1.
Details of individual MRI-guided TMS target selection. (A) Group-averaged functional cortical atlases (n = 1,000) consisting of seven networks (85) were first projected onto individualized cortical surface based on confidence map weights for each network (individual variability in projected confidence maps for three representative subjects is shown). (B) Individualized target coordinates were then determined based on voxels (Left) with the highest confidence in the angular gyrus and superior parietal gyrus for stimulation of the DMN and DAN, respectively (Middle). TMS was performed targeting the DMN (red) and DAN (green), based on confidence maps of RSNs warped onto individual MRI space (Right). (C) The E-field maps were normalized by the maximum E field for each stimulation and for each subject, and then mapped to the fsaverage common template and averaged across subjects for the purpose of visualization. (D) EEG data were also collected simultaneously to TMS. (E) Representative TEPs of all EEG channels after preprocessing, with the topography of sample TEP peak corresponding to brain activity 195 ms after TMS. (F) For source reconstruction of TEPs, digitized EEG channel locations along with anatomical landmarks were registered onto individual MRI space (Left). Forward and inverse modeling of EEG sources were then computed using symmetric boundary element method for forward modeling and minimum norm estimate MNE method for inverse modeling. Resulting EEG source activations were projected onto individual surface space. Confidence maps of DAN (green) and DMN (red) were then projected onto individual surface space (Middle), and used as ROIs to extract baseline normalized (pre-TMS) TMS-EEG source activations (Right).
We hypothesized that TMS of DMN or DAN would reveal distinct spatial−temporal dynamics as measured by source-reconstructed high-density EEG data, and that source-level EEG responses would reflect the spontaneous negative connectivity characterizing DMN and DAN dynamics measured via fMRI (see Fig. 1 D–F for details of source-reconstructed EEG). Moreover, we aimed to document the reproducibility of such network-specific TMS-evoked EEG responses, at both the electrode and source levels, by conducting test−retest visits over a 1-mo period. Finally, given the specific positive association between DMN/DAN negative connectivity and cognition (in particular, fluid intelligence and abstract reasoning) (26), we hypothesized that network-specific TMS-evoked cortical responses would be related to individual variability in cognitive profile, and that the magnitude of DMN/DAN “dissociation” after TMS would be positively correlated with a higher performance on cognitive measures of intelligence.
Results
TMS-Induced Cortical Activations Reveal Network-Specific Spatial−Temporal Dynamics.
The spatial distribution of normalized source activation patterns following DAN and DMN stimulation for a representative subject is provided in Figs. 2 and 3 (see EEG Source Reconstruction for details of EEG source analyses). Following TMS to the parietal node of the DAN, EEG source reconstructions predominantly projected around the superior parietal gyri of both right and left hemispheres, highly overlapping with the parietal topography of DAN (Fig. 2). On the other hand, TMS to the DMN resulted in EEG source reconstructions projected over the frontal midline and parietal regions (precuneus) of the DMN (Fig. 3). Increased activity in the stimulated networks was not just localized to the site of stimulation but propagated specifically into functionally connected distal regions of the same network, revealing that TMS induces network-specific activations as a function of the probed cortical nodes (see Movies S1 and S2 for network activations of DMN and DAN, respectively).
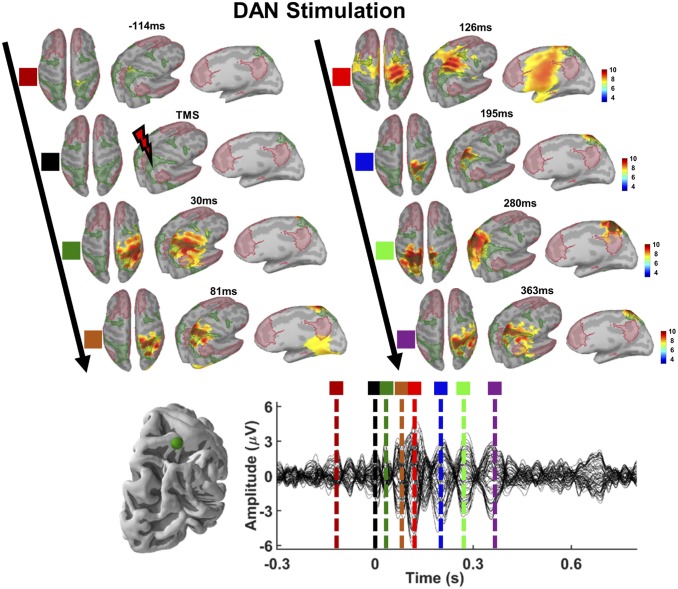
Fig. 2.
Source activation patterns following TMS of DAN. Network maps (DAN, green-shaded areas; DMN, red-shaded areas) and spatiotemporal dynamics of reconstructed EEG sources for a selected time point before, during, and at multiple TEP peaks following TMS are shown (Upper). The corresponding TEP peaks (colored vertical dotted lines) of source activations are shown (Lower). Note that source reconstruction projects locally on the superior parietal gyrus of right hemisphere for earlier TEP peaks (see source activations for 30 ms and 81 ms following TMS), while source reconstruction of a later TEP (280 ms following TMS) maps on the superior parietal gyrus of left hemisphere, suggesting that TMS-induced cortical activations predominantly propagate within the stimulated network.
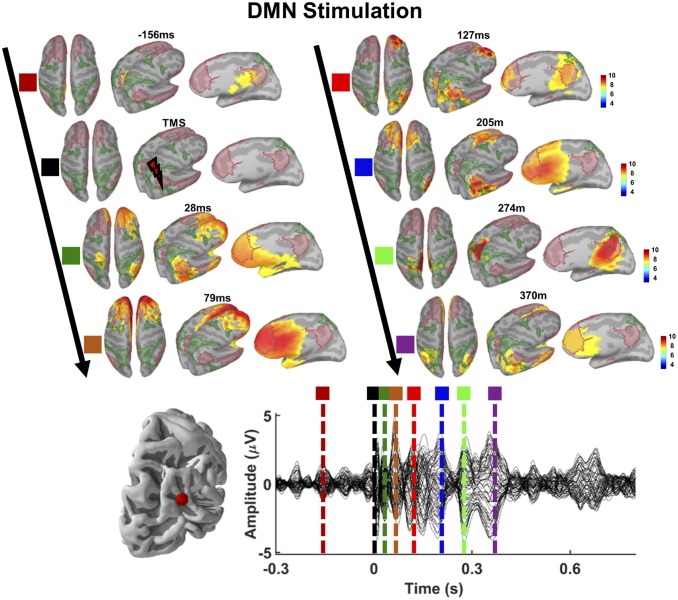
Fig. 3.
Source activation patterns following TMS of DMN. EEG source reconstruction projects over the DMN map, with early TEP peaks localizing around right angular gyrus and ventral medial prefrontal cortex (see source activations for 28 ms), and later TEP peaks localizing around left medial frontal cortex and precuneus (see source activations for 205 ms and 274 ms).
We first performed time−frequency decomposition of average current densities to describe general oscillatory characteristics of each network following stimulation of DAN and DMN. Time−frequency responses and propagation of network activations and group-level statistics are provided in Fig. 4 (see EEG Source Metrics for details). We show that early activations (30 ms to 150 ms) following TMS have broadband oscillation characteristics ranging from theta (4 Hz) to low gamma (∼30 Hz) bands, but significant modulation of evoked responses appears to be primarily in the beta-band (13 Hz to 30 Hz) as a function of the stimulation site (Fig. 4A). Specifically, while beta-band responses are present in both networks across stimulation conditions, significant beta-oscillations are only observed within the stimulated network, suggesting that TMS induces prominent network activations at the “natural frequency” of the stimulated network (27). As for temporal propagation of induced network activations, we observed a clear interaction pattern following TMS such that the stimulation of superior parietal gyrus induced higher cortical activation in the DAN map compared to DMN map, while angular gyrus stimulation resulted in higher cortical activations of DMN compared to DAN (P = 0.01). Additionally, to investigate whether individual network topology would significantly affect the observed propagation patterns, we also performed the same analyses using network maps derived from individual seed-based fMRI connectivity analyses, observing a similar significant interaction pattern (P = 0.025) characterized by higher activation for the stimulated network compared to the nonstimulated network across conditions (SI Appendix, Fig. S4).
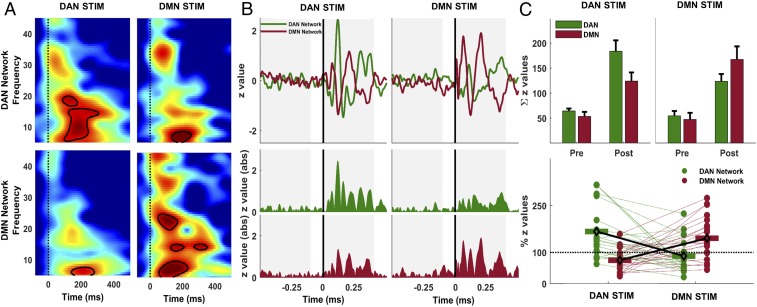
Fig. 4.
Time−frequency responses and propagation of network activations in source space. (A) Time−frequency distribution of ERSPs following stimulation of DAN and DMN in each network. Group-averaged ERSPs for DAN network response are shown following DAN (Upper Left) and DMN (Upper Right) stimulation. DMN responses are shown in the same order (Lower). Vertical dashed lines indicate TMS pulse, and contour lines show significant clusters (P < 0.01) in each panel compared to baseline. (B) Normalized average MNE current density time series in DAN (solid green line) and DMN (solid red line) maps following TMS of DAN and DMN targets are shown (Upper). Colored regions represent rectified current density amplitudes (Lower). Gray shaded regions show 400-ms time windows used to compute area under the curve in each network map for each stimulation. (C) Colored bars in Upper show group-averaged values with SEs in each network map before (Pre) and after (Post) TMS of DAN (green bars) and DMN (red bars). Lower shows individual Post-TMS cortical activation values expressed as the ratio of total cortical activity in the stimulated network relative to the nonstimulated network map. For instance, in the case of DAN stimulation (green dots), individual values show the ratio of z-score sums in the DAN map relative to z-score sums in the DMN map for each subject. Individual trends for the total amount of cortical activity in each network map are shown with colored dotted lines connecting each subject’s ratio scores across stimulation conditions.
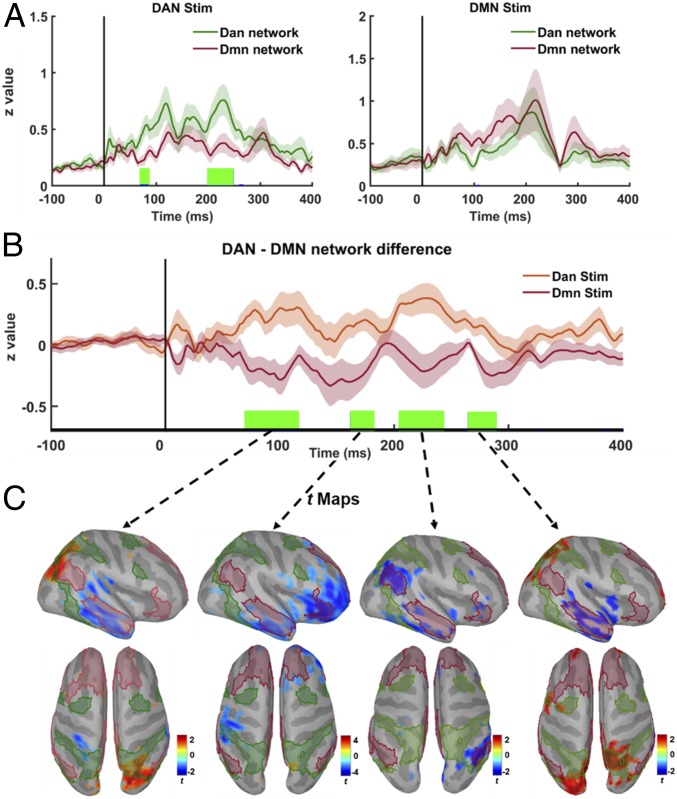
To further examine temporal evolution of network differences and interaction pattern across stimulation conditions, we then computed cluster-based permutation (n = 1,000) paired t test statistics (28) on source activation values, as well as on individual time series representing the difference in activation between DMN and DAN at each time point (see Fig. 5 for details). We found significantly higher activity in DAN around 80 and 200 ms following DAN stimulation. Although DMN time series were higher than DAN time series following DMN stimulation (with significant differences around 100 ms), no time point survived cluster-based permutation correction (Fig. 5 A, Right). As for the DMN−DAN interaction pattern, we computed differences between network-level source activity for all subjects (n = 21) across stimulation conditions, more specifically, by taking DAN as a reference network and subtracting DAN time series from DMN time series for each subject, thus obtaining average network-difference time series. We found significant cluster-based corrected differences around 80- to 120-ms, 160- to 180-ms, 205- to 250-ms, and 270- to 290-ms windows (Fig. 5B). Finally, we computed vertex-wise group average differences to further examine the spatial topography of time windows when significant network differences were observed (Fig. 5C). In the first significant time window around 100 ms, greater activation in the right parietal nodes of the DAN is visible in comparison to activity evoked by DMN stimulation, as well as significantly lower activation of the right inferior temporal node of the DMN. At ∼180 ms, DAN stimulation produces lower activation of both right inferior temporal node and right frontal nodes of the DMN. During the time interval centered around 230 ms, DAN stimulation results in lower activation of the right parietal DMN node. At 280 ms, increased activation in the right parietal DAN node and decreased activation in the right temporal DMN node are observed, altogether demonstrating a spatiotemporal dissociation between DMN and DAN nodes as a function of the stimulation site.
Fig. 5.
Temporal dynamics of TMS-induced network interactions. (A) Comparison of group-averaged time series for each network across stimulation conditions. Solid colored lines show network-level responses for DAN (dark green) and DMN (dark red) averaged across subject and stimulation conditions (DAN: Right; DMN: Left), while shaded regions reflect variation with SE of measurements. Vertical green lines over the x axis show significant time points that survive after cluster-based permutations (P < 0.05). (B) Differences in network-level responses computed by subtracting DAN from DMN current density time series for each subject (n = 21) across stimulation conditions (DAN: orange; DMN: dark red; with shaded regions showing variability in each stimulation condition as SE). Vertical green lines over the x axis show significant time points that survive after cluster-based permutations (P < 0.05). (C) Significant t maps (P < 0.05) averaged over significant time windows in B (green vertical lines) showing differences across the cortex between two stimulation conditions. Higher t values (red) show vertices with significantly higher activity following DAN stimulation, while lower t values (blue) show vertices with significantly higher activity following DMN stimulation.
Network-Targeted TMS Induces Reproducible Cortical Responses.
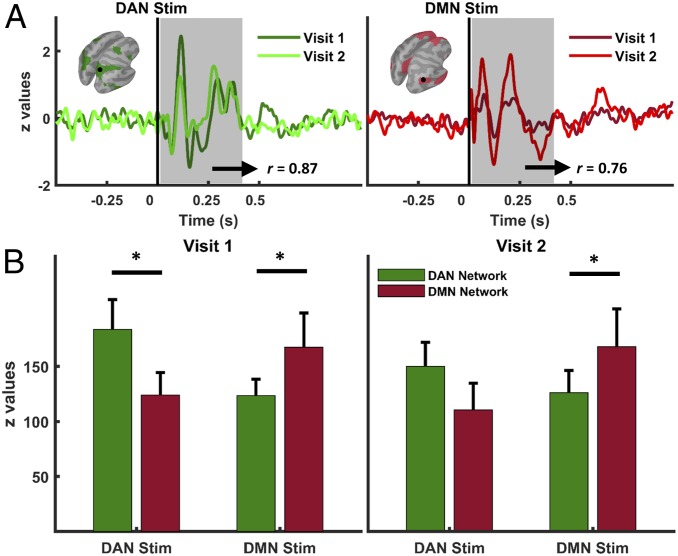
We ran two identical TMS sessions 1 mo apart to test the reproducibility of TMS-induced network activations. We were able to replicate network-specific spatiotemporal dynamics at the source level; source reconstruction results for session 2 revealed a preserved significant interaction pattern (P = 0.02) with increased evoked activity in the stimulated network compared to the nonstimulated one (Fig. 6). Results further confirm that TMS induces network-specific cortical activity dynamics as a function of the probed cortical nodes. No significant effect of time (P > 0.05) was found at the repeated measure ANOVA, indicating that the combination of stimulation conditions remained statistically similar across TMS sessions.
Fig. 6.
Reproducibility of network-level cortical responses to TMS. (A) Normalized average current density time series across TMS visits extracted from DAN (Left) and DMN (Right) maps. (B) Group averages of network activations for each stimulation site (green bars for DAN, and red bars for DMN) are shown for visit 1 (Left) and visit 2 (Right). Asterisks indicate significant differences (DAN Stim visit 1, DAN vs. DMN comparison, *P = 0.02; DMN Stim visit 1, DMN vs. DAN comparison, *P = 0.04; DMN Stim visit 2, DMN vs. DAN comparison, *P = 0.04) between networks after correcting for multiple comparisons (comparisons are shown with horizontal black lines).
Additionally, we ran supplementary correlation analyses in electrode space to further confirm the test−retest reliability of TEPs over the entire scalp. We found highly overlapping scalp topographies within stimulation conditions (i.e., DAN stimulation in visit 1 vs. visit 2) but distinct spatiotemporal dynamics across stimulation conditions (i.e., DAN vs. DMN), corroborating the reproducibility of TMS-induced network-specific activations (see SI Appendix, Results and Fig. S5 for details).
Network Specificity of TMS Activations Is Correlated with Cognitive Performance.
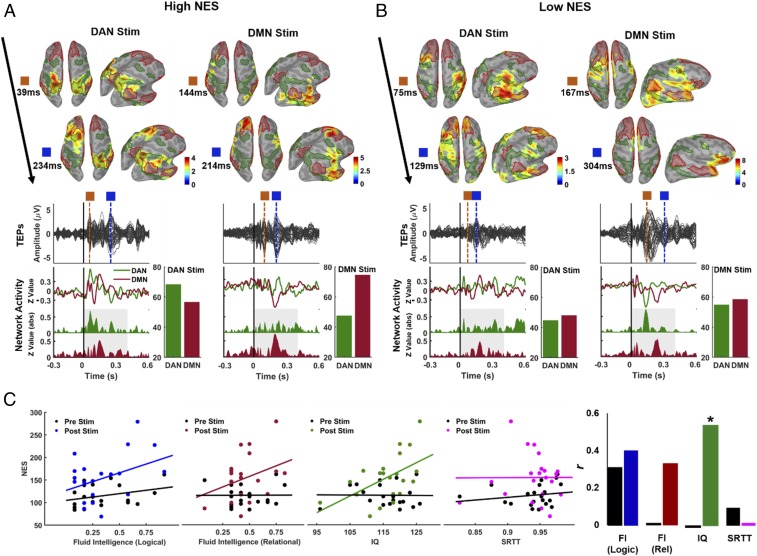
One of our hypotheses was that the magnitude of network dissociation (DMN/DAN) following TMS would be related to individual variability in cognition. We used a network specificity metric (network engagement specificity [NES]), showing the degree of activation patterns within the stimulated network relative to the nonstimulated network, thus quantifying how well TMS-induced activity propagates within the targeted networks (see EEG Source Metrics for details). Given previous reports of an association between cognitive profile and, for example, RSNs dynamics (29), network efficiency (30), and network-to-network interplay (26), our goal was to see whether the specificity of network-level responses to TMS (characterized by high activations in the stimulated networks versus decreased engagement of nonstimulated networks), was related to individual variability in cognition. Higher NES values reflect more specific TMS-induced cortical activity dynamics, possibly capturing brain topological properties related to segregation/integration.
Source activation patterns at selected TEP peaks of representative subjects with high and low NES scores are provided in Fig. 7 A and B, respectively. We found significant and positive correlations between NES values (see Methods for details) and IQ scores (Fig. 7 C, Right, with green-colored scatters: r = 0.53, P = 0.02), showing that individuals with high activity in the stimulated network but relatively low activity in nonstimulated network(s) display a better overall cognitive performance. We also observed positive, but nonsignificant, correlations between logical fluid intelligence scores and NES (Fig. 7 C, Left, with blue-colored scatters: r = 0.4, P = 0.11), and between relational fluid intelligence scores and NES (Fig. 7C, red-colored scatters: r = 0.33, P = 0.17). Notably, no significant correlations were observed between NES and a serial reaction time task (SRTT) (r = 0.01, P = 0.99), suggesting that DMN−DAN dissociation is plausibly more specifically related to high-order cognitive performance than to motor/sensory performance. Additionally, as a control condition, we also computed NES values for cortical activations in the resting-state period (−500 to −100) preceding TMS and examined the correlations between resting-state NES and cognitive performance (Fig. 7C, black-colored scatters). Unlike TMS-induced NES, resting-state EEG NES values were poorly correlated with fluid intelligence (logical: r = 0.29, P = 0.21; relational: r = 0.04, P = 0.80) and IQ scores (r = −0.02, P = 0.84).
Fig. 7.
Relationships between TMS-induced network activation and cognitive performance. (A and B) Spatiotemporal dynamics of reconstructed EEG sources at selected time points (surface images), corresponding TEPs (solid black lines for each EEG channel), current density time series (colored solid lines), and rectified current density amplitudes with the total amount of electrocortical activity (z-score sums) in each network ROI (colored bars) for two representative subjects with (A) a high NES index (see Methods for details) and (B) a low NES index. (C) Scatterplots with regression lines show correlation trends between individual NES scores and cognitive performance at fluid intelligence (Left), IQ (Middle), and SRTT (Right). Black dots/lines show NES index computed before TMS, representing spontaneous network-to-network interplay, while colored dots/lines show NES index computed after perturbation via TMS. Bar plots also show the direction and magnitude of bivariate Pearson correlations (r); *P < 0.05.
Discussion
Here we used fMRI-guided single-pulse TMS to induce a controlled perturbation into parietal nodes of DMN and DAN, and captured fast-evolving electrophysiological dynamics at high temporal and spatial resolution. EEG response dynamics were reconstructed in individual source space using functional network maps as regions of interest (ROIs). We found that targeted stimulation of the DMN node resulted in greater activation of the DMN compared to DAN, whereas stimulation of the DAN resulted in the opposite pattern. Notably, this differential activation was distributed over time and according to stimulated network topography (Fig. 5), rather than just localized to the site of stimulation. Our findings suggest that TMS-evoked activity propagates specifically into functionally connected distal regions of the same network, showing local and distributed effects of TMS over resting-state fMRI networks in humans. Crucially, we replicated our results across two identical sessions 1 mo apart, thus showing that fMRI-guided TMS-induced network activations within RSNs are highly reproducible across sessions. Furthermore, we also provide preliminary evidence that the specificity of network activations across individuals is related to cognitive function. Specifically, we found that cognitive abilities were more strongly correlated with network activations induced by TMS than with resting-state EEG dynamics. Overall, our data revealed several findings with noteworthy implications for future research. First, direct external manipulation of RSN activity by means of TMS combined with high temporal resolution neuroimaging using EEG may provide unique information about fast, millisecond-level interaction patterns between brain networks that are not observable via fMRI or resting-state EEG. External stimulation of a node of a given RSN propagates preferentially within other nodes of the stimulated network and also reveals oscillatory modulations predominantly at the natural frequency of the stimulated network or brain region, thus confirming previous evidence of brain-level effects of TMS (8, 27). Second, TMS-induced network interactions are reproducible such that they can be repeatedly captured across sessions over different days, endorsing their application in future clinical trials focusing on network-level brain dysfunction (31). Finally, preliminary evidence of the correlation between the specificity of network engagement and individual fluid and crystallized intelligence levels suggest perturbation-based propagation patterns as promising physiologic biomarkers of human cognition.
TMS-Induced RSNs Activations.
Originally, the presence of RSNs was first identified by metaanalysis of positron emission tomography scans showing decreased activity during task performance within specific nodes of frontal and parietal brain regions, a network of regions subsequently defined as the DMN (1). Since then, the functional architecture of RSNs in humans has been predominantly studied via fMRI by characterizing spatial dynamics of the BOLD signal in the resting brain. Owing to inherent limitations in the BOLD signal, an increasing number of studies over the years have employed multimodal neuroimaging techniques to better understand the neural origins and spatial−temporal signatures of RSNs (32–40). Combined fMRI−EEG studies have also provided considerable evidence for the electrophysiological basis of RSNs by demonstrating spatially coherent maps in EEG source space that grossly resemble fMRI-based RSNs (36, 39, 41). However, studies relating the spectral dynamics of resting-state EEG to the time course of BOLD activity have revealed heterogeneous findings. In one of the earlier reports, Mantini et al. (38) compared the time course of multiple RSNs identified through independent component analyses (ICA) with broadband EEG spectral power, revealing how EEG oscillations in almost all of the frequency bands are partly correlated with most of the RSN activity, suggesting a link between BOLD activity and millisecond-level EEG activity. However, other studies reported weak or no correlation, or even inverse correlations (42–45). Thus, no consensus has been reached on the nature or direction of correlations between EEG spectral power features and dynamic functional connectivity profiles within or across specific RSNs. Moreover, the correlational nature of these studies is a fundamental limitation for understanding causal dynamics or functional interactions across RSNs. Accordingly, many recent studies have highlighted the importance of moving beyond resting-state correlational studies, and suggested the importance of utilizing direct external manipulation to better infer causal interactions between RSNs (12, 13, 21, 46–49).
Here we employed a network perturbation approach by using fMRI-guided TMS-EEG to examine the extent to which TMS of cortical nodes of the DMN or DAN evokes distinct network-level responses. Our data showed unique TMS-induced activation patterns and oscillatory characteristics as a function of the stimulated cortical node. In particular, we showed that TMS of two cortical nodes with relative anatomical proximity (i.e., both located in the right parietal lobe) generate spatially distinct activation patterns, and that TMS-induced cortical potentials not only endure locally within the stimulated network node but also preferentially propagate following the topography of the same network and at its dominant oscillatory frequency. These results are in line with previous TMS-EEG (15, 18, 27) and TMS-fMRI studies (33, 50–52) showing propagation of TMS-induced activity beyond the stimulation site. Early TMS-EEG studies predominantly focused on the motor system (53, 54) and showed focal responses following TMS that rapidly spread into adjacent ipsilateral motor regions and subsequently propagate into contralateral motor areas, presumably through transcallosal connections. Studies monitoring changes in fMRI connectivity before and after repetitive TMS protocols targeting nonmotor networks have also reported a modulation of network activations within local and distant stimulated networks nodes (51, 55, 56). More recently, reciprocal propagation of evoked responses induced with invasive stimulation has been reported across RSNs following intracranial electrical stimulation of different RSN nodes in neurosurgical patients (57). Our data extend these results by providing evidence of the possibility to 1) engage targeted RSNs following noninvasive perturbation of two neighboring cortical nodes and 2) capture their responses with millisecond resolution, evoking local and distinct distributed cortical propagation patterns resembling network topographies suggested by fMRI data. Previous reports (8, 21, 52) have addressed the clinical and therapeutic value of taking a network perspective on brain stimulation, promoting neuronavigated experimental perturbations to better understand the aberrant network-to-network interplay underlying symptomology in various psychiatric and neurological disorders. Consistent with such notions, our findings support perturbation-based noninvasive neuroimaging as a feasible and reliable multimodal tool to engage and characterize network-level responses in the human brain.
Reproducibility of Network Engagement.
TEPs contain rich neurophysiological information in time, frequency, and source domain, thus serving as a direct measure of both instantaneous and lasting changes in cortical physiological properties (e.g., excitability, inhibition, and connectivity) (14, 58). In addition, TEPs show distinct topographical distribution and component structure across cortical sites in healthy (27) and clinical populations (16, 17, 59–63). To date, although reliability of group-averaged TEPs has been examined at the electrode level (64–66), to the best of our knowledge, no studies have evaluated the reproducibility of TMS-evoked activations at the network level by using RSNs as ROIs in source space. Here, by running two identical sessions 1 mo apart for each study participant, we were able to show that source reconstructions of individual TEPs are highly reproducible both for DAN and DMN (Fig. 5).
There is an ongoing debate regarding the presence of robust nontranscranial peripheral components in TMS-evoked EEG responses as a major factor limiting the interpretation and utility of TEPs. Although the validity and reliability of the present results could be challenged by the contribution of peripherally evoked responses, such as somatosensory and auditory potentials (67), there are several lines of evidence suggesting that our findings reflect transcranial evoked cortical activity. First, we followed state-of-the-art procedures to minimize somatosensory-evoked potentials and auditory-evoked potentials (AEPs), by placing a foam layer underneath the coil and by employing auditory noise masking (68). Second, we applied a two-step ICA to remove TMS-specific artifacts first, as well as remaining muscle, electrode, and cardiac artifacts. This procedure is included to specifically identify and remove components with a spatial topography and time course consistent with AEPs (SI Appendix, Fig. S1). AEPs were clearly present only in five subjects during the second ICA run (less than 25% of our sample), suggesting that noise masking is a useful and effective procedure to suppress unspecific TMS-induced auditory activations. Third, our source analyses showing specificity of TMS-induced activations within the stimulated network strongly argue against the potential contamination of our data with somatosensory or auditory potentials. The presence of temporally stable and high-amplitude sensory responses would dominate the TEP topography and result in relatively uniform activation patterns across stimulation conditions, particularly given that the stimulation sites were anatomically close to each other and would likely result in quite similar nontranscranial effects. Finally, our topographical reliability analyses (SI Appendix, Fig. S5) at the single-electrode level are also in line with source-level network activations, showing topographical specificity for TMS of DAN versus DMN. It should be noted that somatosensory-evoked potentials would be primarily distributed to the somatosensory cortex contralateral to the stimulation site, while auditory-evoked components would be bilaterally distributed over the superior temporal lobes regardless of the stimulation site. The presence of either somatosensory-evoked potentials or AEPs would be expected to dominate the spatial topography (and localized sources) of TEPs, and result in relatively uniform activation patterns across stimulation sites. In particular, we found higher activations over DMN-specific prefrontal regions up to 600 ms after DMN stimulation that are absent for DAN stimulation, suggesting the topographical specificity of evoked responses as a function of the stimulated node/network.
Overall, our reproducibility results for network stimulation expand previous evidence demonstrating that anatomically targeted TEPs can capture brain activity characterizing consciousness and brain plasticity levels, as well as gross cortical physiological differences across brain disorders (15, 17, 27, 60). We believe the reproducibility of network-level responses will provide convincing grounds and motivation for future studies focusing on RSN-guided TMS biomarkers, as well as to better identify therapeutic targets and experimentally modulate connectivity dynamics in abnormally functioning networks.
Specificity of the TMS-Induced Network Activity Is Correlated with Cognition.
The potential for uncovering cortical correlates of variability in human behavior through functional connectivity analysis has generated increasing interest in the study of the human connectome. The majority of studies so far have focused on modeling/correlating data acquired during unconstrained resting state and, to a certain degree, showed that resting-state correlation patterns between various networks can be used to predict individual variability in several cognitive functions (6, 26), personality traits (69), and behavior (70). However, recent evidence suggests that task-based neuroimaging may better delineate the association between individual connectivity patterns and cognitive abilities (46, 47). A recent study by Greene et al. (46), for instance, reported superior performance in predicting fluid intelligence for models built on task fMRI compared to resting-state fMRI ones, suggesting that task-based designs may “perturb” the brain along a particular cognitive dimension and thus provide the opportunity to identify subtle differences in individual patterns of brain connectivity. Although TMS initially perturbs relatively local nodes, induced activity propagates across brain regions over time (71) and provides a snapshot of the connectivity profile of the stimulated node(s). In this context, our NES metric can be considered a proxy of network integrity, as well as a measure of network-to-network integration/segregation. To support this framework, we ran preliminary correlational analyses based on the idea that direct stimulation of functional network nodes (which does not rely on subjects’ attention to and motivation toward a given cognitive task) should generate robust, specific, and reproducible brain states capturing differences in network-to-network dynamics relevant for cognitive performance (48). Our analyses reveal significant positive correlations between the specificity of network activations and fluid/crystallized cognitive abilities, suggesting that the characteristics of brain activity following perturbation within and between networks may constitute a valuable biomarker of human cognition (outperforming resting-state EEG data alone). More specifically, we found that TMS induces an opposing oscillatory pattern between DMN and DAN, highly resembling DMN/DAN fMRI connectivity data (3). The negative functional connectivity pattern between these networks is reported to be critical for optimal cognitive functioning and thought to serve as neural substrate for adaptively directing attentional resources between self-referential and externally oriented tasks (3, 20, 72, 73). Interestingly, and in line with the present results, a recent report by our group has shown that 1) distributed brain regions having robust coactivation during the solution of fluid intelligence (gf) tasks highly overlap with the topography of DAN (74) and, more importantly, 2) the strength of negative correlations between these regions and the DMN is a significant predictor of individual gf performance (26). This suggests that network-targeted TMS-EEG is able to capture the same fMRI dynamics, but at high temporal resolution. More importantly, perturbational approaches like the one adopted in the present investigation could help disentangle the nature of “anticorrelations” in fMRI data (3), by providing a modality-independent, high temporal resolution causal evidence of their existence and potential relevance. Finally, we found that network engagement was not correlated with performance on a control motor learning task, suggesting that the relationship between evoked cortical network dynamics and cognition is not a general and nonspecific finding. Future studies are needed for a comprehensive investigation systematically comparing spontaneous, task-based, and perturbation-based neuromarkers of human cognition.
Limitations of the Study and Future Directions.
Although source localization of EEG responses to TMS of parietal nodes revealed distinct propagation patterns depending on the network being stimulated, more precise spatial mapping of TMS-induced network effects would be better captured by ideally combining TMS-EEG with concurrent fMRI acquisitions (28, 57, 75). While the technology for integrated TMS-EEG−fMRI designs is still in its infancy, such a multimodal neuroimaging approach would allow thorough delineation of fast-evolving dynamics of distributed network activations with high spatial resolution, and better understanding of whether distributed network activations as measured by TMS-EEG are reflected in spatial topography of TMS-evoked BOLD activations as well.
Another important consideration concerns the need to extend the current network perturbation approach with relevant task designs for controlling brain state during stimulation. Given the established dynamic and reciprocal associations between DAN and DMN both at rest and during cognitive performance, it is essential to examine 1) to what extent TMS-induced network activations within and across perturbed networks overlap with network activations during task performance and 2) whether TMS-induced network dynamics prospectively predict task performance in both healthy and neurological/psychiatric populations.
Conclusions
Direct external manipulation of RSN activity may provide valuable information about fast-evolving interactions between functionally opposing brain networks otherwise not observable through low temporal resolution fMRI or resting-state EEG analyses. This corroborates the idea that a controlled perturbation of a complex system provides more unique characterization of its topology and dynamics than measuring its spontaneous behavior, promoting the adoption of perturbation-based biomarkers in cognitive neuroscience and clinical research.
Methods
Participants.
Data collected from 21 right-handed healthy volunteers (mean age = 33.4 ± 8.6 y, ranging from 19 y to 49 y) were analyzed for this study. Experimental protocols and voluntary participation procedures were explained to all participants before they gave their written informed consent to the study which conformed to the Declaration of Helsinki, and had been approved by the Institutional Review Board of the Beth Israel Deaconess Medical Center.
Structural MRI Data Acquisition.
A T1-weighted (T1w) anatomical MRI scan was obtained for all participants and used for neuronavigation. MRI data were acquired on a 3T scanner (GE Healthcare, Ltd.) using a three-dimensional spoiled gradient echo sequence: 166 axial-oriented slices for whole-brain coverage; 240-mm isotropic field-of-view (FOV); 0.937-mm × 0.937-mm × 1-mm native resolution; flip angle = 15°; echo time (TE)/repetition time (TR) ≥ 2.9/6.9 ms; duration ≥ 432 s.
Resting-State fMRI Data and Preprocessing.
Resting-state fMRI data were collected on a 3T GE scanner in three runs of 5 min each with eyes open, with the following parameters: axial plane (bottom-up), FOV = 240 mm, TE = 25 ms, TR = 3,196 ms, slice thickness = 2.5 mm, flip angles = 90°, voxel size of 1.87 × 1.87 × 2.5. We preprocessed the resting-state fMRI data using FMRIPREP (v1.2). To individualize the functional networks, the approach developed by Wang et al., (76) was used. Briefly, this approach includes 18 functional networks using 17 networks from Yeo-2011 group-average functional networks, with an additional network representing the hand area (SI Appendix, Methods). For more details on the procedure for personalization, see the original publication by Wang et al.
Baseline Assessments.
All participants were scheduled for a 5-h baseline visit during which they underwent a careful cognitive and motor performance assessment. The cognitive evaluation included a modified Edinburgh handedness questionnaire (77) and a NEO Personality inventory (NEO-FFI) (78). A total of 14 cognitive tasks were split over two different batteries covering multiple cognitive functions such as attention, abstract reasoning, inhibition, verbal and visuospatial working memory, and switching, presented on a Windows laptop PC (Microsoft) equipped with E-Prime 2.0 (Psychology Software Tools Inc.). As preliminary analyses, we aimed at correlating individual network-targeted TEPs to two broad domains of global human cognition, namely, fluid (gf) and crystallized (gc) intelligence (79, 80). These two domains represent, respectively, 1) abstract reasoning abilities related to hypothesis-testing and reasoning (gf), measured via the Sandia Matrices (SM) (81), and 2) semantic knowledge measured via the Test of Premorbid Functioning (TOPF) (gc) (82). SM and TOPF tests were selected as cognitive tasks representing general intelligence (i.e., the g factor, commonly represented as IQ), both in its fluid (gf) and crystallized (gc) components. As a control task, we also included SRTT. SRTT is a widely used motor task (83) to capture implicit sequence learning (see SI Appendix, Methods for task descriptions).
For each task, accuracy and reaction times (RTs) were recorded and then combined to compute the inverse efficiency scores (IES) (83). IES consider changes in speed−accuracy trade-off typically observed during sustained cognitive testing (83, 84). IES values were used as cognitive task performance scores in the analyses.
TMS.
TMS was delivered using a figure-of-eight−shaped coil with dynamic fluid cooling (MagPro 75-mm Cool B-65; MagVenture A/S) attached to a MagPro X-100 stimulator (MagVenture A/S). Individual high-resolution T1w images were imported into the Brainsight TMS Frameless Navigation system (Rogue Research Inc.), and coregistered to digitized anatomical landmarks for online monitoring of coil positioning.
Motor-evoked potentials (MEPs) were recorded from the right first dorsal interosseous (FDI) and the abductor pollicis brevis (APB) muscles. Ag−AgCl surface electrode pairs were placed on the belly and tendon of the muscles and a ground was placed on the right ulnar styloid process.
EEG.
Whole-scalp 64-channel EEG data were collected with a TMS-compatible amplifier system (actiCHamp system; Brain Products GmbH) and labeled in accordance with the extended 10–20 International System. EEG data were online-referenced to Fp1 electrode. Electrode impedances were maintained below 5 kΩ at a sampling rate of 1,000 Hz. EEG signals were digitized using a BrainCHamp DC amplifier and linked to BrainVision Recorder software (version 1.21) for online monitoring. Digitized EEG electrode locations on the scalp were also coregistered to individual MRI scans using Brainsight TMS Frameless Navigation system.
TMS Targets.
In order to identify individualized TMS targets, group-level resting-state functional networks maps were used, based on a seven-network cortical parcellation (85). The seven networks correspond to visual, somatosensory, limbic, dorsal attention, ventral attention, default mode, and frontoparietal RSNs. Confidence maps for each RSN were used, representing the confidence of each vertex belonging to its assigned network across a sample of 1,000 healthy subjects (expressed as valued between −1 and 1), with larger values indicating higher confidence. By using group-level functional parcellations and confidence maps estimated from 1,000 healthy subjects, we were able to target the most consistent and reliable regions within each network, therefore increasing the generalizability of TMS-EEG findings. We first projected the seven-network functional cortical atlas and the confidence maps onto subject’s cortical surface using the spherical registration implemented in Freesurfer software (Fig. 1A). The resulting maps were then resampled to native structural T1w MRIs. Voxels within each network were weighted by the confidence map, and the voxels with the highest confidence in the right angular gyrus and the right superior parietal were chosen for DMN and DAN stimulations, respectively (Fig. 1E). Individualized DMN and DAN targets mapped to Montreal Neurological Institute (MNI) space are provided to show variability of targeted nodes across individuals (Fig. 1F).
Experimental Procedures.
Throughout the session, participants were comfortably seated in an adjustable chair. At the beginning of each session, the motor hotspot for eliciting MEPs in the FDI muscle was determined by delivering single TMS pulses and moving the TMS coil in small incremental steps after two to three stimulations in each spot, over the hand region of left motor cortex with 45° rotation in relation to the parasagittal plane (inducing posterior-to-anterior current in the underlying cortex) (86). The hotspot was defined as the region where single-pulse TMS elicits larger and more consistent MEPs in the FDI muscle, as compared to the APB muscle, with the minimum stimulation intensity (87). The FDI hotspot was digitized on each participant’s MRI image. Resting motor threshold (RMT) was determined on the FDI hotspot as the minimum stimulation intensity eliciting at least five MEPs (≥50 μV) out of 10 pulses in the relaxed FDI using monophasic current waveforms (86). In compliance with the International Federation of Clinical Neurophysiology safety recommendations, participants were asked to wear earplugs during hotspot and RMT trials to protect their hearing, and to minimize external noise (88). TMS was administered with a thin layer of foam placed under the coil to minimize somatosensory contamination of the TMS-evoked EEG potentials. To minimize AEPs related to the TMS click, auditory white noise masking was used throughout the TMS stimulation (89).
Following determination of RMT, a total of 120 single TMS pulses were delivered to each stimulation target (DMN node target in the angular gyrus and DAN node target in the superior parietal lobule) at an intensity of 120% RMT with randomly jittered (3,000 to 5,000 ms) interstimulus intervals over two repeated blocks of 60 trials each (Fig. 1A). Operators continuously monitored participants during the TMS blocks for their wakefulness, and prompted them to keep their eyes open and stay fully relaxed in case of visible signs of drowsiness or tension. Each participant completed two identical experimental sessions 4 wk apart.
EEG Data Processing.
All EEG data preprocessing was performed offline using EEGLAB functions 14.1 (90−92), and customized scripts running in Matlab R2017b (MathWorks Inc.). All steps of EEG data processing on a sample dataset and resulting signal transformation showing evolution of TEPs, with final scalp topography of a selected TEP are provided in SI Appendix, Methods and Fig. S1. Control analyses examining the impact of TMS-induced muscle artifacts are also provided in SI Appendix, Results and Fig. S3.
EEG Channel Metrics.
Global cortical activation levels following TMS of DMN and DAN were quantified with global mean field power (GMFP) analyses on TEPs (see SI Appendix, Methods and Fig. S2 for details). The GMFP can be used to measure the global brain response to TMS (93). Local mean field power (LMFP) analysis was also performed to identify local evoked activity induced by TMS of DMN and DAN (see SI Appendix, Methods and Fig. S3 for details). TEP time series were also used to test reproducibility of TEPs responses in electrode space.
EEG Source Reconstruction.
All TMS-evoked EEG source reconstruction was performed using Brainstorm (94). First, digitized EEG channel locations and anatomical landmarks of each subject were extracted from Brainsight (nasion, left preauricular, and right preauricular points), and registered onto individual MRI scans in Brainstorm (Fig. 1C). Next, the EEG epochs, −500 ms to 1,000 ms with respect to TMS pulse, for each TMS trial were uploaded, and average epoch time series was generated for each subject (Fig. 1B). Forward modeling of neuroelectric fields was performed using the symmetric boundary element method (95, 96), all with default parameter settings. Noise covariance was estimated from individual trials using the pre-TMS time window as baseline (−500 to 0 ms). Inverse modeling of the cortical sources was performed using the minimum norm estimation (MNE) method with dynamic statistical parametric mapping and constraining source dipoles to the cortical surface. The resulting output of EEG source reconstruction was the MNE current density time series for each cortical voxel.
EEG Source Metrics.
We first extracted average current density time series from DMN (red shaded areas on the cortex in Fig. 1C) and DAN (green shaded areas on the cortex in Fig. 1C), projected on surface space for each individual for both DMN and DAN stimulation. To examine TMS-induced oscillatory characteristics across stimulation conditions, we first computed event-related spectral perturbations (ERSPs) for each network. Using an open source EEGLAB function (newtimef), we implemented Morlet wavelets ranging from 0.5 to 3 cycles with a 500-ms window size sliding 20 ms in each iteration. ERSPs were baseline normalized (−500 to 0 ms), and significant poststimulus activations were determined using bootstrapping.
We then normalized (z score) the current densities to the noise covariance kernel and rectified the resulting time series (Fig. 4 B, Upper) for time domain analyses. We finally computed the total cortical activation within each network in 400-ms windows before (pre-TMS: −500 to −100 ms; Fig. 4 B, Lower) and after (post-TMS: 15 ms to 400 ms) TMS. Resulting current density z-score sums represent the total amount of source reconstructed electrocortical activity in each RSN map for a given time window.
We also examined whether the degree of network-specific activations following TMS were related to individual differences in cognitive abilities. To do so, we quantified an NES by simply computing the average percentage of TMS-induced activity in the stimulated network relative to the nonstimulated network for each stimulation condition, as follows:
NES was used to run bivariate correlations between network specificity and cognition scores.
Statistical Analysis.
All statistical analyses were performed using custom scripts utilizing Matlab statistical toolbox (Version 17A, The MathWorks Inc.). To compare GMFP and LMFP time series across stimulation conditions, we performed cluster-based permutation (permutation n = 10,000) paired t test statistics (28) (see SI Appendix, Results for GMFP and LMFP analyses). Paired sample t tests were used to compare sum of source activation values across conditions both before and after TMS. One-way ANOVA was performed to compare sum GMFP values across stimulation conditions. A repeated measures ANOVA was performed for test−retest comparison with Time (visit 1 vs. visit 2) as within-subject factor. A two-by-two ANOVA (stimulation condition: DAN vs. DMN) × 2 (network maps: DMN vs. DAN) was performed to examine the effect of stimulation condition on network activations both before and after TMS. Pairwise comparisons with Bonferroni corrections (P = 0.05/number of comparisons) were performed to examine significant main effects on network activity differences across stimulation conditions.
Pearson product-moment correlations were used to examine the reproducibility of TEPs across sessions. Simple linear regressions with scatterplots were run to compute bivariate relationships between NES and cognitive performance for gf-relational, gf-logical, and IQ. Significance level was set as (P < 0.05) for all statistical analyses.
Data Availability.
Compressed raw and preprocessed data, as well as code, have been uploaded at the Berenson-Allen Center for Noninvasive Brain Stimulation site (http://www.tmslab.org/santalab.php).
Supplementary Material
Acknowledgments
We thank all participants who took part in the study for their effort. The study was supported by the Harvard-MIT BROAD Institute (Grant ID 6600024-5500000895). A.P.-L. is supported by the Berenson-Allen Foundation, the Sidney R. Baer Jr. Foundation, NIH Grants R01HD069776, R01NS073601, R21 MH099196, R21 NS082870, R21 NS085491, and R21 HD07616, and The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences Grant UL1 RR025758). E.S. is supported by the Beth Israel Deaconess Medical Center via the Chief Academic Officer Award 2017, the Defence Advanced Research Projects Agency via HR001117S0030, and NIH Grants P01 AG031720-06A1, R01 MH117063-01, and R01 AG060981-01. M.M.S. is supported by the Citizens United for Research in Epilepsy foundation, the Football Players Health Study at Harvard University, and NIH Grants R01 MH115949, R01AG060987, R01 NS073601, and P01 AG031720-06A1. M.D.F. was supported by the G. Harold and Leila Y. Mathers Charitable Foundation, the Nancy Lurie Marks Foundation, and the NIH (R01 MH113929). The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of Harvard University and its affiliated academic health care centers, or NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Compressed raw and preprocessed data, as well as code, have been uploaded at the Berenson-Allen Center for Noninvasive Brain Stimulation site (http://www.tmslab.org/santalab.php).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911240117/-/DCSupplemental.
References
- 1.Raichle M. E., et al. , A default mode of brain function. Proc. Natl. Acad. Sci. U.S.A. 98, 676–682 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswal B., Yetkin F. Z., Haughton V. M., Hyde J. S., Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 34, 537–541 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Fox M. D., et al. , The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc. Natl. Acad. Sci. U.S.A. 102, 9673–9678 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox M. D., Snyder A. Z., Zacks J. M., Raichle M. E., Coherent spontaneous activity accounts for trial-to-trial variability in human evoked brain responses. Nat. Neurosci. 9, 23–25 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Buckner R. L., Andrews-Hanna J. R., Schacter D. L., The brain’s default network: Anatomy, function, and relevance to disease. Ann. N. Y. Acad. Sci. 1124, 1–38 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Finn E. S., et al. , Functional connectome fingerprinting: Identifying individuals using patterns of brain connectivity. Nat. Neurosci. 18, 1664–1671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang D., Raichle M. E., Disease and the brain’s dark energy. Nat. Rev. Neurol. 6, 15–28 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Fox M. D., Buckner R. L., White M. P., Greicius M. D., Pascual-Leone A., Efficacy of transcranial magnetic stimulation targets for depression is related to intrinsic functional connectivity with the subgenual cingulate. Biol. Psychiatry 72, 595–603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brookes M. J., et al. , Investigating the electrophysiological basis of resting state networks using magnetoencephalography. Proc. Natl. Acad. Sci. U.S.A. 108, 16783–16788 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Pasquale F., et al. , Temporal dynamics of spontaneous MEG activity in brain networks. Proc. Natl. Acad. Sci. U.S.A. 107, 6040–6045 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hipp J. F., Hawellek D. J., Corbetta M., Siegel M., Engel A. K., Large-scale cortical correlation structure of spontaneous oscillatory activity. Nat. Neurosci. 15, 884–890 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole D. M., Smith S. M., Beckmann C. F., Advances and pitfalls in the analysis and interpretation of resting-state FMRI data. Front. Syst. Neurosci. 4, 8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergmann T. O., Karabanov A., Hartwigsen G., Thielscher A., Siebner H. R., Combining non-invasive transcranial brain stimulation with neuroimaging and electrophysiology: Current approaches and future perspectives. Neuroimage 140, 4–19 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Bortoletto M., Veniero D., Thut G., Miniussi C., The contribution of TMS-EEG coregistration in the exploration of the human cortical connectome. Neurosci. Biobehav. Rev. 49, 114–124 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Casali A. G., et al. , A theoretically based index of consciousness independent of sensory processing and behavior. Sci. Transl. Med. 5, 198ra105 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Shafi M. M., et al. , Physiological consequences of abnormal connectivity in a developmental epilepsy. Ann. Neurol. 77, 487–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosanova M., et al. , Recovery of cortical effective connectivity and recovery of consciousness in vegetative patients. Brain 135, 1308–1320 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massimini M., Boly M., Casali A., Rosanova M., Tononi G., A perturbational approach for evaluating the brain’s capacity for consciousness. Prog. Brain Res. 177, 201–214 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Zhou J., et al. , Divergent network connectivity changes in behavioural variant frontotemporal dementia and Alzheimer’s disease. Brain 133, 1352–1367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corbetta M., Patel G., Shulman G. L., The reorienting system of the human brain: From environment to theory of mind. Neuron 58, 306–324 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox M. D., Greicius M., Clinical applications of resting state functional connectivity. Front. Syst. Neurosci. 4, 19 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greicius M. D., Srivastava G., Reiss A. L., Menon V., Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: Evidence from functional MRI. Proc. Natl. Acad. Sci. U.S.A. 101, 4637–4642 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greicius M. D., Krasnow B., Reiss A. L., Menon V., Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. Proc. Natl. Acad. Sci. U.S.A. 100, 253–258 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheline Y. I., et al. , The default mode network and self-referential processes in depression. Proc. Natl. Acad. Sci. U.S.A. 106, 1942–1947 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitfield-Gabrieli S., Ford J. M., Default mode network activity and connectivity in psychopathology. Annu. Rev. Clin. Psychol. 8, 49–76 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Santarnecchi E., et al. , Network connectivity correlates of variability in fluid intelligence performance. Intelligence 65, 35–47 (2017). [Google Scholar]
- 27.Rosanova M., et al. , Natural frequencies of human corticothalamic circuits. J. Neurosci. 29, 7679–7685 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maris E., Oostenveld R., Nonparametric statistical testing of EEG- and MEG-data. J. Neurosci. Methods 164, 177–190 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Cole M. W., Yarkoni T., Repovs G., Anticevic A., Braver T. S., Global connectivity of prefrontal cortex predicts cognitive control and intelligence. J. Neurosci. 32, 8988–8999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Heuvel M. P., Stam C. J., Kahn R. S., Hulshoff Pol H. E., Efficiency of functional brain networks and intellectual performance. J. Neurosci. 29, 7619–7624 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Etkin A., et al. , Using fMRI connectivity to define a treatment-resistant form of post-traumatic stress disorder. Sci. Transl. Med. 11, eaal3236 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Britz J., Van De Ville D., Michel C. M., BOLD correlates of EEG topography reveal rapid resting-state network dynamics. Neuroimage 52, 1162–1170 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Chang C., Liu Z., Chen M. C., Liu X., Duyn J. H., EEG correlates of time-varying BOLD functional connectivity. Neuroimage 72, 227–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen A. C. N., Feng W., Zhao H., Yin Y., Wang P., EEG default mode network in the human brain: Spectral regional field powers. Neuroimage 41, 561–574 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Feige B., et al. , Distinctive time-lagged resting-state networks revealed by simultaneous EEG-fMRI. Neuroimage 145, 1–10 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Laufs H., Endogenous brain oscillations and related networks detected by surface EEG-combined fMRI. Hum. Brain Mapp. 29, 762–769 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q., Ganzetti M., Wenderoth N., Mantini D., Detecting large-scale brain networks using EEG: Impact of electrode density, head modeling and source localization. Front. Neuroinform. 12, 4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantini D., Perrucci M. G., Del Gratta C., Romani G. L., Corbetta M., Electrophysiological signatures of resting state networks in the human brain. Proc. Natl. Acad. Sci. U.S.A. 104, 13170–13175 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadaghiani S., et al. , Intrinsic connectivity networks, alpha oscillations, and tonic alertness: A simultaneous electroencephalography/functional magnetic resonance imaging study. J. Neurosci. 30, 10243–10250 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tagliazucchi E., von Wegner F., Morzelewski A., Brodbeck V., Laufs H., Dynamic BOLD functional connectivity in humans and its electrophysiological correlates. Front. Hum. Neurosci. 6, 339 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheeringa R., et al. , Neuronal dynamics underlying high- and low-frequency EEG oscillations contribute independently to the human BOLD signal. Neuron 69, 572–583 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Gonçalves S. I., et al. , Correlating the alpha rhythm to BOLD using simultaneous EEG/fMRI: Inter-subject variability. Neuroimage 30, 203–213 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Knyazev G. G., Slobodskoj-Plusnin J. Y., Bocharov A. V., Pylkova L. V., The default mode network and EEG α oscillations: An independent component analysis. Brain Res. 1402, 67–79 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Laufs H., et al. , EEG-correlated fMRI of human alpha activity. Neuroimage 19, 1463–1476 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Wu L., Eichele T., Calhoun V. D., Reactivity of hemodynamic responses and functional connectivity to different states of alpha synchrony: A concurrent EEG-fMRI study. Neuroimage 52, 1252–1260 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greene A. S., Gao S., Scheinost D., Constable R. T., Task-induced brain state manipulation improves prediction of individual traits. Nat. Commun. 9, 2807 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finn E. S., et al. , Can brain state be manipulated to emphasize individual differences in functional connectivity? Neuroimage 160, 140–151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santarnecchi E., Rossi S., Advances in the neuroscience of intelligence: From brain connectivity to brain perturbation. Span. J. Psychol. 19, E94 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Shafi M. M., Westover M. B., Fox M. D., Pascual-Leone A., Exploration and modulation of brain network interactions with noninvasive brain stimulation in combination with neuroimaging. Eur. J. Neurosci. 35, 805–825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amico E., et al. , Tracking dynamic interactions between structural and functional connectivity: A TMS/EEG-dMRI study. Brain Connect. 7, 84–97 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Eldaief M. C., Halko M. A., Buckner R. L., Pascual-Leone A., Transcranial magnetic stimulation modulates the brain’s intrinsic activity in a frequency-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 108, 21229–21234 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fox M. D., et al. , Resting-state networks link invasive and noninvasive brain stimulation across diverse psychiatric and neurological diseases. Proc. Natl. Acad. Sci. U.S.A. 111, E4367–E4375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ilmoniemi R. J., et al. , Neuronal responses to magnetic stimulation reveal cortical reactivity and connectivity. Neuroreport 8, 3537–3540 (1997). [DOI] [PubMed] [Google Scholar]
- 54.Komssi S., et al. , Ipsi- and contralateral EEG reactions to transcranial magnetic stimulation. Clin. Neurophysiol. 113, 175–184 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Hoffman R. E., et al. , Probing the pathophysiology of auditory/verbal hallucinations by combining functional magnetic resonance imaging and transcranial magnetic stimulation. Cereb. Cortex 17, 2733–2743 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hawco C., et al. , Spread of activity following TMS is related to intrinsic resting connectivity to the salience network: A concurrent TMS-fMRI study. Cortex 108, 160–172 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Shine J. M., et al. , Distinct patterns of temporal and directional connectivity among intrinsic networks in the human brain. J. Neurosci. 37, 9667–9674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hill A. T., Rogasch N. C., Fitzgerald P. B., Hoy K. E., TMS-EEG: A window into the neurophysiological effects of transcranial electrical stimulation in non-motor brain regions. Neurosci. Biobehav. Rev. 64, 175–184 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Farzan F., et al. , Characterizing and modulating brain circuitry through transcranial magnetic stimulation combined with electroencephalography. Front. Neural Circuits 10, 73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hallett M., et al. , Contribution of transcranial magnetic stimulation to assessment of brain connectivity and networks. Clin. Neurophysiol. 128, 2125–2139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaskie R. E., Ferrarelli F., Investigating the neurobiology of schizophrenia and other major psychiatric disorders with Transcranial Magnetic Stimulation. Schizophr. Res. 192, 30–38 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Kimiskidis V. K., Transcranial magnetic stimulation (TMS) coupled with electroencephalography (EEG): Biomarker of the future. Rev. Neurol. (Paris) 172, 123–126 (2016). [DOI] [PubMed] [Google Scholar]
- 63.McClintock S. M., Freitas C., Oberman L., Lisanby S. H., Pascual-Leone A., Transcranial magnetic stimulation: A neuroscientific probe of cortical function in schizophrenia. Biol. Psychiatry 70, 19–27 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Casarotto S., et al. , EEG responses to TMS are sensitive to changes in the perturbation parameters and repeatable over time. PLoS One 5, e10281 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kerwin L. J., Keller C. J., Wu W., Narayan M., Etkin A., Test-retest reliability of transcranial magnetic stimulation EEG evoked potentials. Brain Stimul. 11, 536–544 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Lioumis P., Kicić D., Savolainen P., Mäkelä J. P., Kähkönen S., Reproducibility of TMS-evoked EEG responses. Hum. Brain Mapp. 30, 1387–1396 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conde V., et al. , The non-transcranial TMS-evoked potential is an inherent source of ambiguity in TMS-EEG studies. Neuroimage 185, 300–312 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Gordon P. C., Desideri D., Belardinelli P., Zrenner C., Ziemann U., Comparison of cortical EEG responses to realistic sham versus real TMS of human motor cortex. Brain Stimul. 11, 1322–1330 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Adelstein J. S., et al. , Personality is reflected in the brain’s intrinsic functional architecture. PLoS One 6, e27633 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fox M. D., Snyder A. Z., Vincent J. L., Raichle M. E., Intrinsic fluctuations within cortical systems account for intertrial variability in human behavior. Neuron 56, 171–184 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Massimini M., et al. , Breakdown of cortical effective connectivity during sleep. Science 309, 2228–2232 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Broyd S. J., et al. , Default-mode brain dysfunction in mental disorders: A systematic review. Neurosci. Biobehav. Rev. 33, 279–296 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Tavor I., et al. , Task-free MRI predicts individual differences in brain activity during task performance. Science 352, 216–220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santarnecchi E., Emmendorfer A., Pascual-Leone A., Dissecting the Parieto-frontal correlates of fluid intelligence: A comprehensive ALE meta-analysis study. Intelligence 63, 9–28 (2017). [Google Scholar]
- 75.Peters J. C., et al. , On the feasibility of concurrent human TMS-EEG-fMRI measurements. J. Neurophysiol. 109, 1214–1227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang D., et al. , Parcellating cortical functional networks in individuals. Nat. Neurosci. 18, 1853–1860 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ransil B. J., Schachter S. C., Test-retest reliability of the Edinburgh Handedness Inventory and Global Handedness preference measurements, and their correlation. Percept. Mot. Skills 79, 1355–1372 (1994). [DOI] [PubMed] [Google Scholar]
- 78.Costa P. T. Jr, McCrae R. R., Personality in adulthood: A six-year longitudinal study of self-reports and spouse ratings on the NEO Personality Inventory. J. Pers. Soc. Psychol. 54, 853–863 (1988). [DOI] [PubMed] [Google Scholar]
- 79.Geake J. G., Hansen P. C., Functional neural correlates of fluid and crystallized analogizing. Neuroimage 49, 3489–3497 (2010). [DOI] [PubMed] [Google Scholar]
- 80.Gray J. R., Chabris C. F., Braver T. S., Neural mechanisms of general fluid intelligence. Nat. Neurosci. 6, 316–322 (2003). [DOI] [PubMed] [Google Scholar]
- 81.Matzen L. E., et al. , Recreating Raven’s: Software for systematically generating large numbers of Raven-like matrix problems with normed properties. Behav. Res. Methods 42, 525–541 (2010). [DOI] [PubMed] [Google Scholar]
- 82.Nelson H. E., National Adult Reading Test (NART): For the assessment of premorbid intelligence in patients with dementia: Test manual. https://www.academia.edu/2515150/National_Adult_Reading_Test_NART_test_manual_Part_1. Accessed 1 January 2019.
- 83.Townsend J. T., Ashby F. G., Stochastic Modeling of Elementary Psychological Processes (Cambridge University Press Archive, 1983). [Google Scholar]
- 84.Enns J. T., Akhtar N., A developmental study of filtering in visual attention. Child Dev. 60, 1188–1199 (1989). [PubMed] [Google Scholar]
- 85.Yeo B. T., et al. , The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 1125–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rossini P. M., et al. , Non-invasive electrical and magnetic stimulation of the brain, spinal cord, roots and peripheral nerves: Basic principles and procedures for routine clinical and research application. An updated report from an I.F.C.N. Committee. Clin. Neurophysiol. 126, 1071–1107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rothwell J. C., et al. ; The International Federation of Clinical Neurophysiology , Magnetic stimulation: Motor evoked potentials. Electroencephalogr. Clin. Neurophysiol. Suppl. 52, 97–103 (1999). [PubMed] [Google Scholar]
- 88.Rossi S., Hallett M., Rossini P. M., Pascual-Leone A.; Safety of TMS Consensus Group , Safety, ethical considerations, and application guidelines for the use of transcranial magnetic stimulation in clinical practice and research. Clin. Neurophysiol. 120, 2008–2039 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.ter Braack E. M., de Vos C. C., van Putten M. J., Masking the auditory evoked potential in TMS–EEG: A comparison of various methods. Brain Topogr. 28, 520–528 (2015). [DOI] [PubMed] [Google Scholar]
- 90.Delorme A., Makeig S., EEGLAB: An open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods 134, 9–21 (2004). [DOI] [PubMed] [Google Scholar]
- 91.Hyvärinen A., Oja E., A fast fixed-point algorithm for independent component analysis. Neural Comput. 9, 1483–1492 (1997). [Google Scholar]
- 92.Rogasch N. C., et al. , Analysing concurrent transcranial magnetic stimulation and electroencephalographic data: A review and introduction to the open-source TESA software. Neuroimage 147, 934–951 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Esser S. K., et al. , A direct demonstration of cortical LTP in humans: A combined TMS/EEG study. Brain Res. Bull. 69, 86–94 (2006). [DOI] [PubMed] [Google Scholar]
- 94.Tadel F., Baillet S., Mosher J. C., Pantazis D., Leahy R. M., Brainstorm: A user-friendly application for MEG/EEG analysis. Comput. Intell. Neurosci. 2011, 879716 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gramfort A., Papadopoulo T., Olivi E., Clerc M., OpenMEEG: Opensource software for quasistatic bioelectromagnetics. Biomed. Eng. Online 9, 45 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kybic J., Clerc M., Faugeras O., Keriven R., Papadopoulo T., Fast multipole acceleration of the MEG/EEG boundary element method. Phys. Med. Biol. 50, 4695–4710 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Compressed raw and preprocessed data, as well as code, have been uploaded at the Berenson-Allen Center for Noninvasive Brain Stimulation site (http://www.tmslab.org/santalab.php).