Abstract
The classification process of lung nodule detection in a traditional computer-aided detection (CAD) system is complex, and the classification result is heavily dependent on the performance of each step in lung nodule detection, causing low classification accuracy and high false positive rate. In order to alleviate these issues, a lung nodule classification method based on a deep residual network is proposed. Abandoning traditional image processing methods and taking the 50-layer ResNet network structure as the initial model, the deep residual network is constructed by combining residual learning and migration learning. The proposed approach is verified by conducting experiments on the lung computed tomography (CT) images from the publicly available LIDC-IDRI database. An average accuracy of 98.23% and a false positive rate of 1.65% are obtained based on the ten-fold cross-validation method. Compared with the conventional support vector machine (SVM)-based CAD system, the accuracy of our method improved by 9.96% and the false positive rate decreased by 6.95%, while the accuracy improved by 1.75% and 2.42%, respectively, and the false positive rate decreased by 2.07% and 2.22%, respectively, in contrast to the VGG19 model and InceptionV3 convolutional neural networks. The experimental results demonstrate the effectiveness of our proposed method in lung nodule classification for CT images.
1. Introduction
Lung cancer is a malignant disease with poor prognosis, and the average 5-year survival rate of patients is less than 20%. Although there are targeted treatments and various radiotherapy and chemotherapy regimens, the average survival time of advanced lung cancer is only 12 months [1]. Therefore, early detection, early diagnosis, and early treatment of lung cancer can effectively improve the quality of life and survival rate of patients. A spot on the CT of the lung is defined as a lung nodule, which can be benign or malignant. Early lung lesions are mainly characterized by malignant nodules in the lungs. Therefore, it is very important for the clinical treatment of lung cancer to classify lung nodules timely and accurately. In recent years, the improvement of medical and health levels has led to the application of more and more medical digital imaging devices to the clinic. The medical imaging equipment, including X-ray, B-scan ultrasonography, computed tomography (CT), and magnetic resonance imaging (MRI), are improving and optimizing constantly [2]. More and more subtle lesions can be captured by imaging equipment. Among them, computerized tomography is considered to be one of the most effective means of detecting lung cancer early [3]. Doctors need to diagnose malignant nodules accurately by reading the patient's lung CT image; however, reading a large number of CT images is not only time-consuming, and there is also a high probability of misdiagnosis.
With the development of information technology and medical imaging methods, computer-aided detection systems based on CT images have achieved certain research results [4,5]. The use of the CAD system model for the automatic detection and identification of lung nodules not only improves efficiency greatly, it also has higher accuracy and better robustness. The traditional CAD algorithm is divided into two steps: the first step is image preprocessing [6] (pulmonary CT image enhancement, lung parenchymal segmentation, lung ROI extraction, etc.), and the second step is image feature extraction (gray texture features [7], scale-invariant features [8], local binary pattern features [9] and gradient direction histogram features [10], etc.), after which traditional machine learning algorithms (K-nearest neighbors, support vector machines, random forests, etc.) are applied for lung nodule classification. For example, Manikandan and Bharathi [11] extracted grayscale features and the center of gravity of the region of interest for morphological-based target detection classification. Kim et al. [12] extracted 96 nodular morphological features including area, standard deviation, perimeter, and diameter and then extracted 100 ROI deep features with a stacked denoising autoencoder (SDAE) containing 3 hidden layers. Since these features are designed manually, it is difficult to analyze the image comprehensively and deeply. Furthermore, using traditional machine learning algorithms, the previous processing results have a greater impact on the subsequent processing.
As an important branch of machine learning, deep learning has developed rapidly in recent years. Convolutional neural networks (CNNs) have achieved good results in the field of face recognition, object detection, image classification, and other images, due to a large amount of available data and the efficient computing capacity of GPUs, and it has also been applied to medical images [13,14]. Ronneberger et al. [15] reported a new full convolution network (FCN) called U-Net for biomedical image segmentation and achieved promising results. Gao et al. [16] extracted the time domain and spatial domain information features of the echocardiogram by combining two 2D-CNNs and classified the video images of echocardiography to assist diagnosis of heart disease. In 2017, Litjens et al. [17] published a review, which summarizes the research work on deep learning in medical image classification, detection and segmentation, registration, and retrieval.
In the present work, we focus on the research work of lung nodule classification based on a proposed deep residual network. Our two main contributions can be summarized as follows: (1) We have developed a deep residual network based classification method for lung nodules in CT images, abandoning the complex traditional CT image processing method. Our results demonstrate the effectiveness of this approach for lung nodule classification. (2) We also investigated performance comparisons of our approach with two representative deep learning models and one traditional model. It is shown that the performance of our proposed deep learning methods is superior to that of the other two deep learning models and the traditional machine learning method. The presented results indicate the suitability of this approach for lung nodule classification.
The rest of this article is organized as follows. In Section 2, the related work is presented. In Section 3, the related algorithms and method framework are elaborated. The detailed experimental setup, procedure, result comparison, and analysis are presented and discussed in Section 4. The conclusion is given in the last section.
2. Related Work
From the literature, it can be observed that deep learning has achieved a series of satisfactory results in the field of medical imaging, and it also has made great progress in the classification of lung nodules. Hua et al. [18] applied CNNs and a deep belief network (DBN) to distinguish malignant from benign lung nodules, and better discriminative results were achieved with deep learning algorithms. A multicrop CNN was developed by Shen et al. [19], and it is able to automatically extract salient nodule information via cropping different regions from convolutional feature maps and applying max-pooling at varying times. Song et al. [20], respectively, adopted the LeNet (4-layer) CNN structure, a deep neural network (DNN), and a stacked autoencoder (SAE) for lung nodule classification. The CNN network achieved the best performance with an accuracy of 84.2% on the LIDC-IDRI lung image dataset. Hussein et al. [21] proposed an end-to-end deep multiview CNN based on the AlexNet (8-layer) network structure and achieved 92.3% classification accuracy of lung nodules on the LIDC-IDRI dataset. Nibali et al. [22] combined a residual network, course learning, and migration learning to propose the ResNet-18 network structure for lung nodule classification. A classification accuracy of 89.9% was obtained on the test samples collected from the LIDC-IDRI dataset.
Traditional computer-aided detection algorithms based on machine learning have the problem of low classification accuracy due to the uncertainty of artificial feature selection. Most classes handled by CNNs are more obvious classes (such as humans and dogs), and there are only subtle differences between the data in the nodule classification task, so there are certain disadvantages in using CNNs for lung nodule classification tasks. The residual network proposed by He et al. [23] in 2015 won as the champion of the ImageNet Large Scale Visual Recognition Competition (ILSVRC) [24]. As a deep learning model, the residual network has been applied successfully in the fields of text classification and image classification. Lan et al. [25] proposed a new network called Residual U-Net (RUN) to perform the lung nodule detection without the selection of nodule candidates. The idea of a residual network was introduced to improve the traditional U-Net, thus solving the shortcomings of poor results from lacking of network depth. Migration learning is also a very useful machine learning method, which means a pretrained model is reapplied to another task. Using the pretrained CNN models on ImageNet for migration learning has become a common method in medical image analysis. van Ginneken et al. [26] extracted the 4096 off-the-shelf features from the first fully connected layer of a pretrained OverFeat model for lung nodule detection in CT images through a linear support vector machine (SVM).
Inspired by the aforementioned work, in this paper, combined with the theories of residual learning and migration learning, a classification method for lung nodules based on a deep residual network is proposed. The 50-layer ResNet network structure is used as the initial model to reconstruct the global average pooling layer, the fully connected layer, and the classification layer. The experimental results on the LIDC-IDRI dataset show that the proposed method has better performance and adaptability than the SVM method, VGG19 model [27], and InceptionV3 model [28] in the classification of lung nodules.
3. Algorithm and Method Framework
In the present work, the basic principle of a residual network is applied to the classification of pulmonary nodules. The proposed classification method of pulmonary nodules based on a deep residual network includes: (1) on the LIDC-IDRI dataset, the nodule contour is extracted according to the labeling of the experienced radiologists to form the experimental dataset; (2) based on the 50-layer residual network and migration learning idea, the original network weight is preserved, and the global average pooling layer, fully connected layer, and classification layer are constructed; (3) the network model is trained using the extracted experimental dataset to complete the classification task of the pulmonary nodules. Figure 1 depicts the pulmonary nodule classification process based on a deep residual network.
Figure 1.
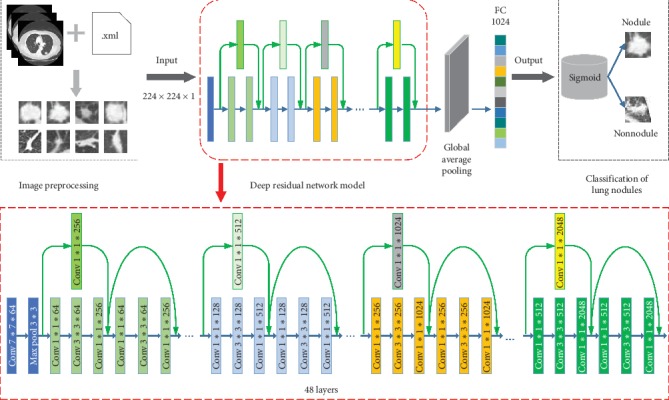
Classification model of pulmonary nodules based on deep residual network.
3.1. Residual Learning
In the process of deep learning, the main problems with an increase in network depth are gradient disappearance and gradient explosion. The traditional solution is the initialization and regularization of data, which solves the gradient problem but results in network performance degradation. The depth has deepened, but the error rate has increased. The purpose of residual learning is to improve the network performance while solving the network gradient problem. If the layers behind the deep network are all identically mapped, the model can be degraded into a shallow network, and then, the problem of network performance degradation caused by the increase in the network depth can be solved.
To achieve identity mapping, only the identity mapping function needs to be used. In residual learning, we design the network as H(x)=F(x)+x, thus, it is converted to learn a residual function F(x)=H(x) − x, and as long as the fitting F(x)=0, it forms an identity map H(x)=x, where x represents the prelayer input, F(x) is the network mapping before the sum, and H(x) is the network mapping from the input to summation. Figure 2 depicts the residual elements in the residual network, which is a basic building block in the residual network.
Figure 2.
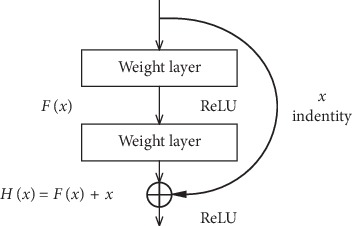
Residual element.
Considering the forward process, the final result represents a direct forward process from the l layer to the L layer and is a continuous operation. The specific calculation process is as follows:
| (1) |
where Wi represents the equivalent mapping method. For residual elements, the forward process is linear, and the subsequent input is equal to the result of the input plus each residual element. The first major feature of the residual network is that the reverse update solves the problem of gradient disappearance. Therefore, when the residual network propagates in the back direction, only the part before the chain law is derived, that is, the gradient from the L layer can be transferred to the l layer stably. The specific derivation process is
| (2) |
Through the structural design of the residual element, the network will avoid the problem of complete gradient disappearance when performing backpropagation training. At the same time, when the performance of the network reaches a bottleneck, the redundant network layer can do identical mapping, which realizes the basic idea of residual learning.
3.2. Deep Residual Network Based Pulmonary Nodule Classification Methodology
As outlined, the lung nodule classification method of deep residual networks proposed in this paper is based on the 50-layer ResNet network, retaining the original weights trained on the ImageNet dataset, removing the original fully connected and classification layers of the network, and adding the global average pooling layer. The global average pooling layer was originally used by Min Lin and Yan in 2014 [29]. They used global average pooling to replace the traditional fully connected layers in the CNN. The idea was to generate one feature map for each corresponding category of the classification task in the last mlpconv layer. They took the average of each feature map, and the resulting vector was fed directly into the softmax layer. Since there are many parameters for the fully connected layer and the fully connected layer are prone to overfitting, thus hampering the generalization ability of the overall network. The number of parameters is reduced greatly after adding the global average pooling layer, which can compress the size of the model well and reduce the occurrence of overfitting. Furthermore, global average pooling sums out the spatial information, thus it is more robust to spatial translations of the input.
After global average pooling layer, we reconstruct the fully connected layer and the classification layer. Since differentiating nodules from nonnodules is a binary classification problem, the sigmoid is used in the classification layer. The ReLU function is employed as an activation function, which makes the feature extraction range of neurons more extensive. The formula is
| (3) |
The design of the deep residual network is embodied by the implementation of the identity mapping in the fast connection mode. The fast connection makes the residual possible, and the identity mapping makes the network deeper. The quick connection can be embodied as our residual network and is composed of multiple stacked layers, each of which uses the residual element as shown in Figure 2. Specifically, the building block of this article is defined as
| (4) |
where x and y represent the input and output vectors and Wi represents the equivalent mapping method. The function F(x, {Wi}) denotes the residual function. As shown in Figure 2, there are two layers, F=W2σ(W1x), in which σ represents ReLU. For simplicity, we temporarily ignore the offset. The operation of F+x is done by a quick connection and an element-by-element addition.
In the design process of this method, the identity maps are implemented by shortcut connections and their outputs are added to the output of the overlay. Shortcut connections neither generate additional parameters nor add computational complexity, and the entire network still uses the backpropagated stochastic gradient descent (SGD) algorithm. At the same time, the residual structures can connect the fully connected layers of the network with the features of each layer of the lung nodule images indirectly, which merge the shallow features and deep features of the image effectively. The residual network can exploit the useful information contained in the images fully, which is expected to improve the accuracy of lung nodule classification [30].
4. Experiment and Result Analysis
4.1. Datasets and Preprocessing
The lung CT images used in our experiment are from the Lung Image Database Consortium collection (LIDC-IDRI) [31–33]. Currently, the LIDC-IDRI dataset is the world's largest public dataset for lung cancer and contains 1,018 cases (a total of 375,590 CT scan images with a scan layer thickness of 1.25 mm∼3 mm and 512 × 512 pixels). For each subject, the nodules are identified by four experienced thoracic radiologists without forced consensus, and the corresponding outline coordinates and characteristic information of the nodules are recorded in an associated XML file. In our experiment, except 8 missing cases and one with damaged images, a total of 1009 instances are adopted to evaluate the effectiveness of the proposed method.
In the process of pulmonary nodule detection, the lung nodule area is initially extracted from the lung CT image. The traditional processing approaches are divided into several procedures such as image preprocessing and image feature extraction. These steps are commonly set by humans, and the result of each step has a direct influence on the subsequent classification performance. Therefore, in the present work, we abandon the traditional method of lung nodule extraction, and according to the radiologist's annotations of the pulmonary nodules, the pulmonary nodule area is extracted directly as the experimental dataset. The size of the lung nodules is in the range of 3 mm∼30 mm, and the nodule area on each CT image is marked by the radiologists. According to the nodule contour coordinate information marked by the radiologist in the associated XML file, the rectangular region of the nodule can be segmented from the CT images, as shown in Figure 3. In addition, the nonnodules ≥3 mm are extracted based on their centroids annotated by the radiologists.
Figure 3.
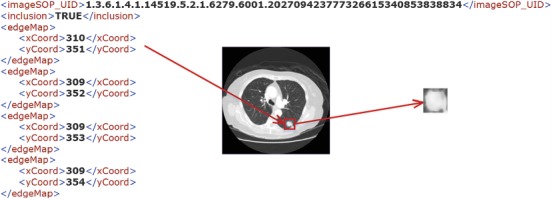
Illustration of extracting lung nodule region from CT images.
In our experiment, a total of 14995 CT slices are collected from 1009 instances in terms of the radiologist's annotations, including 7685 nodule slices and 7310 nonnodule slices. The nodule and nonnodule samples are shown in Figure 4.
Figure 4.
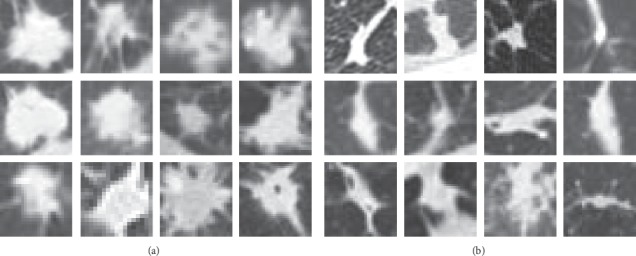
Illustration of extracting lung nodule region from CT images: (a) nodule samples. (b) nonnodule samples.
4.2. Experimental Setup
In order to validate the effectiveness of the network model, the accuracy, precision, specificity, recall, f1-score, false positive rate, and receiver operating characteristic (ROC) curve of the classification of pulmonary nodules are used to evaluate the performance of the algorithm in this paper. All the possible outcomes of a test procedure and the gold standard are listed in Table 1. Table 2 details the formulas to calculate the aforementioned evaluation indexes. Additionally, a statistical significance test is performed to observe the performance differences of different approaches.
Table 1.
All possible outcomes of a test.
| Test result | Gold standard | |
|---|---|---|
| Positive | Negative | |
| Positive | True positive (TP) | False positive (FP) |
| Negative | False negative (FN) | True negative (TN) |
| Total | TP + FN | FP + TN |
Table 2.
Indicators for evaluating algorithm performance.
| Evaluation criteria | Calculation method |
|---|---|
| Accuracy | Accuracy = (TP + TN)/(TP + TN + FP + FN) |
| Precision | Precision = TP/(TP + FP) |
| Specificity | Specificity = TN/(TN + FP) |
| False positive rate | FPR = FP/(FP + TN) |
| Recall | Recall = TP/(TP + FN) |
| F1-score | F1-score = 2 × precision × recall/(precision + recall) |
All the experiments were evaluated using ten-fold cross-validation, and the validation dataset and test dataset were swapped to repeat the experiment. The average result is taken as the final experiment result. The data volume of the nodules and nonnodules is equal basically to confirm the effectiveness of the network. These two hyperparameters of the optimizer SGD method are set to: learning rate (LR) = 0.0001 and momentum = 0.9 [34]. Because the model requires that the input size of the image is 224 × 224 × 1, the extracted nodule images are resized to the same size before being input into our model.
In order to show the effectiveness of our proposed approach, we compare the lung nodule classification performances with different models, namely, the traditional machine learning model and the pretrained CNN models. Specifically, the SVM classification model with curvelet transform features, the VGG19 model and the InceptionV3 model are designed as the comparison algorithms. Among them, the curvelet transform is a multiscale, directional feature extraction method. It has obvious advantages in the description of the contour and texture direction of an image. The fast discrete curvelet transform includes two algorithms. One is the unequispaced FFT transform, in which the curvelet coefficients are found by irregularly sampling the Fourier coefficients of an image. Another one is the Wrapping transform, using a series of translations and a wraparound technique. The unequispaced FFT transform was used in the present work. The first layer (low frequency coefficients) of the curvelet transform coefficients mainly contains the energy of the image and the contour characteristics. The higher frequency coefficients correspond to the image edge and details information. The medium high-level coefficients also describe the edge features. We concatenated all layer coefficients, whose dimension is 4096, as the feature vector. Then, PCA technique was used on it to reduce the feature size by selecting 100 components which explained >95% variances of the original features. The radial basis kernel function is used in the SVM classification method. The penalty parameter c=1 of the error term, and the kernel parameter γ=0.5. The VGG19 model and the InceptionV3 model contain 16 convolutional layers and 47 convolutional layers, respectively. Detailed configurations of the deep residual network, VGG19 model, and InceptionV3 model are listed in Table 3. The global average pooling layer is also employed in VGG19 and InceptionV3 models. Then, the fully connected layer and the sigmoid classification layer are followed. The collected dataset is used to fine-tune these networks. The same training dataset, verification dataset, and test dataset are used in all four models.
Table 3.
Parameter configuration of the deep residual network, VGG19 model, and InceptionV3 model.
| Deep residual network | VGG19 | InceptionV3 | |
|---|---|---|---|
| Input: nodule/nonnodule images | |||
| conv1 7 × 7, 64 | 2 × conv3-64 | conv3-32 | |
| conv3-32 | |||
| conv3-64 | |||
| max pool | max pool | max pool | |
| conv2_x | 2 × conv3-128 | conv1-80 | |
| conv3-192 | |||
| max pool | max pool | ||
| conv3_x | 4 × conv3-256 | block1 | module1 ⟶ concat |
| module2 ⟶ concat | |||
| max pool | module3 ⟶ concat | ||
| conv4_x | 4 × conv3-512 | block2 | module1 ⟶ concat |
| module2 ⟶ concat | |||
| module3 ⟶ concat | |||
| module4 ⟶ concat | |||
| max pool | module5 ⟶ concat | ||
| conv5_x | 4 × conv3-512 | block3 | module1 ⟶ concat |
| module2 ⟶ concat | |||
| max pool | module3 ⟶ concat | ||
| Global average pooling2D | |||
| Fully connected layer-1024 | |||
| Fully connected layer-2 | |||
| Output: sigmoid | |||
4.3. Experimental Results and Analysis
During the training process of the neural network, when all the training datasets are used to train the network once, it is called one epoch. In order to observe the performance relationships between the classification accuracy of the test dataset and the training times, we drew the change curve of the classification accuracy with the increase of the epoch at an interval of 1. Figure 5 shows the change curve of the classification accuracy with the increase of the epoch.
Figure 5.
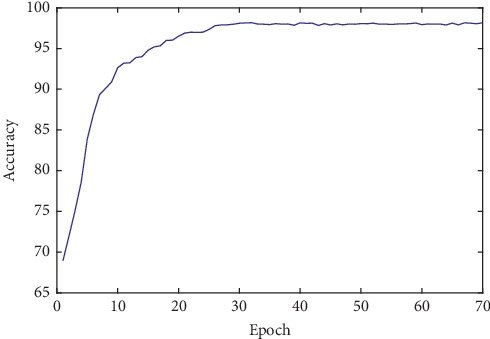
Influence of the classification accuracy with the increase of epoch.
It can be seen from Figure 5 that accuracy increases with the gradual increase of the epoch. When the epoch reaches 30, the highest accuracy is obtained. As the epoch continues to increase, the accuracy rate remains relatively stable. In line with the experimental setup, the SVM algorithm with curvelet transform features, the VGG19 model, and the InceptionV3 model is compared with the proposed method. A comparison of the classification results of the lung nodules using different methods is shown in Table 4.
Table 4.
Comparison of the classification results of lung nodules with different methods.
| Methods | Accuracy (%) | Precision (%) | Recall (%) | Specificity (%) | F1-score (%) | FPR (%) |
|---|---|---|---|---|---|---|
| Curvelet + SVM | 88.27 | 90.12 | 87.69 | 85.94 | 88.89 | 8.60 |
| VGG19 | 96.48 | 97.10 | 95.17 | 96.83 | 96.13 | 3.72 |
| InceptionV3 | 95.81 | 96.35 | 95.30 | 95.76 | 95.85 | 3.87 |
| Deep residual network | 98.23 | 98.46 | 97.70 | 98.35 | 98.06 | 1.65 |
Table 4 shows the results of the four methods. As mentioned, the results are obtained based on ten-fold cross-validation. The traditional machine learning reference method, the SVM algorithm, achieves an accuracy of 88.27%, precision of 90.12%, recall of 87.69%, specificity of 85.94%, f1-score of 88.89%, and a false positive rate of 8.60%. The deep learning method, the VGG19 model, achieves an accuracy of 96.48%, precision of 97.10%, recall of 95.17%, specificity of 96.83%, f1-score of 96.13%, and a false positive rate of 3.72%. InceptionV3 achieves an accuracy of 95.81%, precision of 96.35%, recall of 95.30%, specificity of 95.76%, f1-score of 95.85%, and a false positive rate of 3.87%. The lung nodule classification method based on the proposed deep residual network achieves an accuracy of 98.23%, precision of 98.46%, recall of 97.70%, specificity of 98.35%, f1-score of 98.06%, and a false positive rate of 1.65% in our study. Our proposed method outperforms the SVM algorithm by 9.96%, 8.34%, 10.01%, 12.41%, and 9.17% in terms of accuracy, precision, recall, specificity, and f1-score, and the false positive rate is decreased by 6.95%. Our method outperforms the VGG19 model by 1.75%, 1.36%, 2.53%, 1.52%, and 1.93% in terms of accuracy, precision, recall, specificity, and f1-score, and the false positive rate decreased by 2.07%. Furthermore, our method improved by 2.42%, 2.11%, 2.40%, 2.59%, and 2.21% in terms of accuracy, precision, recall, specificity, and f1-score, respectively, compared to the InceptionV3 model, and the false positive rate decreased by 2.22%. This represents a significant improvement over the SVM model (p < 0.01 in a one-tailed z-test), VGG19 model (p < o.01 in a one-tailed z-test), and InceptionV3 model (p < 0.01 in a one-tailed z-test).
The ROC curve and the area under the ROC curve (AUC) are important indicators for evaluating the performance of the algorithm. The closer the ROC curve is to the upper left, the closer the AUC value is to 1, indicating that the classification result of the algorithm is better. In order to facilitate an intuitive comparison, the ROC curves of the different methods are drawn in a unified coordinate graph, as shown in Figure 6.
Figure 6.
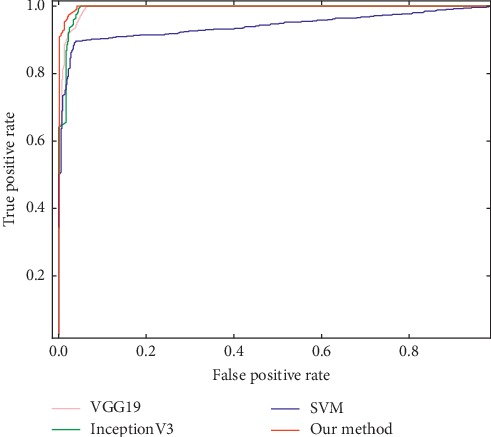
ROC curves of different classification methods.
As can be seen in Figure 6, the ROC curve of our method is closer to the upper left of the graph than the other three methods. Comparable results are achieved by our method, the VGG19 model, and the InceptionV3 model upon the AUC value. A relatively lower result is got by the SVM model. The corresponding AUC values are our method of 0.9971, VGG19 of 0.9935, InceptionV3 of 0.9924, and SVM of 0.9414, respectively. Again, this represents a significant improvement over the SVM (p < 0.01 in a one-tailed z-test) and InceptionV3 model (p < 0.05 in a one-tailed z-test). Although no significant performance differences (p < 0.05) are observed between our method and VGG19 model, it is evident that our method is superior to VGG19 model (p < 0.01 in a one-tailed z-test) in terms of the ROC curve and other evaluation criteria mentioned above. Overall, better results are obtained with our method, which further verifies the feasibility and validity of the proposed method.
Given these experimental results, we can see that the proposed residual network achieves better classification results. The accuracy, precision, recall, f1-score, false positive rate, and AUC values of lung nodule classification are higher than that of the SVM method, the VGG19 model, and the InceptionV3 model. In addition, we compare the experimental results of our method with that of several representative neural network models in the work presented in [20–22]. However, it is difficult to make an objective comparison with the previously published literature due to the variability in the dataset and different validation methods. Nevertheless, it is still important to attempt a relative comparison. For this purpose, we identified several representative methods that have used the same dataset (LIDC-IDRI), employed the same validation procedure (ten-fold cross-validation), and reported better results. Thus, we compared our experimental results with those of literatures [20–22]. 4-layer and 8-layer convolutional neural networks are adopted in [20,21], respectively. Both of them use the ten-fold cross-validation method. An 18-layer ResNet network is adopted in [22], and the literature also uses the randomized splits to evaluate the accuracy of models. A comparison of the classification results of lung nodules in different studies is shown in Table 5.
Table 5.
Comparison of classification results of lung nodules in the literature.
| Methods | Network layer | Accuracy (%) | Recall (sensitivity)(%) | Specificity (%) |
|---|---|---|---|---|
| CNN [20] | 4 | 84.20 | 84.00 | 84.30 |
| High-level attributes + CNN [21] | 8 | 92.30 | — | — |
| ResNet [22] | 18 | 89.90 | 91.10 | 88.60 |
| Deep residual network | 50 | 98.23 | 97.70 | 98.35 |
Note:“—” in the table indicates no data.
From Table 5, we can observe that our network has the best classification performance, with an accuracy of 98.23%, sensitivity of 97.70%, and specificity of 98.35%. The accuracy, sensitivity, and specificity of our method are higher than those of the other three methods. Furthermore, the traditional machine learning method needs different feature extraction methods to select and extract the different features of lung nodules, which increases the complexity and error rate of the operation. Furthermore, it is difficult to reach a deep layer for traditional neural networks due to the problem of gradient disappearance. However, for our proposed method, because the ResNet network applies the theory of residual learning, the problem of network performance degradation caused by the increase of network depth is settled. Our method can learn the characteristics of lung nodule images automatically, and the number of network layers can reach a certain depth, which has better adaptability to lung nodule classification tasks.
5. Conclusion
To address the problems of lung nodule classification, such as a complex classification detection process, low classification accuracy, and high false positive rate, combining the theories of deep learning, migration learning, and residual learning, a novel neural network model for lung nodule classification is proposed. The model is based on a 50-layer residual network model framework, reconstructing the global average pooling layer, the fully connected layer, and the classification layer. The lung nodule image can be used as the input data of the network directly, avoiding complicated feature extraction and selection. The experiment results on the LIDC-IDRI dataset show that the accuracy, precision, specificity, recall, f1-score, false positive rate, and ROC curves of our method outperform the reported results of all the other methods mentioned in this paper, including the neural network models and a traditional machine learning algorithm. This study demonstrates the superior performance of the proposed method in lung nodule classification, which might have the potential to provide a reference for clinical diagnosis.
Although the residual network structure proposed in this paper has better performance in the classification task of lung nodules, it also has a deficiency, this being that a long training time is needed when dealing with a large number of lung CT images. Therefore, it is necessary to further optimize the network model in follow-up work.
Acknowledgments
This work was funded by the National Natural Science Foundation of China (grant nos. 61902282 and 61702370), the Key Program of the Natural Science Foundation of Tianjin (grant no. 18JCZDJC36300), the Technology Plan of Tianjin (grant no: 18ZXRHSY00100), the Open Projects Program of the National Laboratory of Pattern Recognition, Doctoral Foundation of Tianjin Normal University (grant no. 043135202-XB1707), and the Tianjin Municipal Education Commission Project for Scientific Research Plan (grant no. 2018KJ155). This project has also received funding from the European Union's Horizon 2020 Research and Innovation Programme under grant agreement no. 826506 (sustAGE). The authors acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health and their critical role in the creation of the free publicly available LIDC/IDRI database used in this study.
Contributor Information
Ziping Zhao, Email: zhaoziping@tjnu.edu.cn.
Haishuai Wang, Email: haishuai.wang@gmail.com.
Data Availability
Copies of the Lung Image Database Consortium data can be obtained free of charge from https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI/.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Panpan Wu and Xuanchao Sun contributed equally to this work.
References
- 1.Monkam P., Qi S., Xu M., Han F., Zhao X., Qian W. CNN models discriminating between pulmonary micro-nodules and non-odules from CT images. BioMedical Engineering OnLine. 2018;17:p. 16. doi: 10.1186/s12938-018-0529-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sathiya T., Sathiyabhama B. Fuzzy relevance vector machine based classification of lung nodules in computed tomography images. International Journal of Imaging Systems and Technology. 2019;29(3):360–373. doi: 10.1002/ima.22339. [DOI] [Google Scholar]
- 3.Manickavasagam R., Selvan S. Automatic detection and classification of lung nodules in CT image using optimized neuro fuzzy classifier with cuckoo search algorithm. Journal of Medical Systems. 2019;43:p. 9. doi: 10.1007/s10916-019-1177-9. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs C., van Rikxoort E. M., Murphy K., Prokop M., Schaefer-Prokop C. M., van Ginneken B. Computer-aided detection of pulmonary nodules: a comparative study using the public LIDC/IDRI database. European Radiology. 2016;26(7):2139–2147. doi: 10.1007/s00330-015-4030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao X., Liu L., Qi S., Teng Y., Li J., Qian W. Agile convolutional neural network for pulmonary nodule classification using CT images. International Journal of Computer Assisted Radiology and Surgery. 2018;13:1–11. doi: 10.1007/s11548-017-1696-0. [DOI] [PubMed] [Google Scholar]
- 6.Rendon-Gonzalez E., Ponomaryov V. Automatic lung nodule segmentation and classification in CT images based on SVM. Proceedings of the 2016 9th International Kharkiv Symposium on Physics and Engineering of Microwaves, Millimeter and Submillimeter Waves (MSMW); June 2016; Kharkiv, Ukraine. pp. 1–4. [Google Scholar]
- 7.Li S., Xu P., Li B., et al. Predicting lung nodule malignancies by combining deep convolutional neural network and handcrafted features. Physics in Medicine and Biology. 2019;64(17):p. 25. doi: 10.1088/1361-6560/ab326a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farag A. A., Ali A. M., Falk R., Farag A. A., Graham J., Elshazly S. Evaluation of geometric feature descriptors for detection and classification of lung nodules in low dose CT scans of the chest. Proceedings of the 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro; March-April 2011; Chicago, IL, USA. pp. 169–172. [DOI] [Google Scholar]
- 9.Wei G., Ma H., Qian W., et al. Lung nodule classification using local kernel regression models with out-of-sample extension. Biomedical Signal Processing and Control. 2018;40:1–9. doi: 10.1016/j.bspc.2017.08.026. [DOI] [Google Scholar]
- 10.Yang S., Cai W., Zhou Y., Feng D. Feature-based image patch approximation for lung tissue classification. IEEE Transactions on Medical Imaging. 2013;32:797–808. doi: 10.1109/TMI.2013.2241448. [DOI] [PubMed] [Google Scholar]
- 11.Manikandan T., Bharathi N. Lung cancer detection using fuzzy auto-seed cluster means morphological segmentation and SVM classifier. Journal of Medical Systems. 2016;40:181–189. doi: 10.1007/s10916-016-0539-9. [DOI] [PubMed] [Google Scholar]
- 12.Kim B. C., Sung Y. S., Suk H. I. Deep feature learning for pulmonary nodule classification in a lung CT. Proceedings of the 4th International Winter Conference on Brain-Computer Interface (BCI); February 2016; Yongpyong, South Korea. p. p. 4. [DOI] [Google Scholar]
- 13.Setio A. A. A., Ciompi F., Litjens G., et al. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Transactions on Medical Imaging. 2016;35(5):1160–1169. doi: 10.1109/tmi.2016.2536809. [DOI] [PubMed] [Google Scholar]
- 14.Nobrega R. V. M. D., Peixoto S. A., Silva S. P. P. D., Filho P. P. R. Lung nodule classification via deep transfer learning in CT lung images. Proceedings of the 31st International Symposium on Computer-Based Medical Systems (CBMS); June 2018; Karlstad, Sweden. p. p. 6. [DOI] [Google Scholar]
- 15.Ronneberger O., Fischer P., Brox T. U-Net: convolutional networks for biomedical image segmentation. Proceedings of the 18th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI); October 2015; Munich, Germany. pp. 234–241. [Google Scholar]
- 16.Gao X., Li W., Loomes M., Wang L. A fused deep learning architecture for viewpoint classification of echocardiography. Information Fusion. 2017;36:103–113. doi: 10.1016/j.inffus.2016.11.007. [DOI] [Google Scholar]
- 17.Litjens G., Kooi T., Bejnordi B. E., et al. A survey on deep learning in medical image analysis. Medical Image Analysis. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Hua K.-L., Hsu C. H., Hidayati S. C., Cheng W., Chen Y. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets and Therapy. 2015;8:2015–2022. doi: 10.2147/OTT.S80733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen W., Zhou M., Yang F., et al. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognition. 2017;61:663–673. doi: 10.1016/j.patcog.2016.05.029. [DOI] [Google Scholar]
- 20.Song Q., Zhao L., Luo X., Dou X. Using deep learning for classification of lung nodules on computed tomography images. Journal of Healthcare Engineering. August 2017;2017:p. 7. doi: 10.1155/2017/8314740.8314740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hussein S., Gillies R., Cao K., Song Q., Bagci U. Tumornet: lung nodule characterization using multi-view convolutional neural network with Gaussian process. Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017); April 2017; Melbourne, Australia. pp. 1007–1010. [DOI] [Google Scholar]
- 22.Nibali A., He Z., Wollersheim D. Pulmonary nodule classification with deep residual networks. International Journal of Computer Assisted Radiology and Surgery. 2017;12:1799–1808. doi: 10.1007/s11548-017-1605-6. [DOI] [PubMed] [Google Scholar]
- 23.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); June 2016; Las Vegas, NV, USA. pp. 770–778. [DOI] [Google Scholar]
- 24.Russakovsky O., Deng J., Su H., et al. Imagenet large scale visual recognition challenge. International Journal of Computer Vision. 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 25.Lan T., Li Y., Murugi J. K., Ding Y., Qin Z. Run: residual U-Net for computer-aided detection of pulmonary nodules without candidate selection. 2018. https://arxiv.org/abs/1805.11856.
- 26.van Ginneken B., Setio A. A. A., Jacobs C., Ciompi F. Off-the-shelf convolutional neural network features for pulmonary nodule detection in computed tomography scans. Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI); April 2015; New York, NY, USA. pp. 286–289. [DOI] [Google Scholar]
- 27.Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. Proceedings of the 3rd International Conference on Learning Representations (ICLR); 2015; San Diego, CA, USA. p. p. 14. [Google Scholar]
- 28.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Rethinking the inception architecture for computer vision. Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016; Las Vegas, NV, USA. pp. 2818–2826. [Google Scholar]
- 29.Min Lin Q. C., Yan S. Network in network. Proceedings of the 2nd International Conference on Learning Representations (ICLR); 2014; Banff, Canada. p. p. 10. [Google Scholar]
- 30.Huang G., Liu Z., van der Maaten L., Weinberger K. Densely connected convolutional networks. Proceedkngs of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017; Honolulu, HI, USA. p. p. 9. [Google Scholar]
- 31. https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI/
- 32.Armato S. I., McLennan G., Bidaut L., et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Medical Physics. 2011;38:915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark K., Vendt B., Smith K., et al. The cancer imaging archive (TCIA): maintaining and operating a public information repository. Journal of Digital Imaging. 2013;26(6):1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutskever I., Martens J., Dahl G., Hinton G. On the importance of initialization and momentum in deep learning. Proceedings of the 30th International Conference on Machine Learning (ICML); 2013; Atlanta, GA, USA. pp. 1139–1147. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Copies of the Lung Image Database Consortium data can be obtained free of charge from https://wiki.cancerimagingarchive.net/display/Public/LIDC-IDRI/.


