Significance
Shell biorefinery is an emerging concept that upcycles the major component of crustacean shell waste and the like (chitin-rich) into value-added chemicals and materials. To date, the utilization of waste-derived chitin as carbon and nitrogen sources to produce value-added nitrogen-containing chemicals (NCCs) via microbial fermentation has been underexplored. The lack of a compatible pretreatment method in conjunction with a customized NCC-producing strain has placed a major impediment to the valorization effort. This study tackles this challenge by developing an integrated biorefinery process that combines an efficient pretreatment process with a highly customizable microbial engineering platform, opening a door for the better use of crustacean shell waste-derived chitin as a substrate for microbial manufacturing.
Keywords: shrimp shell waste, chitin, metabolic engineering, renewable nitrogenous material, integrated process
Abstract
Chitin is the most abundant renewable nitrogenous material on earth and is accessible to humans in the form of crustacean shell waste. Such waste has been severely underutilized, resulting in both resource wastage and disposal issues. Upcycling chitin-containing waste into value-added products is an attractive solution. However, the direct conversion of crustacean shell waste-derived chitin into a wide spectrum of nitrogen-containing chemicals (NCCs) is challenging via conventional catalytic processes. To address this challenge, in this study, we developed an integrated biorefinery process to upgrade shell waste-derived chitin into two aromatic NCCs that currently cannot be synthesized from chitin via any chemical process (tyrosine and l-DOPA). The process involves a pretreatment of chitin-containing shell waste and an enzymatic/fermentative bioprocess using metabolically engineered Escherichia coli. The pretreatment step achieved an almost 100% recovery and partial depolymerization of chitin from shrimp shell waste (SSW), thereby offering water-soluble chitin hydrolysates for the downstream microbial process under mild conditions. The engineered E. coli strains produced 0.91 g/L tyrosine or 0.41 g/L l-DOPA from 22.5 g/L unpurified SSW-derived chitin hydrolysates, demonstrating the feasibility of upcycling renewable chitin-containing waste into value-added NCCs via this integrated biorefinery, which bypassed the Haber–Bosch process in providing a nitrogen source.
Utilization of renewable feedstock to produce food additives, biofuels, and drugs plays a pivotal role in the chemical industry for the future (1, 2). In particular, the development of sustainable protocols to prepare nitrogen-containing chemicals (NCCs), including a large sum of valuable compounds widely used in pharmaceuticals and beyond, are crucial to sustain a high standard modern lifestyle. Distinct from lignocellulosic biomass that is only enriched with H, C, and O elements, aminopolysaccharides (e.g., chitin) widely found in crustacean species, exoskeleton of insects, and cell wall of fungi contain an additional N element that is more abundant than the fixed N2 in the Haber–Bosch process on earth. Unfortunately, chitin-containing materials at present have been mainly aggregated in the form of waste, such as shellfish waste in the seafood industry. With the utilization of crustacean waste, corresponding to ∼6 to 8 million tons worldwide per year, it is indispensable to hinder odor formation and water pollution caused by improper disposal in coastal regions and to comply with increasingly stringent environmental regulations (3, 4). This crustacean shell waste is cheap feedstock (0.1 to 0.12 US dollars [USD] per kilogram) if it can be upcycled economically into useful chemicals (1, 2).
Crustacean shell waste is composed of 20 to 50 wt% calcium carbonate (CaCO3), 20 to 40 wt% protein, and 15 to 40 wt% chitin (1). Chitin is a linear polymer of N-acetylglucosamine (GlcNAc) linked with β (1 → 4)-glycosidic bonds (5), and is particularly of interest as it is the most abundant natural nitrogen-containing polymer. One-pot catalytic conversion of renewable chitin derived from crustacean shell waste into value-added NCCs has gained considerable attention as the industrial production of NCCs needs nonrenewable fossil fuels and energy-consuming processes (1). However, only a few catalytic processes have been established in several niche areas of industrial chemistry (7). So far, these catalytic processes can only produce a relatively narrow spectrum of products bearing high structural similarity. Harsh and/or energy-intensive conditions such as high temperature and use of concentrated acids are also often required (6). For example, amino sugars, such as hydroxyethyl-2-amino-2-deoxyhexopyranoside and hydroxyethyl-2-acetamido-2-deoxyhexopyranoside, were produced from chitin with a 30 wt% yield at 165 °C with 8 wt% acid as a catalyst (7). N-containing furan derivative with a yield of 7.5 wt% was produced from chitin in a one-step reaction using boric acid at 215 °C (8). The most dominant products from chitin are GlcNAc and glucosamine. Notably, the conventional chemical method to decompose chitin to GlcNAc has typically been carried out by using concentrated HCl (9, 10), which leads to obligated usage of corrosion-proof equipment and generation of a large amount of acidic wastewater (11). Although using cosolvent improved hydrolysis efficiency, the solvent cannot be completely recycled and still incurs high operating costs. Mechanical force-assisted methods (e.g., ball milling) depolymerize H2SO4-impregnated chitin to water-soluble GlcNAc oligomers, and the oligomers can be hydrolyzed into GlcNAc (53% yield) at 170 °C, retaining the acetyl group without additional acid (12). This method represents a more environmentally friendly pretreatment of shell waste for downstream processing.
Microbial fermentation offers a sustainable way to valorize crustacean shell waste-derived chitin into diverse valuable natural NCCs (e.g., amino acids, flavonoids, and alkaloids), which are structurally complex and are difficult to synthesize through one-pot multiple-step catalytic processes. The use of readily accessible chitin from crustacean shell waste as the substrate not only provides carbon but is also a nitrogen source for the formation of cell biomass and NCCs. Complete removal of the protein from shell waste and purification of GlcNAc from depolymerized chitin may not be necessary as the engineered microbe should be able to assimilate GlcNAc and its oligomers in the presence of residual protein and/or its hydrolysates. The pretreatment of crustacean shell waste would only need to remove calcium carbonate and to depolymerize chitin, leading to a mild and energy-efficient process.
Based on the above descriptions, we need to take the following constraints into account to design a cost-effective pretreatment process of crustacean shell waste for downstream microbial assimilation: 1) avoid using concentrated acid/alkali for demineralization and/or deproteinization to lower the capital investment; 2) avoid using a costly cosolvent system for the hydrolysis of chitin; 3) avoid using harsh pretreatment conditions (e.g., high temperature) to prevent degradation of GlcNAc and to lower the operating cost; 4) achieve high and fast recovery of chitin under mild processing conditions from shell waste after demineralization and depolymerization; 5) direct use of unpurified, depolymerized chitin-containing shell waste hydrolysates for microbial fermentation.
Incorporating enzymatic hydrolysis may help develop the process that meets the above criteria. In nature, chitin in crustacean shell waste is enzymatically hydrolyzed by microbes into GlcNAc and its oligomers, thereby providing carbon and nitrogen sources to the microbes. Serratia marcescens has been well studied for degradation of chitin in fungal cell walls as it secretes a large amount of exochitinases, endochitinase, and chitin-active lytic polysaccharide monooxygenase (13–15). These enzymes work simultaneously to degrade chitin into GlcNAc and its oligomers. The use of chitin as the sole carbon and nitrogen sources by Chromobacterium violaceum has also been reported (16). Yet, metabolic engineering of these wild-type microbes to produce value-added NCCs from crustacean shell waste has not yet been achieved due to their slow growth and lack of well-developed genetic tools. As a widely used microbial host in the biotechnological field, Escherichia coli has been designed to produce a variety of natural NCCs (17, 18). E. coli cannot utilize chitin because of its inability to synthesize the enzymes to degrade chitin into soluble and low molecular-weight GlcNAc oligomers, although E. coli was reported to grow on GlcNAc and its dimer (19, 20). Adding chitinase during the fermentation process should be helpful if it depolymerizes larger oligomers in chitin hydrolysates into GlcNAc or (GlcNAc)2. The enzymatic process would not extend the microbial fermentation process time, as the enzymatic hydrolysis of GlcNAc oligomers and the microbial fermentation occur simultaneously at comparable rates.
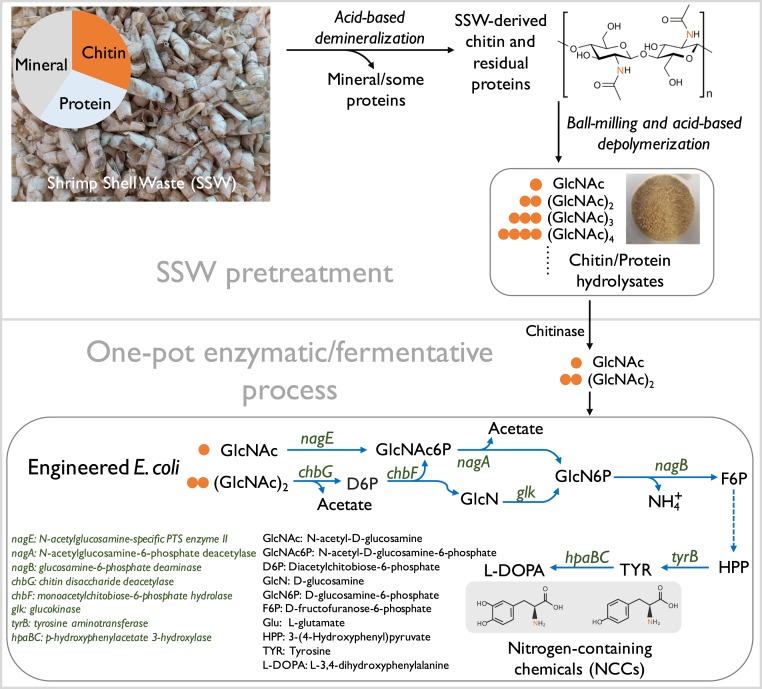
In this study, we developed an integrated biorefinery process to upgrade shrimp shell waste (SSW). The process (Fig. 1) is divided into two stages: 1) pretreatment of SSW; and 2) a one-pot enzymatic/fermentative process. The engineered E. coli strains were able to produce two valuable NCCs, namely tyrosine and l-DOPA, by using unpurified SSW-derived chitin hydrolysates as the sole carbon and nitrogen sources. This integrated biorefinery process links a simple pretreatment of SSW to precisely tailored E. coli strains, thus unlocking an avenue for the valorization of renewable chitin-containing waste into value-added NCCs.
Fig. 1.
An integrated shell biorefinery process for upcycling SSW-derived chitin into NCCs. During pretreatment of SSW into water-soluble chitin hydrolysates, SSW was first demineralized by using diluted acid, which partially removed proteins. The SSW-derived chitin was further depolymerized into water-soluble GlcNAc and its oligomers via a ball-milling H2SO4-catalyzed process, generating chitin hydrolysates mixed with protein hydrolysates. A one-pot enzymatic/fermentative process was performed to produce tyrosine and/or l-DOPA by using engineered E. coli from the unpurified SSW-derived hydrolysates as the sole carbon and nitrogen source. The biosynthetic pathway of tyrosine and l-DOPA from GlcNAc and (GlcNAc)2 is shown. Solid blue lines with arrowhead indicate a one-step enzymatic reaction, while a multiple-step pathway is indicated by the dotted blue line with arrowhead. To enzymatically convert GlcNAc oligomers (n ≥ 4) to fermentable GlcNAc, (GlcNAc)2 or (GlcNAc)3, a small amount of chitinase was added into the culture medium upon inoculation. Gene hapBC coding p-hydroxyphenylacetate 3-hydroxylase was from E. coli W strain (GenBank no. WP_000801473.1).
Results and Discussion
SSW Demineralization and Depolymerization.
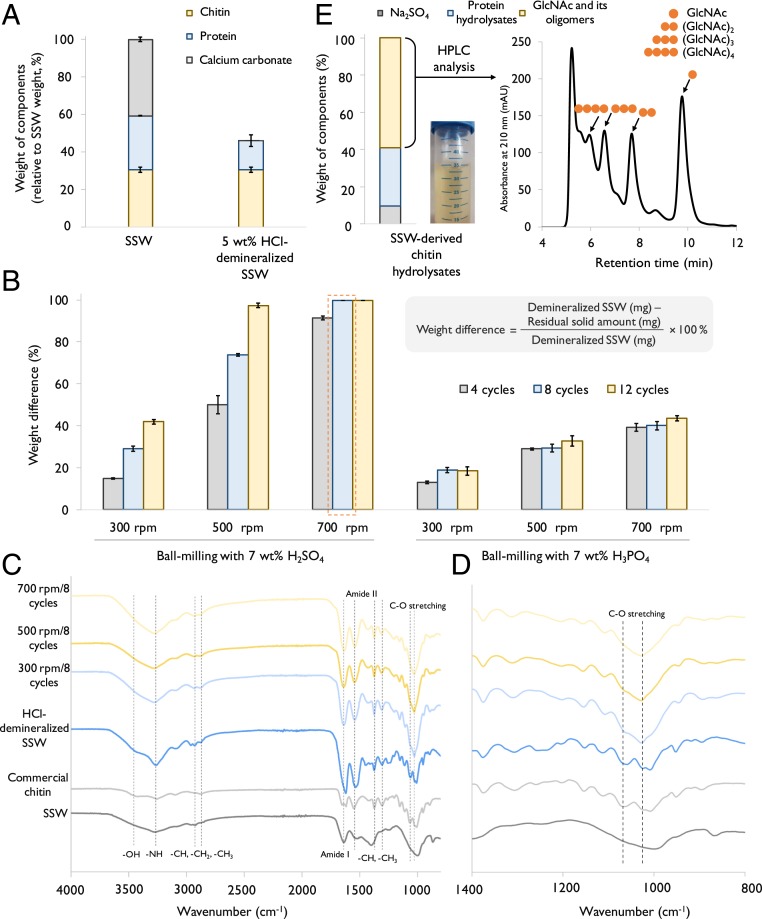
We first analyzed the weight composition of SSW obtained from a local source based on an acid/alkali method (21). The results show that the SSW contained 40.6 wt% of calcium carbonate (CaCO3), 28.7 wt% of protein, and 30.7 wt% of chitin (Fig. 2A). To demineralize SSW, a modified acid-based pretreatment (Fig. 1) was performed to fractionate CaCO3 by using 5.0 wt% of HCl solution according to a previously reported method (21). Based on weight closure, CaCO3 was completely dissolved in an acidic medium and then separated from solid products of SSW. The partial deproteinization also took place concurrently, resulting in 53.9 wt% of protein remaining in the solid demineralized SSW (Fig. 2A).
Fig. 2.
Pretreatment of SSW into water-soluble chitin hydrolysates. (A) Weight of components of SSW before and after HCl demineralization. (B) Optimization of the ball-milling acid-catalyzed hydrolysis process. Cycling number and rotation speed of ball milling were tested systematically with 7.0 wt% of H2SO4 or H3PO4. (C) ATR-IR spectra of solid samples after mechanocatalytic depolymerization with H2SO4 and of control samples. (D) Zoomed in view of C in the range of 800 to 1,400 cm−1. The peaks between 2,875 and 2,935 cm−1 are attributed to −CH stretching, symmetric –CH3 stretching, and asymmetric −CH2 stretching. Amide I and amide II peaks appear at 1,622 to 1,656 cm−1 and 1,552 cm−1, respectively. The peaks ranging from 1,312 to 1,380 cm−1 represent amide III, −CH2 wagging, −CH bending, and symmetric −CH3 deformation. (E) Composition of SSW-derived chitin hydrolysates after ball-milling H2SO4-catalyzed hydrolysis. The SSW-derived chitin hydrolysates were neutralized with sodium hydroxide solution and analyzed by HPLC. Error bars indicate SE (n ≥ 3).
Subsequently, a ball milling-based depolymerization of pretreated SSW was carried out to produce water-soluble GlcNAc and its oligomers for the subsequent enzymatic/fermentative processes (Fig. 1). It is known that an autogenous heat formation (22) during ball milling led to enhancement of organic biopolymers’ reactivity, thereby avoiding the use of harsh reaction media (e.g., cosolvent) owing to the altering of their robust crystalline structures (23). We tuned ball-milling operating parameters, including rotation speed, milling cycle number, and the type of acids (7.0 wt% of H2SO4 or H3PO4) to improve the depolymerization. The yield of the water-soluble products was calculated based on the weight difference before and after ball milling (Fig. 2B). In the case of H2SO4 treatment, the milling cycle number was a crucial factor for improving the yield of water-soluble chitin hydrolysates at low-speed rotations (300 and 500 rpm)—increasing the cycle number from 4 to 12 improved the yield approximately threefold. At 700 rpm, the yield was >90% regardless of milling duration. The rotation speed also had profound influence on the generation of the water-soluble product, and the yields were 29.1%, 73.9%, and 99.8% at 300, 500, and 700 rpm for eight cycles, respectively (Fig. 2B). Apart from H2SO4, we tested the performance of H3PO4 but obtained much worse results (Fig. 2B), which may be partly explained by the encapsulation of more water in H3PO4. It is known that water is able to plasticize biopolymers by reducing the friction and the tensile stress (24). The difference could also be caused by their large difference in pKa (H2SO4: −10; H3PO4: 2.16).
Solid SSW-derived chitin products after ball milling were characterized by attenuated total reflection infrared (ATR-IR) spectroscopy, X-ray diffraction (XRD) analysis, field emission scanning electron microscopy, and solid-state 13C-NMR. The following control samples were included: commercial chitin, untreated SSW, and HCl-demineralized SSW. As shown in the ATR-IR spectrum (Fig. 2 C and D and SI Appendix, Fig. S1), the peaks at 3,261 cm−1 and 3,429 cm−1 are assigned to −NH and −OH stretching, whose changes demonstrate the alteration of hydrogen bonding in the chitin structure (8). The most distinct change was observed for C-O stretching bonds (at 1,029 to 1,068 cm−1), which indicated the existence of glycosidic bonds between GlcNAc units (25). While the peak at 1,068 cm−1 was fairly visible for commercial chitin and HCl-demineralized SSW, its intensity was substantially reduced with increasing ball-milling speed during the H2SO4 treatment. On the other hand, the C-O stretching peak was preserved for the H3PO4-treated sample (SI Appendix, Fig. S1), which is consistent with the low yield obtained by using H3PO4 (SI Appendix, Fig. S2B). The XRD data (SI Appendix, Fig. S2) also suggested that the ball-milling treatment led to the loss of native chitin’s crystalline structure, and that HCl demineralization was successful.
The influence of the mechanocatalytic process on the modified morphology of chitin fiber was displayed (SI Appendix, Fig. S3) at 25,000× magnification. Raw SSW (SI Appendix, Fig. S3A) consisted of small chitin layers without fiber observation and commercial chitin had a partial fibrillation (SI Appendix, Fig. S3B). HCl demineralization created soft organic substances, which was probably due to the partial deproteinization (SI Appendix, Fig. S3C), while the commercial method (NaOH deproteinization and following HCl demineralization) revealed pronounced chitin bundles (SI Appendix, Fig. S3D). Nonetheless, well-defined chitin fibers were disrupted by high mechanical stress during ball milling, which generated small particles (SI Appendix, Fig. S3 E and F). To provide further insights on how mechanocatalytic depolymerization served as a promotional factor for the hydrolysis of chitin into water-soluble GlcNAc and its oligomers, solid-state 13C-NMR was performed (SI Appendix, Fig. S4). The intensities of a few peaks decreased substantially in the ball-milled samples, demonstrating the effect of high mechanical stress on the formation of the amorphous structure. After ball milling (700 rpm), solid chitinous samples featured weaker carbon peaks at 20.3 ppm (C-8) and 170.8 ppm (C-7), which belonged to the acetamido group, and two individual chemical shift peaks belonging to C-3 and C-5 disappeared. Furthermore, the peak of C-1 shifted from 101 ppm to 99 ppm for the mechanocatalytically depolymerized samples caused by the larger amounts of reducing ends, which may be generated due to the partial chain breaking down (26).
After ball milling, SSW-derived chitin hydrolysates were neutralized by using NaOH (1 M) and directly used for downstream microbial fermentation. The chitin hydrolysates after demineralization, partial deproteinization, and depolymerization were composed of GlcNAc and its oligomers (58.9 wt%), protein hydrolysates (31.3 wt%), and Na2SO4 (9.8 wt%, Fig. 2E). The chitin hydrolysates (22.5 g/L) were analyzed by high-performance liquid chromatography (HPLC), and the following compounds were detected (Fig. 2E): GlcNAc (2.8 g/L), (GlcNAc)2 (2.3 g/L), (GlcNAc)3 (1.6 g/L), and larger oligomers (∼5.7 g/L, estimated by using (GlcNAc)3’s standard). The total amount of GlcNAc and its oligomers was ∼54.1 wt% of chitin hydrolysates based on HPLC results, which was close to the weight difference analysis of chitin hydrolysates after SSW demineralization (Fig. 2E). As a comparison, the percentage of GlcNAc and its oligomers in chitin hydrolysates was only 12% when H3PO4-aided mechanocatalytic hydrolysis was used, again confirming that H3PO4 depolymerization was not efficient.
Very recently, Therien et al. reported a new approach to produce GlcNAc from shrimp and crab shell biomass (27). Their process relied on mechanoenzymatic breakdown of chitin instead of using an acid-aided one, and the whole process required over 5 d due to the inherent low activity of the chitinases used. In this study, we focused on obtaining the fermentable GlcNAc and its oligomers instead of only GlcNAc from SSW-derived chitin, and thus the depolymerization can be completed within 2 h by using a mild ball-milling process with a small amount of H2SO4 (7 wt%) without heating and purification. This process almost completely solubilized chitin in SSW, establishing a cost-effective feed stream for the subsequent microbial fermentation.
Engineering E. coli to Produce Tyrosine from GlcNAc.
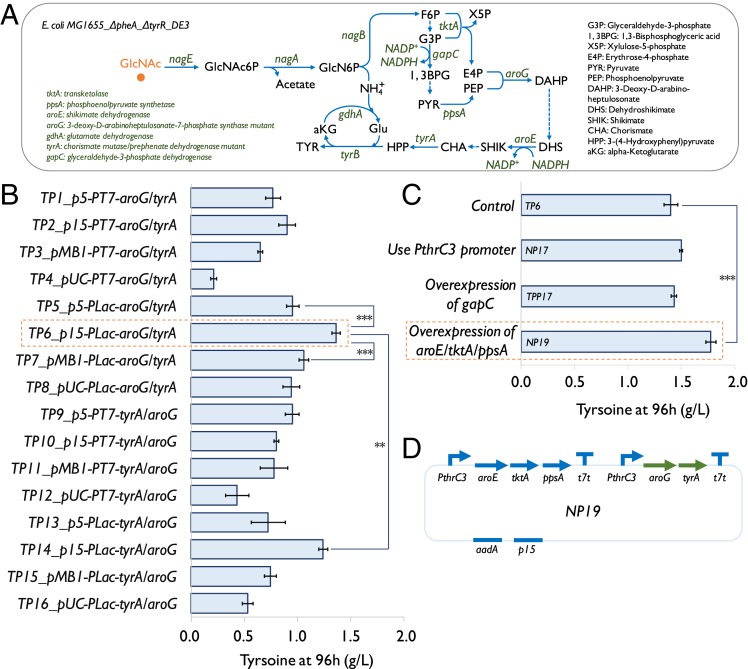
Wild-type E. coli has been demonstrated to grow in the medium using GlcNAc or (GlcNAc)2 as the sole carbon and nitrogen source (19, 20). Both GlcNAc and (GlcNAc)2 derived from SSW-derived chitin hydrolysates would be assimilated intracellularly to form D-glucosamine-6-phosphate, which is further deaminated into D-fructofuranose-6-phosphate. The released ammonium ion (NH4+) can be used as the sole nitrogen source for cell biomass and product formation (Fig. 1B). We selected GlcNAc to provide carbon and nitrogen sources for cell growth and product formation in the development of E. coli strains. Tyrosine, a valuable aromatic amino acid and the precursor for the biosynthesis of many valuable NCCs such as neurotransmitters, hormones, flavonoids, and alkaloids (17, 28, 29), was selected as the target compound. In a previous study (30), 16 strains were constructed in the laboratory to overproduce tyrosine from glucose. All of the strains had two gene deletions (tyrR and pheA) which were known to disrupt the regulation of tyrosine biosynthesis and to eliminate the formation of phenylalanine as a byproduct. All of the strains were also engineered to overexpress a feedback-resistant mutant of 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase (aroGfbr, Asp146 to Asn) and of chorismate mutase/prephenate dehydrogenase (tyrAfbr, Met53 to Ile and Ala354 to Val), both of which are critical enzymes in tyrosine biosynthesis (28). These strains carried plasmids with different replication origins (p5, p15, pMB1, and pUC) and promoters (PT7 and PLac) to express aroG and tyrA. These two genes were arranged in different orders (aroG-tyrA or tyrA-aroG) in some of the plasmids. Such variations led to the different expression level of the two genes and the improvement of tyrosine production from glucose. Here, the 16 strains were tested in a chemically defined medium with 10 g/L of GlcNAc. These E. coli strains exhibited very different capacities to produce tyrosine, with tyrosine titer ranging from 0.2 to 1.4 g/L (Fig. 3B). The best strain (TP6/p15-PLac-A_T) had the plasmid-containing PLac promoter and medium copy replication origin (p15), and produced 1.4 g/L of tyrosine at 96 h after induction (Fig. 3B).
Fig. 3.
Engineering E. coli to overproduce tyrosine from GlcNAc. (A) The schematic drawing of the biosynthetic pathway of producing tyrosine from GlcNAc in an engineered E. coli strain. (B) Screening of 16 tyrosine-producing strains carrying different plasmids with a wide range of combinations of genetic parts using 10 g/L GlcNAc in a chemically defined medium (containing 0.1 M KH2PO4). Replication origins with varied copy number: p5 (pSC101, ∼5), p15 (p15A, ∼10), pMB1 (5–20), and pUC (500 to 700). Inducible promoters: PT7 and PLac. T7 terminator (t7t) and spectinomycin antibiotic resistance (aadA) were used to construct all of the 16 plasmids. Genes aroG and tyrA used in this study were feedback-resistant variants. ***P < 0.001 and **P < 0.01, determined by two-tailed Student’s t test. (C) Characterization of four plasmids that were designed to improve the tyrosine production. Autoinducible promoter: PthrC3. Gene gapC coding glyceraldehyde-3-phosphate dehydrogenase was from C. acetobutylicum. Error bars indicate SE (n ≥ 3, n = 5 for the strains carrying plasmids TP5, TP6, TP7, TP14, and NP19). (D) Schematic diagram of plasmid NP19.
To supply more NADPH that is needed in the biosynthesis of tyrosine (Fig. 3A), we expressed an NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (gapC) from Clostridium acetobutylicum (31) by using plasmid NP17 so that it can generate NADPH instead of NADH in glycolysis (Fig. 3 A and C). The characterization results of the strains carrying these plasmids showed that overexpression of gapC did not lead to the improvement of the tyrosine titer (Fig. 3C), indicating that the NADPH supply was not a limiting step at this stage. We next tested a plasmid (TPP17) constructed before (30), which carried an autoinducible promoter (PthrC3) instead of PLac in the plasmid of TP6. It did not improve the tyrosine production (Fig. 3C), but was still used for the rest of the study, as it simplified the fermentation process by eliminating addition of inducers. Next, to enhance the supply of shikimate (Fig. 3A), we incorporated an upstream module containing three genes (aroE, tktA, and ppsA) driven by the promoter PthrC3 to push carbon flux from glycolysis to tyrosine biosynthesis (Fig. 3 A and C). This plasmid was named NP19. The strain carrying plasmid NP19 (Fig. 3 C and D) produced ∼1.7 g/L of tyrosine in 96 h, which was 20% higher than the titer produced by the strain carrying plasmid TP6 (Fig. 3C), demonstrating that pushing flux to tyrosine biosynthesis was an effective strategy as reported in previous works (28, 32).
We reasoned that overexpression of nagB (coding glucosamine-6-phosphate deaminase) might enhance deamination of D-glucosamine-6-phosphate. We also planned to co-overexpress gdhA (coding glutamate dehydrogenase) whose protein product assimilates NH4+ to produce glutamate that is needed as the amino group donor during tyrosine formation (Fig. 3A). We then constructed four E. coli strains carrying one of the four plasmids (TP6, NP17, TPP17, and NP19) with a nagB-gdhA–overexpressing plasmid (TP20) (SI Appendix, Fig. S5). The overexpression of nagB and gdhA, however, decreased tyrosine production by most strains (SI Appendix, Fig. S5), indicating the relationship between the nitrogen metabolism and tyrosine biosynthesis to be more complicated than what we had hypothesized.
Production of Tyrosine Using GlcNAc- and Glucose-Based Media.
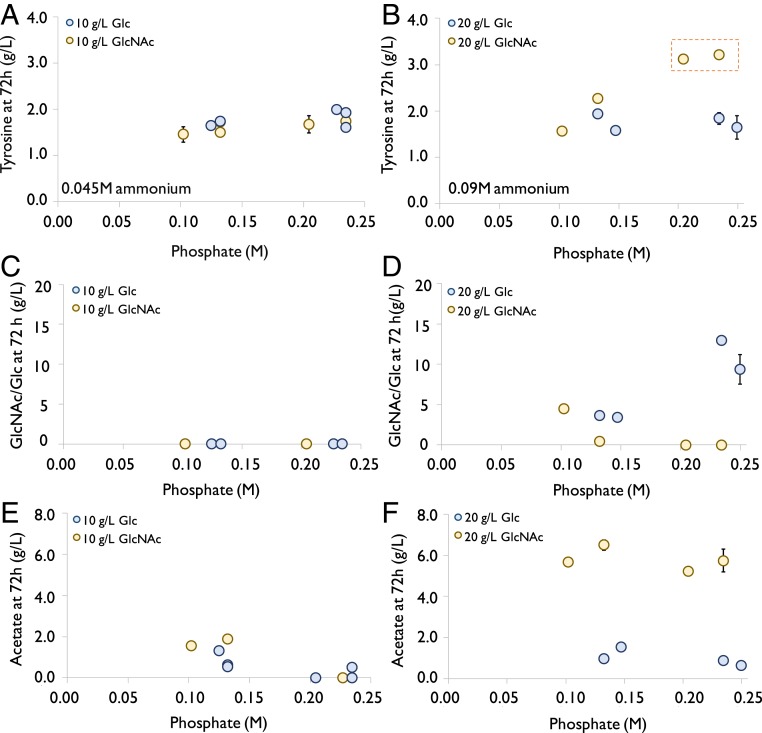
GlcNAc was used as the sole carbon and nitrogen sources for the above experiments of pathway engineering. In literature, glucose has been frequently used as a carbon source for the production of NCCs due to its low price (33). With glucose as the carbon source, the fermentation medium used to produce NCCs required additional nitrogen sources, such as ammonium salts, yeast extract, or peptone (28, 32, 34). Until now, there has been no report on the effect of utilizing GlcNAc as the sole carbon and nitrogen sources on NCC production of engineered E. coli. Hence, we compared tyrosine production using GlcNAc- and glucose-based media at two concentrations (10 and 20 g/L) (Fig. 4 A and B and SI Appendix, Table S1). Since the carbon-to-nitrogen ratio would be fixed if we used GlcNAc as a carbon source without an additional nitrogen source, we matched the nitrogen concentration in the corresponding glucose group by using ammonium salts for fair comparison. We tested a range of phosphate concentrations in all of the groups since phosphate concentration can substantially affect fermentation performance (35). With 10 g/L of the carbon source, a similar titer of tyrosine (∼1.8 g/L) was achieved in all of the tested conditions (Fig. 4A). Interestingly, with 20 g/L of carbon source, increasing phosphate concentration substantially increased the tyrosine titer only in the GlcNAc group. With more than 0.2 M phosphate, 3.2 g/L of tyrosine was produced in the GlcNAc medium, whereas the tyrosine titer did not exceed 2 g/L with the glucose-containing medium in all of the tested conditions (Fig. 4 A and B).
Fig. 4.
Comparison of glucose (Glc) and GlcNAc as a carbon source for producing tyrosine. Two carbon source concentrations were used: 10 g/L (A, C, and E) and 20 g/L (B, D, and F). (A and B) Tyrosine titer at 72 h achieved with different amounts of carbon source and phosphate. (C and D) Concentration of residual carbon source at 72 h. (E and F) Acetate (byproduct) titer at 72 h. The strain carrying plasmid NP19 was used. Ammonium salts were added to the glucose-based media to match the nitrogen concentration in the GlcNAc media in which GlcNAc was the only nitrogen source. The detailed composition of the media used in this part can be found in SI Appendix, Table S1. Error bars indicate SE (n = 3).
Phosphate is a basic component required for the production of phospholipids of microbial cell membranes and of nucleic acids (36). A larger amount of phosphate salts in the medium will also provide a stronger buffering capacity to maintain the pH during fermentation, benefitting cell growth (35). In this experiment, the carbon source was completely utilized in all of the conditions when the initial concentration was 10 g/L (Fig. 4C). A substantial amount of carbon source, however, was left at the end of the experiment when the initial concentration increased to 20 g/L. Increasing phosphate concentration substantially improved uptake of GlcNAc—all of the GlcNAc was consumed when the phosphate concentration was higher than 0.2 M (Fig. 4D). On the contrary, glucose uptake was inefficient with increased phosphate concentration and at least 4 g/L of glucose was left unconsumed at all of the tested conditions (Fig. 4D). The different effect of phosphate concentration on tyrosine production in glucose- and GlcNAc-based media may be due to the fact that cells impose distinct regulations on its metabolism when they use various carbon substrates. Compared with reports in the literature (most of which used glucose and ammonium salts as carbon and nitrogen sources), the titer of tyrosine (3.2 g/L, Fig. 4B) produced in the medium with 20 g/L of GlcNAc and 0.23 M phosphate was among the highest at the test tube/shake flask scale (18, 28, 32, 37) (SI Appendix, Table S2). The tyrosine yield (grams of tyrosine per gram of GlcNAc) at this condition can potentially be further improved, because ∼5 g/L of acetate was produced as a byproduct, which was much more than the one obtained by using glucose as carbon source (Fig. 4 D and E). The overproduced acetate from GlcNAc could be assimilated if the acetate utilization pathway is further improved (38). Nevertheless, the data we have collected so far already serve as strong evidences to support that GlcNAc is a better carbon and nitrogen source than glucose/ammonium salt for the microbial production of tyrosine.
Engineering E. coli to Produce l-DOPA from GlcNAc.
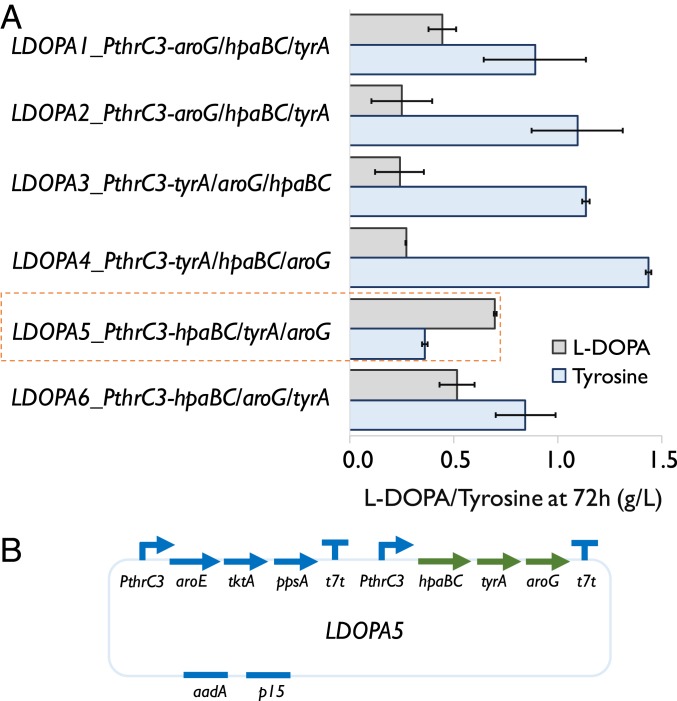
To demonstrate that what we have developed is a platform for microbial production of NCCs, E. coli was subsequently engineered to produce a tyrosine-derived NCC, l-DOPA, a frontline drug treating Parkinson’s disease and the precursor of many important alkaloid natural products (17). We selected a gene (hpaBC) coding 4-hydroxyphenylacetate 3-monooxygenase, a known enzyme capable of converting tyrosine into l-DOPA. We first inserted hpaBC into the downstream module of plasmid NP17 and constructed six plasmids to investigate the effect of gene order of the downstream module on l-DOPA production in E. coli, as the production of l-DOPA could be improved by balancing enzyme expression in the downstream module (28). Screening the six engineered E. coli strains carrying the above-mentioned l-DOPA-producing plasmids was carried out in the medium with 10 g/L of GlcNAc and 0.23 M of phosphate (Fig. 5A). The plasmid carrying the downstream module with a gene order of hpaBC/tyrA/aroG resulted in the highest titer of l-DOPA (0.7 g/L) (Fig. 5 A and B). At this condition, 0.36 g/L of tyrosine was still accumulated in the medium (Fig. 5A), indicating that the expression or the activity of 4-hydroxyphenylacetate 3-monooxygenase can be further enhanced to reach complete conversion of tyrosine to l-DOPA in vivo. It was also observed that the color of the medium became darker over time. The hydroxylase (HapBC) was not supposed to overoxidize l-DOPA, so the darker color could be a result of l-DOPA autooxidation by molecular oxygen into melanin (34).
Fig. 5.
Engineering E. coli to overproduce l-DOPA from GlcNAc. (A) Characterization of six l-DOPA-producing strains, each of which carried a plasmid with aroG, tyrA, and hapBC in different orders. The plasmid also included an operon to overexpress aroE, tktA, and ppsA. The medium containing 0.23 M phosphate and 10 g/L GlcNAc was used for screening l-DOPA-producing strains. Error bars indicate SE (n = 3). (B) Schematic diagram of plasmid LDOPA5.
We next used 20 g/L of GlcNAc to test the production of l-DOPA by the strain carrying plasmid LDOPA5. While the titer of l-DOPA achieved under this condition (0.68 g/L; SI Appendix, Fig. S6) was almost the same as the one obtained with 10 g/L of GlcNAc (Fig. 5A), there was a more than twofold increase in the tyrosine titer (1.03 g/L), which further supported that the step controlled by hpaBC was rate limiting. The titer of l-DOPA obtained with 20 g/L of GlcNAc was two times higher than that obtained using 20 g/L of glucose and ammonium salts (SI Appendix, Fig. S6), again demonstrating that GlcNAc was a better carbon and nitrogen source than glucose with ammonium salts. The titer of l-DOPA produced by the strain carrying LDOPA5 (Fig. 5A) is at the same level of the reported titers achieved by using engineered E. coli strains at the culture tube/shake flask scale (34, 39) (SI Appendix, Table S2). The modularity and flexibility of modifying the strains to overproduce l-DOPA as shown above support that the platform developed in this study is highly customizable in making many tyrosine-derived NCCs.
Production of Tyrosine and l-DOPA from SSW-Derived Hydrolysates.
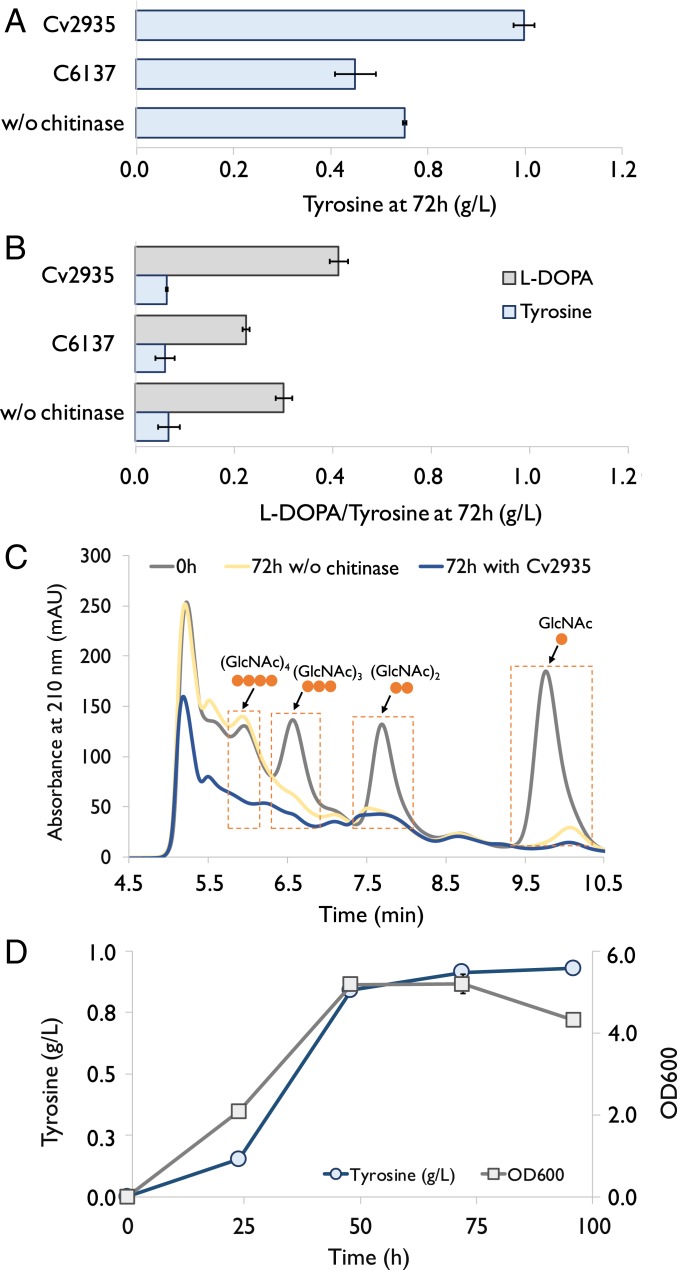
After developing the strains capable of overproducing tyrosine and l-DOPA, we focused on producing them directly from the unpurified SSW-derived chitin hydrolysates. We used a chemically defined medium containing 0.23 M phosphate buffer (SI Appendix, Table S1) with 22.5 g/L SSW-derived chitin hydrolysates, which contained ∼20 g/L chitin and protein hydrolysates (Fig. 2E, ∼10% [wt/wt] of the total weight was sodium sulfate). Fermentation with this medium produced 0.55 g/L tyrosine and 0.3 g/L l-DOPA by using the strains carrying plasmids NP19 and LDOPA5, respectively (Fig. 6 A and B).
Fig. 6.
Production of tyrosine and l-DOPA using SSW-derived chitin hydrolysates as the sole carbon and nitrogen sources. (A) Tyrosine production by using the engineered E. coli strain carrying plasmid NP19 with or without adding chitinases. (B) l-DOPA production by using the engineered E. coli carrying plasmid LDOPA5 with or without adding chitinases. Cv2935 (4 μg/mL): chitinase from C. violaceum. C6137 (8.4 μg/mL): a commercialized chitinase from Streptomyces griseus (Sigma, C6137). (C) HPLC analysis of the consumption of GlcNAc and its oligomers by the engineered E. coli strain carrying plasmid NP19. 0h: at the beginning of the fermentation. “72 h w/o enzyme”: after 72 h since the beginning of the fermentation without adding any chitinase. “72 h with Cv2935”: after 72 h since the beginning of the fermentation with adding Cv2935 (10 μg/mL). (D) Time profile of tyrosine concentration and cell density (OD600) in the one-pot enzymatic/fermentative process. The engineered E. coli strain carrying plasmid NP19 was cultured by using 22.5 g/L SSW-derived chitin hydrolysates. Cv2935 (10 μg/mL) was added into the medium upon inoculation. Error bars indicate SE (n = 3).
Following that, we hypothesized that adding chitinase would be helpful in further breaking down GlcNAc oligomers (n ≥ 3) in chitin hydrolysates into fermentable GlcNAc and (GlcNAc)2, thus improving the product yield. We first selected three chitinases and tested their activities. All of the three chitinases were expressed in E. coli and purified (SI Appendix, Fig. S7). All of them were able to form (GlcNAc)2 and (GlcNAc)3 instead of GlcNAc from the chitin hydrolysates. Among the three, the chitinase (Cv2935) from C. violaceum (40) exhibited the highest activity on GlcNAc oligomers (n ≥ 3), producing more (GlcNAc)2 compared with the other two (SI Appendix, Fig. S8). In addition, the activity of Cv2935 was not inhibited by high substrate concentration (SI Appendix, Fig. S9), making this chitinase a good candidate to develop an enzymatic/fermentative process. We subsequently tested the production of tyrosine and l-DOPA from SSW-derived chitin hydrolysates with Cv2935 or a commercial chitinase (Sigma, C6137) that had been found to produce (GlcNAc)2 and (GlcNAc)3 at a comparable capacity to that of Cv2935 (SI Appendix, Fig. S8). Adding Cv2935 (4.0 μg/mL) indeed increased the titers of tyrosine and l-DOPA, producing 0.8 and 0.41 g/L of tyrosine and l-DOPA, respectively (Fig. 6 A and C). Surprisingly, the addition of the commercial chitinase C6137 (6.8 μg/mL) decreased the production of both tyrosine and l-DOPA (Fig. 6 A and C), possibly because of other components in the commercial enzyme product that could interfere with microbial cell growth. We also employed the plasmids carrying a separate operon to increase hpaBC expression level, but it did not further improve l-DOPA production from SSW-derived chitin hydrolysates with or without chitinase Cv2935 (SI Appendix, Fig. S10).
It was also observed that adding chitinase Cv2935 promoted biomass formation in the first 24 h (SI Appendix, Fig. S11A), further supporting the idea that the chitinase was active in breaking down the GlcNAc oligomers during fermentation. HPLC analysis of the fermentation medium provided concrete evidence that engineered E. coli strains could directly utilize GlcNAc, (GlcNAc)2, and (GlcNAc)3 for biomass formation and NCC production (Fig. 6C). With the addition of Cv2935, the HPLC peak areas of GlcNAc oligomers (n ≥ 3) after fermentation for 72 h were drastically reduced compared with the ones without the addition of chitinase (Fig. 6C and SI Appendix, Fig. S11B), confirming the hydrolysis activity of Cv2935 during fermentation.
We then attempted to add more Cv2935 (10 μg/mL) to accelerate the enzymatic hydrolysis of GlcNAc oligomers and performed the enzymatic/fermentative process. The results indicated that the titer of tyrosine reached 0.91 g/L in 72 h (Fig. 6D), which was slightly higher than the one obtained with less Cv2935 (4.0 μg/mL) (Fig. 6C). Unlike the use of GlcNAc as the sole carbon and nitrogen source, we did not detect any production of acetate when using SSW-derived chitin hydrolysates, suggesting that the slow release of GlcNAc from its oligomers could be beneficial for assimilating acetate by the engineered E. coli cell, similar to substrate-limited fed-batch fermentation. Approximately 70% of total GlcNAc and its oligomers in SSW-derived chitin hydrolysates (22.5 g/L) was consumed to produce biomass and tyrosine, and ∼46% of GlcNAc oligomers (n > 4) remained in the medium with Cv2935 after fermentation for 72 h (Fig. 6C and SI Appendix, Fig. S11 B and D). The hydrolysis of GlcNAc oligomers (n > 4) to (GlcNAc)2 and (GlcNAc)3 almost stopped after 24 h (SI Appendix, Fig. S11 C and D) which may be caused by 1) pH-associated activity loss during acidification of the media in the first 24 h; 2) chitinase was degraded or denatured in the period; and/or 3) a fraction of GlcNAc oligomers cannot be processed by the chitinase due to higher molecular weight or association with other molecules. It was also possible that the HPLC estimation of the remaining GlcNAc oligomers (n ≥ 4) was not accurate and the remaining peak area was from other chemicals in the medium. In such a case, the hydrolysis of the oligomers would be considered to be completed by 24 h during the fermentation.
In an attempt to produce tyrosine with a higher concentration of SSW-derived chitin hydrolysates, 1.3 g/L of tyrosine (SI Appendix, Fig. S11E) was produced from 40 g/L of SSW-derived chitin hydrolysates but with a substantially decreased tyrosine yield (decreased from 0.040 g/g to 0.033 g/g when substrate concentration increased from 22.5 g/L to 40 g/L). Although the titers of these two NCCs were lower than the ones we achieved by using GlcNAc (Fig. 4B), we demonstrated the feasibility of using chitin-containing shell waste to produce value-added NCCs in E. coli via an integrated bioprocess. To improve utilization of SSW-derived GlcNAc oligomers in chitin hydrolysates from SSW, more chitinases can be characterized and the effective ones could be mixed to prepare an enzyme mixture (including exochitinase and endochitinase). Besides, the protein hydrolysates in SSW-derived chitin hydrolysates might also be used as carbon and nitrogen sources once they are enzymatically digested into fermentable short peptides and amino acids.
Summary
The amount of waste from crustacean shell processing industries has increased substantially in recent years, leading to undesired accumulation of chitin-containing waste that has resulted in waste disposal and pollution problems. In this work, an integrated biorefinery process was developed to utilize SSW-derived chitin hydrolysates as the sole carbon and nitrogen sources to produce NCCs, which seamlessly merges the pretreatment of SSW and the microbial fermentation without an additional purification step. We have experimentally demonstrated that two NCCs (tyrosine and l-DOPA) were produced from the unpurified SSW-derived chitin hydrolysates via a one-pot enzymatic/fermentative process, providing a perspective on the production of useful NCCs from renewable nitrogen-containing feedstock such as crustacean shell waste.
Materials and Methods
Materials.
Commercial chitin was purchased from Wako Pure Chemical Industries. Sulphuric acid (98% purity), phosphoric acid (85% purity), and hydrochloric acid (32% purity) were provided by VWR. Sodium hydroxide was purchased from Schedelco. N,N′-diacetylchitobiose (T2912, [GlcNAc]2) and N,N′,N″-triacetylchitotriose (D4215, [GlcNAc]3) were purchased from Tokyo Chemical Industry. All other chemicals used in this study were purchased from Sigma.
SSW Composition Analysis and HCl Demineralization.
Shrimp purchased from a local supermarket in Singapore was used in this study. The body part of the shrimp shell was washed, dried at room temperature, and then ground into powder with a kitchen blender (Tefal). The composition of the shrimp shell was evaluated by a modified alkaline and acidic method (21). For demineralization, the dried SSW (5 g) was stirred with 5 wt% of HCl solution (50 mL) at room temperature for 1 h. After that, the solids (called HCl-demineralized SSW) were separated with centrifugation (8,000 rpm for 3 min), washed with 200 mL of water (five times), and dried (453 K for 24 h). The composition analysis was conducted based on the weight differences before and after each process.
Mechanocatalytic Decomposition of Demineralized SSW.
Dried HCl-treated SSW (450 mg), which contained chitin, residual protein, and H2SO4 (34 mg) was placed into a zirconium oxide ball-mill chamber (45 mL) together with zirconium oxide balls (100 pieces). Ball milling (Fritsch, Pulverisette 7) was performed at 300 to 700 rpm and 4 to 12 cycles. One cycle consisted of 10 min of grinding and 5 min of resting. After grinding, distilled water (10 mL) was added into the ball-mill chambers and rotated at low speed (200 rpm) for 5 min. At this stage, the dissolution of SSW-derived chitin occurred. The undissolved sample was separated and dried in an oven at 353 K overnight. The dissolution was quantified by using the weight change between initial HCl-demineralized SSW and the dried solid sample. SSW-derived chitin hydrolysate dissolved in water was neutralized with 1 M NaOH. For subsequent usage in microbial fermentation, the pH-adjusted SSW-derived chitin hydrolysate was dried by freeze drier to form a solid product to prepare the chitin hydrolysate-based medium.
Ball-milled solid products, HCl-demineralized SSW, SSW, solid products after deproteinization and demineralization, and other pure chemicals were characterized by several techniques, including Fourier-transform infrared spectroscopy equipped with attenuated total reflectance (Thermo Scientific iS50), field emission scanning electron microscopy (JEOL 7610F) coating with platinum before analysis, and X-ray powder diffraction analysis which was operated at 40 kV and 15 mA, and carried out using a Rigaku Miniflex 600 X-Ray Diffractometer with a scan rate of 1.5°/min. Solid state 13C NMR spectroscopy was performed on a Bruker AVNEOO400WB with a cross-polarization magic angle spinning probe.
Plasmids and Strains.
The oligonucleotides used in this study were ordered from Integrated DNA Technologies. A full list of plasmids used in this study is provided in SI Appendix, Table S3, and the construction of these plasmids was carried out by using guanine–thymine standard-based workflow (30). The standard electroporation procedure was used to transfer most constructed plasmids into E. coli MG1655 ΔrecA_ΔendA_ΔpheA_ΔtyrR_DE3 we had developed before (30). Plasmids CPP1, CPP2, and CPP3 were transferred into E. coli BL21 (C2530H, New England Biolabs) via a standard heat-shock method. Colonies appeared on the plate supplemented with proper antibiotic(s) (specified below) after incubation at 37 °C overnight. The Addgene IDs of plasmid NP19, LDOPA5, and LDOPA8 are 140530, 140528, and 140529, respectively.
Growth Media.
Ten milliliters of growth medium was prepared by mixing 5 mL of the basal medium (SI Appendix, Table S1), x mL of carbon substrate stock solution (x depended on the working concentration and the concentration of the stock solution), and 0.017 mL of the master mix. This mixture was topped up to 10 mL by using ultrapure water. The carbon substrate stock solution was 100 g/L GlcNAc stock solution (sterilized by using a 0.22-µm filter), 200 g/L glucose stock solution (autoclaved), or 100 g/L SSW hydrolysates (sterilized by using a 0.22-µm filter). The proper amount of antibiotic solution was added as specified in the relevant sessions. The basal medium containing varied phosphate or ammonium salts was prepared in 1 L of deionized water with 60 mg of l-phenylalanine. Its pH was adjusted to 7 using 400 g/L NaOH. Basal medium 5 (SI Appendix, Table S1) was used with SSW hydrolysates.
The master mix was prepared by mixing 2.5 mL of 0.1 M ferric citrate solution (autoclaved), 1 mL of 4.5 g/L thiamine solution (filtrated through a 0.2-µm nylon filter), 3 mL of 4 mM Na2MoO3 (autoclaved), 1 mL of 1,000× K3 trace elements stock solution (autoclaved), and 1 mL of 1 M MgSO4 solution (autoclaved). The 1,000× K3 trace element stock solution (autoclaved) was prepared by dissolving 5 g of CaCl2∙2H2O, 1.6 g of MnCl2∙4H2O, 0.38 g of CuCl2∙4H2O, 0.5 g of CoCl2∙2H2O, 0.94 g of ZnCl2, 0.0311 g of H3BO3, and 0.4 g of Na2EDTA∙2H2O in 1 L of deionized water.
Production and Purification of Chitinases.
A single colony of an E. coli BL21 strain carrying plasmids CPP1, CPP2, or CPP3 was inoculated into Luria–Bertani broth (LB) with 50 µg/mL spectinomycin, and cultured at 37 °C/250 rpm overnight. Five hundred microliters of the overnight-grown cell suspension was inoculated into 50 mL of the chemically defined medium prepared by using basal medium 9 (SI Appendix, Table S1) with 20 g/L glucose and 50 µg/mL spectinomycin in a 250-mL shake flask, and the culture was incubated at 30 °C/250 rpm until cell density reached 0.5 to 1.0 (optical density [OD600]), at which time the culture was induced by 0.1 mM of isopropyl β-d-1-thiogalactopyranoside (IPTG). The shake flask was incubated at 30 °C/250 rpm for 24 h. The cells were collected by centrifugation at 4,000 rpm for 10 min. The obtained cell pellets were resuspended by using 5 mL B-PER solution (78248, Thermo Fisher Scientific) to extract intracellularly expressed enzymes at 25 °C/160 rpm for 1 h. The expression of the three chitinases was verified by using SDS-polyacrylamide gel electrophoresis (SDS/PAGE) (Mini-PROTEAN TGX Precast Protein Gels, 4561083, Bio-Rad). The Gel Doc EZ Gel Documentation System (Bio-Rad) was used to image the gel, and the results are shown in SI Appendix, Fig. S7A.
The supernatants containing soluble proteins were isolated by centrifugation at 4 °C/12,000 rpm for 10 min (Eppendorf Centrifuge 5424), and then loaded into a HisPur Ni-NTA Spin Column (88224, Thermo Fisher Scientific) containing 1.4 mL of activated nickel–nitrilotriacetic acid resin. The target proteins were purified according to the manufacturer’s instructions. The purification results of the three chitinases can be found in SI Appendix, Fig. S7B. The concentration of purified enzymes was measured by using Bradford assay (Pierce Coomassie Plus Assay Kit, 23236, Thermo Fisher Scientific). Typically, the cells obtained from 50 mL of culture were sufficient to prepare 0.5 mL of concentrated enzyme solution (∼300 µg/mL).
Fermentation and Quantification of Substrates and Products.
A single colony was inoculated into LB with 50 µg/mL spectinomycin or 50 µg/mL spectinomycin plus 50 µg/mL ampicillin, and cultured at 37 °C/250 rpm overnight. Ten microliters of the overnight-grown cell suspension was inoculated into 1 mL of a growth medium with the proper amount of antibiotic(s) in a 14-mL round-bottom Falcon tube, and the culture was incubated at 30 °C/250 rpm until cell density reached 0.5 to 1.0 (OD600), at which point the culture was induced by 0.1 mM IPTG. If PthrC3 promoter was used, IPTG induction was skipped. The commercial and home-made chitinase was added upon inoculation when needed. The tube was incubated at 30 °C/250 rpm for 24 to 96 h. Optical density 600 (OD600) of 10 times-diluted cell suspensions (using ultrapure water) was measured by a microplate reader (Varioskan LUX Multimode Microplate Reader, Thermo Fisher Scientific). The reading was converted into standard OD600 values by using a calibration curve.
For measuring GlcNAc and its oligomers, acetate and glucose, 5 μL of the filtered supernatant obtained from the above protocol was analyzed by HPLC (Agilent 1260 Infinity HPLC). The HPLC conditions are as follows: the column was Aminex HPX-87H Column (Bio-Rad); an isocratic flow was used (the flow rate was 0.7 mL/min); the mobile phase was an aqueous solution containing 5 mM H2SO4; the column temperature was 50 °C; a UV detector (210 nm) was used to quantify GlcNAc and its oligomers; and a refractive index detector was used to quantify acetate and glucose.
One hundred microliters of 6 M HCl was added to 1 mL of cell culture broth for dissolving tyrosine crystals after incubation. The mixture was incubated at 30 °C/200 rpm for 20 min and then centrifuged at 14,000 rpm for 5 min. The supernatant was filtered using a 13 mm, 0.2-µm nylon filter. To measure the concentration of tyrosine and l-DOPA, 2 μL of the filtered supernatant prepared according to the above protocol was analyzed by HPLC (Agilent 1260 Infinity HPLC). The HPLC conditions were as follows: the column was Agilent ZORBAX Eclipse Plus C18 100 mm; an isocratic flow was used (the flow rate was 0.7 mL/min); the mobile phase consisted of acetonitrile (10%, vol/vol), ultrapure water (89.9%, vol/vol), and trifluoroacetic acid (0.1%, vol/vol); the column temperature was 30 °C; and the detector was a UV detector (wavelength: 254 nm for tyrosine and 285 nm for l-DOPA).
Data Availability.
All of the data used to support the claims of this work have been presented in the form of figures and/or tables, which are available in this manuscript or its SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by Singapore Ministry of Education Tier 1 Grants (R-279-000-478-112 and R-279-000-479-112), a Disruptive & Sustainable Technologies for Agricultural Precision Grant, and the National University of Singapore Flagship Green Energy Program (R-279-000-553-646 and R-279-000-553-731).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919862117/-/DCSupplemental.
References
- 1.Yan N., Chen X., Sustainability: Don’t waste seafood waste. Nature 524, 155–157 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Sgobba E., Blöbaum L., Wendisch V. F., Production of food and feed additives from non-food-competing feedstocks: Valorizing N-acetylmuramic acid for amino acid and carotenoid fermentation with Corynebacterium glutamicum. Front. Microbiol. 9, 2046 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osada M., Shoji S., Suenaga S., Ogata M., Conversion of N-acetyl-d-glucosamine to nitrogen-containing chemicals in high-temperature water. Fuel Process. Technol. 195, 106154 (2019). [Google Scholar]
- 4.Zhang J., Yan N., Formic acid-mediated liquefaction of chitin. Green Chem. 18, 5050–5058 (2016). [Google Scholar]
- 5.Bobbink F. D., Zhang J., Pierson Y., Chen X., Yan N., Conversion of chitin derived N-acetyl-D-glucosamine (NAG) into polyols over transition metal catalysts and hydrogen in water. Green Chem. 17, 1024–1031 (2015). [Google Scholar]
- 6.Chen X., Yan N., Novel catalytic systems to convert chitin and lignin into valuable chemicals. Catal. Surv. Asia 18, 164–176 (2014). [Google Scholar]
- 7.Pierson Y., Chen X., Bobbink F. D., Zhang J., Yan N., Acid-catalyzed chitin liquefaction in ethylene glycol. ACS Sustainable. Chem. Eng. 2, 2081–2089 (2014). [Google Scholar]
- 8.Chen X., Chew S. L., Kerton F. M., Yan N., Direct conversion of chitin into a N-containing furan derivative. Green Chem. 16, 2204–2212 (2014). [Google Scholar]
- 9.Bohlmann J. A., et al. , “N-acetyl-D-glucosamine and process for producing N-acetyl-D-glucosamine.” US Patent 6 693 188 (2004).
- 10.Anderson T., et al. , “Glucosamine and N-acetylglucosamine compositions and methods of making the same fungal biomass.” US Patent 8 222 232 (2012).
- 11.Chen J.-K., Shen C.-R., Liu C.-L., N-acetylglucosamine: Production and applications. Mar. Drugs 8, 2493–2516 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yabushita M., Kobayashi H., Kuroki K., Ito S., Fukuoka A., Catalytic depolymerization of chitin with retention of N-acetyl group. ChemSusChem 8, 3760–3763 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Vaaje-Kolstad G., Horn S. J., Sørlie M., Eijsink V. G., The chitinolytic machinery of Serratia marcescens–A model system for enzymatic degradation of recalcitrant polysaccharides. FEBS J. 280, 3028–3049 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Zhang H., Jin Y., Deng Y., Wang D., Zhao Y., Production of chitin from shrimp shell powders using Serratia marcescens B742 and Lactobacillus plantarum ATCC 8014 successive two-step fermentation. Carbohydr. Res. 362, 13–20 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Yan Q., Hong E., Fong S. S., Study of ChiR function in Serratia marcescens and its application for improving 2,3-butanediol from crystal chitin. Appl. Microbiol. Biotechnol. 101, 7567–7578 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Streichsbier F., Utilization of chitin as sole carbon and nitrogen source by Chromobacterium violaceum. FEMS Microbiol. Lett. 19, 129–132 (1983). [Google Scholar]
- 17.Nakagawa A., et al. , A bacterial platform for fermentative production of plant alkaloids. Nat. Commun. 2, 326 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santos C. N., Xiao W., Stephanopoulos G., Rational, combinatorial, and genomic approaches for engineering L-tyrosine production in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 109, 13538–13543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valerio-Lepiniec M., et al. , Analysis of the Escherichia coli glucosamine-6-phosphate synthase activity by isothermal titration calorimetry and differential scanning calorimetry. Arch. Biochem. Biophys. 498, 95–104 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Keyhani N. O., Roseman S., Wild-type Escherichia coli grows on the chitin disaccharide, N,N′-diacetylchitobiose, by expressing the cel operon. Proc. Natl. Acad. Sci. U.S.A. 94, 14367–14371 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shahidi F., Synowiecki J., Isolation and characterization of nutrients and value-added products from snow crab (Chionoecetes opilio) and shrimp (Pandalus borealis) processing discards. J. Agric. Food Chem. 39, 1527–1532 (1991). [Google Scholar]
- 22.James S. L., et al. , Mechanochemistry: Opportunities for new and cleaner synthesis. Chem. Soc. Rev. 41, 413–447 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Margoutidis G., Parsons V. H., Bottaro C. S., Yan N., Kerton F. M., Mechanochemical amorphization of α-chitin and conversion into oligomers of N-acetyl-d-glucosamine. ACS Sustainable. Chem. Eng. 6, 1662–1669 (2018). [Google Scholar]
- 24.Meine N., Rinaldi R., Schüth F., Solvent-free catalytic depolymerization of cellulose to water-soluble oligosaccharides. ChemSusChem 5, 1449–1454 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Pearson F. G., Marchessault R. H., Liang C. Y., Infrared spectra of crystalline polysaccharides. V. Chitin. J. Polym. Sci. 43, 101–116 (1960). [DOI] [PubMed] [Google Scholar]
- 26.Chen X., Gao Y., Wang L., Chen H., Yan N., Effect of treatment methods on chitin structure and its transformation into nitrogen-containing chemicals. ChemPlusChem 80, 1565–1572 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Therien J. P. D., Hammerer F., Friščić T., Auclair K., Mechanoenzymatic breakdown of chitinous material to N-acetylglucosamine: The benefits of a solventless environment. ChemSusChem 12, 3481–3490 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Juminaga D., et al. , Modular engineering of L-tyrosine production in Escherichia coli. Appl. Environ. Microbiol. 78, 89–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lütke-Eversloh T., Stephanopoulos G., L-tyrosine production by deregulated strains of Escherichia coli. Appl. Microbiol. Biotechnol. 75, 103–110 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Ma X., et al. , A standard for near-scarless plasmid construction using reusable DNA parts. Nat. Commun. 10, 3294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martínez I., Zhu J., Lin H., Bennett G. N., San K. Y., Replacing Escherichia coli NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with a NADP-dependent enzyme from Clostridium acetobutylicum facilitates NADPH dependent pathways. Metab. Eng. 10, 352–359 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Kim S. C., Min B. E., Hwang H. G., Seo S. W., Jung G. Y., Pathway optimization by re-design of untranslated regions for L-tyrosine production in Escherichia coli. Sci. Rep. 5, 13853 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wendisch V. F., et al. , The flexible feedstock concept in industrial biotechnology: Metabolic engineering of Escherichia coli, Corynebacterium glutamicum, Pseudomonas, Bacillus and yeast strains for access to alternative carbon sources. J. Biotechnol. 234, 139–157 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Wei T., Cheng B. Y., Liu J. Z., Genome engineering Escherichia coli for L-DOPA overproduction from glucose. Sci. Rep. 6, 30080 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C., et al. , Combining genotype improvement and statistical media optimization for isoprenoid production in E. coli. PLoS One 8, e75164 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh V., et al. , Strategies for fermentation medium optimization: An in-depth review. Front. Microbiol. 7, 2087 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim B., Binkley R., Kim H. U., Lee S. Y., Metabolic engineering of Escherichia coli for the enhanced production of l-tyrosine. Biotechnol. Bioeng. 115, 2554–2564 (2018) [DOI] [PubMed] [Google Scholar]
- 38.Jo M., et al. , Precise tuning of the glyoxylate cycle in Escherichia coli for efficient tyrosine production from acetate. Microb. Cell Fact. 18, 57 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fordjour E., Adipah F. K., Zhou S., Du G., Zhou J., Metabolic engineering of Escherichia coli BL21 (DE3) for de novo production of L-DOPA from D-glucose. Microb. Cell Fact. 18, 74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobo M. D. P., et al. , Expression and efficient secretion of a functional chitinase from Chromobacterium violaceum in Escherichia coli. BMC Biotechnol. 13, 46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data used to support the claims of this work have been presented in the form of figures and/or tables, which are available in this manuscript or its SI Appendix.