Significance
Biological membranes are essential for life. Although bacteria and eukaryotic cells have evolved to produce membranes of different compositions, several bacterial pathogens can hijack and utilize host-synthesized membrane lipids. Here, we show that an obligatory intracellular pathogen, Ehrlichia chaffeensis, deficient in biosynthesis of cholesterol and some glycerophospholipids, actively acquires host-derived membrane components within membrane-bound inclusions (vacuoles). The trafficking of host membrane components to Ehrlichia and membrane-bound inclusions appears to occur via endocytosis and autophagy induced by a bacteria-secreted protein. Numerous intraluminal vesicles were found in Ehrlichia inclusions that may function as a membrane reserve for rapid proliferation of Ehrlichia. Our findings provide insights into host membrane assimilation by an intracellular pathogen, which can be exploited for antibacterial therapy.
Keywords: Ehrlichia chaffeensis, membranes, glycerophospholipids, cholesterol, intraluminal vesicles
Abstract
Ehrlichia chaffeensis, a cholesterol-rich and cholesterol-dependent obligate intracellular bacterium, partially lacks genes for glycerophospholipid biosynthesis. We found here that E. chaffeensis is dependent on host glycerolipid biosynthesis, as an inhibitor of host long-chain acyl CoA synthetases, key enzymes for glycerolipid biosynthesis, significantly reduced bacterial proliferation. E. chaffeensis cannot synthesize phosphatidylcholine or cholesterol but encodes enzymes for phosphatidylethanolamine (PE) biosynthesis; however, exogenous NBD-phosphatidylcholine, Bodipy-PE, and TopFluor-cholesterol were rapidly trafficked to ehrlichiae in infected cells. DiI (3,3′-dioctadecylindocarbocyanine)-prelabeled host-cell membranes were unidirectionally trafficked to Ehrlichia inclusion and bacterial membranes, but DiI-prelabeled Ehrlichia membranes were not trafficked to host-cell membranes. The trafficking of host-cell membranes to Ehrlichia inclusions was dependent on both host endocytic and autophagic pathways, and bacterial protein synthesis, as the respective inhibitors blocked both infection and trafficking of DiI-labeled host membranes to Ehrlichia. In addition, DiI-labeled host-cell membranes were trafficked to autophagosomes induced by the E. chaffeensis type IV secretion system effector Etf-1, which traffic to and fuse with Ehrlichia inclusions. Cryosections of infected cells revealed numerous membranous vesicles inside inclusions, as well as multivesicular bodies docked on the inclusion surface, both of which were immunogold-labeled by a GFP-tagged 2×FYVE protein that binds to phosphatidylinositol 3-phosphate. Focused ion-beam scanning electron microscopy of infected cells validated numerous membranous structures inside bacteria-containing inclusions. Our results support the notion that Ehrlichia inclusions are amphisomes formed through fusion of early endosomes, multivesicular bodies, and early autophagosomes induced by Etf-1, and they provide host-cell glycerophospholipids and cholesterol that are necessary for bacterial proliferation.
The bacterium Ehrlichia chaffeensis causes an emerging tick-borne zoonosis called human monocytic ehrlichiosis, a severe and potentially fatal flu-like systemic disease (1). As a small, obligate intracellular bacterium, E. chaffeensis infects and replicates inside membrane-bound cytoplasmic compartments of monocytes and macrophages. These compartments, known as inclusions or vacuoles, have characteristics of early endosomes and early autophagosomes, but lack lysosomal proteins and NADPH oxidase components, so that ehrlichiae can avoid lysosomal digestion as well as cell death mediated by reactive oxygen species (2–6). Within these compartments, E. chaffeensis utilizes multiple strategies to rapidly obtain essential nutrients from host cells (7, 8). Through fusion of bacteria-containing inclusions with host-derived vesicles produced by the RAB5-regulated autophagosome and endosome pathways, E. chaffeensis can acquire amino acids, metabolic intermediates, iron, and other essential nutrients (4, 9). However, the mechanisms by which ehrlichiae acquire membrane components within membrane-bound inclusions remains unknown.
Bacterial membrane compositions are distinct from those of eukaryotic cells and generally lack cholesterol (10, 11). However, the ehrlichial membrane is rich in cholesterol and ehrlichiae are dependent on host-derived cholesterol for survival and infection (12), as ehrlichiae lack genes for biosynthesis or modification of cholesterol (13). Indeed, unlike Escherichia coli, but similar to eukaryotic cells, host cell-free ehrlichiae are capable of incorporating a considerable amount of exogenous cholesterol (12). Bacteria have phospholipid bilayers of either single membranes (gram-positive bacteria, Mycoplasma) or double membranes (gram-negative bacteria). Phylogenetically and ultrastructurally, ehrlichiae are gram-negative bacteria (i.e., having two lipid bilayers) (14). Unlike most gram-negative bacteria, however, ehrlichiae lack lipopolysaccharide (LPS), including lipid A, peptidoglycan, and capsule components (13).
As ehrlichiae require a large amount of membrane for rapid replication inside host cells [over 600-fold within 3 d (15)], it is possible that they evolved a mechanism to hijack host membrane phospholipids as well. To investigate this possibility, we used multiple independent approaches, including analysis of ehrlichial genomic capacity for phospholipids biosynthesis, inhibition of host glycerolipid biosynthesis, trafficking analysis of fluorescence-labeled cholesterol and phospholipids, immunogold labeling of cryosections, and focused ion beam-scanning electron microscopy (FIB-SEM) in infected cells. Collectively, our results demonstrate unidirectional flow of host membrane phospholipids and cholesterol to ehrlichiae within inclusions, likely mediated by amphisomes and abundant intrainclusion membrane vesicles.
Results
E. chaffeensis Is Partially Defective in Glycerophospholipid Biosynthesis and Dependent on Host-Synthesized Lipids.
The E. chaffeensis genome encodes partial pathways for de novo biosynthesis of fatty acids and phospholipids, including phosphatidylethanolamine (PE), phosphatidylserine, and phosphatidylglycerol, but this organism lacks genes for biosynthesis of phosphatidylcholine (PC) or cardiolipin (SI Appendix, Fig. S1). Pyruvate is the primary substrate for synthesis of acetyl-CoA, a key molecule in the tricarboxylic acid cycle and many biosynthetic pathways, including fatty-acid biosynthesis. Glycerol-3-phosphate is the precursor required for glycerophospholipid biosynthesis. Although E. chaffeensis encodes enzymes that can carry out the tricarboxylic acid cycle, genes encoding the glycolytic pathway are incomplete (SI Appendix, Fig. S1) (13). Consequently, in order to synthesize fatty acids, E. chaffeensis has to import host-cell pyruvate or other glycolysis intermediate metabolites across the inclusion membrane, as well as the bacterial membrane, and utilize them to produce glyceraldehyde-3-phosphate and glycerol-3-phosphate at the expense of bacterial ATP (SI Appendix, Fig. S1).
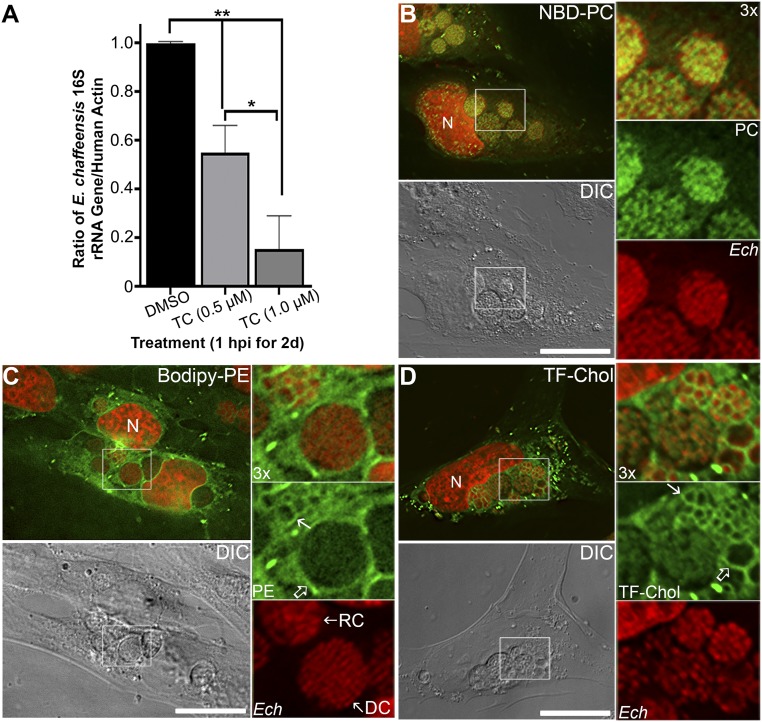
To examine whether E. chaffeensis depends on host-cell phospholipid synthesis, we used triacsin C, a potent inhibitor of host-cell long-chain acyl-CoA synthetases (ACSLs) that are required for de novo synthesis of triacylglycerols and phospholipids from glycerol (16). Treatment of E. chaffeensis-infected THP-1 cells with triacsin C revealed that E. chaffeensis is highly sensitive to inhibition of glycerolipid biosynthesis (Fig. 1). When 0.5 or 1 μM triacsin C was added at 1 h postinfection (hpi), E. chaffeensis infection in THP-1 cells was inhibited by 50 ∼ 90% (Fig. 1A and SI Appendix, Fig. S2A). As majority of E. chaffeensis had been internalized into THP-1 cells within 1 h of incubation with host cells, as shown in previous studies (17, 18), Triacsin C likely blocked E. chaffeensis proliferation within host cells instead of its internalization into the host. To examine whether triacsin C affects E. chaffeensis internalization, host THP-1 cells were pretreated with 0.5 ∼ 1 μM of triacsin C for 1 d, then infected with E. chaffeensis in the absence of triacsin C. Results showed that inhibition of host ASCLs by triacsin C had no effects on Ehrlichia infection at 2 d postinfectoin (dpi) (SI Appendix, Fig. S2 B and C), suggesting that triacsin C did not block E. chaffeensis internalization into the cells and its inhibition on ASCLs was reversible.
Fig. 1.
E. chaffeensis is dependent on host-derived lipids and incorporates exogenous phospholipids and cholesterol. (A) E. chaffeensis-infected THP-1 cells were seeded in a six-well plate. At 1 hpi, cells were incubated with 0, 0.5, or 1 μM triacsin C (TC) for 2 d at 37 °C. DNA was extracted from treated samples, and quantitative PCR was performed for the E. chaffeensis 16S rRNA gene and normalized against human ACTIN. Results are shown as the mean ± SD from three independent experiments. **P < 0.001; *P < 0.01 (ANOVA). (B–D) RF/6A cells were seeded onto cover glasses in a six-well plate for 3 h and then infected with E. chaffeensis (Ech). Cells were incubated with 25 μM NBD-PC for 1 d at 1 dpi (B), or with 5 μM Bodipy-PE for 4 h at 2 dpi (C); Alternatively, infected cells at 1 dpi were washed and replaced with AMEM containing lipoprotein-depleted serum for 8 h, then incubated with 1 μM TF-Chol for 1 d (D). Cells were fixed and DNA was stained with Hoechst 33342 to label host and bacterial DNA (pseudocolored in red). Samples were observed under a DeltaVision microscope. The boxed area in the merged image is enlarged 3× on the Right. DC, dense-core forms with diameter <1 μm and tightly packed chromosomes; DIC, differential interference contrast; RC, reticulate cell forms of E. chaffeensis with larger diameter (≥1 μm) and more loosely-packed chromosome DNAs. Open arrow, inclusion membrane; solid arrow, E. chaffeensis membrane; N, nucleus. Images are representative of at least three independent experiments. (Scale bars, 10 μm.)
Since ehrlichiae (and prokaryotes in general) lack ACSLs, when purified host cell-free E. chaffeensis was pretreated with 0.5 or 1 µM of triacsin C, and then used to infect THP-1 cells, neither Ehrlichia infection nor proliferation in host cells was affected (SI Appendix, Fig. S2D). Similarly, when E. coli was cultured in the presence of 0.5 or 1 µM of triacsin C, its growth curve was not affected (SI Appendix, Fig. S2E), indicating that triacsin C did not act directly on E. chaffeensis or other prokaryotes. Although triacsin C has variable inhibitory effects on proliferation of different mammalian cell types (19), the maximum concentration of triacsin C used in this study (1 µM) had no discernible effect on THP-1 proliferation (SI Appendix, Fig. S2) (as demonstrated by the cell numbers or host actin amount by qPCR). These results indicate that E. chaffeensis is highly susceptible to triacsin C because its proliferation is dependent on host-synthesized lipids (or at least acyl-CoA, the product of ACSLs).
Exogenous Phospholipids and Cholesterol Were Incorporated into E. chaffeensis and Inclusion Membranes in the Host Cells.
Because E. chaffeensis cannot synthesize PC (SI Appendix, Fig. S1), we examined whether a fluorescent, acyl chain-labeled PC analog 7-nitrobenz-2-oxa-1,3-diazol-4-yl-PC (C6-NBD-PC), which can be internalized and transported into yeast vacuoles (20), could be transported into inclusions of E. chaffeensis and across the bacterial membrane. As NBD-labeled lipids can mimic endogenous lipids, they are extensively used as fluorescent analogs of native lipids to study intracellular lipid transport (21, 22). Uninfected RF/6A cells were incubated with NBD-PC, and both the plasma membrane and intracellular vesicles were labeled with NBD-PC (SI Appendix, Fig. S3A). When E. chaffeensis-infected RF/6A cells were incubated with NBD-PC at 1 dpi for 1 d, the membrane of all E. chaffeensis were strongly labeled with NBD-PC (Fig. 1B). Line profile analysis of fluorescence intensity signals of NBD-PC and Hoechst 33342 (Ehrlichia DNA) showed that E. chaffeensis membranes were more strongly labeled than the plasma membrane (∼twofold) or other cytosolic vesicles (∼threefold) (SI Appendix, Fig. S4A). In addition, results suggested the inclusion membranes (SI Appendix, Fig. S4A, solid arrows) were not labeled by NBD-PC.
Bioinformatic analysis suggests that E. chaffeensis can synthesize PE by utilizing metabolic intermediates imported from the host cell using bacteria-produced ATP (SI Appendix, Fig. S1). To investigate whether E. chaffeensis can incorporate exogenous phospholipids that can be synthesized by the bacterium, we incubated the cells with Bodipy-PE [N-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine]. In uninfected RF/6A cells, Bodipy-PE labeled mostly endoplasmic reticulum and mitochondria (SI Appendix, Fig. S3B), most likely because both organelles have a higher percentage of PE in the total membrane lipids than that of plasma membranes (23). However, in E. chaffeensis-infected RF/6A cells, deconvolution microscopy and line profile analysis of fluorescence intensity signals of Bodipy-PE and Hoechst 33342 (Ehrlichia DNA) showed that Bodipy-PE strongly labeled membranes of E. chaffeensis-containing inclusions (Fig. 1C and SI Appendix, Fig. S4B, open arrows), as well as individual bacteria (Fig. 1C and SI Appendix, Fig. S4B, solid arrows). Notably, Bodipy-PE labeled mostly the reticulate cell (RC) forms of E. chaffeensis, which are metabolic active, replicating Ehrlichia with loosely packed chromosomes and larger diameter (≥1 μm), but not the dense-core forms that are metabolically inactive and smaller (<1 μm) (18, 24). These data indicate that E. chaffeensis within inclusions can incorporate host cell-derived phospholipids into their membranes to support bacterial proliferation.
We have shown E. chaffeensis purified from infected host cells readily take up exogenous cholesterol (12); however, how host membrane cholesterol is trafficked to E. chaffeensis in its inclusion is unknown. Dipyrromethene difluoride‐cholesterol (BODIPY- or TopFluor-Cholesterol [TF-Chol]) is a widely used cholesterol analog because it has strong intrinsic fluorescence (bright and photostable) and partitions in membranes similar to natural cholesterol (25, 26). When dissolved in solvent and applied to cells in growth medium containing lipoprotein-deficient serum, TF-Chol diffuses into eukaryotic cells and equilibrates slowly with intracellular membranes (26). We used this approach to visualize the distribution of host membrane cholesterol in E. chaffeensis-infected cells. As shown in SI Appendix, Fig. S3C, uninfected RF/6A cells incubated for 1 d with TF-Chol had a brightly labeled plasma membrane as well as intracellular vesicles. Notably, Ehrlichia infection of cells was not affected by TF-Chol labeling, since percent infected cells and bacterial numbers per cell were similar to those without TF-Chol incubation. Upon incubation of TF-Chol with E. chaffeensis-infected RF/6A cells at 1 dpi for 1 d, TF-Chol extensively labeled membranes of E. chaffeensis inclusions and individual bacteria as shown by fluorescence microscopy (Fig. 1D). Furthermore, line profile analysis confirmed that E. chaffeensis membranes were strongly labeled by TF-Chol (SI Appendix, Fig. S4C, solid arrows), which encircled E. chaffeensis bacteria (SI Appendix, Fig. S4C, red arrows). However, unlike NBD-PC but similar to Bodipy-PE, Ehrlichia inclusion membranes (SI Appendix, Fig. S4C, open arrows) were also strongly labeled by TF-Chol.
Unidirectional Trafficking of the Host Plasma Membrane to Membranes of E. chaffeensis Inclusions and Individual Bacteria.
Given the dependence on host synthesized lipids and incorporation of PC and free cholesterol, we investigated whether the host membrane in bulk, instead of individual components, is trafficked to E. chaffeensis. Here, we adapted a method that has been used for labeling the plasma membrane of cultured endothelial, smooth muscle, and nerve cells (27, 28) with the long alkyl-chain fluorescent indocarbocyanine dye DiI (3,3′-dioctadecylindocarbocyanine, DiIC18) (3). DiI is suitable to track membrane movement because, 1) transfer of DiI between independent membranes is usually negligible (27, 29), 2) DiI and its analogs usually exhibit very low cytotoxicity, and 3) DiI can be retained in nerve cells without leaking to neighboring cells for up to 9 mo (30).
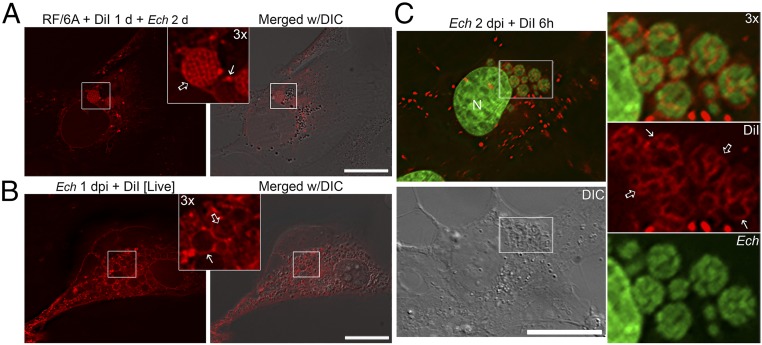
To prevent diffusion of DiI between membranes, cells were fixed with paraformaldehyde (PFA) immediately before visualization and a coverslip sealant containing no organic solvents was used. In uninfected RF/6A cells, periplasmic vesicles were brightly labeled by DiI within 15 min after incubation, and the dye was retained in intracellular vesicles at 1 d postincubation (SI Appendix, Fig. S5). As DiI initially labels the plasma membrane when applied to intact cells, labeling of intracellular vesicles was likely due to endocytosis of plasma membrane-derived vesicles (30). To examine the trafficking of membrane lipids in E. chaffeensis-infected RF/6A cells, RF/6A cells were first labeled with DiI for 1 d, then removed by washing prior to E. chaffeensis infection. At 2 dpi, the membranes of bacterial inclusions and individual Ehrlichia were strongly labeled by DiI in PFA-fixed cells (Fig. 2A, solid and open arrows, respectively), suggesting that DiI-labeled host plasma membranes could be endocytosed and trafficked to ehrlichial inclusions. Alternatively, live-cell imaging was performed in RF/6A cells at early stage of infection (1 dpi) with DiI-labeling for 15 min, and the results showed that DiI-labeled membranes were trafficked to small Ehrlichia inclusions and certain intrainclusional membranes (Fig. 2B, solid and open arrows, respectively), indicating dynamic vesicular trafficking and fusion with bacterial inclusions.
Fig. 2.
DiI-labeled host membranes are trafficked to membranes of E. chaffeensis inclusions, ILVs, and individual bacteria. (A) RF/6A cells were seeded on coverglasses in a six-well plate and incubated with 5 μM DiI for 1 d and washed three times with AMEM to remove excess dye. Cells were then infected with E. chaffeensis for 2 d. After washing, cells were fixed and observed under a DeltaVision microscope. Inlet shows 3× enlargement of the boxed area. Solid arrow, DiI-labeled inclusion membrane; open arrow, individual bacterial membranes. E. chaffeensis inclusions are visible under the DIC image. (B) RF/6A cells seeded on a 35-mm glass-bottom culture dish were infected with E. chaffeensis for 2 d. Cells were incubated with 5 μM DiI in serum-free, phenol red-free AMEM for 15 min and washed three times with AMEM to remove excess dye. Live cells were observed under a DeltaVision microscope in a heated environment. Inlet shows 3× enlargement of the boxed area. Solid arrow, inclusion membrane; open arrow, intrainclusional membranes. E. chaffeensis inclusions are visible under the DIC image. (C) RF/6A cells were seeded onto coverglasses in a 12-well plate for 1 d and then infected with E. chaffeensis for 2 d. Cells were labeled with 5 μM DiI for 6 h, fixed and stained with Hoechst 33342 (pseudocolored green), then observed under a DeltaVision microscope. The boxed area in the merged image is enlarged 3× on the right. Solid arrows, inclusion membrane; open arrows, individual bacterial membranes. Images from above panels are the representative of at least three independent experiments. (Scale bars, 10 μm.)
To confirm DiI-labeled membranes on E. chaffeensis and inclusion, E. chaffeensis-infected RF/6A cells at 1 or 2 dpi were incubated with DiI for 15 min to 1 d, then fixed and stained with DNA dye (Hoechst 33342) to label the bacteria. DiI that was initially incorporated into the host plasma membrane was endocytosed and trafficked to the inclusion membrane, as well as the ehrlichial membrane following 15 min of incubation (SI Appendix, Fig. S6A, solid and open arrows, respectively), similar to those observed by live-cell imaging. Moreover, membranes of nearly all E. chaffeensis inclusions (Fig. 2C, solid arrows) and most individual bacterium (Fig. 2C, open arrows) became labeled by DiI starting at 3 h postincubation, with over 80% of individual bacterial membranes being labeled at 1 d postincubation (Fig. 2C and SI Appendix, Fig. S6 B–D). These data suggested that DiI-labeled host cell membranes were trafficked and incorporated into membranes of Ehrlichia inclusions and individual bacteria in concert with bacterial proliferation. E. chaffeensis infection and proliferation in RF/6A cells were not affected by DiI incubation, based on a similar percentage of infected cells and bacterial numbers per cell compared to those in non-DiI–treated infected cells.
To examine whether E. chaffeensis membranes can be reciprocally transferred to host cells, host cell-free E. chaffeensis was purified and labeled with DiI for 15 min, washed, and then used to infect RF/6A cells. Live-cell imaging revealed that DiI-labeled fluorescent E. chaffeensis were internalized into and proliferated inside RF/6A cells, and the bacterial membranes remained labeled by DiI following infection time course, from 2 h to 3 dpi (SI Appendix, Fig. S7 and Movie S1). However, DiI-labeled E. chaffeensis membranes were not transferred to the membranes of inclusions or host cells (SI Appendix, Fig. S7 and Movie S1). These results indicate that membrane trafficking is unidirectional, from the host cell to Ehrlichia.
Host Membrane Trafficking to E. chaffeensis Depends on Endocytic and Early Autophagic Pathways.
The inclusions of E. chaffeensis have early endosome-like characteristics, including the presence of transferrin, transferrin receptor, the small GTPase RAB5, and its effector EEA1 (5, 9, 31–33). E. chaffeensis inclusions also express early autophagosome markers, such as VPS34, the catalytic subunit of class III phosphatidylinositol 3-kinase (PI3KC3), and ATG5, the autophagy double-membrane initiation protein (4). These vesicular trafficking and fusion pathways are essential for the obligatorily intracellular bacterium to acquire nutrients from the host for the purpose of bacterial growth and proliferation (4, 5, 31–33).
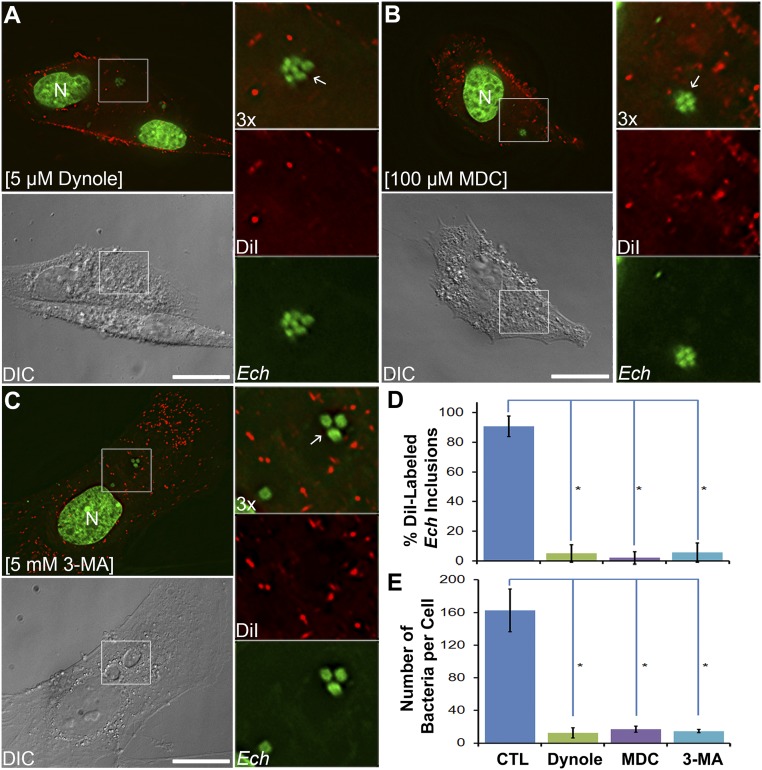
To determine whether endocytic and autophagic pathways are also involved in supplying host-cell membranes to ehrlichiae, we used inhibitors to block these pathways. Monodansylcadaverine (MDC) is an inhibitor of transglutaminase, which is involved in receptor-mediated endocytosis (34–36), and can block the infection of host cells by E. chaffeensis (37). Dynole 34-2 is a potent cell-permeable inhibitor of dynamin I and II, which are required for closing invaginated endosomal membranes (38). 3-Methyladenine (3-MA) inhibits PI3KC3 activity and thus blocks autophagy (39, 40), and it strongly inhibits E. chaffeensis infection of host cells (4). Treatment of infected cells with either of these inhibitors at 1 dpi for 1 d, followed by DiI labeling for 6 h, nearly completely blocked DiI labeling of the membranes of inclusions and bacteria (Fig. 3 A–D). In cells treated with endocytosis inhibitors, most DiI labeling remained at the host-cell plasma membrane (Fig. 3 A and B). Treatment of cells with 3-MA did not affect DiI labeling of endocytosed vesicles but completely blocked the labeling of inclusions and E. chaffeensis cells (Fig. 3 C and D), indicating that autophagy is essential for supplying membranes to E. chaffeensis. In agreement with previous findings (4, 37), all three inhibitors profoundly reduced E. chaffeensis infection of RF/6A cells (Fig. 3 A–C and E) compared with control untreated cells (Fig. 2C and SI Appendix, Fig. S6). These data demonstrate that trafficking of host-cell membranes to ehrlichiae depends on endocytic and early autophagic pathways and is required for ehrlichial proliferation.
Fig. 3.
Lipid trafficking from host membranes to Ehrlichia inclusions depends on endocytic and autophagic pathways. RF/6A cells were seeded on coverslips in a 12-well plate for 1 d and then infected with E. chaffeensis. At 1 dpi, cells were treated for 1 d with 5 μM Dynole 34-2 (A), 100 μM monodansylcadaverine (MDC, B), or 5 mM 3-methyladenine (3-MA, C). Cells were labeled with 5 μM DiI for 6 h, fixed and stained with Hoechst 33342 (pseudocolored green), then observed under a DeltaVision microscope. The boxed area in the merged images is enlarged 3× on the right. Arrows indicate E. chaffeensis inclusions not labeled by DiI. Images are representative of at least three independent experiments. (Scale bars, 10 μm.) (D) Percentage of DiI-labeled membranes of Ehrlichia inclusions or individual bacteria among total inclusions and (E) number of bacteria per host cell were quantified by counting at least 50 cells per group from three independent experiments. Results are shown as the mean ± SD; asterisk (*), significantly different from the control (CTL) group shown in Fig. 2C (P < 0.05, ANOVA).
Host Membrane Trafficking to E. chaffeensis Requires Bacterial Protein Synthesis and Is Driven by a Bacterial Type IV Secretion System Effector.
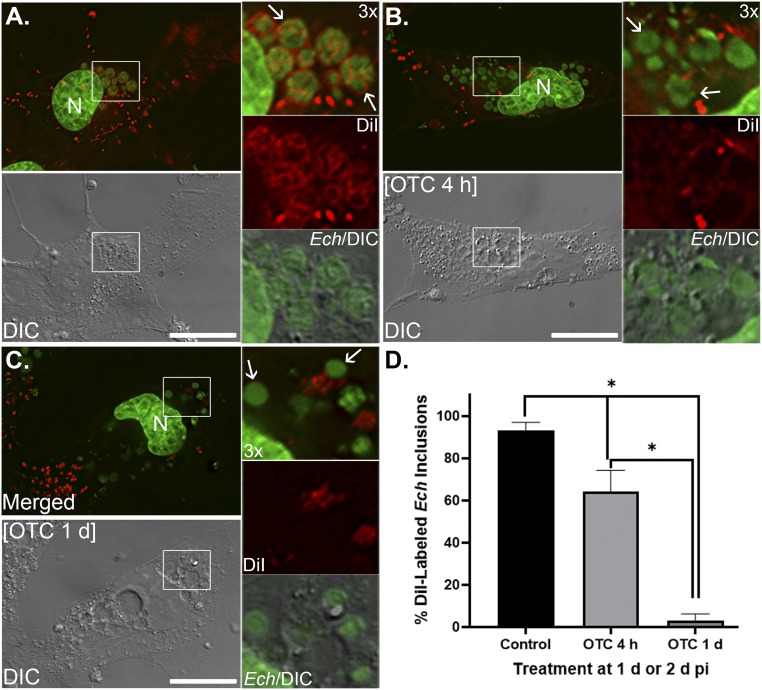
To investigate whether unidirectional trafficking of DiI-labeled host-cell membranes is driven by bacterial proteins, we treated ehrlichiae with a prokaryotic protein synthesis inhibitor, oxytetracycline, then examined the trafficking of DiI-labeled host membrane to Ehrlichia. Results showed that compared to the control groups, DiI-labeling of the membranes of Ehrlichia-containing inclusions was significantly reduced with oxytetracycline treatment for 4 h, and completely abolished at 1 d posttreatment (Fig. 4). In addition, DiI-labeling of E. chaffeensis membranes was not detectable starting at 4 h after oxytetracycline treatment (Fig. 4 B and C). Ehrlichia spp. are susceptible to tetracyclines, and previous studies showed that doxycycline (4 μg/mL) treatment for 1 d reduced the bacterial infection and growth from 62 to ∼20% (41, 42). Our study also indicates that compared to untreated cells (Fig. 4A), bacterial numbers per cell were greatly reduced in E. chaffeensis-infected RF/6A cells treated with oxytetracycline for 1 d (Fig. 4C). In addition, although DNA staining of individual bacteria became less compact, all remaining Ehrlichia inclusions were distinct under differential interference contrast microscopy (Fig. 4 B and C and SI Appendix, Fig. S8). To confirm abrogation of host membrane trafficking to Ehrlichia is due to inhibition of bacterial protein synthesis instead of bacterial death, the viabilities of Ehrlichia in the inclusions were assessed. Since gram-negative bacteria possess high cytoplasmic membrane potentials like mitochondria (43), live-staining of E. chaffeensis-infected cells were performed following oxytetracycline treatment using the membrane-potential–sensitive dye MitoTracker (44, 45). Results showed that most existing E. chaffeensis in the inclusions were still viable at 4 h or 1 d after oxytetracycline treatment (SI Appendix, Fig. S8). These data indicate that newly synthesized E. chaffeensis proteins are required for the trafficking of host membranes to Ehrlichia inclusions.
Fig. 4.
Host membrane trafficking to Ehrlichiae requires bacterial protein synthesis. RF/6A cells were seeded on coverglasses and infected with E. chaffeensis (Ech). Cells were treated with medium control (CTL, A), 5 μg/mL of oxytetracycline (OTC) at 2 dpi for 4 h (B), or at 1 dpi for 1 d (C), then labeled with DiI (5 μM) for 6 h. After fixation and staining with Hoechst 33342, cells were observed under a DeltaVision microscope. Arrows, E. chaffeensis–containing inclusions. (Scale bars, 10 μm.) (D) Percentage of DiI-labeled membranes of Ehrlichia inclusions among total inclusions were quantified by counting at least 10 cells per group, and results are shown as mean ± SD. Asterisk (*), significantly different from the control group and between OTC treatment time courses (P < 0.01, ANOVA).
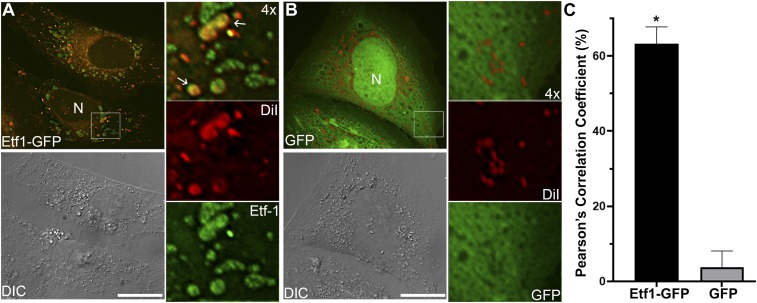
In E. chaffeensis-infected host cells, the bacteria secrete the effector protein Ehrlichia translocated factor-1 (Etf-1) into the host cytoplasm through a type IV secretion system (T4SS) (4, 15), which is essential for ehrlichial proliferation (46). Etf-1–induced early autophagosomes have the characteristics of amphisomes (47), as these vesicles retain RAB5-GTP, VPS34, and ATG5 (4). To investigate whether endosomes derived from the DiI-labeled plasma membrane can fuse with Etf-1–induced autophagosomes, RF/6A cells were transfected with Etf-1–GFP and labeled with DiI at 1 d posttransfection. More than 60% of Etf-1–containing vesicle membranes were labeled with DiI compared with the GFP control as measured by Pearson’s correlation coefficient (Fig. 5), confirming that Etf-1–induced autophagosomes fuse with endosomes. As autophagosomes fuse with morphologically distinct endosomes (47) called multivesicular bodies (MVBs) to form amphisomes, they internally accumulate small membrane vesicles (60 to 80 nm) containing cytoplasmic cargo molecules (48, 49). Because Etf-1–containing vesicles can fuse with Ehrlichia inclusions (4), these results suggest that Etf-1–induced amphisomes can likely deliver intraluminal vesicles (ILVs) into Ehrlichia inclusions to provide membrane lipid components.
Fig. 5.
DiI-labeled host membranes are trafficked to E. chaffeensis T4S effector Etf-1–induced autophagosomes. RF/6A cells were seeded onto coverglasses in a 12-well plate and transfected with Etf-1–GFP (A) or GFP control (B) plasmids. At 1 d posttransfection, cells were labeled with 5 μM DiI for 1 d, then fixed and observed under a DeltaVision microscope. The boxed area in the merged images is enlarged 4× at right. Arrows indicate the DiI-labeled Etf-1–containing vesicles. Images are representative of three independent experiments. (Scale bars, 10 μm.) (C) Colocalization of DiI labeling with Etf1-GFP or GFP was analyzed using Pearson’s correlation coefficient in transfected RF/6A cells by SoftWoRx software. Asterisk (*), significantly different by Student’s unpaired t test (P < 0.01).
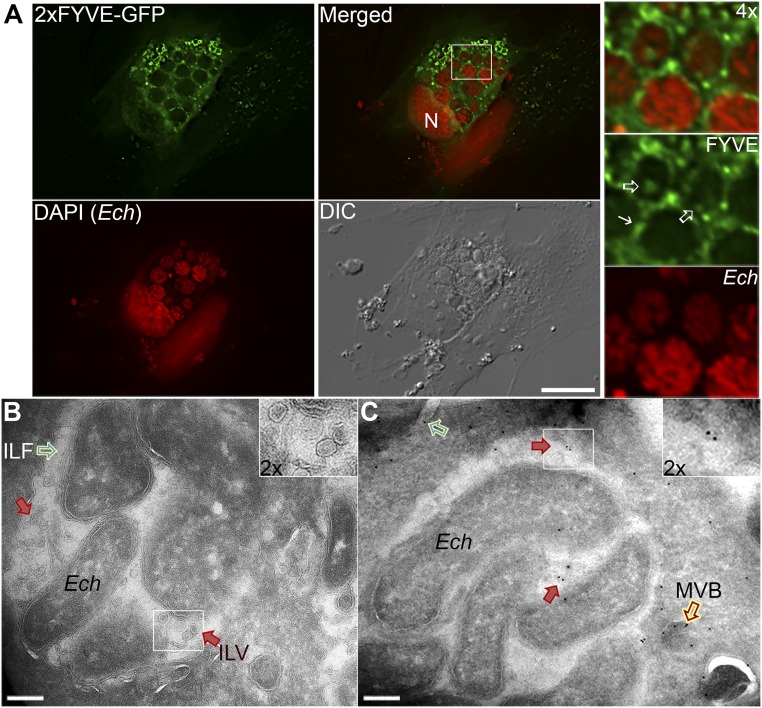
We previously investigated whether Ehrlichia inclusion membranes are enriched with phosphatidylinositol 3-phosphate (PI3P), which is produced by activated PI3KC3 and is highly enriched on early endosomes and in ILVs of MVBs (50). Both VPS34 and EEA1 or RabAnk5, which localize to early endosomal membranes by binding to PI3P, localize to Ehrlichia inclusions (2, 4), indicating the enrichment of PI3P on the inclusion membranes. Indeed, ectopically expressed double-FYVE finger (2×FYVE)-GFP, a fluorescent finger protein probe with unique specificity for PI3P, produced large puncta (vesicles) and was localized and enriched on E. chaffeensis inclusion membranes at 2 dpi (4) (Fig. 5A, solid arrow). Upon closer observation, localization of 2×FYVE-GFP was also detected on ILV membranes within Ehrlichia inclusions (Fig. 6A, open arrows), suggesting that MVBs are a source of intraluminal membranes in inclusions.
Fig. 6.
E. chaffeensis inclusion membranes and ILVs are enriched with PI3P. (A) RF/6A cells were infected with E. chaffeensis for 1 d and transfected with a plasmid encoding 2×FYVE-GFP. At 2 dpi (1 d posttransfection), cells were fixed, permeabilized, and stained with DAPI (pseudocolored red). Images were captured using a DeltaVision microscope. The boxed area in the merged image is enlarged 4× at right. Solid arrow, inclusion membrane; open arrows, intrainclusion vesicles; N, nucleus. (Scale bar, 10 μm.) (B and C) HEK-293 cells were infected with E. chaffeensis for 1 d, then transfected with 2×FYVE-EGFP plasmid by electroporation and cultured for 2 d. Cells were fixed using 4% PFA + 0.2% glutaraldehyde at 37 °C for 1 h. (B) A representative cryoelectron micrograph shows ILV (red arrows) and an intraluminal filament (ILF, green open arrow). (C) For immunogold labeling of 2×FYVE-GFP, frozen/thawed sections through fixed, cryoprotected specimens were incubated with mouse anti-GFP, followed by incubation with colloidal gold (10 nm) conjugation to protein A. Labeling of 2×FYVE-GFP was detected on vesicles inside Ehrlichia inclusions (red solid arrows) and MVBs docked onto ehrlichial inclusions (red open arrow). Another ILF attached to the top left bacterium was also visible (green open arrow). (Scale bars, 0.2 μm.)
To confirm the presence of ILVs within inclusions, HEK293 cells, which can be transfected at high efficiency (>80%) and can be readily infected with E. chaffeensis (4, 17), were used for cryosectioning and immunogold labeling. EM images of cryosections from 2×FYVE-GFP–transfected HEK-293 cells infected with E. chaffeensis had abundant ILVs (Fig. 6B, red arrows) inside inclusions, as well as numerous intraluminal filaments juxtaposed to Ehrlichia surface (Fig. 6 B and C, green open arrow). Immunogold labeling of these cells using a monoclonal antibody against GFP confirmed the presence of vesicles (Fig. 6C, red arrows) within inclusions and MVBs docked onto ehrlichial inclusions (Fig. 6C, red open arrow). Labeling with 2×FYVE-GFP was, however, rarely detected on Ehrlichia. These data supported the idea that E. chaffeensis inclusions contain a large number of intraluminal vesicles derived from MVBs to provide a membrane source for bacterial proliferation. As ILVs of MVBs are cholesterol-rich (50), it is likely that Ehrlichia can also obtain cholesterol from ILVs.
Three-Dimensional Imaging of E. chaffeensis Inclusions by FIB-SEM Substantiates a Large Amount of ILVs.
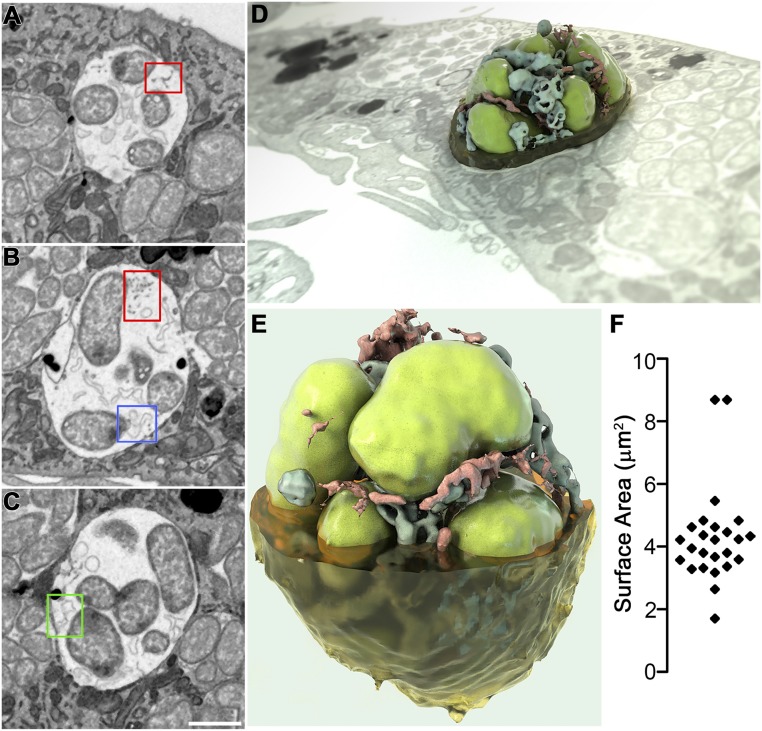
Previous studies with transmission electron microscopy (TEM) revealed E. chaffeensis-inclusions contain membrane vesicles, tubules, and fibrillary materials (7, 24, 51, 52). To further analyze the intraluminal structures, high-resolution 3D images of E. chaffeensis-infected cells were generated by FIB-SEM [also referred to as ion abrasion SEM (53)]. Visualization of the 3D ultrastructure of E. chaffeensis-infected canine DH82 macrophages showed the presence of numerous inclusions that contained large numbers of E. chaffeensis (Fig. 7 and Movies S2–S4). Fusion between inclusions and lysosomes was not observed in any of the infected cells (Fig. 7 and Movies S2 and S3), validating previous results using TEM (7) and immunofluorescence microscopy (2, 5). In each SEM image, numerous ILVs or tubules at 50- to 200-nm diameter were observed, either unattached or attached to the bacterial or inclusion membrane (Fig. 7 A–E and Movies S3 and S4). In addition, bundles of tiny filaments at 10- to 30-nm diameter with connections to bacterial membranes (14) were reproducibly found in every inclusion (Fig. 7 A and B). A 3D reconstruction of an inclusion (Fig. 7 D and E and Movie S4) revealed ILVs and filaments squeezed between tightly packed bacteria.
Fig. 7.
Three-dimensional imaging of intraluminal structures in E. chaffeensis-containing inclusions by FIB-SEM. E. chaffeensis-infected DH82 macrophages at 2 dpi were embedded in resin and subjected to iterative milling with a focused ion beam followed by imaging by SEM. For 3D imaging, a region of interest (red box in Movie S2) through one inclusion was selected and segmented. Image data were binned three times in the xy plane to give a final voxel size of 15 × 15 × 15 nm, and individual 2D images were merged, cropped, aligned, and then reconstructed to produce Movie S3. (A–C) Selected 2D slices through one inclusion demonstrated the presence of filaments or vesicles connected to the inclusion membrane (A) or bacterial (B) membrane (red boxes); branching tubules attached at one edge to either the bacterial membrane (B, blue box), or inclusion membrane (C, green box). (Scale bar, 1 μm.) (D and E) Reconstructed 3D images representing a bacterial vacuole inside a cell or viewed at a different angle (3D ultrastructural volume of the sample is shown in Movie S3). Segmentation shows one vacuole with bacteria (green), filaments (red), and vesicles (blue). (F) Scatter plot of the surface area of each individual bacterium in Movie S3 (4 μm2 average).
Also consistent with the TEM results, most bacteria were ∼1.0 µm in diameter with a mostly oval to round shape with evenly distributed ribosomes and nucleoid DNA fibrils, which were the RC form in the exponential growth phase (18, 24) (Fig. 7 and Movies S2 and S3). A scatter plot depicting the distribution of the estimated membrane surface area of individual bacteria revealed an average surface area of 4 μm2 (Fig. 7F). Despite this small size, Ehrlichia spp. can generate a substantial amount of total membrane when reproducing in infected cells. In in vitro culture of E. chaffeensis in canine DH82 macrophage cells (∼1,200-μm2 plasma membrane area, ∼20-μm diameter), over 200 bacteria are produced in a single host cell within 2 to 3 d (14, 54). Because E. chaffeensis is enveloped by a double phospholipid bilayer, the total bacterial membrane (inner and outer) generated within a single cell is estimated to be ∼1,600 μm2, which is greater than the surface area of the host cell plasma membrane. The presence of a large amount of ILVs in Ehrlichia inclusions, which is derived from host membranes, could provide membrane lipid components to the bacterium and membrane-bound ehrlichial inclusions for its survival and rapid proliferation.
Discussion
The present study reveals that E. chaffeensis uses host-cell membranes as a source for not only cholesterol but also glycerophospholipids for its own survival and proliferation. As the E. chaffeensis genome does not encode biosynthetic pathways for cholesterol (55) and the bacterium is partially defective in de novo glycerophospholipid biosynthesis (SI Appendix, Fig. S1), actively hijacking these lipids from host cells is a remarkable adaptation for survival of this obligate intracellular pathogen. It also allows Ehrlichia to masquerade as a part of the host cell to avoid innate immune recognition. The present study further reveals the involvement of endosomes, autophagosomes, and MVBs activated by bacterial T4SS effectors in ehrlichial acquisition of host-cell membrane lipids. This model of actively formed intrainclusion membranes represents a substantial boost in our understanding of how obligate intracellular pathogens proliferate, and may yield new approaches to overcome infection.
Hijacking and utilization of host lipids are known for intracellular pathogens (reviewed in refs. 56–59). For example, the closely related bacteria in the order Rickettsiales, Anaplasma phagocytophilum, hijacks cholesterol from NPC1-mediated LDL-cholesterol vesicular trafficking pathways (60–62). Membrane extracts of Rickettsia prowazekii contain PC and cardiolipin (63), which cannot be synthesized by this bacterium based on analysis of the recent genome sequence data (64), suggesting that the lipids were derived from host cells and incorporated into rickettsial membranes. Whether or how Ehrlichia utilizes a similar pathway of cholesterol acquisition or acquire cardiolipin remain to be investigated.
Host lipid uptake mechanisms have been extensively studied in an obligatory intravacuolar bacterium Chlamydia trachomatis. C. trachomatis encodes numerous genes for de novo biosynthesis of fatty acids, phosphatidylserine, PE, and phosphatidylglycerol (65), but this obligate intracellular pathogen also recruits and modifies a variety of host-derived lipids, including PC, phosphatidylinositol, sphingomyelin, and cholesterol (66–69). In addition, cytoplasmic lipid droplets (LDs) are translocated into the lumen of Chlamydia-containing vacuoles (70, 71). C. trachomatis utilizes host ceramide-transfer proteins, fusion with MVBs/late endosomes, and coopting of endoplasmic reticulum-Golgi trafficking mechanisms to acquire host lipids (72–74). Chlamydia also recruits host-cell ACSLs to inclusions to synthesize bacterial phospholipids (75). A large group of Chlamydia inclusion membrane proteins, secreted by the bacterial type III secretion systems and localized at Chlamydia inclusion membrane, can modulate host-cell vesicular trafficking, Golgi redistribution, and cytoskeleton dynamics to promote lipid acquisition by the bacterium (reviewed in ref. 76).
Another intravacuolar bacterium, Coxiella burnetii, replicates inside a large acidic-phagolysosome–like parasitophorous vacuole (PV) within macrophages. Host membrane lipids including cholesterol are critical for PV formation and maintenance (77). C. burnetii PV is rich in sterol (77), and the bacterium encodes two sterol reductase enzymes functioning in the very last steps of mammalian cholesterol biosynthesis (56). However, paradoxically PV formation and intracellular replication of C. burnetii don’t require cholesterol, and increasing PV cholesterol, rather, leads to Coxiella death (78, 79). In contrast, LD lipolysis, which is likely regulated by Coxiella T4SS, is critical for bacterial growth (80). Coxiella T4SS effector CvpA involved in the regulation of clathrin-mediated vesicular trafficking events, possibly assisting C. burnetii acquisition of lipids and proteins for PV biogenesis (81).
Although the presence of ILVs in bacterial inclusion compartments is similar to chlamydial inclusions (72), ehrlichial inclusions are distinct from Chlamydia or Coxiella inclusions in that they lack LDs and retain early endosome and early-autophagosome characteristics, and all these aspects are facilitated by its two ehrlichial T4SS effectors (2, 4, 5, 82). The ehrlichial effector Etf-2 binds directly to RAB5-GTP and blocks the RAB5-specific GTPase-activating protein from binding to RAB5-GTP, keeping RAB5-GTP on ehrlichial inclusions and thereby delaying endosomal maturation and blocking fusion between inclusions and lysosomes (5). The T4SS effector Etf-1 nucleates preautophagosome formation in a RAB5-GTP–dependent manner utilizing early endosomal membranes as a scaffold to assemble the early-autophagosomal protein complex Etf-1/RAB5-GTP/VPS34/Beclin1 (4, 8). The PI3KC3 complex VPS34/Beclin1 is the key autophagy nucleation factor (83). Keeping RAB5 in a GTP-bound state is thus essential for ehrlichial survival and proliferation (4, 5). Furthermore, the RAB5 effector VPS34 is recruited to the inclusion membrane, as demonstrated here and previously (4). PI3KC3 activation generates PI3P, and E. chaffeensis infection doubles the level of host-cell PI3P (4). Immunofluorescence labeling showed that PI3P was localized and enriched on E. chaffeensis inclusion membranes, and detected on ILV membranes within Ehrlichia inclusions that were also confirmed by immunogold labeling. However, the current immuno-EM study with cryo-protected but freeze–thawed samples revealed a lesser amount of PI3P on ehrlichial or inclusional membranes; whether this was due to limited accessibility of probes, or PI3P is degraded on, or excluded from ehrlichial membrane remains to be determined. E. chaffeensis encodes a phospholipase (ECH_0935), and it would be interesting to know if it functions in regulating PI3P within Ehrlichia inclusions. PI3P or other phosphoinositides have been identified on the vacuolar membranes containing other intravacuolar bacteria like Mycobacterium, Legionella, and Salmonella, but never inside their inclusions or bacteria themselves (84). Nevertheless, since PI3P is a key player in membrane dynamics and trafficking regulation of endosomes and autophagosomes (85), these data suggest that MVBs in inclusions are a source of intraluminal membranes that provides lipids for rapid proliferation of Ehrlichia.
Membranes of different vesicles and organelles, as well as in cells and bacteria, maintain distinct protein and lipid compositions. This study shows that exogenous lipids traffic to the E. chaffeensis membrane with distinctive localizations: Bodipy-PE and TF-Choll are trafficked to both bacterial and inclusional membranes, whereas NBD-PC is only localized to bacterial membranes (Fig. 1). It is possible that the probe distribution pattern is influenced by the fluorophore. Unlike TopFluor or Bodipy fluorophore that strongly fluoresces in both aqueous and lipid environments, the NBD moiety exhibits remarkable sensitivity to environmental polarity, which is very weakly fluorescent in water but fluoresces brightly in a hydrophobic environment (86). As NBD-PC signal did not overlap with DAPI signal, NBD-PC is likely in ehrlichial membrane, rather than in ehrlichial cytoplasm. However, membrane distribution patterns are distinct: TF-Chol and Bodipy-PE thoroughly envelop individual Ehrlichia DNA, whereas NBD-PC only partially envelops individual Ehrlichia DNA (Fig. 1 and SI Appendix, Fig. S4). Absence of NBD-PC on the ehrlichial inclusion membrane is striking. Since E. chaffeensis encodes a single phospholipase (ECH_0935), whether NBD-PC is modified in the ehrlichial inclusion membrane by this enzyme or NBD-PC is simply excluded from the inclusion membrane remains to be studied. A previous study with purified R. prowazekii reported over 60% of its membrane lipids are PE (63), although it is unknown whether PE is synthesized by Rickettsia or derived from the host cells. Although E. chaffeensis encodes enzymes for PE biosynthesis, exogenous Bodipy-PE is rapidly trafficked to the membranes of Ehrlichia-containing inclusions and individual bacteria during the proliferating RC stage, suggesting that E. chaffeensis may incorporate these membrane lipids from the host for its rapid growth and replication.
The mechanisms by which membrane specificity is maintained are still unclear, but are likely achieved by specific protein–lipid interactions, controlled lateral diffusions, and nonrandom removal of membrane components (87). The extracellular bacterium Borrelia burgdorferi, the agent of tick-borne Lyme disease, also lacks LPS in its membrane (88). Instead, its membranes contain free cholesterol and cholesterol glycolipids that exist as lipid raft-like microdomains, and the growth of this spirochete requires an external cholesterol source (57, 89–91). Studies suggested that Borrelia encodes cholesterol modification enzymes to synthesize cholesterol glycolipids (92, 93), and Borrelia membrane lipoproteins like OspA and OspB can interact with cholesterol lipids (89, 94). However, unlike Borrelia that exchanges cholesterol and phospholipids bidirectionally with host cells through direct contact and outer membrane vesicles (95), trafficking of cholesterol and phospholipids is unidirectional from host cells to Ehrlichia. Given the nature of the obligatory intravacuolar bacteria, it is likely that Ehrlichia maintains the specificities of bacterial and inclusional membranes through specific interactions with host vesicles and nonrandom removal of host components.
In eukaryotes, acyl-CoA is an essential intermediate metabolite for de novo synthesis of phospholipids, triacylglycerol, and cholesterol esters, as well as for degradation pathways that generate ATP, such as β-oxidation (96). Triacsin C, an inhibitor of host-cell ACSLs that are essential for acyl-CoA synthesis (97), significantly blocked E. chaffeensis infection of host cells, suggesting that host de novo synthesized lipids are required for E. chaffeensis proliferation. In contrast to Ehrlichia, Coxiella intracellular growth was enhanced by triacsin C treatment that blocked LD formation (80). E. chaffeensis is more sensitive to triacsin C (∼90% inhibition of growth at 1 µM) than C. trachomatis (56% inhibition at 7.5 µM), or eukaryotic cells (cell type-dependent, but usually IC50 > 1 µM) (16, 75). As the only current choice for treatment of human monocytic ehrlichiosis is the broad-spectrum antibiotic doxycycline, which is only effective when initiated early, exploitation of ehrlichial dependence on host-cell glycerolipid biosynthesis offers potential for development of a novel therapy.
In summary, our study reveals that ehrlichiae actively hijack host intracellular membrane traffic to recycle host membrane lipids to construct their own membranes for rapid intracellular proliferation. Although many aspects of intrainclusion membrane transport remain to be explained, our study allows us to integrate a number of previous observations for E. chaffeensis inclusion assembly and membrane acquisition into a single model. While Ehrlichia employs mechanisms distinct from those of other bacteria to subvert host lipids for its proliferations, the present study provides a better understanding of the roles of lipids in the complex interplay between a pathogen and its host.
Materials and Methods
Bacteria, Cell Culture, and Plasmids Transfection.
Different types of cells were used in this study for specific applications, and all cells can be readily infected with E. chaffeensis with high infectivities (4). Human acute myelocytic leukemia THP-1 cells (ATCC) were used for analysis of E. chaffeensis infection levels and qPCR analysis, since they are human monocytes that are natural hosts of E. chaffeensis infection (98), and are nonadherent, allowing rapid detection of infection levels by light microscopy and DNA extraction for qPCR analysis. RF/6A endothelial cells were used for immunofluorescence labeling assays, because they are adherent cells with thinly spread morphology, and can be infected with E. chaffeensis with high bacterial burden, suitable for immunofluorescence and unambiguous localization analysis (4). Human embryonic kidney HEK293 cells were used for transfection and immunogold-labeling experiments as they can be transfected with high efficiency (4, 99). Canine histiocytic leukemia DH82 cells (14) were used for FIB-SEM studies, since this cell line was used to culture isolate E. chaffeensis from human moncytic ehrlichiosis patient blood samples, and E. chaffeensis is able to reproduce in large quantities (over 200 bacteria per cell) inside 10 to 20 spacious inclusions in the cell, which makes the observation of bacteria inside cells much easier under EM than in other types of cells.
The E. chaffeensis Arkansas strain (100) was cultured in THP-1 cells in RPMI medium 1640 (Mediatech) supplemented with 10% FBS (Atlanta Biologicals) and 2 mM l-glutamine (Gibco). RF/6A cells (ATCC) were cultured in advanced MEM (AMEM, Gibco) supplemented with 5% FBS and 2 mM l-glutamine. DH82 and HEK293 cells (ATCC) were cultured in DMEM (Mediatech) supplemented with 10% FBS and 2 mM l-glutamine. Cultures were incubated at 37 °C under 5% CO2 in a humidified atmosphere. Host cell-free E. chaffeensis was purified from heavily infected THP-1 cells by sonication and subsequent filtration through 5- and 2.7-μm syringe filters, as previously described (4).
Plasmids encoding the Etf-1–GFP fusion protein and 2×FYVE-GFP were described previously (4). RF/6A cells were transfected using Fugene HD (Promega). HEK293 cells (2 × 106 cells in 100 μL OptiMEM medium; Invitrogen) were transfected with 2×FYVE-EGFP plasmid (5 μg) by electroporation (voltage, 100 V; capacitance, 1,000 F; resistance, ∞) in a 0.2-cm cuvette using a Gene Pulser Xcell Electroporation System (Bio-Rad), as previously described (60).
Triacsin C Treatment and qPCR Analysis.
For treatment with the ACSL inhibitor triacsin C (StressMarq Biosciences), E. chaffeensis-infected THP-1 cells at 1 hpi were incubated with 0.5 or 1 µM triacsin C and cultured for an additional 2 d at 37 °C. Alternatively, THP-1 cells were pretreated with 0.5 or 1 μM of triacsin C for 1 d, washed, then infected with E. chaffeensis for 2 d. To examine the effects of triacsin C on bacteria, purified host cell-free E. chaffeensis was treated with 0.5 or 1 µM triacsin C at 37 °C for 30 min, washed twice with RPMI medium 1640, then used to infect THP-1 cells for 3 d. For E. coli treatment, overnight cultures of E. coli DH5α were diluted 1:100 in LB media, and aliquoted 4-mL each into 14-mL tubes in duplicate. Triacsin C (0.5 to 1 μM) were added to E. coli, and the growth curve was measured by OD600 at 37 °C, 275 rpm. As a control, an equal amount of DMSO solvent was used to treat cells or bacteria under the same conditions. Aliquots of cells were cytospun onto slides, and infectivity was assessed by Diff-Quik staining using a Hema 3 stain set (Thermo Fisher Scientific).
For qPCR analysis, DNA samples were purified from the remaining cells and subjected to qCR analysis of the E. chaffeensis 16S rRNA gene with SYBR green qPCR master mix (Thermo Fisher) in an Mx3000P qPCR system (Stratagene). Expression was normalized against human ACTIN, as previously described (5, 6).
Uptake and Trafficking of NBC-PC, TF-Chol, and DiI.
RF/6A cells were seeded onto glass coverslips in a six-well plate and cultured in AMEM supplemented with 5% FBS and 2 mM l-glutamine at 37 °C for 1 d. For exogenous lipid uptake, uninfected or E. chaffeensis-infected RF/6A cells at 1 dpi were incubated with 25 µM NBD-PC (Avanti Polar Lipids) dissolved in DMSO for 1 d, or with 5 μM of Bodipy-PE (Thermo Fisher Scientific) dissolved in DMSO for 4 h at 2 dpi. For TF-Chol (Avanti Polar Lipids), uninfected or infected RF/6A cells at 1 dpi were washed three times with serum-free AMEM, and replaced with AMEM supplemented with 5% lipoprotein-depleted serum (Kalen Biomedical). After culturing for 8 h, TF-Chol (1 μM) dissolved in DMSO was added to cells and incubated for 1 d.
For live-cell imaging, RF/6A cells were grown in a 35-mm chambered glass-bottom culture dish (WillCo Wells) and cultured in phenol red-free AMEM supplemented with 5% FBS and 2 mM l-glutamine. Cells were infected with E. chaffeensis for 2 d and labeled with 5 μM Vybrant DiI cell-labeling solution (Thermo Fisher) for 15 min. Cells were washed three times with AMEM to remove excess dye prior to DeltaVision microscopy. Alternatively, host cell-free E. chaffeensis was incubated with 5 μM DiI for 15 min, washed twice with medium, and used to infect RF/6A cells seeded on a 35-mm glass-bottom culture dish. After 2-h incubation, cells were washed to remove uninternalized bacteria (with time point set as 0 hpi), and cultured in phenol red-free AMEM containing 5% FBS and 2 mM l-glutamine for 3 d.
For time course studies of DiI trafficking, uninfected or E. chaffeensis-infected RF/6A cells at 1 ∼ 2 dpi were incubated with 5 µM DiI for 15 min, 3 h, 6 h, and 1 d. For treatment with inhibitors of endocytic and autophagic pathways, RF/6A cells infected with E. chaffeensis at 1 dpi were incubated with 5 mM 3-methyladenine (Sigma), 5 μM Dynole 34-2 (Abcam), or 100 μM monodansylcadaverine (Sigma). DiI (5 μM) was added at 1 d postinhibitor treatment, and cells were incubated with DiI for 6 h.
For antibiotic oxytetracycline (Sigma) treatment, E. chaffeensis-infected RF/6A cells were treated with 5 μg/mL of oxytetracycline at 1 dpi for 1 d, or at 2 dpi for 4 h, then incubated with DiI for 6 h. To determine the viability of E. chaffeensis within infected RF/6A cells, infected cells following oxytetracycline treatment were incubated with 500 nM MitoTracker Deep Red FM (Thermo Fisher Scientific) in growth media for 30 min at 37 °C. After washing with PBS, cells were fixed in 100% ice-cold methanol for 15 min at -20 °C, and washed three times with PBS for 5 min each.
DeltaVision Deconvolution Microscopy and Image Analysis.
For labeling with fluorescent lipid tracers, no permeabilization or organic solvents were used during fixation, labeling, and mounting to prevent nonspecific diffusion of dyes within host cells. Briefly, cells were washed three times with PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), then fixed in 4% PFA in PBS for 20 min at room temperature. DNA was stained with 1 μg/mL Hoechst 33342 (Invitrogen) in PBS for 15 min.
Alternatively, for Etf-1–GFP or 2×FYVE-GFP transfected RF/6A cells, fixed cells were permeabilized in PGS (PBS supplemented with 0.5% BSA [Sigma], 0.1% gelatin [Sigma], and 0.15% saponin [Sigma]) for 15 min and then incubated with 300 nM DAPI (Invitrogen) to label both host-cell and Ehrlichia DNAs.
Following fixation and labeling, glass coverslips were mounted with SlowFade Diamond Antifade Mountant (Invitrogen) and sealed with a coverslip sealant without organic solvents (Biotum). Fluorescence and differential interference contrast images were captured with a DeltaVision Personal DV Deconvolution microscope system (GE Healthcare). For live-cell imaging, the temperature of the stage and chamber was maintained at 37 °C with humidified air containing 5% CO2 on the DeltaVision system, and time-lapse images were captured using 15-s interval for 10 min.
Line profile and colocalization analyses were performed on a single z-section using SoftWoRx software (GE Healthcare). Percentage colocalization of membrane labeling by various fluorescent lipid tracers in E. chaffeensis inclusions was obtained by counting intrainclusion vesicles or individual bacteria within Ehrlichia inclusions in over 20 cells per experiment from three independent experiments. Statistical analysis was performed by analysis of variance, and P < 0.05 was considered statistically significant. Colocalization of Etf-1–GFP or GFP with DiI was analyzed by SoftWoRx software for the Pearson’s correlation coefficient from 10 to 20 cells per group from three independent experiments.
Immunogold Labeling and Electron Microscopy.
For 2×FYVE-GFP-transfected HEK293 cells with or without E. chaffeensis infection, cells were briefly treated with 0.3% trypsin and mixed with an equal volume of prewarmed (37 °C) 8% formaldehyde + 0.4% glutaraldehyde in 200 mM sodium phosphate buffer (pH 7.4) for 30 min. Cells were centrifuged at 150 × g for 3 min to prevent damage and resuspended in fresh prewarmed 1× fixative (4% formaldehyde + 0.2% glutaraldehyde) in 100 mM sodium phosphate buffer (pH 7.4) at room temperature for an additional 30 min. After centrifugation, each pellet was rinsed three times (5 min each) in sodium phosphate buffer and resuspended in 5% molten gelatin. Cells were immediately centrifuged again, and the tubes were placed on ice to solidify the gelatin. The solidified pellet was cut into cubes that were placed in 2.3 M sucrose containing 15% polyvinylpyrrolidone for 16 h. The cubes were frozen onto Leica specimen stubs and sectioned at −130 °C using a Leica Ultracut UCT cryo-ultramicrotome (Leica Microsystems). Sections of 80- to 120-nm thickness were picked up with a droplet containing methylcellulose and sucrose, as described previously (101), and placed on carbon-coated formvar grids. Sections were sequentially labeled with anti-GFP (Abcam) and 10-nm gold-labeled protein A (Utrecht University, Utrecht, The Netherlands), embedded in 2% methylcellulose, and contrast-stained with 0.3% uranyl acetate. After removing excess fluid, grids were allowed to air dry, and labeling patterns were assessed using a Tecnai-T12 TEM (FEI).
FIB-SEM.
E. chaffeensis-infected DH82 macrophages were prepared according to Ito and Rikihisa’s rickettsial EM protocol (102). The embedded resin block was trimmed and mounted on a specimen stub with conductive epoxy (Chemtronics) and sputter-coated with gold. An NVision 40 dual-beam microscope (Carl Zeiss) equipped with Atlas3D (Fibics Inc.) was used for data collection, as previously described (103). Resin-embedded samples are subjected to an iterative process of milling (slicing) with a focused gallium ion beam and imaging by SEM. A focused gallium ion beam (700 pA) iteratively removed slices every 15 nm. The scanning electron beam was used to record images at pixel sizes of 5 nm in the xy plane for a voxel size of 5 × 5 × 15 nm. SEM images were recorded using an energy-selective backscattered electron detector. Data volumes with a clear presence of E. chaffeensis and sufficient levels of infection were selected. Figures in this work were derived from two datasets; the first set had dimensions of 15 × 30 μm in the xy plane and with 1,181 images making up the z-stack for a final volume of ∼9,975 μm3, and the second set had dimensions of ∼17 × 35 μm in the xy plane and with 1,181 images making up the z-stack for a final volume of 7,735 μm3. For image processing, data were binned three time in the xy plane to give a final voxel size of 15 × 15 × 15 nm. Individual 2D images were merged, cropped, and aligned using customized scripts based on the image processing program IMOD (University of Colorado, Boulder, CO) (104). Features of interest were manually selected with 3DSlicer (https://www.slicer.org/) (105). The sequential stacks of 2D images were converted computationally to produce Movies S2 and S3, and sequential stacks of 2D images in Movie S3 were computationally converted to a 3D ultrastructural volume of the sample (Movie S4). An improved key-frame imaging strategy yielded high-resolution (10 nm) 3D ultrastructural images of local regions of interest (106).
Statistical Analysis.
Statistical analysis was performed with Student’s unpaired t test or analysis of variance, and P < 0.05 was considered statistically significant. All statistical analyses were performed using Prism 8 software (GraphPad).
Data Availability.
All experimental data described in this study are included in the main text and SI Appendix.
Supplementary Material
Acknowledgments
We thank Tim Vojt at College of Veterinary Medicine, The Ohio State University for assistance in the preparation of SI Appendix, Fig. S1 and Movies S2 and S3.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921619117/-/DCSupplemental.
References
- 1.Paddock C. D., Childs J. E., Ehrlichia chaffeensis: A prototypical emerging pathogen. Clin. Microbiol. Rev. 16, 37–64 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mott J., Barnewall R. E., Rikihisa Y., Human granulocytic ehrlichiosis agent and Ehrlichia chaffeensis reside in different cytoplasmic compartments in HL-60 cells. Infect. Immun. 67, 1368–1378 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin M., Rikihisa Y., Degradation of p22phox and inhibition of superoxide generation by Ehrlichia chaffeensis in human monocytes. Cell. Microbiol. 9, 861–874 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Lin M., et al. , Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase. Autophagy 12, 2145–2166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan Q., et al. , Ehrlichia type IV secretion system effector Etf-2 binds to active RAB5 and delays endosome maturation. Proc. Natl. Acad. Sci. U.S.A. 115, E8977–E8986 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teymournejad O., Lin M., Rikihisa Y., Ehrlichia chaffeensis and its invasin EtpE block reactive oxygen species generation by macrophages in a DNase X-dependent manner. MBio 8, e01551-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rikihisa Y., Clinical and biological aspects of infection caused by Ehrlichia chaffeensis. Microbes Infect. 1, 367–376 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Rikihisa Y., Subversion of RAB5-regulated autophagy by the intracellular pathogen Ehrlichia chaffeensis. Small GTPases 10, 343–349 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnewall R. E., Rikihisa Y., Lee E. H., Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect. Immun. 65, 1455–1461 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schäffer C., Messner P., The structure of secondary cell wall polymers: How gram-positive bacteria stick their cell walls together. Microbiology 151, 643–651 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Costerton J. W., Ingram J. M., Cheng K. J., Structure and function of the cell envelope of gram-negative bacteria. Bacteriol. Rev. 38, 87–110 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin M., Rikihisa Y., Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect. Immun. 71, 5324–5331 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunning Hotopp J. C., et al. , Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2, e21 (2006). Correction in: PLoS Genet.2, e213 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rikihisa Y., The tribe Ehrlichieae and ehrlichial diseases. Clin. Microbiol. Rev. 4, 286–308 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H., Bao W., Lin M., Niu H., Rikihisa Y., Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell. Microbiol. 14, 1037–1050 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igal R. A., Wang P., Coleman R. A., Triacsin C blocks de novo synthesis of glycerolipids and cholesterol esters but not recycling of fatty acid into phospholipid: Evidence for functionally separate pools of acyl-CoA. Biochem. J. 324, 529–534 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohan Kumar D., et al. , Ehrlichia chaffeensis uses its surface protein EtpE to bind GPI-anchored protein DNase X and trigger entry into mammalian cells. PLoS Pathog. 9, e1003666 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J. Z., Popov V. L., Gao S., Walker D. H., Yu X. J., The developmental cycle of Ehrlichia chaffeensis in vertebrate cells. Cell. Microbiol. 9, 610–618 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Tomoda H., Igarashi K., Cyong J. C., Omura S., Evidence for an essential role of long chain acyl-CoA synthetase in animal cell proliferation. Inhibition of long chain acyl-CoA synthetase by triacsins caused inhibition of Raji cell proliferation. J. Biol. Chem. 266, 4214–4219 (1991). [PubMed] [Google Scholar]
- 20.Kean L. S., Fuller R. S., Nichols J. W., Retrograde lipid traffic in yeast: Identification of two distinct pathways for internalization of fluorescent-labeled phosphatidylcholine from the plasma membrane. J. Cell Biol. 123, 1403–1419 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koval M., Pagano R. E., Sorting of an internalized plasma membrane lipid between recycling and degradative pathways in normal and Niemann-Pick, type A fibroblasts. J. Cell Biol. 111, 429–442 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wüstner D., Mukherjee S., Maxfield F. R., Müller P., Herrmann A., Vesicular and nonvesicular transport of phosphatidylcholine in polarized HepG2 cells. Traffic 2, 277–296 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Alberts B., et al. , “Membrane structure” in Molecular Biology of the Cell, (Garland Science, New York, NY, ed. 6, 2017), Chap. 10, pp. 565–596. [Google Scholar]
- 24.Popov V. L., Chen S. M., Feng H. M., Walker D. H., Ultrastructural variation of cultured Ehrlichia chaffeensis. J. Med. Microbiol. 43, 411–421 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Hölttä-Vuori M., et al. , BODIPY-cholesterol: A new tool to visualize sterol trafficking in living cells and organisms. Traffic 9, 1839–1849 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Hölttä-Vuori M., Sezgin E., Eggeling C., Ikonen E., Use of BODIPY-cholesterol (TF-Chol) for visualizing lysosomal cholesterol accumulation. Traffic 17, 1054–1057 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honig M. G., Hume R. I., Fluorescent carbocyanine dyes allow living neurons of identified origin to be studied in long-term cultures. J. Cell Biol. 103, 171–187 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ragnarson B., Bengtsson L., Haegerstrand A., Labeling with fluorescent carbocyanine dyes of cultured endothelial and smooth muscle cells by growth in dye-containing medium. Histochemistry 97, 329–333 (1992). [DOI] [PubMed] [Google Scholar]
- 29.Godement P., Vanselow J., Thanos S., Bonhoeffer F., A study in developing visual systems with a new method of staining neurones and their processes in fixed tissue. Development 101, 697–713 (1987). [DOI] [PubMed] [Google Scholar]
- 30.Vidal-Sanz M., Villegas-Pérez M. P., Bray G. M., Aguayo A. J., Persistent retrograde labeling of adult rat retinal ganglion cells with the carbocyanine dye diI. Exp. Neurol. 102, 92–101 (1988). [DOI] [PubMed] [Google Scholar]
- 31.Rikihisa Y., Anaplasma phagocytophilum and Ehrlichia chaffeensis: Subversive manipulators of host cells. Nat. Rev. Microbiol. 8, 328–339 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Rikihisa Y., Molecular events involved in cellular invasion by Ehrlichia chaffeensis and Anaplasma phagocytophilum. Vet. Parasitol. 167, 155–166 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rikihisa Y., Molecular pathogenesis of Ehrlichia chaffeensis infection. Annu. Rev. Microbiol. 69, 283–304 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Abe S., Yamashita K., Kohno H., Ohkubo Y., Involvement of transglutaminase in the receptor-mediated endocytosis of mouse peritoneal macrophages. Biol. Pharm. Bull. 23, 1511–1513 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Davies P. J., et al. , Transglutaminase is essential in receptor-mediated endocytosis of alpha 2-macroglobulin and polypeptide hormones. Nature 283, 162–167 (1980). [DOI] [PubMed] [Google Scholar]
- 36.Levitzki A., Willingham M., Pastan I., Evidence for participation of transglutaminase in receptor-mediated endocytosis. Proc. Natl. Acad. Sci. U.S.A. 77, 2706–2710 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin M., Zhu M. X., Rikihisa Y., Rapid activation of protein tyrosine kinase and phospholipase C-gamma2 and increase in cytosolic free calcium are required by Ehrlichia chaffeensis for internalization and growth in THP-1 cells. Infect. Immun. 70, 889–898 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson M. J., Deane F. M., Robinson P. J., McCluskey A., Synthesis of Dynole 34-2, Dynole 2-24 and Dyngo 4a for investigating dynamin GTPase. Nat. Protoc. 9, 851–870 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Seglen P. O., Gordon P. B., 3-Methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 79, 1889–1892 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindmo K., Stenmark H., Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119, 605–614 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Rikihisa Y., Jiang B. M., In vitro susceptibilities of Ehrlichia risticii to eight antibiotics. Antimicrob. Agents Chemother. 32, 986–991 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brouqui P., Raoult D., In vitro antibiotic susceptibility of the newly recognized agent of ehrlichiosis in humans, Ehrlichia chaffeensis. Antimicrob. Agents Chemother. 36, 2799–2803 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinac B., Saimi Y., Kung C., Ion channels in microbes. Physiol. Rev. 88, 1449–1490 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poot M., et al. , Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J. Histochem. Cytochem. 44, 1363–1372 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Sharp M. D., Pogliano K., An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. U.S.A. 96, 14553–14558 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma P., Teymournejad O., Rikihisa Y., Peptide nucleic acid knockdown and intra-host cell complementation of Ehrlichia type IV secretion system effector. Front. Cell. Infect. Microbiol. 7, 228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fader C. M., Colombo M. I., Autophagy and multivesicular bodies: Two closely related partners. Cell Death Differ. 16, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Berg T. O., Fengsrud M., Strømhaug P. E., Berg T., Seglen P. O., Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J. Biol. Chem. 273, 21883–21892 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Eskelinen E. L., Maturation of autophagic vacuoles in mammalian cells. Autophagy 1, 1–10 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Gillooly D. J., et al. , Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577–4588 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rikihisa Y., “Ultrastructure of Rickettsia with special emphasis on Ehrlichia” in Ehrlichiosis: A Vector-Borne Disease of Animals and Humans, Williams J. C., Kakoma I., Eds. (Kluwer Publishing Co., Norwell, MA, 1990), vol. 54, pp. 22–31. [Google Scholar]
- 52.Popov V. L., et al. , Ultrastructural differentiation of the genogroups in the genus Ehrlichia. J. Med. Microbiol. 47, 235–251 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Drobne D., 3D imaging of cells and tissues by focused ion beam/scanning electron microscopy (FIB/SEM). Methods Mol. Biol. 950, 275–292 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Rikihisa Y., “Ehrlichiae”, in Proceedings of the 5th International Symposium on Rickettsiae and Rickettsial Diseases, Hechemy K., Ed. (International Society for Rickettsiae and Rickettsial Diseases, Slovak Academy of Sciences, Bratislava, Slovak Republic, 1996), pp. 272–286. [Google Scholar]
- 55.Dunning Hotopp J. C., et al. , Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Gilk S. D., “Role of lipids in Coxiella burnetii infection” in Coxiella Burnetii: Recent Advances and New Perspectives in Research of the Q Fever Bacterium, Toman R., Heinzen R. A., Samuel J. E., Mege J.-L., Eds. (Springer Netherlands, Dordrecht, 2012), pp. 199–213. [DOI] [PubMed] [Google Scholar]
- 57.Toledo A., Benach J. L., Hijacking and use of host lipids by intracellular pathogens. Microbiol. Spectr., 10.1128/microbiolspec.VMBF-0001-2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fozo E. M., Rucks E. A., The making and taking of lipids: The role of bacterial lipid synthesis and the harnessing of host lipids in bacterial pathogenesis. Adv. Microb. Physiol. 69, 51–155 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Samanta D., Mulye M., Clemente T. M., Justis A. V., Gilk S. D., Manipulation of host cholesterol by obligate intracellular bacteria. Front. Cell. Infect. Microbiol. 7, 165 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiong Q., Lin M., Huang W., Rikihisa Y., Infection by Anaplasma phagocytophilum requires recruitment of low-density lipoprotein cholesterol by flotillins. MBio 10, e02783-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiong Q., Lin M., Rikihisa Y., Cholesterol-dependent anaplasma phagocytophilum exploits the low-density lipoprotein uptake pathway. PLoS Pathog. 5, e1000329 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiong Q., Rikihisa Y., Subversion of NPC1 pathway of cholesterol transport by Anaplasma phagocytophilum. Cell. Microbiol. 14, 560–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winkler H. H., Miller E. T., Phospholipid composition of Rickettsia prowazeki grown in chicken embryo yolk sacs. J. Bacteriol. 136, 175–178 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Driscoll T. P., et al. , Wholly Rickettsia! Reconstructed metabolic profile of the quintessential bacterial parasite of eukaryotic cells. MBio 8, e00859-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stephens R. S., et al. , Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759 (1998). [DOI] [PubMed] [Google Scholar]
- 66.Hackstadt T., Scidmore M. A., Rockey D. D., Lipid metabolism in Chlamydia trachomatis-infected cells: Directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. U.S.A. 92, 4877–4881 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wylie J. L., Hatch G. M., McClarty G., Host cell phospholipids are trafficked to and then modified by Chlamydia trachomatis. J. Bacteriol. 179, 7233–7242 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Ooij C., et al. , Host cell-derived sphingolipids are required for the intracellular growth of Chlamydia trachomatis. Cell. Microbiol. 2, 627–637 (2000). [DOI] [PubMed] [Google Scholar]
- 69.Carabeo R. A., Mead D. J., Hackstadt T., Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc. Natl. Acad. Sci. U.S.A. 100, 6771–6776 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cocchiaro J. L., Kumar Y., Fischer E. R., Hackstadt T., Valdivia R. H., Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. U.S.A. 105, 9379–9384 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elwell C. A., Engel J. N., Lipid acquisition by intracellular Chlamydiae. Cell. Microbiol. 14, 1010–1018 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beatty W. L., Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J. Cell Sci. 119, 350–359 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Elwell C. A., et al. , Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathog. 7, e1002198 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Agaisse H., Derré I., Expression of the effector protein IncD in Chlamydia trachomatis mediates recruitment of the lipid transfer protein CERT and the endoplasmic reticulum-resident protein VAPB to the inclusion membrane. Infect. Immun. 82, 2037–2047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Recuero-Checa M. A., et al. , Chlamydia trachomatis growth and development requires the activity of host long-chain Acyl-CoA Synthetases (ACSLs). Sci. Rep. 6, 23148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bugalhão J. N., Mota L. J., The multiple functions of the numerous Chlamydia trachomatis secreted proteins: The tip of the iceberg. Microb. Cell 6, 414–449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Howe D., Heinzen R. A., Coxiella burnetii inhabits a cholesterol-rich vacuole and influences cellular cholesterol metabolism. Cell. Microbiol. 8, 496–507 (2006). [DOI] [PubMed] [Google Scholar]
- 78.Gilk S. D., et al. , Bacterial colonization of host cells in the absence of cholesterol. PLoS Pathog. 9, e1003107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mulye M., Samanta D., Winfree S., Heinzen R. A., Gilk S. D., Elevated cholesterol in the Coxiella burnetii intracellular niche is bacteriolytic. MBio 8, e02313-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mulye M., Zapata B., Gilk S. D., Altering lipid droplet homeostasis affects Coxiella burnetii intracellular growth. PLoS One 13, e0192215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Larson C. L., Beare P. A., Howe D., Heinzen R. A., Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, E4770–E4779 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rikihisa Y., Role and function of type IV secretion systems in Anaplasma and Ehrlichia species. Curr. Top. Microbiol. Immunol. 413, 297–321 (2018) [DOI] [PubMed] [Google Scholar]
- 83.Itakura E., Kishi C., Inoue K., Mizushima N., Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19, 5360–5372 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weber S. S., Ragaz C., Hilbi H., Pathogen trafficking pathways and host phosphoinositide metabolism. Mol. Microbiol. 71, 1341–1352 (2009). [DOI] [PubMed] [Google Scholar]
- 85.Nascimbeni A. C., Codogno P., Morel E., Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J. 284, 1267–1278 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Haldar S., Chattopadhyay A., “Application of NBD-labeled lipids in membrane and cell biology” in Fluorescent Methods to Study Biological Membranes, Mély Y., Duportail G., Eds. (Springer Berlin Heidelberg, Berlin, Heidelberg, 2013), pp. 37–50. [Google Scholar]
- 87.Palade G. E., Membrane biogenesis: An overview. Methods Enzymol. 96, xxix–lv (1983). [DOI] [PubMed] [Google Scholar]
- 88.Takayama K., Rothenberg R. J., Barbour A. G., Absence of lipopolysaccharide in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 55, 2311–2313 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.LaRocca T. J., et al. , Cholesterol lipids of Borrelia burgdorferi form lipid rafts and are required for the bactericidal activity of a complement-independent antibody. Cell Host Microbe 8, 331–342 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.LaRocca T. J., et al. , Proving lipid rafts exist: Membrane domains in the prokaryote Borrelia burgdorferi have the same properties as eukaryotic lipid rafts. PLoS Pathog. 9, e1003353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Toledo A., Monzón J. D., Coleman J. L., Garcia-Monco J. C., Benach J. L., Hypercholesterolemia and ApoE deficiency result in severe infection with Lyme disease and relapsing-fever Borrelia. Proc. Natl. Acad. Sci. U.S.A. 112, 5491–5496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fraser C. M., et al. , Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390, 580–586 (1997). [DOI] [PubMed] [Google Scholar]
- 93.Ostberg Y., Berg S., Comstedt P., Wieslander A., Bergström S., Functional analysis of a lipid galactosyltransferase synthesizing the major envelope lipid in the Lyme disease spirochete Borrelia burgdorferi. FEMS Microbiol. Lett. 272, 22–29 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Toledo A., et al. , Selective association of outer surface lipoproteins with the lipid rafts of Borrelia burgdorferi. MBio 5, e00899-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Crowley J. T., et al. , Lipid exchange between Borrelia burgdorferi and host cells. PLoS Pathog. 9, e1003109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grevengoed T. J., Klett E. L., Coleman R. A., Acyl-CoA metabolism and partitioning. Annu. Rev. Nutr. 34, 1–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tomoda H., Igarashi K., Omura S., Inhibition of acyl-CoA synthetase by triacsins. Biochim. Biophys. Acta 921, 595–598 (1987). [PubMed] [Google Scholar]
- 98.Barnewall R. E., Rikihisa Y., Abrogation of gamma interferon-induced inhibition of Ehrlichia chaffeensis infection in human monocytes with iron-transferrin. Infect. Immun. 62, 4804–4810 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miura K., et al. , Ehrlichia chaffeensis induces monocyte inflammatory responses through MyD88, ERK, and NF-κB but not through TRIF, interleukin-1 receptor 1 (IL-1R1)/IL-18R1, or toll-like receptors. Infect. Immun. 79, 4947–4956 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dawson J. E., et al. , Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 29, 2741–2745 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Butan C., et al. , Spiral architecture of the nucleoid in Bdellovibrio bacteriovorus. J. Bacteriol. 193, 1341–1350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ito S., Rikihisa Y., “Techniques for electron microscopy of rickettsiae” in Rickettsiae and Rickettsial Diseases, Burgdorfer W., Anacker R. L., Eds. (Academic Press, New York, 1981), pp. 213–227. [Google Scholar]
- 103.Do T., et al. , Three-dimensional imaging of HIV-1 virological synapses reveals membrane architectures involved in virus transmission. J. Virol. 88, 10327–10339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 105.Fedorov A., et al. , 3D Slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging 30, 1323–1341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Narayan K., et al. , Multi-resolution correlative focused ion beam scanning electron microscopy: Applications to cell biology. J. Struct. Biol. 185, 278–284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All experimental data described in this study are included in the main text and SI Appendix.