Abstract
Respiratory viruses are the most frequent causative agents of disease in humans, with significant impact on morbidity and mortality worldwide. Common respiratory agents from several virus families are well adapted to efficient person-to-person transmission and circulate in a global scale, and community-based studies conducted over the past five decades or so confirm that these viruses are the predominant etiological agents of acute respiratory infections (ARIs). The respiratory viruses that most commonly circulate in all continents as endemic or epidemic agents are influenza virus, respiratory syncytial virus, parainfluenza viruses, metapneumovirus, rhinovirus, coronaviruses, adenoviruses, and bocaviruses. Although vaccines and effective antiviral drugs are not yet available for most of these viruses, much progress has been made in the understanding of their biology and fundamental issues of host–parasite relationship. This article is a summary of the current knowledge about these viruses.
Keywords: Bocavirus, common cold, Coronavirus, Metapneumovirus, parainfluenza virus, respiratory adenovirus, respiratory syncytial virus, respiratory viral infections, rhinovirus, SARS
Keywords: ACE2, angiotensin-converting enzyme 2; ARDS, acute respiratory distress syndrome; ARI, acute respiratory infection; CAR, Coxsackie B and adenovirus receptor; ECMO, extracorporeal membrane oxygenation; HBD-2, human β-defensin 2; HBoV, human bocavirus; HCoV, human coronavirus; HE, hemagglutinin; HMPV, human metapneumovirus; HPIV, human parainfluenza virus; HRSV, human respiratory syncytial virus; HRV, human rhinovirus; ICAM, intercellular adhesion molecule; IRES, internal ribosomal entry site; LDLR, low-density lipoprotein receptor; LRI, lower respiratory infection; LRT, lower respiratory tract; LRTI, lower respiratory tract infection; SARS, severe acute respiratory syndrome; SARS-CoV, SARS coronavirus; URI, upper respiratory infection; UTR, untranslated region; VLP, virus-like particle
Defining Statement
This article presents an overview of the most important respiratory viruses, with the exception of influenza virus, covered in a separate article. This overview includes a description of the agents, their epidemiology, pathogenesis, clinical features, and management and control. The viruses covered are respiratory syncytial virus, parainfluenza viruses, metapneumovirus, and coronaviruses, including the agents of SARS, respiratory adenoviruses, rhinovirus, and bocavirus.
Introduction
Respiratory viruses are the most frequent causative agents of disease in humans, with significant impact on morbidity and mortality worldwide, mainly in children. Approximately one-fifth of all childhood deaths worldwide are related to acute respiratory infections (ARIs), particularly in impoverished populations of tropical regions, where ARI case-to-fatality ratios can be remarkably higher than in temperate regions of the world. Eight human respiratory viruses circulate commonly in all age groups and are recognized as adapted to efficient person-to-person transmission (Table 1). In addition to these, SARS coronavirus (SARS-CoV) and avian influenza virus H5N1 have emerged in recent years as threats to public health. SARS-CoV has been out of circulation since 2003, and avian influenza virus H5N1 has caused limited outbreaks of human infections.
Table 1.
Common respiratory viruses, their classification, principal syndromes, and main detection methods
| Virus | Classificationa | Principal syndromes | Virus detection methods |
|---|---|---|---|
| HRSV | Groups A and B | URI, bronchiolitis, croup, bronchitis, pneumonia | Culture, Ag detection, RT–PCR |
| HPIV | Types 1, 2, 3, 4 | URI, croup, bronchiolitis, bronchitis, pneumonia | Culture, Ag detection, RT–PCR |
| HRV | Species A, B, and C With 100 serotypes | URI; asthma and COPD exacerbation | Culture, RT–PCR |
| ADV | 51 serotypes | URI, PCF, bronchitis, pneumonia | Culture, Ag detection, PCR |
| HCoV | Types OC43, 229E, NL(NH), HKU1 | URI, bronchitis, pneumonia | Culture, RT–PCR |
| SARS-CoV | 1 type | SARS | Culture, RT–PCR |
| HMPV | Groups A and B | URI, bronchitis, pneumonia | Culture, RT–PCR |
| HBoV | 2 lineages | URI, bronchiolitis, asthma exacerbation,bronchitis, pneumonia | PCR |
ADV, adenovirus; Ag, antigen; HBoV, human bocavirus; HCoV, human coronavirus; HMPV, human metapneumovirus; HPIV, human parainfluenza virus; HRSV, human respiratory syncytial virus; HRV, human rhinovirus; PCF, pharyngoconjunctival fever; SARS, severe acute respiratory syndrome; SARS-CoV, Coronavirus associated with SARS; URI, upper respiratory infection; RT–PCR, reverse transcription–polymerase chain reaction, which includes both conventional and real-time methods.
Classification in species, subgroups, serotypes, or lineages.
As the currently known respiratory viruses still do not account for all clinically relevant human viral respiratory illnesses, systematic searches for new agents using molecular tools are expected to discover previously unidentified agents. Although respiratory viruses cause a great burden of diseases, only a few preventive or therapeutic interventions are currently available. However, recent advances in molecular and cell biology of respiratory viruses will hopefully result in the development of useful interventions. In this article, we focus on the common respiratory viruses, with the exception of influenza virus, covered in a separate article.
Human Respiratory Syncytial Virus
The Agent
Human respiratory syncytial virus (HRSV) is the only known human pathogen of the genus Pneumovirus, family Paramyxoviridae, first isolated in 1956 from a chimpanzee with coryza. One year later, the agent was recovered from two children with respiratory illness. HRSV virions are heterogeneous in size and shape, and consist of a helical nucleocapsid containing a 15 220 nt negative-sense single-stranded RNA, tightly bound to the nucleoprotein (N). The virus particle is surrounded by a lipid bilayer containing the glycoproteins G (receptor binding), F (membrane fusion), and SH (unknown function). The virions also contain an RNA-dependent RNA polymerase (L), a phosphoprotein (P), and a 22K protein (M2-1) (Figure 1). In addition, three nonstructural proteins (NS1, NS2, and M2-2) are produced in infected cells.
Figure 1.
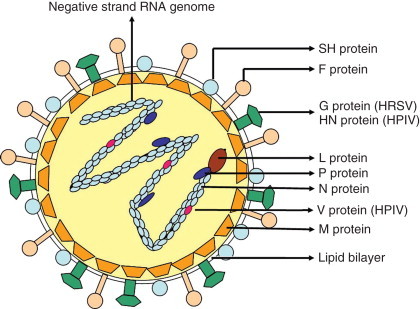
Schematic structure of a paramyxovirus. The virus envelope contains the F, SH, and the G/HN proteins. Underlying the envelope is the M (matrix). The nucleocapsid consists of a negative-strand RNA encapsidated by the N (nucleocapsid) protein. Associated with the nucleocapsid are L (large-polymerase), P (phosphoprotein), and V (cysteine-rich) proteins.
Two HRSV groups, A and B, were originally distinguished based on the antigenic differences in the attachment glycoprotein G. Later, this separation was confirmed in other genes and phylogenetic analysis of G gene sequences allowed further division of groups into HRSV genotypes. While new genotypes continue to emerge, others that circulated previously tend to disappear.
HRSV binds to cell surface glycosaminoglycans through the viral glycoprotein G, although G protein-independent binding also exists. Upon virus–receptor interaction, fusion of virus and cell membranes takes place through the binding of viral protein F to cell GTPase RhoA. HRSV internalization is considered to be pH-independent and may happen either in plasma or in endosomal membranes. Expression of the viral protein F on the plasma membrane of infected cells promotes fusion between adjacent cells, resulting in the formation of syncytia, the hallmark of paramyxovirus cytopathic effect. Once in the cytoplasm, the negative-strand viral RNA is transcribed by the coordinated action of the proteins L, N, P, and M2-1; resulting in a set of mRNAs, which then direct the synthesis of viral proteins. The transcription of mRNAs occurs in a gradient with decreasing amounts of transcripts being generated from the 3′ end to the 5′ end of the genome template (Figure 2). At some point, the polymerase complex switches from mRNA transcription to viral-RNA replication, generating a full-length RNA of polarity opposite to that of the genome (antigenome). Antigenome serves as a template for the polymerase complex to generate negative-sense RNA genomes of the progeny. The viral protein M2-2 seems to alter the polymerase complex in a way that may direct preference of replication over transcription. The viral matrix (M) protein mediates the association of nucleocapsids with lipid rafts containing HRSV envelope proteins in the plasma membrane, giving rise to virus budding. Viral proteins NS1 and NS2 are suppressors of type 1 interferon induction.
Figure 2.
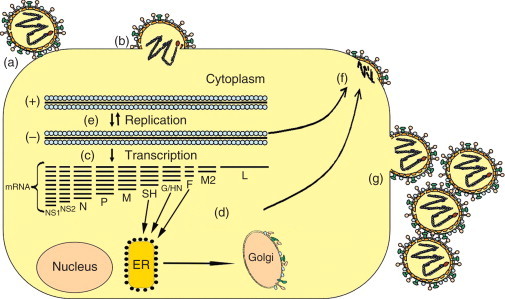
Schematic representation of a paramyxovirus replication cycle. (a) The G/HN protein binds to cell receptors and (b) promotes viral adsorption that results in the fusion of viral envelope with cell membrane mediated by F protein. Then genome is released into the cytoplasm. (c) A set of mRNAs is transcribed using the genome as template. (d) These mRNAs are then translated in specific proteins, some of which are glycosylated in association with the endoplasmic reticulum and the Golgi apparatus. (e) Envelope glycoproteins direct the points where new virions are formed. (f) New genomes produced by RNA replication are incorporated in newly formed virus particles. (g) The mature virus particles are released by budding from the plasma membrane.
Epidemiology
HRSV is the single most frequent cause of lower respiratory tract infection (LRTI) leading to morbidity and mortality in children worldwide. Most children have been infected by HRSV at the age of 2, and reinfections are common throughout life. Annual seasonal outbreaks of HRSV occur in winter/early spring in temperate regions and during rainy seasons in tropical areas, likely because of increased indoor crowding. More than one genotype of HRSV groups A and B cocirculate in the same seasonal outbreak, with the group predominance changing from year to year, with no apparent correlation of group or genotype with clinical manifestations or disease severity. HRSV transmission requires close contact, either by large particle aerosols or by contamination of hands followed by inoculation into the eye or nose, and secondary infections in family contacts of an index case are common.
An estimated 30% of all infants will have HRSV infection requiring medical attention and 2% of them will be hospitalized. Approximately 10% of all children will have bronchiolitis in the first year of life, and 60–90% of them are caused by HRSV. In certain regions, LRTIs caused by HRSV are responsible for up to 85% of hospitalizations in infants during seasonal outbreaks.
Most of what is known about HRSV evolution is based on phylogenetic studies of the G protein gene, which is quite variable among clinical isolates. Rates of accumulation of nucleotide substitutions in this gene have been calculated at 1.83 × 10−3 and 1.95 × 10−3 substitutions per site per year, respectively, for HRSV A and B. Steady accumulation of amino acid changes over time leads to the emergence of new lineages, and therefore HRSV genotypes predominant in an outbreak tend to be replaced by others in subsequent years. This is likely because of the development of herd immunity toward immunodominant epitopes. A mutant HRSV B containing 20 amino acid reduplication in the variable region of the G protein ectodomain was recently identified and made it possible to track the dissemination of novel HRSV strains. This insertion, first detected in Argentina in 1999, was isolated in the following years in Belgium, United Kingdom, Japan, and North America, indicating that a new HRSV strain can spread rapidly worldwide. Remarkably, after a period of 1–2 years, this new variant became the predominant HRSV B strain with further accumulation of variation in the G gene.
Pathogenesis and Clinical Features
HRSV replicates in respiratory epithelia, reaching high titers in nasal secretions and causing virus shedding for up to 3 weeks after the end of symptoms. Cell-to-cell spread leads to involvement of the entire respiratory tree by HRSV, reaching bronchioles 1–3 days after the onset of symptoms, inducing necrosis of ciliated cells, syncytia formation, peribronchiolar inflammation with abundant lymphocytes and macrophages, and impairment of secretion clearance, resulting in small airway obstruction and lung hyperinflation typical of bronchiolitis. HRSV pneumonia with interstitial mononuclear infiltrate, eosinophilic cytoplasmic inclusions in epithelial cells, and multinucleated giant cells, frequently coexist with bronchiolitis. HRSV disease is particularly severe in young babies, whose immature airways are unable to compensate for the virus-induced damage.
Naturally acquired immunity to RSV is incomplete and short-lived, but disease severity is reduced on reinfections. Secretory IgA correlates better with protection than serum antibody level and patient age, and preexisting HRSV-specific maternal antibodies influence the development of disease. Cell-mediated immune response is central to recovery from infection, and immunosuppressed patients are at risk of severe HRSV disease and fatal outcome. The type of immune response to HRSV is a major factor in the development of wheezing and asthma exacerbations. A deviation toward Th2 cytokine response is associated with more severe disease, whereas a Th1 response leads to effective viral clearance and milder illness. A preexisting Th1 deficiency may be associated with severe disease in some children, which may be a marker of predisposition to wheezing and asthma later in life. The HRSV G protein, which is produced both as membrane-anchored and as soluble forms, is a type II glycoprotein with two highly variable extracellular mucin-like domains separated by a cysteine-rich, much conserved central region that inhibits innate immunity and modulates cytokine production. HRSV infection of epithelial cells triggers the release of Th1 cytokines (IFN-γ, IL-2, IL-12), Th2 cytokines (IL-4, IL-5, IL-6, IL-10), IFN-α, IFN-β, and chemokines that amplify the immune response and inflammatory reaction.
Children vaccinated in the 1960s with a formalin-inactivated HRSV vaccine had severe disease when exposed to natural infection. This was apparently due to the imbalance between protective and immunopathologic T-cell responses elicited by the vaccine that would favor a CD4+ Th2 pattern upon subsequent HRSV infections, whereas a previous natural infection would favor a CD4+ Th1 pattern in response to reinfection. Following a 2–8 days incubation period, HRSV infection causes illness ranging from mild upper respiratory infection (URI) to severe lower respiratory infection (LRI), including pneumonia, bronchiolitis, tracheobronchitis, and croup. In infants and young children, URI is frequently accompanied by otitis media and fever. The most frequent manifestation of LRI due to HRSV in infants is bronchiolitis, which is usually preceded by 2–3 days of symptoms of the upper respiratory tract, and is characterized by tachypnea, dyspnea, cough, expiratory wheezing, air trapping, and in more severe cases, intercostal muscle retractions and cyanosis. Fever is present in 50% of infants, with lung hyperaeration and sometimes segment atelectasis on chest X-ray. Blood count usually shows lymphocytosis, and an increase in neutrophils can be associated with bacterial superinfection. Acute otitis media is found in up to 60% of children with bronchiolitis. Patients with underlying conditions, such as congenital heart disease, prematurity or cystic fibrosis, and bronchopulmonary dysplasia, as well as immunocompromised hosts of any age, are at risk for severe and fatal HRSV infections.
In children above 3 years of age and adults, HRSV illness is usually mild to moderate, with upper respiratory tract symptoms, including coryza, cough, sore throat, and hoarseness, often accompanied by low-grade fever. Exacerbations of chronic pulmonary diseases and wheezing can be seen in adults with HRSV, and severe pneumonia can also occur, particularly in the elderly. The role of HRSV infections in wheezing and asthma exacerbations in infants, especially in those with family history of allergy, has been established in studies in several different parts of the world, including temperate as well as tropical regions. In the elderly, HRSV infections in the lower respiratory tract (LRT) are mainly characterized by interstitial pneumonia, prolonged cough, and dyspnea in persons with chronic pulmonary conditions, and HRSV should be considered in the differential diagnosis of flu-like illness.
Diagnosis
Nasopharyngeal aspirates, nasal washings, and swabs, as well as LRT samples, can be used as specimens for HRSV detection. Sample storage and freeze–thawing significantly reduce HRSV recovery in culture. HRSV can be isolated in several human cell lines, such as HEp-2, HeLa, and A549, and syncytia formation can be noted in HEp-2 cells in 3–5 days.
Rapid HRSV antigen detection methods, including immunofluorescence of sloughed respiratory cells, EIA, and membrane-based chromatography, are sensitive and specific. Commercially available point-of-care simple tests require no equipment and are ideal for emergency room and primary care HRSV diagnosis.
Detection of HRSV RNA by RT–PCR, both conventional and based on real-time techniques, has become widely accepted as the most sensitive diagnostic assay. In addition to sensitive qualitative results, real-time methods are applicable to quantitation of viral load and could simultaneously detect the subtype RSV directly in clinical specimens. Other amplification methods not based on PCR, such as NASBA, have become commercially available for HRSV diagnosis. HRSV serology has limited value for case management, but may be useful for epidemiologic surveys.
Management and Control
HRSV URIs require no specific treatment, and antibiotics may be considered only in the presence of bacterial complications, such as acute otitis media. In infants with HRSV bronchiolitis, supportive treatment consists of bronchodilators and prevention of hypoxemia and electrolyte imbalance. Measures to prevent hypoxemia may vary from simple aspiration of respiratory secretions to mechanical ventilatory support, or even extracorporeal membrane oxygenation (ECMO).
The only antiviral drug currently approved for the treatment of infants with HRSV is the synthetic nucleoside ribavirin, delivered by small-particle aerosol via mist tent, mask, oxygen hood, or ventilator. Although ribavirin trials have lacked power to provide definitive recommendation, the antiviral has been recommended for infants and young children with underlying conditions, such as congenital heart disease, cystic fibrosis, or immunosuppression. This therapy can also be considered for premature babies, infants younger than 6 weeks of age, or severely ill. Ribavirin therapy may reduce the duration of hospital stay and requirement for mechanical ventilation. Considering that the chemokine/receptor system is central to the inflammatory cell recruitment in HRSV bronchiolitis, effective chemokine receptor blockers under development may also become useful to reduce inflammation and disease severity.
No vaccine is currently licensed for HRSV prophylaxis. The disease enhancement related to a Th2 bias, caused by formalin-inactivated vaccine in the 1960s, significantly slowed progress toward an HRSV vaccine. Immune response to naturally acquired HRSV is poor, and developing an effective vaccine faces the challenge of improving over the natural infection to elicit a balanced immune response, including Th1/Th2 CD4 T cells, CD8 T cells, and hopefully HRSV-neutralizing antibodies, including secretory IgA. Intranasally delivered genetically engineered attenuated or vectored vaccines are the most promising alternative for newborns, whereas subunit vaccine candidates combined with adjuvants to promote a protective response could be an alternative to previously exposed children.
Passive immunization of high-risk infants with monthly applications of HRSV immunoglobulin during the seasonal outbreaks reduces the incidence and severity of HRSV infections in high-risk children. This costly intervention is the only available means of protecting high-risk children against serious HRSV disease. Monthly intramuscular injections of humanized monoclonal antibody (palivizumab) should be considered for passive immunoprophylaxis during HRSV season for high-risk infants, such as preterm babies less than 6 months old, children with congenital heart disease, and children less than 2 years of age with bronchopulmonary dysplasia. A second-generation mAb (motavizumab) with increased affinity is on phase III clinical trials as of this writing, and a third-generation mAb (numax-YTE) has been developed to increase the antibody serum half-life, thus obviating the need for monthly dosing.
Hospitalized infants with RSV infection should be isolated or grouped to prevent nosocomial cross-infection. Hand washing, use of eye–nose goggles, gowns, gloves, and decontamination of surfaces and fomites are additional control measures.
Human Parainfluenza Viruses
Agents
Human parainfluenza viruses (HPIVs) are frequent causes of LRTIs in infants and children worldwide. HPIVs share structural and biological characteristics with HRSV and are distributed in two genera of the family Paramyxoviridae. HPIVs are classified antigenically into types 1–4, and HPIV-4 has subtypes A and B. HPIV types 1 and 3 are classified in the genus Respirovirus, whereas HPIV types 2 and 4 are in the genus Rubulavirus. The virions are pleomorphic, with single-stranded negative-sense RNA, ranging from 150 to 200 nm in diameter (Figure 1). The glycoprotein HN, with hemagglutinin (HE) and neuraminidase activities, promotes virus–cell attachment by binding to sialic acid present on the cell surface proteins. Upon cleavage by proteolytic enzymes, viral protein F mediates the fusion of viral and cell membranes, with consequent release of the nucleocapsid in the cytoplasm. The virus life cycle occurs in a way similar to other Paramyxoviridae (Figure 2). Nonstructural proteins equivalent to M2, NS1, and NS2 of HRSV are not present in HPIV. HPIV zinc-binding protein V is found in high intracellular levels, but in little quantity in viral particles, and plays roles controlling viral-RNA synthesis and counteracting host cell interferon type 1 response.
Epidemiology
Primary HPIV infection occurs early in childhood and by age 5 virtually all children are seropositive. Reinfections are frequent, but disease severity in reinfections is inversely proportional to the titer of serum-neutralizing antibody elicited by the previous infection. HPIV-1 and -3 are the types most frequently associated with LRI in children, the immunocompromised, the chronically ill, and the elderly, whereas HPIV-4 causes mostly URI in children and adults. In most temperate regions of both hemispheres, HPIV-1 causes outbreaks in the fall of alternate years, either alone or in cocirculation with HPIV-2. HPIV-3 causes spring and summer outbreaks in temperate areas, but circulates endemically throughout the year, especially in immunocompromised or chronically ill, whereas HPIV-4 occurs sporadically throughout the year in children and adults. In tropical regions, higher HPIV activity occurs during rainy seasons. HPIV spreads mainly within families and closed communities, such as nurseries, day care centers, and pediatric wards, with high secondary attack rates. The virus does not persist long in the environment and is transmitted mainly by large droplets and fomites. Viral shedding usually lasts 3–10 days, but prolonged shedding for months has been reported in very young children and immunosuppressed hosts. The rate of HPIV infection in infants is similar to that of HRSV, but HPIV causes less hospitalizations than HRSV.
Pathogenesis and Clinical Features
HPIV replicates in ciliated cells causing cytolysis of the respiratory mucosa. The infection begins in the upper respiratory tract and disseminates down the respiratory tree. The larynx and trachea are mostly involved in the croup syndrome, and extensive involvement of the lower respiratory tree may be present in tracheobronchitis, bronchopneumonia, and bronchiolitis. Similar to what occurs with HRSV, amplified inflammatory response induced by viral infection of epithelial cells causes mononuclear interstitial infiltrate, epithelial necrosis, inflammatory exudate into the alveoli, and hyaline membrane formation in the lungs. In cases of croup, mononuclear inflammatory cell infiltrate is seen in the subglotic area.
Host immunity is largely mediated by antibodies to the two surface proteins HN and F, and secretory antibody is the best marker of protection against HPIV. However, this protection is limited and reinfections are frequent. T-cell immune response is involved in the clearance of virus, and immunocompromised hosts may develop progressive and even lethal disease.
In addition to the host immune response, factors inherent to the virus may be central for pathogenicity, such as the varying susceptibility of the HPIV F protein to cleavage by proteolytic enzymes present in infected tissues.
After a 2–4 days incubation period, patients with HPIV infections may develop rhinitis, pharyngitis, laryngotracheobronchitis (croup), bronchiolitis, or pneumonia. Approximately, two-thirds of all HPIV infections in children result in febrile URI with associated otitis media in 10–34% of cases. The remaining one-third of HPIV infections causes croup, bronchiolitis, or pneumonia. HPIV bronchopneumonia and bronchiolitis are usually less severe than those caused by HRSV.
HPIV-1 and -2 cause up to 74% of all cases of croup, the most striking clinical presentation of HPIV infection, commonly seen in children younger than 3 years. Barking or brassy cough, dysphonia, inspiratory stridor, and suprasternal retraction due to subglotic edema are cardinal features. Most children recover in 2–5 days, but some may develop a bronchopneumonia–croup syndrome. Other less common manifestations are apnea, sudden infant death syndrome, parotitis, and myopericarditis, and there are suggestions that certain HPIV strains may become neuroinvasive.
Since immunity to HPIVs is incomplete, reinfections occur throughout life. Adults with HPIV infection generally have nonspecific URI, commonly with hoarseness. HPIVs can cause particularly severe diseases in immunocompromised hosts, especially children with severe combined immunodeficiency, interferon-γ deficiency, patients on chemotherapy, and bone marrow or solid organ transplant recipients. Mortality in bone marrow transplant patients with HPIV infection can reach 44%.
Diagnosis
HPIV is present in respiratory secretions up to 8 days from the onset of symptoms and can be isolated from nasopharyngeal washes/aspirates, swabs, or bronchoalveolar lavage in cultures of monkey kidney primary cells or several continuous cell lines, such as HEp-2, Vero, MDCK, LLC-MK2, BHK, and HeLa. The presence of HPIV in monolayers can be confirmed by hemadsorption with guinea pig erythrocytes and immunofluorescence. Isolation of HPIV in the shell vial assay format has produced mixed results. If for HPIV on exfoliated respiratory epithelial cells has been used for decades, but its sensitivity is moderate to low. Sensitive assays for the detection of HPIV RNA by RT–PCR, including multiplex format, and real-time PCR-based tests have become widely accepted. Serologic assays for HPIV IgM and IgG are highly specific, but are not useful for acute patient management.
Management and Control
At present, only supportive and symptomatic treatment is available for HPIV infections. In patients with croup, nebulization with racemic epinephrine or budesonide has rapid effect, and short-term, high-dose systemic corticosteroids may reduce the need for intubation. Besides anecdotal reports of aerosolized ribavirin treatment of immunocompromised patients with HPIV LRT infections, no specific antiviral treatment is licensed for HPIV. Inhibitors of the protein HN are effective in vitro and in animal models, but have not reached clinical use.
No vaccines are currently available for the prevention of HPIV infections. Early trials with inactivated HPIV vaccine in the 1960s were unsuccessful. Reverse genetics has provided means for identifying the basis of attenuation of vaccine candidates. Live attenuated cold-adapted HPIV-3 vaccines were immunogenic for children and still hold promise for clinical application. Also, candidate vaccines using murine Sendai virus expressing HPIV-3 F or HN proteins, elicited neutralizing antibodies and IFN-γ production in cotton rats, were protective against challenge and encourage development for future human use.
As in the case of HRSV infections, hand washing, use of eye–nose goggles, gowns, and gloves, as well as decontamination of fomites, should be used to prevent nosocomial spread of HPIV.
Human Metapneumovirus
The Agent
Human metapneumovirus (HMPV) is now recognized as a frequent cause of community-acquired ARI in children and adults worldwide. This agent was discovered in the Netherlands in 2001, through the study of previously unidentified viral isolates that induced CPE in cultures of LLC-MK2 cells. These viral isolates had been recovered from respiratory secretions of 28 children with ARI occurring in winter time, over a period of 20 years. Electron microscopy of cell cultures showing CPE revealed paramyxovirus-like particles, and sequencing of randomly primed PCR products revealed genome sequence and organization consistent with a paramyxovirus of the subfamily Pneumovirinae, related to avian pneumovirus of the genus Metapneumovirus. Rather than an avian virus that can infect humans, the new agent is a primary human pathogen, and screening of banked serum samples for HMPV antibodies in the Netherlands indicates that this virus has been in circulation for at least 5 decades.
HMPV particles are enveloped, pleomorphic, spherical, or filamentous particles, of about 209 nm in diameter. Like other paramyxoviruses, HMPV has a negative-sense, single-stranded RNA genome, and the viral replication occurs in a gradient manner. Complete genome sequencing revealed that, in contrast to pneumoviruses, HMPV has different positioning of the genes between M and L, and lacks NS1 and NS2 genes. Similar to HRSV, HMPV genes G and SH are not essential for virus replication. The protein M2-2 appears to control the switch from transcription to replication, and removal of the M2-2 gene leads to the accumulation of viral mRNAs and attenuation of viral replication.
Genetic and antigenic studies indicate that, similar to HRSV, HMPV isolates cluster into two main genetic subgroups, A and B, with 93–95% similarity of F gene sequences, and each subgroup includes at least two genogroups (A1, A2, B1, and B2). In contrast, the G protein gene sequence is only 30–35% conserved between subgroups. There is no apparent correlation between subgroups and disease severity.
Epidemiology
In temperate climates, HMPV circulates predominantly in late winter and spring, with peak activity closely following, or overlapping, that of HRSV. Different strains of both subgroups A and B cocirculate in the same outbreak, but vary from location to location and from year to year. By the age of 5 years, more than 90% of children have antibodies to HMPV, and the seroprevalence among adults is virtually 100%.
Although the incidence of HMPV disease may vary significantly from year to year in one location, this agent is generally detected in 5–10% of samples from ARI patients. Of note, this percentage can be much higher during peak months, when it reached up to 45% in some studies.
The peak age of hospitalization for HMPV is from 6 to 12 months and males seem to be at greater risk of LRT involvement. HMPV is rarely detected in samples from asymptomatic patients and has been associated with adult ARI, mainly in the elderly and in those with debilitating underlying conditions.
Pathogenesis and Clinical Features
Little is known about HMPV-specific mechanisms of pathogenesis. Animal studies show disruption of the respiratory epithelium, epithelial cell sloughing, and inflammatory infiltrates in the lung. In pathologic studies of humans with underlying diseases and HMPV infection, the main findings are acute and organizing lung injury, diffuse alveolar damage, sloughed epithelial cells with eosinophilic cytoplasmic inclusions, multinucleated giant cells, histiocytes, and hyaline membrane formation.
Clinically, HMPV infections closely resemble those caused by HRSV, ranging from mild upper ARI to severe bronchiolitis and pneumonia in children of the same age group at risk for HRSV. However, the median age of children hospitalized with HMPV infection is greater than that for children with HRSV. HMPV is now considered second only to HRSV as a cause of bronchiolitis, with chest X-rays revealing infiltrates, hyperinflation, and peribronchial cuffing. The most frequent symptoms of HMPV infections are fever, tachypnea, dyspnea, cough, hypoxia, wheezing, stridor, rhinitis, and sore throat, and otitis media is frequently present. HRSV in hospitalized infants and young children may require intensive care and mechanical ventilation, and coinfection by HMPV appears to increase the likelihood of severe HRSV disease by up to tenfold. In addition to LRTIs, HMPV also causes upper respiratory symptoms, indistinguishable from those caused by other common respiratory viruses.
HMPV may cause more serious infections in patients with comorbid or immunosuppressive conditions, as well as in the very young and the elderly. In one study, all individuals older than 65 with LRTIs caused by HMPV had at least one underlying chronic or debilitating condition, including lymphoma, leukemia, or neurologic or cardiovascular diseases.
HMPV has been recognized as a frequent cause of acute wheezing in children. A study found that 47% of the children with HMPV in Brazil had wheezing and 31% had chest indrawing. Conversely, 8% of the wheezing children in Finland had HMPV. In addition, a previous history of asthma has been more frequently associated with HMPV than with HRSV, and HMPV-infected patients are more often treated with bronchodilators and corticosteroids than HRSV-infected patients.
Diagnosis
HMPV can be isolated from nasopharyngeal washes/aspirates in LLC-MK2 cells with medium containing trypsin. The virus grows poorly and the cytopathic effect, characteristically negative on hemadsorption testing, develops usually late after inoculation (up to 23 days). RT–PCR is currently the most widely adopted method for HMPV detection. Primer sets targeting the N and L genes are reported to be the most sensitive to detect HMPV of both genetic groups. Real-time PCR assay for HMPV has now become the gold standard, being more sensitive than conventional RT–PCR. There has been a report of the circulation of strains of HMPV genetically distinct from the four already identified lineages, detectable by RT–PCR only with primers for the N gene. If such strains become widespread, primer sets for RT–PCR may have to be modified.
HMPV-specific antibodies have been developed and are now commercialized for HMPV immunofluorescence assays. While this method is not as sensitive as real-time RT–PCR, it is readily applicable in diagnostic laboratories, where IF for respiratory viruses is routinely done.
Management and Control
Other than supportive measures, oxygen therapy, bronchodilators, corticosteroids, and mechanical ventilation, there is no specific antiviral treatment for HMPV. Ribavirin and NMSO3 are inhibitory for HMPV in vitro.
Although an HMPV vaccine is not available at this time, the demonstration that hamsters, ferrets, and African green monkeys are susceptible to infection, and that hamsters vaccinated with HMPV A were protected from challenge with HMPV of both A and B genetic groups, opens possibilities for HMPV vaccine design.
Humanized neutralizing monoclonal antibody to the F protein is active in experimentally infected animals and is likely to become available for prophylaxis of HMPV in the future.
Rhinovirus
The Agent
Human rhinoviruses (HRVs) are the most frequent respiratory pathogens of humans, and the most commonly detected viruses in samples from common cold sufferers. HRVs are small, nonenveloped, positive-stranded RNA viruses in the family Picornaviridae, genus Rhinovirus, distributed in two species, A (75 serotypes) and B (25 serotypes). Recently described new strains containing deletions in the VP1 region may constitute a new species C. HRV atomic structure was solved and revealed an icosahedral particle composed of 12 pentamers with a diameter of about 30 nm (Figure 3). Surrounding a fivefold vertex of VP1 capsid proteins from adjacent pentamers, there is a 1.2–3.0 nm wide canyon that contains the receptor binding site for the HRVs of the major receptor group. The HRV genome is a 7.4 kb monocistronic, single-stranded, uncapped RNA, with a small genomic protein (VPg) covalently linked to the 5′ end and a poly(A) tail at the 3′ end. The first 600–700 nt at the 5′ end constitute a highly structured untranslated region (UTR), where an internal ribosomal entry site (IRES) permits initiation of translation. Following receptor binding, the viral positive-strand RNA is released into the cytoplasm, and translation of the single ORF produces one polyprotein that is cleaved cotranslationally to generate three precursors – P1 that originates capsid proteins, P2 and P3, originates nonstructural proteins and VPg. Further cleavages of these precursors generate 11 end-products, and the last cleavage of VP0 into VP2 and VP4 occurs only at the final stages of virus maturation (Figure 4). One of the P3 cleavage products, 3D, is an RNA-based RNA polymerase that will produce a negative-stranded full-length copy of the genome to be used as a template for the production of an expanding pool of positive-stranded RNAs. The positive-stranded RNAs can be either translated into viral proteins or packaged as genome into newly assembled virions. The HRV replication cycle takes place in association with vesicle membranes in the cytoplasm and mature virions are released upon cell in lysis (Figure 5).
Figure 3.
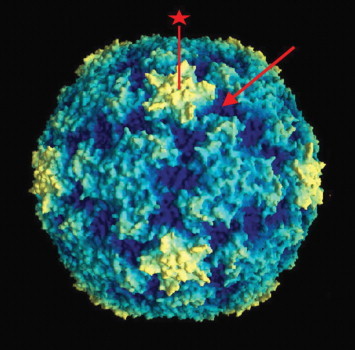
Molecular graphics image of HRV 14. The star-shaped region corresponds to the pentamers formed by convergence of five adjacent VP1 units (star symbol). ICAM-1, the receptor for the majority of HRV serotypes, binds within the canyon, off each tip of the star (arrow). Photo courtesy of Jean-Yves Sgro, University of Wisconsin, Madison, WI. Reproduced from Rossmann et al. (1985) Nature 317: 145–153.
Figure 4.
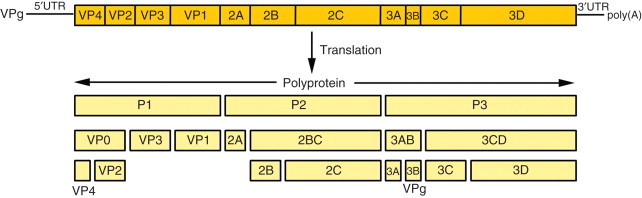
Schematic representation of HRV genome and polyprotein organization. Diagram of the RNA genome with VPg protein, the 5′ untranslated region (5′UTR), the protein coding region, the 3′UTR, and the poly(A) tail (top). Processing pattern of rhinovirus polyprotein. Proteases 2Apro and 3Cpro cleave the polyprotein, generating P1, P2, and P3 precursors (bottom). All intermediate and final cleavages are carried out by 3Cpro and its precursor, 3CDpro, except for the VP0 peptide cleavage into VP4 and VP1, which is done by an as yet unknown protease.
Figure 5.
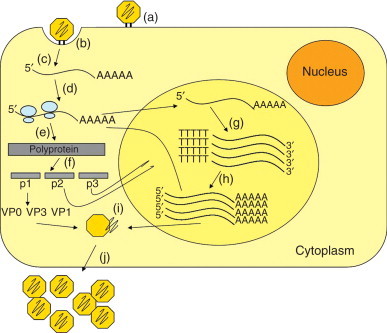
Schematic overview of HRV replication cycle. (a) Virus binds to cell receptor, (b) triggering endocytosis, and (c) genome uncoating. (d) The released positive-strand genome directs the immediate translation (e) of the polyprotein. (f) The nascent polyprotein is cleaved cotranslationally to produce the virus proteins. The RNA synthesis occurs anchored on vesicle membranes. (g) The positive genome is copied by the viral RNA polymerase to form the full-length negative RNA replicative intermediate, which then (h) serves as a template to produce additional positive RNA genomes. (i) The capsid proteins and the newly synthesized RNA genomes are assembled into virions, (j) which are released by cell lysis.
Until recently, the recognized 100 HRV serotypes were classified according to receptor specificity into three groups: The major group with 90 serotypes, whose receptor is intercellular adhesion molecule-1 (ICAM-1); the minor group with 10 serotypes, whose receptor is the low-density lipoprotein receptor (LDLR); and HRV-87 that shares properties with human enterovirus 68 and utilizes sialic acid residues on cell proteins as receptor. The range of HRV serotypes and species by the currently accepted classification system, which is based on sequencing and phylogenetic comparisons rather than on receptor usage or antiviral susceptibility, is expected to expand as more sequence information is obtained from field strains. In fact, a new genetic clade has been recently detected in Hong Kong, consisting of strains associated mostly with LRTI in children with wheezing that contained important deletions in the VP1 region, probably representing a new species of HRV, named HRV C.
HRV is stable for days on environmental surfaces and is resistant to ethanol, ether, chloroform, and nonionic detergents, but is sensitive to UV light, pH lower than 5, and to halogens, such as chlorine, bromine, iodine, and phenolic disinfectants.
Epidemiology
HRV is the predominant agent of ARI in the world and infections occur in people from all continents, including remotely located population groups, such as Bushmen from the Kalahari Desert, native Alaskans, and isolated Amazon Indian tribes. Rhinoviruses have been clearly shown as frequent cause of colds in the United States and in Western Europe and are frequently associated with ARI in children throughout the world. HRV has been estimated to cause up to 80% of all autumn colds in adults in the United States and around 50% of all respiratory infections in ambulatory children in tropical areas.
Evidence suggests that indoor HRV transmission is favored by high relative humidity and crowding of young children, as occurs in the United States at the beginning of the school term, which may explain the autumn seasonal peak of HRV. In tropical regions, however, where relative humidity remains above 70%, reaching 90% during the rainy season, longitudinal studies have found no obvious HRV seasonality.
HRV transmission requires close exposure and occurs mainly by hand-to-hand contact, followed by self-inoculation into the eye or nose, but can also happen by airborne spread. Once HRV reaches the nasal cavity, infection occurs in virtually 100% of susceptible subjects, and approximately 75% of those infected develop illness after 1–2 days of incubation. Children play a central role in spreading the virus in the household.
Pathogenesis and Clinical Features
HRV replication is restricted to the respiratory epithelium, taking place in scattered ciliated cells of the nose and in nonciliated cells of the nasopharynx, and this tropism seems to be a consequence of receptor availability. Infection of a limited number of cells triggers the nuclear translocation of NF-κB and gene expression of cytokines, chemokines, and inflammatory mediators. These, in association with the stimulation of local parasympathetic nerve endings, result in the development of cold symptoms. Kinins, prostaglandins, proinflammatory cytokines, and chemokines may contribute to vasodilation, increased vascular permeability, influx of polymorphonuclear leukocytes, exocrine gland secretion, and nerve ending stimulation, resulting in nasal obstruction, rhinorrhea, sneezing, cough, and sore throat.
Serotype-specific neutralizing IgM, IgG, and IgA antibodies develop in most infected persons in 7–21 days and persist for years. Protection from infection is partially attributed to the presence of IgA antibody in nasal secretions, and recovery from illness is more dependent on cell-mediated immunity.
HRV-induced blastogenesis, natural killer cell activity, mitogen-stimulated cell production of IL-2 and IFN-γ have been documented during HRV infection. HRV induces the expression of human β-defensin 2 (HBD-2) in the respiratory epithelium, which supports a role for HBD-2 in host defense against this agent.
HRV-induced colds are clinically indistinguishable from colds of other viral etiologies and the main symptoms are nasal discharge, nasal obstruction, sneezing, sore or scratchy throat, hoarseness, cough, and headache. Facial and ear pressure may be present, but fever and malaise are uncommon. These symptoms last approximately 7 days, but may persist for up to 2 weeks in 25% of cases. Infants and toddlers may display only nasal discharge and be otherwise asymptomatic. There is no clear association between distinct clinical outcomes and any particular serotypes or species of HRV.
The majority of patients have obstruction and mucosal abnormalities of the sinus cavities, eustachian tubes, and the middle ear, which predispose to secondary bacterial sinusitis and otitis media. HRV RNA may be detected by RT–PCR in maxillary sinus brushings in 40% of adults presenting with acute sinusitis, and in 24% of the samples of middle ear fluid from children less than 7 years of age with diagnosis of acute otitis media. HRV is frequently associated with exacerbations of chronic obstructive pulmonary disease and asthma attacks in children over 2 years of age and in adults.
In addition to colds, HRV has been increasingly recognized as a major cause of LRTI in children and immunocom- promised hosts, and has been detected over 3 times more often than HRSV in association with wheezing in the first year of life.
A major recent advance in HRV research has been the development of experimental animal models of infection, using LDLR-binding minor serotypes in BALB/c mice or ICAM-1 binding major serotypes in human ICAM-1 transgenic mice. Such models will boost the research on pathogenesis and antiviral therapies.
Diagnosis
HRV can be detected in respiratory secretions by isolation in cultures of susceptible cell lines. Cell lines of primate origin support HRV propagation, but certain strains of HeLa cells and human embryonic fibroblasts provide higher sensitivity for HRV isolation from clinical specimens. HRV shedding peaks around 48 h after infection and declines rapidly, but may remain at low levels for up to 3 weeks. The optimal growth temperature for HRV is 33 °C–35 °C in a roller drum and cultures should be examined for 10–14 days. The presence of HRV, indicated by the typical CPE, can be confirmed by the acid sensitivity of the isolate. Unlike other picornaviruses, HRVs are acid-labile, a property that distinguishes them from enteroviruses.
Rapid assays for HRV detection, like immunofluorescence and other antigen detection methods, are not available, because of the large number of serotypes. The homotypic nature of HRV antibodies restricts serology to experimental settings.
RT–PCR in clinical samples is the most sensitive method for HRV detection. Real-time PCR-based assays have been developed in several laboratories, using mostly primers directed to the conserved 5′UTR of the genome, and have the potential to detect serotypes of both HRV species. Real-time PCR multiplex assays directed to conserved sequences of different viral species and genera, as well as recently developed methods such as MultiCode-PLx and Mass-Tag, can detect several viral pathogens in a single run and are expected to become methods of choice for large-scale sample testing in the near future.
Management and Control
Several trials of antiviral agents for HRV have been conducted, but no specific treatment has been licensed, mainly because of the lack of potency, untoward side effects, and drug delivery problems. Symptomatic treatment can be done with a variety of nonprescription medications. Systemic sympathomimetic decongestants may reduce nasal obstruction, first-generation antihistamines may reduce sneezing and rhinorrhea, and nonsteroidal anti-inflammatory drugs may reduce headache, cough, and systemic symptoms.
The large number of HRV serotypes with minimal cross-antigenicity has hampered the development of an HRV vaccine. It may be possible to reduce the exposure to HRV by hand washing after contact with a cold sufferer or after handling objects that may have been contaminated with respiratory secretions. Application of the virucidal agents, salicylic acid or pyroglutamic acid, to the hands reduced recovery of rhinovirus from the hand skin of treated persons.
Short-term postexposure prophylaxis with intranasal IFN-α significantly reduced the incidence of HRV colds in household contacts of an index case. However, the cost and the local side effects associated with the difficulty to make the drug available to homes in a timely fashion reduce the utility of this approach.
Respiratory Adenoviruses
Agents
Adenoviruses are nonenveloped, icosahedral DNA viruses of the genus Mastadenovirus, family Adenoviridae. The adenovirus capsid consists of three morphologically, antigenically, and functionally distinct types of capsomere: hexons, penton bases, and penton fibers that project from the penton bases (Figure 6). The hexon and penton bases contain complement fixing, group-specific antigens common to all human adenoviruses, whereas the fibers have primarily neutralizing and hemagglutination-inhibiting, type-specific antigens. Serum neutralization permits the classification of human adenoviruses in 51 distinct serotypes, distributed in six species, A–F. Adenoviruses are commonly accompanied by small, single-stranded DNA parvoviruses known as adeno-associated viruses, which do not seem to cause any specific disease.
Figure 6.
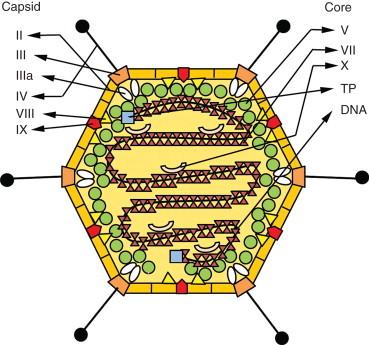
Schematic organization of the adenovirus virion. A naked icosahedral capsid contains a double-stranded DNA genome. The main capsid proteins are the hexon (II), penton base (III), and the penton fiber (IV), which projects from each vertex and binds to cell receptor. In addition to these, other capsid proteins are depicted (IIIa, VIII, and IX). The viral core contains the genome and the proteins V, VII, X, and TP – terminal protein).
The fiber protein binds to the host cell through the protein Coxsackie B and adenovirus receptor (CAR), a protein of the immunoglobulin superfamily that serves as high-affinity receptor for the attachment of adenovirus species A, C, D, E, and F. Plasma membrane protein CD46 is the ligand for the fiber of adenovirus species B. Following the fiber/receptor binding, the interaction of the penton base with cell surface integrins triggers endocytosis and the exposure of the virus to the cytosol, with microtubule-mediated transport to the nucleus and final disassembly at the nuclear pore. In the nucleus, a set of ‘early’ mRNAs is transcribed that will direct translation of proteins that modulate cell cycle, block apoptosis, shut down the host mRNA transport from the nucleus and DNA replication. Then the ‘late’ mRNAs are transcribed and their products include viral structural proteins. Virus assembly takes place in the nucleus, and the infectious cycle is completed by the release of up to 1 million virions upon cell lysis (Figure 7).
Figure 7.
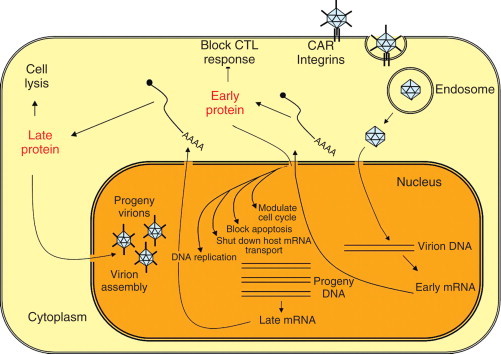
Adenovirus replication cycle. The virus binds to the receptor CAR (Coxsackie B and adenovirus receptor) and integrins. After adsorption, virus is internalized by receptor-mediated endocytosis and directed to the nuclear pore where final disassembly occurs. The viral DNA genome is released into the nucleus and the early set of genes are expressed. Early viral gene products mediate further viral gene expression and DNA replication. Then the late viral genes are expressed, generating structural proteins, and assembly of progeny virions occurs. New viruses are released by cell lysis.
Adenoviruses are stable over a wide pH range (5–9), resistant to isopropyl alcohol, ether, and chloroform. They are stable for weeks at room temperature and for years at approximately 20 °C or colder, and can even be lyophilized. They are inactivated by sodium hypochlorite and by exposure to 60 °C for 2 min.
Epidemiology
Respiratory diseases are among the most frequent manifestations of infections by adenoviruses, particularly in children under the age of 5 years. Respiratory infections by adenoviruses occur worldwide and with no apparent seasonality. Outbreaks can occur, especially in crowding conditions, and are more frequent in late winter, spring, and early summer. The most common serotypes associated with respiratory disease are serotypes 1–7 and 21, and adenovirus of low-numbered serotypes (1, 2, 3, and 5) are more frequent before the age of 5 years, accounting for 5–20% of cases of URI and approximately 5% of cases of LRI in children. In adults, adenoviruses occur sporadically and cause mostly URI.
Infections by adenoviruses types 4 and 7 are usually epidemic, with attack rates of 6–16% per week in newly assembled military recruits, whose adenovirus carriage rate may be as high as 18%. In this group, the adenoviral syndromes vary from mild colds to severe LRI, but overall attack rates may reach 80%, with 20–40% of the individuals needing hospitalization. In tropical areas, the incidence of adenovirus infections in military recruits is lower, and different serotypes may be involved.
Pharyngoconjunctival fever caused by adenoviruses types 3 and 7 may be epidemic or endemic among children during the summer in temperate climates, and has been associated with inadequate chlorination or filtration of swimming pools. Ocular transmission has also been associated with physician offices where sterilization or hand washing was inadequate. Asymptomatic adenoviral infections and prolonged carrier state are common.
Pathogenesis and Clinical Features
Up to 50% of nonepidemic adenoviral infections are asymptomatic, and, in fact, adenoviruses were discovered because of their propensity for latency in adenoidal tissue. Symptomatic infections may involve all parts of the respiratory tract and generally initiate in the upper respiratory epithelium.
Adenovirus infection results in necrosis of cells of airway epithelia and may cause viremia by systemic virus dissemination in immunocompromised persons. Bronchiolitis, interstitial pneumonitis, and mononuclear cell infiltrates are part of the inflammatory process in the lungs. In addition to lytic infection, adenoviruses may become latent in epithelial and lymphoid cells, which is probably important to maintain the virus in populations. Protection from adenovirus infection and disease is mainly due to type-specific neutralizing antibody, but reinfections, mostly asymptomatic, may occur. A long-lived T-cell immune response develops in most infected immunocompetent persons and is responsible not only for recovery from infection, but also for tissue pathologic changes.
The incubation period of adenovirus infections averages 10 days and the usual symptoms are those of a febrile cold. In children, the fever may be high and long-lasting. Pharyngitis is common and may be associated with fever, pharyngeal exudate, granular appearance of the mucosa, and anterior cervical adenopathy, similarly to streptococcal pharyngitis. The most frequent complication of adenoviral colds is acute otitis media, which occurs in up to 30% of cases.
Adenoviruses can be recovered from up to 20% of cases of pharyngitis in small children and symptoms may be concurrent with pharyngoconjunctival fever, a syndrome caused mostly by adenovirus types 3 and 7, characterized by conjunctivitis, frequently unilateral, which may last for 1–2 weeks, preauricular adenopathy, cough, rhinitis, malaise, and fever.
Adenovirus LRIs, mainly bronchitis and pneumonia, may represent over 10% of childhood LRIs in temperate areas. Permanent lung parenchymal damage may occur, especially when adenoviral infection is concurrent with measles. Clinical manifestations of epidemic adenoviral infections in military recruits may range from colds to severe pneumonia. Typically, the manifestations are fever, pharyngeal symptoms, cough, chest pain, headache, and malaise. Overwhelming pneumonitis may be part of disseminated adenoviral infections in newborn infants and patients with immunodeficiencies, including AIDS. However, the frequent concomitance of other respiratory pathogens in AIDS patients and the high prevalence of asymptomatic adenovirus infection shed doubt on the causal role of the adenovirus in these patients.
Diagnosis
Adenoviruses can be detected in respiratory, ocular, or ear secretions, but clinical correlation is required, because asymptomatic virus shedding is common. Isolation of adenoviruses in cell culture with identification by IF has been in use for decades, but direct detection of viral antigens or viral DNA by PCR in clinical samples is a sensitive and rapid alternative.
Adenoviruses replicate well in continuous cell lines of epithelial origin, such as HEp-2, HeLa, and A549, and can be adapted to grow in human embryonic lung fibroblasts. In culture, they cause a characteristic CPE and maintenance of cultures for 2 weeks combined with blind passage may increase adenovirus recovery. Inoculation of cells by centrifugation in the shell vial format followed by immunostaining may shorten the detection time.
Rapid antigen detection by immunochromatography is around 95% sensitive in comparison with cell culture and can be easily used in point-of-care diagnosis of adenovirus. However, both conventional and real-time PCR are currently accepted as the most sensitive diagnostic methods. Positive results by PCR should be interpreted with caution, given the propensity of adenoviruses to cause latency. Quantitative real-time PCR can also be used to monitor viral load in transplant recipients or immunosuppressed patients, allowing for appropriate interventions to be initiated, such as immunosuppressive regimen adjustment.
Several serologic tests can detect antibodies to the common hexon antigen. However, their clinical utility is restricted.
Management and Control
At present, there is no routine effective antiviral treatment for adenovirus infections. Successful therapy of severe adenoviral infections in immunocompromised patients with IV ribavirin has been reported. Trifluridine and cidofovir have shown antiviral effect and in vitro and can potentially be used for treatment.
A live vaccine consisting of wild-type adenovirus packaged in enterically coated capsules induces immunity by ensuring enteric replication without infection of the respiratory tree. Proper sterilization, hand washing, and chlorination can prevent adenovirus spread via tonometers, hands, and swimming pools.
Human Coronaviruses Unrelated to SARS
Agents
Human coronavirus (HCoV) subtypes 229E and OC43 were the only coronaviruses identified in humans until 2003, when identification of the severe acute respiratory syndrome (SARS) coronavirus led to a renewed research on HCoVs. Following that, novel coronaviruses have been discovered. In 2004, investigators in the Netherlands isolated a distinct HCoV, subtype NL63 (also known as NL or NH), from a child with bronchiolitis and conjunctivitis. This agent, closely related to coronavirus 229E, has subsequently been detected in children and adults worldwide. A second novel coronavirus, subtype HKU1, was detected in 2005 from adults with pneumonia in Hong Kong. On the basis of antigenic and genetic studies, the known HCoVs are distributed in two of the three coronavirus groups so far identified. HCoV-229E and -NL63 belong to group I; and HCoV-OC43, HKU1, and SARS-CoV belong to group II, while group III contains no known human agents, but only avian coronaviruses.
Coronaviruses are enveloped viruses with distinct virion morphology, displaying widely spaced, long petal-shaped spikes at the surface, that confer to the virus a crownlike appearance, origin of the name corona (Figure 8). The viral envelope contains a long helical nucleocapsid with single-, positive-stranded RNA 27–32 kb in size, the largest known viral-RNA genome.
Figure 8.
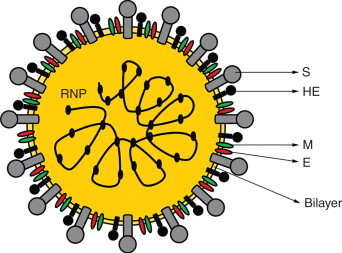
Schematic coronavirus structure. The virions are spherical, with an envelope containing a prominent crown (‘corona’) of peplomers of S (spike) glycoprotein. HE (hemagglutinin), E (small envelope protein), and M (membrane glycoprotein). The genome is a positive-stranded RNA associated with the N (nucleocapsid phosphoprotein), composing the helical RNP (ribonucleoprotein).
Coronavirus RNA synthesis occurs in the cytoplasm via a negative-strand RNA intermediate (Figure 9). The viral RNA is capped at the 5′ end , where there is a ‘leader’ sequence followed by an UTR. At the 3′ end, there is a terminal UTR, followed by a poly(A) tail. The ORF 1 of the genomic RNA is translated into a polyprotein that is processed to yield the proteins that form the transcriptase–helicase complex. The genomic RNA is used as a template to synthesize negative-sense RNAs, which are in turn used to synthesize full-length genomic RNA and subgenomic mRNAs. The mRNAs direct translation of the viral structural and nonstructural proteins. Progeny viruses assemble and bud in vesicles between the endoplasmic reticulum and the Golgi apparatus, later released by exocytosis.
Figure 9.
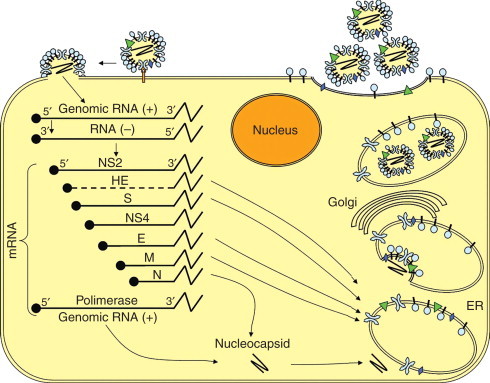
Schematic representation of the life cycle of a coronavirus. Virion binds receptors on the plasma membrane by the S glycoproteins resulting in fusion of viral envelope and plasma or endosomal membrane. ORF 1 of the genomic RNA is translated generating the RNA-dependent RNA polymerase that synthesizes negative-sense RNAs, which serve as templates to generate full-length genomic RNA and subgenomic (NS2, HE, S, NS4, E, M, and N). mRNAs are translated in structural and nonstructural proteins. The N protein and newly synthesized genomic RNA form the nucleocapsid. Other structural proteins are inserted in the endoplasmic reticulum (ER) where they are cotranslationally glycosylated and trimerized. The nucleocapsids are then enclosed by these proteins in the ER and transported to the Golgi apparatus. Mature virions are apparently released by exocytosis-like fusion of smooth-walled vesicles with the plasma membrane.
The viral envelope contains the structural proteins S (spike), M (membrane), E (envelope), and only in the case of some group II coronaviruses, the HE. The S glycoprotein contains neutralizing and T cell epitopes and functions as the cell receptor ligand. The M protein is embedded in the envelope and interacts with the N (nucleocapsid) protein during maturation. In addition to the nucleocapsid and envelope proteins, a replicase is present in cells infected by all coronaviruses. Except for HCoV-OC43, which binds the cell receptor via the HE, the other HCoVs bind via the S (spike) protein. The S protein of HCoV-229E binds to the metalloprotease human aminopeptidase N, or CD13, at the cell surface, and entry is independent of enzymatic activity of the receptor. The HE of HCoV-OC43 binds to the sialic acid present in glycoproteins on the cell surface, and this interaction facilitates infection. HCoV-NL63, like SARS-CoV, utilizes the angiotensin-converting enzyme 2 (ACE2) as cell receptor.
Epidemiology
HCoVs have been found throughout the world and are considered to be the second most frequent cause of common colds, accounting for an average rate of 15% of ARI in the general population in the United States. However, the rates may be quite variable from year to year, ranging from 1% to 35% in years of peak activity. HCoV-229E and -OC43 cause common colds with variable frequency, depending mainly on the detection method and the season of the study. HCoV infections occur mainly in the winter and spring months, but summer activity has also been documented. HCoV-229E and -OC43 have caused winter outbreaks at 2–4-year intervals in temperate regions. Little is known about the prevalence of HCoV-229E and -OC43 in tropical countries. Data on the frequency and impact of HCoV-NL63 and -HKU1 infections are still scarce.
Pathogenesis and Clinical Features
There is no convenient small animal model to study the pathogenesis of HCoV, and humans naturally or experimentally infected are the only source of information. HCoV-229E is known to infect airway epithelial cells from the apical surface, where the receptor is constitutively expressed, and to exit productively infected cells through the same route. Ultrastructural studies of nasal epithelium of volunteers experimentally infected with HCoV-229E revealed epithelial cell damage, ciliary loss, and cytolysis on day 3 postinfection.
In the United States, seropositivity to HCoV-OC43 and -229E rises during the first 5 years of life, and around 40% of adults are seropositive. Symptomatic reinfections are possible, despite the presence of antibodies, suggesting rapidly waning immune response or circulation of closely related but antigenically different viruses.
HCoVs are transmitted by the respiratory route, and experimentally infected volunteers shed virus for approximately 5 days, beginning 48 h after infection, which is approximately the time of onset of symptoms. The peak of symptoms occurs 2–4 days postinoculation. The usual manifestations of HCoV infection are typical common colds, but more severe disease can be seen in the very young, the elderly, and immunocompromised hosts. The incubation period tends to be 1 day longer than that for HRV colds, with illness duration of 6–7 days. Low-grade fever may occur in up to 20% of the patients, and in addition to nasal symptoms, cough and sore throat occur frequently. More serious infections of the LRT caused by HCoV have also been documented, either sporadically in infants with pneumonia and immunocompromised patients, or in up to 33% of previously healthy Marine Corps recruits with pneumonia. In addition, HCoV-229E and -OC43 have been recognized in association with influenza-like illnesses in frail elderly patients.
HCoV-NL63 seems to play a major role as a cause of LRTI in children, and up to 45% of children younger than 3 years with infection by HCoV-NL63 in Germany had croup. HCoV-HKU1 has also been associated with LRTI both in children and in elderly patients with underlying diseases.
HCoV infections have also been associated with exacerbations of asthma, chronic bronchitis, and recurrent wheezing in children. Similar to HRV, HCoV infections have been frequently recognized in association with otitis media and maxillary sinusitis in children and adults. HCoV was detected by RT–PCR in the middle ear effusion or nasopharyngeal aspirate from 16 of 92 (17%) children with acute otitis media in Finland and in nasal swabs from 3 of 20 adults with acute maxillary sinusitis.
Some studies indicate that respiratory HCoVs are able to reach the central nervous system, but the real importance of these agents as cause of diseases outside the respiratory tract remains to be determined yet.
Diagnosis
Laboratory diagnosis of HCoVs in clinical samples by isolation in cell cultures is difficult. RT–PCR-based assays for HCoVs utilizing primers for relatively conserved regions of the genome are currently the best alternative for virus detection. More recently, quantitative real-time PCR for HCoVs has been developed in several laboratories, providing a rapid and sensitive way for detection and determination of viral load, with potential applications in pathogenesis studies. Serologic diagnosis of HCoVs is sensitive and specific, but has limited application in case management.
Management and Control
Intranasal interferon protects against experimental infection with HCoV-229E, but there is no antiviral drug therapy currently available for non-SARS HCoVs. Considering the usually self-limited course of infection, supportive care and symptomatic relief are sufficient. No vaccines are currently available for HCoV.
SARS Coronavirus
The Agent
SARS was first reported to the WHO by Carlo Urbani, a physician working in Hanoi, Vietnam, in February 2003. However, retrospective analysis revealed that cases compatible with this disease had occurred in China, in the province of Guangdong, starting in November 2002. In February 2003, a single infected person spent 1 day at a hotel in Hong Kong and transmitted the virus to 16 other people, who in turn seeded outbreaks of SARS in other countries in Asia and Canada. In a matter of weeks, SARS had affected thousands of people in 25 countries in all continents, causing enormous global impact. SARS-CoV was isolated and characterized in record time after the onset of the 2002–03 outbreak.
SARS-CoV shares structural features and genome organization with other Coronaviridae, and the recognition of the peculiar coronavirus morphology by electron microscopy of Vero E6 cells inoculated with oropharyngeal material from a patient was the initial clue to the identification of the agent. The viral genome is 29 727 nt in length, with 14 ORFs coding for 28 putative proteins. It is known that SARS-CoV nsp1 protein increases cellular RNA degradation and thus might facilitate SARS-CoV replication or block immune responses. Trimers of the S protein form the peplomers and bind the cell receptor, being the main target for neutralizing antibodies. M and N proteins of SARS-CoV can induce apoptosis of host cells and may play an important role in pathogenesis.
SARS-CoV is phylogenetically different from previously known coronaviruses, but isolates from different origins are relatively similar. Genome analysis reveals that SARS-CoV is neither a host-range mutant nor a recombinant of previously known coronaviruses but rather an independently emerged virus. SARS-CoV seems to have evolved from an animal SARS-like virus, acquiring greater fitness in humans during the course of the outbreaks, probably through the appearance of nucleotide deletions in ORF8.
During the infection the N-terminal portion of the spike glycoprotein binds to the virus receptor, identified as the metallopeptidase ACE2 homologue, triggering pH-dependent endocytosis. SARS-CoV spike protein can also bind the dendritic cell-specific C-type lectin ICAM-3 grabbing nonintegrin (DC-SIGN), which does not result in dendritic cell infection by the agent but allows for SARS-CoV to be transported to susceptible target cells elsewhere. Upon entry, SARS-CoV replication cycle is probably similar to that of other coronaviruses.
Epidemiology
SARS-CoV has a zoonotic origin and horseshoe bats seem to be its natural reservoir. At the beginning of the 2002–03 outbreak, probably animal-to-human interspecies transmission was involved providing the source of an agent that later adapted to efficient human-to-human transmission. Palm civets and perhaps other mammals served as amplification hosts. Interestingly, shortly after a wildlife trade ban was imposed to control the SARS outbreak, there were no further naturally acquired human cases of SARS in Guangdong. SARS-CoV is mainly transmitted among humans by the deposition of infected droplets or aerosols on the respiratory epithelium. In addition, transmission is infrequent during the first 5 days of illness, partly because of the low viral load in respiratory secretions during that phase. Excretion of SARS-CoV in sputa and stools may average 21–27 days after symptom onset, respectively.
Transmission of SARS-CoV among health-care workers and between patients in the hospital setting played a pivotal role in outbreak propagation. Assisting during intubation, suctioning, and manipulating ventilatory apparatuses were high-risk activities. SARS-CoV may persist for up to 2 days on environmental surfaces and 4 days in diarrheal stools.
Approximately, 8500 cases were reported in the outbreak, with fatality rate of at least 10%. At present no circulation of SARS-CoV is registered.
Pathogenesis and Clinical Features
The mechanism of how SARS-CoV damages cells and tissues is not completely understood. The virus evades innate immune response defenses, interferes with interferon production, degrades cell mRNAs, and modulates ubiquitination pathway. Cell damage occurs by necrosis, apoptosis, and syncytium formation.
SARS-CoV has been detected in respiratory tissue using immunohistochemistry, in situ hybridization, and electron microscopy of pneumocytes, as well as on the apical surface of enterocytes. Marked inflammatory infiltrates have not been observed in the intestine, and the pathogenesis of the SARS-CoV-related diarrhea remains largely unknown. Pulmonary tissue shows diffuse alveolar damage, mixed infiltrate, lung edema, hyaline membrane, abundant macrophages in alveoli and interstitium, and syncytia formation. Higher viral loads of SARS-CoV can be detected in the LRT than in the upper airways.
Besides respiratory secretions and stools, SARS-CoV can be detected in urine in up to 30% of patients. The effect of SARS-CoV infection on the immune system is highlighted by pronounced T-cell lymphopenia and elevation of several inflammatory cytokines (IL-1β, IL-6, and IL-12) and chemokines (MCP-1 and IP-10) observed in patients. Since immunologic markers in the peripheral blood may not reflect what happens in the microenvironment of the lung, the pathogenic importance of these findings is not clear. Seroconversion was documented in 93% of the patients at around 20 days, and the rise in IgG titers correlates with decrease in viral load. Paradoxically, clinical worsening also occurs during this phase, suggesting that, rather than unchecked viral replication, immunopathologic factors may be responsible for the lung lesions.
The incubation period of SARS is 4–6 days and initial clinical symptoms and signs are systemic symptoms, such as fever, chills, myalgia, and malaise. Respiratory symptoms appear 2–7 days later, most frequently nonproductive cough, dyspnea, chest pain, headache, and sore throat. Diarrhea and vomiting may occur. Chest radiograms frequently reveal infiltrates consistent with viral pneumonitis, consisting mostly of consolidations and ground-glass opacifications. Fever generally subsides in 48 h, but one or two relapses within 8–15 days may be observed. Watery diarrhea with an average of six evacuations per day is common.
Radiologic worsening of the pulmonary lesions seen at admission then occurs, and development of diffuse ground-glass changes frequently heralds the development of acute respiratory distress syndrome (ARDS). Hypoxemia is noted in approximately half of the patients at around 9 days after the onset of symptoms, and a high proportion of those admitted to the intensive care unit (ICU), especially older males, require mechanical ventilation around day 13. Prognosis is related to the level of viral replication in tissues, and patients with high viral loads in serum, nasopharyngeal aspirates, or feces tend to have poor clinical outcome. Old age and severe underlying diseases have been identified as predictors of poor outcome. Fatality rates based on cases admitted to hospitals were 13% for patients younger than 60 years and 43% for those older than 60 years.
Diagnosis
The ability to grow SARS-CoV in Vero E6 cell cultures was critical to identifying the agent. SARS-CoV can be recovered by isolation from respiratory secretions, feces, and urine, but this procedure can only be done in biosafety level 3 laboratories not available in most hospitals of the world. Although not useful for early diagnosis, seroconversion determined by IFA or EIA remains the gold standard for confirming SARS diagnosis. IgG seroconversion is detectable in over 90% of patients at around day 28.
Rapid diagnosis by antigen detection is feasible, but less sensitive than RT–PCR. Real-time quantitative RT–PCR of nasopharyngeal aspirates is the method of choice with around 80% sensitivity. Because of low viral loads in the upper respiratory tract in the first few days of illness, SARS-CoV is detectable by RT–PCR in nasopharyngeal aspirates in only one-third of patients at presentation and in two-thirds just at day 14. RT–PCR may be positive for SARS-CoV in stools from as much as 97% of patients at day 14 and in urine in 42% of samples at day 15. The sensitivity of RT–PCR could be increased by testing multiple specimens, including nasopharyngeal, serum, and fecal samples.
Management and Control
The main component of treatment of SARS patients is supportive therapy, chiefly the management of hypoxemia and ARDS. SARS-CoV is only modestly susceptible to ribavirin in vitro, and therapeutic doses are difficult to achieve clinically. Many potential antiviral compounds have been evaluated in vitro, but just a few have been tested in animal models and even fewer in patients. Interferons (IFN-αn1/n3, leukocytic IFN-α, IFN-β) and protease inhibitors were consistently active in vitro and may be considered for treatment. ACE2 analogues, antiviral peptides against the S protein, and small interfering RNA (siRNA) have also been effective in vitro. Pegylated IFN-α2a was shown to be useful in a monkey model. Convalescent plasma has also been tested in the treatment of SARS patients and reduced the frequency of poor outcome when given before 14 days of illness.
It is impossible to predict whether naturally reemerging SARS-CoV would cause a global outbreak. At present, no SARS-CoV vaccine is available for human use. Nevertheless, a vaccine for this agent would be relevant for high-risk individuals, such as workers in laboratories, hospitals, and game animal farming. Therefore, considerable effort has been directed at developing such vaccine. It has been shown that SARS-CoV S protein expressed on chimeric parainfluenza virus or in vaccinia virus, as well as spike protein – encoding DNA-induced neutralizing antibodies and protected experimental animals from challenge with live virus. Several other types of vaccine candidates have been developed, including inactivated virus, subunit vaccines, virus-like particles (VLPs), DNA vaccines, heterologous expression systems, and vaccines derived from SARS-CoV by reverse genetics.
Human Bocavirus
The Agent
Human bocavirus (HBoV) was recently discovered by means of an elegant metagenomic approach and has now been detected in samples from children with ARI in studies around the world. HBoV was described in 2005 after a screening of a pool of respiratory clinical samples that had tested negative for other viral agents. DNA extracted from a gradient purified virus-enriched fraction of the pool of clinical samples was randomly amplified by PCR, then cloned and sequenced, revealing the presence of a previously unknown parvovirus. Bioinformatic analysis of the sequences then revealed it to be a novel member of the family Parvoviridae, provisionally classified by sequence homology and genome organization in the genus Bocavirus, which already included two other viruses: the bovine parvovirus 1 and canine minute virus.
HBoV virions consist of small nonenveloped icosahedral particles with a single-stranded DNA genome of approximately 5300 nt, organized in a way similar to that of other known bocaviruses, with three ORFs, two encoding the nonstructural proteins NS1 and NP-1, and a third nesting the two capsid proteins, VP1 and VP2 (Figure 10). NS1 is likely to be the viral polymerase and the function of NP-1 is still unknown. HBoV replication cycle has not been studied and is likely to be similar to that of other autonomous Parvoviridae. Two genetically distinct clusters have been detected and the degree of variability is higher in the gene of capsid proteins. HBoV has not been propagated in cell cultures and this has hampered cell biology studies and production of virus stocks for pathogenesis studies.
Figure 10.
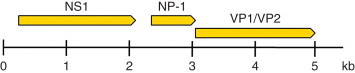
Schematic map of HBoV genome. The single-stranded DNA genome contains three ORFs: NS1, NP-1, and VP1/VP2. VP1 and VP2 are alternative splicing products of the same ORF.
Epidemiology
HBoV circulates worldwide and prevalence studies, done mostly by PCR in respiratory samples from children, have shown detection rates of 1.5–19%, depending on the regions where studies were conducted and the sensitivity of the assays. So far, there has been no clear HBoV seasonality.
The fact that HBoV may persist for a long time after primary infection and positive PCR results may be found in healthy carriers. This asymptomatic persistence may seriously affect incidence estimates. Seroprevalence in children in the United States, using VP2 VLPs as antigen, revealed that over 85% of children older than 4 years have already been exposed to HBoV. While studies have reported HBoV detection in all age groups, children younger than 2 years represent the vast majority of patients investigated to date.
Pathogenesis and Clinical Features
Several studies have detected HBoV as the sole virus in association with respiratory symptoms, sometimes with high viral loads, consistent with disease causation. However, HBoV has been frequently detected by PCR in healthy subjects, what may represent shedding of unknown origin. In addition to this, the lack of culture system and animal models makes it difficult to fulfill Koch’s postulates and even molecular causality criteria. Case–control studies have found significant difference in the prevalence of HBoV in patients with ARI as compared to healthy controls. However, up to 76% of patients with detectable HBoV also shed other respiratory viruses at the same time.
Parvoviruses require proliferating cells for their replication and animal bocaviruses infect respiratory and intestinal epithelium, and lymphoid organs. The sites of HBoV replication have not been determined, but the virus is apparently not confined to the respiratory tract, since it has been detected in stools and serum, suggesting systemic infection. Importantly, HBoV can be found in 45% of the stool samples from patients with respiratory symptoms. This, however, is not enough to support a role of HBoV as an agent of gastroenteritis.
Even though HBoV’s causative role in disease remains to be confirmed, the agent has been detected in association with URI symptoms, including cough, rhinitis, and fever, as well as in patients with bronchitis, bronchiolitis, pneumonia, and asthma exacerbation. The range of clinical symptoms is similar to that observed for HRSV and HMPV infections, and the patient may remain symptomatic for 1–2 weeks. Acute otitis media may be present in 42% and gastrointestinal symptoms have been reported in up to 25% of patients.
Diagnosis
The only diagnostic method available for HBoV is PCR on nasopharyngeal aspirates, washes, and swabs. Primers directed to the more conserved NS1 gene are favored. In addition to its diagnostic importance, real-time PCR assays have the potential for quantitation of viral loads, and therefore can contribute to HBoV pathogenesis investigation. Serology has been described using recombinant VP2 and baculovirus-expressed VP2 VLPs as antigens, but as for other respiratory viruses, its application to case management would be limited.
Management and Control
No specific treatment exists for HBoV infections and, if HBoV is confirmed to contribute significantly to disease in children, vaccine development will become a possibility.
See also:
Influenza
Further Reading
- Bartlett N.W., Walton R.P., Edwards M.R. Mouse models of rhinovirus-induced disease and exacerbation of allergic airway inflammation. Nature Medicine. 2008;14:199–204. doi: 10.1038/nm1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cane P. Respiratory syncytial virus. In: Zuckerman Ajm I.K., editor. 1st edn. vol. 14. Elsevier; Amsterdam: 2007. p. 340. (Perspectives in Medical Virology). [Google Scholar]
- Cheng V.C., Lau S.K., Woo P.C.Y., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clinical Microbiology Reviews. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn J.S. Epidemiology of human metapneumovirus. Clinical Microbiology Reviews. 2006;19:546–557. doi: 10.1128/CMR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn J.S., Kesebir D., Cotmore S.F. Seroepidemiology of human bocavirus defined using recombinant virus-like particles. The Journal of Infectious Diseases. 2008;198:41–50. doi: 10.1086/588674. [DOI] [PubMed] [Google Scholar]
- Knipe D.H., Howley P.M., editors. 1st edn. vol. 1 and 2. Lippincott Williams & Wilkins; Philadelphia: 2007. (Fields Virology). [Google Scholar]
- Kusel M.M., de Klerk N.H., Holt P.G. Role of respiratory viruses in acute upper and lower respiratory tract illness in the first year of life: A birth cohort study. The Pediatric Infectious Disease Journal. 2006;25:680–686. doi: 10.1097/01.inf.0000226912.88900.a3. [DOI] [PubMed] [Google Scholar]
- Lau S.K., Yip C.C., Tsoi H.W. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. Journal of Clinical Microbiology. 2007;45:3655–3664. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Holloway B., Dare R.K. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. Journal of Clinical Microbiology. 2008;46:533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IM (2008) Human rhinoviruses: The cold wars resume. Journal of Clinical Virology. (in press). [DOI] [PMC free article] [PubMed]
- Pyrc K., Berkhout Bvan der Hoek L. The novel human coronaviruses NL63 and HKU1. Journal of VIirology. 2007;81:3051–3057. doi: 10.1128/JVI.01466-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. Journal of Virology. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildgen O., Muller A., Allander T. Human bocavirus: Passenger or pathogen in acute respiratory tract infections? Clinical Microbiology Reviews. 2008;21:291–304. doi: 10.1128/CMR.00030-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Pfarr D.S., Losonsky G.A., Kiener P.A. Immunoprophylaxis of RSV infection: Advancing from RSV-IGIV to palivizumab and motavizumab. Current Topics in Microbiology and Immunology. 2008;317:103–123. doi: 10.1007/978-3-540-72146-8_4. [DOI] [PubMed] [Google Scholar]
- Zhan X., Slobod K.S., Krishnamurthy S. Sendai virus recombinant vaccine expressing hPIV-3 HN or F elicits protective immunity and combines with a second recombinant to prevent hPIV-1, hPIV-3 and RSV infections. Vaccine. 2008;26:3480–3488. doi: 10.1016/j.vaccine.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


