History of Virus Classification and Virus Nomenclature
Humans feel the need to classify natural entities and the viruses are no exception. As in other biological systems, virus classification is an approximate and imperfect exercise. Like any other type of classification, it is a totally artificial and human-driven activity without any natural base. However, science requires workable descriptions of living systems and their constituent parts, and, when achieved properly, classifications are extremely useful for showing similar characteristics and properties across populations. Unfortunately for virus taxonomy no fossil record exists and so evolutionary relationships are very speculative, meaning that only a logical and precise virus classification can provide indications of the evolution of viruses. Appropriately chosen classification criteria are also informative in the case of newly discovered viruses. In theory, nomenclature and classification are totally independent, but for viruses both issues are often considered at the same time. As a result, taxonomic names for the viruses have always been the subject of passionate discussions and the taxonomic status of viruses is a sensitive and critical issue.
Virus classification is a relatively new exercise, as the first evidence for existence of a virus was only presented at the end of the nineteenth century by Beijerinck in 1898. It was not until 1927 that Johnson, a plant virologist, drew attention to the need for a system of virus nomenclature and classification. First efforts to classify viruses utilized a range of ecological and biological criteria, including pathogenic properties in the case of human and animal viruses, and symptoms for plant viruses. For example, viruses sharing the pathogenic properties causing hepatitis (e.g. hepatitis A virus, hepatitis B virus, yellow fever virus, and Rift Valley fever virus) were grouped together as ‘the hepatitis viruses’. Virology developed substantially in the 1930s and early classifications for the viruses reflected these advances. In 1939, Holmes published a classification of plant viruses dependent on host reactions and differential host species, using a binomial-trinomial nomenclature based on the name of the infected plant; however, only 89 viruses were described and classified in this way. With the development of electron microscopy and biochemical studies in the 1950s, the first virus groupings based on common virion properties emerged: like the Herpesvirus group described by Andrewes in 1954, the Myxovirus group by Andrewes et al, in 1955, and the Poxvirus group by Fenner and Burnet in 1957. During this period there was also an explosion of newly discovered viruses; in response, several individuals and committees independently proposed virus classification systems but none was widely adopted by the scientific community. It became obvious that only an international association of virologists could propose a comprehensive and universally acceptable system of virus classification.
At the 1966 International Congress for Microbiology held in Moscow, the International Committee on Nomenclature of Viruses (ICNV) was established by an international group of 43 virologists. An international organization was set up with the aim of developing a taxonomy and nomenclature system for all viruses that would be recognized worldwide. The name of the ICNV was changed in 1974 to the more appropriate International Committee on Taxonomy of Viruses (ICTV), which remains active today. ICTV, the unique official committee of the Virology Division, is now considered the international official body for all matters related to taxonomy and nomenclature of viruses.
Since the founding of the ICTV, all virologists have agreed that the hundreds of viruses isolated from different organisms should be classified together in a unique system, but separate from other microorganisms such as fungi, bacteria and mycoplasma. However, there was much controversy on the way to do it. Lwoff, Horne and Tournier argued for the adoption of a system classifying viruses into subphyla, classes, orders, suborders and families. Descending hierarchical divisions would have been based on nucleic acid type (DNA or RNA), strandedness (single or double), presence or absence of an envelope, capsid symmetry, and so on. The hierarchy of this system has never been recognized by the ICTV; nevertheless, the types of criteria used became the basis of the universal taxonomy system now in place, and all ICTV reports have used this scheme. Until 1990, no hierarchical classification level higher than the family was used, however, the system has recently begun to move in this direction. A first order, Mononegavirales, was accepted in 1990, and another two, Caudovirales and Nidovirales, were adopted in 1996. In its nonlinnean structure, the scheme is quite different from that used in the taxonomy of bacteria and other organisms. Nevertheless, the usefulness of the scheme is being demonstrated by its wide application. It has replaced all competing classification schemes for all viruses and no one would now dispute with the ICTV the international mandate to name and classify viruses.
Since its establishment, a total of seven virus taxonomic reports (also known by the names of the ICTV Presidents acting as Editors in Chief of the reports) have been published by the ICTV: Wildy in 1971; Fenner in 1976; Matthews in 1979; Matthews in 1982; Francki et al in 1991; Murphy et al in 1995; and van Regenmortel et al in 1999. At the first meeting in Mexico City in 1970, two families with a corresponding two genera and 24 floating genera were adopted to begin the grouping of the vertebrate, invertebrate and bacterial viruses. In addition, 16 plant virus groups were designated, as reported by Matthews in 1983. The fifth ICTV report, edited by Francki et al in 1991, described one order, 40 families, nine subfamilies, 102 genera, two floating genera and two subgenera for vertebrate, invertebrate, bacterial and fungal viruses, and 32 groups and seven subgroups for plant viruses. While most virologists shifted to placing viruses in families and genera, plant virologists retained the term ‘groups’ until 1993. It was only in 1995, as described in the sixth ICTV report, that the ICTV proposed a uniform system for all viruses, with two orders, 50 families, nine subfamilies, 126 genera, 23 floating genera and four subgenera encompassing 2644 assigned viruses. Most recently, at the 28th meeting of the ICTV in March 1998 in San Diego, California, the Universal Virus Classification was adopted; this comprises three orders, 56 families, nine subfamilies, 203 genera, 30 floating genera and a total of 3954 species, strains and/or serotypes of species and tentative species. It is a general trend that the number of described taxa and the number of species of viruses is increasing steadily, easily explained by the increasing complexity of the virus classification and by the amount of data available to demarcate viruses.
With precise and complete descriptions available for a large number of virus families, this classification now constitutes a valuable source of information for new ‘unknown’ viruses. Therefore, the ICTV classification is not only a taxonomic exercise for virus evolutionists but also a valuable diagnostic tool and educational system for virologists, teachers, medical doctors and epidemiologists.
How does the ICTV operate?
The ICTV is a committee of the Virology Division, which is in turn part of the International Union of Microbiological Societies. The ICTV is a nonprofit-making organization composed of prominent virologists representing countries from throughout the world who work to designate virus names and taxa through a democratic process. The ICTV operates through a number of committees, subcommittees and study groups consisting of more than 492 eminent virologists with expertise in viruses infecting humans, animals, insects, protozoa, archaea, bacteria, mycoplasma, fungi, algae, yeasts and plants. Taxonomic proposals are initiated and formulated by individuals or by the study groups. These proposals are revised and accepted by the corresponding subcommittees and presented for executive committee approval. All decisions are then ratified at a plenary session (or also now by postal vote) held at each Virology Congress where all members of ICTV and more than 50 representatives of national microbiological societies are represented. At present, there are 47 study groups working in concert with six subcommittees – namely, the vertebrate, invertebrate, plant, bacteria, fungus and virus data subcommittees. The ICTV does not impose any taxonomic terms or taxa but ensures that all propositions are compatible with ICTV rules for homogeneity and consistency. The ICTV regularly publishes reports describing all existing virus taxa with a list of classified viruses as well as descriptions of virus families and genera. An Internet web site, where the most important information relative to virus taxonomy is made available, is updated regularly. The sixth report was published by Murphy et al (1995) and the seventh by van Regenmortel et al (1999).
The increasing number of virus species and virus strains being identified, together with the explosion of data on many descriptive aspects of viruses and viral diseases, and particularly sequence data, has led the ICTV to launch an international virus database project. This project, termed ICTVdB, is scheduled to be fully operational and accessible to the scientific community around the year 2000. The ICTVdB, in addition to the taxonomic descriptions of all the taxa, will comprise all the information available about each virus species, and later each virus strain, for all the descriptors necessary to identify and recognize all viruses.
A Universal System for Virus Classification
There are currently two systems in use for classifying organisms: the linnean and the adansonian systems. The former is the monothetic hierarchical classification applied by Linnaeus to plants and animals, while the adansonian is a polythetic hierarchical system initially proposed by Adanson in 1763. In 1984 Maurin and collaborators suggested applying the linnean classification system to the viruses. Although convenient to use, this system has shortcomings when applied to the classification of viruses. Firstly, it is difficult to appreciate the validity of a particular criteria. For example, it may not be appropriate to use the number of genomic components as a hierarchical criteria. Secondly, there are no obvious reasons for prioritizing criteria, and in consequence it is difficult to rank all the available criteria. For instance, is the nature of the genome (DNA/RNA) more important than the sense of the coding sequence of the genome or the shape of the virus particles?
The adansonian system considers all available criteria at once and makes several classifications, taking the criteria into consideration successively. The criteria leading to the same classifications are considered as correlated and are therefore not discriminatory. Subsequently, a subset of criteria are considered, and the process is repeated until all criteria can be ranked to provide the best discrimination of the species. This system has not been used frequently in the past owing to its labor-intensive nature, but this situation has changed as a result of the power and availability of today's computer technology. Furthermore, qualitative and quantitative data can be simultaneously considered when generating such a classification. In the case of viruses, it was determined by Harrison and collaborators in 1971 that at least 60 characters could be used for a complete virus description (Table 1 ). Thus, the limiting factor for applying the adansonian system is now not its labor-intensive nature but the lack of data for many of the viruses.
Table 1.
Virus family descriptors used in virus taxonomy
| I Virion properties |
| A Morphology properties of virions |
| 1 Size |
| 2 Shape |
| 3 Presence or absence of an envelope and peplomers |
| 4 Capsomeric symmetry and structure |
| B Physical properties of virions |
| 1 Molecular mass |
| 2 Buoyant density |
| 3 Sedimentation coefficient |
| 4 pH stability |
| 5 Thermal stability |
| 6 Cation (Mg2+, Mn2+) stability |
| 7 Solvent stability |
| 8 Detergent stability |
| 9 Radiation stability |
| C Properties of genome |
| 1 Type of nucleic acid DNA or RNA |
| 2 Strandedness: single-stranded or double-stranded |
| 3 Linear or circular |
| 4 Sense: positive, negative or ambisense |
| 5 Number of segments |
| 6 Size of genome or genome segments |
| 7 Presence or absence and type of 5′ terminal cap |
| 8 Presence or absence of 5′ terminal covalently linked polypeptide |
| 9 Presence or absence of 3′ terminal poly (A) tract (or other specific tract) |
| 10 Nucleotide sequence comparisons |
| D Properties of proteins |
| 1 Number |
| 2 Size |
| 3 Functional activities (especially virion transcriptase, virion reverse transcriptase, virion hemagglutinin, virion neuraminidase, virion fusion protein) |
| 4 Amino acid sequence comparisons |
| E Lipids |
| 1 Presence or absence |
| 2 Nature |
| F Carbohydrates |
| 1 Presence or absence |
| 2 Nature |
| II Genome organization and replication |
| 1 Genome organization |
| 2 Strategy of replication of nucleic acid |
| 3 Characteristics of transcription |
| 4 Characteristics of translation and post-translational processing |
| 5 Site of accumulation of virion proteins, site of assembly, site of maturation and release |
| 6 Cytopathology, inclusion body formation |
| III Antigenic properties |
| 1 Serological relationships |
| 2 Mapping epitopes |
| IV Biological properties |
| 1 Host range, natural and experimental |
| 2 Pathogenicity, association with disease |
| 3 Tissue tropisms, pathology, histopathology |
| 4 Mode of transmission in nature |
| 5 Vector relationships |
| 6 Geographic distribution |
Adapted from ICTV guidelines for family descriptions.
In addition, the increasing number of viral nucleic acid sequences being reported, in combination with the appropriate computer software, allows the comparison of viruses to generate different phylogenetic trees, according to the gene or set of genes used, as for example proposed by Koonin in 1991, Dolja and Koonin in 1991 and Dolja et al in 1991. However, to date, none of them has satisfactorily provided a clear classification of all viruses. A multidimensional classification, taking into account all the criteria necessary to describe viruses, would probably be the most appropriate way of representing a virus classification, but again the shortcomings of data for some viruses would prevent the use of this system in the foreseeable future.
For almost 25 years, the ICTV has been classifying viruses essentially at the family and genus levels using a nonsystematic polythetic approach. Viruses were clustered first in genera and then in families. A subset of characters, including physicochemical, structural, genomic and biological criteria, is then used to compare and group viruses. This subset of characters may change from one family to another, according to the availability of the data and the importance of a particular character for a particular family. It is obvious that there is no homogeneity in this respect throughout the virus classification and that virologists weigh the criteria differently in this subjective process, leading to the generation of a nonhomogeneous classification. Nevertheless, over time we can see stability of the current ICTV classification at the genus and family level. When sequence, genomic organization and replicative cycle data are subsequently used for taxonomic purposes, they usually confirm the actual classification. It is also obvious that hierarchical classifications above the family level will encounter conflicts between phenotypic and genotypic criteria and that virologists will have to consider the entire classification process in order to progress in this direction.
Currently, and for practical reasons only, virus classification is structured according to the presentation indicated in Table 2, Table 3 . This ‘Order of Presentation of the Viruses’ does not reflect any hierarchical or phylogenetic classification but only a convenient order of presentation of the virus taxa. Since a taxonomic structure above the level of family (with the exception of the orders Mononegavirales, Caudovirales and Nidovirales) has not been developed extensively, any listing must be arbitrary. The order of presentation of virus families and genera follows four criteria: (1) the nature of the viral nucleic acid; (2) the strandedness of the nucleic acid; (3) the use of a reverse transcription process (DNA or RNA); and (4) the positive or negative sense of gene coding on the encapsidated genome. These four criteria give rise to six clusters comprising the 86 families and floating genera of viruses. In the past, two other criteria were also taken in account: the presence or absence of a lipid envelope and the segmentation of the genome as mono-, bi-, tri-, tetra- or multipartite. However, it has become clear that the presence of an envelope was entirely related to the nature of the host and that families could comprise genera having viruses with segmented or nonsegmented genomes, but sharing all other properties, including genome organization and sequence homology. These criteria have been therefore abandoned.
Table 2.
Order of presentation of the viruses
| Order | Family | Subfamily | Genus | Type species | Host |
|---|---|---|---|---|---|
| The DNA viruses | |||||
| The dsDNA viruses | |||||
| Caudovirales | Myoviridae | ||||
| “T4-like viruses”a | Enterobacteria phage T4 | Bacteria | |||
| “P1-like viruses” | Enterobacteria phage P1 | Bacteria | |||
| “P2-like viruses” | Enterobacteria phage P2 | Bacteria | |||
| “Mu-like viruses” | Enterobacteria phage Mu | Bacteria | |||
| “SP01-like viruses” | Bacillus phage SP01 | Bacteria | |||
| “ϕH-like viruses” | Halobacterium virus ϕH | Archaea | |||
| Siphoviridae | |||||
| “λ-like viruses” | Enterobacteria phage λ | Bacteria | |||
| “T1-like viruses” | Enterobacteria phage T1 | Bacteria | |||
| “T5-like viruses” | Enterobacteria phage T5 | Bacteria | |||
| “L5-like viruses” | Mycobacterium phage L5 | Bacteria | |||
| “c2-like viruses” | Lactococcus phage c2 | Bacteria | |||
| “ψM1-like viruses” | Methanobacterium virus ψM1 | Archaea | |||
| Podoviridae | |||||
| “T7-like viruses” | Enterobacteria phage T7 | Bacteria | |||
| “P22-like viruses” | Enterobacteria phage P22 | Bacteria | |||
| “ϕ29-like viruses” | Bacillus phage ϕ29 | Bacteria | |||
| Tectiviridae | Tectivirus | Enterobacteria phage PRD1 | Bacteria | ||
| Corticoviridae | Corticovirus | Alteromonas phage PM2 | Bacteria | ||
| Plasmaviridae | Plasmavirus | Acholeplasma phage L2 | Mycoplasma | ||
| Lipothrixviridae | Lipothrixvirus | Thermoproteus virus 1 | Archaea | ||
| Rudiviridae | Rudivirus | Sulfolobus virus SIRV1 | Archaea | ||
| Fuselloviridae | Fusellovirus | Sulfolobus virus SSV1 | Archaea | ||
| “SNDV-like viruses” | Sulfolobus virus SNDV | Archaea | |||
| Poxviridae | |||||
| Chordopoxvirinae | |||||
| Orthopoxvirus | Vaccinia virus | Vertebrates | |||
| Parapoxvirus | Orf virus | Vertebrates | |||
| Avipoxvirus | Fowlpox virus | Vertebrates | |||
| Capripoxvirus | Sheeppox virus | Vertebrates | |||
| Leporipoxvirus | Myxoma virus | Vertebrates | |||
| Suipoxvirus | Swinepox virus | Vertebrates | |||
| Molluscipoxvirus | Molluscum contagiosum virus | Vertebrates | |||
| Yatapoxvirus | Yaba monkey tumor virus | Vertebrates | |||
| Entomopoxvirinae | |||||
| Entomopoxvirus A | Melolontha melolontha entomopoxvirus | Invertebrates | |||
| Entomopoxvirus B | Amsacta moorei entomopoxvirus | Invertebrates | |||
| Entomopoxvirus C | Chironomus luridus entomopoxvirus | Invertebrates | |||
| Asfarviridae | Asfivirus | African swine fever virus | Vertebratesb | ||
| Iridoviridae | |||||
| Iridovirus | Chilo iridescent virus | Invertebrates | |||
| Chloriridovirus | Mosquito iridescent virus | Invertebrates | |||
| Ranavirus | Frog virus 3 | Vertebrates | |||
| Lymphocystivirus | Flounder virus | Vertebrates | |||
| Phycodnaviridae | |||||
| Chlorovirus | Paramecium bursaria Chlorella virus 1 | Algae | |||
| Prasinovirus | Micromonas pusilla virus SP1 | Algae | |||
| Prymnesiovirus | Chrysochromulina brevifilum virus | Algae | |||
| Phaeovirus | Ectocarpus siliculosus virus 1 | Algae | |||
| Baculoviridae | |||||
| Nucleopolyhedrovirus | Autographa californica nucleopolyhedrovirus | Invertebrates | |||
| Granulovirus | Cydia pomonella granulovirus | Invertebrates | |||
| Herpesviridae | |||||
| Alphaherpesvirinae | |||||
| Simplexvirus | Human herpesvirus 1 | Vertebrates | |||
| Varicellovirus | Human herpesvirus 3 | Vertebrates | |||
| “Marek's disease-like viruses” | Marek's disease virus | Vertebrates | |||
| “ILTV-like viruses” | Infectious laryngotracheitis virus | Vertebrates | |||
| Betaherpesvirinae | |||||
| Cytomegalovirus | Human herpesvirus 5 | Vertebrates | |||
| Muromegalovirus | Mouse cytomegalovirus 1 | Vertebrates | |||
| Roseolovirus | Human herpesvirus 6 | Vertebrates | |||
| Gammaherpesvirinae | |||||
| Lymphocryptovirus | Human herpesvirus 4 | Vertebrates | |||
| Rhadinovirus | Ateline herpesvirus 2 | Vertebrates | |||
| “Ictalurid herpes-like viruses” | Ictalurid herpesvirus 1 | Vertebrates | |||
| Adenoviridae | |||||
| Mastadenovirus | Human adenovirus 2 | Vertebrates | |||
| Aviadenovirus | Fowl adenovirus 1 | Vertebrates | |||
| Rhizidiovirus | Rhizidiomyces virus | Fungi | |||
| Polyomaviridae | |||||
| Polyomavirus | Murine polyomavirus | Vertebrates | |||
| Papillomaviridae | |||||
| Papillomavirus | Cottontail rabbit papillomavirus | Vertebrates | |||
| Polydnaviridae | |||||
| Ichnovirus | Campoletis sonorensis virus | Invertebrates | |||
| Bracovirus | Cotesia melanoscela virus | Invertebrates | |||
| Ascoviridae | Ascovirus | Spodoptera frugiperda ascovirus | Invertebrates | ||
| The ssDNA viruses | |||||
| Inoviridae | |||||
| Inovirus | Coliphage fd | Bacteria | |||
| Plectrovirus | Acholeplasma phage L51 | Mycoplasma | |||
| Microviridae | |||||
| Microvirus | Coliphage ϕX174 | Bacteria | |||
| Spiromicrovirus | Spiroplasma phage 4 | Spiroplasma | |||
| Bdellomicrovirus | Bdellovibrio phage MAC1 | Bacteria | |||
| Chlamydiamicrovirus | Chlamydia phage 1 | Bacteria | |||
| Geminiviridae | |||||
| Mastrevirus | Maize streak virus | Plants | |||
| Curtovirus | Beet curly top virus | Plants | |||
| Begomovirus | Bean golden mosaic virus | Plants | |||
| Circoviridae | Circovirus | Chicken anemia virus | Vertebrates | ||
| Nanovirus | Subterranean clover stunt virus | Plants | |||
| Parvoviridae | |||||
| Parvovirinae | |||||
| Parvovirus | Mice minute virus | Vertebrates | |||
| Erythrovirus | B19 virus | Vertebrates | |||
| Dependovirus | Adeno-associated virus 2 | Vertebrates | |||
| Densovirinae | |||||
| Densovirus | Junonia coenia densovirus | Invertebrates | |||
| Iteravirus | Bombyx mori densovirus | Invertebrates | |||
| Brevidensovirus | Aedes aegypti densovirus | Invertebrates | |||
| The DNA and RNA reverse transcribing viruses | |||||
| Hepadnaviridae | |||||
| Orthohepadnavirus | Hepatitis B virus | Vertebrates | |||
| Avihepadnavirus | Duck hepatitis B virus | Vertebrates | |||
| Caulimoviridae | |||||
| Caulimovirus | Cauliflower mosaic virus | Plants | |||
| “PVCV-like viruses” | Petunia vein-clearing virus | Plants | |||
| “SbCMV-like viruses” | Soybean chlorotic mottle virus | Plants | |||
| “CsVMV-like viruses” | Cassava vein mosaic virus | Plants | |||
| Badnavirus | Commelina yellow mottle virus | Plants | |||
| “RTBV-like viruses” | Rice tungro bacilliform virus | Plants | |||
| Pseudoviridae | |||||
| Pseudovirus | Saccharomyces cerevisiae Ty1 virus | Yeast, Plants | |||
| Hemivirus | Drosophila melanogaster copia virus | Yeast, Invertebrates | |||
| Metaviridae | |||||
| Metavirus | Saccharomyces cerevisiae Ty3 virus | Yeast, Plants, Invertebrates | |||
| Errantivirus | Drosophila melanogaster gypsy virus | Invertebrates | |||
| Retroviridae | |||||
| Alpharetrovirus | Avian leukosis virus | Vertebrates | |||
| Betaretrovirus | Mason-Pfizer monkey virus | Vertebrates | |||
| Gammaretrovirus | Mouse mammary tumor virus | Vertebrates | |||
| Deltaretrovirus | Bovine leukemia virus | Vertebrates | |||
| Epsilonretrovirus | Walleye dermal sarcoma virus | Vertebrates | |||
| Lentivirus | Human immunodeficiency virus 1 | Vertebrates | |||
| Spumavirus | Human spumavirus | Vertebrates | |||
| The RNA viruses | |||||
| The dsRNA viruses | |||||
| Cystoviridae | Cystovirus | Pseudomonas phage ϕ6 | Bacteria | ||
| Reoviridae | |||||
| Orthoreovirus | Reovirus 3 | Vertebrates | |||
| Orbivirus | Bluetongue virus 1 | Vertebrates | |||
| Rotavirus | Simian rotavirus SA11 | Vertebrates | |||
| Coltivirus | Colorado tick fever virus | Vertebrates | |||
| Aquareovirus | Golden shiner virus | Vertebrates | |||
| Cypovirus | Bombyx mori cypovirus 1 | Invertebrates | |||
| Fijivirus | Fiji disease virus | Plants | |||
| Phytoreovirus | Wound tumor virus | Plants | |||
| Oryzavirus | Rice ragged stunt virus | Plants | |||
| Birnaviridae | |||||
| Aquabirnavirus | Infectious pancreatic necrosis virus | Vertebrates | |||
| Avibirnavirus | Infectious bursal disease virus | Vertebrates | |||
| Entomobirnavirus | Drosophila X virus | Invertebrates | |||
| Totiviridae | |||||
| Totivirus | Saccharomyces cerevisiae virus L-A | Fungi | |||
| Giardiavirus | Giardia lamblia virus | Protozoa | |||
| Leishmaniavirus | Leishmania RNA virus 1-1 | Protozoa | |||
| Partitiviridae | |||||
| Partitivirus | Gaeumannomyces graminis virus 019/6-A | Fungi | |||
| Chrysovirus | Penicillium chrysogenum virus | Fungi | |||
| Alphacryptovirus | White clover cryptic virus 1 | Plants | |||
| Betacryptovirus | White clover cryptic virus 2 | Plants | |||
| Hypoviridae | Hypovirus | Cryphonectria hypovirus 1-EP713 | Fungi | ||
| Varicosavirus | Lettuce big-vein virus | Plants | |||
| The negative-stranded ssRNA viruses | |||||
| Mononegavirales | |||||
| Bornaviridae | |||||
| Bornavirus | Borna disease virus | Vertebrates | |||
| Filoviridae | |||||
| “Ebola-like viruses” Zaïre | Ebola virus | Vertebrates | |||
| “Marburg-like viruses” | Marburg virus | Vertebrates | |||
| Paramyxoviridae | |||||
| Paramyxovirinae | |||||
| Respirovirus | Human parainfluenza virus 1 | Vertebrates | |||
| Morbillivirus | Measles virus | Vertebrates | |||
| Rubulavirus | Mumps virus | Vertebrates | |||
| Pneumovirinae | |||||
| Pneumovirus | Human respiratory syncytial virus | Vertebrates | |||
| Metapneumovirus | Turkey rhinotracheitis virus | Vertebrates | |||
| Rhabdoviridae | |||||
| Vesiculovirus | Vesicular stomatitis Indiana virus | Vertebrates | |||
| Lyssavirus | Rabies virus | Vertebrates | |||
| Ephemerovirus | Bovine ephemeral fever virus | Vertebrates | |||
| Novirhabdovirus | Infectious hematopoietic necrosis virus | Vertebrates | |||
| Cytorhabdovirus | Lettuce necrotic yellows virus | Plants | |||
| Nucleorhabdovirus | Potato yellow dwarf virus | Plants | |||
| Orthomyxoviridae | |||||
| Influenzavirus A | Influenza A virus | Vertebrates | |||
| Influenzavirus B | Influenza B virus | Vertebrates | |||
| Influenzavirus C | Influenza C virus | Vertebrates | |||
| Thogotovirus | Thogoto virus | Vertebrates | |||
| Bunyaviridae | |||||
| Bunyavirus | Bunyamwera virus | Vertebrates | |||
| Hantavirus | Hantaan virus | Vertebrates | |||
| Nairovirus | Nairobi sheep disease virus | Vertebrates | |||
| Phlebovirus | Sandfly fever Sicilian virus | Vertebrates | |||
| Tospovirus | Tomato spotted wilt virus | Plants | |||
| Tenuivirus | Rice stripe virus | Plants | |||
| Ophiovirus | Citrus psorosis virus | Plants | |||
| Arenaviridae | Arenavirus | Lymphocytic choriomeningitis virus | Vertebrates | ||
| Deltavirus | Hepatitis delta virus | Vertebrates | |||
| The positive-stranded ssRNA viruses | |||||
| Leviviridae | |||||
| Levivirus | Enterobacteria phage MS2 | Bacteria | |||
| Allolevivirus | Enterobacteria phage Qβ | Bacteria | |||
| Narnaviridae | |||||
| Narnavirus | Saccharomyces cerevisiae 20S narnavirus | Yeast | |||
| Mitovirus | Cryphonectria parasitica NB631 virus | Yeast | |||
| Picornaviridae | |||||
| Enterovirus | Poliovirus 1 | Vertebrates | |||
| Rhinovirus | Human rhinovirus 1A | Vertebrates | |||
| Hepatovirus | Hepatitis A virus | Vertebrates | |||
| Cardiovirus | Encephalomyocarditis virus | Vertebrates | |||
| Aphthovirus | Foot-and-mouth disease virus O | Vertebrates | |||
| Parechovirus | Human echovirus 22 | Vertebrates | |||
| “Cricket paralysis-like viruses” | Cricket paralysis virus | Invertebrates | |||
| Sequiviridae | |||||
| Sequivirus | Parsnip yellow fleck virus | Plants | |||
| Waïkavirus | Rice tungro spherical virus | Plants | |||
| Comoviridae | |||||
| Comovirus | Cowpea mosaic virus | Plants | |||
| Fabavirus | Broad bean wilt virus 1 | Plants | |||
| Nepovirus | Tobacco ringspot virus | Plants | |||
| Potyviridae | |||||
| Potyvirus | Potato virus Y | Plants | |||
| Rymovirus | Ryegrass mosaic virus | Plants | |||
| Macluravirus | Maclura mosaic virus | Plants | |||
| Ipomovirus | Sweet potato mild mottle virus | Plants | |||
| Bymovirus | Barley yellow mosaic virus | Plants | |||
| Tritimovirus | Wheat streak mosaic virus | Plants | |||
| Caliciviridae | |||||
| Vesivirus | Swine vesicular exanthema virus | Vertebrates | |||
| Lagovirus | Rabbit hemorrhagic disease virus | Vertebrates | |||
| “Norwalk-like viruses” | Norwalk virus | Vertebrates | |||
| “Sapporo-like viruses” | Sapporo virus | Vertebrates | |||
| “Hepatitis E-like viruses” | Hepatitis E virus | Vertebrates | |||
| Astroviridae | Astrovirus | Human astrovirus 1 | Vertebrates | ||
| Nodaviridae | |||||
| Alphanodavirus | Nodamura virus | Invertebrates | |||
| Betanodavirus | Striped jack nervous necrosis virus | Vertebrates | |||
| Tetraviridae | |||||
| Betatetravirus | Nudaurelia capensis β virus | Invertebrates | |||
| Omegatetravirus | Nudaurelia capensis ω virus | Invertebrates | |||
| Sobemovirus | Southern bean mosaic virus | Plants | |||
| Marafivirus | Maize rayado fino virus | Plants | |||
| Luteoviridae | |||||
| Luteovirus | Barley yellow dwarf virus – MAV | Plants | |||
| Polerovirus | Potato leafroll virus | Plants | |||
| Enamovirus | Pea enation mosaic virus 1 | Plants | |||
| Umbravirus | Carrot mottle virus | Plants | |||
| Tombusviridae | |||||
| Avenavirus | Oat chlorotic stunt virus | Plants | |||
| Aureusvirus | Pothos latent virus | Plants | |||
| Carmovirus | Carnation mottle virus | Plants | |||
| Dianthovirus | Carnation ringspot virus | Plants | |||
| Machlomovirus | Maize chlorotic mottle virus | Plants | |||
| Necrovirus | Tobacco necrosis virus | Plants | |||
| Panicovirus | Panicum mosaic virus | Plants | |||
| Tombusvirus | Tomato bushy stunt virus | Plants | |||
| Nidovirales | |||||
| Coronaviridae | |||||
| Coronavirus | Avian infectious bronchitis virus | Vertebrates | |||
| Torovirus | Berne virus | Vertebrates | |||
| Arteriviridae | |||||
| Arterivirus | Equine arteritis virus | Vertebrates | |||
| Flaviviridae | |||||
| Flavivirus | Yellow fever virus | Vertebrates | |||
| Pestivirus | Bovine diarrhea virus | Vertebrates | |||
| Hepacivirus | Hepatitis C virus | Vertebrates | |||
| Togaviridae | |||||
| Alphavirus | Sindbis virus | Vertebrates | |||
| Rubivirus | Rubella virus | Vertebrates | |||
| Tobamovirus | Tobacco mosaic virus | Plants | |||
| Tobravirus | Tobacco rattle virus | Plants | |||
| Hordeivirus | Barley stripe mosaic virus | Plants | |||
| Furovirus | Soil-borne wheat mosaic virus | Plants | |||
| Pomovirus | Potato mop-top virus | Plants | |||
| Pecluvirus | Peanut clump virus | Plants | |||
| Benyvirus | Beet necrotic yellow vein virus | Plants | |||
| Bromoviridae | |||||
| Alfamovirus | Alfalfa mosaic virus | Plants | |||
| Bromovirus | Brome mosaic virus | Plants | |||
| Cucumovirus | Cucumber mosaic virus | Plants | |||
| Ilarvirus | Tobacco streak virus | Plants | |||
| Oleavirus | Olive latent virus 2 | Plants | |||
| Ourmiavirus | Ourmia melon virus | Plants | |||
| Idaeovirus | Rasberry bushy dwarf virus | Plants | |||
| Closteroviridae | |||||
| Closterovirus | Beet yellows virus | Plants | |||
| Crinivirus | Lettuce infectious yellows virus | Plants | |||
| Capillovirus | Apple stem grooving virus | Plants | |||
| Trichovirus | Apple chlorotic leaf spot virus | Plants | |||
| Vitivirus | Grapevine virus A | Plants | |||
| Tymovirus | Turnip yellow mosaic virus | Plants | |||
| Carlavirus | Carnation latent virus | Plants | |||
| Potexvirus | Potato virus X | Plants | |||
| Allexivirus | Shallot virus X | Plants | |||
| Foveavirus | Apple stem pitting virus | Plants | |||
| Barnaviridae | Barnavirus | Mushroom bacilliform virus | Fungi | ||
| Unassigned viruses | |||||
| The subviral agents: viroids, satellites and agents of spongiform encephalopathies (prions) | |||||
| Subviral agent | Family | Genus | Type species | Host | |
| Viroids | |||||
| Pospiviroidae | |||||
| Pospiviroid | Potato spindle tuber viroid | Plants | |||
| Hostuviroid | Hop stunt viroid | Plants | |||
| Cocadviroid | Coconut cadang-cadang viroid | Plants | |||
| Apscaviroid | Apple scar skin viroid | Plants | |||
| Coleviroid | Coleus blumei viroid 1 | Plants | |||
| Avsunviroidae | |||||
| Avsunviroid | Avocado sunblotch viroid | Plants | |||
| Pelamoviroid | Peach latent mosaic virus | Plants | |||
| Satellites | Plants | ||||
| Invertebrates | |||||
| Fungi | |||||
| Prions | Vertebrates | ||||
| Fungi | |||||
Quotes are used to denote taxon names that are not approved ICTV international names, and are thus temporary until formal names are approved.
Vertebrate arthropod-borne viruses are listed according to their vertebrate hosts.
Table 3.
Orders, families and floating genera of viruses according to the seventh ICTV report (1999)
| Criteria | Order | Family | Floating genus | Morphology | Genome configuration | Genome size (kb) | Virus host |
Number of species |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Strains/serotypes | Tentative | Total | ||||||||
| dsDNA | Caudovirales | Myoviridae | Tailed phage | 1 linear | 336 | Bacteria, archaea | 15 | 23 | 117 | 155 | |
| Siphoviridae | Tailed phage | 1 linear | 53 | Bacteria, archaea | 7 | 0 | 137 | 144 | |||
| Podoviridae | Tailed phage | 1 linear | 40 | Bacteria, archaea | 8 | 12 | 66 | 86 | |||
| Tectiviridae | Isometric | 1 linear | 16 | Bacteria | 4 | 0 | 38 | 42 | |||
| Corticoviridae | Isometric | 1 circular supercoiled | 10 | Bacteria | 1 | 0 | 2 | 3 | |||
| Plasmaviridae | Pleomorphic | 1 circular | 12 | Mycoplasma | 1 | 0 | 7 | 8 | |||
| Lipothrixviridae | Rod | 1 linear | 16 | Archaea | 2 | 0 | 0 | 2 | |||
| Rudiviridae | Rod | 1 linear | 33–36 | Archaea | 2 | 0 | 1 | 3 | |||
| Fuselloviridae | Lemon-shape | 1 circular supercoiled | 15 | Archaea | 1 | 0 | 0 | 1 | |||
| “SNDV-like viruses” | Droplet-shape | 1 circular | 20 | Archaea | 1 | 0 | 0 | 1 | |||
| Poxviridae | Ovioid | 1 linear | 130–375 | Vertebrate, invertebrate | 62 | 8 | 23 | 93 | |||
| Asfarviridae | Isometric | 1 circular | 170–190 | Vertebrate | 1 | 0 | 0 | 1 | |||
| Iridoviridae | Isometric | 1 linear | 160–400 | Vertebrate, invertebrate | 17 | 4 | 3 | 24 | |||
| Phycodnaviridae | Isometric | 1 linear | 250–350 | Algae | 27 | 0 | 38 | 65 | |||
| Baculoviridae | Bacilliform | 1 circular supercoiled | 90–230 | Invertebrate | 17 | 6 | 7 | 30 | |||
| Herpesviridae | Isometric | 1 linear | 120–220 | Vertebrate | 56 | 0 | 65 | 121 | |||
| Adenoviridae | Isometric | 1 linear | 32–48 | Vertebrate | 26 | 102 | 35 | 163 | |||
| Rhizidiovirus | Isometric | 1 linear | 27 | Fungus | 1 | 0 | 0 | 1 | |||
| Polyomaviridae | Isometric | 1 circular | 5 | Vertebrate | 12 | 4 | 0 | 16 | |||
| Papillomaviridae | Isometric | 1 circular | 6.8–8.4 | Vertebrate | 7 | 0 | 88 | 95 | |||
| Polydnaviridae | Rod, fusiform | 1 circular supercoiled | 2–28 | Invertebrate | 59 | 0 | 0 | 59 | |||
| Ascoviridae | Ovoid and bacilliform | 1 circular | 100–180 | Invertebrate | 3 | 0 | 1 | 4 | |||
| 330 | 159 | 628 | 117 | ||||||||
| ssDNA | Inoviridae | Rod | 1 circular | 7–20 | Bacteria, mycoplasma | 36 | 7 | 5 | 48 | ||
| Microviridae | Isometric | 1 circular | 6 | Bacteria, spiroplasma | 7 | 0 | 33 | 40 | |||
| Geminivirus | Isometric | 1 or 2 circular | 3–6 | Plant | 94 | 2 | 10 | 106 | |||
| Circoviridae | Isometric | 1 circular | 1.7–2.3 | Vertebrate | 3 | 0 | 1 | 4 | |||
| Nanovirus | Isometric | 6–9 circular | 6–9 | Plant | 4 | 0 | 1 | 5 | |||
| Parvoviridae | Isometric | 1 − strand | 6–8 | Vertebrate, invertebrate | 38 | 0 | 16 | 54 | |||
| 182 | 9 | 66 | 257 | ||||||||
| ssDNA RT | Hepadnaviridae | Isometric | 1 circular – strand | 3 | Vertebrate | 5 | 0 | 2 | 7 | ||
| dsDNA RT | Caulimoviridae | Isometric, bacilliform | 1 circular | 8 | Plant | 26 | 0 | 8 | 34 | ||
| ssRNA RT | Pseudoviridae | Ovoid | 1 linear | 5–8 | Yeast, plant | 15 | 0 | 0 | 15 | ||
| ssRNA RT | Metaviridae | Isometric | 1 linear | 4–10 | Yeast, fungus, Invertebrate | 18 | 0 | 1 | 19 | ||
| ssRNA RT | Retroviridae | Spherical | dimer 1 + segment | 7–10 | Vertebrate | 59 | 44 | 2 | 105 | ||
| 123 | 44 | 13 | 180 | ||||||||
| dsRNA | Cystoviridae | Isometric | 3 segments | 17 | Bacteria | 1 | 0 | 0 | 1 | ||
| Reoviridae | Isometric | 10–12 segments | 19–62 | Vertebrate, invertebrate, plant | 62 | 256 | 39 | 357 | |||
| Birnaviridae | Isometric | 2 segments | 6 | Vertebrate, invertebrate | 4 | 21 | 1 | 26 | |||
| Totiviridae | Isometric | 1 segment | 5–7 | Fungus, protozoa | 18 | 0 | 5 | 23 | |||
| Partitiviridae | Isometric | 2 segments | 3–10 | Fungus, plant | 30 | 0 | 15 | 45 | |||
| Hypoviridae | Pleomorphic | 1 segment | 9–13 | Fungus | 3 | 0 | 2 | 5 | |||
| Varicosavirus | Rod | 2 segments | 14 | Plant | 1 | 0 | 3 | 4 | |||
| 119 | 277 | 65 | 461 | ||||||||
| Negative | Mononegavirales | Bornaviridae | Spherical | 1 − segment | 9 | Vertebrate | 1 | 0 | 1 | 2 | |
| ssRNA | |||||||||||
| Filoviridae | Bacilliform | 1 − segment | 13 | Vertebrate | 5 | 19 | 0 | 24 | |||
| Paramyxoviridae | Helical | 1 − segment | 15–16 | Vertebrate | 31 | 5 | 2 | 38 | |||
| Rhabdoviridae | Bacilliform | 1 − segment | 10–13 | Vertebrate, plant | 37 | 0 | 142 | 179 | |||
| Orthomyxoviridae | Helical | 8 − segments | 13–14 | Vertebrate | 5 | 1 | 0 | 6 | |||
| Bunyaviridae | Spherical | 3 − segments | 12–23 | Vertebrate, plant | 93 | 236 | 66 | 395 | |||
| Tenuivirus | Filaments | 4 −?segments | 15–19 | Plant | 6 | 0 | 5 | 11 | |||
| Ophiovirus | Filaments | 3 − segments | 12 | Plant | 3 | 0 | 0 | 3 | |||
| Arenaviridae | Spherical | 2 − segments | 11 | Vertebrate | 19 | 27 | 2 | 48 | |||
| Deltavirus | Spherical | 1 circular − strand | 1.7 | Vertebrate | 1 | 0 | 0 | 1 | |||
| 201 | 288 | 218 | 707 | ||||||||
| Positive ssRNA | Leviviridae | Isometric | 1 + segment | 3–4 | Bacteria | 4 | 18 | 35 | 57 | ||
| Narnaviridae | Ribonucleic complex | 1 + segment | 2.5 | Yeast | 3 | 0 | 0 | 3 | |||
| Picornaviridae | Isometric | 1 + segment | 7–8.5 | Vertebrate | 16 | 105 | 137 | 258 | |||
| “CrPV-like viruses” | Isometric | 1 + segment | 9–10 | Invertebrate | 5 | 0 | 0 | 5 | |||
| Sequiviridae | Isometric | 1 + segment | 9–12 | Plant | 5 | 0 | 0 | 5 | |||
| Comoviridae | Isometric | 2 + segments | 9–16 | Plant | 50 | 0 | 9 | 59 | |||
| Potyviridae | Rod | 1 or 2 + segments | 8–12 | Plant | 106 | 0 | 92 | 198 | |||
| Caliciviridae | Isometric | 1 + segment | 8 | Vertebrate | 6 | 40 | 8 | 54 | |||
| “HEV-like viruses” | Isometric | 1 + segment | 7 | Vertebrate | 1 | 0 | 0 | 1 | |||
| Astroviridae | Isometric | 1 + segment | 7–8 | Vertebrate | 6 | 13 | 0 | 19 | |||
| Nodaviridae | Isometric | 2 + segments | 5 | Vertebrate, invertebrate | 14 | 0 | 0 | 14 | |||
| Tetraviridae | Isometric | 1 + segment | 5 | Invertebrate | 9 | 0 | 0 | 9 | |||
| Sobemovirus | Isometric | 1 + segment | 4 | Plant | 11 | 0 | 3 | 14 | |||
| Marafivirus | Isometric | 1 + segment | 6–7 | Plant | 3 | 0 | 0 | 3 | |||
| Luteoviridae | Isometric | 1 or 2 + segment | 6–9 | Plant | 8 | 0 | 11 | 9 | |||
| Umbravirus | No particles | 1 + segment | 4 | Plant | 7 | 0 | 15 | 22 | |||
| Tombusviridae | Isometric | 1 or 2 + segment | 4–5.5 | Plant | 38 | 0 | 11 | 49 | |||
| Nidovirales | Coronaviridae | Pleomorphic | 1 + segment | 28–33 | Vertebrate | 16 | 5 | 1 | 22 | ||
| Arteriviridae | Spherical | 1 + segment | 13–16 | Vertebrate | 4 | 0 | 0 | 4 | |||
| Flaviviridae | Isometric | 1 + segment | 10–12 | Vertebrate | 57 | 47 | 6 | 110 | |||
| Togaviridae | Isometric | 1 + segment | 10–13 | Vertebrate | 23 | 6 | 0 | 29 | |||
| ssRNA | Tobamovirus | Rod | 1 + segment | 6 | Plant | 16 | 0 | 3 | 19 | ||
| Positive sense | Tobravirus | Rod | 2 + segments | 9–11 | Plant | 3 | 0 | 0 | 3 | ||
| Hordeivirus | Rod | 3 + segments | 10 | Plant | 4 | 0 | 0 | 4 | |||
| Furovirus | Rod | 2 + segments | 9–11 | Plant | 1 | 0 | 4 | 5 | |||
| Pomovirus | Rod | 3 + segments | 12 | Plant | 4 | 0 | 0 | 4 | |||
| Pecluvirus | Rod | 2 + segments | 10 | Plant | 2 | 0 | 0 | 2 | |||
| Benyvirus | Rod | 4 (or 5) + segments | 14–16 | Plant | 2 | 0 | 0 | 2 | |||
| Bromoviridae | Isometric, bacilliform | 3 + segments | 8–9 | Plant | 28 | 0 | 0 | 28 | |||
| Ourmiavirus | Bacilliform | 3 + segments | 4–5 | Plant | 3 | 0 | 0 | 3 | |||
| Idaeovirus | 3 + segments | 8 | Plant | 1 | 0 | 1 | |||||
| Closteroviridae | Rod | 1 or 2 + segments | 15–19 | Plant | 18 | 0 | 16 | 34 | |||
| Capillovirus | Rod | 1 + segment | 7 | Plant | 3 | 0 | 1 | 4 | |||
| Trichovirus | Rod | 1 + segment | 7.5 | Plant | 3 | 0 | 1 | 4 | |||
| Vitivirus | Rod | 1 + segment | 7.5 | Plant | 4 | 0 | 1 | 5 | |||
| Tymovirus | Isometric | 1 + segment | 6 | Plant | 20 | 0 | 1 | 21 | |||
| Carlavirus | Rod | 1 + segment | 7–8 | Plant | 31 | 0 | 29 | 60 | |||
| Potexvirus | Rod | 1 + segment | 6 | Plant | 26 | 0 | 18 | 44 | |||
| Allexivirus | Rod | 1 + segment | 9 | Plant | 6 | 0 | 3 | 9 | |||
| Foveavirus | Rod | 1 + segment | 8–9 | Plant | 2 | 0 | 1 | 3 | |||
| Barnaviridae | Bacilliform | 1 + segment | 4 | Fungus | 1 | 0 | 1 | 2 | |||
| 565 | 234 | 403 | 1202 | ||||||||
| Unassigned viruses | — | — | — | All | 30 | 0 | 0 | 30 | |||
| 1550 | 1011 | 1393 | 3954 | ||||||||
| Viroids | — | — | — | Plant | 27 | 0 | 8 | 35 | |||
| Satellites | — | — | — | Plant | 33 | 0 | 6 | 39 | |||
The Virus Species Concept and its Application
In 1991 the ICTV accepted the concept that viruses exist as species, in a similar manner to other organisms, and adopted a definition for a virus species proposed by van Regenmortel in 1990: ‘A virus species is a polythetic class of viruses that constitutes a replicating lineage and occupies a particular ecological niche.’ This simple definition and the position taken by the ICTV has already had, and will continue to have, a profound effect on virus classification. Effectively, in the sixth ICTV report virus names were indicated in ‘List of species’ but they were in fact a ‘List of virus names’ with undefined taxonomic status. In the seventh ICTV report, according to the polythetic nature of the species definition, a ‘List of species-demarcating criteria’ is provided for each genus, indicating how virus species can be identified in this particular genus. Viruses are then differentiated in species and tentative species according to this list of criteria and the availability of information to demarcate the species.
First, it is intended to define for each genus the criteria demarcating a virus species, and, second, to compare these criteria from one genus to the next, searching for homogeneity throughout the virus classification. Naturally this list of criteria should follow the polythetic nature of the species definition and more than one criteria should be used to determine a new species. It is obvious that most of the criteria in the list of demarcating criteria are shared amongst the different genera, within and across families; namely, host range, serological relationships, vector transmission type, tissue tropism, genome rearrangement and sequence homology (Table 4 ). However, if the types of criteria are similar, the levels of demarcation clearly differ from one family to another. This may reflect differences in appreciation from one family to another but also the differential ranking of a particular criterion in different families. The huge differences (up to 30%) in sequences among nucleoproteins of species of lentiviruses does not have the same biological significance as small differences in capsid protein sequences (1–10%) of species of potyviruses, and therefore universal levels of sequence identity for similar genes may not exist for viruses! The levels of demarcation may even change from one gene to another within the same family. Homogenization of the application of the species definition concept throughout the virus definition will be the next challenge of ICTV for the eighth report to be published by 2002. This, in turn, will contribute to homogeneity of the genus and family demarcation criteria (Table 4) and will permit creation of new families or merging of existing families. However, it is important to note that the nature of the demarcating criteria at the genus level will probably not change as these have passed the test of time. Despite the fact that they were mostly established using biochemical and structural criteria, they remained valid when correlated with genome organization and sequence data.
Table 4.
List of criteria demarcating different virus taxa
| I Order |
|---|
| Common properties between several families including: |
| Biochemical composition |
| Virus replication strategy |
| Particle structure (to some extent) |
| General genome organization |
| II Family |
| Common properties between several genera including: |
| Biochemical composition |
| Virus replication strategy |
| Nature of the particle structure |
| Genome organization |
| III Genus |
| Common properties within a genus including: |
| Virus replication strategy |
| Genome size, organization and/or number of segments |
| Sequence homologies (hybridization properties) |
| Vector transmission |
| IV Species |
| Common properties within a species including: |
| Genome rearrangement |
| Sequence homologies (hybridization properties) |
| Serological relationships |
| Vector transmission |
| Host range |
| Pathogenicity |
| Tissue tropism |
| Geographical distribution |
The Universal Virus Classification
The present universal system of virus taxonomy is set arbitrarily at hierarchical levels of order, family (in some cases subfamily), genus and species. Lower hierarchical levels, such as subspecies, strain, serotype, variant, pathotype and isolate are established by international specialty groups and/or by culture collections, but not by the ICTV. However some of them may be indicated in the ICTV report for information or because in the past these names were listed as ‘viruses’ in previous reports.
Virus species
The species taxon is always regarded as the most important taxonomic level in classification but it has proved to be the most difficult to apply to the viruses. ICTV definition of a virus species was long considered to be ‘a concept that will normally be represented by a cluster of strains from a variety of sources, or a population of strains from a particular source, which have in common a set or pattern of correlating stable properties that separates the cluster from other clusters of strains’ as stated by Matthews in 1982 and by Francki et al in 1991. This was a general definition, which was in fact not very useful for practically delineating species in a particular family. Furthermore, this definition directly addressed the definition of a virus strain, which had never been attempted in the history of virus taxonomy. In 1991, the ICTV Executive Committee accepted a definition proposed by van Regenmortel in 1990 (see above). This definition states: ‘A virus species is a polythetic class of viruses that constitutes a replicating lineage and occupies a particular ecological niche.’ The major advantage in this definition is that it can accommodate the inherent variability of viruses and is not dependent on the existence of a unique characteristic. Members of a polythetic class are defined by more than one property and no single property is absolutely essential and necessary. Thus in each family it might be possible to determine the set of properties of the class ‘species’ and to check if the family members are species of this family or if they belong to a lower taxonomic level. The ICTV is currently conducting this exercise throughout all virus families. This exercise should ultimately result in an excellent evaluation of a precise definition of each virus species in the entire classification.
Several practical matters are related to the definition of a virus species with the goal of improving the usefulness of virus classification. These include: (1) homogeneity of the different taxa; (2) diagnosis-related matters; (3) virus collections; (4) evolution studies; (5) biotechnology; (6) sequence database projects; (7) virus database projects; and now (8) intellectual property rights.
Virus families and genera
There is no formal definition for a genus, but it is commonly considered as: ‘a population of virus species that share common characteristics and are different from other populations of species’. Although this definition is somewhat elusive, this level of classification seems enduring and useful; some genera have been moved from one family to another over the years, but the composition and description of the genera has remained stable. The characters defining a genus differ from one family to another and there is a tendency to create genera with fewer differences between them. Upon examination, there is more and more evidence that the members of a genus have a common evolutionary origin. The use of subgenera has been abandoned in current virus classification.
Notwithstanding the creation of the ICTV, plant virologists continued to classify plant viruses in ‘groups’, refusing to place them in genera and families. However, owing to obvious similarities, plant reoviruses and rhabdoviruses had been integrated into the families Reoviridae and Rhabdoviridae (Table 2). This position was mostly due to plant virologists’ refusal to accept binomial nomenclature. Since this form of nomenclature was withdrawn from the ICTV classification rules in 1995, they subsequently accepted the placing of plant viruses into species, genera and families as shown in the sixth ICTV report. However, there are still 30 of so-called ‘floating genera’ that do not pertain to any family. This is mostly due to the fact that plant virologists prefer to accumulate data on virus species and genera before clustering appropriate genera in families. It is remarkable that this attitude has also been adopted by other virologists as a convenient way of classifying viruses, without having to move genera out of families when it becomes apparent that they are part of a distinct family. For example, the members of the floating genus ‘cricket paralysis-like viruses’ share enough properties with picornaviruses to be included in the family Picornaviridae; however, they also possess properties that would justify their classification in a separate family. Only new data or new viruses will permit a definitive position, therefore for the time being it remains a floating genus. Similarly the same strategy is used to create a floating genus ‘ictalurid herpes-like viruses’, within the family Herpesviridae, although in this case it is a floating genus within the family because of uncertainty as to whether the members of this genus should be classified in one of the existing subfamilies or to a new subfamily.
Virus orders
As mentioned above, the higher hierarchy levels for virus classification are extremely difficult to establish. Despite several propositions in the past, only three have been accepted: Caudovirales, Mononegavirales and Nidovirales. The first virus order, Mononegavirales, was established in 1990 and comprises the nonsegmented single-stranded RNA negative-sense viruses, namely the families Bornaviridae, Filoviridae, Paramyxoviridae and Rhabdoviridae. This decision was taken because of the great similarity of many criteria between these families, including their replication strategy. A second order, Caudovirales, contains all the families of double-stranded DNA phages possessing a tail, including the families Myoviridae, Podoviridae and Siphoviridae. A third order, Nidovirales, comprising the families Coronaviridae and Arteriviridae, was accepted in 1996 because of the impossibility of grouping together these two taxonomic entities, which share many properties and yet are so different, as a single family. Many members of the ICTV advocate the creation of many more orders, but it has been decided to proceed cautiously to avoid creation of short-lived orders. The creation of formal taxa higher than the orders, for example, kingdoms, classes and subclasses, has not been considered by the ICTV.
Virus Taxa Descriptions
Virus classification continues to evolve with the technologies available for describing viruses. The first wave of descriptions, those before 1940, mostly took into account the visual symptoms of the diseases caused by viruses, along with their modes of transmission. A second wave, between 1940 and 1970, brought together an enormous amount of information from studies of virion morphology (electron microscopy, structural data), biology (serology and virus properties) and physicochemical properties of viruses (nature and size of genome, number and size of viral proteins). Since 1970, the third wave of virus descriptions has included genome and replicative information as well as molecular relationships with virus hosts. There is a correlative modification of the list of virus descriptors and Table 1 lists the family and genera descriptors which are used in the current ICTV report. Figure 1, Figure 2, Figure 3, Figure 4, Figure 5 are diagrammatic representations of families and genera of viruses infecting vertebrates, invertebrates, plants, fungi, yeasts, protozoa and bacteria. The most recent wave of information used to classify viruses is naturally nucleotide and amino acid sequences. It is becoming more and more prevalent in virus taxonomy, as exemplified by the presence of a significant number of ‘phylogenetic trees’ in the seventh ICTV report, and by the huge number of scientific publications on this topic. Some scientists promote the concept of ‘quantitative taxonomy’ aimed at demonstrating that virus sequences contain all the coding information required for all the biological properties of the viruses. This is in complete agreement with the polythetic concept of the virus species definition, as demonstrated for example by Padidam et al in 1995, van Regenmortel et al in 1997, Hyppia et al in 1998, and Aleman et al in 1999.
Figure 1.
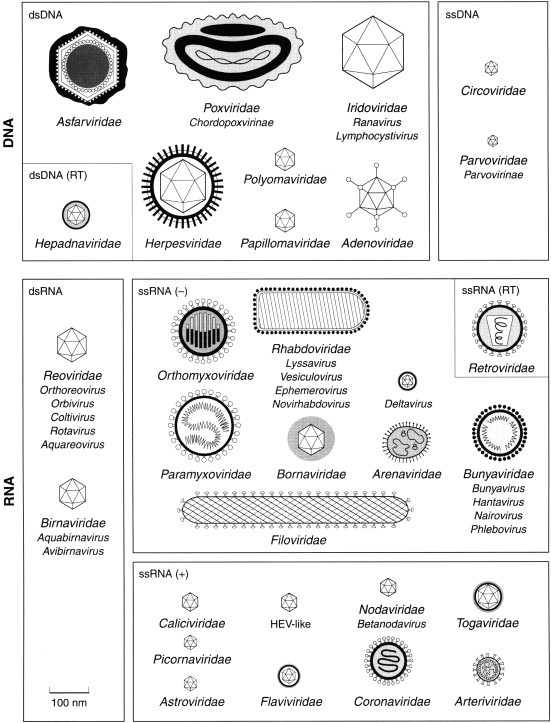
Families and genera of viruses infecting vertebrates.
Figure 2.
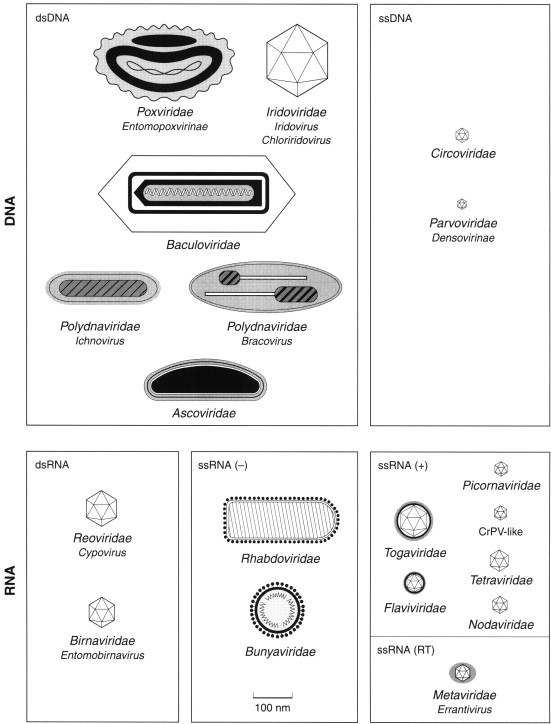
Families and genera of viruses infecting invertebrates.
Figure 3.
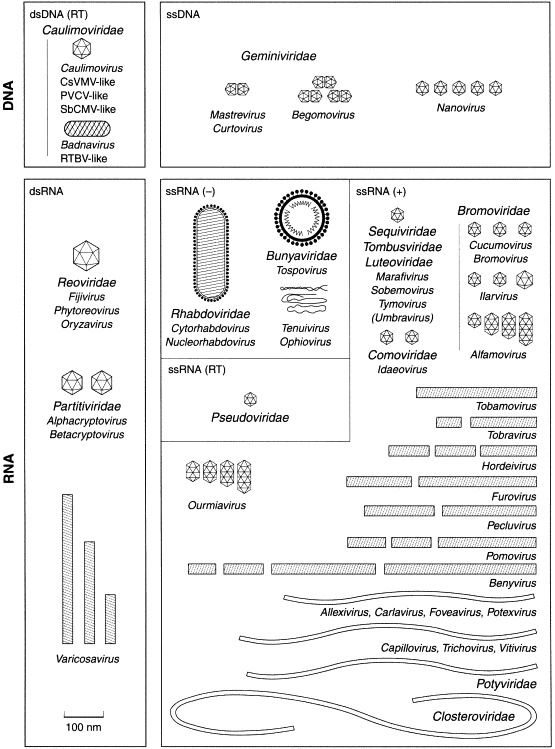
Families and genera of viruses infecting plants.
Figure 4.
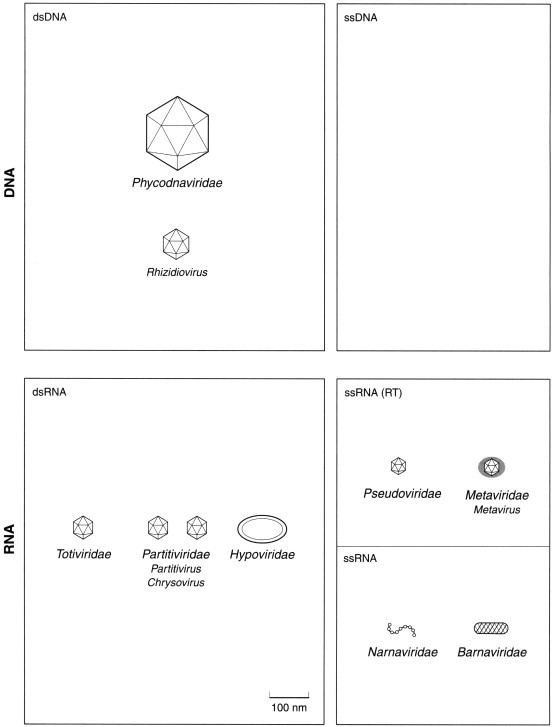
Families of viruses infecting algae, fungi, yeasts and protozoa.
Figure 5.
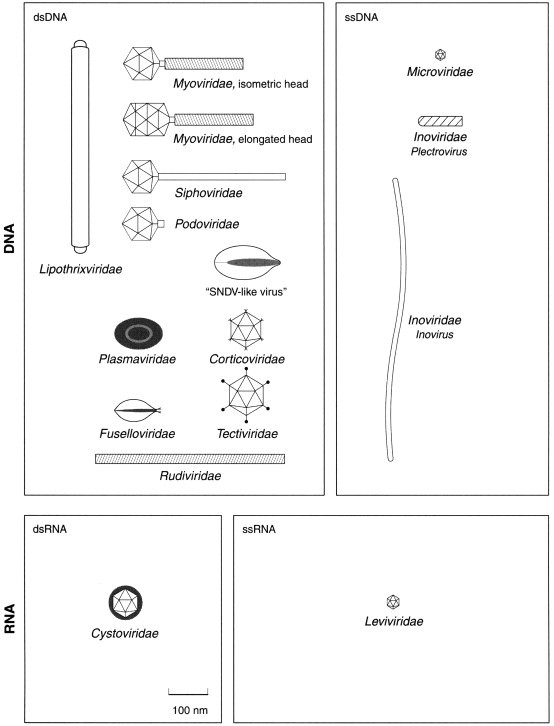
Families and genera of viruses infecting bacteria.
The impact of descriptions on virus classification has been particularly influenced by electron microscopy and of the negative staining technique for virions. This technique had an immediate influence on diagnostics and classification of viruses. With negative staining, viruses could be identified from poorly purified preparations of all tissue types, and information about size, shape, structure and symmetry could be quickly provided. As a result, virology progressed simultaneously for all viruses infecting animals, insects, plants and bacteria. Thin sections of infected tissues brought a new dimension to virus classification by providing information about virion morphogenesis and cytopathogenic effects. These techniques, in conjunction with the determination of the nature of the genome, provided a major source of information for the system of virus classification established in the 1980s, as shown by the large number of viruses listed in the fifth ICTV report in 1989.
In many instances the properties of viruses belonging to the same genus are correlated. Thus, the classification of a few of them will likely be sufficient to allow the classification of a new virus into an established genus. For example, a plant virus with filamentous particles of 700–850 nm and transmitted by aphids is likely to be a member of the genus Potyvirus. Establishment of new genera in the future will require more information. Most of the properties listed in Table 3 will have to be precisely analyzed to warrant the formation of a new genus.
Table 3 lists 45 different categories of properties but each category includes many items. Lists of virus descriptors usually comprise 1000–2000 descriptors. The establishment of a universal list of virus descriptors is under way and should be adopted by ICTV around 2000 with the establishment of the ICTVdB. It will contain a common set of descriptors for all viruses and subsets for specific viruses in relation to their specific hosts (human, animal, insect, plant and bacterial).
A Uniform Nomenclature of Viral Taxa
When a genus is approved by ICTV, a type species is designated. However, none of these type species have received a new international name and only English names are used. Latinized binomial names for virus names have been supported by animal and human virologists of ICTV for many years, but have never been implemented. This suggestion was in fact withdrawn from ICTV nomenclature rules in 1990 and consequently such names as Herpesvirus varicella or Polyomavirus hominis should not be used. For several years, plant virologists have adopted a different nomenclature, using the vernacular name of a virus but replacing the word ‘virus’ by the genus name; for example, Cucumber mosaic cucumovirus and Tobacco mosaic tobamovirus. Though this usage is favored by many scientists, and examples of such practice can be found for human, animal and insect viruses (e.g. Human rhinovirus, Canine calicivirus, Acheta densovirus…), it has not been universally adopted by the ICTV.
The ICTV has set rules for virus nomenclature and orthography of taxonomic names that are regularly revisited and improved. The last word of international virus species names is ‘virus’, the international genus names universally end in ‘…virus’, the international subfamily names end in ‘…virinae’, the international family names end with ‘…viridae’, and the international order names are ending in ‘…virales’. In formal taxonomic usage, the virus order, family, subfamily, genus and species names are all printed in italics (or underlined) and the first letter is capitalized. For all taxa except the species names, new names are created de novo following ICTV guidelines, but in the case of virus names English vernacular form is used. In formal usage, the name of the taxon precedes the name of the taxonomic unit; for example, ‘the family Picornaviridae’ or ‘the genus Rhinovirus’. In informal vernacular usage, order, family, subfamily, genus and species names are written in lower case Roman script; they are not capitalized nor italicized (or underlined). Additionally, in informal usage, the name of the taxon should not include the formal suffix, and it should follow the term for the taxonomic unit; for example, ‘the mononegavirales order’, ‘the adenovirus family’, ‘the avihepadnavirus genus’ or ‘the tobacco mosaic virus’ species. Virus names are often abbreviated for convenient reasons, but ICTV has not set up guidelines to generate such abbreviations. The ICTV reports list abbreviations most commonly used by specialists and the ICTV reports help virologists to identify duplicates of abbreviations in order to decrease the number of such duplicates. In 1988 plant virologists initiated the publication of such lists and have indicated guidelines for the creation of new virus names and new abbreviations. These guidelines were last published in 1991 by Fauquet and Martelli and will be updated again in 1999.
To avoid ambiguous virus identifications, it has been recommended to journal editors that published papers follow ICTV guidelines for proper virus identification and nomenclature, and that viruses should be cited with their full taxonomic terminology when they are first mentioned in an article. For example:
-
•
Order Caudovirales, family Podoviridae, genus ‘T7-like viruses’, species Enterobacteria phage T7.
-
•
Order Mononegavirales, family Paramyxoviridae, subfamily Paramyxovirinae, genus Rubulavirus, species Mumps virus.
-
•
Order Nidovirales, family Coronaviridae, genus Coronavirus, species Avian infectious bronchitis virus.
-
•
Family Iridoviridae, genus Iridovirus, species Chilo iridescent virus.
-
•
Family Picornaviridae, genus Enterovirus, species Poliovirus, serotype Human poliovirus 1.
-
•
Genus Tobamovirus, species Tobacco mosaic virus.
See also:
PHAGE TAXONOMY AND CLASSIFICATION; VIRUS STRUCTURE | Atomic Structure; VIRUS STRUCTURE | Principles of Virus Structure.
Further Reading
- Fauquet C.M., Martelli G.P. Up-dated ICTV list of names and abbreviations of viruses, viroids and satellites infecting plants. Arch. Virol. 1995;140:393. doi: 10.1007/BF01309874. [DOI] [PubMed] [Google Scholar]
- Francki R.I.B., Milne R.G., Hatta T. Atlas of Plant Viruses. CRC Press; Boca Raton: 1985. [Google Scholar]
- Francki R.I.B., Fauquet C.M., Knudson D.L., Brown F. Fifth Report of the International Committee on Taxonomy of Viruses. Springer; Vienna: 1991. Classification and Nomenclature of Viruses. [Google Scholar]
- Lwoff A., Horne R., Tournier P. A system of viruses. 1962. Cold Spring Harb. Symp. Quant. Biol. [DOI] [PubMed] [Google Scholar]
- Matthews R.E.F. The History of Viral Taxonomy. A Critical Appraisal of Viral Taxonomy. CRC Press; Boca Raton: 1983. pp. 1–35. [Google Scholar]
- Murphy F.A., Fauquet C.M., Bishop D.H.L., editors. Virus Taxonomy. Sixth Report of the International Committee on Taxonomy of Viruses. Springer; Vienna: 1995. [Google Scholar]
- van Regenmortel M.H.V., Bishop D.H.L., Fauquet C.M. Guidelines to the demarcation of virus species. Arch. Virol. 1997;142:1505. [PubMed] [Google Scholar]
- van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., editors. Virus Taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press; New York: 1999. [Google Scholar]


