I. GENERAL INTRODUCTION
Chemiluminescence, the emission of light from a chemical reaction, has been studied extensively for many decades. Chemiluminescent processes constitute a very special class of chemical reactions in which products (or intermediates) are produced in electronically excited states that are very short-lived and rapidly decay with concomitant emission of light. Similar chemiluminescent reactions, called bioluminescence, occur in nature in species as diverse as the firefly (Photinus pyralis), marine bacteria (Vibrio harveyi), and others. Most chemiluminescence reactions involve oxidations of a variety of organic compounds as well as naturally occurring materials, resulting in the generation of light-emitting excited states. This phenomenon was first described with synthetic organic compounds in 1877 (Radziszewski, 1877).
Chemiluminescent reactions do not produce very high intensity light signals because of many efficient quenching processes that compete with the radiative decay of the excited states. Nevertheless, chemiluminescence has been used effectively as a very sensitive detection system in many applications (Carter and Kricka, 1982; Harber, 1982; Kricka and Carter, 1982), largely because no background light signals are generated since the emitting excited state is created in a dark chemical reaction (compared to scattered excitation light in fluorescence.) Therefore, in theory, every photon detected is a true signal of the assay. This feature of chemiluminescent molecules—coupled with long shelf-life, elimination of hazards associated with the use of radioisotopes, and their detectability at 10−21 moles (detection of alkaline phosphatase with chemiluminescent dioxetane substrate; Kricka, 1992)—makes them ideal as a reporter system for immunoassays and DNA probe hybridization assays.
In this chapter we describe the use of various chemiluminescence methodologies for the detection of viruses in DNA hybridization assays. A short discussion of instrumentation used in chemiluminescence measurement is also included.
II. CHEMILUMINESCENCE METHODS
A. Dioxetanes
Dioxetanes are four-membered cyclic peroxides that have been implicated as short-lived unstable intermediates in oxidation reactions that result in chemiluminescence (McCapra, 1966). Thus, 1,2-dioxetanes differ from most other chemiluminescence systems because these compounds do not require oxidation to emit light. Recently developed 1,2-dioxetanes that can be activated to luminesce by enzymes have been used successfully for bioanalyte detection. Dephosphorylation of adamantyl- and derivatized adamantyl-l,2-dioxetane phosphate substrates, such as AMPPD® [disodium 3-(4-methoxyspiro{l,2-dioxetane-3,2′-tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate] and CSPD® [disodium 3-(4-methoxyspiro{l,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate] by alkaline phosphatase results in the formation of a destablized anion that fragments further to form an excited state of methyl meta-oxybenzoate anion that emits light at 477 nm (Fig. 1 ; Bronstein et al., 1989a, 1991; Bronstein and Dimond, 1990; Bronstein and Sparks, 1992).
Figure 1.
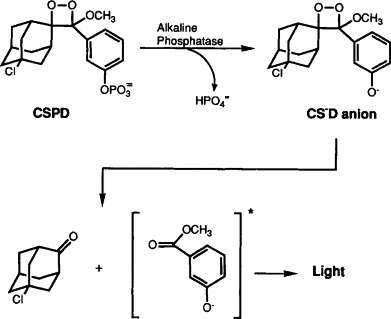
Chemiluminescent decomposition of CSPD® 1,2-dioxetane triggered by enzymatic dephosphorylation.
1,2-Dioxetane substrates for alkaline phosphatase are widely used in DNA hybridization assays (Bronstein, et al., 1990; Pollard-Knight et al., 1990b; Tumolo et al., 1992). DNA probes are labeled with alkaline phosphatase either indirectly, with a biotin or hapten label followed by binding streptavidin- or antibody-alkaline phosphatase conjugates, or directly by covalent bonding to enzyme (oligonucleotide probes). Biotin has been the most popular ligand for indirect labeling, but hapten labels other than biotin have also been employed including digoxigenin, fluorescein, and 2,4-dinitrophenyl. Dioxetane-based chemiluminescent indirect labeling and detection systems for DNA hybridization assays, as well as for immunoassays and DNA sequencing, are widely available from many commercial suppliers. With a nick-translated biotinylated DNA probe, as little as 380 fg (7.9 × 104 copies) of target pBR322 DNA can be detected on a Southern blot (Bronstein et al., 1990). Using digoxigenin-labeled random-primed DNA probes with a membrane-based assay and photographic film detection, a sensitivity level of 10–50 fg of target DNA was obtained for the detection of purified cytomegalovirus (CMV) or parvovirus B19 DNA (Musiani et al., 1991a).
Direct labeling of oligonucleotide probes with alkaline phosphatase (Jablonski et al., 1986) is possible with systems from Promega (Madison, WI) and Cambridge Research Biochemicals (Wilmington, DE). The detection of a single copy gene in 0.25 μg human genomic DNA with Southern blot analysis has been achieved with an alkaline phosphatase-labeled oligonucleotide probe (Cate et al., 1991).
Comparisons have shown that the sensitivity of alkaline phosphatase-dioxetane chemiluminescence detection is comparable to or better than that of 32P-based detection. In a human genomic Southern blot analysis of the tissue plasminogen activator gene, the sensitivity achieved with an alkaline phosphatase-labeled oligonucleotide probe was 12-fold higher than that achieved with the same 32P-5′-end-labeled probe, and the speed of detection was enhanced 40-fold with the alkaline phosphatase-labeled probe (Cate et al., 1991). Slot blot hybridization of human serum samples with the alkaline phosphatase-labeled AmpliProbe® system (ImClone Systems, New York) showed a higher sensitivity in the detection of hepatitis B virus (HBV) relative to 32P-labeled nick-translated probes (Yang et al., 1991). Similar sensitivities were obtained with an indirect digoxigenin-labeled probe and a random-primed 32P-labeled probe in a dot blot hybridization assay for amplified human immunodeficiency virus type 1 (HIV-1) DNA (Zachar et al., 1991).
The sensitivity of alkaline phosphatase-dioxetane chemiluminescence detection has been shown to be superior to other nonisotopic systems based on colorimetric detection in membrane-based hybridization assays (Bronstein and Kricka, 1989; Bronstein and Voyta, 1989; Bronstein et al., 1989c; Musiani et al., 1991a,1992). Finally, alkaline phosphatase-dioxetane detection has been demonstrated to be two to five times more sensitive than enhanced luminol chemiluminescent detection (described subsequently) in a solution hybridization assay system (Clyne et al., 1989; Urdea et al., 1990). Furthermore, the alkaline phosphatase-dioxetane detection system consists of fewer components, which are more stable than those required for an enhanced luminol chemiluminescent reaction (Beck and Köster, 1990). Although several other alkaline phosphatase-based chemiluminescent assays also exist, involving alternative substrates and coupled reactions, the most sensitive and widely used assays are those with 1,2-dioxetane substrates (for review of alternative systems, see Kricka, 1991).
B. Luminol
Luminol and other cyclic diacylhydrazide derivatives can be oxidized in the presence of peroxide and peroxidase to generate an unstable intermediate in the excited state that chemiluminesces. Luminols can be used as direct chemiluminescent labels or as the chemiluminescent detectors of a peroxidase enzyme label (Kricka, 1991). Activation of luminol chemiluminescence with horseradish peroxidase (HRP) using an enhanced luminol system (enhanced chemiluminescence, ECL) has been done in DNA hybridization assays (Matthews et al., 1985; Durrani et al., 1990; Durrant, 1992) and immunoassays (reviewed by Bronstein and Sparks, 1992; Whitehead et al., 1983; Thorpe et al., 1985; Kricka et al., 1987).
DNA probes can be labeled indirectly with HRP by binding streptavidin-HRP or anti-hapten-HRP conjugates or covalently by direct enzyme conjugation with oligonucleotides and longer double-stranded DNAs. The detection of single copy genes in 0.5 μg human genomic DNA has been reported with indirectly labeled probes (Simmonds et al., 1991). Direct HRP-labeled DNA probes have been used for both membrane-based DNA hybridization assays (Pollard-Knight et al., 1990a; Simmonds et al., 1991) and solution-phase hybridization assays (Urdea et al., 1990). Detection of a single-copy gene on a Southern blot of <2 μg human genomic DNA, with a sensitivity of <1 amol target DNA, has been demonstrated (Pollard-Knight et al., 1990a) using direct HRP-labeled probes 0.3–5.1 kb in length. Similar sensitivity (1 amol target DNA) was also reported by Durrant et al. (1990). Both indirect and direct HRP labeling systems for nucleic acids and detection systems for HRP-catalyzed chemiluminescent reactions (ECL gene detection system) are available from Amersham (Arlington Heights, IL).
C. Acridinium Esters
Acridinium esters (AE) are direct chemiluminescent labels for antibodies (Weeks et al., 1983) and DNA probes (Septak, 1989; Nelson and Kacian, 1990; Nelson et al., 1992), in contrast to dioxetane and luminol systems, in which an enzyme label catalyzes the chemiluminescent reaction. N-Methyl acridinium esters react with hydrogen peroxide under basic conditions to yield an excited state N-methylacridone which emits light at 430 nm (reviewed by Nelson and Kacian, 1990). Oligonucleotide DNA probes can be labeled convalently with AEs by reaction of modified N-hydroxysuccinimide-AE with a primary alkyl amine on a linker arm that was previously incorporated during oligonucleotide synthesis (Nelson and Kacian, 1990). Preparation of AE-labeled oligonucleotide probes has also been described by Septak (1989). The AE label does not affect probe hybridization characteristics; relatively large amounts of clinical specimen material may be used without interfering with hybridization and detection of AE-labeled probes.
Probe hybridization and detection reactions are performed in solution, using either separation or nonseparation formats. In a separation or heterogeneous assay, hybridized probe may be separated and detected by selective binding to microspheres, which can be separated from solution magnetically. In a nonseparation or homogeneous format, also termed a hybridization protection assay (H?α), the ester bond of the unhybridized probe can be hydrolyzed by differential chemical hydrolysis, thus rendering its AE label nonchemiluminescent, whereas the AE label of the hybridized probe is minimally affected (Nelson and Kacian, 1990). This type of assay is possible because hybridization provides an intercalation site for the AE label, thereby protecting the AE molecule residing in the hybridized region from hydrolysis (Arnold et al., 1989).
The sensitivity of this detection system is approximately 5 × 10−19 mol AE-labeled oligonucleotide, and the linear dynamic range is greater than four orders of magnitude (Nelson and Kacian, 1990). Similar sensitivities for the detection of an amplified gag sequence (4 HIV proviral copies per 150,000 cells) were achieved with colorimetric, chemiluminescence and 32P-labeling methods (Ou et al., 1990; Rapier et al., 1993). Schmidt (1991) was able to detect 0.05 fmol target HIV-1 DNA with AE-labeled gag probes and obtained greater sensitivity with chemiluminescence than with the same 32P-end-labeled probe in a dot blot hybridization assay.
D. Electrochemilutninescence
Electrochemiluminescence is a process in which the excited state products are generated via an electrochemical reaction (Faulkner and Glass, 1982). Electrochemiluminescence occurs when specific metal chelates such as ruthenium (II) tris(bipyridyl) [Ru(bpy)], utilized as labels, undergo a series of chemical reactions at an electrode surface. Electrochemiluminescent labels for DNA hybridization assays have been utilized in a highly sensitive, simple, and versatile assay system. Oligonucleotide probes, synthesized with a free 5′-amino group, are readily labeled with Ru(bpy)-NHS ester (Blackburn et al., 1991; Kenten et al., 1991). Alternatively, oligonucleotide probes may be labeled during synthesis by incorporating labeled phosphoramidites (Kenten et al., 1992; DiCesare et al., 1993).
Electrochemiluminescent labels are relatively small molecules (∼1000 dalton) that are extremely stable and may be coupled to nucleic acids, haptens, or proteins without affecting immunoreactivity or hybridization characteristics. The dynamic range for detection of these labels has been reported to be over six orders of magnitude (Blackburn et al., 1991). These advantages, compared with other nonisotopic detection methods, provide potential wide utility in automated nonradioactive clinical diagnostic assays, including both DNA hybridization and immunoassay formats. A disadvantage of electrochemiluminescence, however, is a need for specialized instrumentation that can induce generation of electrochemically-excited states coupled with sensitive light detection.
Blackburn et al. (1991) used electrochemiluminescence detection with a DNA probe assay to quantify polymerase chain reaction (PCR)-amplified HIV-1 gag sequences. Double-stranded biotinylated PCR product was captured on streptavidin-coated microparticles and treated with alkali. Ru(bpy)3 2+-labeled oligonucleotide probe was then hybridized to the particle-bound DNA, washed, and quantified. A linear response was generated over the range of 50 to 2000 gene copies, and the detection of less than 10 copies of the HIV-1 gag was attained. An automated system for electrochemiluminescence quantification of PCR products (QPCR System 5000; Perkin-Elmer Corporation, Norwalk, CT) has been developed (DiCesare et al., 1993) and is used for detection of viral disease. This system provides detection limits of 10–200 amol and a linear dynamic range greater than three orders of magnitude. The system has been used for the detection of HIV-1 over a range of 3 to 106 copies of target DNA (Wages et al., 1993).
Because of the electrogeneration of the emitting species, which requires contact of the metal chelate label with an electrode, it is difficult to envision that simple membrane-based blotting assays that can be imaged on film could be designed using electrochemiluminescence.
E. Bioluminescence
Bioluminescent reactions, a special class of chemiluminescent reactions that occur in nature and are catalyzed by a luciferase or photoproteins, offer an alternative method for luminescence detection of protein and DNA (Kricka, 1991). Two bioluminescent reaction systems have been used for DNA hybridization assays, both of which are coupled enzymatic reactions. One system, used for membrane-based DNA hybridization, couples the production of d-luciferin from d-luciferin-O-phosphate, catalyzed by alkaline phosphatase (as a direct or indirect label) and the oxidation of d-luciferin, catalyzed by firefly luciferase, with concomitant light emission (Hauber and Geiger, 1987,1988; Hauber et al., 1988,1989; Geiger, 1992). The other system, used with both membrane-based and solution hybridization assays, couples reactions catalyzed by glucose-6-phosphate dehydrogenase (G6PDH), NAD(P)H: FMN oxidoreductase, and marine bacterial luciferase to produce the light (Balaguer et al., 1989a,b,1991a,b; Nicolas et al., 1990,1992). Although bioluminescence-based DNA detection systems have not become as widely used as chemiluminescence systems for DNA hybridization assays, they do offer another alternative for sensitive nonradioactive biomolecule detection.
III. INSTRUMENTATION FOR CHEMILUMINESCENCE ASSAYS
A wide spectrum of instruments is currently available for recording and quantifying chemiluminescent signal intensities. These instruments, known as luminometers, use a light detector that consists of a photomultiplier tube in photon counting mode, positioned close to the light source (microtiter plate or tube) to maximize photon collection efficiency. Among commercially available luminometers, semi-automated tube instruments such as the AutoClinilumat LB952T (Berthold/EG&G, Wallac, Inc., Gaithersburg, MD) and microtiter plate readers such as the ML 1000 (Dynatech Laboratories, Chantilly, VA) are most popular (reviewed by Bronstein and Kricka, 1990; Stanley, 1992a,b, 1993b).
Chemiluminescence signals originating from blotting experiments performed on membranes can be detected by imaging on X-ray or instant photographic films. These films offer simple, convenient, and inexpensive detectors of chemiluminescence that can be used successfully for qualitative determinations and some signal quantification. Camera luminometers that house instant photographic film are suitable for the detection of light emission from blots and microtiter plate wells, and are available from Amersham, Analytical Luminescence Laboratory (San Diego, CA), Dynatech Laboratories, and Tropix, Inc. (Bedford, MA).
Finally, photon-counting cameras are available and are most suitable for the detection and accurate quantification of low-light signals. This instrumentation usually consists of a light detector such as a silicon target, silicon diode array, or charge-coupled device (CCD) coupled to a lens system, a controller, and a digital image processor. Since most of these camera systems are capable of imaging in two dimensions, micro- and macroscopic luminescent specimens can be analyzed spatially and temporally. The Argus-100/CL (Hamamatsu Corporation, Photonic Microscopy, Inc., Oak Brook, IL) is a photon-counting imaging device that has been used in the detection of blotted proteins (Hauber et al., 1988). The Star I CCD cooled camera system (Photometrics Ltd., Tucson, AZ) exhibits very low dark current background and a wide dynamic range and has been used successfully to detect protein and nucleic acid analytes in solution and on membranes (Martin and Bronstein, 1993, 1994.).
IV. CHEMILUMINESCENCE ASSAYS FOR VIRUS DETECTION
The combination of DNA hybridization assays with chemiluminescence detection methods has enabled the development of rapid, sensitive, quantitative, nonradioactive assays that are amenable to automation. DNA hybridization technology is becoming accepted as a reliable clinical laboratory technique for the identification of infectious organisms and has fueled the need for more rapid, sensitive, and automated assay formats. Culture assay methods are laborious, time-consuming, and costly, and sometimes impossible to use. Antigen-based detection assays including fluorescent antibody and immunoassay techniques, although faster and automatable, are often less sensitive than culture techniques. With the advent of technologies such as PCR, DNA probe methods offer rapid, easy, and highly sensitive assay formats. DNA hybridization assays using radioactive labels are sensitive and are easily quantified, but health, environmental, disposal, and cost concerns render these systems less than ideal as widely used clinical assays.
Chemiluminescence methods for the detection of viral agents as well as other microorganisms have become widely used (Table 1 ), and continued development will certainly expand their applications in research and clinical diagnostic tests. More traditional immunoassays have also been developed and used with chemiluminescence for the detection of various viral antigens and the assessment of immune status with respect to viruses (selected references in Table 1). A survey of commercially available products that incorporate chemiluminescence or bioluminescence techniques and reagents for specific assays and nonspecific detection systems is available (Stanley, 1993a,b).
TABLE 1.
Selected Studies That Have Used Chemiluminescence to Detect Viruses, Chlamydia trachomatis, and Other Microorganisms
Assay formats include: BH, bead-based hybridization; H, hybridization; IA, immunoassay; IM, immunity; ISH, in situ hybridization; MH, membrane-based hybridization; PCR, polymerase chain reaction; RT, reverse transcriptase; SD, strand displacement; SH, solution hybridization.
Chemiluminescent (CL) methods employed (if known): AE, acridinium ester; AP, alkaline phosphatase (most likely with dioxetane substrate); BL, bioluminescence; DX, dioxetane; EL, electrochemiluminescence; ILU, isoluminol; LU, enhanced luminol.
A. DNA Hybridization Assay Formats
Several DNA hybridization assay formats including membrane-based, solution, and in situ hybridization have been coupled with chemiluminescence for the detection of viruses and other infectious agents. Membrane-based chemiluminescent hybridization assays have employed either 1,2-dioxetane substrates for alkaline phosphatase or the enhanced chemiluminescence reaction of luminol and HRP, and are imaged on X-ray or photographic films or imaged directly and quantified using a CCD camera system. Solution hybridization assays are performed with 1,2-dioxetanes, luminol, and AE labels, and the emitted light signal is measured in a luminometer. Electrochemiluminescent labels are also used for solution hybridization assays and are detected with an instrument combining an electrochemical flow cell, a potentiostat, and a photomultiplier tube. In situ hybridization has been performed using both 1,2-dioxetanes and enhanced luminol with either photographic film detection or a CCD camera system.
B. Chemiluminescence Detection Systems
1. Dioxetanes
Alkaline phosphatase-dioxetane chemiluminescence systems have been used in a wide variety of DNA hybridization assays for detection of infectious agents. Membrane-based hybridization assays have been used for the detection of HBV (Bronstein et al., 1989c; Yang et al., 1991; Escarceller et al., 1992), herpes simplex virus (HSV-1) (Bronstein and Voyta, 1989), CMV (Musiani et al., 1991a,1992; Yang et al., 1991), HIV-1 (Zachar et al., 1991), and other viral agents (Fouly et al., 1992; Tham and Stanislawek, 1992a,b; Fuchs et al., 1993). Solution hybridization assays include those for HBV (Urdea et al., 1990), HIV-1 (Suzuki et al., 1992), and Chlamydia (Clyne et al., 1989; Urdea et al., 1989). In situ hybridization assays have been performed with both HSV-1 infected cells (Bronstein and Voyta, 1989) and HIV-infected cells (Bronstein et al., 1989b). Finally, assays for retroviruses based on the detection of reverse transcriptase activity can be coupled with chemiluminescence detection by measuring the enzymatic incorporation of digoxigenin-labeled nucleotides with anti-digoxigenin alkaline phosphatase and a dioxetane substrate (Suzuki et al., 1993).
Commercially available detection systems incorporating dioxetanes include the AmpliProbe® system (ImClone Systems) for membrane-based hybridization assays for HBV, CMV, and EBV (Yang et al., 1991), the Hybrid Capture™ System HBV DNA Assay (Murex Diagnostics Ltd., Kent, UK), and solution hybridization assay systems for Chlamydia trachomatis and HBV detection (Chiron Corporation; Clyne et al., 1989; Urdea et al., 1989,1990).
2. Luminol
DNA hybridization assays using the ECL system with direct HRP-labeled probes include detection of bovine enteric coronavirus in a slot blot hybridization assay (Collomb et al., 1992) and a solution-phase hybridization assay for HBV DNA (Urdea et al., 1987,1990). In situ hybridization for detection of human papillomavirus (HPV) type 16 has been performed with an indirect labeled probe (Hawkins and Cumming, 1990). ECL systems have also been used for the immunoassay detection of several viruses, including grapevine closterovirus (Pollini et al., 1993) and parvovirus B 19 (O'Neill and Coyle, 1992).
3. Acridinium Esters
DNA hybridization assays incorporating AE-labeled probes have been developed for detection of several infectious agents from clinical samples, including C. trachomatis, Neisseria gonorrhoeae, fungal pathogens, mycobacteria, and several common bacterial pathogens (Nelson and Kacian, 1990). These assay systems, called PACE 2™ and ACCUPROBE™, are available commercially through Gen-Probe, Inc. (San Diego, CA). The Gen-Probe system for screening for Chlamydia has been compared with both culture and nonculture antigen detection methods including enzyme immunoassays and immunofluorescent antibody tests (Gratton et al., 1990; Mercer et al., 1990; Iwen et al., 1991). The PACE 2™ system can provide a rapid, reliable alternative to culture and immunoassay methods for the detection of Chlamydia from cervical samples (Iwen et al., 1991). Solution hybridization (hybridization protection) assays with AE-labeled probes have been used for the detection of PCR-amplified HIV-1 DNA (Ou et al., 1990; Schmidt, 1991; Rapier et al., 1993).
In addition, AEs have also been used to label antibodies that have been incorporated into automated immunoassay formats for the detection of infectious agents and antibody screening from clinical samples (Khalil et al., 1991a,b).
4. Electrochemiluminescence
Electrochemiluminescence detection has been used in both manual (Blackburn et al., 1991; Gudibande et al., 1992; Kenten et al., 1992) and automated (QPCR System 5000; Wages et al., 1993) post-PCR amplification DNA hybridization assays for the detection of HIV-1 and HPV (Kenten et al., 1991).
5. Bioluminescence
Detection of asymmetric amplified papillomavirus sequences using solution-phase hybridization with a G6PDH-labeled oligonucleotide and solid-phase capture has been performed using a bioluminescence assay (Balaguer et al., 1991b).
V. CHEMILUMINESCENCE DETECTION PROTOCOLS
A. Hepatitis B Virus
Hepatitis B “core antigen” DNA, immobilized on nylon membrane, is hybridized with an alkaline phosphatase-labeled oligonucleotide probe. Hybridized probe is then detected with the 1,2-dioxetane substrate AMPPD (Bronstein et al., 1989c).
1. Materials
Hepatitis B core antigen plasmid DNA and alkaline phosphatase-labeled probe, included in a SNAP® Hybridization System, and GeneScreen Plus™ nylon membrane were obtained from NEN/DuPont (Boston, MA). AMPPD and CSPD are from Tropix.
2. Target DNA Preparation and Probe Hybridization
HBV “core antigen” (HBVc) plasmid DNA (100 ng; 1.2 × 1010 copies) was dissolved in 25 μl sterile deionized H2O and serially diluted with 0.3 M NaOH to produce target DNA samples ranging in concentration from 4.88 × 103 to 0.98 × 108 copies/μl. Blots were prepared as described here:
-
1.
Incubate diluted DNA samples at room temperature for 15 min to denature, and spot 1 μl of each dilution onto dry membrane strips (1 × 8 cm).
-
2.
Rinse blots with 2 M NH4OAc and then with 0.6 M NaCl, 0.08 M sodium citrate, pH 7.0.
-
3.
Prehybridize with 3 ml hybridization buffer [0.75 M NaCl, 0.075 M sodium citrate (5 X SSC), 0.5% bovine serum, 0.5% polyvinylpyrrolidone, 1% sodium dodecyl sulfate (SDS), pH 7.0] for 15 min at 55°C.
-
4.
Hybridize with hybridization buffer containing 1.0 nM alkaline phosphatase-labeled oligonucleotide probe for 30 min at 55°C.
-
5.
Wash sequentially for 5 min each in:
1 X SSC, pH 7.0, 1% SDS at room temperature
1 X SSC, pH 7.0, 1% Triton X-100 at 55°C
1 X SSC, pH 7.0, at room temperature
3. Chemiluminescence Detection
-
1.
Wash hybridized blots with 0.1% bovine serum albumin (BSA), 0.05 M sodium carbonate, pH 9.5.
-
2.
Saturate blot with 100 μl 1.6 mM AMPPD in 0.1% BSA, 0.05 M sodium carbonate, 1.0 mM MgCl2, pH 9.5.
NOTE: Alternatively, an improved buffer (0.1 M diethanolamine, 1.0 mM MgCl2, pH 10.0) can be substituted for this wash, using 0.25 mM AMPPD or CSPD in this buffer for substrate incubation.
-
3.
Place blots in a plastic pouch and image light emission in a camera luminometer with Polaroid Instant Black and White Type 612 (ASA 20,000) photographic film.
NOTE: Alternatively, blots can be imaged on standard X-ray film.
-
4.
Digitize photographic film image using a black and white RBP Reflectance Densitometer (Tobias Associates, Inc., Ivyland, PA).
4. Results
Figure 2 shows a time course of the chemiluminescent DNA hybridization assay for HBVc antigen DNA. Serial dilutions of plasmid DNA were hybridized with alkaline phosphastase-labeled oligonucleotide probe, incubated with chemiluminescent substrate, and imaged on photographic film. Each photograph corresponds to a 30-min exposure. With this chemiluminescence assay, 1.18 × 106 copies of HBVc DNA can be detected within 30 min of substrate incubation. After a 2-hr incubation, 4.39 × 104 copies can be detected. In contrast, with the colorimetric bromochloroindolyl phosphate/nitro blue tetrazolium (BCIP/NBT) substrate system, 9.8 × 107 and 1.07 × 107 copies can be detected after 30 min or 2 hr of substrate incubation, respectively (results not shown). Quantitative results were obtained by measuring reflection densities from the imaged photographic film using a black and white reflection densitometer (Fig. 3 ). These values could be used to establish a dose-response curve for the reflection densities as a function of HBVc plasmid concentration, from which HBVc DNA levels in clinical specimens could be determined. Use of the improved chemiluminescence detection protocol, incorporating the diethanolamine substrate buffer and CSPD chemiluminescent substrate, results in even greater sensitivity for DNA hybridization assays and would increase the sensitivity of this HBV DNA assay. Imaging and quantification of this membrane-based assay with rapidly evolving CCD camera systems will likely provide even greater sensitivity and a greater linear dynamic range than that achieved with densitometry.
Figure 2.

Chemiluminescent detection of hepatitis B “core antigen” plasmid DNA with AMPPD substrate in alkaline phosphatase-based DNA hybridization assay.
Reprinted with permission from Bronstein et al. (1989c).
Figure 3.

Hepatitis B virus “core antigen” plasmid DNA hybridization assay. Reflection density vs. number of copies of target DNA. Densitometric analysis of the Polaroid instant black and white photographic film image [0.00 (white)–2.00 (black)].
Reprinted with permission from Bronstein et al. (1989c).
5. Summary
Chemiluminescent detection of HBV DNA has also been performed with the AmpliProbe® system (ImClone Systems). This signal amplification probe system incorporates multiple target-specific primary and multiple secondary probes, alkaline phosphatase-labeled oligonucleotides that hybridize to the primary probes, in a two-step hybridization system (Yang et al., 1991). Chemiluminescence detection is performed with a dioxetane substrate. With this system, 0.4 pg (1 × 105 copies) purified target HBV genomic DNA can be detected in a chemiluminescent slot blot assay (Farmar and Castaneda, 1991; Yang et al., 1991). Identical assays performed with serum samples (25 μl) demonstrated that this chemiluminescent DNA hybridization system has the same specificity and sensitivity as immunoassays and is more sensitive than a 32P-labeled nick-translated probe (Yang et al., 1991).
Escarceller et al. (1992) report the use of digoxigenin-labeled probes, anti-digoxigenin alkaline phosphatase, and AMPPD for the direct detection of HBV sequences in human serum samples. These investigators achieved a limit of sensitivity of 2–5 pg, which was equivalent to that obtained with both colorimetric detection and a 32P-labeled probe. These researchers also used digoxigenin-labeled oligonucleotide primers for PCR amplification of HBV DNA purified from human serum, followed by immunological detection of the digoxigenin label (as described), a method that can be used in conjunction with alternatively labeled primers for multiple amplifications.
A chemiluminescent assay incorporating a solution-phase hybridization of synthetic oligonucleotides to target DNA, followed by solid-phase capture, labeling, and detection with either HRP or alkaline phosphatase-labeled oligonucleotides and chemiluminescent substrates has been used to achieve the detection of 0.2 pg (6 × 104 copies) HBV DNA in human serum samples in 4 hr. This solution DNA hybridization method includes novel labeling and amplification schemes and has been performed with both polystyrene bead and microtiter well capture systems (Urdea et al., 1987,1990).
Chemiluminescence techniques have also been used in the development of automated enzyme immunoassay systems for the detection of HBV in human sera (Khalil et al., 1991a,b; Bouveresse and Bourgeois, 1992).
B. Herpes Simplex Virus
Two chemiluminescent DNA hybridization assays for HSV, dot blot hybridization and in situ hybridization, are described here as originally reported by Bronstein and Voyta (1989). In these assays, HSV-1 plasmid DNA, immobilized on nylon membrane, or HSV-1-infected Vero cells, fixed and mounted on microscope slides, were hybridized with an alkaline phosphatase-labeled HSV-1 oligonucleotide probe and detected with AMPPD.
1. Materials
HSV-1 plasmid DNA and alkaline phosphatase-labeled oligonucleotide probe, included in a SNAP® Hybridization System, and GeneScreen Plus nylon membrane were obtained from NEN/DuPont. HSV-l-infected Vero cells were provided by Drs. J. Kershner and E. Jablonski (Molecular Biosystems, San Diego, CA). AMPPD and Emerald™ luminescence-amplifying material are from Tropix.
2. Dot Blot Hybridization and Chemiluminescence Detection
This membrane hybridization protocol is similar to that described for HBV detection.
-
1.
Serially dilute HSV-1 plasmid DNA in 0.3 M NaOH, denature, and spot l-μl aliquots onto dry membrane strips.
-
2.
Prehybridize blots with hybridization buffer (0.5% BSA, 0.5% polyvinylpyrrolidone, 1% SDS) for 15 min at 55°C.
-
3.
Hybridize with hybridization solution (containing alkaline phosphatase-labeled HSV-1 oligonucleotide probe) for 30 min at 55°C.
-
4.
Wash sequentially for 5 min each in:
2× SSC, 1% SDS at room temperature
1× SSC, 1% Triton X-100 at 55°C
1× SSC, 1% Triton X-100 at room temperature
1× SSC at room temperature
-
5.
Wash hybridized blots with 0.05 M sodium carbonate/bicarbonate, 1 mM MgCl2, pH 9.5 (substrate buffer).
-
6.
Saturate blot with 1.6 mM AMPPD (in substrate buffer) for 5 min.
NOTE: As described for HBV detection, the diethanolamine buffer and 0.25 mM AMPPD or CSPD can be substituted in Steps 5 and 6 for increased sensitivity.
-
7.
Image blots with Polaroid Type 612 Instant Black and White film.
NOTE: Alternatively, blots can be imaged on X-ray film.
3. In Situ Hybridization and Chemiluminescence Detection
-
1.
Infect Vero cells with HSV-1 (MacIntyre strain) for 1 hr at room temperature.
-
2.
Harvest cells with trypsin/versene after the addition of 2% fetal calf serum at 0, 2, 4, 6, 8, 10, 12, 24, and 48 hr.
-
3.
Pellet cells, fix in 95% ethanol, and mount on glass microscope slides.
-
4.
Treat mounted slides with 0.2 M HCl for 2 min, rinse with deionized water, and immerse in 70% ethanol. Prior to hybridization, remove slides from ethanol and dry.
-
5.
Immerse slides in 0.1% BSA, 5X SSC for 15 min at 70°C. Treat with 0.3 M NaOH for 1 min at room temperature. Rinse with phosphate-buffered saline (PBS).
-
6.
Hybridize cells with the alkaline phosphatase-labeled HSV-1 oligonucleotide probe at a concentration of 5 nM in 0.1% BSA, 5X SSC for 20 min at 60°C.
-
7.
Wash slides briefly in hybridization buffer at 60°C, and then extensively with 1X SSC at 50°C.
-
8.
Wash with 0.05 M sodium carbonate/bicarbonate, 1 m M MgCl2, pH 9.5 (substrate buffer).
-
9.
Incubate with 0.8 mM AMPPD, 10% Emerald in substrate buffer for 5 min.
-
10.
Place slides in a camera luuminometer and expose to Polaroid Type 612 Instant Black and White film.
4. Results
With the dot blot hybridization assay for HSV-1 plasmid DNA, detection limits achieved with the chemiluminescent substrate AMPPD are 1.3 × 105 and 1.4 × 104 copies of target HSV-1 DNA, with a 30-min exposure performed 1 hr after substrate addition and a 45-min exposure performed 4 hr after substrate addition, respectively (results not shown). The sensitivity achieved with AMPPD is 25- to 100-fold higher than that obtained with the colorimetric BCIP/NBT substrate system (results not shown). Fig. 4 shows the time course of viral infection assayed by in situ DNA hybridization with chemiluminescence detection. Use of the AMPPD chemiluminescent substrate enables the detection of HSV-1-infected cells within 6 hr postinfection. Again, with this assay format, CCD detection and imaging may provide even greater sensitivity than that achieved with photographic film.
Figure 4.

Chemiluminescent detection of in situ DNA hybridization of herpes simplex virus I-infected Vero cells: time course of infection.
Reprinted from Bronstein and Voyta, Clinical Chemistry (1989), 35, 1856–1857, Courtesy of the American Association for Clinical Chemistry, Inc.
5. Summary
In situ hybridization with chemiluminescence detection has also been used to detect HIV-infected cells (Bronstein et al., 1989b) and HPV type 16 in a cervical carcinoma cell line (Hawkins and Cumming, 1990). The latter protocol involved the use of biotinylated HPV 16 DNA probes (Enzo Diagnostics, New York), a streptavidin-HRP conjugate, and ECL detection reagents coupled with a CCD imaging system. Detection of fewer than 10 HPV-positive cells (containing 600 copies of HPV 16 DNA per cell) among 10,000 HPV-negative cells on a single slide was achieved. However, this detection level is not necessarily the limit of sensitivity; with improved optical instrumentation, in situ hybridization coupled with CCD detection may provide a valuable diagnostic tool for the rapid and automated identification of viral sequences within cells.
VI. CONCLUSION
Chemiluminescence detection technologies combined with DNA hybridization methods provide rapid, sensitive, nonradioactive, automatable assay formats for the clinical diagnosis of infectious agents, as well as for research use. Rapidly evolving chemiluminescent enzyme substrates and labels, techniques, and assay and detection instrumentation, coupled with continued advances in DNA hybridization technologies, will further refine and improve the specificity and sensitivity of chemiluminescent DNA detection methods, bringing them into more widespread use.
ACKNOWLEDGMENTS
We are very grateful to Larry Kricka, Chris Martin, John Voyta, and Alison Sparks for editorial assistance.
REFERENCES
- Akin A., Wu C.C., Lin T.L., Keirs R.W. Chemiluminescent detection of infectious bursal disease virus with a PCR-generated nonradiolabeled probe. J. Vet. Diagn. Invest. 1993;5:166–173. doi: 10.1177/104063879300500205. [DOI] [PubMed] [Google Scholar]
- Akita G.Y., Glenn J., Castro A.E., Osburn B.I. Detection of bluetongue virus in clinical samples by polymerase chain reaction. J. Vet. Diagn. Invest. 1993;5:154–158. doi: 10.1177/104063879300500202. [DOI] [PubMed] [Google Scholar]
- Arenkov P.I., Berezin V.A., Starodub N.F. Chemiluminescence fiber optic immunosensor for detecting antibodies to the influenza virus. Ukr. Biokhim. 1991;63:99–103. [PubMed] [Google Scholar]
- Arnold L.J., Hammond P.W., Wiese W.A., Nelson N.C. Assay formats involving acridinium-ester-labeled DNA probes. Clin. Chem. 1989;35:1588–1594. [PubMed] [Google Scholar]
- Balaguer P.T., Térouanne B., Boussioux A.-M., Nicolas J.-C. Use of bioluminescence in nucleic acid hybridization reactions. J. Biolumin. Chemilumin. 1989;4:302–309. doi: 10.1002/bio.1170040142. [DOI] [PubMed] [Google Scholar]
- Balaguer P., Térouanne B., Eliaou J.F., Humbert M., Boussioux A.M., Nicolas J.C. Use of glucose-6-phosphate dehydrogenase as a new label for nucleic acid hybridization reactions. Anal. Biochem. 1989;180:50–54. doi: 10.1016/0003-2697(89)90085-7. [DOI] [PubMed] [Google Scholar]
- Balaguer P., Térouanne B., Boussioux A., Nicolas J. Quantification of DNA sequences obtained by polymerase chain reaction using a bioluminescent adsorbent. Anal. Biochem. 1991;195:105–110. doi: 10.1016/0003-2697(91)90303-b. [DOI] [PubMed] [Google Scholar]
- Balaguer P., Térouanne B., Boussioux A.M., Nicolas J.C. Papillomavirus quantification using asymmetric amplification and a rapid bioluminescent assay. In: Stanley P.E., Kricka L.J., editors. Bioluminescence & Chemiluminescence: Current Status. John Wiley & Sons; Chichester: 1991. pp. 143–146. [Google Scholar]
- Barker R.H., Jr., Banchongaksorn T., Courval J.M., Suwonkerd W., Rimwungtragoon K., Wirth D.F. A simple method to detect Plasmodium falciparum directly from blood samples using the polymerase chain reactions. Am. J. Trop. Med. Hyg. 1992;46:416–426. doi: 10.4269/ajtmh.1992.46.416. [DOI] [PubMed] [Google Scholar]
- Beck S., Köster H. Applications of dioxetane chemiluminescent probes to molecular biology. Anal. Chem. 1990;62:2258–2270. doi: 10.1021/ac00220a003. [DOI] [PubMed] [Google Scholar]
- Bettens F., Pichler W.J., de Weck A.L. Incorporation of biotinylated nucleotides for the quantification of PCR-amplified HIV-1 DNA by chemiluminescence. Eur. J. Clin. Chem. Clin. Biochem. 1991;29:685–688. [PubMed] [Google Scholar]
- Blackburn G.F., Shah H.P., Kenten J.H., Leland J., Kamin R.A., Link J., Peterman J., Powell M.J., Shah A., Talley D.B., Tyagi S.K., Wilkins E., Wu T.-G., Massey R.J. Electrochemiluminescence detection for development of immunoassays and DNA probe assays for clinical diagnostics. Clin. Chem. 1991;37:1534–1539. [PubMed] [Google Scholar]
- Bouveresse E., Bourgeois J.P.J. Chemiluminescent enzyme immunoassay for the detection of hepatitis B surface antigen (HBsAg) Clin. Chem. 1992;38:1090. [Google Scholar]
- Boxall E.H. Enhanced luminescent assays for hepatitis markers: Assesssment of post vaccine responses. Arch. Virol. Suppl. 1992;4:156–159. doi: 10.1007/978-3-7091-5633-9_34. [DOI] [PubMed] [Google Scholar]
- Bronstein I., Dimond P. Chemiluminescent compounds for diagnostic tests. Diagn. Clin. Test. 1990;28:36–39. [Google Scholar]
- Bronstein I., Kricka L.J. Clinical applications of luminescent assays for enzymes and enzyme labels. J. Clin. Lab. Anal. 1989;3:316–322. doi: 10.1002/jcla.1860030511. [DOI] [PubMed] [Google Scholar]
- Bronstein I., Kricka L.J. Instrumentation for luminescent assays. Am. Clin. Lab. 1990;9(1):33–37. [PubMed] [Google Scholar]
- Bronstein I., Sparks A. Sensitive enzyme immunoassays with chemiluminescent detection. In: Nakamura R.M., Kasahara Y., Rechnitz G.A., editors. Immunochemical Assays and Biosensor Technology for the 1990s. American Society for Microbiology; Washington, D.C.: 1992. pp. 229–250. [Google Scholar]
- Bronstein I., Voyta J.C. Chemiluminescent detection of herpes simplex virus I DNA in blot and in-situ hybridization assays. Clin. Chem. 1989;35:1856–1857. [PubMed] [Google Scholar]
- Bronstein I., Edwards B., Voyta J.C. 1,2-Dioxetanes: Novel chemiluminescent enzyme substrates. Applications to immunoassays. J. Biolumin. Chemilumin. 1989;4:99–111. doi: 10.1002/bio.1170040116. [DOI] [PubMed] [Google Scholar]
- Bronstein I., Kerschner J.H., Voyta J.C., Jablonski E.G. Chemiluminescent detection of HIV infected cells using in-situ hybridization assay technique. Presented at the V International Conference on AIDS; Montreal, Quebec, Canada, June 4–9; 1989. [Google Scholar]
- Bronstein I., Voyta J.C., Edwards B. A comparison of chemiluminescent and colorimetric substrates in a hepatitis B virus DNA hybridization assay. Anal. Biochem. 1989;180:95–98. doi: 10.1016/0003-2697(89)90093-6. [DOI] [PubMed] [Google Scholar]
- Bronstein I., Voyta J.C., Lazzari K.G., Murphy O., Edwards B., Kricka L.J. Rapid and sensitive detection of DNA in Southern blots with chemiluminescence. Bio Techniques. 1990;8:310–313. [PubMed] [Google Scholar]
- Bronstein I., Juo R.-R., Voyta J.C., Edwards B. Novel chemiluminescent adamantyl 1,2-dioxetane enzyme substrates. In: Stanley P., Kricka L.J., editors. Bioluminescence and Chemiluminescence: Current Status. John Wiley; Washington, D.C.: 1991. pp. 73–82. [Google Scholar]
- Bull T., Shanson D. Evaluation of a commercial chemiluminescent gene probe system “AccuProbe” for the rapid differentiation of mycobacteria, including “MAIC X,” isolated from blood and other sites, from patients with AIDS. J. Hosp. Infect. 1992;21:143–149. doi: 10.1016/0195-6701(92)90034-j. [DOI] [PubMed] [Google Scholar]
- Carter T.J.N., Kricka L.J. Analytical applications of chemiluminescence. In: Kricka L.J., Carter T.J.N., editors. Clinical and Biochemical Luminescence. Marcel Dekker; Chichester: 1982. pp. 135–151. [Google Scholar]
- Cate R.L., Ehrenfels C.W., Wysk M., Tizard R., Voyta J.C., Murphy O.J., Bronstein I. Genomic Southern analysis with alkaline-phosphatase-conjugated oligonucleotide probes and the chemiluminescent substrate AMPPD. Genetic Analysis, Techniques Applications. 1991;8:102–106. doi: 10.1016/1050-3862(91)90044-r. [DOI] [PubMed] [Google Scholar]
- Chanteloup E., Clement A., Payne J. Chemiluminescent enzyme immunoassay for the evaluation of the immune status to rubella virus. Clin. Chem. 1992;38:1085. [Google Scholar]
- Clyne J.M., Running J.A., Stempien M., Stephens R.S., Akhavan-Tafti H., Schaap A.P., Urdea M.S. A rapid chemiluminescent DNA hybridization assay for the detection of Chlamydia trachomatis. 1989;4:357–366. doi: 10.1002/bio.1170040149. [DOI] [PubMed] [Google Scholar]
- Collomb J., Finance C., Alabouch S., Laporte J. Radioactive and enzymatic cloned cDNA probes for bovine enteric coronavirus detection by molecular hybridization. Arch. Virol. 1992;125:25–37. doi: 10.1007/BF01309626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway B., Adler K.E., Bechtel L.J., Kaplan J.C., Hirsch M.S. Detection of HIV-1 DNA in crude cell lysates of peripheral blood mononuclear cells by the polymerase chain reaction and nonradioactive oligonucleotide probes. J. Acq. Immune Def. Syndr. 1990;3:1059–1064. [PubMed] [Google Scholar]
- Cook R.F., Cook S.J., Issel C.J. A nonradioactive micro-assay for released reverse transcriptase activity of a lentivirus. Bio Techniques. 1992;13:380–386. [PubMed] [Google Scholar]
- Dalessio J., Ashley R. Highly sensitive enhanced chemiluminescence immunodetection methods for herpes simplex virus type-2 Western immunoblot. J. Clin. Microbiol. 1992;30:1005–1010. doi: 10.1128/jcm.30.4.1005-1007.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J., Clifton N., Seshkin K., Gooch W. Use of rapid, nonradioactive DNA probes in culture confirmation tests to detect Streptococcus agalactiae, Haemophilus influenzae, and Enterococcus spp. from pediatric patients with significant infections. J. Clin. Microbiol. 1991;29:80–82. doi: 10.1128/jcm.29.1.80-82.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCesare J., Grossman B., Katz E., Picozza, Ragusa R., Woudenberg T. A high-sensitivity electrochemiluminescence-based detection system for automated PCR product quantification. Bio Techniques. 1993;15:152–157. [PubMed] [Google Scholar]
- Donahue C., Jurgensen S., Nycz C., Schram J.L., Shank D., Vonk G.P., Walker G.T. Detection of mycobacterial DNA using strand displacement amplification and a chemiluminescent microwell assay. J. Biolumin. Chemilumin. 1993;8:77. [Google Scholar]
- Dumornay W., Roblin P., Gelling M., Hammerschlag M., Worku M. Comparison of a chemiluminometric immunoassay with culture for diagnosis of chlamydial infections in infants. J. Clin. Microbiol. 1992;30:1867–1869. doi: 10.1128/jcm.30.7.1867-1869.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant I. Detection of horseradish peroxidase by enhanced chemiluminescence. In: Kricka L.J., editor. Nonisotopic DNA Probe Techniques. Academic Press; New York: 1992. pp. 167–183. [Google Scholar]
- Durrant I., Benge L.C.A., Sturrock C., Devenish A.T., Howe R., Roe S., Moore M., Scozzafava G., Proudfoot L.M.F., Richardson T.C., McFarthing K.G. The application of enhanced chemiluminescence to membrane-based nucleic acid detection. Bio Techniques. 1990;8:564–570. [PubMed] [Google Scholar]
- Eis-Hubinger A.M., Kaiser R., Kleim J.P., Dlugosch D., Estor A., Kleeman E., Lange C.E., Schneweis K.E. Detection of varicella zoster virus infections using polymerase chain reaction. Hautarzt. 1992;43:767–771. [PubMed] [Google Scholar]
- Escarceller M., Rodriguez-Frias F., Jardi R., San-Segundo B., Eritja R. Detection of hepatitis B virus DNA in human serum samples: Use of digoxigenin-labeled oligonucleotides as modified primers for the polymerase chain reaction. Anal. Biochem. 1992;206:36–42. doi: 10.1016/s0003-2697(05)80007-7. [DOI] [PubMed] [Google Scholar]
- Farmar J.G., Castaneda M. An improved preparation and purification of oligonucleotide-alkaline phosphatase conjugates. Bio Techniques. 1991;11:588–589. [PubMed] [Google Scholar]
- Faulkner L.R., Glass R.S. Electrochemiluminescence. In: Adam W., Cilento G., editors. Chemical and Biological Generation of Excited States. Academic Press; San Diego: 1982. pp. 191–227. [Google Scholar]
- Fouly H.M., Domier L.L., D'Arcy C.J. A rapid chemiluminescent detection method for barley yellow dwarf virus. J. Virol. Meth. 1992;39:291–298. doi: 10.1016/0166-0934(92)90102-j. [DOI] [PubMed] [Google Scholar]
- Fuchs F., Leparc I., Kopecka H., Garin D., Aymard M. Use of cRNA digoxigenin-labelled probes for detection of enteroviruses in humans and in the environment. J. Virol. Meth. 1993;42:217–226. doi: 10.1016/0166-0934(93)90034-o. [DOI] [PubMed] [Google Scholar]
- Geiger C.P., Caselmann W.H. Non-radioactive hybridization with hepatitis C virus-specific probes created during polymerase chain reaction: A fast and simple procedure to verify hepatitis C virus infection. J. Hepatol. 1992;15:387–390. doi: 10.1016/0168-8278(92)90074-y. [DOI] [PubMed] [Google Scholar]
- Geiger R.E. Detection of alkaline phosphatase by bioluminescence. In: Kricka L.J., editor. Nonisotopic DNA Probe Techniques. Academic Press; New York: 1992. pp. 113–126. [Google Scholar]
- Gratton C.A., Lim-Fong R., Prasad E., Kibsey P.C. Comparison of a DNA probe with culture for detecting Chlamydia trachomatis directly from genital specimens. Mol. Cell. Probes. 1990;4:25–31. doi: 10.1016/0890-8508(90)90036-y. [DOI] [PubMed] [Google Scholar]
- Gudibande S.R., Kenten J.H., Link J., Friedman K., Massey R.J. Rapid non-separation electrochemiluminescent DNA hybridization assays for PCR products, using 3′-labelled oligonucleotide probes. Mol. Cell. Probes. 1992;6:495–503. doi: 10.1016/0890-8508(92)90046-z. [DOI] [PubMed] [Google Scholar]
- Gustaferro C.A., Persing D.H. Chemiluminescent universal probe for bacterial ribotyping. J. Clin. Microbiol. 1992;30:1039–1041. doi: 10.1128/jcm.30.4.1039-1041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harber M.J. Applications of luminescence in medical microbiology and hematology. In: Kricka L.J., Carter T.J.N., editors. Clinical and Biochemical Luminescence. Marcel Dekker; San Diego: 1982. pp. 189–218. [Google Scholar]
- Hauber R., Geiger R. A new, very sensitive, bioluminescence-enhanced detection system for protein blotting. Ultrasensitive detection systems for protein blotting and DNA hybridization, I. J. Clin. Chem. Clin. Biochem. 1987;25:511–514. doi: 10.1515/cclm.1987.25.8.511. [DOI] [PubMed] [Google Scholar]
- Hauber R., Geiger R. A sensitive, bioluminescence-enhanced detection method for DNA dot-hybridization. Nucleic Acid Res. 1988;16:1213. doi: 10.1093/nar/16.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber R., Miska W., Schleinkofer L., Geiger R. The application of a photon-counting camera in very sensitive, bioluminescence-enhanced detection systems for protein blotting. Ultrasensitive detection systems for protein blotting and DNA hybridization, II. J. Clin. Chem. Clin. Biochem. 1988;26:147–148. [PubMed] [Google Scholar]
- Hauber R., Miska W., Schleinkofer L., Geiger R. New, sensitive, radioactive-free bioluminescence-enhanced detection system in protein blotting and nucleic acid hybridization. J. Biolumin. Chemilumin. 1989;4:367–372. doi: 10.1002/bio.1170040150. [DOI] [PubMed] [Google Scholar]
- Hawkins E., Cumming R. Enhanced chemiluminescence for tissue antigen and cellular viral DNA detection. J. Histochem. Cytochem. 1990;38:415–419. doi: 10.1177/38.3.1689340. [DOI] [PubMed] [Google Scholar]
- Henchal E.A., Polo S.L., Vorndam V., Yaemsiri C., Innis B.L., Hoke C.H. Sensitivity and specificity of a universal primer set for the rapid diagnosis of dengue virus infections by polymerase chain reaction and nucleic acid hybridization. Am. J. Trop. Med. Hyg. 1991;45:418–428. doi: 10.4269/ajtmh.1991.45.418. [DOI] [PubMed] [Google Scholar]
- Hornsleth A., Aaen K., Gundestrup M. Detection of respiratory syncytial virus and rotavirus by enhanced chemiluminescence enzyme-linked immunosorbent assay. J. Clin. Microbiol. 1988;26:630–635. doi: 10.1128/jcm.26.4.630-635.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland D., Samuel D. Enhanced chemiluminescence ELISA for the detection of antibody to hepatitis B virus surface antigen. J. Biolumin. Chemilumin. 1989;4:159–163. doi: 10.1002/bio.1170040124. [DOI] [PubMed] [Google Scholar]
- Ishii J.K., Ghosh S.S. Bead-based sandwich hybridization characteristics of oligonucleotide-alkaline phosphatase conjugates and their potential for quantitating target RNA sequences. Bioconjugate Chem. 1993;4:34–41. doi: 10.1021/bc00019a005. [DOI] [PubMed] [Google Scholar]
- Iwen P.C., Blair T.M., Woods G.L. Comparison of the Gen-Probe PACE 2 system, direct fluorescent-antibody, and cell culture for detecting Chlamydia trachomatis in cervical specimens. Am. J. Clin. Pathol. 1991;95:578–582. doi: 10.1093/ajcp/95.4.578. [DOI] [PubMed] [Google Scholar]
- Jablonski E., Moomaw E.W., Tullis R.H., Ruth J.L. Preparation of oligodeoxynucleotide-alkaline phosphatase conjugates and their use as hybridization probes. Nucleic Acids Res. 1986;14:6115–6128. doi: 10.1093/nar/14.15.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs R.M., Smith H.E., Gregory B., Valli V.E., Whetstone C.A. Detection of multiple retroviral infections in cattle and cross-reactivity of bovine immunodeficiency-like virus and human immunodeficiency virus type 1 proteins using bovine and human sera in a western blot assay. Can. J. Vet. Res. 1992;56:353–359. [PMC free article] [PubMed] [Google Scholar]
- Jang D., Sellors J.W., Mahony J.B., Pickard L., Chernesky M.A. Effects of broadening the gold standard on the performance of a chemiluminometric immunoassay to detect Chlamydia trachomatis antigens in centrifuged first void urine and urethral swab samples from men. Sex. Trans. Dis. 1992;19:315–319. [PubMed] [Google Scholar]
- Kenten J.H., Casadei J., Link J., Lupolid S., Willey J., Powell M., Rees A., Massey R. Rapid electrochemiluminescence assays of polymerase chain reaction products. Clin. Chem. 1991;37:1626–1632. [PubMed] [Google Scholar]
- Kenten J.H., Gudibande S., Link J., Willey J.J., Curfman B., Major E.O., Massey R.J. Improved electrochemiluminescent label for DNA probe assays: Rapid quantitative assays of HIV-1 polymerase chain reaction products. Clin. Chem. 1992;38:873–879. [PubMed] [Google Scholar]
- Khalil O.S., Hanna C.F., Huff D., Zurek T.F., Murphy B., Pepe C., Genger K. Reaction tray and noncontract transfer method for heterogeneous chemiluminescence immunoassays. Clin. Chem. 1991;37:1612–1617. [PubMed] [Google Scholar]
- Khalil O.S., Zurek T.F., Tryba J., Hanna C.F., Hollar R., Pepe C., Genger K., Brentz C., Murphy B., Abunimeh N., Carver R., Harder P., Coleman C., Robertson E., Wolf-Rogers J. Abbott Prism: A multichannel heterogeneous chemiluminescence immunoassay analyzer. Clin. Chem. 1991;37:1540–1547. [PubMed] [Google Scholar]
- Kricka L.J. Chemiluminescent and bioluminescent techniques. Clin. Chem. 1991;37:1472–1481. [PubMed] [Google Scholar]
- Kricka L.J. Nucleic acid hybridization test formats: Strategies and applications. In: Kricka L.J., editor. Nonisotopic DNA Probe Techniques. Academic Press; New York: 1992. pp. 3–28. [Google Scholar]
- Kricka L.J., Carter T.J.N. Luminescent immunoassays. In: Kricka L.J., Carter T.J.N., editors. Clinical and Biochemical Luminescence. Marcel Dekker; San Diego: 1982. pp. 153–178. [Google Scholar]
- Kricka L.J., Thorpe G.H.G., Stott R.A.W. Enhanced chemiluminescence enzyme immunoassay. Pure Appl. Chem. 1987;59:651–654. [Google Scholar]
- Kuroda N., Nakashima K., Akiyama S., Shiraki H., Maeda Y. Photographic chemiluminescent ELISA for detection of anti-human T-cell leukemia virus type-I antibodies by using synthetic peptides as antigents. Clin. Chim. Acta. 1992;211:113–119. doi: 10.1016/0009-8981(92)90110-c. [DOI] [PubMed] [Google Scholar]
- Martin C.S., Bronstein I. Imaging of chemiluminescent signals with cooled CCD camera systems. Presented at the 2nd European Seminars on Low Light Imaging; Florence, Italy, September 1–4; 1993. [Google Scholar]
- Martin C.S., Bronstein I. Imaging of chemiluminescent signals with cooled CCD camera systems. J. Biolumin. Chemilumin. 1994;9:145–153. doi: 10.1002/bio.1170090308. [DOI] [PubMed] [Google Scholar]
- Martinez M.L., Weiss R.C. Detection of feline infectious peritonitis virus infection in cell cultures and peripheral blood mononuclear leukocytes of experimentally infected cats using a biotinylated cDNA probe. Vet. Microbiol. 1993;34:259–271. doi: 10.1016/0378-1135(93)90016-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J.A., Batki A., Hynds C., Kricka L.J. Enhanced chemiluminescent method for the detection of DNA dot-hybridization assays. Anal. Biochem. 1985;151:205–209. doi: 10.1016/0003-2697(85)90073-9. [DOI] [PubMed] [Google Scholar]
- McCapra F. The chemiluminescence of organic compounds. Q. Rev. Chem. Soc. 1966;20:485–510. [Google Scholar]
- McCartney R.A., Harbour J., Roome A.P.C.H., Caul E.O. Comparison of enhanced chemiluminescence and microparticle enzyme immunoassay for the measurement of hepatitis-B surface antibody. Vaccine. 1993;11:941–945. doi: 10.1016/0264-410x(93)90383-9. [DOI] [PubMed] [Google Scholar]
- McKimm-Breschkin J.L. Rapid treatment of whole cells and RNA viruses for analysis of RNA slot blot hybridization. Virus Res. 1992;22:199–206. doi: 10.1016/0168-1702(92)90051-a. [DOI] [PubMed] [Google Scholar]
- Mercer L.J., Robinson D.C., Sahm D.F., Lawrie M.J., Hajj S.N. Comparison of chemiluminescent DNA probe to cell culture for the screening of Chlamydia trachomatis in a gynecology clinic population. Obstet. Gynecol. 1990;76:114–117. [PubMed] [Google Scholar]
- Miliukiene V., Dikiniene N., Vidziunaite R., Mikalauskiene G., Veleckaite A., Mikulskis P. The comparative evaluation of spectrophotometric and chemiluminescent detection in an immunoenzyme test of antibodies to the antigens of the bovine leukosis virus. Zh. Mikrobiol. Epidemiol. Immunobiol. 1991;7:64–66. [PubMed] [Google Scholar]
- Musiani M., Zerbini M., Gibellini D., Gentilomi G., La Placa M., Ferri E., Girotti S. Chemiluminescent assay for the detection of viral and plasmid DNA using dogoxigenin-labeled probes. Anal. Biochem. 1991;194:394–398. doi: 10.1016/0003-2697(91)90247-q. [DOI] [PubMed] [Google Scholar]
- Musiani M., Zerbini M., Gibellini D., Gentilomi G., Venturoli S., Gallinella G., Ferri E., Girotti S. Chemiluminescence dot blot hybridization assay for detection of B19 parvovirus DNA in human sera. J. Clin. Microbiol. 1991;29:2047–2050. doi: 10.1128/jcm.29.9.2047-2050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiani M., Zerbini M., Gentilomi G., Gibellini D., Gallinella G., Venturoli S., La Placa M. Detection of CMV DNA in clinical samples of AIDS patients by chemiluminescence by hybridization. J. Virol. Meth. 1992;38:1–9. doi: 10.1016/0166-0934(92)90164-9. [DOI] [PubMed] [Google Scholar]
- Nelson N.C., Kacian D.L. Chemiluminescent DNA probes: A comparison of the acridinium ester and dioxetane detection system and their use in clinical diagnostic assays. Clin. Chim. Acta. 1990;194:73–90. doi: 10.1016/0009-8981(90)90304-b. [DOI] [PubMed] [Google Scholar]
- Nelson N.C., Reynolds M.A., Arnold L.J., Jr. Detection of acridinium esters by chemiluminescence. In: Kricka L.J., editor. Nonisotopic DNA Probe Techniques. Academic Press; New York: 1992. pp. 275–310. [Google Scholar]
- Neman-Simha V., Delmas-Beauvieux M.C., Geniaux M., Bebear C. Evaluation of a chemiluminometric immunoassay and a direct immunofluorescence test for detecting Chlamydia trachomatis in urogenital specimens. Eur. J. Clin. Microbiol. Infect. Dis. 1991;10:662–665. doi: 10.1007/BF01975822. [DOI] [PubMed] [Google Scholar]
- Nicolas J.C., Térouanne B., Balaguer P., Boussioux A.M., Crastes de Paulet A. A bioluminescent solid phase for immunoassays and DNA probes. Ann. Biol. Clin. (Paris) 1990;48:573–579. [PubMed] [Google Scholar]
- Nicolas J.-C., Balaguer P., Térouanne B., Villebrun M.A., Boussioux A.-M. Detection of glucose 6-phosphate dehydrogenase by bioluminescence. In: Kricka L.J., editor. Nonisotopic DNA Probe Techniques. Academic Press; San Diego: 1992. pp. 203–225. [Google Scholar]
- O'Neill H.J., Coyle P.V. Two anti-parvovirus B19 IgM capture assays incorporating a mouse monoclonal antibody specific for B19 viral capsid proteins Vp1 and Vp2. Arch. Virol. 1992;123:125–134. doi: 10.1007/BF01317143. [DOI] [PubMed] [Google Scholar]
- Ou C.-Y., McDonough S.H., Cabanas D., Ryder T.B., Harper M., Moore J., Schochetman G. Rapid and quantitative detection of enzymatically amplified HIV-1 DNA using chemiluminescent oligonucleotide probes. AIDS Res. Hum. Retroviruses. 1990;6:1323–1329. doi: 10.1089/aid.1990.6.1323. [DOI] [PubMed] [Google Scholar]
- Papsidero L.D., Dittmer R.P., Vaickus L., Poiesz B.J. Monoclonal antibodies and chemiluminescence immunoassay for detection of the surface protein of human T-cell lumphotropic virus. J. Clin. Microbiol. 1992;30:351–358. doi: 10.1128/jcm.30.2.351-358.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podleckis E.V., Hammond R.W., Hurtt S.S., Hadidi A. Chemiluminescent detection of potato and pome fruit viroids by dogoxigenin-labeled dot blot and tissue blot hybridization. J. Virol. Meth. 1993;43:147–158. doi: 10.1016/0166-0934(93)90072-y. [DOI] [PubMed] [Google Scholar]
- Pollard-Knight D., Read C.A., Downes M.J., Howard L.A., Leadbetter M.R., Pheby S.A., McNaughton E., Syms A., Brady M.A.W. Nonradioactive nucleic acid detection by enhanced chemiluminescence using probes directly labeled with horseradish peroxidase. Anal. Biochem. 1990;185:84–89. doi: 10.1016/0003-2697(90)90259-c. [DOI] [PubMed] [Google Scholar]
- Pollard-Knight D., Simmonds A.C., Schaap A.P., Akhavan H., Brady M.A.W. Nonradioactive DNA detection on Southern blots by enzymatically triggered chemiluminescence. Anal. Biochem. 1990;185:353–358. doi: 10.1016/0003-2697(90)90307-u. [DOI] [PubMed] [Google Scholar]
- Pollini C.P., Giunchedi L., Credi R. A chemiluminescent immunoassay for the diagnosis of grapevine closteroviruses on nitrocellulose membrane. J. Virol. Meth. 1993;42:107–116. doi: 10.1016/0166-0934(93)90182-q. [DOI] [PubMed] [Google Scholar]
- Pronovost A.D., Baumgarten A., Hsiung G.D. Sensitive chemiluminescent enzyme-linked immunosorbent assay for quantification of human immunoglobulin G and detection of herpes simplex virus. J. Clin. Microbiol. 1981;13:97–101. doi: 10.1128/jcm.13.1.97-101.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchhammerstoeckl E., Heinz F.X., Kunz C. Evaluation of 3 nonradioactive DNA detection systems for identification of herpes simplex DNA amplified from cerebrospinal fluid. J. Virol. Meth. 1993;43:257–266. doi: 10.1016/0166-0934(93)90081-2. [DOI] [PubMed] [Google Scholar]
- Radziszewski B. Untersuchungen über Hydrobenzamid, Amarin und Lophin. Chem. Berlin. 1877;10:70–75. [Google Scholar]
- Rapier J.M., Villamarzo Y., Schochetman G., Ou C.Y., Brakel C.L., Donegan J., Maltzman W., Lee S., Kirtikar D., Gatica D. Nonradioactive colorimetric microplate hybridization assay for detecting amplified human immunodeficiency virus DNA. Clin. Chem. 1993;39:244–247. [PubMed] [Google Scholar]
- Robertson E.F., Weare J.A., Randell R., Holland P.V., Madsen G., Decker R.H. Characterization of a reduction-sensitive factor from human plasma responsible for apparent false activity in competitive assays for antibody to hepatitis B core antigen. J. Clin. Microbiol. 1991;29:605–610. doi: 10.1128/jcm.29.3.605-610.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar F.H., Sakr W.A., Li Y.W., Sreepathi P., Crissman J.D. Detection of human papillomavirus (HPV) DNA in human prostatic tissues by polymerase chain reaction (PCR) Prostate. 1993;22:171–180. doi: 10.1002/pros.2990220210. [DOI] [PubMed] [Google Scholar]
- Schmidt B.L. A rapid chemiluminescence detection method for PCR-amplified HIV-1 DNA. J. Virol. Meth. 1991;32:233–244. doi: 10.1016/0166-0934(91)90054-4. [DOI] [PubMed] [Google Scholar]
- Schmidt B.L., Gschnait F. Detection of causal agents of AIDS using polymerase chain reaction and chemiluminescence measurement. Hautarzt. 1991;42:754–758. [PubMed] [Google Scholar]
- Scieux C., Bianchi A., Henry S., Brunat N., Abdennader S., Vexiau D., Janier M., Morel P., Lagrange P.H. Evaluation of a chemiluminometric immunoassay for detection of Chlamydia trachomatis in the urine of male and female patients. Eur. J. Clin. Microbiol. Infect. Dis. 1992;11:704–708. doi: 10.1007/BF01989974. [DOI] [PubMed] [Google Scholar]
- Scieux C., Bianchi A., Vassias I., Meouchy R., Felten A., Morel P., Perol Y. Evaluation of a new chemiluminometric immunoassay, Magic Lite Chlamydia, for detecting Chlamydia trachomatis antigen from urogenital specimens. Sex. Transm. Dis. 1992;19:161–164. [PubMed] [Google Scholar]
- Septak M. Acridinium ester-labelled DNA oligonucleotide probes. J. Biolumin. Chemilumin. 1989;4:351–356. doi: 10.1002/bio.1170040148. [DOI] [PubMed] [Google Scholar]
- Simmonds A.C., Cunningham M., Durrant I., Fowler S.J., Evans M.R. Enhanced chemiluminescence in filter-based DNA detection. Clin. Chem. 1991;37:1527–1528. [PubMed] [Google Scholar]
- Sritharan V., Barker R.H., Jr. A simple method for diagnosing M. tuberculosis infection in clinical samples using PCR. Mol. Cell. Probes. 1991;5:385–395. doi: 10.1016/s0890-8508(06)80011-3. [DOI] [PubMed] [Google Scholar]
- Stanley P.E. A survey of more than 90 commercially available luminometers and imaging devices for low light measurements of chemiluminescence and bioluminescence, including instruments for manual, automatic and specialized operation, for HPLC, LC, GLC and microtitre plates. Part 1: Descriptions. J. Biolumin. Chemilumin. 1992;7:77–108. doi: 10.1002/bio.1170070202. [DOI] [PubMed] [Google Scholar]
- Stanley P.E. A survey of more than 90 commercially available luminometers and imaging devices for low light measurements of chemiluminescence and bioluminescence, including instruments for manual, automatic and specialized operation, for HPLC, LC, GLC and microtitre plates. Part 2: Photographs. J. Biolumin. Chemilumin. 1992;7:157–169. doi: 10.1002/bio.1170070302. [DOI] [PubMed] [Google Scholar]
- Stanley P.E. A survey of some commercially available kits and reagents which include bioluminescence or chemiluminescence for their operation—including immunoassays, hybridization, labels, probes, blots and ATP-based rapid microbiology: Products from more than 40 companies. J. Biolumin. Chemilumin. 1993;8:51–63. doi: 10.1002/bio.1170080202. [DOI] [PubMed] [Google Scholar]
- Stanley P.E. Commercially available luminometers and imaging devices for low-light measurements and kits and reagents utilizing bioluminescence or chemiluminescence: Survey update 1. J. Biolumin. Chemilumin. 1993;8:237–240. doi: 10.1002/bio.1170080502. [DOI] [PubMed] [Google Scholar]
- Stauber S., Siegel G., Janitschke K., Schulze-Forster K., Simon D. Detection of polymerase chain reaction (PCR) products of Toxoplasma gondii by enhanced chemiluminescence (ECL) J. Biolumin. Chemilumin. 1991;6:283. [Google Scholar]
- Suzuki K., Okamoto N., Watanabe S., Kano T. Chemiluminescent microtiter method for detecting PCR amplified HIV-1 DNA. J. Virol. Meth. 1992;38:113–122. doi: 10.1016/0166-0934(92)90174-c. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Craddock B.P., Kano T., Steigbigel R.T. Chemiluminescent enzyme-linked immunoassay for reverse transcriptase, illustrated by detection of HIV reverse transcriptase. Anal. Biochem. 1993;210:277–281. doi: 10.1006/abio.1993.1196. [DOI] [PubMed] [Google Scholar]
- Tham K.M., Stanislawek W.L. Detection of chicken anaemia agent DNA sequences by the polymerase chain reaction. Arch. Virol. 1992;127:245–255. doi: 10.1007/BF01309588. [DOI] [PubMed] [Google Scholar]
- Tham K.M., Stanislawek W.L. Polymerase chain reaction amplification for direct detection of chicken anemia virus DNA in tissues and sera. Avian Dis. 1992;36:1000–1006. [PubMed] [Google Scholar]
- Thorpe G.H.G., Kricka L.J., Moseley S.B., Whitehead T.P. Phenols as enhancers of the chemiluminescent horseradish peroxidase-luminol-hydrogen peroxide reaction: Application in luminescence-monitored enzyme immunoassays. Clin. Chem. 1985;31:1335–1341. [PubMed] [Google Scholar]
- Tumolo A., Nguyen Q., Witney F., Murphy O.J., Voyta J.C., Bronstein I. Detection of DNA on membranes with alkaline phosphatase-labeled probes and chemiluminescent AMPPD substrate. In: Kricka L.J., editor. Nonisotopic DNA Probe Techniques. Academic Press; San Diego: 1992. pp. 127–145. [Google Scholar]
- Urdea M.S., Running J.A., Horn T., Clyne J., Ku L., Warner B.D. A novel method for the rapid detection of specific nucleotide sequences in crude biological samples without blotting or radioactivity; application to the analysis of hepatitis B virus in human serum. Gene. 1987;61:253–264. doi: 10.1016/0378-1119(87)90189-2. [DOI] [PubMed] [Google Scholar]
- Urdea M.S., Kolberg J., Clyne J., Running J.A., Besemer D., Warner B., Sanchez-Pescador R. Application of a rapid non-radioisotopic nucleic acid analysis system to the detection of sexually transmitted disease-causing organisms and their associated antimicrobial resistances. Clin. Chem. 1989;35:1571–1575. [PubMed] [Google Scholar]
- Urdea M.S., Kolberg J., Warner B.D., Horn T., Clyne J., Ku L., Running J.A. A novel method for the rapid detection of hepatitis B virus in human serum samples without blotting or radioactivity. In: van Dyke K., van Dyke R., editors. Luminescence Immunoassay and Molecular Applications. CRC Press; San Diego: 1990. pp. 275–292. [Google Scholar]
- Vlaspolder F., Matsaers J.A., Blog F., Notowicz A. Value of a DNA probe assay (Gen-Probe) compared with that of culture for diagnosis of gonococcal infection. J. Clin. Microbiol. 1993;31:107–110. doi: 10.1128/jcm.31.1.107-110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieger A.M., Medenblik A.M., van-Gijlswijk R.P., Tanke H.J., van-der-Ploeg M., Gratama J.W., Raap A.K. Quantitation of polymerase chain reaction products by hybridization-based assays with fluorescent, colorimetric, or chemiluminescent detection. Anal. Biochem. 1992;205:1–7. doi: 10.1016/0003-2697(92)90570-w. [DOI] [PubMed] [Google Scholar]
- Wages J.M., Dolenga L., Fowler A.K. Electrochemiluminescence detection and quantitation of PCR-amplified DNA. Amplifications. 1993;10:1–3. [Google Scholar]
- Weare J.A., Robertson E.F., Madsen G., Hu R., Decker R.H. Improvement in the specificity of assays for detection of antibody to hepatitis B core antigen. J. Clin. Microbiol. 1991;29:600–604. doi: 10.1128/jcm.29.3.600-604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks I., Behesti I., McCapra F., Campbell A.K., Woodhead J.S. Acridinium esters as high-specific-activity labels in immunoassay. Clin. Chem. 1983;29:1474–1479. [PubMed] [Google Scholar]
- Welnicki M., Hiruki C. Chemiluminescent assay for the detection of viral and viroid RNAs using digoxigenin labelled probes. J. Biolumin. Chemilumin. 1993;8:127. [Google Scholar]
- Whitehead T.P., Thorpe G.H.C., Carter T.J.N., Groucutt C., Kricka L.J. Enhanced luminescence procedure for sensitive determination of peroxidase-labelled conjugates in immunoassay. Nature. 1983;305:158–159. [Google Scholar]
- Yang J.Q., Tata P.V., Park-Turkel H.S., Waksal H.W. The application of AmpliProbe in diagnostics. BioTechniques. 1991;11:392–397. [PubMed] [Google Scholar]
- Zachar V., Mayer V., Aboagye-Mathiesen G., Norskov-Laruitsen N., Ebbesen P. Enhanced chemiluminescence-based hybridization analysis for PCR-mediated HIV-1 DNA detection offers an alternative to 32P-labelled probes. J. Virol. Meth. 1991;33:391–395. doi: 10.1016/0166-0934(91)90039-3. [DOI] [PubMed] [Google Scholar]


