Background
Imaging mass spectrometry enables in situ label-free detection of thousands of metabolites from intact tissue samples. However, automated steps for multi-omics analyses and interpretation of histological images have not yet been implemented in mass spectrometry data analysis workflows. The characterization of molecular properties within cellular and histological features is done via time-consuming, non-objective, and irreproducible definitions of regions of interest, which are often accompanied by a loss of spatial resolution due to mass spectra averaging.
Methods: We developed a new imaging pipeline called Spatial Correlation Image Analysis (SPACiAL), which is a computational multimodal workflow designed to combine molecular imaging data with multiplex immunohistochemistry (IHC). SPACiAL allows comprehensive and spatially resolved in situ correlation analyses on a cellular resolution. To demonstrate the method, matrix-assisted laser desorption-ionization (MALDI) Fourier-transform ion cyclotron resonance (FTICR) imaging mass spectrometry of metabolites and multiplex IHC staining were performed on the very same tissue section of mouse pancreatic islets and on human gastric cancer tissue specimens. The SPACiAL pipeline was used to perform an automatic, semantic-based, functional tissue annotation of histological and cellular features to identify metabolic profiles. Spatial correlation networks were generated to analyze metabolic heterogeneity associated with cellular features.
Results: To demonstrate the new method, the SPACiAL pipeline was used to identify metabolic signatures of alpha and beta cells within islets of Langerhans, which are cell types that are not distinguishable via morphology alone. The semantic-based, functional tissue annotation allows an unprecedented analysis of metabolic heterogeneity via the generation of spatial correlation networks. Additionally, we demonstrated intra- and intertumoral metabolic heterogeneity within HER2/neu-positive and -negative gastric tumor cells.
Conclusions: We developed the SPACiAL workflow to provide IHC-guided in situ metabolomics on intact tissue sections. Diminishing the workload by automated recognition of histological and functional features, the pipeline allows comprehensive analyses of metabolic heterogeneity. The multimodality of immunohistochemical staining and extensive molecular information from imaging mass spectrometry has the advantage of increasing both the efficiency and precision for spatially resolved analyses of specific cell types. The SPACiAL method is a stepping stone for the objective analysis of high-throughput, multi-omics data from clinical research and practice that is required for diagnostics, biomarker discovery, or therapy response prediction.
Keywords: Imaging mass spectrometry, Multiplex immunohistochemistry, In situ metabolomics, Tissue annotation, Pixel-accurate analysis
Graphical abstract
Highlights
-
•
Novel method enables phenotype-guided in situ metabolomics on intact tissue sections.
-
•
Metabolic heterogeneity can be objectified under (patho-)physiological conditions.
-
•
Innovative approach for tissue-based, preclinical, and clinical research.
1. Introduction
Computational automation of routine tasks and artificial intelligence guided analyses rapidly gain significance with the increasing amount of data generated from single tissue samples [1]. With the rise of digital pathology, a major objective for precision medicine is the integration of morphological and molecular imaging data from multi-omics studies [2]. Matrix-assisted laser desorption-ionization (MALDI) imaging mass spectrometry (IMS) can be used for in situ imaging of metabolites from frozen or formalin-fixed, paraffin-embedded (FFPE) tissue samples [3]. Providing spatially resolved and label-free detection of hundreds to thousands of molecules within a single tissue section, MALDI imaging has proven to be an invaluable tool for digital histopathology. However, the spatial resolution of molecule distributions is often not fully utilized. For instance, cell- and tissue-specific structures, such as tumors, are frequently analyzed by manually annotating the respective areas on whole-slide images to mark so-called regions of interest (ROIs) [4]. The spatially resolved mass spectra in these regions are lost because only the mean or representative spectrum of each ROI is used for subsequent calculations. Such segmentation approaches fail to preserve molecular heterogeneity and spatial distribution, while whole-sample-based classifications fail for tissue sections comprised of cells belonging to different classes [5]. With the availability of tissue samples from large-scale clinical cohort studies, manual preprocessing steps can easily take weeks or months of work. Furthermore, manually annotating tumor regions is not only time-consuming, non-objective, and irreproducible, but also requires extensive histology knowledge that only expert pathologists possess. Additionally, it only permits the annotation of regions and cell types that are histologically distinguishable, while molecular alterations often do not manifest morphologically. Current efforts to utilize machine learning algorithms to automatically distinguish cell types [6] based on nothing more than hematoxylin and eosin tissue staining may work, for example, on tumor and stroma cells, but are not able to identify morphologically indistinguishable tumor subtypes.
Traditionally, when morphology alone is not sufficient, even clinical pathologists resort to immunohistochemistry (IHC) to localize proteins or peptides in a single tissue section [7]. Compared to efforts to define ROIs or distinguish cell types based on molecular distributions [[8], [9], [10]], immunostainings represent a method to precisely label specific cell types [11]. The technique is currently used to classify tumors or to perform structural tissue analyses to help pathologists establish a diagnosis [7]. IHC staining is commonly used for cell type labeling [12], but their potential for automated, semantic-based, functional tissue annotation and spatially resolved molecular analyses of heterogeneity is not fully utilized. In recent years, imaging mass spectrometry data and immunohistochemical staining have been successfully combined to increase the resolution of MALDI images [13] or to characterize individual dissociated cells [14], but no in situ tissue analysis with automatic identification of ROIs and data integration has been presented. While there is some software available for tissue image analysis, there currently is no method that integrates and analyzes the comprehensive molecular data from imaging mass spectrometry in combination with morphological, proteomic, and genetic information from other omics fields. The translation of imaging mass spectrometry into experimental clinical applications requires time-efficient data post-processing and comprehensive analyses of spatially resolved molecular information by avoiding expensive manual annotations or loss of resolution due to the generation of mean or representative spectra. In research, the use of immunostainings in combination with molecular data represent a significant improvement in scientific quality by solving the problem of time-consuming and irreproducible user-defined ROIs. In particular, the analysis of metabolic heterogeneity is hampered by pseudoheterogeneity originating from inaccuracies during manual annotation or by the use of consecutive tissue sections. The analysis of metabolic heterogeneity in tissues requires a strict coherence to consistent tissue and data preprocessing.
Here, we present our Spatial Correlation Image Analysis (SPACiAL) pipeline, a computational multimodal workflow to integrate molecular imaging mass spectrometry data with multiplex IHC staining from the very same tissue section to provide automated and reliable annotations and allow comprehensive and pixel-accurate correlation analyses of heterogeneity to combine data from multi-omics studies. The pipeline represents a starting point for the objective analysis of high-throughput data from clinical research and practice, which is required for tissue-based diagnostics and research.
To demonstrate the versatility and analytical power of the SPACiAL method, we deliberately chose two examples of molecular heterogeneity in both a physiological and a pathophysiological application: First, we performed a high-resolution analysis of islets of Langerhans in mouse pancreases. Phenotypic and functional beta cell heterogeneity has been shown to provide pancreatic islets with functional flexibility to adapt to physiological changes in the environment [15]. The metabolomic analysis of islet and islet cell heterogeneity requires in situ analyses of intact islets within tissue slices [16,17] and it has been insufficiently studied in their natural histological context. With SPACiAL, we distinguish alpha and beta cells and investigate the heterogeneity of different islets within one animal. Second, an analysis of tissue samples from patients with gastric cancer was carried out. The metabolomic, intratumoral, heterogeneous nature of the human epidermal growth factor receptor 2 (HER2/neu) is insufficiently studied in situ–especially in relation to gastric cancer–even though it is highly relevant for diagnostics and response to HER2/neu-based treatment. The SPACiAL pipeline was applied on tissue resection specimens and on a tissue microarray to distinguish HER2/neu-positive and -negative tumor cells and to investigate the molecular intra- and intertumoral heterogeneity. The multimodal approach utilizes pixel-wise molecular information to investigate metabolic heterogeneity via spatial correlation networks from cell populations automatically identified by multiplex immunohistochemical analysis.
2. Methods
2.1. Tissue specimens and Fourier-transform ion cyclotron resonance (FTICR) MALDI IMS analysis
Pancreas/islets of Langerhans were obtained from a C57BL/6 N mouse and the sample was flash frozen in liquid nitrogen until measurement. The animal was provided ad libitum access to food and water. All animal studies were conducted in accordance with German animal welfare legislation and approved by the government of Upper Bavaria. FFPE tissue patient samples of gastric cancer were collected between 1995 and 2018 at the University of Leipzig and at the Department of Surgery, Klinikum Rechts der Isar, Munich, Germany. The resection specimens were processed in a highly standardized manner and fixed for 12–24 h in 10% neutral buffered formalin, followed by tissue dehydration and paraffin embedding with fully automated systems. The study was approved by the local Ethics Committees. All patients provided informed, signed consent.
Tissue preparation steps for MALDI imaging analysis was performed as previously described [3,18,19]. In brief, frozen (12 μm, Leica Microsystems, CM1950, Germany) and FFPE sections (4 μm, Microm, HM340E, Thermo Fisher Scientific, USA) were mounted onto indium-tin-oxide (ITO)-coated glass slides (Bruker Daltonik, Bremen, Germany) pretreated with 1:1 poly-l-lysine (Sigma Aldrich, Munich, Germany) and 0.1% Nonidet P-40 (Sigma). The air-dried tissue sections were spray-coated with 10 mg/ml of 9-aminoacridine hydrochloride monohydrate matrix (Sigma–Aldrich, Munich, Germany) in 70% methanol using the SunCollect™ sprayer (Sunchrom, Friedrichsdorf, Germany). Prior matrix application, FFPE tissue sections were incubated additionally for 1 h at 70 °C and deparaffinized in xylene (2 × 8 min). Spray-coating of the matrix was conducted in eight passes (ascending flow rates 10 μl/min, 20 μl/min, 30 μl/min for layers 1–3, and layers 4–8 with 40 μl/min), utilizing 2 mm line distance and a spray velocity of 900 mm/min.
Metabolites were detected in negative-ion mode on a 7 T Solarix XR FTICR mass spectrometer (Bruker Daltonik) equipped with a dual ESI-MALDI source and a SmartBeam-II Nd: YAG (355 nm) laser. Data acquisition parameters were specified in ftmsControl software 2.2 and flexImaging (v. 5.0) (Bruker Daltonik). Mass spectra were acquired in negative-ion mode covering 75–1,100, with a 1 M transient (0.367 s duration) and an estimated resolving power of 49,000 at m/z 200,000. The laser operated at a frequency of 1,000 Hz utilizing 200 laser shots per pixel with a pixel resolution of 15 μm (islets of Langerhans) and 60 μm (gastric cancer), respectively. l-Arginine was used for external calibration in the ESI mode. On-tissue double mass spectrometry (MS/MS) was conducted on islets of Langerhans from the consecutive mouse pancreatic tissue section using continuous accumulation of selected ions’ mode and collision-induced dissociation (CID) in the collision cell (Supplementary Figure 17). MS/MS spectra were analyzed by Bruker Compass DataAnalysis 5.0 (Build 203.2.3586).
2.2. Multiplex fluorescent immunohistochemical staining
After MALDI IMS analysis, 9-aminoacridine matrix was removed with 70% ethanol for 5 min from tissue sections followed by immunohistochemical staining. Pancreatic islets were analyzed by double staining for insulin [Insulin-monoclonal rabbit anti-insulin (1:800), catalog no. 3014, Cell Signaling Technology, Germany; AF750-goat anti-rabbit (1:100), catalog no. A21039, Thermo Fisher Scientific, US] and glucagon [polyclonal guinea pig anti-glucagon (1:3,000), catalog no. M182, Takara, USA; biotinylated goat anti-guinea pig immunoglobulin G (IgG, 1:100), catalog no. BA-7000, Vector Laboratories, US; streptavidin-Cy3, catalog no. SA1010, Thermo Fisher Scientific].
Double staining of human gastric cancer tissue specimens and a tissue microarray was performed using HER2 [polyclonal rabbit anti-human c-erbB-2 oncoprotein (1:300), catalog no. A0485, DAKO, CiteAb Ltd., UK] and pan-cytokeratin [monoclonal mouse pan cytokeratin plus [AE1/AE3+8/18] (1:75), catalog no. CM162, Biocare Medical, US]. Signal detection was conducted using fluorescence-labeled secondary antibodies [Goat Anti-rabbit IgG (H + L)-Cross-Adsorbed Secondary Antibody-DyLight 633 (1:200), catalog no. 35563; and Goat Anti-Mouse IgG (H + L)-Cross-Adsorbed Secondary Antibody-Alexa Fluor 750 (1:100), catalog no. A-21037, both Thermo Fisher Scientific]. Nuclei were identified with Hoechst 33342 in all stainings. Fluorescence stainings were scanned with an AxioScan.Z1 digital slide scanner (Zeiss) equipped with a 20x magnification objective and visualized with the software ZEN 2.3 blue edition (Zeiss). Multi-images were exported as TIF files. Additionally, tissue sections were stained with hematoxylin and eosin after MALDI and IHC for internal visual validation.
2.3. Peak picking
The Bruker software flexImaging (v. 5.0) was used to export all root mean square normalized mass spectra as processed imzML files. An in-house python 3 pipeline was written to perform pixel-wise and parallelized peak picking. For each coordinate (i.e., spectrum), the peak picking pipeline began by resampling the mass () and intensity values between 75 and 1,100 Dalton (Da) with a step size of 0.0005 Da. Intensity values were resampled by choosing the maximum intensity per window. Noise levels were estimated for windows of 10 Da, and all peaks falling below their respective noise level were filtered. The noise level was calculated as 2.2 times the 85th percentile of the intensity values within the window. If fewer than 200 intensities fell within 1 window, which frequently happens in the higher mass range, then their neighboring windows are considered until at least 200 intensities can be used for the calculation. Since the noise level is expected to increase with the value and to avoid extreme noise level fluctuations, the level of the first and last window were used as upper bounds. After noise-filtering, only local maxima were kept as preliminary peaks. Preliminary peaks within each spectrum were merged as previously described [18]. The merged peaks of all coordinates were then aligned, if their distance did not exceed with . Peaks that occur in less than 0.5% of the spectra were filtered. Picked peaks were saved as an imzML file. Noise levels and the peak pickings were verified by manual inspection of random sample coordinates.
2.4. Metabolite annotation
The Human Metabolome Database [20] (HMDB, v. 4.0) was used to functionally annotate values. The metabolite XML file was downloaded for offline use and a local PostgreSQL (v. 11) database was set up. Molecules were annotated by allowing M−H, M–H2O–H, M + Na–2H, M + Cl and M + K–2H as negative adducts with a mass tolerance of 4 ppm. A keyword search was performed on the description text to filter compounds with multiple annotations. Specifically, compounds with indications of being drug-, plant-, food-, or bacteria-specific were filtered stringently.
2.5. Image co-registration
The imzML file of picked peaks was used to create a master image of the MALDI measurement region (imzML-grid). All additional images were precisely co-registered onto this image, allowing an exact integration and correlation of molecular MALDI data with immunostainings. The co-registration was done with the Landmark Correspondences plugin of FIJI ImageJ [21] (v. 1.52p). Alternatively, co-registration is also feasible with Adobe Photoshop CC 2019 or the GNU Image Manipulation Program (GIMP, v.2.10.8). A gray-scale tissue overview image and measurement points were exported with flexImaging (v. 5.0) and then fitted onto the master image. The integration of mass spectra and image data is done by co-registering the tissue scanned subsequent to MALDI imaging mass spectrometry and mapping the matrix ablation marks to the imzML-grid. The 4′,6-diamidino-2-phenylindole (DAPI) staining and all other stainings were finally fitted onto the precisely co-registered tissue image.
2.6. Region inclusion/exclusion criteria
After co-registration, all images had the exact same dimension and resolution. SPACiAL now offers the option to create a mask, where the user can manually label regions that should be excluded from subsequent analyses. Such regions may comprise tissue folds, swept away tissue, artifacts, or regions that are generally of no interest.
To integrate the data from all images, they have to be scaled to the exact MALDI measurement resolution by averaging the color values per x/y-coordinate. The IHC images are then converted into numerical matrices comprised of values corresponding to the lightness values for each pixel. SPACiAL can create images to allow validation of automatically defined ROIs (e.g., HER2/neu-positive tumor regions).
2.7. Pixel-accurate definition of HER2/neu-positive tumor regions
FFPE tissue sections of human gastric cancer samples were used to analyze the metabolic heterogeneity within HER2/neu-positive tumor regions. Tumor cells were annotated via the pan-cytokeratin staining. They were then classified as HER2/neu positive, if they also exhibited a positive signal in the HER2/neu staining. Otherwise, they were classified as HER2/neu-negative.
2.8. Networks
Correlation networks were created with Cytoscape [22] (v. 3.7.1). In all networks, nodes represent metabolites with node sizes corresponding to the mean intensity. Edges represent spatial correlations with line thickness and opacity increasing with the correlation coefficient. Nodes were colored red if their metabolites take part in glycolysis or they were colored depending on the molecule super class defined in HMDB [lipids and lipid-like molecules (yellow); nucleosides, nucleotides, and analogs (light red); organic acids and derivatives (green); organoheterocyclic compounds (lime green); alkaloids and derivatives (pink); organic oxygen compounds (blue); benzenoids (violet); phenylpropanoids and polyketides (orange); others (gray)]. All networks were either visualized using the yFiles circular layout or edge-weighted spring-embedded layout using the absolute value of the correlation coefficient. For the pancreatic islet cells, circular networks were generated by filtering edges with a coefficient smaller than 0.7 and by only visualizing direct neighbors of glucose 6-phosphate. All islets were located on the same tissue slide and were analyzed concurrently. The multiplex staining of the complete tissue is shown in Supplementary Figure 1. All remaining networks were generated by showing metabolites with at least one correlation coefficient larger than 0.5, but without filtering edges. The complete networks are shown in Supplementary Figures 15 and 16. The multiplex staining of the complete tissues are shown in Supplementary Figures 8, 10, 12, and 14.
2.9. Statistical analyses
For the networks, pairwise Spearman rank-order correlations (Python 3.7, SciPy 1.2.0) were calculated between annotated metabolites using their intensities, and the resulting p-values were adjusted with Benjamini/Hochberg correction (Python 3.7, StatsModels 0.9.0). For the pancreatic islet cells, circular networks were generated by filtering edges with a coefficient smaller than 0.7. Network metrics (Supplementary Tables 1–3) were calculated using Cytoscape's plugin NetworkAnalyzer.
Metabolites localized predominantly on alpha or beta cells in islets of Langerhans were identified by using the Mann–Whitney U-test (Python 3.7, SciPy 1.2.0). The p-values were adjusted with Benjamini/Hochberg correction (Python 3.7, StatsModels 0.9.0). The number of cell-type specific pixels per islet ranges between 59 and 194 for alpha cells and between 112 and 228 for beta cells. The Python 3.7 package NumPy 1.15.4 was used to calculate statistics for the intensity distributions of ADP, cholesterol sulfate, and 3-O-sulfogalactosylceramide in the islets of Langerhans (Supplementary Figures 5–7).
3. Results and discussion
3.1. The SPACiAL workflow for immunohistochemistry-guided imaging mass spectrometry
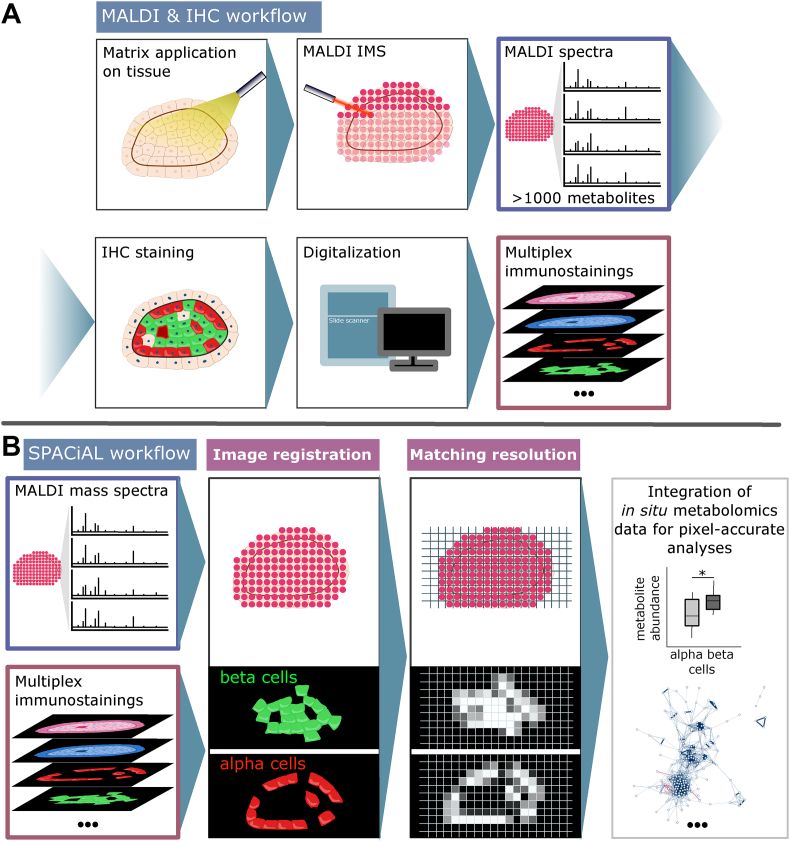
The SPACiAL pipeline comprises a series of MALDI data and image processing steps to combine molecular data with morphological and immunophenotypic information from IHC stainings or other imaging data. Immunostaining following MALDI imaging has previously been shown to be feasible [23,24]; hence the entire workflow works on the very same tissue section. Here, we have demonstrated that even multiplex immunostainings are entirely possible after MALDI imaging of the very same tissue section, which allows automatic data integration of morphological and spatially resolved in situ data of thousands of molecules via the SPACiAL method. The entire tissue and data pre-processing workflow preceding the application of the SPACiAL algorithm includes matrix coating of tissue sections, MALDI imaging, peak picking, matrix removal, IHC staining, and image digitalization, which is shown schematically for an islet of Langerhans with glucagon, insulin and DAPI staining (Figure 1A). SPACiAL then uses MALDI imaging files to create a reference image for subsequent co-registration of the molecular data with other image information (Figure 1B). The digitized and co-registered immunostaining images are scaled to match the exact MALDI resolution and then converted into numerical data without loss of spatial resolution. This ultimately allows pixel-accurate, objective tissue annotations based on semantics and function, which is shown here as an example for alpha and beta cells stained with glucagon (red) and insulin (green), respectively, and one metabolite (yellow) co-localizing with alpha cells (Figure 2). The SPACiAL pipeline paves the way for further statistical calculations and for the analysis of tissue heterogeneity and previously infeasible molecular in situ analyses of cell subpopulations within intact tissue sections. To illustrate the versatility and analytical power of the SPACiAL pipeline, it was applied on two datasets; a physiological use case and a pathophysiological use case.
Figure 1.
Workflow of immunohistochemistry-guided in situ metabolomics using the example of an islet of Langerhans. A: MALDI and IHC workflow starting with matrix application on tissue sections, MALDI imaging and data processing including peak picking and annotation. The matrix is then removed for subsequent multiplex immunohistochemical staining of the very same tissue section using DAPI, glucagon, and insulin. The individual stainings are digitized with a slide scanner. B: The SPACiAL pipeline integrates molecular MALDI data and immunohistochemical data. The IHC images need to be co-registered to the coordinates of the mass spectra per pixel. The MALDI data file is used to generate an image of the measurement region, which can be used for precise co-registration with the tissue image and tissue stainings. Once co-registered, the staining images are scaled to match the resolution of the measurement and color values per pixel are used for the definition of regions or for pixel-accurate analyses of metabolic correlations or heterogeneity.
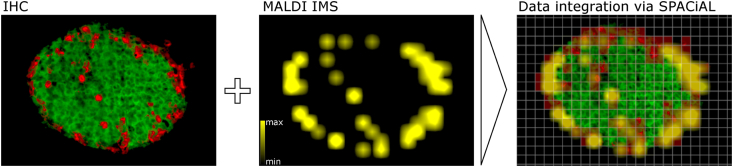
Figure 2.
Multi-omics data integration via the SPACiAL method. Left: Islet of Langerhans with immunohistochemical staining (glucagon in red, insulin in green). Middle: Spatial distribution of 3-O Sulfogalactosylceramide (m/z 778.5147). Right: Data integration via SPACiAL, utilizing the IHC stainings to automatically identify alpha cells (semi-transparent, pixelated staining in red) and correlating metabolites. Lateral MALDI resolution (pixel): 15 μm.
3.2. SPACiAL analysis of metabolic heterogeneity within and between islets of Langerhans
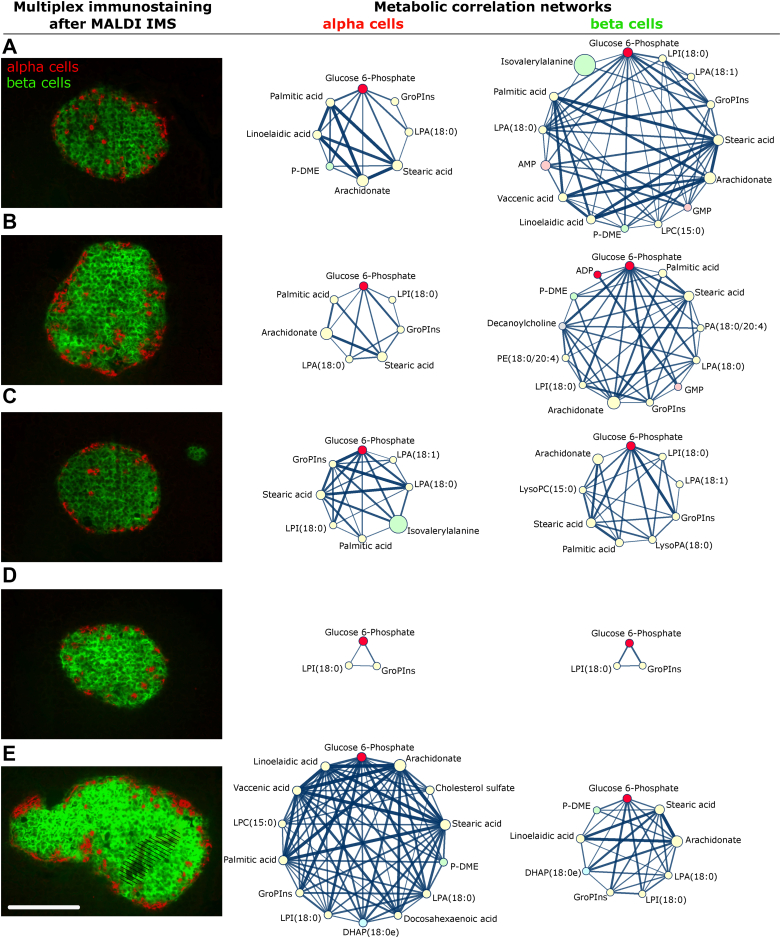
To demonstrate the SPACiAL pipeline, it was applied on islets of Langerhans in the pancreas of a wild-type mouse to distinguish the glucagon-releasing alpha and insulin-releasing beta cells and to investigate the heterogeneity of different islets within one animal. Previous studies highlight heterogeneity as a fundamental characteristic of pancreatic islets [25]. Beta cells are functionally heterogeneous and display different activity patterns in response to glucose stimulation or the ability to secret insulin [26]. The metabolic heterogeneity within automatically detected alpha and beta cells was analyzed in detail for the glucose metabolism. The islets of Langerhans – originating from one tissue section (Supplementary Figures 1–4) – were imaged with both high lateral (15 μm) and high mass resolution. Correlation networks were created to identify functional relationships of metabolites with glucose 6-phosphate and to assess metabolic heterogeneity within and between individual islets of Langerhans (Figure 3, Supplementary Table 1). Glucose 6-phosphate was chosen as a relevant example because it is an important intermediate in the glycolysis, gluconeogenesis, and pentose phosphate pathways.
Figure 3.
Metabolic heterogeneity within and between islets of Langerhans in a pancreatic tissue section of one mouse (A–E). The column on the left shows multiplex immunostainings after MALDI imaging mass spectrometry. Alpha cells (red) and beta cells (green) are stained with glucagon and insulin, respectively. A tissue fold in the fifth islet (E) was excluded from analyses (dashed). The second and third columns show spatial correlation networks for metabolites in alpha and beta cells, respectively. Nodes and edges represent compounds and their spatial correlation. The networks shown here only include direct neighbors of the glucose 6-phosphate node and edges representing a correlation coefficient of at least 0.7. Scale bar, 150 μm. Abbreviations: adenosine monophosphate (AMP), guanosine monophosphate (GMP), phosphatidic acid (PA), phosphatidylethanolamine (PE), lysophosphatidic acid (LPA), lysophospholipid (LPC), lysophosphatidylinositol (LPI), dihydroxyacetone phosphate (DHAP), glycerophosphoinositol (GroPIns), phosphodimethylethanolamine (P-DME).
Clear differences regarding network size were found between the islets and islet cell populations, reflecting differential metabolic states (Figure 3). For example, the alpha and beta cell network of the islet in Figure 3D indicates a low dependency on glucose metabolism, with only two metabolites showing a significant correlation to glucose 6-phosphate. Within networks from other islets, a variety of metabolites, including lipids, nucleotides, amino acid, and analogs, correlate with glucose 6-phosphate (Figure 3). The highest number of correlations were found in beta and alpha cell populations of the islets A and E, respectively, indicating a high dependency on glucose metabolism. The spatial distribution of lipid-associated compounds, such as palmitic acid, stearic acid, lysophosphatidylinositol (LPI), and lysophosphatidic acid (LPA) were found to be correlated almost consistently. Other compounds, such as phosphodimethylethanolamine (P-DME) or glycerophosphoinositol (GroPIns), were found to inconsistently correlate with glucose 6-phosphate.
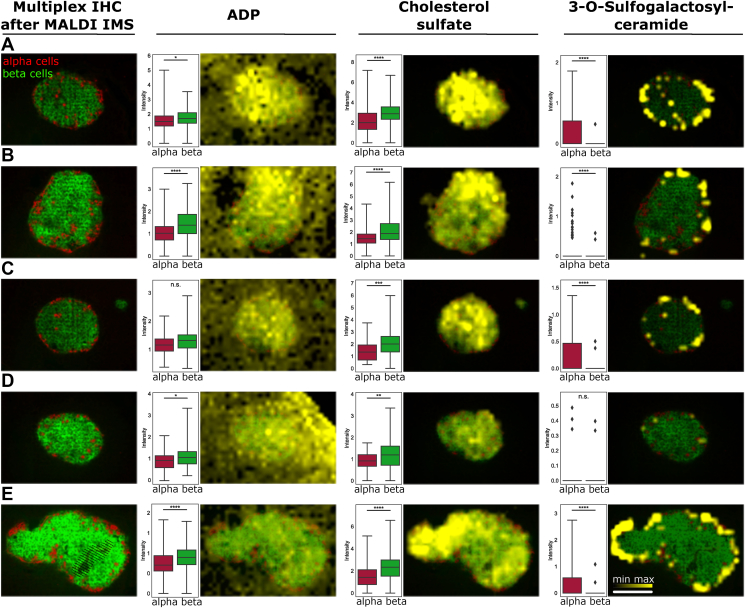
Metabolic signatures related to specific cell types and subpopulations can now easily be extracted with SPACiAL. Alpha and beta cells were defined automatically as ROIs, and metabolic differences between alpha and beta cells were assessed. Significant differences were detected for adenosine diphosphate (ADP), cholesterol sulfate, and 3-O-sulfogalactosylceramide (Figure 4). The presence of ADP, cholesterol sulfate, and 3-O-sulfogalactosylceramide was validated via MALDI FTICR on-tissue MS/MS using quadrupole collision-induced dissociation and comparison to standard compounds (Supplementary Figure 17). Not all islets reveal similar significant changes, also reflecting inter- and intra-islet metabolic heterogeneity. For instance, across four of the five measured islets, significantly higher ADP levels were detected in beta cells in comparison to alpha cells. Thus, the SPACiAL pipeline paves the way for in situ analyses of individual energy conditions of alpha and beta cells in each islet due to adenine nucleotide measurements. Cholesterol sulfate was found abundantly in beta cells, but it also exhibits a strong heterogeneous distribution between islets and even within cells of the same islet. Cholesterol sulfate is a component of the cell membrane [27] and in pancreatic beta cells, elevated intracellular cholesterol levels have been associated with reduced insulin secretion in mice [27,28]. Correlating with alpha cells, we found the 3-O-sulfogalactosylceramide. Sulfatides are glycosphingolipids which have been described in pancreatic islets with different abundances in alpha and beta cells [[29], [30], [31]].
Figure 4.
Multiplex immunohistochemistry-guided imaging mass spectrometry on islets of Langerhans to automatically distinguish morphologically indistinguishable cell types (A–E). Alpha and beta cells were stained with glucagon (red) and insulin (green), respectively. The spatial distributions of ADP, cholesterol sulfate and 3-O-sulfogalactosylceramide (sulfatide) are visualized (yellow). Pixel-wise intensity distributions are shown for alpha (red) and beta cells (green), respectively. See the methods section for details about the statistical analysis. Scale bar, 150 μm.
Finally, pronounced molecular heterogeneity, both within single and between different islets of Langerhans, is reflected by a varying distribution of metabolite abundances (Supplementary Figures 5–7). Between islets and between cell types, the standard deviation of metabolite intensities differs by a factor of between 2.28 and 14.25 (ADP 0.32–0.73, cholesterol sulfate 0.5–1.52; 3-O-sulfogalactosylceramide 0.04–0.57). Hence, even individual cells within one islet exhibit different metabolite compositions, possibly reflecting different metabolic or cell differentiation states.
The in situ analysis of metabolic heterogeneity within pancreatic islets is just one potential field of application for the SPACiAL pipeline. Metabolic data together with detailed spatial information can be exploited to assess the extent and modulation of alpha and beta cells in situ. Because both insulin and glucagon are dysregulated in pathophysiological conditions, such as diabetes, our pipeline is valuable for future studies. The analysis of different subpopulations of islets of Langerhans can help to illuminate underlying phenotypic mechanisms to expand our knowledge of cell function and develop new therapeutic strategies.
3.3. Intratumoral metabolic heterogeneity in gastric cancer
The SPACiAL strategy has been shown to be powerful for close-to single-cell analyses of the metabolome in tissues of animal models, but it is also valuable for clinically relevant tissue analyses regarding diagnostics, prognosis, and therapy response prediction. For this reason, we applied the SPACiAL pipeline for the analysis of intra- and intertumoral heterogeneity in gastric cancer. While we used glucagon and insulin to stain alpha and beta cells within a frozen pancreatic tissue section, here we used pan-cytokeratin as an epithelial marker to stain tumor cells and HER2/neu for tumor cell classification within human FFPE tissue sections.
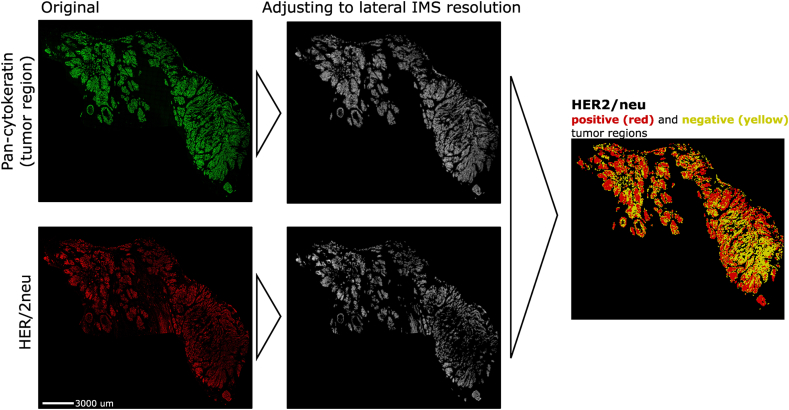
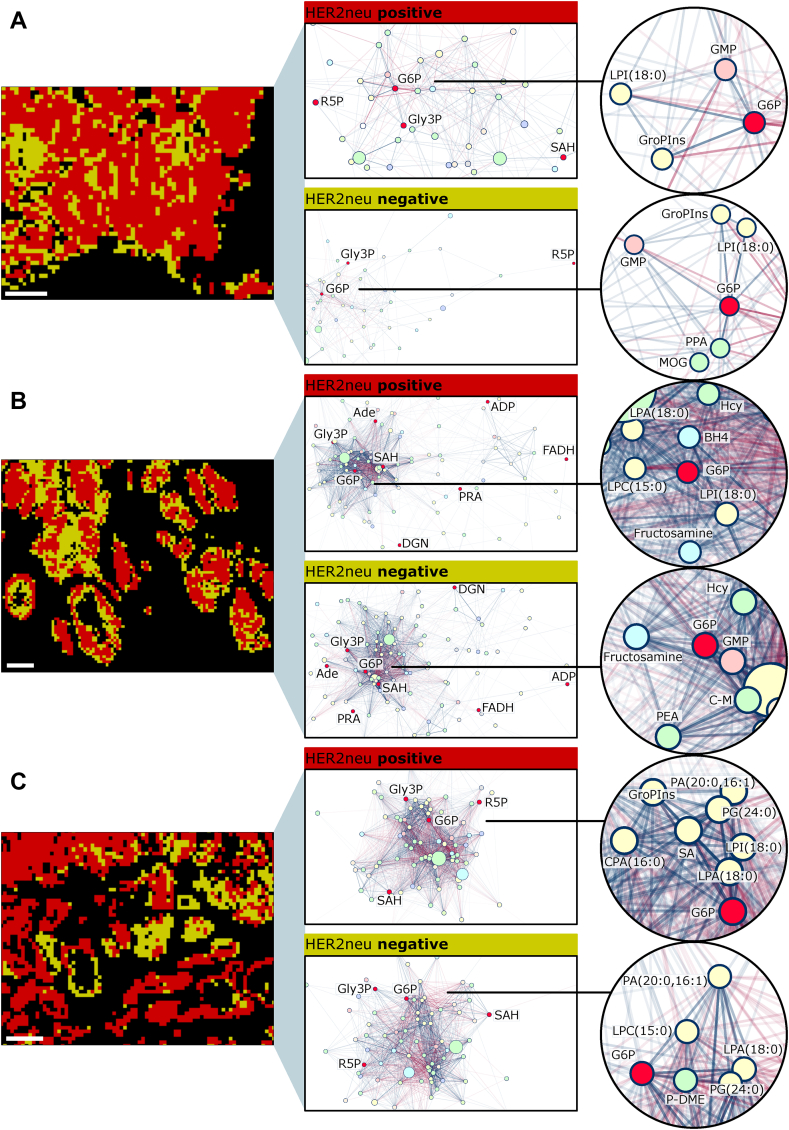
In gastric cancer, intratumoral HER2/neu heterogeneity is frequent, but its clinical significance remains open in terms of treatment with trastuzumab-chemotherapy [[32], [33], [34], [35], [36], [37], [38]]. The investigation of HER2/neu heterogeneity in gastric cancer in relation to the metabolic state of tumors is completely unexplored and may contribute to the improvement of treatment success. The SPACiAL pipeline was applied on tissue samples from three patients with gastric cancer to evaluate metabolic heterogeneity depending on the HER2/neu state. SPACiAL automatically determines the HER2/neu-positive and -negative tumor regions in a standardized way by evaluating expression values both in quantity on the basis of pixel intensity and localization by pixel co-localization (Figure 5). Regions displaying both pan-cytokeratin and HER2/neu signals are defined as HER2/neu-positive tumor regions, while regions displaying only a pan-cytokeratin signal are classified as HER2/neu-negative. Whole slide immunohistochemical stainings and regions defined as HER2/neu-positive (red) and -negative (yellow) are shown in Supplementary Figures 8–13. The pixel-accurate annotation allows an unprecedented analysis of metabolic heterogeneity within tumor cells based on metabolic correlation networks that were calculated for annotated metabolites detected and stringently filtered from gastric cancer tissue sections (Figure 6). For visualization purposes, a zoom-in of HER2/neu-positive and -negative tumor regions of Supplementary Figures 9, 11, and 13 is shown.
Figure 5.
Image processing workflow to define HER2/neu positive and negative tumor regions. Pan-cytokeratin (green) as an epithelial marker to stain tumor cells. HER2/neu positive cells are shown in red. Both stainings are adjusted to match the lateral imaging mass spectrometry resolution (60 μm) and combined to classify HER2/neu positive and negative tumor cells. Scale bar, 3000 μm.
Figure 6.
Intratumoral heterogeneity of spatially correlating metabolites in three human gastric cancer tissue sections, visualized via spatial correlation networks (A–C). Left: Close-up of the HER2/neu positive (red) and negative (yellow) tumor regions. Middle: Spatial correlation networks for metabolites. Edges represent positive (blue) and negative (red) spatial correlations between metabolites. Line thickness and transparency correspond to the correlation coefficient. Right: Zoom-in to glucose 6-phosphate. Abbreviations: glucose 6-phosphate (G6P), glycerol 3-phosphate (Gly3P), ribose 5-phosphate (R5P), S-adenosylhomocysteine (SAH), glycerophosphoinositol (GroPIns), adenosine diphosphate (ADP), guanosine monophosphate (GMP), adenine (Ade), 5-phosphoribosylamine (PRA), reduce flavin adenine dinucleotide (FADH), d-glutamine (DGN), cysteinyl-methionine (C–M), homocysteine (Hcy), phosphatidic acid (PA), phosphatidylglycerol (PG), cyclic phosphatidic acid (CPA), lysophosphatidic acid (LPA), lysophospholipid (LPC), lysophosphatidylinositol (LPI), phosphodimethylethanolamine (P-DME), phosphopantothenate (PPA), dimethyl-2-oxoglutarate (MOG), tetrahydrobiopterin (BH4), O-phosphoethanolamine (PEA), stearic acid (SA). Scale bar, 600 μm.
Since glucose plays a major role in altered energy metabolism in cancer [39], focusing on captured glucose as glucose 6-phosphate in the analysis of correlation networks provides insight into the complexity of the tumor biology regarding HER2/neu status. The spatial correlation networks comprise 67 to 171 metabolites (Supplementary Table 2) in HER2/neu-positive or -negative tumor regions, revealing intratumoral heterogeneity (Figure 6A–C). The number of correlating metabolites was identified in samples A and C, but the number and strength of pairwise spatial correlations differed, leading to different network structures; in particular, the network density is higher in C by a factor of approximately four (Supplementary Table 2). A majority of the correlating metabolites belong to the class of lipids and lipid-like molecules. In HER2/neu-positive regions of all patients, lysophosphatidylinositole (LPI) abundance correlates positively with glucose 6-phosphate. LPI is a bioactive lipid produced by the phospholipase A family, which is believed to play an essential role in several physiological and pathological processes [40]. As a ligand for the G-protein-coupled receptor GPR55, LPI may increase the glycolytic activity, since a GPR55 antagonist was shown to decrease glycolytic activity in cancer cell lines [41]. In one sample, glucose 6-phosphate formed a cluster together with numerous lipids (Figure 6C), while in the other two samples, the neighboring nodes belonged to different metabolic classes – including carbohydrates, dipeptides and glycosylamines (Figure 6A,B). The intratumoral heterogeneity was most prominent in tumor sample C, reflected by the difference in degree, average shortest path length, and clustering coefficient of HER2/neu-positive and -negative metabolic networks (Figure 6 and Table 1). Overall, the degree and clustering coefficient of the glucose 6-phosphate node varies more strongly between patient samples, than between HER2/neu-positive and -negative tumor regions within individual patient samples, reflecting intertumoral heterogeneity (Table 1).
Table 1.
Network metrics for the glucose 6-phosphate node. Sample identifiers (A, B and C) correspond to the samples in Figure 6. The degree of a node represents the number of neighbors in the network. The average shortest path length is the average, minimum number of edges between glucose 6-phosphate node and any other node. The clustering coefficient is a measure for the connections between neighboring nodes.
| Sample | HER2/neu state | Degree | Average shortest path length | Clustering coefficient |
|---|---|---|---|---|
| A | positive | 27 | 1.697 | 0.285 |
| A | negative | 22 | 1.942 | 0.398 |
| B | positive | 54 | 2.012 | 0.721 |
| B | negative | 55 | 1.964 | 0.689 |
| C | positive | 62 | 1.294 | 0.733 |
| C | negative | 41 | 1.5 | 0.472 |
3.4. Intertumoral metabolic heterogeneity in gastric cancer
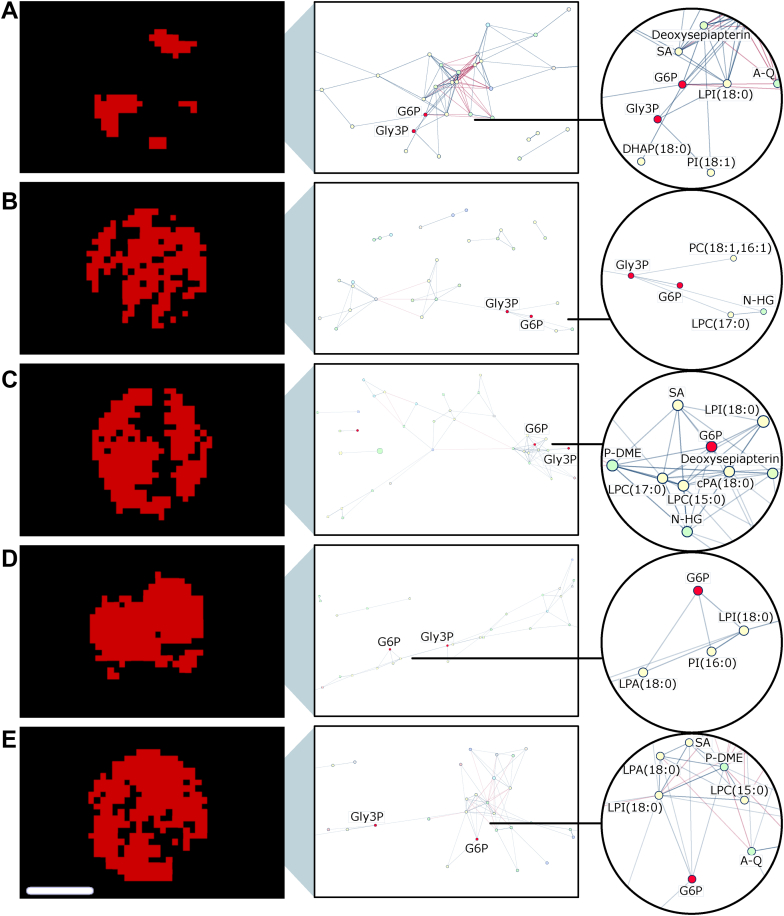
To additionally demonstrate the compatibility of SPACiAL for high-throughput multiplex phenotyping, metabolic correlation networks were created for gastric cancer patient tissues from an FFPE tissue microarray (Figure 7A–E). Networks on the extracted HER2/neu-positive tumor regions of 5 gastric cancer patients comprised 30 to 39 metabolites and exhibited diverse correlation patterns. Similar to the results from whole gastric cancer resection specimens, most of the correlating metabolites are lipids. An altered lipid metabolism has been described previously in a HER2/neu-positive breast cancer model [42]. Thus, a changed lipid metabolism may be associated with a high positive correlation of individual lipids to glucose 6-phosphate in human gastric cancer patients.
Figure 7.
Intertumoral heterogeneity of metabolites in five tissue cores from HER2/neu positive patients with gastric cancer, visualized via spatial correlation networks (A–E). Edges represent positive (blue) and negative (red) spatial correlations between metabolites. Line thickness and transparency correspond to the correlation coefficient. Right: Zoom-in to glucose 6-phosphate. Abbreviations: glucose 6-phosphate (G6P), glycerol 3-phosphate (Gly3P), phospholipid (PC), phosphatidylinositol (PI), cyclic phosphatidic acid (CPA), lysophosphatidic acid (LPA), lysophospholipid (LPC), lysophosphatidylinositol (LPI), phosphodimethylethanolamine (P-DME), stearic acid (SA), dihydroxyacetone phosphate (DHAP), alanylglutamine (A–Q), histidinyl-glycine (N-HG). Scale bar, 600 μm.
The diversity between metabolic correlation networks in individual patients demonstrates high intertumoral heterogeneity of HER2/neu-positive gastric cancer tissue (also see network metrics in Supplementary Table 3). The novel pipeline is a starting point for intra- and intertumoral heterogeneity analyses, enabling simultaneous analysis of distinct tissue and cellular compartments. The spatially resolved information from the molecular analysis has been used in our study to generate correlation networks between metabolites within ROIs that are automatically defined by immunohistochemical staining.
4. Conclusions
Our SPACiAL workflow integrates molecular MALDI imaging mass spectrometry data with IHC staining to facilitate automatic, reliable, and pixel-accurate annotation of specific cell types. In this context, the phenotypical information provided by IHC complements in situ molecular information for cell type-specific evaluation. The pipeline was demonstrated for both physiological and pathophysiological applications to investigate metabolic heterogeneity in alpha and beta cells from islets of Langerhans of a mouse model and in HER2/neu-positive tumor cells from patients with gastric cancer.
Glucagon-releasing alpha and insulin-releasing beta cells of different pancreatic islets within one animal were automatically annotated, demonstrating the basic functionality of the SPACiAL pipeline as a tool for objective IHC-guided annotation of otherwise histologically indistinguishable cell types. The pixel-accurate annotation and analysis of metabolites allows previously infeasible assessments of metabolomic heterogeneity between islets of Langerhans. Additionally, tissue samples from patients with gastric cancer were chosen to demonstrate the methodological advantages of SPACiAL for the analysis of intra- and intertumoral heterogeneity. The SPACiAL strategy can be extended by integrating other in situ datasets from tissue analytic platforms, since all spatially resolved information of a tissue section can be integrated in this pipeline (e.g., morphometrics, fluorescence in situ hybridization, and imaging mass cytometry). Prospectively, the application can also be useful for the automatic readout of regions of interest for metabolite quantification on an absolute, rather than on a relative scale. Quantification is a major topic of investigation in the targeted MALDI IMS field concentrating on the analysis of a subset of metabolites. Furthermore, the workflow was demonstrated to be compatible with both frozen and FFPE tissue samples. With SPACiAL, hundreds of distinct samples within tissue microarrays can be analyzed simultaneously. In contrast to the traditional analysis of mean spectra per ROI, SPACiAL allows in-depth and full use of available data without loss of resolution. With the spatial correlation networks of metabolites and the comparative approach to investigate islet cell heterogeneity, we demonstrate one of the many possibilities to utilize MALDI data. Combining the data from multi-omics studies, the pipeline represents an important starting point for the objective analysis of high-throughput data from large-scale clinical cohort studies, which are required for artificial intelligence-guided diagnostics, biomarker discovery, or therapy prediction.
We expect that further development of computational techniques for automatic feature identification based on multi-parameter molecular imaging data will remain an important area of ongoing investigation, and the results we present provide a useful framework and resource for advancing future studies.
Funding
The study was funded by the Ministry of Education and Research of the Federal Republic of Germany (BMBF; Grant Nos. 01ZX1610B and 01KT1615), the Deutsche Forschungsgemeinschaft (Grant Nos. SFB 824TP C04, CRC/TRR 205 S01) and the Deutsche Krebshilfe (No. 70112617) to A. Walch. Funding was provided through the Impulse and Networking Fund of the Helmholtz Association and the Helmholtz Zentrum München (Helmholtz Enterprise-2018-6) to A. Buck.
Data availability
Imaging mass spectrometry data for mouse pancreatic islets of Langerhans are provided at Zenodo.org (DOI:10.5281/zenodo.3607915) [43].
Acknowledgments
We thank Ulrike Buchholz, Claudia-Mareike Pflüger, Andreas Voss, Cristina Hübner Freitas, and Elenore Samson for excellent technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.01.017.
Contributor Information
Achim Buck, Email: achim.buck@helmholtz-muenchen.de.
Axel Walch, Email: axel.walch@helmholtz-muenchen.de.
Declaration of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Colling R., Pitman H., Oien K., Rajpoot N., Macklin P., Snead D. Artificial intelligence in digital pathology: a roadmap to routine use in clinical practice. The Journal of Pathology: Path. 2019:5310. doi: 10.1002/path.5310. [DOI] [PubMed] [Google Scholar]
- 2.Huang S., Chaudhary K., Garmire L.X. More is better: recent progress in multi-omics data integration methods. Frontiers in Genetics. 2017;8(JUN):1–12. doi: 10.3389/fgene.2017.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck A., Ly A., Balluff B., Sun N., Gorzolka K., Feuchtinger A. High-resolution MALDI-FT-ICR MS imaging for the analysis of metabolites from formalin-fixed, paraffin-embedded clinical tissue samples. The Journal of Pathology. 2015;237:123–132. doi: 10.1002/path.4560. [DOI] [PubMed] [Google Scholar]
- 4.Aeffner F., Zarella M., Buchbinder N., Bui M., Goodman M., Hartman D. Introduction to digital image analysis in whole-slide imaging: a white paper from the digital pathology association. Journal of Pathology Informatics. 2019;10(1):9. doi: 10.4103/jpi.jpi_82_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inglese P., Correia G., Pruski P., Glen R.C., Takats Z. Colocalization features for classification of tumors using desorption electrospray ionization mass spectrometry imaging. Analytical Chemistry. 2019;91(10):6530–6540. doi: 10.1021/acs.analchem.8b05598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulten W., Bándi P., Hoven J., van de Loo R., Lotz J., Weiss N. Epithelium segmentation using deep learning in H&E-stained prostate specimens with immunohistochemistry as reference standard. Scientific Reports. 2019;9(1):1–10. doi: 10.1038/s41598-018-37257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed M., Broeckx G., Baggerman G., Schildermans K., Pauwels P., Van Craenenbroeck A.H. 2019. Next-generation protein analysis in the pathology department; pp. 1–6. [DOI] [PubMed] [Google Scholar]
- 8.Alexandrov T. MALDI imaging mass spectrometry: statistical data analysis and current computational challenges. BMC Bioinformatics. 2012;13(Suppl 1) doi: 10.1186/1471-2105-13-S16-S11. (Suppl 16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansson E.T., Comi T.J., Rubakhin S.S., Sweedler J.V. Single cell peptide heterogeneity of rat islets of Langerhans. ACS Chemical Biology. 2016;11(9):2588–2595. doi: 10.1021/acschembio.6b00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prentice B.M., Hart N.J., Phillips N., Haliyur R., Judd A., Armandala R. Imaging mass spectrometry enables molecular profiling of mouse and human pancreatic tissue. Diabetologia. 2019;62(6):1036–1047. doi: 10.1007/s00125-019-4855-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feuchtinger A., Stiehler T., Jütting U., Marjanovic G., Luber B., Langer R. Image analysis of immunohistochemistry is superior to visual scoring as shown for patient outcome of esophageal adenocarcinoma. Histochemistry and Cell Biology. 2014;143(1):1–9. doi: 10.1007/s00418-014-1258-2. [DOI] [PubMed] [Google Scholar]
- 12.Brieu N., Caie P., Gavriel C., Schmidt G., Harrison D.J. 2018. Context-based interpolation of coarse deep learning prediction maps for the segmentation of fine structures in immunofluorescence images (March 2018) p. 24. [Google Scholar]
- 13.Van De Plas R., Yang J., Spraggins J., Caprioli R.M. Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping. Nature Methods. 2015;12(4):366–372. doi: 10.1038/nmeth.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann E.K., Comi T.J., Rubakhin S.S., Sweedler J.V. Lipid heterogeneity between astrocytes and neurons revealed by single-cell MALDI-MS combined with immunocytochemical classification. Angewandte Chemie International Edition. 2019;58(18):5910–5914. doi: 10.1002/anie.201812892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roscioni S.S., Migliorini A., Gegg M., Lickert H. Impact of islet architecture on β-cell heterogeneity, plasticity and function. Nature Reviews Endocrinology. 2016;12(12):695–709. doi: 10.1038/nrendo.2016.147. [DOI] [PubMed] [Google Scholar]
- 16.Carrano A.C., Mulas F., Zeng C., Sander M. Interrogating islets in health and disease with single-cell technologies. Molecular Metabolism. 2017;6(9):991–1001. doi: 10.1016/j.molmet.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farack L., Golan M., Egozi A., Dezorella N., Bahar Halpern K., Ben-Moshe S. Transcriptional heterogeneity of beta cells in the intact pancreas. Developmental Cell. 2019;48(1):115–125. doi: 10.1016/j.devcel.2018.11.001. e4. [DOI] [PubMed] [Google Scholar]
- 18.Ly A., Buck A., Balluff B., Sun N., Gorzolka K., Feuchtinger A. High-mass-resolution MALDI mass spectrometry imaging of metabolites from formalin-fixed paraffin-embedded tissue. Nature Protocols. 2016;11(8):1428–1443. doi: 10.1038/nprot.2016.081. [DOI] [PubMed] [Google Scholar]
- 19.Aichler M., Borgmann D., Krumsiek J., Buck A., MacDonald P.E., Fox J.E.M. N-acyl taurines and acylcarnitines cause an imbalance in insulin synthesis and secretion provoking β cell dysfunction in type 2 diabetes. Cell Metabolism. 2017;25(6):1334–1347. doi: 10.1016/j.cmet.2017.04.012. e4. [DOI] [PubMed] [Google Scholar]
- 20.Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Rosa V. HMDB 4.0 : the human metabolome database for 2018. Nucleic Acids Research. 2018;46:608–617. doi: 10.1093/nar/gkx1089. (November 2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kriegsmann K., Longuespée R., Hundemer M., Zgorzelski C., Casadonte R., Schwamborn K. Combined immunohistochemistry after mass spectrometry imaging for superior spatial information. Proteomics - Clinical Applications. 2019;13(1):1–8. doi: 10.1002/prca.201800035. [DOI] [PubMed] [Google Scholar]
- 24.Kaya I., Michno W., Brinet D., Iacone Y., Zanni G., Blennow K. Histology-compatible MALDI mass spectrometry based imaging of neuronal lipids for subsequent immunofluorescent staining. Analytical Chemistry. 2017;89(8):4685–4694. doi: 10.1021/acs.analchem.7b00313. [DOI] [PubMed] [Google Scholar]
- 25.Da Silva Xavier G., Rutter G.A. Metabolic and functional heterogeneity in pancreatic β cells. Journal of Molecular Biology. 2019 doi: 10.1016/j.jmb.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Van Schravendijk C.F.H., Kiekens R., Pipeleers D.G. Pancreatic β cell heterogeneity in glucose-induced insulin secretion. Journal of Biological Chemistry. 1992;267(30):21344–21348. [PubMed] [Google Scholar]
- 27.Strott C.A., Higashi Y. Cholesterol sulfate in human physiology: what's it all about? Journal of Lipid Research. 2003;44(7):1268–1278. doi: 10.1194/jlr.R300005-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Dirkx R., Solimena M. Cholesterol-enriched membrane rafts and insulin secretion. Journal of Diabetes Investigation. 2012;3(4):339–346. doi: 10.1111/j.2040-1124.2012.00200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi T., Suzuki T. Role of sulfatide in normal and pathological cells and tissues. Journal of Lipid Research. 2012;53(8):1437–1450. doi: 10.1194/jlr.R026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boslem E., Meikle P.J., Biden T.J. Roles of ceramide and sphingolipids in pancreatic β-cell function and dysfunction. Islets. 2012;4(3):177–187. doi: 10.4161/isl.20102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm L.J., Krogvold L., Hasselby J.P., Kaur S., Claessens L.A., Russell M.A. Abnormal islet sphingolipid metabolism in type 1 diabetes. Diabetologia. 2018;61(7):1650–1661. doi: 10.1007/s00125-018-4614-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee H.E., Park K.U., Yoo S.B., Nam S.K., Park D.J., Kim H.-H. Clinical significance of intratumoral HER2 heterogeneity in gastric cancer. European Journal of Cancer. 2013;49(6):1448–1457. doi: 10.1016/j.ejca.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 33.Kurokawa Y., Matsuura N., Kimura Y., Adachi S., Fujita J., Imamura H. Multicenter large-scale study of prognostic impact of HER2 expression in patients with resectable gastric cancer. Gastric Cancer. 2015;18(4):691–697. doi: 10.1007/s10120-014-0430-7. [DOI] [PubMed] [Google Scholar]
- 34.Ahn S., Ahn S., van Vrancken M., Lee M., Ha S.Y., Lee H. Ideal number of biopsy tumor fragments for predicting HER2 status in gastric carcinoma resection specimens. Oncotarget. 2015;6(35):38372–38380. doi: 10.18632/oncotarget.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yagi S., Wakatsuki T., Yamamoto N., Chin K., Takahari D., Ogura M. Clinical significance of intratumoral HER2 heterogeneity on trastuzumab efficacy using endoscopic biopsy specimens in patients with advanced HER2 positive gastric cancer. Gastric Cancer. 2019;22(3):518–525. doi: 10.1007/s10120-018-0887-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H.J., Kim J.Y., Park S.Y., Park I.A., Song I.H., Yu J.H. Clinicopathologic significance of the intratumoral heterogeneity of HER2 gene amplification in HER2-positive breast cancer patients treated with adjuvant trastuzumab. American Journal of Clinical Pathology. 2015;144(4):570–578. doi: 10.1309/AJCP51HCGPOPWSCY. [DOI] [PubMed] [Google Scholar]
- 37.Wakatsuki T., Yamamoto N., Sano T., Chin K., Kawachi H., Takahari D. Clinical impact of intratumoral HER2 heterogeneity on trastuzumab efficacy in patients with HER2-positive gastric cancer. Journal of Gastroenterology. 2018;53(11):1186–1195. doi: 10.1007/s00535-018-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaito A., Kuwata T., Tokunaga M., Shitara K., Sato R., Akimoto T. HER2 heterogeneity is a poor prognosticator for HER2-positive gastric cancer. World Journal of Clinical Cases. 2019;7(15):1964–1977. doi: 10.12998/wjcc.v7.i15.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanahan D., Weinberg R.a. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Arifin S.A., Falasca M. Lysophosphatidylinositol signalling and metabolic diseases. Metabolites. 2016;6(1):1–11. doi: 10.3390/metabo6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernier M., Catazaro J., Singh N.S., Wnorowski A., Boguszewska-Czubara A., Jozwiak K. GPR55 receptor antagonist decreases glycolytic activity in PANC-1 pancreatic cancer cell line and tumor xenografts. International Journal of Cancer. 2017;141(10):2131–2142. doi: 10.1002/ijc.30904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai C., Arceo J., Arnold J., Sreekumar A., Dovichi N.J., Li J. Metabolomics of oncogene-specific metabolic reprogramming during breast cancer. Cancer & Metabolism. 2018;6(5):1–17. doi: 10.1186/s40170-018-0175-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prade V.M., Kunzke T., Feuchtinger A., Rohm M., Luber B., Lordick F. 2020. MALDI FTICR MS imaging data of pancreatic mouse tissue. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Imaging mass spectrometry data for mouse pancreatic islets of Langerhans are provided at Zenodo.org (DOI:10.5281/zenodo.3607915) [43].