Abstract
Objective
Compromised brain cholesterol turnover and altered regulation of brain cholesterol metabolism have been allied with some neurodegenerative diseases, including Huntington’s disease (HD). Following our previous studies in HD, in this study we aim to investigate in vitro in a neuroblastoma cellular model of HD, the effect of CYP46A1 overexpression, an essential enzyme in cholesterol metabolism, on huntingtin aggregation and levels.
Results
We found that CYP46A1 reduces the quantity and size of mutant huntingtin aggregates in cells, as well as the levels of mutant huntingtin protein. Additionally, our results suggest that the observed beneficial effects of CYP46A1 in HD cells are linked to the activation of autophagy. Taken together, our results further demonstrate that CYP46A1 is a pertinent target to counteract HD progression.
Keywords: CYP46A1, Cholesterol, Neuroblastoma cells, Huntingtin, Autophagy
Introduction
Huntington’s disease (HD), is an autosomal dominant inherited neurodegenerative disorder [1, 2], resulting from an abnormal CAG-repeat expansion in the coding region of the HTT gene, resulting in an expanded polyglutamine (polyQ) tract of the huntingtin protein (HTT) [3]. HTT protein gains a toxic function, and abnormally accumulates in the cells leading to their death [4, 5]. Clinically, HD is associated with degeneration of the striatum and cerebral cortex [6–8], and symptoms such as uncontrolled movements, chorea, dystonia and oculomotor impairments [9]. Until now there is no therapy available to delay or stop HD progression [10].
Several studies have demonstrated that cholesterol metabolism impairment in the brain could be implicated in different neurodegenerative diseases [11, 12]. Brain cholesterol is crucial for brain homeostasis and physiology, throughout development and in the course of neuronal lifespan [13, 14]. Brain cholesterol is nearly absolutely synthesized in situ [15], given that the blood-brain-barrier (BBB) precludes its efflux/influx [16]. 24-hydroxylase (CYP46A1), a key enzyme in cholesterol turn over and clearance is mostly expressed in neurons and hydroxylates cholesterol into 24S-hydroxycholesterol (24S-OHC) that freely crosses the BBB reaching peripheral circulation [17]. CYP46A1 also play an important role in neural homeostasis [17–20]. It has been shown that CYP46A1 deficiency leads to neuronal dysfunction, neuronal death, motor and cognitive impairments and that CYP46A1 overexpression improved neurodegenerative HD-related traits [21–24]. Previously, we and others showed that cholesterol metabolism is impaired in HD, and strategies to reactivate it, such as CYP46A1 expression restoration, show beneficial effects in different HD model mice, including an improvement in neuronal function, a prevention in neuronal degeneration and the recovery of motor deficits [22, 24–29]. Here, we investigated the impact of CYP46A1 overexpression in huntingtin aggregates and soluble mutant HTT (HTT-MUT) levels and characterized a potential molecular mechanism.
Main text
Materials and methods
Neuroblastoma cells culture and transfections
Mouse neuroblastoma cell line (N2a cells; ATCC® CCL-131) were maintained at 37 °C, 5% CO2, in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% foetal bovine serum, 100 U/ml penicillin and 100 mg/ml streptomycin (Gibco). The different plasmids used were transfected in the cells using PEI (Polyethylenimine, #274651, Polysciences Inc.) in total concentration of 500 ng for each DNA. Cells were lysed for Western blot processing, harvested for flow cytometry analysis or fixed for fluorescence microscopy analysis, 48 h post-transfection.
Plasmids
Plasmids used: pEGFP-HTTQ74 (Addgene #40262, exon 1 of HTT with 319 bp) [30], pAAV-HA-CYP46A1 (provided by Brainvectis), ptfLC3-RFP-GFP (Addgene #21074) [31]. The LacZ gene was cloned in our laboratory under the control of a PGK promoter [32].
Western blot
The Western blot processing was performed as described previously [33]. Antibodies used for immunoblotting: mouse anti-GFP (1:1000, Biolegends, #668205), rabbit anti-LC3B (1:1000, Novus, #NB100-2220), rabbit anti-p62/SQSTM1 (1:1000, Cell Signaling, #5114), mouse anti-ubiquitin (1:1000, Cell Signaling, #3936), mouse anti-β-tubulin, (1:10,000, Sigma-Aldrich, #T7816) and mouse anti-actin (1:10,000, Sigma-Aldrich, #A5316).
Immunocytochemistry
Immunocytochemistry was performed based on protocols described previously [33]. Primary antibodies were incubated overnight at 4 °C (haemagglutinin (HA) tag, 1:1000, Abcam, #ab9110). Secondary antibodies were incubated for 2 h at room temperature (Alexa 594 or Alexa 647, Thermo Fisher).
Fluorescence microscopy analysis
Mutant HTT aggregates were directly counted in a blind fashion way in 100 random cells with 40X objective in a Zeiss Axio Imager Z2. For the aggregate’s areas assessment, the CellProfiler software was used [34]. Images were acquired with 40X objective in a Zeiss Axio Imager Z2. For the immunocytochemistry experiments, the LC3-GFP-RFP puncta was blindly counted in 100 random cells with 40X objective in a Zeiss Axio Imager Z2.
Chloroquine treatment
We performed the chloroquine (ChQ) treatment (SIGMA, 100 μm), an inhibitor of autolysosomal degradation, as previously described [33].
Ubiquitin-proteasome system (UPS) inhibition in neuroblastoma cells
Six hours prior to collection (and 42 h post-transfection), transfected N2a cells were treated with MG-132 (Sigma-Aldrich, 5 μM), an UPS inhibitor. After this treatment N2a cells were collected for western blot analysis.
Flow cytometry
Transfected cells were twice washed with PBS and then carefully harvested in PBS on ice. The different conditions were then processed in a flow cytometer (Becton–Dickinson FACSCalibur) using the adequate lasers, as previously described [35]. Briefly, for each condition 50,000 cells were acquired in the selected gate, and positive cells were plotted as GFP fluorescence intensity (FL1 channel, 530 ± 30 nm).
Statistical analysis
Statistical analyses were performed using One-way ANOVA analysis with Bonferroni’s multiple comparisons test or unpaired t-Student test. Results are expressed as mean ± SEM. Significant thresholds were set at *P < 0.05, **P < 0.001, ***P < 0.0001, ****P < 0.00001. All analyses were performed using GraphPad Prism (GraphPad Software v6, La Jolla, USA).
Results
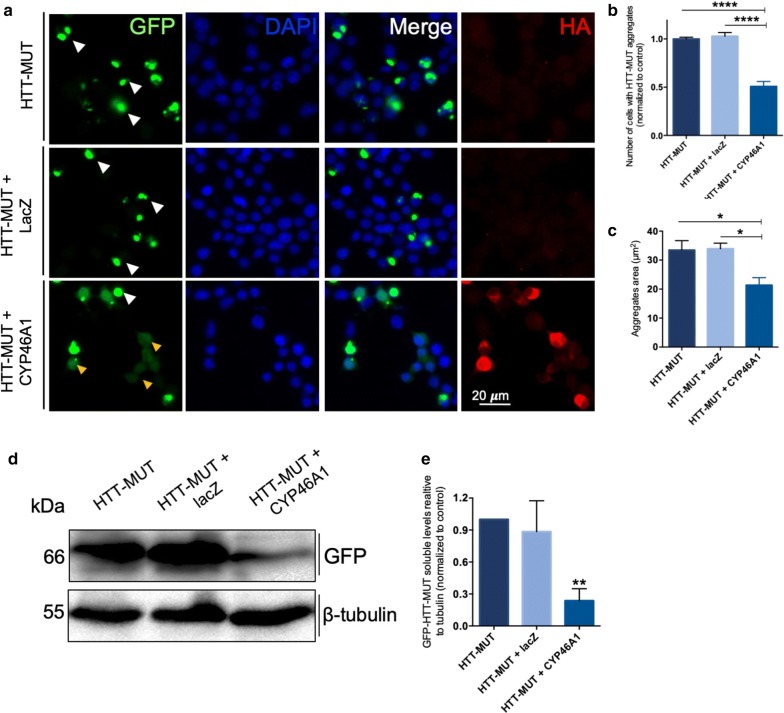
We analyzed by immunocytochemistry the impact of CYP46A1 overexpression in neuroblastoma (N2a) cells overexpressing the exon 1 of HTT-MUT carrying 74 glutamine (Q74) fused with GFP. N2a cells were co-transfected with plasmids encoding HTT-MUT and either the plasmid encoding CYP46A1 or as control the plasmid for lacZ (Fig. 1a). Approximately 90% of all transfected cells express both plasmids, without difference in cell viability. We observed that the overexpression of CYP46A1 (labeled with HA tag) significantly reduced the number of cells with HTT-MUT aggregates compared to HTT-MUT condition and to HTT-MUT and lacZ (Fig. 1b). The aggregates area (µm2) was also significantly reduced upon CYP46A1 expression (Fig. 1c), as compared to HTT-MUT, and to HTT-MUT + lacZ. Next, we evaluated the impact of CYP46A1 overexpression in the protein levels of HTT-MUT. Western blot analysis of N2a lysates using anti-GFP antibody showed a statistically significant reduction in the levels of soluble HTT-MUT in the condition with CYP46A1, as compared to both control conditions (Fig. 1d–e). Altogether, these results show a robust effect of CYP46A1 in reducing the levels of HTT-MUT aggregates and protein.
Fig. 1.
CYP46A1 overexpression decreases the number of aggregates and the levels of soluble mutant HTT. a N2a cells were transfected with pEGFP-HttQ74, co-transfected with pEGFP-HttQ74 and pAAV-HA-CYP46A1, and co-transfected with pEGFP-HttQ74 and LacZ, highlighting the fusion protein with GFP, the nuclei labeled with DAPI and the expression of CYP46A1 by the labeling with HA tag. For each condition, 100 cells were randomly counted. b The number of cells with aggregates (white arrows) upon CYP46A1 overexpression was significantly reduced compared to control conditions (note the yellow arrows with expression of HTT-MUT without aggregates). c The size of HTT-MUT aggregates was smaller upon CYP46A1 overexpression relatively to both control conditions. d Western blot analysis probed with mouse anti-GFP depicting the soluble HTT-MUT (66 kDa) and tubulin as loading control. Densitometric analysis showed that CYP46A1 expression leads to a significant reduction of the soluble HTT-MUT (e). Values are expressed as mean ± SEM. (a–c, n = 5 independent experiments; d–e, n = 4 independent experiments; One-way ANOVA with Bonferroni’s multiple comparisons test, *P < 0.05; **P < 0.001; ****P < 0.00001)
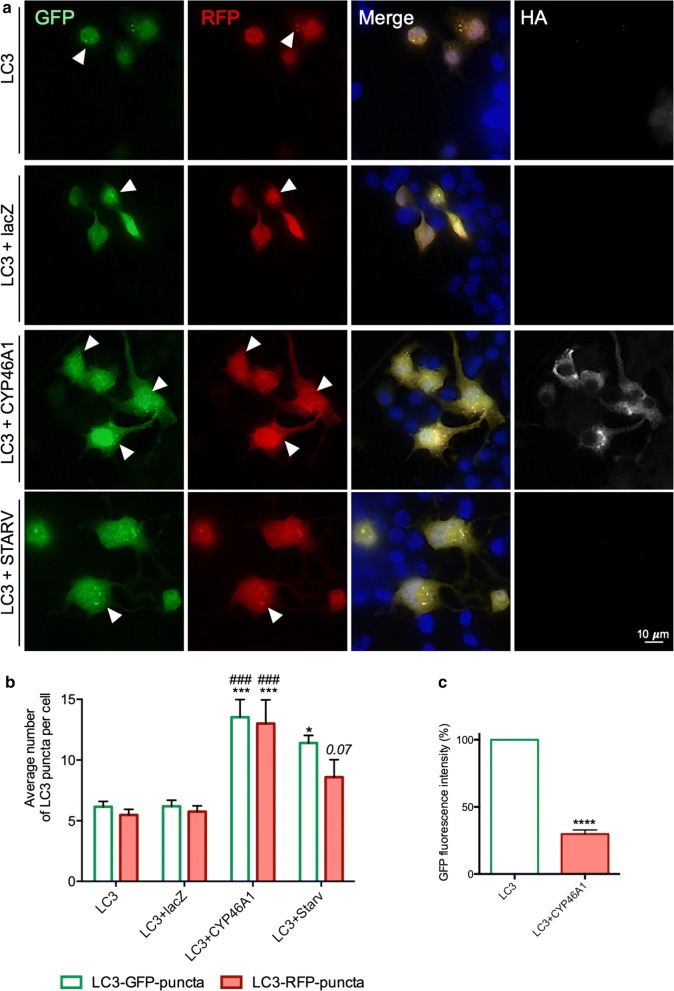
To further investigate the mechanisms of CYP46A1-mediated effects in the clearance of HTT-MUT via autophagic pathway experiments were preformed using a plasmid expressing the autophagic protein LC3B fused with RFP or GFP, and using an autophagy inhibitor (chloroquine—ChQ; Fig. 2a) [31]. We found that CYP46A1 significantly increased both LC3-GFP and LC3-RFP puncta, as compared to control conditions (Fig. 2b;). This increase was even more pronounced than the one observed in the starvation condition, which is a positive control for autophagy activation, thus suggesting that CYP46A1 is activating autophagy. We also performed a cytometry analysis using cells transfected with the LC3-GFP-RFP plasmid. We observed that CYP46A1 overexpression leads to a significant reduction in GFP fluorescence intensity compared to the control condition (Fig. 2c). As this experiment was performed in the absence of ChQ, the results suggest that CYP46A1 is activating autophagy, and promoting autophagy degradation, which could explain the observed reduction in the levels of HTT-MUT aggregates and protein.
Fig. 2.
CYP46A1 overexpression significantly increases the number of LC3 puncta upon autophagy inhibition. a N2a cells were transfected with ptfLC3-RFP-GFP, co-transfected with ptfLC3-RFP-GFP and CYP46A1, and co-transfected with ptfLC3-RFP-GFP and lacZ. An additional condition was used, promoting starvation, as a positive control for autophagy activation. The visualization of LC3-puncta is possible upon autophagy inhibition with chloroquine in all the experimental conditions. The presence of LC3-RFP-GFP puncta (co-localized) refers to the presence of mature autophagosomes, whereas LC3-RFP puncta (without GFP) refers to autolysosomes. Representative microscopy images. b For each condition 100 cells were randomly counted in different microscopy fields. The number LC3-puncta per cell upon CYP46A1 overexpression was significantly increased compared to both control conditions. c The total fluorescence intensity (GFP) was significantly reduced upon CYP46A1 overexpression, compared to control conditions, thus suggesting an increase in the autophagic clearance. Values expressed as mean ± SEM. (a, b, n = 5 independent experiments, two-way ANOVA with Bonferroni’s multiple comparisons test, *P < 0.05; ***P < 0.0001;###P < 0.0001 comparing to LC3 + lacZ; c, n = 3 independent experiments Unpaired Student’s t test, ****P < 0.00001)
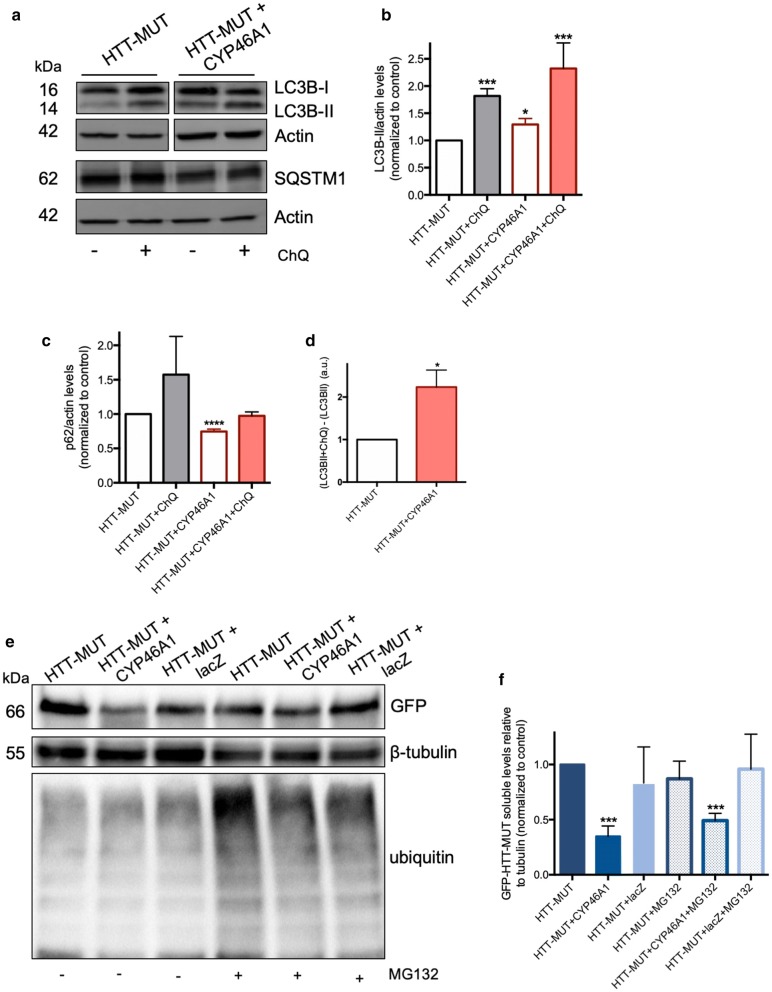
Next, we analyzed the impact of CYP46A1 in several autophagy markers by western blot (Fig. 3a). Overexpression of CYP46A1 significantly increases LC3B-II levels compared to control condition (Fig. 3b), thus suggesting an activation of autophagy. Moreover, in the presence of ChQ the results follow the same trend. SQSTM1/p62 is a protein involved in the degradation of autophagy via a specific interaction between p62 and LC3 [44]. Importantly, a significant reduction in the SQSTM1/p62 levels was observed upon CYP46A1 overexpression, as compared to the control condition (Fig. 3c), suggesting an increase in autophagic activity. Next, we calculated the autophagic flux by measuring the LC3B turnover assay, which is used to quantify the amount of LC3B-II that is delivered to the lysosomes and thus the autophagy dynamics. We found that CYP46A1 overexpression leads to a significant increase in the autophagic flux, as compared to the control condition (Fig. 3d). To assess selective autophagy activation by CYP46A1, we performed an experiment inhibiting the ubiquitin–proteasome system (UPS) using MG132, which leads to the accumulation of polyubiquitinated proteins (Fig. 3e). The CYP46A1 overexpression reduced the levels of HTT-MUT independently of UPS inhibition (Fig. 3f). Altogether, all these results suggest that CYP46A1 is able to activate autophagy pathway in a cellular model of HD.
Fig. 3.
CYP46A1 overexpression promotes the activation of autophagy, independently of the UPS system. a N2a cells were transfected with pEGFP-HTTQ74, co-transfected with pEGFP-HTTQ74 and CYP46A1. Western blots were probed with rabbit anti-LC3B, rabbit anti-p62/SQSTM1, and mouse anti-actin. b The densitometric analysis showed that CYP46A1 expression leads to a significant increase in the LC3B-II levels and c to a significant reduction in the SQSTM1/p62 levels, both compared to the experimental control. The results with chloroquine inhibition of autophagy follow the same trend, suggesting a robust autophagy activation upon CYP46A1 overexpression. d This activation is also supported by a significant increase in the autophagic net flux. e Representative western blot probed for GFP of protein lysates from N2a cells of the different experimental conditions, with and without the proteasome inhibitor MG132 (5 M). The western blot was also probed for tubulin, and poly-ubiquitin to highlight the UPS inhibition. f The densitometric analysis showed that CYP46A1 expression decreases GFP levels (HTT-MUT), with and without ubiquitin–proteasome system inhibition. (n = 4 independent experiments, Unpaired Student’s t-test; one-way ANOVA with Bonferroni’s multiple comparisons test, *P < 0.05; **P < 0.001; ***P < 0.0001)
Discussion
Impaired brain cholesterol metabolism has been broadly associated with neurodegenerative disorders. We and others showed that strategies aiming at restoring cholesterol metabolism using different strategies are beneficial in AD, SCA and HD mouse models [22, 24, 36, 37]. Local synthesis/efflux of cholesterol is essential for the structural regulation of cells and membranes within the brain, being implicated in signal transduction, release of neurotransmitters and membrane trafficking [38]. Cholesterol has not the ability to cross the blood–brain-barrier (BBB). In order to be eliminated by the brain, therefore allowing brain cholesterol homeostasis, the excess of cholesterol is converted by the 24-hydroxylase (exclusively expressed in neurons) into 24-OHC that crosses the BBB, allowing brain cholesterol efflux [39, 40].
We have previously shown that CYP46A1 expression was able to reduce the number and size of intranuclear protein aggregates within the striatum of HD mouse models and improve motor impairment [22], emphasizing the neuroprotective effect conferred by CYP46A1 in HD mice [22, 24]. Here, we further show that CYP46A1 is able to decrease the number and size of HTT-MUT aggregates within a neuroblastoma cellular model of HD. Furthermore, reduction of soluble HTT-MUT was decreased upon CYP46A1 overexpression. Autophagy is impaired in HD [41] and activating autophagy by expressing CYP46A1, may in part explain how inclusions and protein levels are decreased. We previously showed in a SCA3 in vitro and in vivo models that CYP46A1 robustly increased the autophagic flux, i.e., augmented the levels of LC3-II and decreased the p62/SQSTM1 levels [36].
It is important to point that several studies have previously shown that the activation of autophagy, both using molecular or pharmacological approaches, is effective in removing HTT-MUT, and other mutated polyglutamine proteins, being therefore a privileged target for the development of effective therapeutic approaches [42–44, 33, 45–49].
Altogether, our data suggest that CYP46A1 degrades HTT-MUT via autophagy and that CYP46A1 may be a good target to alleviate HD progression. Therefore, our findings and our previous studies suggest that CYP46A1 is implicated in HTT-MUT degradation via autophagy.
Limitations
Cellular models display several limitations and in vivo studies must be perform as robust platforms for disease modelling supporting CYP46A1 in the activation of autophagy. Furthermore, additional studies of autophagy could be performed in this cellular model, including with additional controls, to further strengthen the observed results.
Acknowledgements
We like to acknowledge the Light Microscopy Unit of CBMR-UAlg.
Abbreviations
- HD
Huntington’s disease
- HTT
Huntingtin
- BBB
Blood–brain-barrier
- DMEM
Dulbecco’s modified Eagle’s medium
- PEI
Polyethylenimine
- HA
Haemagglutinin
- ChQ
Chloroquine
- UPS
Ubiquitin–proteasome system
- N2a
Neuroblastoma cells
Authors’ contributions
CN designed and drafted of the work; AC, substantively revised the work; RGC acquired and analysed the results; RK acquired and analysed the results; RS acquired and analysed the results; RN acquired and analysed the results; SCS acquired and analysed the results, AM acquired and analysed the results, CAM substantively revised it, SB substantively revised it, JC substantively revised it, NC, designed and drafted of the work, SA, designed and drafted of the work. All authors read and approved the final manuscript.
Funding
This work was supported by Brainvectis and E.rare: E-Rare Joint Transnational Call for Proposals 2017 “Transnational Research Projects for Innovative Therapeutic Approaches for Rare Diseases”. CN laboratory is supported by the French Muscular Dystrophy Association (AFM-Téléthon), the Ataxia UK, and the Fundação para a Ciência e Tecnologia (project ALG-01-0145-FEDER-29480 “SeGrPolyQ”). AM is supported by a Ph.D. fellowship from FCT (SFRH/BD/133192/2017).
Data availability
All the raw data generated are available upon reasonable request to corresponding author.
Ethics approval and consent to participate
The manuscript does not contain any individual person’s data in any form.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nathalie Cartier and Sandro Alves contributed equally to this work
Contributor Information
Clévio Nóbrega, Email: cdnobrega@ualg.pt.
Nathalie Cartier, Email: nathalie.cartier@inserm.fr.
Sandro Alves, Email: sandropfalves@gmail.com, Email: sandro.alves@brainvectis.com.
References
- 1.Ross CA, Aylward EH, Wild EJ, Langbehn DR, Long JD, Warner JH, Scahill RI, Leavitt BR, Stout JC, Paulsen JS, Reilmann R, Unschuld PG, Wexler A, Margolis RL, Tabrizi SJ. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol. 2014;10:204–216. doi: 10.1038/nrneurol.2014.24. [DOI] [PubMed] [Google Scholar]
- 2.McColgan P, Tabrizi SJ. Huntington’s disease: a clinical review. Eur J Neurol. 2018;25:24–34. doi: 10.1111/ene.13413. [DOI] [PubMed] [Google Scholar]
- 3.Kim SD, Fung VS. An update on Huntington’s disease: from the gene to the clinic. Curr Opin Neurol. 2014;27:477–483. doi: 10.1097/WCO.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 4.Jansen AH, van Hal M, Op den Kelder IC, Meier RT, de Ruiter AA, Schut MH, Smith DL, Grit C, Brouwer N, Kamphuis W, Boddeke HW, den Dunnen WF, van Roon WM, Bates GP, Hol EM, Reits EA. Frequency of nuclear mutant huntingtin inclusion formation in neurons and glia is cell-type-specific. Glia. 2017;65:50–61. doi: 10.1002/glia.23050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 6.Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP., Jr Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence AD, Hodges JR, Rosser AE, Kershaw A, Ffrench-Constant C, Rubinsztein DC, Robbins TW, Sahakian BJ. Evidence for specific cognitive deficits in preclinical Huntington’s disease. Brain J Neurol. 1998;121(Pt 7):1329–1341. doi: 10.1093/brain/121.7.1329. [DOI] [PubMed] [Google Scholar]
- 8.Kassubek J, Gaus W, Landwehrmeyer GB. Evidence for more widespread cerebral pathology in early HD: an MRI-based morphometric analysis. Neurology. 2004;62:523–524. doi: 10.1212/WNL.62.3.523-a. [DOI] [PubMed] [Google Scholar]
- 9.Roos RA. Huntington’s disease: a clinical review. Orphanet J Rare Dis. 2010;5:40. doi: 10.1186/1750-1172-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabrizi SJ, Ghosh R, Leavitt BR. Huntingtin lowering strategies for disease modification in Huntington’s disease. Neuron. 2019;101:801–819. doi: 10.1016/j.neuron.2019.01.039. [DOI] [PubMed] [Google Scholar]
- 11.Vance JE. Dysregulation of cholesterol balance in the brain: contribution to neurodegenerative diseases. Dis Models Mech. 2012;5:746–755. doi: 10.1242/dmm.010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korade Z, Kenworthy AK. Lipid rafts, cholesterol, and the brain. Neuropharmacology. 2008;55:1265–1273. doi: 10.1016/j.neuropharm.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin MG, Pfrieger F, Dotti CG. Cholesterol in brain disease: sometimes determinant and frequently implicated. EMBO Rep. 2014;15:1036–1052. doi: 10.15252/embr.201439225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camargo N, Smit AB, Verheijen MH. SREBPs: SREBP function in glia-neuron interactions. FEBS J. 2009;276:628–636. doi: 10.1111/j.1742-4658.2008.06808.x. [DOI] [PubMed] [Google Scholar]
- 15.Dietschy JM, Turley SD. Control of cholesterol turnover in the mouse. J Biol Chem. 2002;277:3801–3804. doi: 10.1074/jbc.R100057200. [DOI] [PubMed] [Google Scholar]
- 16.Bjorkhem I, Meaney S. Brain cholesterol: long secret life behind a barrier. Arterioscler Thromb Vasc Biol. 2004;24:806–815. doi: 10.1161/01.ATV.0000120374.59826.1b. [DOI] [PubMed] [Google Scholar]
- 17.Bjorkhem I, Lutjohann D, Diczfalusy U, Stahle L, Ahlborg G, Wahren J. Cholesterol homeostasis in human brain: turnover of 24S-hydroxycholesterol and evidence for a cerebral origin of most of this oxysterol in the circulation. J Lipid Res. 1998;39:1594–1600. [PubMed] [Google Scholar]
- 18.Meaney S, Hassan M, Sakinis A, Lutjohann D, von Bergmann K, Wennmalm A, Diczfalusy U, Bjorkhem I. Evidence that the major oxysterols in human circulation originate from distinct pools of cholesterol: a stable isotope study. J Lipid Res. 2001;42:70–78. [PubMed] [Google Scholar]
- 19.Lund EG, Guileyardo JM, Russell DW. cDNA cloning of cholesterol 24-hydroxylase, a mediator of cholesterol homeostasis in the brain. Proc Natl Acad Sci USA. 1999;96:7238–7243. doi: 10.1073/pnas.96.13.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sodero AO, Trovo L, Iannilli F, Van Veldhoven P, Dotti CG, Martin MG. Regulation of tyrosine kinase B activity by the Cyp46/cholesterol loss pathway in mature hippocampal neurons: relevance for neuronal survival under stress and in aging. J Neurochem. 2011;116:747–755. doi: 10.1111/j.1471-4159.2010.07079.x. [DOI] [PubMed] [Google Scholar]
- 21.Ayciriex S, Djelti F, Alves S, Regazzetti A, Gaudin M, Varin J, Langui D, Bieche I, Hudry E, Dargere D, Aubourg P, Auzeil N, Laprevote O, Cartier N. Neuronal cholesterol accumulation induced by Cyp46a1 down-regulation in mouse hippocampus disrupts brain lipid homeostasis. Front Mole Neurosci. 2017;10:211. doi: 10.3389/fnmol.2017.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boussicault L, Alves S, Lamaziere A, Planques A, Heck N, Moumne L, Despres G, Bolte S, Hu A, Pages C, Galvan L, Piguet F, Aubourg P, Cartier N, Caboche J, Betuing S. CYP46A1, the rate-limiting enzyme for cholesterol degradation, is neuroprotective in Huntington’s disease. Brain J Neurol. 2016;139:953–970. doi: 10.1093/brain/awv384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djelti F, Braudeau J, Hudry E, Dhenain M, Varin J, Bieche I, Marquer C, Chali F, Ayciriex S, Auzeil N, Alves S, Langui D, Potier MC, Laprevote O, Vidaud M, Duyckaerts C, Miles R, Aubourg P, Cartier N. CYP46A1 inhibition, brain cholesterol accumulation and neurodegeneration pave the way for Alzheimer’s disease. Brain J Neurol. 2015;138:2383–2398. doi: 10.1093/brain/awv166. [DOI] [PubMed] [Google Scholar]
- 24.Kacher R, Lamaziere A, Heck N, Kappes V, Mounier C, Despres G, Dembitskaya Y, Perrin E, Christaller W, Sasidharan Nair S, Messent V, Cartier N, Vanhoutte P, Venance L, Saudou F, Neri C, Caboche J, Betuing S. CYP46A1 gene therapy deciphers the role of brain cholesterol metabolism in Huntington’s disease. Brain J Neurol. 2019;142:2432–2450. doi: 10.1093/brain/awz174. [DOI] [PubMed] [Google Scholar]
- 25.Valenza M, Rigamonti D, Goffredo D, Zuccato C, Fenu S, Jamot L, Strand A, Tarditi A, Woodman B, Racchi M, Mariotti C, Di Donato S, Corsini A, Bates G, Pruss R, Olson JM, Sipione S, Tartari M, Cattaneo E. Dysfunction of the cholesterol biosynthetic pathway in Huntington’s disease. J Neurosci. 2005;25:9932–9939. doi: 10.1523/JNEUROSCI.3355-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valenza M, Leoni V, Tarditi A, Mariotti C, Bjorkhem I, Di Donato S, Cattaneo E. Progressive dysfunction of the cholesterol biosynthesis pathway in the R6/2 mouse model of Huntington’s disease. Neurobiol Dis. 2007;28:133–142. doi: 10.1016/j.nbd.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Valenza M, Carroll JB, Leoni V, Bertram LN, Bjorkhem I, Singaraja RR, Di Donato S, Lutjohann D, Hayden MR, Cattaneo E. Cholesterol biosynthesis pathway is disturbed in YAC128 mice and is modulated by huntingtin mutation. Hum Mol Genet. 2007;16:2187–2198. doi: 10.1093/hmg/ddm170. [DOI] [PubMed] [Google Scholar]
- 28.Valenza M, Leoni V, Karasinska JM, Petricca L, Fan J, Carroll J, Pouladi MA, Fossale E, Nguyen HP, Riess O, MacDonald M, Wellington C, DiDonato S, Hayden M, Cattaneo E. Cholesterol defect is marked across multiple rodent models of Huntington’s disease and is manifest in astrocytes. J Neurosci. 2010;30:10844–10850. doi: 10.1523/JNEUROSCI.0917-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valenza M, Chen JY, Di Paolo E, Ruozi B, Belletti D, Ferrari Bardile C, Leoni V, Caccia C, Brilli E, Di Donato S, Boido MM, Vercelli A, Vandelli MA, Forni F, Cepeda C, Levine MS, Tosi G, Cattaneo E. Cholesterol-loaded nanoparticles ameliorate synaptic and cognitive function in Huntington’s disease mice. EMBO Mol Med. 2015;7:1547–1564. doi: 10.15252/emmm.201505413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narain Y, Wyttenbach A, Rankin J, Furlong RA, Rubinsztein DC. A molecular investigation of true dominance in Huntington’s disease. J Med Genet. 1999;36:739–746. doi: 10.1136/jmg.36.10.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 32.Nobrega C, Carmo-Silva S, Albuquerque D, Vasconcelos-Ferreira A, Vijayakumar UG, Mendonca L, Hirai H, de Almeida LP. Re-establishing ataxin-2 downregulates translation of mutant ataxin-3 and alleviates Machado-Joseph disease. Brain J Neurol. 2015;138:3537–3554. doi: 10.1093/brain/awv298. [DOI] [PubMed] [Google Scholar]
- 33.Marcelo A, Brito F, Carmo-Silva S, Matos CA, Alves-Cruzeiro J, Vasconcelos-Ferreira A, Koppenol R, Mendonca L, de Almeida LP, Nobrega C. Cordycepin activates autophagy through AMPK phosphorylation to reduce abnormalities in Machado-Joseph disease models. Hum Mol Genet. 2019;28:51–63. doi: 10.1093/hmg/ddy328. [DOI] [PubMed] [Google Scholar]
- 34.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. Cell Profiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nobrega C, Nascimento-Ferreira I, Onofre I, Albuquerque D, Conceicao M, Deglon N, de Almeida LP. Overexpression of mutant ataxin-3 in mouse cerebellum induces ataxia and cerebellar neuropathology. Cerebellum (London, England) 2013;12:441–455. doi: 10.1007/s12311-012-0432-0. [DOI] [PubMed] [Google Scholar]
- 36.Nobrega C, Mendonca L, Marcelo A, Lamaziere A, Tome S, Despres G, Matos CA, Mechmet F, Langui D, den Dunnen W, de Almeida LP, Cartier N, Alves S. Restoring brain cholesterol turnover improves autophagy and has therapeutic potential in mouse models of spinocerebellar ataxia. Acta Neuropathol. 2019;138:837–858. doi: 10.1007/s00401-019-02019-7. [DOI] [PubMed] [Google Scholar]
- 37.Hudry E, Van Dam D, Kulik W, De Deyn PP, Stet FS, Ahouansou O, Benraiss A, Delacourte A, Bougneres P, Aubourg P, Cartier N. Adeno-associated virus gene therapy with cholesterol 24-hydroxylase reduces the amyloid pathology before or after the onset of amyloid plaques in mouse models of Alzheimer’s disease. Mol Ther. 2010;18:44–53. doi: 10.1038/mt.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bjorkhem I. Crossing the barrier: oxysterols as cholesterol transporters and metabolic modulators in the brain. J Intern Med. 2006;260:493–508. doi: 10.1111/j.1365-2796.2006.01725.x. [DOI] [PubMed] [Google Scholar]
- 39.Russell DW, Halford RW, Ramirez DM, Shah R, Kotti T. Cholesterol 24-hydroxylase: an enzyme of cholesterol turnover in the brain. Annu Rev Biochem. 2009;78:1017–1040. doi: 10.1146/annurev.biochem.78.072407.103859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez DM, Andersson S, Russell DW. Neuronal expression and subcellular localization of cholesterol 24-hydroxylase in the mouse brain. J Comp Neurol. 2008;507:1676–1693. doi: 10.1002/cne.21605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Son JH, Shim JH, Kim K-H, Ha J-Y, Han JY. Neuronal autophagy and neurodegenerative diseases. Exp Mol Med. 2012;44:89. doi: 10.3858/emm.2012.44.2.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin DD, Ladha S, Ehrnhoefer DE, Hayden MR. Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci. 2015;38:26–35. doi: 10.1016/j.tins.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Sittler A, Muriel MP, Marinello M, Brice A, den Dunnen W, Alves S. Deregulation of autophagy in postmortem brains of Machado-Joseph disease patients. Neuropathology. 2018;38:113–124. doi: 10.1111/neup.12433. [DOI] [PubMed] [Google Scholar]
- 44.Alves S, Cormier-Dequaire F, Marinello M, Marais T, Muriel MP, Beaumatin F, Charbonnier-Beaupel F, Tahiri K, Seilhean D, El Hachimi K, Ruberg M, Stevanin G, Barkats M, den Dunnen W, Priault M, Brice A, Durr A, Corvol JC, Sittler A. The autophagy/lysosome pathway is impaired in SCA7 patients and SCA7 knock-in mice. Acta Neuropathol. 2014;128:705–722. doi: 10.1007/s00401-014-1289-8. [DOI] [PubMed] [Google Scholar]
- 45.Sarkar S, Krishna G, Imarisio S, Saiki S, O’Kane CJ, Rubinsztein DC. A rational mechanism for combination treatment of Huntington’s disease using lithium and rapamycin. Hum Mol Genet. 2008;17:170–178. doi: 10.1093/hmg/ddm294. [DOI] [PubMed] [Google Scholar]
- 46.Pierzynowska K, Gaffke L, Hać A, Mantej J, Niedziałek N, Brokowska J, Węgrzyn G. Correction of Huntington’s disease phenotype by genistein-induced autophagy in the cellular model. Neuromol Med. 2018;20:112–123. doi: 10.1007/s12017-018-8482-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, O’Kane CJ, Floto RA, Rubinsztein DC. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nascimento-Ferreira I, Nóbrega C, Vasconcelos-Ferreira A, Onofre I, Albuquerque D, Aveleira C, Hirai H, Déglon N, Pereira de Almeida L. Beclin 1 mitigates motor and neuropathological deficits in genetic mouse models of Machado-Joseph disease. Brain. 2013;136:2173–2188. doi: 10.1093/brain/awt144. [DOI] [PubMed] [Google Scholar]
- 49.Matos CA, Pereira de Almeida L, Nóbrega C. Machado-Joseph disease/spinocerebellar ataxia type 3: lessons from disease pathogenesis and clues into therapy. J Neurochem. 2019;148:8–28. doi: 10.1111/jnc.14541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the raw data generated are available upon reasonable request to corresponding author.