Abstract
Background
Chest pain is one of the most common complaints among patients presenting to the emergency department (ED). Causes of chest pain can be benign or life threatening, making accurate risk stratification a critical issue in the ED. In addition to the use of established clinical scores, prior studies have attempted to create predictive models with heart rate variability (HRV). In this study, we proposed heart rate n-variability (HRnV), an alternative representation of beat-to-beat variation in electrocardiogram (ECG), and investigated its association with major adverse cardiac events (MACE) in ED patients with chest pain.
Methods
We conducted a retrospective analysis of data collected from the ED of a tertiary hospital in Singapore between September 2010 and July 2015. Patients > 20 years old who presented to the ED with chief complaint of chest pain were conveniently recruited. Five to six-minute single-lead ECGs, demographics, medical history, troponin, and other required variables were collected. We developed the HRnV-Calc software to calculate HRnV parameters. The primary outcome was 30-day MACE, which included all-cause death, acute myocardial infarction, and revascularization. Univariable and multivariable logistic regression analyses were conducted to investigate the association between individual risk factors and the outcome. Receiver operating characteristic (ROC) analysis was performed to compare the HRnV model (based on leave-one-out cross-validation) against other clinical scores in predicting 30-day MACE.
Results
A total of 795 patients were included in the analysis, of which 247 (31%) had MACE within 30 days. The MACE group was older, with a higher proportion being male patients. Twenty-one conventional HRV and 115 HRnV parameters were calculated. In univariable analysis, eleven HRV and 48 HRnV parameters were significantly associated with 30-day MACE. The multivariable stepwise logistic regression identified 16 predictors that were strongly associated with MACE outcome; these predictors consisted of one HRV, seven HRnV parameters, troponin, ST segment changes, and several other factors. The HRnV model outperformed several clinical scores in the ROC analysis.
Conclusions
The novel HRnV representation demonstrated its value of augmenting HRV and traditional risk factors in designing a robust risk stratification tool for patients with chest pain in the ED.
Keywords: Heart rate variability (HRV), Heart rate n-variability (HRnV), Electrocardiogram, Chest pain, Risk stratification, Emergency department
Background
Chest pain, which may be caused by life-threatening myocardial infarction (MI) or benign musculoskeletal pain, is one of the most common presenting complaints in the emergency department (ED) [1–3]. Majority of chest pain patients are subjected to extensive diagnostic tests to rule out acute coronary syndrome (ACS), resulting in oftentimes, prolonged and costly ED admission, with only a small proportion of these patients eventually receiving a diagnosis of ACS [3]. This can strain crowded EDs and reduce availability of resources for patients who need urgent medical attention. Hence, early identification of chest pain patients who are at high-risk of developing adverse cardiac events has been a pressing issue to contend with in the ED. Several established clinical scores have been used for risk stratifying chest pain patients in the ED [4, 5], including the History, ECG, Age, Risk factors and Troponin (HEART) [6], the Thrombolysis in Myocardial Infarction (TIMI) [7], and the Global Registry of Acute Coronary Events (GRACE) [8] scores. Of these scores, the HEART score is the most accurate and widely used [5, 9–12], with recent studies focusing on the development of risk score-based clinical pathways for rapid, yet safe discharge of low-risk patients [1, 3, 13, 14].
In a recent review of clinical scores for ED patients with chest pain [5], heart rate variability (HRV) has demonstrated its capability in building predictive models for accurate risk stratification [15–17]. HRV is a widely adopted tool for evaluating changes in cardiac autonomic regulation, and has been shown to be strongly associated with the autonomic nervous system (ANS) [18–20]. HRV analysis characterizes the beat-to-beat variation in an electrocardiogram (ECG) by utilizing time and frequency domains, and nonlinear analyses [19]. Reduced HRV has been found to be a significant predictor of adverse cardiac outcomes [21]. Given the complexity of quantifying HRV representation, several tools such as the PhysioNet Cardiovascular Signal Toolbox [22] and Kubios HRV [23] have been developed to standardize HRV analyses.
Based on the principle of parameter calculation on normal R-R intervals (RRIs; in this paper, RRIs are equivalent to normal-to-normal [NN] intervals, in which abnormal beats have been removed), HRV analysis generates only one set of parameters from a fixed length of ECG record. This limits the amount of information that can be extracted from raw ECG signals. In this paper, we proposed a novel representation of beat-to-beat variation, named as heart rate n-variability (HRnV) [24] to characterize RRIs from a different perspective. With the use of HRnV measures, multiple sets of parameters can be calculated from the same ECG record, which significantly increases the amount of extracted information. Our study is the first clinical application and evaluation of the HRnV representation in risk stratification of chest pain patients in the ED. We hypothesized that HRnV, while closely related to conventional HRV, can provide supplementary information associated with adverse cardiac events. We also investigated the potential use of HRnV parameters to develop risk prediction tools.
Methods
Study design and setting
We conducted a retrospective analysis of data collected in our previous study on risk stratification of chest pain patients in the ED [9]. A convenience sample of patients was recruited at the ED of Singapore General Hospital, a tertiary hospital with around-the-clock primary percutaneous coronary intervention capabilities and a median door-to-balloon time of 101 min [25], between September 2010 and July 2015. At ED triage, patients are classified using the Patient Acuity Category Scale (PACS), with PACS 1 patients being the most critically ill and requiring immediate medical attention and PACS 4 patients being non-urgent cases. In this study, patients > 20 years old who presented to the ED with chief complaint of chest pain and with PACS of 1 or 2 were included. Patients were excluded from the study if they had ST-elevation myocardial infarction (STEMI) or an obvious non-cardiac etiology of chest pain diagnosed by the primary emergency physician. Patients were also excluded if their ECGs had high level of noise or if they were in non-sinus rhythm; these criteria were applied to ensure the quality of HRV and HRnV analyses. Ethical approval was obtained from the Centralized Institutional Review Board (CIRB, Ref: 2014/584/C) of SingHealth, the largest public healthcare system in Singapore that includes the Singapore General Hospital as a key partner. Patient consent was waived for this study.
Data collection
During the data collection period, five to six-minute single-lead (lead II) ECG recordings were retrieved from the X-Series Monitor (ZOLL Medical Corporation, Chelmsford, MA). The first set of vital signs and troponin values from the recruited patients were extracted from the hospital’s electronic health records (EHR). In this study, high-sensitivity troponin-T was used, and an abnormal value was defined as > 0.03 ng/mL [26]; it was further stratified into three groups and coded as 0 if the value was ≤0.03 ng/mL, 1 if the value was between 1 and 3 times the normal limit, and 2 if the value was > 3 times the normal limit. Additionally, patients’ first 12-lead ECGs were interpreted by two independent clinical reviewers. Pathologic ST-elevation, ST-depression, T-wave inversions, and Q-waves were recorded. Patient demographics, medical history, and information required for computing the HEART, TIMI, and GRACE scores were retrospectively reviewed and obtained from EHR.
Proposed HRnV representation of beat-to-beat variation in ECG
HRnV: a novel measure with non-overlapping RRIs
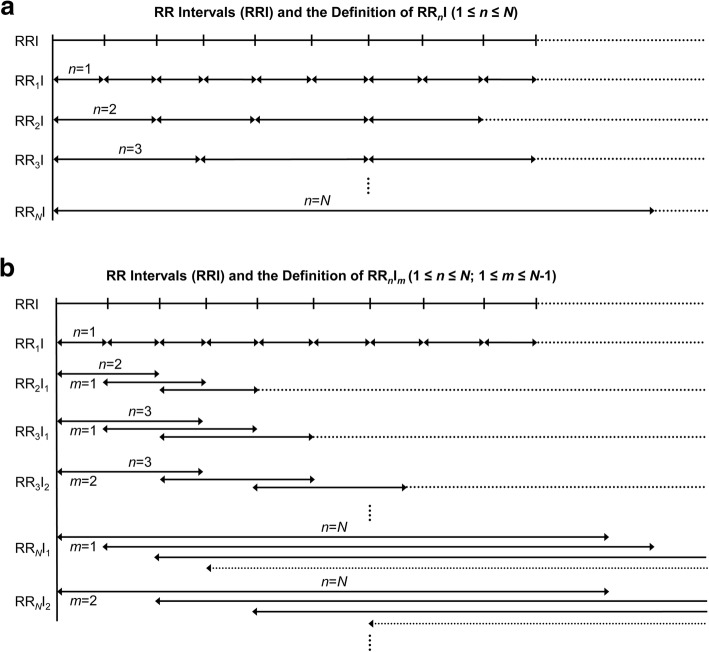
Prior to introducing the new HRnV measure, we define a new type of RRI called RRnI, where 1 ≤ n ≤ N, and ; is the total number of RRIs. The definition of RRnI is illustrated in Fig. 1a. When n = 1, RRnI is equivalent to conventional RRI. When n > 1, every n adjacent RRI is connected to form a new sequence of RRnIs. By using this strategy, we can create a maximum number of (N − 1) new RRnI sequences from conventional single RRI sequence. With these newly generated RRnI sequences, the calculation of HRnV parameters is straightforward and can be accomplished by applying established quantitative methods including time and frequency domain analyses and nonlinear analysis [18, 19]. In describing this new measure, we use the term “HRnV” prior to parameter names to indicate that these parameters are calculated from RRnI sequences. As noted in the above, HRnV is a novel measure based on newly generated, non-overlapping RRnIs. The computed HRnV parameters include but are not limited to the following: the average of RRnIs (HRnV mean NN), standard deviation of RRnIs (HRnV SDNN), square root of the mean squared differences between RRnIs (HRnV RMSSD), the number of times that the absolute difference between two successive RRnIs exceeds 50 ms (HRnV NN50), HRnV NN50 divided by the total number of RRnIs (HRnV pNN50), the integral of the RRnI histogram divided by the height of the histogram (HRnV triangular index), low frequency power (HRnV LF power), high frequency power (HRnV HF power), approximate entropy (HRnV ApEn), sample entropy (HRnV SampEn), and detrended fluctuation analysis (HRnV DFA), among others. Notably, two new parameters NN50n and pNN50n are created, where 50 × n ms is set as the threshold to assess the difference between pairs of consecutive RRnIs.
Fig. 1.
a Illustration of R-R intervals (RRIs) and the definition of RRnI where 1 ≤ n ≤ N and . is the total number of RRIs; b Illustration of RRIs and the definition of RRnIm where 1 ≤ n ≤ N, 1 ≤ m ≤ N − 1, and . is the total number of RRIs and m indicates the non-overlapping portion between two consecutive RRnIm sequences
HRnVm: a novel measure with overlapping RRIs
Like RRnI that is used in HRnV, to define HRnVm measure, we introduce another type of RRI called RRnIm, where 1 ≤ n ≤ N, 1 ≤ m ≤ N − 1, and . In the RRnIm sequence, m is used to indicate the level of overlap between consecutive RRnIm sequences. As illustrated in Fig. 1b, (n − m) RRIs form the overlapping portions. When m = n, RRnIm becomes RRnI; therefore, the upper limit of m is N − 1. By controlling the overlap among these newly generated RRnIm sequences, we can create a maximum number of (N × (N − 1)/2) RRnIm sequences (excluding the RRnI sequence) from conventional single RRI sequence. For each of the newly created RRnIm sequences, we apply time and frequency domain analyses, and nonlinear analysis to calculate HRnVm parameters. We add the term “HRnVm” prior to the parameters to denote that they are computed from RRnIm sequences. For example, the average RRnIm intervals and the sample entropy are written as HRnVm mean NN and HRnVm SampEn, respectively. The HRnVm measure extracts more information than HRnV, by adopting a strategy of controlling sequence overlap.
HRnV analysis and parameter calculation
We developed the HRnV-Calc software suite (https://github.com/nliulab/HRnV) to calculate HRnV parameters. The HRnV-Calc software integrates functions from the PhysioNet Cardiovascular Signal Toolbox [22] to perform standardized ECG signal processing and QRS complex detection. Given the short ECG records in this study, the upper limit of n was set as three; thus, six sets of parameters were calculated, namely HRV, HR2V, HR2V1, HR3V, HR3V1, and HR3V2.
Clinical outcomes
The primary endpoint in this study was a composite outcome of major adverse cardiac events (MACE) [27], including all-cause death, acute myocardial infarction (AMI), and revascularization (coronary artery bypass graft [CABG] or percutaneous coronary intervention [PCI]) within 30 days of ED presentation.
Statistical analysis
Continuous variables were presented as mean and standard deviation and compared between two categories of the primary outcome (MACE) using two-sample t-test. Categorical variables were presented as frequency and percentage and compared between two categories of the primary outcome (MACE) using chi-square test. A statistically significant difference was defined as p < 0.05. To evaluate the HRnV parameters and other risk factors, we conducted univariable and multivariable analyses and subsequently developed simple prediction models using traditional logistic regression. In building the HRnV prediction model, we selected candidate variables with p < 0.2 in the univariable analysis and fed them into the multivariable stepwise logistic regression. To evaluate the predictive performance, we used leave-one-out cross-validation (LOOCV) to conduct the analysis.
Receiver operating characteristic (ROC) analysis [28] was performed to compare prediction performances among the HRnV model, HEART, TIMI and GRACE scores. The area under the ROC curve (AUC), sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were reported as predictive measures. Data preparation, descriptive analysis, and predictive model development were performed in R version 3.6.0 (R Foundation, Vienna, Austria); ROC analysis was conducted in MATLAB R2019a (MathWorks, Natick, MA).
Results
A total of 795 patients were selected from the originally recruited 922 patients [9]. Twenty-eight patients were excluded for ECG recording issues, four were excluded for obvious non-cardiac chest pain, and 95 were excluded for irregular rhythm/artifacts. Among the included 795 patients, 247 (31%) had the primary outcome of 30-day MACE. Table 1 shows patient baseline characteristics. Patients with the primary outcome were older (mean age 61 years vs. 59 years, p = 0.035), with a higher proportion being males (76.1% vs. 64.6%, p = 0.002). There was no statistically significant difference between MACE and non-MACE groups in terms of patient ethnicity. Factors such as history of diabetes and current smoking status were significantly more prevalent in the group with MACE.
Table 1.
Patient baseline characteristics
| Total (n = 795) | MACE (n = 247) | Non-MACE (n = 548) | p-value | |
|---|---|---|---|---|
| Age, mean (SD) | 59.63 (12.88) | 61.06 (11.38) | 58.99 (13.47) | 0.035 |
| Male gender, n (%) | 542 (68.2) | 188 (76.1) | 354 (64.6) | 0.002 |
| Race, n (%) | 0.623 | |||
| Chinese | 492 (61.9) | 159 (64.4) | 333 (60.8) | |
| Indian | 129 (16.2) | 34 (13.8) | 95 (17.3) | |
| Malay | 150 (18.9) | 46 (18.6) | 104 (19.0) | |
| Other | 24 (3.0) | 8 (3.2) | 16 (2.9) | |
| Medical history, n (%) | ||||
| Ischemic heart disease | 343 (43.1) | 115 (46.6) | 228 (41.6) | 0.22 |
| Diabetes | 278 (35.0) | 106 (42.9) | 172 (31.4) | 0.002 |
| Hypertension | 509 (64.0) | 161 (65.2) | 348 (63.5) | 0.707 |
| Hypercholesterolemia | 476 (59.9) | 151 (61.1) | 325 (59.3) | 0.683 |
| Stroke | 58 (7.3) | 15 (6.1) | 43 (7.8) | 0.458 |
| Cancer | 29 (3.6) | 7 (2.8) | 22 (4.0) | 0.537 |
| Respiratory disease | 31 (3.9) | 5 (2.0) | 26 (4.7) | 0.102 |
| Chronic kidney disease | 87 (10.9) | 26 (10.5) | 61 (11.1) | 0.32 |
| Congestive heart failure | 38 (4.8) | 9 (3.6) | 29 (5.3) | 0.407 |
| History of PCI | 199 (25.0) | 68 (27.5) | 131 (23.9) | 0.316 |
| History of CABG | 71 (8.9) | 26 (10.5) | 45 (8.2) | 0.355 |
| History of AMI | 133 (16.7) | 48 (19.4) | 85 (15.5) | 0.288 |
| Active smoker | 197 (24.8) | 73 (29.6) | 124 (22.6) | 0.003 |
MACE Major adverse cardiac events, SD Standard deviation, PCI Percutaneous coronary intervention, CABG Coronary artery bypass graft, AMI Acute myocardial infarction
Descriptive analyses of HRV and HRnV parameters are tabulated in Table 2. In this clinical case study, N was set as 3, thus HR2V, HR2V1, HR3V, HR3V1 and HR3V2 parameters were calculated. Among time domain parameters such as mean NN, SDNN and RMSSD, the HRnV and HRnVm values were generally incremental with an increase in n. Notably, HR2V NN50 and HR3V NN50 were much lower than conventional HRV NN50. Moreover, NN50n and pNN50n are parameters specifically applicable to the HRnV representation. Like time domain parameters, the same trend of changes in frequency domain parameters were observed. The magnitude of increment in VLF power and LF power was larger than that of HF power with increasing n. One exception, however, was the normalized HF power, where HRnV and HRnVm parameters were smaller than that of HRV. In nonlinear analysis, there were marked differences in Poincaré SD2 values between HRV and HRnV parameters. HR2V SampEn and HR3V SampEn were considerably larger compared to SampEn parameters of HRV, HR2V1, HR3V1, and HR3V2, as their confidence intervals (CIs) were wide. The wide CI was due to insufficient data points of less than 200 [19], as our ECG recordings were only five to six minutes long. HR2V1, HR3V1 and HR3V2 were free from this issue as they were calculated from overlapping RRnIm sequences where more data points were available.
Table 2.
Descriptive analyses of heart rate variability (HRV) and heart rate n-variability (HRnV) parameters
| HRV | HR2V | HR2V1 | HR3V | HR3V1 | HR3V2 | |
|---|---|---|---|---|---|---|
| Mean NN (s) | 829.40 (169.49) | 1656.65 (339.85) | 1658.81 (338.99) | 2484.80 (509.33) | 2488.22 (508.50) | 2485.02 (509.84) |
| SDNN (s) | 38.16 (25.49) | 62.28 (45.45) | 68.81 (47.00) | 82.06 (62.47) | 97.79 (67.46) | 87.77 (64.52) |
| RMSSD (s) | 30.04 (23.07) | 32.61 (26.68) | 33.79 (25.67) | 34.83 (28.86) | 36.27 (26.50) | 34.98 (27.43) |
| Skewness | −0.65 (2.34) | −0.41 (1.66) | −0.59 (1.95) | − 0.29 (1.29) | −0.55 (1.69) | − 0.38 (1.42) |
| Kurtosis | 14.59 (26.83) | 7.33 (13.58) | 10.17 (17.90) | 5.15 (8.13) | 8.06 (12.92) | 5.98 (9.75) |
| Triangular index | 7.68 (4.19) | 10.38 (5.10) | 12.60 (6.45) | 11.47 (5.29) | 16.25 (7.94) | 13.06 (6.04) |
| NN50 (count) | 21.08 (33.98) | 14.46 (20.35) | 29.35 (40.03) | 11.57 (15.05) | 35.29 (44.34) | 17.41 (22.51) |
| pNN50 (%) | 6.31 (11.08) | 8.66 (13.18) | 8.75 (12.97) | 10.31 (14.27) | 10.38 (13.95) | 10.28 (14.20) |
| NN50n (count) | – | 4.16 (9.72) | 8.45 (18.76) | 1.37 (3.72) | 4.37 (10.72) | 2.08 (5.48) |
| pNN50n (%) | – | 2.60 (6.67) | 2.64 (6.47) | 1.32 (3.95) | 1.39 (3.86) | 1.33 (3.87) |
| Total power (ms2) | 2518.30 (4797.05) | 7797.46 (16,947.44) | 9156.26 (17,970.75) | 13,904.78 (37,182.24) | 18,714.67 (37,620.26) | 15,706.11 (34,845.52) |
| VLF power (ms2) | 985.18 (1991.52) | 3401.42 (6569.37) | 3922.74 (7987.46) | 6503.53 (14,205.11) | 8772.26 (17,986.63) | 7567.79 (14,666.32) |
| LF power (ms2) | 732.36 (1841.88) | 2626.83 (7593.16) | 2782.48 (7212.62) | 5091.49 (18,402.20) | 5740.99 (15,243.38) | 5397.76 (16,001.18) |
| HF power (ms2) | 527.27 (1232.69) | 1328.86 (4033.96) | 1361.53 (3433.55) | 1661.69 (7237.55) | 1762.45 (4851.11) | 1761.05 (6477.63) |
| LF power norm (nu) | 56.76 (19.20) | 66.82 (18.17) | 66.42 (17.35) | 76.53 (15.32) | 77.65 (14.55) | 77.93 (14.95) |
| HF power norm (nu) | 43.24 (19.20) | 33.18 (18.17) | 33.58 (17.35) | 23.47 (15.32) | 22.35 (14.55) | 22.07 (14.95) |
| LF/HF | 1.99 (1.93) | 3.24 (2.95) | 3.04 (2.73) | 5.60 (5.21) | 5.79 (4.99) | 6.06 (5.18) |
| Poincaré SD1 (ms) | 21.27 (16.34) | 23.12 (18.93) | 23.92 (18.18) | 24.72 (20.50) | 25.68 (18.77) | 24.80 (19.46) |
| Poincaré SD2 (ms) | 48.82 (33.29) | 84.47 (62.15) | 93.88 (64.58) | 112.87 (86.62) | 135.55 (94.02) | 121.20 (89.72) |
| SampEn | 1.57 (0.51) | 83.84 (2324.24) | 1.33 (0.48) | 248.48 (4020.64) | 1.06 (0.41) | 1.14 (0.45) |
| ApEn | 0.99 (0.20) | 0.72 (0.18) | 0.91 (0.17) | 0.60 (0.15) | 0.84 (0.17) | 0.70 (0.15) |
| DFA, α1 | 0.99 (0.31) | 1.24 (0.29) | 1.23 (0.27) | 1.41 (0.27) | 1.42 (0.23) | 1.42 (0.25) |
| DFA, α2 | 0.95 (0.22) | 0.98 (0.35) | 0.98 (0.22) | 0.86 (0.65) | 1.01 (0.22) | 1.02 (0.36) |
HRV Heart rate variability, mean NN Average of R-R intervals, SDNN Standard deviation of R-R intervals, RMSSD Square root of the mean squared differences between R-R intervals; NN50, the number of times that the absolute difference between 2 successive R-R intervals exceeds 50 ms; pNN50, NN50 divided by the total number of R-R intervals; NN50n, the number of times that the absolute difference between 2 successive RRnI/RRnIm sequences exceeds 50 × n ms; pNN50n, NN50n divided by the total number of RRnI/RRnIm sequences; VLF Very low frequency, LF Low frequency, HF High frequency, SD Standard deviation, SampEn Sample entropy, ApEn Approximate entropy, DFA Detrended fluctuation analysis
Table 3 presents the results of univariable analyses of HRnV and HRnVm parameters. Eleven out of 21 conventional HRV parameters were statistically significant. Additionally, 13 HR2V, six HR3V, 11 HR2V1, seven HR3V1 and 11 HR3V2 parameters were also significant. Overall, additional 115 HRnV parameters were derived, among which 48 showed statistical significances between patients with 30-day MACE and those without. Among all HRV and HRnV parameters, mean NN, SDNN, RMSSD, NN50, pNN50, HF power, Poincaré SD1 and SD2 were statistically significant in at least five out of six measures (i.e., HRV, HR2V, HR2V1, HR3V, HR3V1, and HR3V2). Furthermore, skewness, LF power, SampEn, and ApEn, which did not demonstrate statistical significance in conventional HRV analysis, were statistically significant in HRnV representation. Table 4 presents the results of the multivariable analyses of HRnV and HRnVm parameters by adjusting for age and sex. After adjustment, several parameters such as NN50 of HR3V and HR3V2, and triangular index of HRV, HR2V, and HR3V2, became statistically non-significant, while parameters such as ApEn of HR2V, HR2V1, and HR3V2 became statistically significant.
Table 3.
Univariable analysis of HRnV and HRnVm parameters
| HRV | HR2V | HR3V | ||||
| OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | |
| Mean NN | 0.999 (0.998–1.000) | 0.023* | 0.999 (0.999–1.000) | 0.023* | 1.000 (0.999–1.000) | 0.023* |
| SDNN | 0.992 (0.986–0.999) | 0.023* | 0.996 (0.992–1.000) | 0.028* | 0.997 (0.995–1.000) | 0.060 |
| RMSSD | 0.990 (0.982–0.998) | 0.010* | 0.992 (0.985–0.998) | 0.011* | 0.994 (0.988–0.999) | 0.030* |
| Skewness | 1.059 (0.991–1.132) | 0.088 | 1.079 (0.981–1.186) | 0.118 | 1.139 (1.006–1.290) | 0.040* |
| Kurtosis | 1.006 (1.000–1.011) | 0.038* | 1.009 (0.998–1.019) | 0.113 | 1.011 (0.993–1.029) | 0.242 |
| Triangular index | 0.961 (0.925–0.998) | 0.039* | 0.967 (0.938–0.997) | 0.032* | 0.978 (0.950–1.007) | 0.133 |
| NN50 | 0.993 (0.987–0.998) | 0.008* | 0.989 (0.981–0.998) | 0.012* | 0.988 (0.977–0.999) | 0.031* |
| pNN50 | 0.978 (0.962–0.995) | 0.009* | 0.984 (0.971–0.997) | 0.014* | 0.987 (0.976–0.999) | 0.027* |
| NN50n | – | – | 0.982 (0.964–1.001) | 0.065 | 0.952 (0.905–1.002) | 0.059 |
| pNN50n | – | – | 0.974 (0.946–1.002) | 0.069 | 0.951 (0.903–1.001) | 0.054 |
| Total power | 1.000 (1.000–1.000) | 0.031* | 1.000 (1.000–1.000) | 0.021* | 1.000 (1.000–1.000) | 0.072 |
| VLF power | 1.000 (1.000–1.000) | 0.132 | 1.000 (1.000–1.000) | 0.070 | 1.000 (1.000–1.000) | 0.133 |
| LF power | 1.000 (1.000–1.000) | 0.077 | 1.000 (1.000–1.000) | 0.023* | 1.000 (1.000–1.000) | 0.063 |
| HF power | 1.000 (0.999–1.000) | 0.002* | 1.000 (1.000–1.000) | 0.014* | 1.000 (1.000–1.000) | 0.074 |
| LF power norm | 1.001 (0.994–1.009) | 0.738 | 0.999 (0.99–1.007) | 0.733 | 0.994 (0.985–1.004) | 0.248 |
| HF power norm | 0.999 (0.991–1.007) | 0.738 | 1.001 (0.993–1.01) | 0.733 | 1.006 (0.996–1.015) | 0.248 |
| LF/HF | 1.034 (0.959–1.116) | 0.381 | 1.014 (0.964–1.066) | 0.592 | 1.001 (0.973–1.031) | 0.923 |
| Poincaré SD1 | 0.986 (0.975–0.997) | 0.010* | 0.988 (0.979–0.997) | 0.011* | 0.991 (0.983–0.999) | 0.029* |
| Poincaré SD2 | 0.995 (0.990–1.000) | 0.032* | 0.997 (0.994–1.000) | 0.032* | 0.998 (0.996–1.000) | 0.063 |
| SampEn | 0.813 (0.604–1.095) | 0.173 | 0.730 (0.545–0.977) | 0.035* | 1.000 (1.000–1.000) | 0.932 |
| ApEn | 1.645 (0.752–3.598) | 0.213 | 2.319 (1.003–5.357) | 0.049* | 1.241 (0.463–3.327) | 0.667 |
| DFA, α1 | 0.953 (0.585–1.552) | 0.846 | 1.031 (0.611–1.741) | 0.908 | 0.968 (0.560–1.672) | 0.907 |
| DFA, α2 | 1.532 (0.773–3.034) | 0.221 | 1.202 (0.782–1.848) | 0.401 | 1.184 (0.934–1.500) | 0.163 |
| HR2V1 | HR3V1 | HR3V2 | ||||
| OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | |
| Mean NN | 0.999 (0.999–1.000) | 0.023* | 1.000 (0.999–1.000) | 0.023* | 1.000 (0.999–1.000) | 0.023* |
| SDNN | 0.996 (0.993–1.000) | 0.034* | 0.997 (0.995–1.000) | 0.042* | 0.997 (0.995–1.000) | 0.034* |
| RMSSD | 0.991 (0.984–0.998) | 0.010* | 0.992 (0.986–0.999) | 0.016* | 0.993 (0.986–0.999) | 0.016* |
| Skewness | 1.061 (0.980–1.149) | 0.144 | 1.072 (0.978–1.176) | 0.139 | 1.098 (0.982–1.227) | 0.100 |
| Kurtosis | 1.007 (0.999–1.015) | 0.082 | 1.006 (0.994–1.017) | 0.333 | 1.010 (0.995–1.025) | 0.195 |
| Triangular index | 0.981 (0.958–1.005) | 0.119 | 0.982 (0.963–1.001) | 0.065 | 0.974 (0.949–0.999) | 0.040* |
| NN50 | 0.995 (0.991–0.999) | 0.018* | 0.996 (0.993–1.000) | 0.052 | 0.992 (0.985–0.999) | 0.035* |
| pNN50 | 0.984 (0.972–0.997) | 0.020* | 0.988 (0.977–1.000) | 0.049* | 0.988 (0.976–0.999) | 0.035* |
| NN50n | 0.989 (0.979–1.000) | 0.043* | 0.982 (0.964–1.000) | 0.054 | 0.974 (0.943–1.007) | 0.118 |
| pNN50n | 0.969 (0.939–0.999) | 0.046* | 0.947 (0.895–1.002) | 0.058 | 0.960 (0.914–1.009) | 0.109 |
| Total power | 1.000 (1.000–1.000) | 0.048* | 1.000 (1.000–1.000) | 0.072 | 1.000 (1.000–1.000) | 0.029* |
| VLF power | 1.000 (1.000–1.000) | 0.139 | 1.000 (1.000–1.000) | 0.145 | 1.000 (1.000–1.000) | 0.074 |
| LF power | 1.000 (1.000–1.000) | 0.084 | 1.000 (1.000–1.000) | 0.092 | 1.000 (1.000–1.000) | 0.027* |
| HF power | 1.000 (1.000–1.000) | 0.005* | 1.000 (1.000–1.000) | 0.010* | 1.000 (1.000–1.000) | 0.022* |
| LF power norm | 1.000 (0.991–1.008) | 0.937 | 0.995 (0.985–1.006) | 0.382 | 0.995 (0.986–1.005) | 0.356 |
| HF power norm | 1.000 (0.992–1.009) | 0.937 | 1.005 (0.994–1.015) | 0.382 | 1.005 (0.995–1.015) | 0.356 |
| LF/HF | 1.024 (0.970–1.080) | 0.387 | 1.003 (0.973–1.033) | 0.863 | 0.999 (0.971–1.029) | 0.966 |
| Poincaré SD1 | 0.987 (0.978–0.997) | 0.010* | 0.989 (0.980–0.998) | 0.016* | 0.989 (0.981–0.998) | 0.016* |
| Poincaré SD2 | 0.997 (0.995–1.000) | 0.039* | 0.998 (0.996–1.000) | 0.045* | 0.998 (0.996–1.000) | 0.037* |
| SampEn | 0.854 (0.623–1.171) | 0.328 | 0.802 (0.553–1.161) | 0.242 | 0.709 (0.500–1.005) | 0.053 |
| ApEn | 2.065 (0.842–5.064) | 0.113 | 1.207 (0.499–2.922) | 0.677 | 2.558 (0.906–7.222) | 0.076 |
| DFA, α1 | 0.888 (0.514–1.537) | 0.672 | 1.039 (0.547–1.971) | 0.907 | 1.004 (0.549–1.835) | 0.991 |
| DFA, α2 | 1.557 (0.782–3.098) | 0.208 | 1.554 (0.780–3.093) | 0.210 | 1.169 (0.764–1.789) | 0.472 |
HRV Heart rate variability, OR Odds ratio, CI Confidence interval, mean NN Average of R-R intervals, SDNN Standard deviation of R-R intervals, RMSSD Square root of the mean squared differences between R-R intervals, NN50 The number of times that the absolute difference between 2 successive R-R intervals exceeds 50 ms, pNN50, NN50 divided by the total number of R-R intervals; NN50n, the number of times that the absolute difference between 2 successive RRnI/RRnIm sequences exceeds 50 × n ms; pNN50n, NN50n divided by the total number of RRnI/RRnIm sequences; VLF Very low frequency, LF Low frequency, HF High frequency, SD Standard deviation, SampEn Sample entropy, ApEn Approximate entropy, DFA Detrended fluctuation analysis
* p < 0.05
Table 4.
Multivariable analysis of HRnV and HRnVm parameters by adjusting for age and sex
| HRV | HR2V | HR3V | ||||
| OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | |
| Mean NN | 0.999 (0.998–1) | 0.005* | 0.999 (0.999–1.000) | 0.005* | 1.000 (0.999–1.000) | 0.005* |
| SDNN | 0.993 (0.986–0.999) | 0.035* | 0.996 (0.992–1.000) | 0.040* | 0.998 (0.995–1.000) | 0.093 |
| RMSSD | 0.990 (0.982–0.998) | 0.011* | 0.992 (0.985–0.999) | 0.016* | 0.994 (0.988–1.000) | 0.047* |
| Skewness | 1.064 (0.995–1.138) | 0.068 | 1.082 (0.983–1.191) | 0.109 | 1.140 (1.005–1.293) | 0.042* |
| Kurtosis | 1.005 (1.000–1.011) | 0.047* | 1.008 (0.997–1.019) | 0.139 | 1.011 (0.993–1.030) | 0.238 |
| Triangular index | 0.967 (0.93–1.006) | 0.093 | 0.971 (0.940–1.002) | 0.070 | 0.982 (0.953–1.013) | 0.256 |
| NN50 | 0.993 (0.988–0.999) | 0.013* | 0.991 (0.982–0.999) | 0.030* | 0.990 (0.979–1.001) | 0.078 |
| pNN50 | 0.979 (0.963–0.996) | 0.015* | 0.986 (0.972–0.999) | 0.033* | 0.989 (0.977–1.001) | 0.063 |
| NN50n | – | – | 0.983 (0.964–1.002) | 0.081 | 0.954 (0.906–1.005) | 0.077 |
| pNN50n | – | – | 0.975 (0.947–1.004) | 0.086 | 0.952 (0.903–1.004) | 0.069 |
| Total power | 1.000 (1.000–1.000) | 0.042* | 1.000 (1.000–1.000) | 0.026* | 1.000 (1.000–1.000) | 0.104 |
| VLF power | 1.000 (1.000–1.000) | 0.167 | 1.000 (1.000–1.000) | 0.082 | 1.000 (1.000–1.000) | 0.152 |
| LF power | 1.000 (1.000–1.000) | 0.093 | 1.000 (1.000–1.000) | 0.033* | 1.000 (1.000–1.000) | 0.105 |
| HF power | 1.000 (0.999–1.000) | 0.003* | 1.000 (1.000–1.000) | 0.016* | 1.000 (1.000–1.000) | 0.101 |
| LF power norm | 1.002 (0.994–1.011) | 0.589 | 0.999 (0.990–1.007) | 0.769 | 0.994 (0.984–1.003) | 0.202 |
| HF power norm | 0.998 (0.989–1.006) | 0.589 | 1.001 (0.993–1.010) | 0.769 | 1.006 (0.997–1.016) | 0.202 |
| LF/HF | 1.039 (0.961–1.124) | 0.336 | 1.013 (0.962–1.066) | 0.620 | 0.999 (0.970–1.028) | 0.928 |
| Poincaré SD1 | 0.986 (0.975–0.997) | 0.011* | 0.989 (0.980–0.998) | 0.016* | 0.992 (0.983–1.000) | 0.047* |
| Poincaré SD2 | 0.995 (0.990–1.000) | 0.050* | 0.997 (0.994–1.000) | 0.046* | 0.998 (0.996–1.000) | 0.098 |
| SampEn | 0.852 (0.630–1.152) | 0.297 | 0.752 (0.559–1.010) | 0.058 | 1.000 (1.000–1.000) | 0.956 |
| ApEn | 1.669 (0.754–3.693) | 0.207 | 2.668 (1.139–6.246) | 0.024* | 1.507 (0.555–4.096) | 0.421 |
| DFA, α1 | 0.991 (0.593–1.654) | 0.971 | 1.072 (0.622–1.848) | 0.802 | 0.962 (0.550–1.682) | 0.891 |
| DFA, α2 | 1.499 (0.750–2.993) | 0.252 | 1.204 (0.782–1.853) | 0.400 | 1.193 (0.941–1.512) | 0.146 |
| HR2V1 | HR3V1 | HR3V2 | ||||
| OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | |
| Mean NN | 0.999 (0.999–1.000) | 0.005* | 1.000 (0.999–1.000) | 0.005* | 1.000 (0.999–1.000) | 0.005* |
| SDNN | 0.996 (0.993–1.000) | 0.052 | 0.998 (0.995–1.000) | 0.064 | 0.997 (0.995–1.000) | 0.049* |
| RMSSD | 0.992 (0.985–0.998) | 0.015* | 0.993 (0.986–0.999) | 0.023* | 0.993 (0.987–0.999) | 0.023* |
| Skewness | 1.066 (0.984–1.156) | 0.118 | 1.079 (0.983–1.185) | 0.108 | 1.099 (0.982–1.229) | 0.099 |
| Kurtosis | 1.007 (0.999–1.015) | 0.096 | 1.005 (0.994–1.017) | 0.377 | 1.010 (0.994–1.025) | 0.218 |
| Triangular index | 0.985 (0.960–1.010) | 0.234 | 0.985 (0.965–1.005) | 0.137 | 0.977 (0.951–1.003) | 0.088 |
| NN50 | 0.996 (0.991–1.000) | 0.047* | 0.997 (0.993–1.001) | 0.130 | 0.993 (0.986–1.001) | 0.084 |
| pNN50 | 0.986 (0.973–1.000) | 0.046* | 0.990 (0.979–1.002) | 0.111 | 0.989 (0.978–1.001) | 0.076 |
| NN50n | 0.990 (0.980–1.000) | 0.059* | 0.982 (0.963–1.001) | 0.064 | 0.975 (0.943–1.008) | 0.142 |
| pNN50n | 0.971 (0.941–1.002) | 0.063 | 0.947 (0.893–1.004) | 0.067 | 0.962 (0.915–1.012) | 0.131 |
| Total power | 1.000 (1.000–1.000) | 0.064 | 1.000 (1.000–1.000) | 0.096 | 1.000 (1.000–1.000) | 0.035* |
| VLF power | 1.000 (1.000–1.000) | 0.173 | 1.000 (1.000–1.000) | 0.180 | 1.000 (1.000–1.000) | 0.086 |
| LF power | 1.000 (1.000–1.000) | 0.100 | 1.000 (1.000–1.000) | 0.108 | 1.000 (1.000–1.000) | 0.037* |
| HF power | 1.000 (1.000–1.000) | 0.006* | 1.000 (1.000–1.000) | 0.014* | 1.000 (1.000–1.000) | 0.025* |
| LF power norm | 1.000 (0.991–1.009) | 0.960 | 0.995 (0.984–1.005) | 0.324 | 0.995 (0.985–1.005) | 0.329 |
| HF power norm | 1.000 (0.991–1.009) | 0.960 | 1.005 (0.995–1.016) | 0.324 | 1.005 (0.995–1.015) | 0.329 |
| LF/HF | 1.023 (0.968–1.081) | 0.428 | 0.999 (0.969–1.030) | 0.940 | 0.996 (0.967–1.026) | 0.786 |
| Poincaré SD1 | 0.988 (0.979–0.998) | 0.015* | 0.990 (0.981–0.999) | 0.023* | 0.990 (0.981–0.999) | 0.023* |
| Poincaré SD2 | 0.997 (0.995–1.000) | 0.059 | 0.998 (0.997–1.000) | 0.068 | 0.998 (0.996–1.000) | 0.052 |
| SampEn | 0.870 (0.632–1.197) | 0.393 | 0.842 (0.578–1.227) | 0.371 | 0.716 (0.504–1.019) | 0.064 |
| ApEn | 2.520 (1.009–6.298) | 0.048* | 1.413 (0.575–3.471) | 0.451 | 3.461 (1.201–9.971) | 0.021* |
| DFA, α1 | 0.898 (0.508–1.587) | 0.710 | 1.068 (0.555–2.058) | 0.843 | 1.005 (0.543–1.838) | 0.988 |
| DFA, α2 | 1.507 (0.751–3.025) | 0.249 | 1.500 (0.746–3.014) | 0.255 | 1.172 (0.764–1.798) | 0.467 |
HRV Heart rate variability, OR Odds ratio, CI Confidence interval, mean NN average of R-R intervals, SDNN Standard deviation of R-R intervals, RMSSD Square root of the mean squared differences between R-R intervals, NN50, the number of times that the absolute difference between 2 successive R-R intervals exceeds 50 ms; pNN50, NN50 divided by the total number of R-R intervals; NN50n, the number of times that the absolute difference between 2 successive RRnI/RRnIm sequences exceeds 50 × n ms; pNN50n, NN50n divided by the total number of RRnI/RRnIm sequences; VLF Very low frequency, LF Low frequency, HF High frequency, SD Standard deviation, SampEn Sample entropy, ApEn Approximate entropy, DFA Detrended fluctuation analysis
* p < 0.05
Table 5 lists the 16 variables that were selected through multivariable stepwise logistic regression, among which there were one conventional HRV parameter and seven HRnV parameters. In addition to traditional predictors of adverse cardiac outcomes such as ST segment changes and troponin, HR2V ApEn (OR = 0.095; 95% CI 0.014–0.628), HR2V1 ApEn (OR = 19.700; 95% CI 2.942–131.900) and HR3V skewness (1.560; 95% CI 1.116–2.181) also demonstrated strong predictive power in assessing the risk of 30-day MACE. Figure 2 shows the ROC curves and Table 6 presents the results of ROC analysis in evaluating the predictive performance of the HRnV model (based on LOOCV), HEART, TIMI, and GRACE scores. The HRnV model achieved the highest AUC value and outperformed HEART, TIMI, and GRACE scores in terms of specificity, PPV, and NPV at the optimal cut-off scores, defined as the points nearest to the upper-left corner of the ROC curves.
Table 5.
Multivariable analysis with stepwise logistic regression (backward selection) on all variables
| Variable | Adjusted OR | 95% CI |
|---|---|---|
| Age | 1.021 | 1.002–1.041 |
| Diastolic BP | 1.018 | 1.003–1.034 |
| Pain score | 1.082 | 1.003–1.168 |
| ST-elevation | 6.449 | 2.762–15.059 |
| ST-depression | 4.827 | 2.511–9.277 |
| Q wave | 3.383 | 1.668–6.860 |
| Cardiac historya | 7.838 | 5.192–11.832 |
| Troponin | 4.406 | 3.218–6.033 |
| HRV NN50 | 0.981 | 0.970–0.991 |
| HR2V skewness | 0.806 | 0.622–1.045 |
| HR2V SampEn | 0.600 | 0.348–1.035 |
| HR2V ApEn | 0.095 | 0.014–0.628 |
| HR2V1 ApEn | 19.700 | 2.942–131.900 |
| HR3V RMSSD | 1.024 | 1.008–1.040 |
| HR3V skewness | 1.560 | 1.116–2.181 |
| HR3V2 HF power | 1.000 | 1.000–1.000 |
BP Blood pressure, HRV Heart rate variability, OR Odds ratio, CI Confidence interval; mean NN, average of R-R intervals; RMSSD, square root of the mean squared differences between R-R intervals; NN50, the number of times that the absolute difference between 2 successive R-R intervals exceeds 50 ms; LF Low frequency, HF High frequency, SampEn Sample entropy, ApEn Approximate entropy
aCardiac history was a numeric value that was derived from the narrative in the hospital charts. Its value was zero if the patient history contained characteristics of atypical cardiac chest pain; Its value was two if the history contained characteristics of typical cardiac chest pain; Its value was one if the history contained characteristics of both atypical and typical cardiac chest pain
Fig. 2.
The receiver operating characteristic (ROC) curves produced by heart rate n-variability (HRnV) model (performance was based on leave-one-out cross-validation), the History, ECG, Age, Risk factors and Troponin (HEART) score, the Thrombolysis in Myocardial Infarction (TIMI) score, and the Global Registry of Acute Coronary Events (GRACE) score
Table 6.
Comparison of performance of the HRnV model (based on leave-one-out cross-validation), HEART, TIMI, and GRACE scores in predicting 30-day major adverse cardiac events (MACE)
| AUC (95% CI) | Cut-off | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) | |
|---|---|---|---|---|---|---|
| HRnV Model | 0.888 (0.860–0.917) | 0.3611a | 77.3% (72.1–82.5%) | 81.8% (78.5–85.0%) | 65.6% (60.2–71.1%) | 88.9% (86.1–91.6%) |
| – | 0.0352 | 99.2% (98.1–100.0%) | 39.6% (35.5–43.7%) | 42.5% (38.5–46.6%) | 99.1% (97.8–100.0%) | |
| HEART | 0.841 (0.808–0.874) | 5a | 78.9% (73.9–84.0%) | 72.8% (69.1–76.5%) | 56.7% (51.4–61.9%) | 88.5% (85.5–91.4%) |
| – | 3 | 99.6% (98.8–100.0%) | 35.8% (31.8–39.8%) | 41.1% (37.2–45.1%) | 99.5% (98.5–100.0%) | |
| TIMI | 0.681 (0.639–0.723) | 2a | 63.6% (57.6–69.6%) | 58.4% (54.3–62.5%) | 40.8% (35.9–45.7%) | 78.0% (74.0–82.1%) |
| – | 0 | 98.4% (96.8–100.0%) | 19.3% (16.0–22.7%) | 35.5% (31.9–39.1%) | 96.4% (92.9–99.9%) | |
| GRACE | 0.665 (0.623–0.707) | 107a | 64.0% (58.0–70.0%) | 60.8% (56.7–64.9%) | 42.4% (37.3–47.4%) | 78.9% (75.0–82.8%) |
| – | 60 | 98.8% (97.4–100.0%) | 8.0% (5.8–10.3%) | 32.6% (29.3–36.0%) | 93.6% (86.6–100.0%) |
AUC Area under the curve, CI Confidence interval, PPV Positive predictive value, NPV Negative predictive value, HEART History, ECG, Age, Risk factors and Troponin, TIMI Thrombolysis in Myocardial Infarction, GRACE Global Registry of Acute Coronary Events
aOptimal cut-off values, defined as the points nearest to the upper-left corner on the ROC curves
Discussion
HRV has generated significant research interest in the past decades [18, 19, 29], with majority of studies focusing on development of advanced nonlinear techniques to derive novel parameters [30, 31]. There is, however, a paucity of research on alternative approaches to analyze RRIs. Vollmer [32] used relative RRIs to describe the relative variation of consecutive RRIs, with which HRV was analyzed. Likewise, we proposed a novel HRnV representation, providing more HRnV parameters than conventional HRV analysis. In this paper, we introduced two measures of HRnV, namely HRnV and HRnVm. HRnV was calculated based on non-overlapping RRnI sequences, while HRnVm was computed from overlapping RRnIm sequences. HRnV was not developed to replace the conventional HRV but to augment it. It enables the creation of additional parameters from raw ECGs, and thus empowers the extraction of supplementary information.
In our clinical case study, we investigated the predictive value of HRnV parameters in assessing the risk of 30-day MACE for chest pain patients in the ED. In addition to 21 HRV parameters, 115 HRnV parameters were derived, of which 48 were found to be statistically significant in their association with the primary outcome. Notably, even with a small n (three in our study), newly generated HRnV parameters greatly boosted the number of candidate predictors. When longer ECG records are available, more HRnV parameters can be calculated. With HRnV parameters, HRV parameters, vital signs, and several established risk factors, we conducted multivariable logistic regression analysis and selected age, diastolic BP, pain score, ST-elevation, ST-depression, Q wave, cardiac history, troponin, HRV NN50, and seven HRnV parameters. In addition to traditional risk factors such as ST segment changes, HR2V ApEn, HR2V1 ApEn, and HR3V skewness were found to be strong predictors for 30-day MACE. Compared to the HEART, TIMI, and GRACE scores, the HRnV model achieved the highest AUC, specificity, PPV, and NPV values at the optimal cut-off points in ROC analysis. This demonstrated the clinical utility of HRnV in determining the risk of 30-day MACE for ED patients with chest pain.
Due to the wide differential diagnosis for chest pain, accurate stratification is vital, particularly for preventing low-risk patients from obtaining expensive and unnecessary medical testing and intervention [3]. Although the TIMI and GRACE scores have been validated for risk prediction of patients with chest pain in the ED [4, 33, 34], some criteria used in these scores may be inappropriate for undifferentiated chest pain cohorts in the ED, as they were originally developed for post-acute myocardial infarction patients [1]. In comparison, the HEART score was derived from a population of ED patients with chest pain, and has been extensively validated worldwide [10, 13, 27, 35]. It has demonstrated its utility in identifying both low-risk patients for possible early discharge and high-risk patients for urgent intervention. Built upon established scores, several chest pain pathways [14, 36–38] have been implemented and tested, particularly for the management of low-risk patients. Than et al. [38] evaluated a TIMI score-based accelerated diagnostic protocol (ADP) with a reported sensitivity of 99.3% and NPV of 99.1%. Similarly, a systematic review by Laureano-Phillips et al. [39] reported that the HEART score achieved both sensitivity and NPV of 100% in several validation studies. Furthermore, a cost-effectiveness study conducted in Brisbane, Australia reported economic benefits by adopting an ADP in the ED, with reduction in expected cost and length of stay amongst patients with chest pain [40].
Most established clinical scores use conventional risk factors such as biomarkers, medical history, and presenting vital signs. However, patient history can sometimes be subjective and blood tests, such as troponin, require waiting time. HRV, as a noninvasive measure, can be easily calculated from ECGs; it is an objective tool to assess the activities of the ANS [19]. It also has the advantage of requiring only several minutes to acquire (five to six minutes in our protocol), which is much faster than serum biomarkers. Over the past decades, HRV has been widely investigated in a broad range of clinical applications, particularly in cardiovascular research. Apart from being associated with sudden cardiac death [18], HRV also showed significant correlations with adverse clinical outcomes in prehospital setting [41] and with MACE outcomes in ED patients with chest pain [17]. HRV parameters have been integrated with other risk factors into machine learning algorithms to predict adverse outcomes [42, 43]. These promising results motivated the use of HRV to develop objective and computerized risk stratification tools for chest pain patients [44, 45]. In an updated review of clinical scores for chest pain, Liu et al. [5] summarized several studies which aimed to develop alternative techniques for risk stratification.
This study aimed to present novel HRnV representation and its measures and investigate their association with clinical outcomes. Although HRnV parameters showed promising performance in identifying high-risk chest pain patients, this study was not intended to create a ready-to-use clinical tool. Instead, we demonstrated the feasibility of utilizing HRnV parameters to augment conventional HRV and risk factors in designing a prediction tool/score. These HRnV parameters can be readily calculated without the collection of supplementary data. In this study, with five to six-minute ECG recording and n = 3, five-fold more HRnV parameters were calculated compared to HRV alone. When longer ECG recordings are available and parameter n is larger, more HRnV parameters can be derived. To build a HRnV-based risk stratification tool, a systematic approach is needed to derive a point-based, consistent score to ease its clinical application and practical implementation.
As a natural extension of conventional HRV, HRnV representation creates the opportunity to generate additional parameters. This representation could also serve as a smoother for RRIs, making them less sensitive to sudden changes caused by abnormal heart beats (e.g. very short or very long RRI). However, since HRnV is a novel representation of beat-to-beat variations in ECG, many technical issues need to be addressed in future research. For instance, as shown in Table 2, SampEn became larger when the available number of data points was less than 200 [19], suggesting that additional research is required to investigate its applicability to short ECG records. Moreover, parameters NN50n and pNN50n are newly introduced in HRnV representation only. They characterize the number of times that the absolute difference between two successive RRnI sequences exceeds 50 × n ms, by assuming that the absolute difference may be magnified when the corresponding RRnI is n times longer than RRI. Thus, in-depth investigations are required in the selection of appropriate thresholds. More importantly, physiological interpretations of the HRnV parameters and their normal values [29] need to be determined through numerous research. One example is the identification of frequency bands that correlate with certain physiological phenomenon. In the current analysis, the conventional cut-off values were adopted (i.e., ≤0.04 Hz as very low frequency range; 0.04–0.15 Hz as low frequency range; 0.15–0.4 Hz as high frequency range). With the increase in n, frequency domain analysis may need to be changed accordingly.
Beyond its use in risk stratification of ED patients with chest pain, HRnV can potentially be used in other clinical domains, where conventional HRV has been extensively investigated [46–49]. With the augmented RRnI and RRnIm sequences, HRnV could possibly capture more dynamic changes in cardiac rhythms than HRV. This capability enables the extraction of additional information from limited raw ECGs. This study utilized HRnV parameters as independent risk factors and analyzed them with traditional biostatistical methods. There are multiple ways to use HRnV parameters, e.g. each set of HRnV parameters can be analyzed individually and subsequently combined with an ensemble learning [50] (a special type of machine learning algorithm) architecture to reach a decision. However, artificial intelligence and machine learning methods generally create black-box predictive models, making interpretation a challenge [51].
Limitations
This study has several limitations. First, we did not develop a scoring tool for practical clinical use. The primary aim of this study was to demonstrate the feasibility of using HRnV parameters and common risk factors to build predictive models. Second, the HRnV model was evaluated with LOOCV strategy due to the small sample size. Ideally, separate patient cohorts are needed to train and test prediction models. When a new scoring tool is developed, it is necessary to conduct external validations on cohorts with diverse patient characteristics. Furthermore, properly designed clinical pathways are needed as well. Third, the patients included in this study were mainly from the high acuity group, resulting in a higher 30-day MACE rate (i.e., 31%) compared to other similar studies [10, 39]. As a result, the generalizability of the HRnV model developed in this study may be uncertain in other patient cohorts. Fourth, the calculated HRnV and HRV parameters depended on the choice of tools and methods for ECG signal analysis. Thus, the values of these parameters may vary across studies. Last, the physiology of HRnV and interpretations of its measures are mostly unknown; calculation of some parameters also needs to be standardized. All these require future collaborative research efforts between clinicians and scientists to address.
Conclusions
In this study, we proposed a novel HRnV representation and investigated the use of HRnV and established risk factors to develop a predictive model for risk stratification of patients with chest pain in the ED. Multiple HRnV parameters were found to be statistically significant predictors, which effectively augmented conventional HRV, vital signs, troponin, and cardiac risk factors in building an effective model with good discrimination performance. The HRnV model outperformed the HEART, TIMI, and GRACE scores in the ROC analysis. It also demonstrated its capability in identifying low-risk patients, which could potentially be used to build a new clinical pathway. Moving forward, we suggest further development of a point-based, ready-to-use HRnV risk stratification tool. Although some issues remain to be addressed, we hope to stimulate a new stream of research on HRnV. We believe that future endeavors in this field will lead to the possibility of in-depth evaluation of the associations between HRnV measures and various human diseases.
Acknowledgements
We would like to thank and acknowledge the contributions of doctors, nurses, and clinical research coordinators from the Department of Emergency Medicine, Singapore General Hospital.
Abbreviations
- ACS
Acute coronary syndrome
- ADP
Accelerated diagnostic protocol
- AMI
Acute myocardial infarction
- ANS
Autonomic nervous system
- AUC
Area under the curve
- ApEn
Approximate entropy
- CABG
Coronary artery bypass graft
- CI
Confidence interval
- DFA
Detrended fluctuation analysis
- ED
Emergency department
- EHR
Electronic health records
- GRACE
Global registry of acute coronary events
- HEART
History, ECG, age, risk factors and troponin
- HF
High frequency
- HRnV
Heart rate n-variability
- HRV
Heart rate variability
- LF
Low frequency
- LOOCV
Leave-one-out cross-validation
- MACE
Major adverse cardiac events
- Mean NN
Average of R-R intervals
- MI
Myocardial infarction
- NN50
The number of times that the absolute difference between 2 successive R-R intervals exceeds 50 ms
- NN50n
The number of times that the absolute difference between 2 successive RRnI/RRnIm sequences exceeds 50 × n ms
- NPV
Negative predictive value
- PACS
Patient acuity category scale
- PCI
Percutaneous coronary intervention
- pNN50
NN50 divided by the total number of R-R intervals
- pNN50n
NN50n divided by the total number of RRnI/RRnIm sequences
- PPV
Positive predictive value
- RMSSD
Square root of the mean squared differences between R-R intervals
- ROC
Receiver operating characteristic
- RRI
R-R interval
- SampEn
Sample entropy
- SD
Standard deviation
- SDNN
Standard deviation of R-R intervals
- STEMI
ST-elevation myocardial infarction
- TIMI
Thrombolysis in Myocardial Infarction
- VLF
Very low frequency
Authors’ contributions
NL invented the HRnV representation, conceived the study, supervised the project, and wrote the first draft of the manuscript. NL, DG, ZXK, and FX performed the analyses. NL, DG, ZXK, AFWH, FX, TT, JTS, PPP, BC, SHL, JWCT, and MEHO contributed to evaluation of the HRnV measures, interpretation of the results, and revision of the manuscript. NL, DG, ZXK, AFWH, FX, TT, JTS, PPP, BC, SHL, JWCT, and MEHO approved the final manuscript.
Funding
This work was supported by the SHF Foundation Research Grant (SHF/FG652P/2017). The study sponsor was not involved in the study design; in the collection, analysis and interpretation of data; in the writing of the manuscript; and in the decision to submit the manuscript for publication.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The ethical approval was obtained from the Centralized Institutional Review Board (CIRB, Ref: 2014/584/C) of SingHealth, in which patient consent was waived.
Consent for publication
Not applicable.
Competing interests
NL and MEHO hold patents related to using heart rate variability and artificial intelligence for medical monitoring. NL, DG, ZXK, and MEHO are currently advisers to TIIM SG. The other authors report no conflicts.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Long B, Koyfman A. Best clinical practice: current controversies in the evaluation of Low-risk chest pain with risk stratification aids. Part 2. J Emerg Med. 2017;52(1):43–51. doi: 10.1016/j.jemermed.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Long B, Koyfman A. Best clinical practice: current controversies in evaluation of Low-risk chest pain-part 1. J Emerg Med. 2016;51(6):668–676. doi: 10.1016/j.jemermed.2016.07.103. [DOI] [PubMed] [Google Scholar]
- 3.Hollander JE, Than M, Mueller C. State-of-the-art evaluation of emergency department patients presenting with potential acute coronary syndromes. Circulation. 2016;134(7):547–564. doi: 10.1161/CIRCULATIONAHA.116.021886. [DOI] [PubMed] [Google Scholar]
- 4.Backus BE, Six AJ, Kelder JH, Gibler WB, Moll FL, Doevendans PA. Risk scores for patients with chest pain: evaluation in the emergency department. Curr Cardiol Rev. 2011;7(1):2–8. doi: 10.2174/157340311795677662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu N, Ng JCJ, Ting CE, Sakamoto JT, Ho AFW, Koh ZX, Pek PP, Lim SH, Ong MEH. Clinical scores for risk stratification of chest pain patients in the emergency department: an updated systematic review. J Emerg Crit Care Med. 2018;2:16.
- 6.Six AJ, Cullen L, Backus BE, Greenslade J, Parsonage W, Aldous S, Doevendans PA, Than M. The HEART score for the assessment of patients with chest pain in the emergency department: a multinational validation study. Crit Pathw Cardiol. 2013;12(3):121–126. doi: 10.1097/HPC.0b013e31828b327e. [DOI] [PubMed] [Google Scholar]
- 7.Antman EM, Cohen M, Bernink PJ, McCabe CH, Horacek T, Papuchis G, Mautner B, Corbalan R, Radley D, Braunwald E. The TIMI risk score for unstable angina/non-ST elevation MI: a method for prognostication and therapeutic decision making. Jama. 2000;284(7):835–842. doi: 10.1001/jama.284.7.835. [DOI] [PubMed] [Google Scholar]
- 8.Granger CB, Goldberg RJ, Dabbous O, Pieper KS, Eagle KA, Cannon CP, Van De Werf F, Avezum A, Goodman SG, Flather MD, et al. Predictors of hospital mortality in the global registry of acute coronary events. Arch Intern Med. 2003;163(19):2345–2353. doi: 10.1001/archinte.163.19.2345. [DOI] [PubMed] [Google Scholar]
- 9.Sakamoto JT, Liu N, Koh ZX, Guo D, Heldeweg MLA, Ng JCJ, Ong MEH. Integrating heart rate variability, vital signs, electrocardiogram, and troponin to triage chest pain patients in the ED. Am J Emerg Med. 2018;36(2):185–192. [DOI] [PubMed]
- 10.Poldervaart JM, Langedijk M, Backus BE, Dekker IMC, Six AJ, Doevendans PA, Hoes AW, Reitsma JB. Comparison of the GRACE, HEART and TIMI score to predict major adverse cardiac events in chest pain patients at the emergency department. Int J Cardiol. 2017;227:656–661. doi: 10.1016/j.ijcard.2016.10.080. [DOI] [PubMed] [Google Scholar]
- 11.Fernando SM, Tran A, Cheng W, Rochwerg B, Taljaard M, Thiruganasambandamoorthy V, Kyeremanteng K, Perry JJ. Prognostic accuracy of the HEART score for prediction of major adverse cardiac events in patients presenting with chest pain: a systematic review and meta-analysis. Acad Emerg Med Off J Soc Acad Emerg Med. 2019;26(2):140–151. doi: 10.1111/acem.13649. [DOI] [PubMed] [Google Scholar]
- 12.Reaney PDW, Elliott HI, Noman A, Cooper JG. Risk stratifying chest pain patients in the emergency department using HEART, GRACE and TIMI scores, with a single contemporary troponin result, to predict major adverse cardiac events. Emerg Med J. 2018;35(7):420–427. doi: 10.1136/emermed-2017-207172. [DOI] [PubMed] [Google Scholar]
- 13.Van Den Berg P, Body R. The HEART score for early rule out of acute coronary syndromes in the emergency department: a systematic review and meta-analysis. Eur Heart J Acute Cardiovasc Care. 2018;7(2):111–119. doi: 10.1177/2048872617710788. [DOI] [PubMed] [Google Scholar]
- 14.Mahler SA, Riley RF, Hiestand BC, Russell GB, Hoekstra JW, Lefebvre CW, Nicks BA, Cline DM, Askew KL, Elliott SB, et al. The HEART pathway randomized trial: identifying emergency department patients with acute chest pain for early discharge. Circ Cardiovasc Qual Outcomes. 2015;8(2):195–203. doi: 10.1161/CIRCOUTCOMES.114.001384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu N, Lin Z, Cao J, Koh ZX, Zhang T, Huang G-B, Ser W, Ong MEH. An intelligent scoring system and its application to cardiac arrest prediction. IEEE Trans Inf Technol Biomed. 2012;16(6):1324–1331. doi: 10.1109/TITB.2012.2212448. [DOI] [PubMed] [Google Scholar]
- 16.Liu N, Koh ZX, Chua ECP, Tan LML, Lin Z, Mirza B, Ong MEH. Risk scoring for prediction of acute cardiac complications from imbalanced clinical data. IEEE J Biomed Health Inform. 2014;18(6):1894–1902. doi: 10.1109/JBHI.2014.2303481. [DOI] [PubMed] [Google Scholar]
- 17.Ong MEH, Goh K, Fook-Chong S, Haaland B, Wai KL, Koh ZX, Shahidah N, Lin Z. Heart rate variability risk score for prediction of acute cardiac complications in ED patients with chest pain. Am J Emerg Med. 2013;31(8):1201–1207. doi: 10.1016/j.ajem.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 18.Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology Heart rate variability: standards of measurement, physiological interpretation and clinical use. Circulation. 1996;93(5):1043–1065. doi: 10.1161/01.CIR.93.5.1043. [DOI] [PubMed] [Google Scholar]
- 19.Rajendra Acharya U, Paul Joseph K, Kannathal N, Lim CM, Suri JS. Heart rate variability: a review. Med Biol Eng Comput. 2006;44(12):1031–1051. doi: 10.1007/s11517-006-0119-0. [DOI] [PubMed] [Google Scholar]
- 20.Billman GE. Heart rate variability - a historical perspective. Front Physiol. 2011;2:86. doi: 10.3389/fphys.2011.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carpeggiani C, L'Abbate A, Landi P, Michelassi C, Raciti M, Macerata A, Emdin M. Early assessment of heart rate variability is predictive of in-hospital death and major complications after acute myocardial infarction. Int J Cardiol. 2004;96(3):361–368. doi: 10.1016/j.ijcard.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 22.Vest AN, Da Poian G, Li Q, Liu C, Nemati S, Shah AJ, Clifford GD. An open source benchmarked toolbox for cardiovascular waveform and interval analysis. Physiol Meas. 2018;39(10):105004. doi: 10.1088/1361-6579/aae021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarvainen MP, Niskanen JP, Lipponen JA, Ranta-Aho PO, Karjalainen PA. Kubios HRV–heart rate variability analysis software. Comput Methods Prog Biomed. 2014;113:210–220. doi: 10.1016/j.cmpb.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 24.Liu N, Guo D, Koh ZX, Ho AFW, Ong MEH. Heart rate n-variability (HRnV): A novel representation of beat-to-beat variation in electrocardiogram. bioRxiv. 2018:449504. 10.1101/449504.
- 25.Ho AF, Fook-Chong S, Pek PP, Ng YY, Wong AS, Ong ME. Prehospital presentation of patients with ST-segment elevation myocardial infarction in Singapore. Int J Cardiol. 2013;168(4):4273–4276. doi: 10.1016/j.ijcard.2013.04.204. [DOI] [PubMed] [Google Scholar]
- 26.Januzzi JL, Jr, Bamberg F, Lee H, Truong QA, Nichols JH, Karakas M, Mohammed AA, Schlett CL, Nagurney JT, Hoffmann U, et al. High-sensitivity troponin T concentrations in acute chest pain patients evaluated with cardiac computed tomography. Circulation. 2010;121(10):1227–1234. doi: 10.1161/CIRCULATIONAHA.109.893826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakamoto JT, Liu N, Koh ZX, Fung NX, Heldeweg ML, Ng JC, Ong ME. Comparing HEART, TIMI, and GRACE scores for prediction of 30-day major adverse cardiac events in high acuity chest pain patients in the emergency department. Int J Cardiol. 2016;221:759–764. doi: 10.1016/j.ijcard.2016.07.147. [DOI] [PubMed] [Google Scholar]
- 28.Fawcett T. An introduction to ROC analysis. Pattern Recogn Lett. 2006;27(8):861–874. doi: 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- 29.Shaffer F, Ginsberg JP. An overview of heart rate variability metrics and norms. Front Public Health. 2017;5:258. doi: 10.3389/fpubh.2017.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng CK, Havlin S, Stanley HE, Goldberger AL. Quantification of scaling exponents and crossover phenomena in nonstationary heartbeat time series. Chaos. 1995;5(1):82–87. doi: 10.1063/1.166141. [DOI] [PubMed] [Google Scholar]
- 31.Barbieri R, Matten EC, Alabi AA, Brown EN. A point-process model of human heartbeat intervals: new definitions of heart rate and heart rate variability. Am J Phys Heart Circ Phys. 2005;288(1):H424–H435. doi: 10.1152/ajpheart.00482.2003. [DOI] [PubMed] [Google Scholar]
- 32.Vollmer M. Computing in Cardiology Conference (CinC): 6–9 Sept. 2015. 2015. A robust, simple and reliable measure of heart rate variability using relative RR intervals; pp. 609–612. [Google Scholar]
- 33.Chase M, Robey JL, Zogby KE, Sease KL, Shofer FS, Hollander JE. Prospective validation of the thrombolysis in myocardial infarction risk score in the emergency department chest pain population. Ann Emerg Med. 2006;48(3):252–259. doi: 10.1016/j.annemergmed.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 34.Pollack CV, Sites FD, Shofer FS, Sease KL, Hollander JE. Application of the TIMI risk score for unstable angina and non-ST elevation acute coronary syndrome to an unselected emergency department chest pain population. Acad Emerg Med Off J Soc Acad Emerg Med. 2006;13(1):13–18. doi: 10.1197/j.aem.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 35.Marcoon S, Chang AM, Lee B, Salhi R, Hollander JE. HEART score to further risk stratify patients with low TIMI scores. Crit Pathw Cardiol. 2013;12(1):1–5. doi: 10.1097/HPC.0b013e31827377e1. [DOI] [PubMed] [Google Scholar]
- 36.Long B, Oliver J, Streitz M, Koyfman A. An end-user's guide to the HEART score and pathway. Am J Emerg Med. 2017;35(9):1350–1355. doi: 10.1016/j.ajem.2017.03.047. [DOI] [PubMed] [Google Scholar]
- 37.Than MP, Pickering JW, Aldous SJ, Cullen L, Frampton CM, Peacock WF, Jaffe AS, Goodacre SW, Richards AM, Ardagh MW, et al. Effectiveness of EDACS versus ADAPT accelerated diagnostic pathways for chest pain: a pragmatic randomized controlled trial embedded within practice. Ann Emerg Med. 2016;68(1):93–102. doi: 10.1016/j.annemergmed.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Than M, Cullen L, Reid CM, Lim SH, Aldous S, Ardagh MW, Peacock WF, Parsonage WA, Ho HF, Ko HF, et al. A 2-h diagnostic protocol to assess patients with chest pain symptoms in the Asia-Pacific region (ASPECT): a prospective observational validation study. Lancet (London, England) 2011;377(9771):1077–1084. doi: 10.1016/S0140-6736(11)60310-3. [DOI] [PubMed] [Google Scholar]
- 39.Laureano-Phillips J, Robinson RD, Aryal S, Blair S, Wilson D, Boyd K, Schrader CD, Zenarosa NR, Wang H. HEART score risk stratification of Low-risk chest pain patients in the emergency department: a systematic review and meta-analysis. Ann Emerg Med. 2019;74(2):187–203. doi: 10.1016/j.annemergmed.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 40.Cheng Q, Greenslade JH, Parsonage WA, Barnett AG, Merollini K, Graves N, Peacock WF, Cullen L. Change to costs and lengths of stay in the emergency department and the Brisbane protocol: an observational study. BMJ Open. 2016;6(2):e009746. doi: 10.1136/bmjopen-2015-009746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ong ME, Padmanabhan P, Chan YH, Lin Z, Overton J, Ward KR, Fei DY. An observational, prospective study exploring the use of heart rate variability as a predictor of clinical outcomes in pre-hospital ambulance patients. Resuscitation. 2008;78(3):289–297. doi: 10.1016/j.resuscitation.2008.03.224. [DOI] [PubMed] [Google Scholar]
- 42.Liu N, Zhang Z, Ho AFW, Ong MEH. Artificial intelligence in emergency medicine. J Emerg Crit Care Med. 2018;2:82. doi: 10.21037/jeccm.2018.10.08. [DOI] [Google Scholar]
- 43.Chiew CJ, Liu N, Tagami T, Wong TH, Koh ZX, Ong MEH. Heart rate variability based machine learning models for risk prediction of suspected sepsis patients in the emergency department. Medicine. 2019;98(6):e14197. doi: 10.1097/MD.0000000000014197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu N, Sakamoto JT, Cao J, Koh ZX, Ho AFW, Lin Z, Ong MEH. Ensemble-based risk scoring with extreme learning machine for prediction of adverse cardiac events. Cogn Comput. 2017;9(4):545–554. doi: 10.1007/s12559-017-9455-7. [DOI] [Google Scholar]
- 45.Liu N, Koh ZX, Goh J, Lin Z, Haaland B, Ting BP, Ong MEH. Prediction of adverse cardiac events in emergency department patients with chest pain using machine learning for variable selection. BMC Med Inform Decis Mak. 2014;14:75. doi: 10.1186/1472-6947-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quintana DS, Alvares GA, Heathers JA. Guidelines for reporting articles on psychiatry and heart rate variability (GRAPH): recommendations to advance research communication. Transl Psychiatry. 2016;6:e803. doi: 10.1038/tp.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kloter E, Barrueto K, Klein SD, Scholkmann F, Wolf U. Heart rate variability as a prognostic factor for Cancer survival - a systematic review. Front Physiol. 2018;9:623. doi: 10.3389/fphys.2018.00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schroeder EB, Chambless LE, Liao D, Prineas RJ, Evans GW, Rosamond WD, Heiss G. Diabetes, glucose, insulin, and heart rate variability: the atherosclerosis risk in communities (ARIC) study. Diabetes Care. 2005;28(3):668–674. doi: 10.2337/diacare.28.3.668. [DOI] [PubMed] [Google Scholar]
- 49.Buchman TG, Stein PK, Goldstein B. Heart rate variability in critical illness and critical care. Curr Opin Crit Care. 2002;8(4):311–315. doi: 10.1097/00075198-200208000-00007. [DOI] [PubMed] [Google Scholar]
- 50.Polikar R. Ensemble based systems in decision making. IEEE Circuits Syst Mag. 2006;6(3):21–44. doi: 10.1109/MCAS.2006.1688199. [DOI] [Google Scholar]
- 51.Lundberg SM, Nair B, Vavilala MS, Horibe M, Eisses MJ, Adams T, Liston DE, King-Wai Low D, Newman SF, Kim J, et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng. 2018;2(10):749–760. doi: 10.1038/s41551-018-0304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.